Abstract
Three strains of Xenorhabdus nematophilus showed insecticidal activity when fed to Pieris brassicae (cabbage white butterfly) larvae. From one of these strains (X. nematophilus PMFI296) a cosmid genome library was prepared in Escherichia coli and screened for oral insecticidal activity. Two overlapping cosmid clones were shown to encode insecticidal proteins, which had activity when expressed in E. coli (50% lethal concentration [LC50] of 2 to 6 μg of total protein/g of diet). The complete sequence of one cosmid (cHRIM1) was obtained. On cHRIM1, five genes (xptA1, -A2, -B1, -C1, and -D1) showed homology with up to 49% identity to insecticidal toxins identified in Photorhabdus luminescens, and also a smaller gene (chi) showed homology to a putative chitinase gene (38% identity). Transposon mutagenesis of the cosmid insert indicated that the genes xptA2, xptD1, and chi were not important for the expression of insecticidal activity toward P. brassicae. One gene (xptA1) was found to be central for the expression of activity, and the genes xptB1 and xptC1 were needed for full activity. The location of these genes together on the chromosome and therefore present on a single cosmid insert probably accounted for the detection of insecticidal activity in this E. coli clone. Although multiple genes may be needed for full activity, E. coli cells expressing the xptA1 gene from the bacteriophage lambda PL promoter were shown to have insecticidal activity (LC50 of 112 μg of total protein/g of diet). This is contrary to the toxin genes identified in P. luminescens, which were not insecticidal when expressed individually in E. coli. High-level gene expression and the use of a sensitive insect may have aided in the detection of insecticidal activity in the E. coli clone expressing xptA1. The location of these toxin genes and the chitinase gene and the presence of mobile elements (insertion sequence) and tRNA genes on cHRIM1 indicates that this region of DNA represents a pathogenicity island on the genome of X. nematophilus PMFI296.
Currently, the most successful microbial insecticide is based on the bacterium Bacillus thuringiensis (16, 21) that produces insecticidal crystalline toxins during sporulation (20). The recent use of B. thuringiensis on a large scale and the development and use of transgenic plants expressing these toxin genes (3, 23) may enhance the development of resistant insect populations. New protein toxins are therefore required to provide a greater diversity of genes for use in pest control (14, 15, 31).
In the search for new genes, we have identified, by testing nonluminescent bacterial strains (Xenorhabdus species) isolated from insect parasitic nematodes (IPNs) for their ability to kill insects, a group of orally active protein toxins (22). Both IPNs and even their associated bacteria and toxins when injected into the insect hemocoel are well known for their ability to kill insects (2, 4, 12, 27, 32, 33). These properties are important for pathogenicity; however, for exploitation a toxin that is active when fed to insects is important. Ensign et al. (13) have identified protein toxins from the bacterium Photorhabdus luminescens with oral insecticidal activity which have been studied in detail (8, 9, 13, 18). However, when the toxin genes from P. luminescens were cloned and expressed in Escherichia coli they did not display insecticidal activity (9). Since both Xenorhabdus and Photorhabdus spp. cannot survive in water or soil for long, their use as a biopesticide may be limited. It is therefore important that the toxicity detected in these bacteria can be genetically moved to other bacteria, microorganisms, or plants for their exploitation. To enable this and to study the toxins in greater detail, the ability to express these proteins in a heterologous host and maintain their insecticidal activity is essential. We describe here the discovery of a strain of X. nematophilus and the isolation of an E. coli clone expressing a region of its genomic DNA which, when fed to Pieris brassicae larvae, causes a rapid cessation in feeding and mortality. The sequence of the insert and the identification of the genes involved in insecticidal activity are presented.
MATERIALS AND METHODS
Bacterial strains and media.
A strain of X. nematophilus 9965 was obtained from the NCIMB culture collection, Aberdeen, United Kingdom. Strains X. bovenii (UQM2872), X. beddingii (UQM2211), X. poinarii (ATCC 4921), and X. redingi were a gift from Noel Boemare, INRA-CNRS, Paris, France. The other Xenorhabdus strains were isolated from infective nematodes present in United Kingdom soil by using an insect entrapment method (4). Samples of hemolymph containing characteristic rod-shaped bacteria were plated onto NBTA (1) composed of nutrient agar (NA; Difco, Detroit, Mich.) containing bromophenol blue (25 mg liter−1) and triphenyl 235 tetrazolium chloride (4 mg liter−1) and incubated at 25°C for 72 h. Plates where the bacterial colonies were dark blue were subcultured on NBTA and incubated at 25°C for 72 h to ensure purity. Nonluminescent strains were presumptively identified as Xenorhabdus species and used in this work. The strains were routinely maintained on NA.
Insect bioassay.
The insecticidal activity of bacterial isolates was tested with neonate P. brassicae larvae. Individual colonies were cultured in Luria broth (LB; Difco) at 28°C for 18 h at 200 rpm. A 50-μl sample was spread onto an agar-based artificial diet (11) which contained streptomycin (20 μg ml−1) and cefataxime and tetracycline (each at 100 μg ml−1). To each container (4.5 cm in diameter), 10 larvae were added, and the assay pots were incubated at 25°C (16-h day length period), with a relative humidity of 80% for 96 h. Daily recordings of larval mortality and the size of the surviving larvae were made. Insecticidal strains were selected when reduced larval size was observed and >50% of the insects had been killed.
The potency of active bacteria was tested using a multidose incorporation assay. Assays were performed with a series of five dilutions of the original cultures made in phosphate-buffered saline (0.05 mM phosphate buffer with 0.125 M NaCl) using a minimum of 40 larvae per concentration. Assays were incubated at 25°C, with a relative humidity of 80%, for 6 days with a 16-h day length period. The mortality was recorded, and the 50% lethal concentration (LC50) was determined by probit analysis (17). In the assays, negative controls included treatments with just water and with E. coli. The protein concentration of samples was determined using the BCA reagent kit (Pierce).
Strain characterization.
PCR amplification of DNA was performed on purified DNA using an Omnigene Thermocycler (Hybaid, Teddington, United Kingdom). DNA was isolated from bacterial cultures grown at 30°C for 18 h on NA using a Qiagen kit (Qiagen, Dorking, United Kingdom). A loopful of each culture was resuspended in 1 ml of buffer B1; to this, 0.35 ml of buffer B2 and 0.4-g glass beads (0.1 mm in diameter) were added. The sample was vortexed at maximum speed for 10 min and centrifuged at 13,000 × g for 5 min. The supernatant was applied to the top of a Qiawell 8 column (Qiagen), and the DNA was purified according to the manufacturer's instructions. After ethanol precipitation the sample was resuspended in 100 μl of 1 mM Tris-HCl (pH 8). To a 100-μl PCR reaction, 4 to 20 μl of DNA was added. The reaction mix contained 1× buffer (Flowgen, Lichfield, United Kingdom), 100 pmol of each deoxynucleoside triphosphate, 200 μmol of each primer, and 1 U of Dynazyme (Flowgen). The samples were overlaid with 50 μl of mineral oil. For amplification of the 16S rRNA gene, the primers AAGGAGGTGATCCAGCCGCA and GGAGAGTTAGATCTTGGCTC were used (10). Following amplification, a 5-μl aliquot was run on a standard 1% (wt/vol) agarose gel set at 100 V for 2.5 h to identify PCR products. The 16S rRNA gene PCR products (5 to 17 μl) were digested with the restriction enzymes AluI, CfoI, DdeI, HaeI, HinfI, RsaI, and Sau3A in a final volume of 20 μl in 1× buffer at 37°C for 3 h. All comparisons from a single digest were made on one gel to ensure the reliability of any comparisons.
Cloning of genomic DNA fragments encoding insecticidal activity.
Total DNA was isolated from a culture of PMFI296 grown in LB (Sigma, Poole, United Kingdom) at 28°C and 200 rpm to an optical density of 1.5 (600 nm) using a genomic DNA purification kit (Qiagen). Genomic DNA was partially digested with Sau3A to give a dominant fragment size between 30 to 50 kb. The genomic DNA was ligated to Supercos DNA (Stratagene, Amsterdam, The Netherlands) cut with BamHI and XbaI using standard procedures (25). The ligated DNA was packaged (Stratagene) and transformed into E. coli DH5α cells. Any colonies that grew on LB agar supplemented with 50 μg of kanamycin ml−1 at 30°C within 30 h were tested for insecticidal activity by small-scale toxicity tests. Subcloning was performed using standard techniques as outlined in the appropriate sections.
DNA sequencing.
A system of subcloning larger fragments of cHRIM1 in pUC19, designing new sequencing primers and walking out from known sequences, and random sequencing of small EcoRV DNA fragments of cHRIM1 in pUC19 was used to obtain the sequence of the cosmid insert. All sequencing reactions were performed using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Liverpool, United Kingdom) and analyzed on an automated DNA sequencer (Applied Biosystems). Sequences were edited and assembled using Sequencher 3.1 (Gene Codes Corp.) and DNA∗ (DNAStar, Inc.) software. Sequence analysis was performed using the packages FASTA (6.0), PILEUP (GCG), PLOTSIMILARITY (GCG), and the EBI internet system. DNA mapping and translation were performed using the programs Clone Manager and Enhance (Scientific & Educational Software).
Transposon mutagenesis.
Transposon mutagenesis using Tn3 was carried out essentially by the method of Seifert et al. (30). cHRIM1 DNA was cut with PstI (sites only within the cosmid) and cloned into the plasmid pBRMCS-2 cut with NsiI to create plasmid pHRIM555 (kanamycin resistant). This removed the ampicillin resistance gene from the cosmid and created a mobilizable plasmid system. Plasmid pHRIM555 was transformed into E. coli (pLBIO1) selecting for kanamycin and chloramphenicol resistance. The resulting strain was mated with E. coli RDP146 (pOX38:mTn3-HIS3), and the cointegrates were resolved by mating this strain with E. coli NS2115Sm and selection with ampicillin, kanamycin, and streptomycin. Artificial transposon mutagenesis was also carried out using the transposon AT2 on purified DNA (Qiagen-Midi) following the manufacturer's protocols (Applied Biosystems). To locate the transposon insertion positions, plasmid DNA was purified (Qiagen Qiawell 8 Miniprep), and the primers Tn3Pw3 (24) (TACTCATATATACTTTAGAT), Tn3Pw6 (24) (ATACGCTCACGTACATGCTA), and AT2+ and AT2− (Applied Biosystems) were used to sequence outward from the transposon ends.
SDS-PAGE of cell proteins.
X. nematophilus PMFI296 was grown in 50 ml of LB at 30°C at 150 rpm for 2 days. E. coli, E. coli (cHRIM1), and E. coli (clone 338) were grown at 30°C for 1 day in LB and LB containing 50 μg of kanamycin or 20 μg of chloramphenicol ml−1, respectively. The cell cultures were centrifuged at 10,000 × g for 15 min, and the cell pellet was washed twice with phosphate-buffered (10 mM) saline (125 mM). The cells were disrupted by three rounds of sonication (18 μ peak to peak for 30 s), and the cell debris was removed by centrifugation at 13,000 × g for 15 min. Samples (5 μl) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 3 to 8% precast gradient gel (Novex, San Diego, Calif.) run at 150 V for 1.5 h. The gels were either stained with 0.25% (wt/vol) Coomassie brilliant blue in 40% (vol/vol) methanol–10% (vol/vol) acetic acid for 1 h and destained for 3 h or else stained for 45 min with SYPRO Orange (Bio-Rad, Hemel Hampstead, United Kingdom) in 7.5% (vol/vol) acetic acid. N′-terminal protein sequencing was performed on peptides transferred to a polyvinylidene difluoride membrane after SDS-PAGE using the manufacturer's recommended conditions (Novex). Sequencing was carried out at the Protein Sequencing Service of the Babraham Institute.
Induction of expression from PL promoter.
High-level expression of proteins was achieved by subcloning the xptA1 gene into the vector pLEX (Invitrogen, Groningen, The Netherlands). E. coli and E. coli carrying pHRIM801 were grown in RMG media (2% [wt/vol] amicase, 5% [wt/vol] glucose, and 1 μM MgCl2 in M9 minimal salts solution) containing ampicillin (50 μg ml−1), where appropriate, at 30°C and 200 rpm for 18 h. An aliquot (1 ml) was transferred to LB containing tryptophan (100 μg ml−1) and, where appropriate, ampicillin (50 μg ml−1) to induce expression from the bacteriophage lambda PL promoter. After incubation at either 37 or 30°C, the cells were collected at various intervals by centrifugation at 8,000 × g for 10 min. The cells were analyzed directly by SDS-PAGE for the presence of a 280-kDa protein. A soluble fraction and a fraction containing inclusion bodies were prepared as outlined in Sambrook et al. (29), using sonication to disrupt cells (three 30-s bursts at 18 μ peak to peak) and 50 mM phosphate buffer (pH 7.2) containing 0.5 M urea to wash the inclusion bodies. The two fractions (soluble and inclusion bodies) were analyzed by SDS-PAGE.
Nucleotide sequence.
The complete sequence of the cosmid cHRIM1 indicating the positions of genes described here has been deposited in the EMBL database under accession number AJ308438.
RESULTS
Identification of insecticidal Xenorhabdus strains.
The type strains and new isolates obtained by insect entrapment were tested for toxicity when fed to insects. The type strain X. nematophilus 9965 killed 100% of the insects at a dose of 106 cells/cm2. When a total of 50 isolates obtained by entrapment from soil was screened for insecticidal activity, two showed equivalent activity toward P. brassicae. This indicated that most of the strains obtained were not as insecticidal to this insect under the conditions used. The three active isolates were characterized by PCR-restriction fragment length polymorphism (RFLP) analysis of their 16S rRNA gene and compared to the type strains and a small selection of uncharacterized isolates. The 16S rRNA RFLP patterns of the active strains were identical and included the type strain of X. nematophilus 9965. Since more than four enzymes were used to generate these patterns, the active strains were therefore identified as subspecies of X. nematophilus (7, 10). One of these strains, PMFI296, was studied in greater detail.
Cloning and expression of the insecticidal activity of X. nematophilus PMFI296.
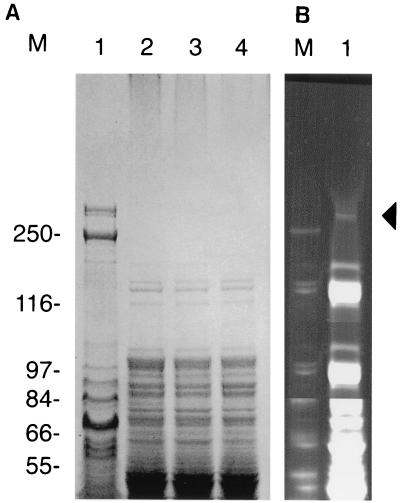
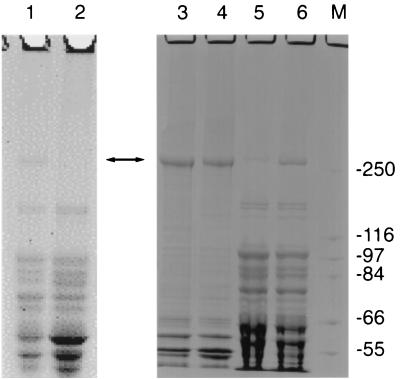
In a screen of more than 500 E. coli clones containing cosmids with inserts of X. nematophilus PMFI296 genomic DNA (ca. 40 kb), 2 were found to have insecticidal activity (Table 1). Both clones (cHRIM1 and cHRIM2) were mapped with the enzymes BamHI, EcoRI, HindIII, SacI, and SalI. Cosmid cHRIM1 was found to have an insert of approximately 39 kb, while cosmid cHRIM2 had a larger insert with an approximate size of 43 kb. The restriction pattern of the clones indicated that the insert in cHRIM1 was a slightly smaller internal fragment of the insert in cHRIM2. E. coli (cHRIM1) grown at 30°C had the greatest insecticidal activity of these cosmids; however, the LC50 of these cells was ca. 1/50 of that obtained from the original strain (Table 1). This loss in activity may be attributed to the presence on cHRIM1 of only some of the toxins of strain PMFI296, the poor expression of these genes in E. coli, or the degradation of the toxins after expression. The protein profiles of X. nematophilus cells and E. coli clones are shown in Fig. 1. One large protein of ca. 280 kDa was present in X. nematophilus PMFI296 cells. Its size (>250 kDa) and N′-terminal sequence (MIKVNELLDK) indicated that this protein was XptA1, a toxin coded by the xptA1 gene on cHRIM1 and shown here to be essential for insecticidal activity. In X. nematophilus this protein is expressed at a reasonable level and is clearly seen in the total cell protein profile when the gel is stained with Coomassie blue. In E. coli (cHRIM1) this protein cannot be detected using Coomassie blue staining. This indicates that the XptA1 protein is present at a lower level in the E. coli cells compared to X. nematophilus cells. When a more sensitive protein stain is used (SYPRO), a protein of the correct size can be seen in E. coli (cHRIM1) cells. Insufficient protein was present to obtain any protein sequence from this band; however, a protein of similar size was not detected in E. coli cells (data not shown). Therefore, the expression or stability of this protein would seem to account for a significant proportion of the reduced insecticidal activity of the E. coli cosmid clone.
TABLE 1.
Insecticidal activity of E. coli clones to P. brassicae
| Strain | Clone | LC50 (μg of protein/g of diet) |
|---|---|---|
| E. coli | cHRIM1 | 2.4 |
| E. coli | cHRIM2 | 6.2 |
| E. coli | pHRIM801 | 112 |
| E. coli | >260 (no mortality) | |
| X. nematophilus | 0.043 | |
| PMFI296 |
FIG. 1.
SDS-PAGE analysis of cell extracts from X. nematophilus pMFI296 and E. coli clones. (A) Lane 1, X. nematophilus pMFI296; lane 2, E. coli; lane 3, E. coli (cHRIM1); and lane 4, E. coli (338/2) stained with coomassie brilliant blue. (B) Lane 1, E. coli (cHRIM1) stained with SYPRO Orange. The position of XptA1 is marked with an arrow. Size markers (M) are presented (in kilodaltons).
Sequence analysis of cHRIM1.
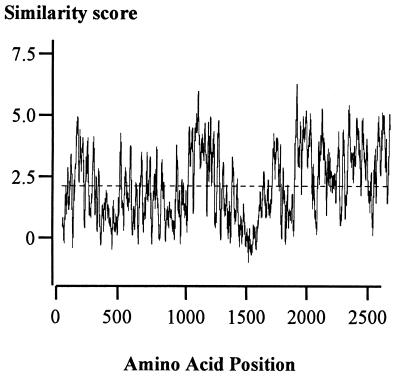
The complete sequence of cHRIM1 was obtained. The insert size is 38.4 kb and has an average G+C composition of 34.6%, which is low but equivalent to other members of the Enterobacteriaceae. Similarity searches have revealed four complete and one incomplete predicted open reading frames that illustrate differing levels of homology to insecticidal protein toxins that have been identified in P. luminescens (9, 13). Figure 2 displays a map of cHRIM1 indicating their position, and Table 2 summarizes the names given to these genes and the homologies detected. The genes were designated xpt, for Xenorhabdus protein toxin, grouped A to D, and further numbered according to the level of DNA homology and overall size. Two similar genes were identified: xptA1 and xptA2. Both of these genes and their predicted proteins showed homology to each other and to the tcb and tcd genes (40 to 45% identity) and the tcaB and tccA genes (26 to 40% identity) present in P. luminescens. The arrangement of xptA1 and xptA2 indicates that they are convergently transcribed. Since they are homologous genes and close, this arrangement may prevent homologous recombination and rearrangement of the DNA in this region. The greatest similarity of the XptA1 and XptA2 proteins was to the Tcb and Tcd proteins. A multiple sequence alignment of these proteins indicates areas with different levels of similarity (Fig. 3). The last 700 amino acids of these proteins (C′-terminal region) showed the greatest overall levels of protein similarity (above their average similarity). Two other smaller regions before this area also showed higher levels of similarity. These regions with greater overall similarity may serve a similar function and may be unable to vary to the same extent as other areas of the protein.
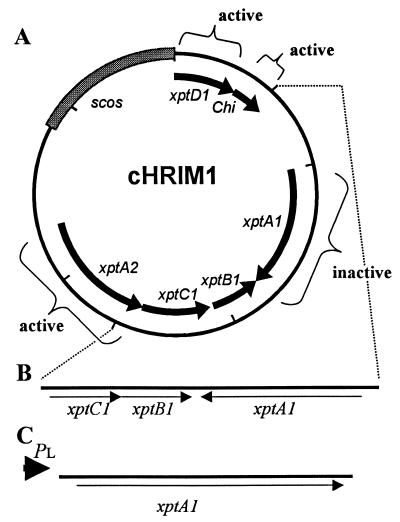
FIG. 2.
Characterization of the contribution of genes on cHRIM1 to insecticidal activity. (A) Map indicating the effect of transposon insertions in genes, including insertions that did not alter insecticidal activity (active) and those that resulted in the loss of insecticidal activity (inactive). Scos, cosmid vector. (B) Map of the insert in the smallest active clone 338/2 constructed using the SalI site at the end of the chitinase gene (chi) and one transposon insertion obtained at the end of the xptA2 gene (dotted lines). (C) Map of insert in plasmid pHRIM801, where the xptA1 gene was expressed from the bacteriophage PL promoter (PL).
TABLE 2.
Similarity of predicted proteins and regions of DNA on cosmid cHRIM1 to sequences on the SWISSPROT and EMBL databases as determined by using the program FASTA (6.0)a
| Homology and X. nematophilus gene or region | Start of comparison (X. nematophilus gene) | Homologous protein or gene | EMBL accession no. | Start of comparison (target protein or gene) | Region length (amino acids) | % Identity |
|---|---|---|---|---|---|---|
| Protein homology | ||||||
| xptA1 | 23 | TcbA | 085160 | 23 | 2,545 | 44.9 |
| 10 | TcdA | Q9RN45 | 1 | 2,576 | 44.5 | |
| 1,775 | TcaB | 085152 | 442 | 1,213 | 38.9 | |
| 945 | TccB | 085156 | 30 | 1,645 | 28.5 | |
| 40 | TccA | 085151 | 40 | 141 | 29.7 | |
| xptA2 | 23 | TcbA | 085160 | 23 | 2,554 | 40.6 |
| 1 | TcdA | Q9RN45 | 25 | 2,560 | 43.2 | |
| 1,786 | TcaB | 085152 | 440 | 768 | 35.5 | |
| 965 | TccB | 085156 | 40 | 1,635 | 26.4 | |
| 100 | TccA | 085151 | 116 | 71 | 46.4 | |
| xptB1 | 5 | TccC | 085157 | 8 | 902 | 48.2 |
| xptC1 | 1 | TcaC | 085153 | 1 | 1,435 | 48.8 |
| xptD1 | 730 | TccB | 085156 | 260 | 847 | 38.9 |
| 317 | TcbA | 085160 | 1,752 | 795 | 32.8 | |
| 207 | TcaB | 085152 | 350 | 899 | 31.3 | |
| chi | 98 | Exochitinase | 052863 | 1 | 553 | 37.7 |
| IS | 1 | IS630 | X05955 | 1 | 343 | 58.1 |
| Ptrans | 10 | Transposase | AAC82732 | 25 | 273 | 47.3 |
| Phage | 1 | P4-helicase | P10277 | 626 | 152 | 47.3 |
| DNA homology | ||||||
| mit1 | Mitochondrial | A1034559 | 60.0 | |||
| DNA (AT rich) | D86630 | 55.1 | ||||
| Aq026159 | 60.3 | |||||
| Retrovirus | Q03276 | 30.7 | ||||
| R1 | J01459 | 60.0 | ||||
| 15S rRNA | X03240 | 57.0 | ||||
| mit2 | AT rich | X54011 | 55.0 | |||
| tRNA | Y13767 | 65.0 | ||||
| AT rich | J01459 | 65.1 |
The starting point and length over which the protein comparisons were made between X. nematophilus and homologous target proteins (P. luminescens toxins or other proteins). The positions of these genes and the regions of DNA on cHRIM1 are outlined in Fig. 1, and the details are listed in the EMBL submission.
FIG. 3.
Multiple sequence comparison of the protein sequences XptA1 and XptA2 from X. nematophilus and TcbA and TcdA from P. luminescens using the package Plotsimilarity. Scores at amino acid positions along the alignment are presented (Henikoff-Henikoff amino acid similarity score, which ranges from −4 to +11), with the average similarity score represented by a dashed line.
Between the xptA1 and xptA2 genes, two further predicted open reading frames are present, xptB1 and xptC1, which show the greatest similarity to the tccC (48% identity) and tcaC (49% identity) genes of P. luminescens, respectively (8, 13). An additional gene that is not complete on the cosmid cHRIM1, xptD1, was found to have similarity to the tccB, tcbA, and tcaB genes (31 to 39% identity) in P. luminescens (8, 13). Therefore, on cHRIM1 four complete genes and one incomplete gene with homology to others implicated in insecticidal activity have been identified. As Table 1 illustrates, these genes also show a degree of homology to one and another between the different groups.
In addition to these toxin genes, a small region of DNA upstream of the xptA1 gene encodes for a predicted protein (648 amino acids) that shows similarity to chitinase protein sequences. The greatest similarity to the exochitinase protein (EMBL accession no. 052863, 695 amino acids) from the Glossina morsitans S-endosymbiont was detected (38% identity); this bacterium is also a member of the gamma subdivision of the proteobacteria. The close proximity of the predicted chitinase gene and the insecticidal toxin genes further indicates that this is a cluster of pathogenicity factors on the chromosome of PMFI296, all of which may be involved to some degree in the overall insecticidal activity observed for this cosmid clone.
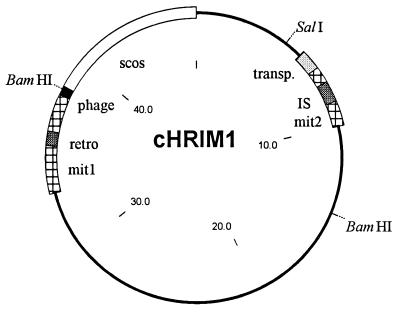
Other gene and predicted protein sequence homologies were also identified on cHRIM1 that relate to DNA mobility; these are illustrated in Fig. 4. An insertion sequence (IS) was located 1,328 bp upstream of the xptA1 gene, which transcribes toward the start of this gene. The IS is homologous to IS630 detected in Salmonella species and other Enterobacteriaceae (26). This insertion sequence has an imperfect inverted repeat of the sequence ATTATGAAAACTTATTTAA and has a single 1,041-bp open reading frame coding for a 347-amino-acid transposase (58% sequence identity to other transposases). This structure is typical of insertion sequences of this class (28). Just 814 bp upstream of the insertion sequence is an open reading frame that shows similarity (47% identity) to a putative transposase present on plasmid PMT-1 from Yersinia pestis (25), and a direct repeat of the sequence ATAAAATTTTCCGG on either side of this gene was detected. Finally, a small area of DNA showed similarity (47% identity) to the C′-terminal half of a bacteriophage P2 primase protein (34). This area of similarity lies at the extreme edge of the cosmid clone, and it is possible that other areas of a complete phage genome lie upstream of this region. The presence of an IS element, a putative transposase, and bacteriophage DNA sequences adjacent to the toxin genes highlights the potential mobility of the DNA within this region.
FIG. 4.
Map of cHRIM1 indicating the position of mobile elements and areas not related to known insecticidal genes. Restriction sites for SalI and BamHI are shown. transp., hypothetical transposase, retro., retrotransposon; phage, bacteriophage DNA; mit1 and mit2, AT-rich regions similar to mitochondrial DNA; Scos, cosmid vector (size in kilobase pairs).
Unusually, areas of the cHRIM1 sequence show considerable similarity (up to 60% identity) to mitochondrial DNA sequences from nematodes and other eukaryotic organisms (Fig. 4, mit1 and mit2). Upstream of the xptA2 gene, before the phage primase region, the DNA shows similarity (up to 60% identity) to mitochondrial rRNA, the mitochondrial retrotransposase (R1), and AT-rich intergenic regions with possible replication functions (mit1). For bacteria, the presence of this unusual region of DNA further indicates greater DNA mobility than expected. In addition, DNA homology to a tRNA gene (65% identity) and regions of similarity to mitochondrial and eukaryotic DNA (55 to 65% identity) was detected in the regions surrounding the insertion sequence and the putative bacterial transposase gene (mit2). The separation of these two regions may indicate that the block of DNA from the start of the xptA2 gene through to the start of the xptA1 gene has integrated as one unit into this area. The chitinase and truncated xptD1 gene, although linked to this region by sharing homology to genes implicated with insecticidal activity, may represent a separate unit.
Identification of the genes on cHRIM1 involved in insecticidal activity.
To characterize the contribution of different genes and DNA sequence(s) to the toxic activity of E. coli cells containing cHRIM1, transposon mutagenesis of the insert was performed. The PstI insert in pHRIM555, comprising the whole of the insert in cHRIM1 and a small region of the original cosmid (Supercos1) was mutated with the transposon Tn3. A total of 110 transposon mutants in pHRIM555 were generated. A total of 16 mutants were mapped by digestion with HindIII and EcoRI, and in all cases a different restriction pattern was obtained, indicating that a diversity of insertion points were obtained. Of the 110 mutants, only 6 showed a loss of insecticidal activity. The insertion points of the six nonactive strains were identified, and these were located within the xptA1 gene. A number of other transposon insertions were sequenced to determine their location. Transposon insertions were found in the xptA2, xptD1, and chi genes, and no loss in insecticidal activity toward P. brassicae was detected (Fig. 2). These results indicated that the expression of these genes did not contribute to the overall insecticidal activity of E. coli (cHRIM1). Two deletions of the cosmid were made. Using the SalI sites in the cosmid and at the end of the predicted chitinase gene (chi), a deletion of cHRIM1 removing the xptD1 and chi genes was made. Also, using an AT2 transposon mutant (Tn116), with an insertion at the end of the xptA2 gene, a deletion of the mit1 region and xptA2 gene was made. The resulting clone (338/2) when expressed in E. coli was as insecticidal as E. coli (cHIRM1). On this clone the xptA1, xptB1, and xptC1 genes are complete (Fig. 2B). Transposon mutants in the xptC1 and xptB1 genes were not obtained with the Tn3 transposon, and their individual contributions to insecticidal activity could not be determined. It is possible that an insertion in these genes resulted in DNA rearrangements or produced a clone that was lethal to the host cell and was not represented in the transposon library. Plasmid clones with smaller inserts than that present in clone 338 were not insecticidal (data not shown).
Insecticidal activity of xptA1.
The xptA1 gene that encodes for a protein with a predicted size of 287 kDa and upstream from the ATG start (−8 bp) is a predicted ribosomal binding site (RBS; AGGA). To insert the xptA1 gene under the control of the bacteriophage lambda PL promoter, the cosmid cHRIM1 was cut with NotI and the 14-kb fragment was inserted into the NotI site of pLEX to create plasmid pHRIM800. Plasmid pHRIM800 was cut with KpnI and AflII and religated in the presence of the linker 5′-TTAAGTAC-3′ (0.5 pmol/μl) to create plasmid pHRIM801. In this plasmid the RBS site and ATG start of the xpt1A gene was brought close to the PL promoter. When expression from the PL promoter was induced in E. coli, a large protein greater than 250 kDa was seen, which was not present in E. coli cells (Fig. 5). The N′-terminal sequence of this protein (10 amino acids) was determined and was identical to the predicted xptA1 gene product. With induction at 37°C for 18 h, most of this protein was found in inclusion bodies and no insecticidal activity was detected. With shorter periods of induction at 30°C, more of this protein was found in the soluble fraction (Fig. 5). E. coli (pHRIM801) cells were insecticidal (Table 1), reducing larval size and killing when applied at a high concentration. The best results were obtained with induction at 30°C for 18 h. At the levels used, the control E. coli cells did not kill P. brassicae larvae, and therefore the LC50 value was greater than the detection limit (Table 1). No difference between the insecticidal activity of the soluble fraction and the inclusion body sample from E. coli (pHRIM801) was detected. The results indicated that the overall insecticidal activity of the XptA1 protein was low.
FIG. 5.
SDS-PAGE analysis of the expression of xptA1 from the PL promoter in E. coli cells. Whole-cell extracts after 18 h of growth at 37°C. Lane 1, E. coli (pHRIM600); lane 2, E. coli. Cell fractions after growth for 4 h at 30°C (lanes 3 and 5) and for 18 h at 30°C (lanes 4 and 6). Samples were separated into inclusion bodies (lanes 3 and 4) and soluble fractions (lanes 5 and 6). Markers (M) are presented (in kilodaltons).
DISCUSSION
A few bacterial isolates from insect parasitic nematodes when fed to P. brassicae quickly stopped larval feeding and caused mortality within 24 h. The highly active isolates were all identified as strains of X. nematophilus. From one of these a region of DNA encoding proteins with insecticidal activity toward P. brassicae was cloned. The smallest cosmid clone (cHRIM1) was found to have a 39-kb insert, which initially could not be reduced in size without the loss of insecticidal activity when expressed in E. coli. The way the insects were killed and the physical properties of the insecticidal activity expressed in the clone (including its high molecular weight, its sensitivity to heat treatment, and the degree of proteinase K resistance) were similar to those found in the original Xenorhabdus strain (22). A lower level of insecticidal activity was detected in this E. coli clone compared to that obtained for the original X. nematophilus PMFI296 strain. When the growth temperature for E. coli cells was reduced to 30°C, the insecticidal activity was improved. However, at best E. coli (cHRIM1) cells had approximately 1/50 of the activity of the original strain (Table 1). By comparing the levels of one known protein toxin (XptA1) in E. coli (cHRIM1) to that detected in X. nematophilus PMFI296, at least part of the reduced insecticidal activity of the clone could be attributed to poor expression or stability of this protein in E. coli. By taking into account the differences in the protein levels where at least a fivefold drop in expression is evident (Fig. 1) and the respective insecticidal activities of the cells (Table 1), it is estimated that the specific activity of the cloned toxins is unlikely to exceed a tenfold reduction compared to the wild-type strain. However, additional toxins on the chromosome of X. nematophilus not present on the cosmid cHRIM1 could also contribute to the total insecticidal activity of the original strain. A transposon mutagenesis study on cHRIM1 identified one open reading frame (xptA1) that was central to insecticidal activity. This gene was similar in structure and had homology to two genes (tcb and tcd) identified in P. luminescens (10). A second gene on cHRIM1 (xptA2) was a similar size and shared homology with xptA1. This indicates that a close family of toxin genes, similar in size and sequence, exist between Xenorhabdus and Photorhabdus species. The two predicted proteins (XptA1 and XptA2) also shared a degree of similarity to other insecticidal toxins identified in P. luminescens, i.e., TcaB and TccB (10). This also indicates that a degree of similarity exists between the different toxin groups. Using DNA and protein similarity, four other genes (xptB1, -C1, and -D1 and chi) previously implicated with insecticidal activity were identified on cHRIM1.
Transposon mutagenesis and subcloning were used to assess the involvement of these genes in the expression of insecticidal activity in E. coli (cHRIM1). Three of the genes (xptA2, xptD1, and chi) did not contribute to the insecticidal activity seen toward P. brassicae. The individual contribution of the two genes, xptB1 and xptC1, could not be determined since Tn3 transposons in these genes were not detected. Since the smallest active clone (338/2) contains both these genes and the xptA1 gene and since the overall level of insecticidal activity of the toxin XptA1 is poor, one or both of these genes (xptB1 and -C1) are needed for full activity. The isolation of cosmid clones expressing proteins with insecticidal activity has provided this insight into the gene interactions for toxin activity. This region can now be introduced into other bacteria to assess any changes in the expression of activity, or to complement different toxins such as those from B. thuringiensis.
P. luminescens toxin genes when expressed individually in E. coli were not insecticidal (9). In contrast, the xptA1 gene from X. nematophilus was insecticidal. The high level of expression of the xptA1 gene from the bacteriophage PL promoter may be responsible for our ability to detect insecticidal activity for this single toxin. In addition, a greater sensitivity of our target insect may also have been partly responsible for this. Certainly, the XptA1 protein is insecticidal, and it is also central for the activity of E. coli (cHRIM1) cells. However, the combination of the different toxins on the cosmid is important for overall activity. The cosmid genomic cloning strategy adopted here may provide a useful way for obtaining active proteins that are insecticidal when expressed in heterologous hosts. This may provide a faster solution to the expression of active toxins than studying purified toxins. For example, if one or more of the gene products on cHRIM1 interacts with XptA1 to activate it then, unless the activation step becomes obvious during the purification of the active protein, it may become difficult to determine the changes that have taken place in this protein. The activated protein may have an altered structure, processed at either end, or it may become protected in some way from inactivation in the insect gut. These processes may be difficult to detect in a purified peptide. Thus, in an E. coli strain expressing proteins from a cosmid clone that has insecticidal activity, at least some of these factors would have to be present on the clone. These interactions can then be studied at the genetic level. However, strain PMFI296 may be unusual and the key toxin genes may not be close together on the chromosome in other strains, and thus a cloning approach would fail. Different protein interactions may also be responsible for differences in insecticidal activity seen toward other insect species. As such, the xptA2 and chitinase genes may be important for full activity toward insects other than P. brassicae.
The fact that genes similar to those identified here in X. nematophilus have been found in distantly related strains of P. luminescens indicates that a common set of insect toxins exist in bacteria associated with IPNs. This suggests that either of these strains have had these genes since they diverged from a common ancestor in which they may have been required to fulfill a similar function or, alternatively, that gene exchange has taken place either between these strains or between these strains and a common, as-yet-unidentified strain in more recent times. The presence of an insertion sequence, phage DNA, and mitochondrial DNA near the toxin genes on cHRIM1 indicates that gene exchange between strains is likely to have been involved in the distribution of toxin genes. However, it is possible that this area on the chromosome may act as a hot spot for the insertion of DNA coming from other sources. A better understanding of these processes may be gained from the analysis of a greater diversity of strains. The dependence of this bacteria on a nematode host and its growth within an insect may explain how this strain has acquired DNA that is similar to eukaryotic mitochondrial DNA sequences through lateral DNA transfer in this close relationship. It is interesting that the presence of homology to a tRNA gene and the prediction of three tRNA genes near to the potential point of insertion of the toxin genes shows similarity with the other mobile virulence regions in E. coli, Salmonella species, Dichelobacter nodosus, and other bacteria (5, 6, 19). In these instances, large segments of DNA also insert into tRNA loci, and a number of independent insertion events may have led to the mosaic structure now seen in the genomes of these strains. As well as the presence of virulence determinants, the presence of IS elements, tRNA genes, and phage genes are all hallmarks of a pathogenicity island (19). We therefore conclude that that the insert in cHRIM1 represents a pathogenicity island on the genome of X. nematophilus PMFI296.
ACKNOWLEDGMENTS
This work was funded by the Ministry of Agriculture Fisheries and Food (United Kingdom) and the Biological and Biotechnological Sciences Research Council (United Kingdom).
We thank Umesh Patel at Cambridge Biosciences and Ruth Finch at Horticulture Research International for their help in the DNA sequencing work. We also thank Pat Barker at the protein sequencing unit, Babraham Institute (United Kingdom), for help with this work.
REFERENCES
- 1.Akhurst R J. Morphological and functional dimorphism in Xenorhabdus spp. bacteria symbiotically associated with insect pathogenic nematodes Neoaplectana and Heterorhabditis. J Gen Microbiol. 1980;121:303–309. doi: 10.1099/00221287-128-12-3061. [DOI] [PubMed] [Google Scholar]
- 2.Akhurst R J, Dunphy G B. Symbiotically associated entomopathogenic bacteria, nematodes, and their insect hosts. In: Beckage N, Thompson S, Federici B, editors. Parasites and pathogens of insects. Vol. 2. New York, N.Y: Academic Press, Inc.; 1993. pp. 1–23. [Google Scholar]
- 3.Augustyniak J, Dabert M, Wypijewski K. Transgenes in plants:protection against viruses and insects. Acta Physiol Planta. 1997;19:561–569. [Google Scholar]
- 4.Bedding R A, Akhurst R J. Nematodes and their biological control of insect pests. Nematologia. 1975;21:109–110. [Google Scholar]
- 5.BlancPortard A B, Solomon F, Kayser J, Groisman E A. The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol. 1999;181:998–1004. doi: 10.1128/jb.181.3.998-1004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloomfield G A, Whittle G, McDonagh M B, Katz M E, Cheetham B F. Analysis of sequences flanking the vap regions of Dichelobacter nodosus: evidence for multiple integration events, a killer system, and a new genetic element. Microbiology. 1997;143:553–562. doi: 10.1099/00221287-143-2-553. [DOI] [PubMed] [Google Scholar]
- 7.Boemare N E, Akhurst R J, Mourant R G. DNA relatedness between Xenorhabdus spp. (Enterobacteriacea), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdusgen. nov. Int J Syst Bacteriol. 1993;43:249–255. [Google Scholar]
- 8.Bowen D J. Characterization of a high molecular weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. M.S. thesis. University of Wisconsin, Madison; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowen D, Rocheleau T A, Blackburn M, Andreev O, Golubeva E, Bhartia R, Ffrench-Constant R H. Novel insecticidal toxins from the bacterium Photorhabdus luminescens. Science. 1998;28:2129–2134. doi: 10.1126/science.280.5372.2129. [DOI] [PubMed] [Google Scholar]
- 10.Brunel B, Givaudan A, Lanois A, Akhurst R J, Boemare N. Fast and accurate identification of Xenorhabdus and Photorhabdusspecies by restriction analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1997;63:574–580. doi: 10.1128/aem.63.2.574-580.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David W A L, Gardiner B O C. Rearing Pieris brassicae L. larvaeon a semi-synthetic diet. Nature. 1965;207:882–883. [Google Scholar]
- 12.Dunphy G B, Webster J M. Lipopolysaccharides of Xenorhabdus nematophilus (Enterobacteriacea) and their haemocyte toxicity in non-immune Galleria mellonella(Insecta:Lepidoptera) larvae. J Gen Microbiol. 1988;134:1017–1028. [Google Scholar]
- 13.Ensign, J. C., et al. 1997. U.S. patent WO97/17432.
- 14.Estruch J J, Warren G W, Mullins M A, Nye G J, Craig J A, Koziel M G. Vip3A, a novel Bacillus thuringiensisvegetative insecticidal protein with a wide spectrum of activity against lepidopteran insects. Proc Natl Acad Sci USA. 1996;93:5389–5394. doi: 10.1073/pnas.93.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estruch J J, Carozzi N B, Desai N, Duck N B, Warren G W, Koziel M G. Transgenic plants: an emerging approach to pest control. Nat Biotechnol. 1997;15:137–141. doi: 10.1038/nbt0297-137. [DOI] [PubMed] [Google Scholar]
- 16.Feitelson J S, Payne J, Kim M. Bacillus thuringiensis: insects and beyond. Biotechnology. 1992;10:271–275. [Google Scholar]
- 17.Finney D J. Probit analysis. Cambridge, United Kingdom: Cambridge University Press; 1971. [Google Scholar]
- 18.Guo L, Fatig R O, Orr G L, Schafer B W, Strickland J A, Sukhapinda K, Woodsworth A T, Petell J K. Photorhabdus luminescensW-14 insecticidal activity consists of at least two similar but distinct proteins. J Biol Chem. 1999;274:9836–9842. doi: 10.1074/jbc.274.14.9836. [DOI] [PubMed] [Google Scholar]
- 19.Hare J M, Wagner A K, McDonough K A. Independent acquisition and insertion into different chromosomal locations of the same pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis. Mol Microbiol. 1999;31:291–303. doi: 10.1046/j.1365-2958.1999.01172.x. [DOI] [PubMed] [Google Scholar]
- 20.Hofte H, Whiteley H R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaquet F, Hutter R, Luthy P. Specificity of Bacillus thuringiensisdelta-endotoxin. Appl Environ Microbiol. 1987;53:500–504. doi: 10.1128/aem.53.3.500-504.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarrett, P., D. Ellis, and J. A. W. Morgan. 1997. U.S. patent GB97/02284.
- 23.Jouanin L, Bonadebottino M, Girard C, Morrot G, Giband M. Transgenic plants for insect resistance. Plant Sci. 1998;131:1–11. [Google Scholar]
- 24.Kang W, Crook N E, Winstanley D, O'Reilly D R. Complete sequence and transposon mutagenesis of the BamHI J fragment of Cydia pomonellagranulosis virus. Virus Genes. 1997;14:131–136. doi: 10.1023/a:1007917317770. [DOI] [PubMed] [Google Scholar]
- 25.Lindler L E, Plano G V, Burland V, Mayhew G F, Blattner F R. Complete DNA sequence and detailed analysis of the Yersinia pestisKIM5 plasmid encoding murine toxin and capsular antigen. Infect Immun. 1998;66:5731–5742. doi: 10.1128/iai.66.12.5731-5742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsutani S, Ohtsubo H, Maeda Y, Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987;196:445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- 27.Paul V J, Frautschy S, Fenical W, Nealson K H. Antibiotics in microbial ecology: isolation and structure assignment of several new anti-bacterial compounds for insect-symbiotic bacteria Xenorhabdusspp. J Chem Ecol. 1981;7:589–597. doi: 10.1007/BF00987707. [DOI] [PubMed] [Google Scholar]
- 28.Rezsohazy R, Hallet B, Delcour J, Mahillon J. The IS4family of insertion sequences evidence for a conserved transposase motif. Mol Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Seifert H S, Chen E Y, So M, Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83:735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Z, Corbin D R, Greenplate J T, Grenbenok R J, Galbraith D W, Pucell J P. Studies on the mode of action of cholesterol oxidase on insect midgut membranes. Arch Insect Biochem Physiol. 1997;34:429–442. [Google Scholar]
- 32.Tachibana M, Hori H, Suzuki N, Uechi T, Kobayashi D, Iwahana H, Kaya H K. Larvicidal activity of the symbiotic bacterium Xenorhabdus japonicus from the entomopathogenic nematode Steinernema kushidia against Anomala cuprea(Coleoptera:Scarabaeidae) J Invert Pathol. 1996;68:152–159. doi: 10.1006/jipa.1996.0073. [DOI] [PubMed] [Google Scholar]
- 33.Yamanaka S, Hagiwara A, Nishimura Y, Tanabe H, Ishibashi N. Biochemical and physiological characteristics of Xenorhabdus species symbiotically associated with entomopathogenic nematodes including Stemernema kushidai and their pathogenicity against Spodoptera litura(Lepidoptera:Noctuidae) Arch Microbiol. 1992;158:387–393. [Google Scholar]
- 34.Ziegelin G, Scherzinger E, Lurz R, Lanka E. Phage P4 alpha protein is multifunctional with origin recognition, helicase and primase activities. EMBO J. 1993;12:3703–3708. doi: 10.1002/j.1460-2075.1993.tb06045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]