Abstract
Cisplatin is effective against many types of carcinoma. However, a high rate of renal damage is a clinical problem. Thus, there is a need to establish a method to prevent it. Although various compounds have been reported to be effective against cisplatin‐induced renal injury, there are no examples of their clinical application. Therefore, we attempted to search for prophylactic agents with a high potential for clinical application. We used Cascade Eye to identify genes that are altered during cisplatin‐induced renal injury, Library of Integrated Network‐based Cellular Signatures (LINCS) to identify drugs that inhibit changes in gene expression, and a large database of spontaneous adverse drug reaction reports to identify drugs that could prevent cisplatin‐induced kidney injury in clinical practice. In total, 10 candidate drugs were identified. Using the US Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS), we identified drugs that reduce cisplatin‐induced kidney injury. Fenofibrate was selected as a candidate drug to prevent cisplatin‐induced kidney injury based on the FAERS analysis. A model was used to evaluate the efficacy of fenofibrate against cisplatin‐induced renal injury. Studies using HK2 cells and mouse models showed that fenofibrate significantly inhibited cisplatin‐induced renal injury but did not inhibit the antitumor effect of cisplatin. Fenofibrate is a candidate prophylactic drug with high clinical applicability for cisplatin‐induced renal injury. Analysis of data from multiple big databases will improve the search for novel prophylactic drugs with high clinical applicability. For the practical application of these findings, evaluation in prospective controlled trials is necessary.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Current coping strategies in clinical practice do not completely prevent cisplatin‐induced renal injury, warranting development of supportive care.
WHAT QUESTION DID THIS STUDY ADDRESS?
Many drugs and compounds have been reported to be effective against cisplatin‐induced renal injury in basic in vitro and in vivo studies, but none have been applied clinically. In this study, we used medical big data to identify candidate prophylactic drugs with high potential for clinical application.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study used an artificial intelligence tool called Cascade Eye and two large medical information databases, Library of Integrated Network‐based Cellular Signatures (LINCS) and US Food and Drug Administration Adverse Event Reporting System (FAERS), to identify candidate drugs that reduce cisplatin‐induced kidney injury and found that the candidate drug fenofibrate has the potential to prevent cisplatin‐induced kidney injury.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
The developed methods can be applied to the development of prophylactic drugs against various adverse drug reactions and will have an impact on future clinical pharmacology and translational research.
INTRODUCTION
Cisplatin is a platinum‐based anticancer drug used in the chemotherapy of several malignant solid tumors, including lung, bladder, and head and neck cancer. 1 It binds to purine bases in intracellular DNA and forms cross‐links to inhibit DNA replication and transcription, thereby exerting an antitumor effect. However, cisplatin causes numerous side effects, such as acute kidney injury, hearing impairment, nausea and vomiting, and myelosuppression. In particular, acute kidney injury occurs in ~30% of patients treated with cisplatin, 2 restricting further cisplatin administration. Acute renal injury may lead to chronic tubulointerstitial fibrosis or irreversible chronic tubulopathy, possibly resulting in chronic renal failure. 3 Additionally, hypomagnesemia is frequently observed during cisplatin administration because of increased renal excretion and gastrointestinal toxicity. 4 Although various platinum‐based drugs have been developed to reduce side effects, cisplatin remains widely used in clinical practice. To prevent cisplatin‐induced nephrotoxicity, the administration of supplemental fluids such as saline (more than 3 L/day) and diuretic medication is recommended. 4 However, renal injury cannot be completely prevented, and further development of supportive care is required. Although basic experiments have identified a number of candidates for treating cisplatin‐induced renal injury, none have been commercialized. 5 In addition to pharmacodynamic safety, the drug must be safe for use in combination with cisplatin or other anticancer drugs used with cisplatin without affecting efficacy. Therefore, a drug that is determined to be effective in basic experiments may not be useful in clinical practice. Reverse translational research can be used to search for effective drugs in the clinical setting and verify their efficacy and mechanism of action in basic experiments.
Drug repositioning, a method of discovering new effects in an existing approved drug and developing that drug as a treatment for a different disease, was used to find a preventive medication for cisplatin‐induced kidney injury. As information on safety and pharmacokinetics in humans has been accumulated, repositioned drugs can be rapidly applied clinically. Even when basic research has shown some degree of safety and efficacy, ~90% of drugs are abandoned for further analysis at the clinical trial stage. 6 Thus, drug development is a time‐consuming and labor‐intensive process. Drug repositioning is gaining worldwide attention to resolve this problem. The number of papers related to drug repositioning has increased considerably (~160 times) over the past 20 years and has tripled since 2016. Large‐scale medical information databases have been constructed to provide information on changes in gene expression and adverse drug reactions, making it possible to reuse drugs. We used the US Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) to identify prophylactic drugs against cisplatin‐induced renal injury among existing approved drugs. FAERS collects and publishes voluntary reports of adverse events reported worldwide. With over 14 million reports, it is the largest database of its kind. Additionally, the Library of Integrated Network‐based Cellular Signatures (LINCS) program is a drug discovery tool that simulates the changes in the expression of ~20,000 genes in response to chemical substances using human cell lines. It has been applied to research on the development of novel therapeutics. 7 , 8 Specifically, it has been used in many studies, including our own, to identify drugs that can prevent adverse events of high‐risk drugs. 9 , 10 , 11 However, the analysis of big data alone is insufficient as the evidence of efficacy. In this study, we used big data analysis to identify candidate drugs to prevent cisplatin‐induced renal injury and examined their efficacy and safety in cells and animals.
MATERIALS AND METHODS
Data mining‐based selection to identify a network of genes involved in preventing kidney damage
Cascade Eye, developed by FRONTEO Inc., was used to extract a list of genes with altered expression upon renal injury. Cascade Eye is a natural language processing‐based artificial intelligence (AI) tool that can extract all genes related to a particular disease or condition that appears in published papers. It can also identify genes that are causative or responsive to the disease or condition. Cascade Eye identified 20 genes that were responsive to cisplatin‐induced renal injury (Table S1).
Selection of prophylactic drug candidates using the National Institutes of Health gene expression database
In Cascade Eye, we identified 20 genes that respond to cisplatin‐induced renal injury and further analyzed these 20 genes with LINCS L1000CDS2. Among these, genes associated with the promotion of cisplatin‐induced acute kidney injury were classified as “upregulated genes,” and genes associated with the suppression of cisplatin‐induced acute kidney injury were classified as “downregulated genes” based on a literature search. These genes were entered into LINCS L1000CDS2, a web‐based search application. The top 50 compounds causing gene expression opposite to the entered pattern were searched in the order of the similarity score of the expression pattern. Among the top 50 compounds in the search results, compounds that were approved as drugs by the FDA were selected as candidate prophylactic drugs.
Effect of concomitant drugs using FAERS
From January 2007 to March 2017, there were 7,738,415 spontaneous adverse event reports that were submitted to the FAERS, which were downloaded from the FDA website (https://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/ucm082193.htm; data accessed on September 2020). 12 Duplicate data were excluded in accordance with the FDA recommendations, and the remaining 6,994,117 reports were used for analysis. MySQL software (version 5.7.21) was used to build a database that integrated the FAERS data, and R version 3.2.1 (R Foundation for Statistical Computing, Vienna, Austria) was used for statistical analyses.
Adverse events were designated according to the 47 extracted terms from the “acute renal failure (SMQ 20000003)” group, which is based on the Medical Dictionary for Regulatory Activities/J version 23.1, International Glossary of Pharmaceutical Terms of International Council for Harmonization of Technical Requirements for Pharmaceuticals for Human Use (Table S1).
The risk of adverse events was assessed using the reporting odds ratio (ROR) and 95% confidence interval (CI). Patients administered with cisplatin were classified into four groups: patients who (1) used drug A and reported acute renal failure, (2) used drug A and did not report acute renal failure, (3) did not use drug A and reported acute renal failure, and (4) did not use drug A and did not report acute renal failure. Based on the following equation, the ROR and 95% CI were calculated.
All tests were two‐tailed, and results with p values < 0.05 were considered statistically significant.
Animal model of cisplatin‐induced nephrotoxicity
All animal experiments were performed in accordance with the ARRIVE guidelines. All experimental procedures were performed in accordance with the guidelines of the Animal Research Committee of Tokushima University Graduate School, and the protocol was approved by the Institutional Review Board of Tokushima University Graduate School for Animal Protection (Permit Number: T30‐85). Nine‐to‐10‐week‐old C57BL/6J male mice (weighing 24–27 g) were purchased from Nippon CLEA (Tokyo, Japan) and maintained with ad libitum access to water and food (type NMF; Oriental Yeast, Tokyo, Japan). The relative humidity in the breeding room was 50 ± 10%, and the room temperature was 26 ± 1°C, with a 12‐h light/dark cycle (lights on at 8:00, lights off at 20:00). The mouse model of cisplatin‐induced renal injury was generated following a previously described method. 13
The mice were randomly divided into eight groups (n = 7–9 per group): vehicle‐injected, cisplatin‐injected, and fenofibrate (30, 100, and 300 mg/kg) or bezafibrate (30, 100, and 300 mg/kg) administered to the cisplatin‐injected group. The mice were intraperitoneally injected with cisplatin (15 mg/kg) or vehicle (saline). Thirty minutes before cisplatin injection, the mice were administered the prophylactic drug candidates or vehicle (0.1% tween20 aqueous solution); 72 h after cisplatin injection, experimental mice were anesthetized, and samples (serum, urine, and kidney) were collected for subsequent analysis. Anesthesia was induced by the inhalation of isoflurane under a 4% diluted vaporizer setting (vaporized in oxygen 1 L/min) and maintained with isoflurane under a 2% vaporizer setting. The isoflurane was delivered via a small face mask. The doses were based on those used in previous animal studies of these candidates as dyslipidemia agents. 14 , 15 , 16 , 17
Determination of blood and urine creatinine and blood urea nitrogen levels
The levels of blood urea nitrogen (BUN), serum creatinine (serum Cr), and urine creatinine (urine Cr) in the serum and urine samples collected 72 h after cisplatin administration were determined by Oriental Yeast Industries (Shiga, Japan). Creatinine clearance (Ccr) was calculated as follows:
 |
Real‐time polymerase chain reaction using kidney tissues from mice with cisplatin‐induced kidney injury
The kidneys were collected from mice with cisplatin‐induced kidney injury. RNA was extracted from the kidney samples using an RNA extraction solution (NIPPON GENE, Tokyo, Japan) according to the manufacturer’s instructions. cDNA was reverse‐transcribed using the PrimeScript RT Reagent kit (Takara Bio, Shiga, Japan) and a PCR Thermal Cycler Dice (Takara Bio). The cDNA from each sample was mixed with forward and reverse primers and THUNDERBIRD SYBR qPCR Mix (Toyobo, Osaka, Japan); polymerase chain reaction (PCR) was performed using an Applied Biosystems StepOnePlus system (Applied Biosystems, Foster City, CA). Fold changes in gene expression relative to those of the control group were evaluated using mouse Gapdh as an internal standard. The primer sets used for PCR are presented in Table S1. All experiments were performed in triplicate. All data were analyzed using CFX Manager software (Bio‐Rad Laboratories, Hercules, CA). 18
Histological analysis
Renal tubular injury was assessed as previously described. 19 Kidney samples were fixed with 4% paraformaldehyde and embedded in paraffin. The samples were cut into 4‐μm sections, which were stained with hematoxylin and eosin. Tubular damage was scored blindly by three or more researchers trained in tissue assessment, other than the experimenter, according to the percentage of damage (tubular necrosis, brush border loss, cast formation, tubular dilation, and tubular degeneration), as follows: 0, normal; 1, <25%; 2, 25–50%; 3, 50–75%; and 4, >75%. Ten random microscopic fields per kidney section were used for quantification using the BX53 microscope (Olympus, Tokyo, Japan).
Cell culture
We used HK2 cells (ATCC CRL‐2190; human proximal tubule cell: obtained from American Type Culture Collection, Manassas, VA) to investigate the effect of fenofibrate on cisplatin‐induced cell death. To test the effect of fenofibrate on the anticancer effect of cisplatin, we used mouse Lewis lung carcinoma (LLC) cells and Colon 26 murine colon carcinoma cells. The cells were maintained and subcultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin in an atmosphere of 95% air/5% CO2 at 37°C, according to the culture protocol. The cells were passaged at 80% confluence, and experiments were performed using cells with passages between 5 and 15.
Cell viability assay
Cell viability was assessed using a Cell Counting Kit‐8 (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. The cells were seeded into 96‐well plates at a concentration of 5 × 104 cells per well and incubated at 37°C for 24 h. Then, they were cultured in media with or without 50 μM cisplatin for 24 h. Fenofibrate (1, 10, and 100 μM) or bezafibrate (1, 10, and 100 μM) was used for treatment with or without cisplatin administration. Additionally, 2 μM PPARα inhibitor GW6471 was used. Fenofibrate, bezafibrate, and GW6471 were dissolved in dimethylsulfoxide (DMSO) and adjusted to a final concentration of 0.2% DMSO. Cell viability was assessed by measuring the absorbance of WST‐8 formazan at 450 nm using a plate reader (Model680 microplate reader; Bio‐Rad Laboratories).
Statistical analysis
To compare three or more groups, one‐way analysis of variance (ANOVA) was performed. Tukey’s test was performed as a post hoc analysis. R version 3.2.1 for Windows was used for statistical analyses. Results with a two‐tailed p < 0.05 were considered statistically significant.
RESULTS
Identification of agents capable of preventing cisplatin‐induced kidney injury
First, we extracted genes whose expression was altered by cisplatin administration. Using an AI tool called Cascade Eye, we identified 20 genes whose expression was altered during cisplatin‐induced kidney injury. Among these, we classified seven associated with the promotion of acute kidney injury as “upregulated genes” and 13 associated with the suppression of acute kidney injury as “downregulated genes” by literature search (Table S1).
We then used the L1000CDS2 search engine to identify 10 approved drugs that counteract the altered gene expression in cisplatin (Table 1).
TABLE 1.
Drug candidate selection by LINCS analysis
| Cyclosporin A | Menadione |
| Diflorasone diacetate | Metoprolol |
| Diltiazem | Niclosamide |
| Fenofibrate | Vinpocetine |
| Gemcitabine | Vorinostat |
Note: We identified compounds that promote the expression of suppressor genes and inhibit the expression of accelerator genes in cisplatin‐induced renal injury by LINCS analysis. Ten approved drugs were extracted.
Abbreviation: LINCS, Library of Integrated Network‐based Cellular Signatures.
FAERS analysis results
We used FAERS to examine the effects of these 10 drugs on cisplatin‐induced renal injury. We included 28,755 patients treated with cisplatin in the analysis after excluding duplicate data from 6,994,117 reports. The RORs for fenofibrate, diltiazem, metoprolol, vorinostat, and cyclosporine were 0.39, 0.93, 1.02, 1.10, and 1.90, respectively (Table 2). Of the 10 drugs extracted by LINCS, only the five drugs mentioned above had reports of concomitant use of cisplatin. Based on these results, fenofibrate was identified as a novel candidate for prophylaxis.
TABLE 2.
Effect of prophylactic drug candidates on the occurrence of cisplatin‐induced ARF using the FAERS data analysis
| Drug | ARF (%) without the drug | ARF (%) with the drug | ROR (95% CI) | p Value |
|---|---|---|---|---|
| Fenofibrate | 8.81 (2324/26379) | 3.45 (2/58) | 0.39 (0.05–1.48) | 0.236 |
| Diltiazem | 7.54 (2009/26662) | 6.98 (6/86) | 0.93 (0.33–2.10) | 1 |
| Metoprolol | 7.53 (1994/26474) | 7.66 (21/274) | 1.02 (0.33–2.10) | 0.909 |
| Vorinostat | 8.80 (2320/26375) | 9.68 (6/62) | 1.10 (0.39–2.54) | 0.822 |
| Cyclosporine | 8.78 (2315/26371) | 16.67 (11/66) | 1.90 (0.90–3.63) | 0.056 |
Note: Among the 10 drugs extracted by the LINCS analysis, there were five drugs for which FAERS analysis was possible. Among these, fenofibrate had an ROR <1 in the FAERS analysis, and the number of reports of cisplatin‐induced renal injury was significantly lower than reports without fenofibrate. Statistical analysis was conducted using Fisher’s exact test.
Abbreviations: ARF, acute renal failure; CI, confidence interval; FAERS, US Food and Drug Administration Adverse Event Reporting System; LINCS, Library of Integrated Network‐based Cellular Signatures; ROR, reporting odds ratio.
Effects of fenofibrate or bezafibrate on cisplatin‐induced kidney injury in mice
The effect of fenofibrate or bezafibrate on cisplatin‐induced renal injury was investigated in mice. In cisplatin‐treated mice, body weight was lower than that in solvent‐treated mice; however, kidney weight was unchanged (Table 3). Cisplatin‐treated mice showed renal damage with increased mRNA expression of Kim‐1 and Lcn2, markers of renal tubular damage, increased plasma BUN, and significantly decreased creatinine clearance compared with the solvent‐treated group (Table 3, Figure 1a,b). The combination of cisplatin and fenofibrate significantly ameliorated the cisplatin‐induced renal impairment in a dose‐dependent manner (Table 1, Figure 1a,b). In contrast, the combination with bezafibrate did not improve cisplatin‐induced renal injury (Figure 1a,b).
TABLE 3.
Body weight, kidney weight, and renal function in vehicle‐treated mice and cisplatin‐treated mice with or without fenofibrate or bezafibrate
| Vehicle | Feno (300 mg/kg) | Beza (300 mg/kg) | Cisplatin | Cisplatin + Feno (30 mg/kg) | Cisplatin + Feno (100 mg/kg) | Cisplatin + Feno (300 mg/kg) | Cisplatin + Beza (30 mg/kg) | Cisplatin + Beza (100 mg/kg) | Cisplatin + Beza (300 mg/kg) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Initial body weight, g | 24.4 ± 0.5 | 27.8 ± 0.5 | 24.4 ± 0.7 | 23.9 ± 0.5 | 26.3 ± 0.6 | 26.7 ± 0.5 | 26.8 ± 0.5 | 25.8 ± 0.5 | 25.4 ± 0.5 | 26.1 ± 0.6 |
| Post body weight, g | 24.5 ± 0.5 | 26.6 ± 0.5 | 23.1 ± 0.8 | 18.9 ± 0.2† | 20.5 ± 0.7 | 20.9 ± 0.4 | 22.1 ± 0.8 | 22.2 ± 0.8 | 21.4 ± 0.8 | 21.3 ± 0.5 |
| Kidney weight, mg | 167.5 ± 8.4 | 170.8 ± 17.2 | 163.8 ± 6.9 | 140.0 ± 10. | 154.4 ± 5.8 | 155.6 ± 3.9 | 164.3 ± 6.4 | 155.1 ± 6.0 | 164.3 ± 5.1 | 155.6 ± 4.8 |
| Kidney weight/body weight, mg/g | 6.86 ± 0.38 | 6.44 ± 0.73 | 7.11 ± 0.28 | 7.41 ± 0.49 | 7.54 ± 0.32 | 7.45 ± 0.23 | 7.45 ± 0.37 | 6.98 ± 0.17 | 7.73 ± 0.28 | 7.30 ± 0.14 |
| Urine volume, ml | 1.66 ± 0.15 | 1.67 ± 0.17 | 1.52 ± 0.20 | 0.74 ± 0.11† | 0.43 ± 0.05 | 0.63 ± 0.08 | 0.81 ± 0.14 | 0.96 ± 0.12 | 0.89 ± 0.13 | 0.69 ± 0.10 |
| BUN, mg/dl | 19.2 ± 2.3 | 19.3 ± 1.0 | 23.4 ± 1.5 | 132.4 ± 13.2† | 80.9 ± 5.8 | 66.4 ± 5.3* | 43.3 ± 1.6** | 79.2 ± 14.8 | 104.0 ± 28.7 | 83.5 ± 14.4 |
| Ccr, ml/min/kg | 11.59 ± 1.82 | 9.80 ± 1.53 | 8.67 ± 0.53 | 2.14 ± 0.36† | 2.47 ± 0.30 | 3.75 ± 0.72 | 5.53 ± 0.57** | 4.29 ± 0.51* | 4.95 ± 0.59* | 3.94 ± 0.43 |
| BUN Ccr−1 ratio | 2.2 ± 0.4 | 2.1 ± 0.3 | 2.7 ± 0.3 | 86.4 ± 25.9† | 35.6 ± 6.2 | 22.2 ± 6.1* | 8.5 ± 1.0** | 26.4 ± 8.8* | 23.1 ± 5.8* | 22.8 ± 4.0* |
Note: Data are presented as mean ± SEM. † p < 0.05 versus vehicle mice, *p < 0.05 and **p < 0.01 versus cisplatin mice. n = 4–9 in each group.
Abbreviations: Beza, bezafibrate; BUN, blood urea nitrogen; Ccr, creatinine clearance; Feno, fenofibrate.
FIGURE 1.
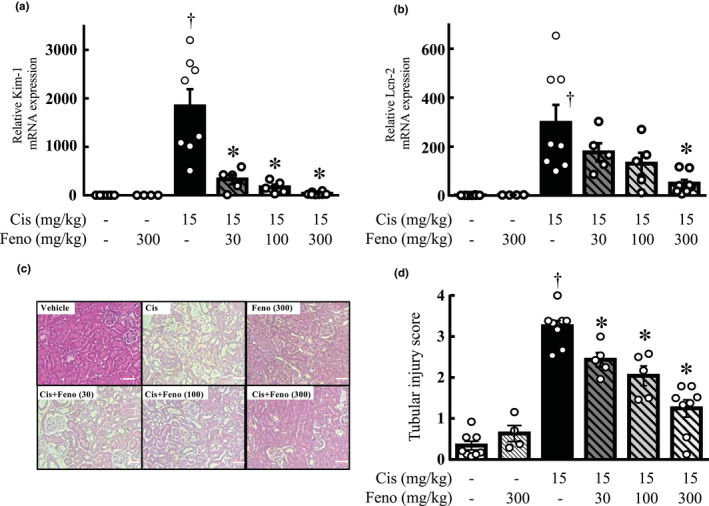
Effect of fenofibrate on cisplatin‐induced nephrotoxicity. (a, b) The mRNA expression levels of kidney injury markers Kim‐1 (a) and Lcn‐2 (b) in the kidneys of mice in each group. (c) Representative hematoxylin and eosin staining (HE) of the kidney section of the control mice, cisplatin‐injected mice with vehicle or fenofibrate. The scale bar indicates 100 μm. (d) Quantitative analysis of renal damage scores. Values are expressed as mean ± SEM. Cis, cisplatin; Feno, fenofibrate. † p < 0.05 versus vehicle mice, *p < 0.01 versus cisplatin mice, n = 4–9 in each group
Histological evaluation
Histological changes in renal tubules of mice were observed by hematoxylin and eosin staining. In cisplatin‐treated mice, the degeneration and destruction of renal tissue were observed, including loss of nuclei, hollowing out of proximal tubular cells, and dilatation of tubules. Mice treated with fenofibrate exhibited less tubular dilation and necrosis of tubular cells than those treated with cisplatin alone (Figure 1c). Tubular dysfunction in kidney sections was scored blindly from 0 to 4 by several researchers. The fenofibrate combination group scored significantly higher than the cisplatin alone group in a dose‐dependent manner (Figure 1d).
Influence of inflammatory cytokines
Quantitative real‐time PCR confirmed that the mRNA expression levels of IL‐1β, IL‐6, and TNF‐α in the kidneys were increased by cisplatin treatment; however, they were significantly lower in the kidneys of the cisplatin and fenofibrate combination group than in those of the cisplatin group (Figure 2).
FIGURE 2.
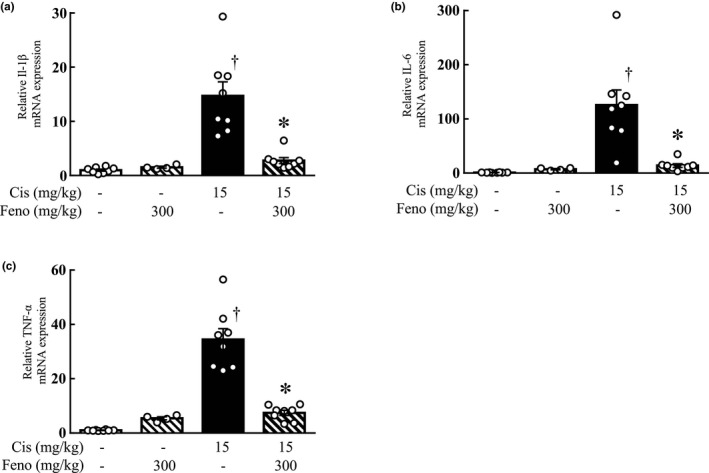
Effect of fenofibrate on inflammatory cytokine expression in the kidneys. The mRNA expression levels of inflammatory cytokines IL‐1β (a), IL‐6 (b), and TNF‐α (c) in the kidneys of mice in each group. Values are expressed as mean ± SEM. Cis, cisplatin; Feno, fenofibrate. † p < 0.05 versus vehicle mice, *p < 0.01 versus cisplatin mice, n = 4–9 in each group
Involvement of PPARα in the inhibitory effect of fenofibrate on cisplatin‐induced cytotoxicity
The effect of fenofibrate and bezafibrate on cisplatin‐induced cell death was investigated using HK2 cells. Twenty‐four hour after cisplatin (50 μM) treatment, the cell viability of HK2 cells was significantly lower than that in the vehicle‐treated group. A combination of fenofibrate (1, 10, and 100 μM) significantly increased the cell viability in a dose‐dependent manner compared with the cisplatin‐treated group (Figure 3a). In contrast, the combination of bezafibrate (1, 10, and 100 μM) exhibited no change in cell viability (Figure 3b). We further investigated the involvement of PPARα in the inhibitory effect of fenofibrate on cisplatin‐induced cytotoxicity; the addition of GW6471, a PPARα inhibitor, suppressed the improvement of cell viability by fenofibrate (Figure 3c).
FIGURE 3.
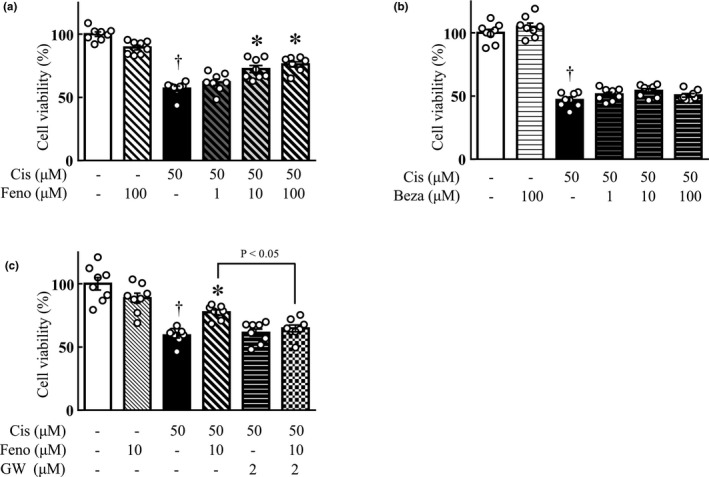
Effect of fibrates on cisplatin cytotoxicity using HK2 cells. Cell viability after 24 h of incubation in medium with or without 50 μM cisplatin was calculated as 100% for the vehicle group. (a, b) Fenofibrate (1, 10, 100 μM) a or bezafibrate (1, 10, 100 μM) b was administered simultaneously with cisplatin. (c) The 2 μM of GW6471, a PPARα inhibitor, was used. Values are expressed as mean ± SEM. Cis, cisplatin; Beza, bezafibrate; Feno, fenofibrate; GW, GW6471. † p < 0.05 versus vehicle, *p < 0.01 versus cisplatin, n = 8 in each group
Effect of fenofibrate on the anticancer action of cisplatin
The effect of fenofibrate on the anticancer effect of cisplatin was examined using LLC cells, a mouse lung cancer cell line, and colon‐26 cells, a mouse colon cancer cell line. Twenty‐four h after cisplatin (50 μM) treatment, the cell viability of LLC and colon‐26 cells was significantly lower than that in the vehicle‐treated group. The combined use of fenofibrate (100 μM) did not change the cell viability as compared with the cisplatin‐treated group (Figure 4).
FIGURE 4.
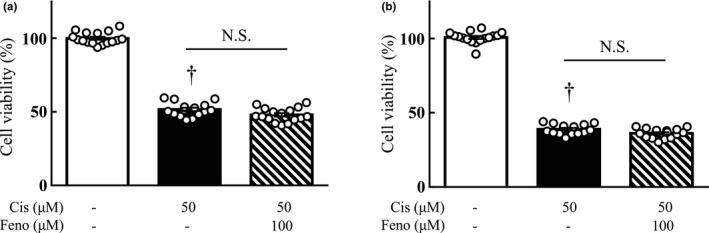
Effect of fibrates on the anticancer effect of cisplatin on tumor cells, LLC (a), Colon‐26 (b). Cell viability after 24 h of incubation in medium with or without 50 μM cisplatin was calculated as 100% for the vehicle group. Fenofibrate (100 μM) was administered simultaneously with cisplatin. Values are expressed as mean ± SEM. Cis, cisplatin; Feno, fenofibrate. † p < 0.05 versus vehicle. N.S. indicates not significant. n = 16 in each group
DISCUSSION
In this study, we searched for candidates for drugs with high clinical potential to reduce cisplatin‐induced renal injury by analyzing using an AI tool called Cascade Eye and two large medical information databases, LINCS and FAERS. Furthermore, basic research using animal models and cells suggest that fenofibrate, not bezafibrate, is a potential prophylactic drug for the prevention of cisplatin‐induced renal injury.
Adverse effects of cisplatin, including renal impairment, are a major clinical challenge as they interfere with the continuation of treatment. Using an AI tool called Cascade Eye, we extracted a group of genes that have been reported to be involved in the mechanism of cisplatin‐induced kidney injury development. Furthermore, using LINCS, we searched for drugs that can reverse the gene expression changes induced by cisplatin and extracted 10 existing approved drugs. We also used FAERS to compare the reported frequency of cisplatin‐induced kidney injury in patients treated with cisplatin with and without the candidate drugs. Fenofibrate achieved an ROR < 1, suggesting that it reduces the incidence of cisplatin‐induced renal injury (Table 2). Although fenofibrate is used as a treatment for dyslipidemia, in the FAERS database, we searched for statins that treat dyslipidemia as well as a similar drug, bezafibrate. Fenofibrate had the smallest reported odds ratio for cisplatin‐induced kidney injury (data not shown). A number of other drugs had odds ratios >1, indicating that dyslipidemia drugs do not uniformly reduce the risk of developing renal injury. Therefore, the improvement of dyslipidemia may not contribute to the improvement of renal impairment by cisplatin. Furthermore, analysis of rhabdomyolysis, a known side effect of fenofibrate, showed that the ROR for rhabdomyolysis was smaller than 1 (data not shown) in patients who used fenofibrate in combination with cisplatin compared to those who used fenofibrate alone. Our data suggest that the combination of cisplatin and fenofibrate may not increase side effects. These results suggest that fenofibrate has a high potential for clinical application as a prophylactic agent against cisplatin‐induced renal injury. It has been reported that PPARα stimulators and fibrates are effective against cisplatin‐induced renal injury in basic in vivo and in vitro studies (PMID: 26536032 and 27193727). However, in vivo studies using mouse models of cisplatin‐induced kidney injury have not compared fenofibrate and bezafibrate, which are fibrates frequently used in clinical practice, and their mechanisms of action are controversial. As fenofibrate was extracted as a candidate drug through big data analysis, we also examined the effect of bezafibrate, a known fibrate drug. The doses of fenofibrate and bezafibrate were determined based on those used in previous studies to treat mice. 20 , 21 , 22 , 23 In addition to BUN and Ccr, the expression levels of Kim‐1 and Lcn‐2 in the kidneys were measured and evaluated histologically as indicators of renal damage. Cisplatin‐induced renal injury is assumed to be caused by proximal tubular cell injury, and just as BUN and Ccr values reflect GFR, Kim‐1, and Lcn‐2 in the kidneys are considered to be suitable indicators of renal injury. In animal experiments, Kim‐1 and Lcn‐2 in the kidneys are significantly increased in cisplatin‐induced kidney injury models. 24 , 25 IL‐6, IL‐1β, and TNF‐α are pro‐inflammatory cytokines; TNF‐α plays an important role in the progression of cisplatin‐induced renal injury and is involved in the release of other pro‐inflammatory cytokines. Previous studies have shown that the release of TNF‐α in renal cells, but not immune cells, is involved in the worsening of renal injury. 26 , 27
Cisplatin‐induced renal injury occurs when cisplatin is taken up by proximal tubular cells in the kidney via the high‐affinity copper transporter 1 and organic cation transporter (OCT2) on the basement membrane side of the cell. 4 Because fenofibrate has been reported to downregulate the renal OCT2 transporter via a PPARα‐independent pathway, 15 renal platinum levels were measured to investigate the effect of fenofibrate on cisplatin accumulation in the kidneys. Consequently, there was no change in the amount of platinum in the kidneys, whole blood, or urine (data not shown).
Cisplatin‐induced renal injury is believed to be caused by cisplatin‐induced oxidative stress and DNA damage, which inhibit the nuclear translocation of PPARα, resulting in ATP depletion, lipotoxicity, and tubular cell damage. The beta‐oxidation of free fatty acids is a major energy‐producing pathway in the kidneys and is inhibited in acute renal failure. The inactivation of PPARα and decreased expression of PPARγ are considered the main causes of the inhibition of fatty acid oxidation. 17 Additionally, a PPARα agonist (WY‐14643) has been reported to inhibit cisplatin‐induced renal injury via PPARα. 14 Based on these reports, we conducted cell experiments to investigate whether PPARα is involved in the mechanism of the inhibitory effect of fenofibrate on cisplatin‐induced renal injury. The results showed that fenofibrate ameliorated cisplatin‐induced cytotoxicity; however, the addition of GW6471, a PPARα inhibitor, tended to decrease cell viability. In addition, pretreatment with fenofibrate had the same effect as simultaneous treatment (data not shown). Therefore, the mechanism of the inhibition of cisplatin‐induced renal injury by fenofibrate may partly involve the activation of PPARα, which has been reported to have anti‐inflammatory effects. 28
The difference in the effects of fenofibrate and bezafibrate on cisplatin‐induced renal injury may be partly related to the difference in the affinity of fenofibrate and bezafibrate for PPARα. Reportedly, the half‐maximal effective concentration (EC50) values of fenofibrate and bezafibrate for human PPARα are 30 and 50 μM, respectively. 29 However, cell experiment results suggest that the activation of PPARα is only part of the mechanism of the nephroprotective action of fenofibrate. Another possible pathway of fenofibrate is the inhibition of oxidative stress. 30 Previous studies using oxidative stress‐induced ischemic kidney injury models have reported that PPARα agonists reduce the levels of BUN, Ccr, inflammatory cytokines, and serum creatinine, which are exacerbated in mouse models of ischemic kidney injury. 31 , 32 , 33 Fenofibrate has been reported to have antitumor effects in numerous cell and animal experiments alone. 16 , 34 The present cell experiments suggest that fenofibrate does not interfere with the antitumor effect of cisplatin. Previous reports have shown that fenofibrate has a direct antitumor effect in vitro (PMID: 29760790) and an inhibitory effect on tumor growth by inhibiting angiogenesis in vivo (PMID: 18199835). Therefore, it is suggested that in patients with cancer, fenofibrate itself is unlikely to interfere with the therapeutic effect of cisplatin.
There are certain limitations to this study. First, the gene expression recorded in LINCS is based on cell line data. Therefore, rather than using LINCS alone to determine candidate drugs, FAERS was used to evaluate the efficacy of LINCS on the reported rate of cisplatin‐induced renal injury. In fact, FAERS analysis results showed that none of the existing approved drugs extracted from the LINCS except fenofibrate were effective against cisplatin‐induced renal injury events, and these were subsequently excluded as candidate drugs. Second, although FAERS is a database for reporting spontaneous adverse events, it does not include all patients who have used cisplatin or concomitant medications because patients who have not experienced adverse events are not registered. Additionally, some patients who experienced adverse events were not reported. Therefore, the reported incidence of renal injury owing to cisplatin calculated in the FAERS analysis may be different from the actual incidence in clinical practice. Third, the FAERS analysis does not consider the effect of confounding factors; the ROR is calculated based on the number of reports on the use of concomitant medications but is not adjusted for age, gender, or medical history. Moreover, medical history could not be evaluated because it was not listed in FAERS. Because of these limitations, the results of the FAERS review may not accurately reflect the actual patients. However, considering the difficulty of collecting case reports of inconsistent adverse effects, such as renal impairment caused by cisplatin, FAERS analysis results, which contains a large amount of data from actual patients, may be useful in investigating actual clinical practice. Therefore, further evidence, such as multicenter randomized clinical trials, is needed to determine whether fenofibrate is effective in preventing renal injury events in patients receiving cisplatin.
Furthermore, it has recently been reported that the adverse effect of fenofibrate, an elevation in serum creatinine tested at the time of diagnosis of rhabdomyolysis, is transient and reversible. Fenofibrate has also been reported to inhibit proteinuria and may have a renoprotective effect in the long term. However, when high doses of fenofibrate are used in patients using drugs that decrease renal blood flow or in patients with impaired renal function, monitoring of serum creatinine is necessary. 35 Therefore, as fenofibrate‐associated increase in serum creatinine level has been reported, the use of fenofibrate to prevent cisplatin‐induced acute kidney injury should be preceded by the risk–benefit analysis for patients with normal and impaired renal function, and the need for monitoring serum creatinine should be evaluated.
To summarize, this study analyzed an AI tool called Cascade Eye and two large medical information databases, LINCS and FAERS, to identify candidate drugs that reduce cisplatin‐induced kidney injury and found that the candidate drug, fenofibrate, is a drug with high potential for clinical application as a preventive agent for cisplatin‐induced kidney injury events.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
M.G., M.K., and A.M. designed the research. A.M., T.Y., A.Y., T.N., and H.H. performed the research. H.H., K.M., F.A., N.O., T.S., M.C., K.Y., Y.I.I., H.Y., Y.Z., and K.I. analyzed the data; M.K. and M.G. wrote the manuscript.
Supporting information
Table S1
ACKNOWLEDGEMENTS
This research was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant numbers 20K07132, 20H05799, and 20K16077). This study was supported by FRONTEO Inc. and Support Center for Advanced Medical Sciences, Tokushima University Graduate School of Biomedical Sciences.
Kanda M, Goda M, Maegawa A, et al. Discovery of preventive drugs for cisplatin‐induced acute kidney injury using big data analysis. Clin Transl Sci. 2022;15:1664‐1675. doi: 10.1111/cts.13282
Funding information
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant numbers 20 K07132 and 20H05799). The funders had no involvement in the study design, collection, analysis, and interpretation of data; preparation of the report; or the decision to submit the manuscript for publication.
REFERENCES
- 1. Chovanec M, Abu Zaid M, Hanna N, El‐Kouri N, Einhorn LH, Albany C. Long‐term toxicity of cisplatin in germ‐cell tumor survivors. Ann Oncol. 2017;28:2670‐2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Isnard‐Bagnis C, Moulin B, Launay‐Vacher V, Izzedine H, Tostivint I, Deray G. Anticancer drug‐induced nephrotoxicity. Nephrol Ther. 2005;1:101‐114. [DOI] [PubMed] [Google Scholar]
- 3. Arany I, Safirstein RL. Cisplatin nephrotoxicity. Semin Nephrol. 2003;23:460‐464. [DOI] [PubMed] [Google Scholar]
- 4. Karasawa T, Steyger PS. An integrated view of cisplatin‐induced nephrotoxicity and ototoxicity. Toxicol Lett. 2015;237:219‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ali BH, Al Moundhri MS. Agents ameliorating or augmenting the nephrotoxicity of cisplatin and other platinum compounds: a review of some recent research. Food Chem Toxicol. 2006;44:1173‐1183. [DOI] [PubMed] [Google Scholar]
- 6. Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates. Nat Rev Drug Discov. 2004;3:711‐715. [DOI] [PubMed] [Google Scholar]
- 7. Keenan AB, Jenkins SL, Jagodnik KM, et al. The library of integrated network‐based cellular signatures NIH program: system‐level cataloging of human cells response to perturbations. Cell Syst. 2018;6:13‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duan Q, Reid SP, Clark NR, et al. L1000CDS2: LINCS L1000 characteristic direction signatures search engine. NPJ Syst Biol Appl. 2016;2:16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zamami Y, Niimura T, Koyama T, et al. Search for therapeutic agents for cardiac arrest using a drug discovery tool and large‐scale medical information database. Front Pharmacol. 2019;10:1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Izawa‐Ishizawa Y, Imanishi M, Zamami Y, et al. Development of a novel aortic dissection mouse model and evaluation of drug efficacy using in‐vivo assays and database analyses. J Hypertens. 2019;37:73‐83. [DOI] [PubMed] [Google Scholar]
- 11. Okada N, Niimura T, Zamami Y, et al. Pharmacovigilance evaluation of the relationship between impaired glucose metabolism and BCR‐ABL inhibitor use by using an adverse drug event reporting database. Cancer Med. 2019;8:174‐181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sasaoka S, Matsui T, Abe J, et al. Evaluation of the association of hand‐foot syndrome with anticancer drugs using the US Food and Drug Administration adverse event reporting system (FAERS) and Japanese adverse drug event report (JADER) databases. Yakugaku Zasshi. 2016;136:507‐515. [DOI] [PubMed] [Google Scholar]
- 13. Goda M, Kanda M, Yoshioka T, et al. Effects of 5‐HT₃ receptor antagonists on cisplatin‐induced kidney injury. Clin Transl Sci. 2021;14:1906‐1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li S, Gokden N, Okusa MD, Bhatt R, Portilla D. Anti‐inflammatory effect of fibrate protects from cisplatin‐induced ARF. Am J Physiol Renal Physiol. 2005;289:F469‐F480. [DOI] [PubMed] [Google Scholar]
- 15. Asavapanumas N, Kittayaruksakul S, Meetam P, Muanprasat C, Chatsudthipong V, Soodvilai S. Fenofibrate down‐regulates renal OCT2‐mediated organic cation transport via PPARα‐independent pathways. Drug Metab Pharmacokinet. 2012;27:513‐519. [DOI] [PubMed] [Google Scholar]
- 16. Lian X, Wang G, Zhou H, Zheng Z, Fu Y, Cai L. Anticancer properties of fenofibrate: a repurposing use. J Cancer. 2018;9:1527‐1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jang HS, Noh MR, Jung EM, et al. Proximal tubule cyclophilin D regulates fatty acid oxidation in cisplatin‐induced acute kidney injury. Kidney Int. 2020;97:327‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C[T]) method. Methods. 2001;25:402‐408. [DOI] [PubMed] [Google Scholar]
- 19. Nojiri T, Hosoda H, Kimura T, et al. Protective effects of ghrelin on cisplatin‐induced nephrotoxicity in mice. Peptides. 2016;82:85‐91. [DOI] [PubMed] [Google Scholar]
- 20. Nagayama T, Tsuchiya A, Arakawa R, et al. Hypolipidemic action of fenofibrate and bezafibrate in normo‐ and hyper‐lipidemic animals. Japanese Pharmacol Ther. 1995;23:1047‐1054. [Google Scholar]
- 21. Tsuchiya A, Kasai H, Nagayama T, Saitoh K. Comparison of hypolipidemic action of fenofibrate, bezafibrate, clinofibrate and clofibrate in rats. Japanese Pharmacol Ther. 1995;23:1041‐1046. [Google Scholar]
- 22. Jiang B, Wang YJ, Wang H, et al. Antidepressant‐like effects of fenofibrate in mice via the hippocampal brain‐derived neurotrophic factor signalling pathway. Br J Pharmacol. 2017;174:177‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferguson LB, Most D, Blednov YA, Harris RA. PPAR agonists regulate brain gene expression: relationship to their effects on ethanol consumption. Neuropharmacology. 2014;86:397‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu X, Meng X, Xu M, et al. Celastrol ameliorates cisplatin nephrotoxicity by inhibiting NF‐κB and improving mitochondrial function. EBioMedicine. 2018;36:266‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mori K, Nakao K. Neutrophil gelatinase‐associated lipocalin as the real‐time indicator of active kidney damage. Kidney Int. 2007;71:967‐970. [DOI] [PubMed] [Google Scholar]
- 26. Zhang B, Ramesh G, Norbury CC, Reeves WB. Cisplatin‐induced nephrotoxicity is mediated by tumor necrosis factor‐alpha produced by renal parenchymal cells. Kidney Int. 2007;72:37‐44. [DOI] [PubMed] [Google Scholar]
- 27. Faubel S, Lewis EC, Reznikov L, et al. Cisplatin‐induced acute renal failure is associated with an increase in the cytokines interleukin (IL)‐1beta, IL‐18, IL‐6, and neutrophil infiltration in the kidney. J Pharmacol Exp Ther. 2007;322:8‐15. [DOI] [PubMed] [Google Scholar]
- 28. Kawahito Y. The roles of PPARs in immune and inflammatory system: application for arthritic joint disease. Ensho Saisei. 2003;23:74‐83. [Google Scholar]
- 29. Ghonem NS, Assis DN, Boyer JL. Fibrates and cholestasis. Hepatology. 2015;62:635‐643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Helmy MM, Helmy MW, El‐Mas MM. Additive renoprotection by pioglitazone and fenofibrate against inflammatory, oxidative and apoptotic manifestations of cisplatin nephrotoxicity: modulation by PPARs. PLoS One. 2015;10:e0142303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang FJ, He YH, Zhou JH. Fenofibrate pre‐treatment suppressed inflammation by activating phosphoinositide 3 kinase/protein kinase B (PI3K/Akt) signaling in renal ischemia‐reperfusion injury. J Huazhong Univ Sci Technol Med Sci. 2015;35:58‐63. [DOI] [PubMed] [Google Scholar]
- 32. Portilla D, Dai G, Peters JM, Gonzalez FJ, Crew MD, Proia AD. Etomoxir‐induced PPARalpha‐modulated enzymes protect during acute renal failure. Am J Physiol Renal Physiol. 2000;278:F667‐F675. [DOI] [PubMed] [Google Scholar]
- 33. Lv J, Wang X, Liu SY, et al. Protective effect of fenofibrate in renal ischemia reperfusion injury: involved in suppressing kinase 2 (JAK2)/transcription 3 (STAT3)/p53 signaling activation. Pathol Biol (Paris). 2015;63:236‐242. [DOI] [PubMed] [Google Scholar]
- 34. Li T, Zhang Q, Zhang J, Yang G, et al. Fenofibrate induces apoptosis of triple‐negative breast cancer cells via activation of NF‐κB pathway. BMC Cancer. 2014;14:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kostapanos MS, Florentin M, Elisaf MS. Fenofibrate and the kidney: an overview. Eur J Clin Invest. 2013;43:522‐531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1


