Abstract
Actionable drug–gene pairs relevant to depression treatment include CYP2D6 and CYP2C19 with specific antidepressants. While clinical use of pharmacogenetic testing is growing, little is known about pharmacogenetic testing for depression treatment in managed care. We determined the incidence of single‐gene CYP2D6 and CYP2C19 testing following a new depression episode among US managed care patients, and described characteristics and antidepressant use of patients receiving tests. We used paid medical and pharmacy claims for patients from commercial health plans in the US. For adult patients with a new depression episode from January 1, 2013 to June 30, 2018, we identified covered claims for single‐gene CYP2D6 and CYP2C19 pharmacogenetic tests and antidepressant fills. Fewer than 1% (n = 1795) of the depressed cohort (n = 438,534) received a single‐gene CYP2D6 or CYP2C19 test through their insurance within 365 days of their earliest depression episode. The percentage of patients who received a test nearly tripled from 0.2% in 2013 to 0.5% in 2014 before plateauing at 0.4% from 2014 to 2017. Among the patients who received a single‐gene CYP2D6 or CYP2C19 test and filled an antidepressant within 365 days of their depression diagnosis, up to 30% may have had their initial antidepressant informed by the test result. Our findings describe the use of antidepressants before and after pharmacogenetic testing, which is clinically relevant as pharmacogenomic testing becomes more common in clinical practice. Our study also emphasizes the need for procedure and billing codes that capture multiple‐gene panel tests to be more widely implemented in administrative databases.
Study Highlights
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Major depressive disorder is common and many patients do not respond to first‐line treatment. Examples of actionable drug–gene pairs relevant to antidepressants include CYP2D6 and CYP2C19, yet little is known about the extent of pharmacogenetic testing for depression treatment in managed care.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study sought to address the incidence of single‐gene pharmacogenetic testing following a depression episode among a managed care population, and the characteristics of antidepressant use before and after testing is received.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
Single‐gene CYP2D6 or CYP2C19 testing through insurance was rare. However, among patients who received a single‐gene CYP2D6 or CYP2C19 test and filled an antidepressant after their depression diagnosis, up to 30% may have had their initial antidepressant informed by the test result.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
As precision medicine research in the mental health field continues to evolve, these findings illustrate how real‐world insurance claims data can be leveraged to answer important questions about the clinical utility of pharmacogenetic testing in depression.
INTRODUCTION
Major depressive disorder (MDD) affects over 260 million individuals worldwide, with an estimated lifetime prevalence of 16% in the US. 1 , 2 Treatment of MDD is challenging; a comparative effectiveness review showed that 37% of patients with acute‐phase MDD did not achieve response within 6–12 weeks, and 53% did not achieve remission, following first‐line treatment with a second‐generation antidepressant. 3 In this current era of genomic medicine, there has been increased interest in the extent to which drug response and adverse effects are governed by genetic variation (i.e., pharmacogenetics). 4 , 5
Genetic variants are thought to explain up to 42% of the variability in antidepressant drug response. 6 CYP2D6 and CYP2C19 are key drug metabolizing enzyme genes that play a role in antidepressant pharmacology. CYP2D6 and CYP2C19 variants have been associated with altered metabolism and plasma exposure of certain selective serotonin reuptake inhibitors (SSRIs), serotonin and norepinephrine reuptake inhibitors (SNRIs), and tricyclic antidepressants (TCAs). 7 , 8 , 9 , 10 As a result, the Clinical Pharmacogenetics Implementation Consortium (CPIC) and the Dutch Pharmacogenetics Working Group (DPWG) have published evidence‐based dosing guidelines for several antidepressants. 7 , 8 , 9 , 10 In addition, over 10 antidepressants have pharmacogenetic information related to CYP2D6 or CYP2C19 in US Food and Drug Administration (FDA) labeling. 11 , 12 , 13
The annual proportion of patients taking an antidepressant with published CYP2D6‐ or CYP2C19‐based pharmacogenetic guidelines is high, ranging from 45% to 84%, in the adult primary care setting. 13 In some clinical studies, the use of pharmacogenetic‐guided treatment in depression has been associated with increased treatment efficacy (i.e., symptom remission and treatment response) and increased medication tolerability. 14 , 15 , 16 , 17 , 18 , 19 , 20 However, other findings are inconsistent, with some trials not finding sustained responses or significant improvements in primary outcomes. 17 , 21 Regardless, the International Society of Psychiatric Genetics issued a genetic testing statement which included: “pharmacogenetic testing should be viewed as a decision‐support tool to assist in thoughtful implementation of good clinical care” and “genetic information for CYP2C19 and CYP2D6 would likely be most beneficial for individuals who have experienced an inadequate response or adverse reaction to a previous antidepressant…trial.” 22 , 23 At the same time, clinicians and patients are becoming increasingly familiar with pharmacogenetic testing, particularly for mental health disorders. 24 , 25 , 26
While clinical use of pharmacogenetic testing is growing, little is known about how many patients receive single‐gene pharmacogenetic testing related to depression treatment through their insurance. The objectives of this study were to determine the incidence of single‐gene CYP2D6 and CYP2C19 testing following a new depression episode among a cohort from a US managed care population, and to describe the characteristics and antidepressant fills of patients who received testing.
METHODS
Data source
IQVIA PharMetrics® Plus for Academics was used for this retrospective cohort study. PharMetrics Plus for Academics is a health plan claims database that includes longitudinal paid medical and pharmacy claims for over 100 million unique enrollees across the US. The University of Colorado Skaggs School of Pharmacy and Pharmaceutical Sciences holds a license for a depression extract that includes all patients from PharMetrics Plus for Academics with a diagnosis code indicating depression, or a procedure code or medication fill indicating depression treatment from January 1, 2013 through June 30, 2018. Available information for these patients includes service types and dates, diagnoses, procedures, medications filled, and demographics such as year of birth, gender, region of residence, and type of insurance. Race/ethnicity information is not available. The data are nationally representative in terms of age and gender distributions of patients in the US commercially insured population. 27 This study was approved as exempt by the Colorado Multiple Institutional Review Board.
Study population
We identified a cohort of adult (age ≥ 18 years) patients with a new depression episode based on criteria specified by the National Committee for Quality Assurance's Health Plan and Employer Data and Information Set: (1) ICD‐9 or ICD‐10 diagnosis code indicating MDD (single or recurrent episode), depression not otherwise specified, or dysthymia; (2) no depression diagnoses during the 120 days prior to the depression episode; and (3) no antidepressant fills during the 90 days prior to the depression episode. 28 , 29 Diagnoses could have been recorded during any inpatient, emergency department, or outpatient visit and were not restricted to any specialty or to primary care. Diagnosis codes indicating MDD in partial or full remission were not included in our definition of a new episode because they may indicate the current depression diagnosis is related to a recent episode. For patients with more than one depression episode meeting inclusion criteria, only the earliest episode was included in analyses. We further required patients to have continuous health plan enrollment and continuous medication benefits during the 4 months prior to the new depression episode and the 12 months following the new depression episode. This depressed cohort served as the denominator for our incidence calculations.
We then identified the sub‐cohort of patients from the depressed cohort who received a single‐gene CYP2D6 and/or CYP2C19 pharmacogenetic test through their insurance following their index depression episode. We used Current Procedural Terminology (CPT) procedure codes to determine whether patients received a single‐gene CYP2D6 (CPT code 81226) and/or CYP2C19 (CPT code 81225) pharmacogenetic test within 365 days of their index depression episode. We did not require a patient to have a depression‐related diagnosis on the same claim as the pharmacogenetic test. This sub‐cohort served as the numerator for incidence calculations.
Measures
Receipt of antidepressants
We determined whether patients filled an antidepressant within 365 days of their index depression episode using the Generic Product Identifier (GPI) code and date the medication was filled. The earliest antidepressant filled on or after the new depression episode was considered the initiating antidepressant. Classes of antidepressants included SSRI (GPI code 5816*); SNRI (GPI code 5818*); TCA (GPI code 5820*); monoamine oxidase inhibitors (MAOI; GPI code 5810*); tetracyclic antidepressants (GPI code 5803*); and newer‐generation antidepressants such as bupropion, nefazodone, and trazodone (GPI codes 5830*, 5812*). We also identified fills of specific antidepressants with CPIC guidelines indicating a drug–gene interaction with CYP2D6 or CYP2C19 (see Table 3).
TABLE 3.
Medication switching or discontinuing after a single‐gene CYP2D6 or CYP2C19 test among patients who filled an antidepressant prior to the CYP2D6 (n = 921) or CYP2C19 test (n = 1026)
| Antidepressant medication | Filled antidepressant prior to test | Switched or discontinued after the test a |
|---|---|---|
| N (%) | N (%) | |
| Single‐gene CYP2D6 (N = 921) | ||
| SSRIs | ||
| Paroxetine | 46 (5.0) | 31 (67.4) |
| Fluvoxamine | 5 (0.5) | 4 (80.0) |
| TCAs b | ||
| Amitriptyline | 36 (3.9) | 27 (75) |
| Clomipramine | 1 (0.1) | 1 (100) |
| Doxepin | 16 (1.7) | 13 (81.3) |
| Nortriptyline | 20 (2.2) | 12 (60) |
| Desipramine | 1 (0.1) | 0 (0) |
| Single‐gene CYP2C19 (N = 1026) | ||
| SSRIs | ||
| Citalopram | 125 (12.8) | 76 (60.8) |
| Escitalopram | 98 (9.6) | 65 (66.3) |
| Sertraline | 162 (15.8) | 105 (64.8) |
| TCAs b | ||
| Amitriptyline | 40 (3.9) | 29 (72.5) |
| Clomipramine | 2 (0.2) | 2 (100) |
| Doxepin | 14 (1.4) | 11 (78.6) |
Abbreviations: CPIC, Clinical Pharmacogenetics Implementation Consortium; SNRI, serotonin and norepinephrine reuptake inhibitor; SSRI, selective serotonin reuptake inhibitor; TCA, tricyclic antidepressant.
Switch or discontinue defined as a patient filling the medication at least once prior to their single‐gene CYP2D6 or CYP2C19 test and not filling the same medication following the singe‐gene CYP2D6 or CYP2C19 test; percentage is calculated as the number who switched or discontinued divided by the number of patients who filled that medication prior to the test.
Imipramine is a TCA with CPIC guidelines indicating a drug–gene interaction with CYP2D6 or CYP2C19 but no patients filled imipramine prior to their singe‐gene CYP2D6 or CYP2C19 test.
Pharmacogenetic‐informed antidepressant initiation
For patients with at least one antidepressant fill and a single‐gene CYP2D6 and/or CYP2C19 pharmacogenetic test following their index depression episode, we inferred whether the antidepressant was initiated after availability of the earliest single‐gene CYP2D6 and/or CYP2C19 test result, to be used as a proxy for pharmacogenetic‐informed antidepressant initiation. Based on typical external laboratory turnaround times of 1–3 weeks, we assumed test results would be available 14 days after receipt of the single‐gene CYP2D6 or CYP2C19 test. 30 Therefore, antidepressants filled prior to the date of the earliest single‐gene CYP2D6 and/or CYP2C19 test plus 14 days were considered to have been initiated prior to the test results being available and not informed by the pharmacogenetic test results; antidepressants filled after the earliest single‐gene CYP2D6 and/or CYP2C19 test plus 14 days were considered initiated after the availability of the test results and could have been informed by the pharmacogenetic test results.
Antidepressant switching
To examine potential antidepressant switching after a pharmacogenetic test, we identified and compared classes and specific antidepressants filled prior to and following presumed receipt of single‐gene CYP2D6 and CYP2C19 test results. We considered the following specific antidepressant medications with CPIC guidelines indicating a potential drug–gene interaction: SSRIs paroxetine, fluvoxamine, citalopram, escitalopram, and sertraline; and TCAs amitriptyline, clomipramine, doxepin, nortriptyline, and desipramine.
Patient characteristics
We calculated age in years using year of birth and the index depression episode date. Gender, region of residence (East, Midwest, South, West), and insurance type (commercial, Medicare Risk and Medicaid administered through private carriers [i.e., Managed Medicare/Medicaid], or other/unknown) were also available.
Statistical analysis
We characterized the patients in the depression cohort using descriptive statistics such as counts and percentages for categorical variables, and means, medians, and ranges for continuous variables. We calculated the annual incidence of CYP2D6 and CYP2C19 testing among the depression cohort to observe time trends. Annual incidence was calculated as the total number of patients receiving a CYP2D6 or CYP2C19 test in that year divided by the total number of patients with a new depression episode in that year. SAS version 9.4 was used for data management and statistical calculations. 31
RESULTS
Patient characteristics
We identified almost half a million adults (n = 438,534) with a new depression episode from 2013 to 2017. The percentage of patients in the depression cohort who received a CYP2D6 or CYP2C19 test nearly tripled from approximately 0.2% in 2013 to almost 0.5% in 2014, before attaining a general plateau around 0.4% from 2014 to 2017 (Figure 1). Among the entire depression cohort during the study period, fewer than 1% (n = 1795) received a single‐gene CYP2D6 or CYP2C19 test through their insurance within 365 days of their earliest depression episode (Table 1). More patients received a CYP2C19 test (n = 1726, 0.39%) than a CYP2D6 test (n = 1536, 0.35%). The majority of those who received at least one of these tests (n = 1795) received both (n = 1467, 81.7%). Nearly half of the depression cohort (n = 210,129) filled an antidepressant within 365 days of their earliest depression episode; 59% of patients who received a single‐gene CYP2D6 or CYP2C19 test filled an antidepressant within 365 days of their earliest depression episode. The majority of patients (63%) receiving a single‐gene CYP2D6 or CYP2C19 test had Managed Medicaid/Medicare at the time of their depression episode; among the entire depression cohort, 37% had Managed Medicaid/Medicare.
FIGURE 1.
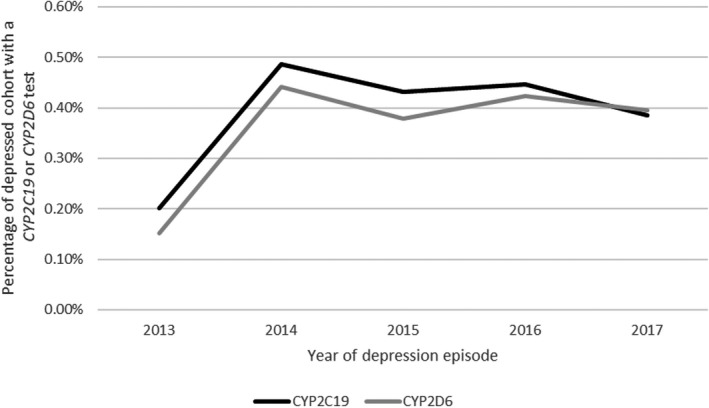
Annual incidence of single‐gene CYP2C19 and CYP2D6 tests; denominators include all patients with an index depression diagnosis within each year and numerators include all patients with a single‐gene CYP2C19 or CYP2D6 test within 365 days of their index depression episode
TABLE 1.
Characteristics of the depressed cohort (N = 438,534) and sub‐cohort who received a single‐gene CYP2D6 or CYP2C19 test within 365 days of their index depression episode (n = 1795)
| Characteristic | Depression cohort (n = 438,534) | Received a single‐gene CYP2D6 or CYP2C19 test within 365 days (n = 1795) |
|---|---|---|
| Age (years) | ||
| Mean (SD), median | 43.2 (15.8), 43 | 42.2 (14.3), 41 |
| Min–max | 18–84 | 18–83 |
| 18–44, n (%) | 158,884 (36.2) | 664 (37.0) |
| 45–64, n (%) | 173,353 (39.5) | 759 (42.3) |
| 65+, n (%) | 106,297 (24.3) | 372 (20.7) |
| Women, n (%) | 291,772 (66.5) | 1159 (64.6) |
| Region, n (%) | ||
| East | 78,980 (18.0) | 139 (7.7) |
| Midwest | 141,898 (32.4) | 585 (32.6) |
| South | 84,476 (19.0) | 540 (30.1) |
| West | 134,183 (30.6) | 531 (29.6) |
| Insurance type, n (%) | ||
| Commercial | 265,696 (60.6) | 638 (35.4) |
| Managed Medicare/Medicaid | 164,588 (37.5) | 1134 (63.2) |
| Other/unknown | 8250 (1.9) | 23 (1.3) |
| Depression treatment within 365 days | ||
| Filled an antidepressant, n (%) | 210,129 (47.9) | 1059 (59.0) |
| Filled another psychotropic, n (%) | 118,466 (27.0) | 959 (53.4) |
| Anxiolytic | 90,094 (20.5) | 750 (41.8) |
| Antipsychotic/mood stabilizer | 27,417 (6.2) | 389 (21.7) |
| Stimulant | 19,719 (4.5) | 143 (8.0) |
Receipt of antidepressants
For patients who received a single‐gene CYP2D6 or CYP2C19 test through their insurance and filled an antidepressant (n = 1059), the average time to first antidepressant fill was 80 days (Table 2). The mean days to first antidepressant fill was 165 days for patients who received their single‐gene CYP2D6 or CYP2C19 test before filling their first antidepressant, in contrast to a mean of 43 days to first antidepressant fill for patients who received their single‐gene CYP2D6 or CYP2C19 test after filling their first antidepressant (results not reported in Table 2). Nearly half (49%) the patients who filled an antidepressant and received a single‐gene CYP2D6 or CYP2C19 test initiated an antidepressant with a CPIC guideline. The most commonly filled class of antidepressants was SSRIs (53%), followed by newer‐generation antidepressants (21%) and SNRIs (14%).
TABLE 2.
Depression treatment within 365 days after an index depression episode among patients who received a single‐gene CYP2D6 or CYP2C19 test (n = 1795)
| Depression treatment characteristic | Received a single‐gene CYP2D6 or CYP2C19 test within 365 days of index depression diagnosis (n = 1795) |
|---|---|
| Filled an antidepressant, n (%) | 1059 (59.0) |
| Days to earliest fill, mean (SD), median | 80.1 (100.6), 29 |
| Filled an antidepressant with a CPIC guideline indicating a potential drug–gene interaction with CYP2D6 or CYP2C19, n (%) a | 519 (49.0) |
| Class of index AD fill, n (%) | |
| SSRIs | 567 (53.5) |
| SNRIs | 153 (14.5) |
| Tricyclic ADs | 69 (6.5) |
| Tetracyclic ADs | 45 (4.2) |
| MAOIs | 1 (0.09) |
| Newer‐generation ADs | 224 (21.1) |
| Timing of antidepressant fill relative to pharmacogenetic test, n (%) | |
| Prior to receipt of the pharmacogenetic test | 601 (56.8) |
| Within 0–14 days after the pharmacogenetic test | 140 (13.2) |
| >14 days after the pharmacogenetic test | 318 (30.0) |
Abbreviations: AD, antidepressant; CPIC, Clinical Pharmacogenetics Implementation Consortium; MAOI, monoamine oxidase inhibitor; SNRI, serotonin and norepinephrine reuptake inhibitor; SSRI, selective serotonin reuptake inhibitor.
Antidepressants with a CPIC guideline indicating a drug–gene interaction with CYP2D6 or CYP2C19 include amitriptyline, clomipramine, doxepin, imipramine, trimipramine, nortriptyline, desipramine, trimipramine, protriptyline, paroxetine, and fluvoxamine, citalopram, escitalopram, sertraline.
Among the patients who received a single‐gene CYP2D6 or CYP2C19 test and filled an antidepressant within 365 days of their depression diagnosis, 30% may have had their initial antidepressant informed by the test result because they did not fill their initial antidepressant until after the earliest single‐gene CYP2D6 or CYP2C19 test results were presumed to have been available (i.e., date of CYP2D6 or CYP2C19 test +14 days). More than half (57%) filled their antidepressant prior to the pharmacogenetic test and 13% filled their antidepressant within 0–14 days following the pharmacogenetic test (Table 2).
Antidepressant switching at the class level
Figure 2 presents potential switching between classes of antidepressants following receipt of the single‐gene pharmacogenetic tests for patients who filled their initial antidepressant prior to their single‐gene CYP2D6 or CYP2C19 test (n = 741). In general, 58% (n = 432) of patients who filled an antidepressant before their single‐gene CYP2D6 or CYP2C19 test filled the same class of antidepressant after their single‐gene CYP2D6 or CYP2C19 test. About half of the patients who received a single‐gene CYP2D6 or CYP2C19 test and filled an SSRI, SNRI, or newer‐generation antidepressant prior to the test filled the same class of antidepressant after the test (52%, 50%, and 51%, respectively). One‐third of the patients who received a test and initiated on a TCA or tetracyclic antidepressant filled the same class of antidepressant after the test (33% and 36%, respectively). However, some movement was seen between classes of antidepressants: 14% of patients initiated on an SSRI switched to an SNRI after the single‐gene CYP2D6 or CYP2C19 test; 15% of patients initiated on an SNRI switched to a TCA after the single‐gene CYP2D6 or CYP2C19 test; almost 30% of patients initiated on a tetracyclic antidepressant switched to an SSRI after the single‐gene CYP2D6 or CYP2C19 test; and almost 30% of patients initiated on a newer‐generation antidepressant switched to an SSRI after the single‐gene CYP2D6 or CYP2C19 test. Regardless of the initial class of antidepressant, 30–46% of patients did not fill any antidepressants after the single‐gene CYP2D6 or CYP2C19 test.
FIGURE 2.
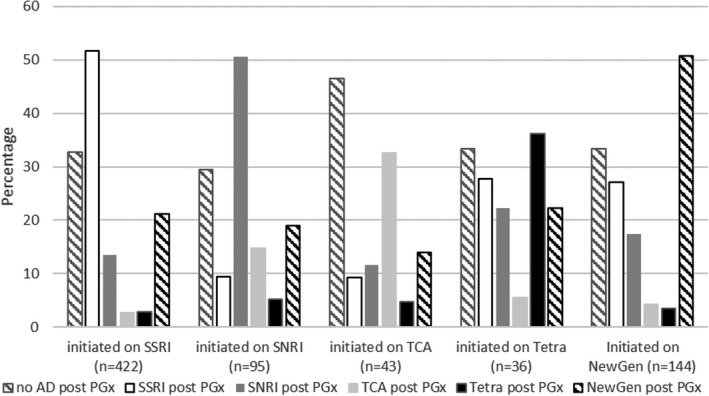
Class of antidepressants filled prior to and following the earliest receipt of a single‐gene CYP2D6 or CYP2C19 test. Note one patient who filled a monoamine oxidase inhibitor (MAOI) prior to their single‐gene CYP2D6 or CYP2C19 test and filled no antidepressant after their test is not represented in the figure. AD, antidepressant; PGx, pharmacogenetic test; NewGen, newer‐generation antidepressant; SNRI, serotonin and norepinephrine reuptake inhibitor; SSRI, selective serotonin reuptake inhibitor; Tetra, tetracyclic antidepressant; TCA, tricyclic antidepressant
Of the patients who initiated an antidepressant prior to their single‐gene CYP2D6 or CYP2C19 test and did not fill any antidepressant after the single‐gene CYP2D6 or CYP2C19 test (n = 247), 32% filled some other type of psychotropic medication: 23% filled an anxiolytic, 9% filled a mood stabilizer, and 5% filled a stimulant (results not presented here in a table or figure).
Antidepressant switching at the medication level
In addition to examining potential changes between antidepressant classes, we also examined potential switching or discontinuation of specific antidepressants with CPIC guidelines related to CYP2D6 and CYP2C19 (Table 3). Switching/discontinuation was defined as at least one fill of the specific medication prior to the single‐gene CYP2D6 or CYP2C19 test and no fills of the same medication after the test. Regardless of the initial medication, at least 60% of patients did not fill the same medication after their single‐gene CYP2D6 or CYP2C19 test. Among patients who received a single‐gene CYP2D6 test (n = 921), 67% of those who filled paroxetine prior to the test did not fill paroxetine again after the test. Among patients who received a single‐gene CYP2C19 test (n = 1026), 66% of those who filled escitalopram prior to the test did not fill escitalopram again after the test, and 61% of those who filled citalopram prior to the test did not fill citalopram again after the test.
DISCUSSION
Using a large database of paid medical and pharmacy claims in the US, we identified a cohort of adult depressed patients and determined whether they received single‐gene CYP2D6 and CYP2C19 pharmacogenetic tests through insurance within 1 year after their index depression episode. Single‐gene testing for these drug metabolizing enzyme genes with the potential to interact with certain antidepressants was not common, seen in fewer than 1% of the nearly half a million adults with a new episode of depression. While low, prevalence of CYP2D6 or CYP2C19 testing nearly tripled from 2013 to 2014, before attaining a plateau from 2014 to 2017. Among patients who received one of these single‐gene pharmacogenetic tests through their insurance, over half (59%) filled an antidepressant during the 1 year following their depression episode.
The low rates of single‐gene CYP2D6 and CYP2C19 testing through insurance in this depressed cohort is likely due to a range of factors, including limited insurance coverage and reimbursement of pharmacogenetic testing during the time period evaluated in this study. 32 , 33 , 34 In 2012, Hresko et al 33 identified 27 pharmacogenomic tests and their corresponding drug indication(s) covered by the top 12 US insurers (including Aetna, Cigna, Humana, UnitedHealth, and Kaiser Foundation Group). While these 27 tests did include CYP2D6 and CYP2C19, none of the coverage was for an antidepressant drug indication. A similar but more recent study reported private health insurance coverage for 34 common gene–drug‐indication groups – no coverage was reported for any gene–drug‐indication groups including CYP2D6 or CYP2C19 for an indication of depression. 34 However, the insurance coverage landscape has changed in recent years. In October 2019, UnitedHealthcare issued coverage rationale indicating the use of pharmacogenetic multigene panel testing to guide therapy decisions is “proven and medically necessary for antidepressants and antipsychotics medication” when a patient has a diagnosis of MDD or generalized anxiety disorder, fails at least one prior medication to treat their condition, and the multigene panel includes no more than 15 relevant genes. 35
From our sub‐cohort of patients who filled an antidepressant and received a single‐gene CYP2D6 or CYP2C19 test, 57% filled their antidepressant before the pharmacogenetic test and 13% filled the initial antidepressant after the test but before the results were likely to have been available. These results suggest their clinicians did not use pharmacogenetic testing to guide initial depression treatment but rather waited until the patients had tried at least one antidepressant. The fact that nearly one‐third of the patients who initiated an antidepressant prior to their single‐gene CYP2D6 or CYP2C19 test then filled some other type of psychotropic medication rather than another antidepressant after the test may be an indication that those patients had more severe or complex depression. We are unaware of any observational studies that inform the real‐world timing of pharmacogenetic testing for depression treatment in practice. However, our findings are consistent with the International Society of Psychiatric Genetics testing statement that “genetic information for CYP2C19 and CYP2D6 would likely be most beneficial for individuals who have experienced an inadequate response or adverse reaction to a previous antidepressant,” and with the UnitedHealthcare coverage requirement that patients fail at least one prior mediation to treat their depression before receiving a pharmacogenetic test. 22 , 35
We also detected switching or discontinuation at the medication level among patients who received a single‐gene CYP2D6 or CYP2C19 test and had filled at least one prior antidepressant. Regardless of the initial antidepressant filled, at least 60% of patients did not fill the same medication after their single‐gene CYP2D6 or CYP2C19 test. Given the limitations of our data, we could not identify whether a patient was on a medication compatible with their genotype/predicted phenotype or whether the result of the single‐gene CYP2D6 or CYP2C19 test guided the treatment choice. However, published clinical trials comparing a pharmacogenomic‐guided depression treatment group to standard care (i.e., not guided by pharmacogenomic test results), such as the GUIDED trial, found the proportion of patients in the intervention group (i.e., pharmacogenomic‐guided treatment) taking a medication compatible with their genotype/predicted phenotype increased significantly from 80% at baseline to 92% at 8 weeks, while the control group (treatment as usual) stayed relatively stable with 76% taking a medication compatible with their genotype/predicted phenotype throughout the study. 16
It is important to acknowledge our study did not capture all patients who received pharmacogenetic testing so we likely underestimated the incidence of single‐gene CYP2D6 or CYP2C19 testing. In addition to those who pay out‐of‐pocket, patients are increasingly using direct‐to‐consumer genetic testing or receiving multiple‐gene panel tests, which have become more common in recent years and are not captured in our insurance claims data. 36 , 37 , 38 , 39 When using insurance claims data for secondary research we are dependent on certain codes being available in the data. Pharmacogenetic testing is captured using the CPT coding system, which until recently did not include specific codes to capture panel testing. While procedure codes now exist for multiple‐gene panel testing, 40 there were none recorded in the depression extract from which our study cohort was drawn. We also could not identify patients who received a single‐gene CYP2D6 or CYP2C19 test and had it coded using a general CPT code (e.g., 81479, Unlisted molecular pathology procedure). Patients in our depressed cohort could have received CYP2D6 or CYP2C19 tests that were not available in our claims data source, resulting in misclassifying those patients as having not received a test; therefore, comparisons between patients labeled as receiving testing and those labeled as not receiving testing were not appropriate.
We could also not determine the timing of testing in relation to medication prescribing (e.g., testing performed before or at the same time a pharmacogenetically‐relevant medication was prescribed) because we only know the date the medication was filled. We were also not able to capture antidepressants filled outside of insurance (i.e., self‐pay). Finally, the PharMetrics Plus for Academics is a Health Plan Claims database nationally representative in terms of age and gender distributions of patients in the US commercially insured population, but we cannot generalize our findings to populations outside US managed health care systems.
This is the first study we are aware of that has examined demographic and antidepressant treatment characteristics of depressed patients who have received single‐gene CYP2D6 or CYP2C19 tests covered by their insurance. While we were only able to consider single‐gene pharmacogenetic tests, we were able to include a large cohort of patients with a depression episode and evaluate those who received a test that could have been used to inform their depression treatment. Our findings describe the use of antidepressants before and after pharmacogenetic testing, which is clinically relevant as pharmacogenomic testing becomes more common in clinical practice. Our study also emphasizes the need for procedure and billing codes that capture multiple‐gene panel tests to be more widely implemented in administrative databases. As precision medicine research in the mental health field continues to evolve, insurance claims are a source of real‐world data that can be leveraged to answer important questions about the clinical utility of pharmacogenetic testing in depression.
CONFLICT OF INTEREST
The authors declare no competing interests for this work.
AUTHOR CONTRIBUTIONS
H.D.A., T.M.T., D.P.K., K.R.C., N.M., and C.L.A. wrote the manuscript. H.D.A., T.M.T., and C.L.A. designed the research. H.D.A. performed the research. H.D.A. analyzed the data.
ACKNOWLEDGMENTS
The statements, findings, conclusions, views, and opinions contained and expressed in this article are based in part on data obtained under license from IQVIA™. Source: IQVIA PharMetrics® Plus for Academics (formerly known as Legacy PharMetrics), January 1, 2007–September 30, 2017, IQVIA™. All Rights Reserved. The statements, findings, conclusions, views, and opinions contained and expressed herein are not necessarily those of IQVIA™ or any of its affiliated or subsidiary entities.
Anderson HD, Thant TM, Kao DP, et al. Pharmacogenetic testing among patients with depression in a US managed care population. Clin Transl Sci. 2022;15:1644‐1653. doi: 10.1111/cts.13279
Funding information
No funding was received for this work.
REFERENCES
- 1. James SL, Abate D, Abate KH, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet. 2018;392(10159):1789‐1858. doi: 10.1016/S0140-6736(18)32279-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kessler RC, Berglund P, Demler O, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS‐R). JAMA. 2003;289(23):3095‐3105. doi: 10.1001/jama.289.23.3095 [DOI] [PubMed] [Google Scholar]
- 3. Gartlehner G, Hansen RA, Morgan LC, et al. Comparative benefits and harms of second‐generation antidepressants for treating major depressive disorder: an updated meta‐analysis. Ann Intern Med. 2011;155(11):772‐785. doi: 10.7326/0003-4819-155-11-201112060-00009 [DOI] [PubMed] [Google Scholar]
- 4. Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526(7573):343‐350. doi: 10.1038/nature15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. Lancet. 2019;394(10197):521‐532. doi: 10.1016/s0140-6736(19)31276-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tansey KE, Guipponi M, Hu X, et al. Contribution of common genetic variants to antidepressant response. Biol Psychiatry. 2013;73(7):679‐682. doi: 10.1016/j.biopsych.2012.10.030 [DOI] [PubMed] [Google Scholar]
- 7. Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacy Promotion (KNMP) . Accessed 29 October 2020. https://www.knmp.nl/downloads/pharmacogenetic‐recommendations‐august‐2020.pdf
- 8. Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127‐134. doi: 10.1002/cpt.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37‐44. doi: 10.1002/cpt.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte‐an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662‐673. doi: 10.1038/clpt.2011.34 [DOI] [PubMed] [Google Scholar]
- 11. Hicks JK, Bishop JR, Gammal RS, et al. A call for clear and consistent communications regarding the role of pharmacogenetics in antidepressant pharmacotherapy. Clin Pharmacol Ther. 2020;107(1):50‐52. doi: 10.1002/cpt.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. U.S. Food and Drug Administration . Accessed 29 October 2020. https://www.fda.gov/drugs/science‐and‐research‐drugs/table‐pharmacogenomic‐biomarkers‐drug‐labeling
- 13. Jessel CD, Mostafa S, Potiriadis M, Everall IP, Gunn JM, Bousman CA. Use of antidepressants with pharmacogenetic prescribing guidelines in a 10‐year depression cohort of adult primary care patients. Pharmacogenet Genomics. 2020;30(7):145‐152. doi: 10.1097/fpc.0000000000000406 [DOI] [PubMed] [Google Scholar]
- 14. Bousman CA, Arandjelovic K, Mancuso SG, Eyre HA, Dunlop BW. Pharmacogenetic tests and depressive symptom remission: a meta‐analysis of randomized controlled trials. Pharmacogenomics. 2019;20(1):37‐47. doi: 10.2217/pgs-2018-0142 [DOI] [PubMed] [Google Scholar]
- 15. Bradley P, Shiekh M, Mehra V, et al. Improved efficacy with targeted pharmacogenetic‐guided treatment of patients with depression and anxiety: a randomized clinical trial demonstrating clinical utility. J Psychiatr Res. 2018;96:100‐107. doi: 10.1016/j.jpsychires.2017.09.024 [DOI] [PubMed] [Google Scholar]
- 16. Greden JF, Parikh SV, Rothschild AJ, et al. Impact of pharmacogenomics on clinical outcomes in major depressive disorder in the GUIDED trial: a large, patient‐ and rater‐blinded, randomized, controlled study. J Psychiatr Res. 2019;111:59‐67. doi: 10.1016/j.jpsychires.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 17. Pérez V, Salavert A, Espadaler J, et al. Efficacy of prospective pharmacogenetic testing in the treatment of major depressive disorder: results of a randomized, double‐blind clinical trial. BMC Psychiatry. 2017;17(1):250. doi: 10.1186/s12888-017-1412-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rosenblat JD, Lee Y, McIntyre RS. The effect of pharmacogenomic testing on response and remission rates in the acute treatment of major depressive disorder: a meta‐analysis. J Affect Disord. 2018;241:484‐491. doi: 10.1016/j.jad.2018.08.056 [DOI] [PubMed] [Google Scholar]
- 19. Singh AB. Improved antidepressant remission in major depression via a pharmacokinetic pathway polygene pharmacogenetic report. Clin Psychopharmacol Neurosci. 2015;13(2):150‐156. doi: 10.9758/cpn.2015.13.2.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winner JG, Carhart JM, Altar CA, Allen JD, Dechairo BM. A prospective, randomized, double‐blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16(89):219‐227. [PubMed] [Google Scholar]
- 21. Thase ME, Parikh SV, Rothschild AJ, et al. Impact of pharmacogenomics on clinical outcomes for patients taking medications with gene–drug interactions in a randomized controlled trial. J Clin Psychiatry. 2019;80(6):19m12910. [DOI] [PubMed] [Google Scholar]
- 22. International Society for Psychiatric Genetics (ISPG) . Genetic testing and psychiatric disorders: a Statement from the International Society of Psychiatric Genetics. Accessed 29 October 2020. https://ispg.net/genetic‐testing‐statement/
- 23. Bousman CA, Bengesser SA, Aitchison KJ, et al. Review and consensus on pharmacogenomic testing in psychiatry. Pharmacopsychiatry. 2021;54(01):5‐17. [DOI] [PubMed] [Google Scholar]
- 24. Haga SB, Mills R, Moaddeb J, Allen Lapointe N, Cho A, Ginsburg GS. Patient experiences with pharmacogenetic testing in a primary care setting. Pharmacogenomics. 2016;17(15):1629‐1636. doi: 10.2217/pgs-2016-0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lemke AA, Hutten Selkirk CG, Glaser NS, et al. Primary care physician experiences with integrated pharmacogenomic testing in a community health system. Pers Med. 2017;14(5):389‐400. doi: 10.2217/pme-2017-0036 [DOI] [PubMed] [Google Scholar]
- 26. Liko I, Lai E, Griffin RJ, Aquilante CL, Lee YM. Patients' perspectives on psychiatric pharmacogenetic testing. Pharmacopsychiatry. 2020;53:256‐261. doi: 10.1055/a-1183-5029 [DOI] [PubMed] [Google Scholar]
- 27. IQVIA . IQVIA PharMetrics Plus for Academics Users Guide & Data Dictionary . 2017.
- 28. National Committee for Quality Assurance . HEDIS® Volume 2: Technical Specifications . 2004.
- 29. Scholle S. NCQA behavioral health measurement efforts. J Manag Care Pharm. 2005;11(3):S9‐S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bousman CA, Hopwood M. Commercial pharmacogenetic‐based decision‐support tools in psychiatry. Lancet Psychiatry. 2016;3(6):585‐590. doi: 10.1016/s2215-0366(16)00017-1 [DOI] [PubMed] [Google Scholar]
- 31. The SAS System for Windows Version 9.4. SAS Institute; 2016. [Google Scholar]
- 32. Cohen J, Wilson A, Manzolillo K. Clinical and economic challenges facing pharmacogenomics. Pharmacogenomics J. 2013;13(4):378‐388. doi: 10.1038/tpj.2011.63 [DOI] [PubMed] [Google Scholar]
- 33. Hresko A, Haga SB. Insurance coverage policies for personalized medicine. J Pers Med. 2012;2(4):201‐216. doi: 10.3390/jpm2040201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park SK, Thigpen J, Lee IJ. Coverage of pharmacogenetic tests by private health insurance companies. J Am Pharm Assoc. 2020;60:352‐356. doi: 10.1016/j.japh.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 35.UnitedHealthcare Pharmacogenetic Testing Policy Number: 2020T0587H. Accessed March 3, 2020. https://www.uhcprovider.com/content/dam/provider/docs/public/policies/comm‐medical‐drug/pharmacogenetic‐testing.pdf
- 36. Allyse MA, Robinson DH, Ferber MJ, Sharp RR. Direct‐to‐consumer testing 2.0: emerging models of direct‐to‐consumer genetic testing. Mayo Clin Proc. 2018;93(1):113‐120. doi: 10.1016/j.mayocp.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 37. Bousman CA, Zierhut H, Muller DJ. Navigating the labyrinth of pharmacogenetic testing: a guide to test selection. Clin Pharmacol Ther. 2019;106(2):309‐312. doi: 10.1002/cpt.1432 [DOI] [PubMed] [Google Scholar]
- 38. Phillips KA, Deverka PA, Hooker GW, Douglas MP. Genetic test availability and spending: where are we now? Where are we going? Health Affairs (Project Hope). 2018;37(5):710‐716. doi: 10.1377/hlthaff.2017.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vo TT, Bell GC, Owusu Obeng A, Hicks JK, Dunnenberger HM. Pharmacogenomics implementation: considerations for selecting a reference laboratory. Pharmacotherapy. 2017;37(9):1014‐1022. doi: 10.1002/phar.1985 [DOI] [PubMed] [Google Scholar]
- 40.UnitedHealthcare Pharmacogenetic Testing Policy Number: 2022T0587J. Accessed May 19, 2021. https://www.uhcprovider.com/content/dam/provider/docs/public/policies/comm‐medical‐drug/pharmacogenetic‐testing.pdf


