Abstract
We expressed cDNAs coding for manganese peroxidases (MnPs) from the basidiomycetes Ceriporiopsis subvermispora (MnP1) and Phanerochaete chrysosporium (H4) under control of the α-amylase promoter from Aspergillus oryzae in Aspergillus nidulans. The recombinant proteins (rMnP1 and rH4) were expressed at similar levels and had molecular masses, both before and after deglycosylation, that were the same as those described for the MnPs isolated from the corresponding parental strains. Isoelectric focusing (IEF) analysis of rH4 revealed several isoforms with pIs between 4.83 and 4.06, and one of these pIs coincided with the pI described for H4 isolated from P. chrysosporium (pI 4.6). IEF of rMnP1 resolved four isoenzymes with pIs between 3.45 and 3.15, and the pattern closely resembled the pattern observed with MnPs isolated from C. subvermispora grown in solid-state cultures. We compared the abilities of recombinant MnPs to use various substrates and found that rH4 could oxidize o-dianisidine and p-anisidine without externally added manganese, a property not previously reported for this MnP isoenzyme from P. chrysosporium.
Ceriporiopsis subvermispora and Phanerochaete chrysosporium are among the most widely used filamentous fungi in laboratory studies of lignin biodegradation. Both of these species produce manganese peroxidase (MnP), a heme protein that catalyzes the H2O2-dependent oxidation of Mn(II) to Mn(III) (5, 16). Mn(III), chelated by organic acids, can oxidize a wide variety of phenolic compounds (28, 29). MnP is oxidized by H2O2 to generate a two-electron deficient intermediate known as compound I. Compound I can oxidize either Mn(II) or phenolic substrates by one electron, giving rise to compound II. The MnP cycle is completed when compound II gains an electron, yielding the resting enzyme. MnPs from P. chrysosporium (28), but not MnPs from C. subvermispora (24), have been reported to have an absolute requirement for Mn(II) as a reductant in the final step.
In most fungi, MnP appears to be produced as a family of isoenzymes, which may be encoded by structurally related genes. In the case of C. subvermispora, the isoenzyme pattern depends on the growth conditions. For example, in liquid cultures in salt medium, up to seven MnP isoenzymes with a molecular mass of 52.5 kDa and pIs ranging from 4.13 to 5.58 can be detected (12, 24). However, when grown on wood chips, this fungus produces four isoenzymes, all of which have a molecular mass of 62.5 kDa and more acidic pIs (12). To date, four genes coding for MnP in C. subvermispora have been cloned and sequenced (13, 23). In contrast, P. chrysosporium produces three MnP isoenzymes encoded by distinct genes that are differentially regulated at the transcriptional level (for a review, see reference 4).
It is difficult to isolate individual MnPs for characterization, as the isoenzymes often have similar physical properties. This problem is particularly evident in C. subvermispora, in which MnP isoenzymes can be purified only by preparative isolectric focusing (IEF) (24). However, this problem can be circumvented by using Aspergillus expression systems, which can produce fully active, secreted peroxidases (19, 21; H. D. Andersen, E. B. Jensen, and K. G. Welinder, 1992, European Patent Office).
The objective of this work was to determine if Aspergillus nidulans is a suitable host for expression of MnPs from C. subvermispora free of cross contamination with similar isoenzymes. For comparative purposes, we also transformed this strain with the cDNA from P. chrysosporium coding for MnP isoenzyme H4, which had been expressed previously in Aspergillus oryzae (21). We used MnP1 from C. subvermispora (13) and H4 from P. chrysosporium produced in A. nidulans to identify novel properties of both enzymes.
MATERIALS AND METHODS
Strains and culture conditions.
C. subvermispora FP-105752 was obtained from the Center for Mycology Research, Forest Products Laboratory, Madison, Wis. A. nidulans A722 (pyrG89 pabaA1 fwA1 uaY9) was obtained from the Fungal Genetic Stock Center (Kansas City, Kans.).
Plasmids and genetic construction.
Several expression vectors were prepared with MnP1 cDNA from C. subvermispora (formerly MnP13-1) (14). All constructs were made by the PCR overlap extension technique (7) by using proofreading polymerase Pfu (Stratagene, La Jolla, Calif.), and the junctions of fusions were sequenced. Plasmids pmCsMnP1 and pssCsMnP1 were similar in that expression of the C. subvermispora cDNA was under control of a 680-bp fragment containing the TAKA amylase promoter (Andersen et al., European Patent Office) and a 199-bp fragment containing the glucoamylase terminator from Aspergillus awamori (9). The vectors differed in their secretion signals. In pmCsMnP1 the TAKA amylase signal peptide was used, whereas in pssCsMnP1 the C. subvermispora MnP1 signal was used. Plasmid pTAAMnP1 contains the MnP1 cDNA from P. chrysosporium coding for isoenzyme H4 fused to the TAKA amylase signal sequence and to the regulatory elements mentioned above (21). Plasmids pmCsMnP1, pssCsMnP1, and pTAAMnP1 all lack a selectable marker and were cotransformed into a pyrG recipient with plasmid ppyrG (Fungal Genetic Stock Center). ppyrG-mCsMnP1, which could be transformed directly into A722, was created by cloning the pmCsMnP1 expression cassette into ppyrG.
Transformation.
Protoplasts were obtained from A. nidulans A722 as described by Ballance et al. (1), except that 0.25% Novozyme 234 (Calbiochem, San Diego, Calif.) was used. Cotransformation of protoplasts was performed by the procedure of Oakley et al. (15) by using 5 μg of ppyrG and 5 μg of the plasmid harboring the foreign cDNA. Alternatively, transformation was performed with 10 μg of ppyrG-mCsMnP1. Transformants were confirmed by DNA dot blot hybridization. For each transformation, five to seven colonies were analyzed.
Growth of transformants.
Twenty-five milliliters of Aspergillus minimal medium (3) supplemented with 5% maltose was inoculated with 107 spores and incubated for 2 to 3 days at 30°C in an orbital shaker (250 rpm). The mycelium was harvested by filtration through Miracloth (Calbiochem).
Nucleic acid isolation and analysis.
Mycelial pellets were snap frozen in liquid nitrogen and incubated at 65°C for 1 h in a solution containing 50 mM Tris-HCl buffer (pH 7.2), 50 mM EDTA, 3% sodium dodecyl sulfate (SDS), and 1% β-mercaptoethanol (TE buffer). Samples were extracted twice with phenol-chloroform, and the aqueous phase was precipitated with ethanol. DNA was resuspended in TE buffer. RNA was extracted as previously described (7). For Southern blot hybridization, 10 μg of genomic DNA was digested with an appropriate restriction enzyme, size fractionated in a 1% agarose gel, transferred to a Nytran membrane, and probed with 32P-labeled MnP cDNA fragments. For Northern blot hybridization, total RNA was fractionated by formaldehyde-agarose gel electrophoresis and analyzed for the presence of MnP mRNA (7). DNA probes were labeled with [α-32P]dCTP by nick translation (Gibco BRL).
Screening for rMnP production.
We evaluated cultures for recombinant MnP (rMnP) production by performing a colorimetric assay in a 16-well culture dish. Thirty-microliter samples of culture medium were taken from each culture and added to 300 μl of a solution containing 100 mM sodium tartrate (pH 5.0), 200 μM o-dianisidine, 50 μM H2O2, and 100 μM Mn(II). Color development at 460 nm was assessed visually after 15 min of incubation.
Enzyme purification.
Extracellular fluids from cultures of transformants pssCs-MnP1-4 and pTAAMnP1-3 were concentrated 10-fold by filtration in a 185-ml Amicon cell containing a 10-kDa-cutoff membrane. The extracts were then dialyzed twice against 500 ml of 25 mM sodium acetate (pH 6.0) and fractionated by chromatography on Q-Sepharose (12).
Zymograms.
SDS-polyacrylamide gel electrophoresis (PAGE) was performed as described by Laemmli (11). Samples were applied in nonreducing denaturing 1× loading buffer without boiling and electrophoresed at 4°C. The gels were incubated in 100 mM sodium tartrate buffer (pH 5.0) containing 100 μM MnSO4 and 50 μM CaCl2 for 1 h and then stained for activity with a solution containing the same solution supplemented with 200 μM o-dianisidine plus 50 μM H2O2. After the gels were photographed, they were stained with Coomassie blue.
Enzymatic incubation.
Enzyme activity was measured at 30°C with a model 160 UV-visible recording spectrophotometer (Shimadzu, Kyoto, Japan). The reaction mixtures (1 ml) contained 100 mM sodium tartrate (pH 5.0), 100 μM MnSO4, 50 μM H2O2, and various amounts of substrate in the millimolar range. The wavelength used to monitor the progress of the reaction depended on the substrate. Initial velocities were recorded between 0 and 15 s. Km values for o-dianisidine and p-anisidine were determinated with 0.03 and 0.007 U of rMnP, respectively, by using Eadie-Hofstee plots. One unit of MnP activity was defined as the amount of enzyme required to oxidize 1 mmol of vanillylacetone in 1 min.
Endoglycosidase treatment.
Thirty microliters (5 μg) of each rMnP was combined with 2 μl of 10% SDS and with 25 μl of a solution containing 50 mM Tris-HCl (pH 7.5), 4% β-mercaptoethanol, and 200 mM EDTA. The resulting mixture was boiled for 5 min. Then 1 U (20 μl) of endoglycosidase F–N-glycosidase F (Sigma Chemical Co., St. Louis, Mo.) was added, and the reaction mixture was incubated for 30 h at 37°C.
Other methods.
Analytical IEF was performed by using Servalyt Precotes 3–6 polyacrylamide gels (Serva Fine Biochemicals Inc., Westbury, N.Y.) (12). Extracellular protease contents were determined by the method of Sarath et al. (20). Protein concentrations were measured by the method of Bradford using bovine serum albumin as the standard (2).
RESULTS
Transformation.
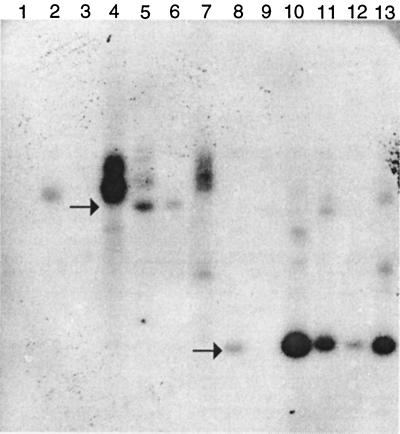
Southern blots revealed that complex integration events occurred among transformants (Fig. 1). Genomic DNAs of selected pyrG transformants were digested with BamHI or PstI and probed with 32P-labeled MnP1 cDNA. In all four vectors used, PstI sites flanked a 1.7-kb region comprising the promoter, cDNA, and terminator. A single BamHI site was located outside this expression cassette in pmCsMnP1 and pssCsMnP1. In ppyrG-mCsMnP1, BamHI digestion yielded a 4.5-kb fragment containing the expression cassette and portions of pyrG. Typical of pyrG-based A. nidulans transformation, the copy numbers and integration context varied among the transformants. The intense signals obtained with BamHI-digested pssCsMnP1-9 (Fig. 1, lane 4) and pssCsMnP1-4 (lane 7) were consistent with tandem duplications at multiple loci. The multicopy nature of the transformants also was supported by the relative band intensity after PstI digestion of ppyrG-mCsMnP1-4 (lane 11). Transformant pmCsMnP1-9 contained no intact expression cassette (lanes 3 and 9).
FIG. 1.
Southern blot analysis of transformants probed with MnP1 cDNA. Ten-microgram samples of total DNA from A. nidulans transformants were digested with BamHI (lanes 1 to 7) or PstI (lanes 8 to 13) and subjected to Southern blot analysis. The lanes contained transformants pmCsMnP1-5 (lanes 2 and 8), pmCsMnP1-9 (lanes 3 and 9), pssCsMnP1-9 (lanes 4 and 10), ppyrG-mCsMnP1-4 (lanes 5 and 11), ppyrG-mCsMnP1-6 (lanes 6 and 12), and pssCsMnP1-4 (lanes 7 and 13). Lane 1 contained DNA from A. nidulans transformed only with ppyrG. The arrows indicate the positions of a 4.5-kb fragment (upper arrow) and a 1.7-kb fragment (lower arrow).
Production of rMnPs.
Transformants confirmed by Southern hybridization were screened for extracellular MnP activity when they were grown in minimal medium. Preliminary determinations showed that pTAAMnP1-containing transformants exhibited the highest levels of extracellular MnP activity and that pssCsMnP1 transformants secreted more MnP than pmCsMnP1 transformants.
Conditions were partially optimized to obtain higher titers of extracellular MnP activity. The spore concentration of the inoculum did not significantly affect enzyme production. Addition of 0.5 mg of hemin per ml at the time of inoculation resulted in the highest enzyme yield, but we routinely used 0.25 mg/ml to facilitate the purification process (Fig. 2). MnP activity decreased dramatically on day 3 or 4, which coincided with an increase in the extracellular protease levels (data not shown). Both transformants were strongly induced by maltose at a concentration of 5% (wt/vol), although glucose gave significant residual activities (Fig. 3A). After the culture conditions were refined, enzyme production by transformants pTAAMnP1-3 and pssCsMnP1-4 varied between 5 and 7 mg/liter and between 3 and 6 mg/liter, respectively (between 1 and 1.4 mg/g [dry weight] of mycelia and between 0.5 and 1 mg/g [dry weight] of mycelia, respectively). Transformants containing pmCsMnP1 or ppyrG-mCsMnP1 never produced more than 0.6 mg of rMnP per liter. rMnP Northern blots gave strong signals for RNA derived from the maltose-containing medium (Fig. 3B). These results are consistent with the results of recent studies of TAKA amylase transcriptional regulation in A. nidulans (22).
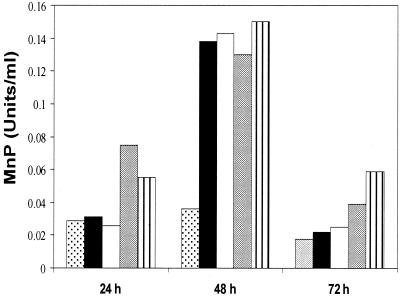
FIG. 2.
Effect of hemin concentration on rMnP production. Cultures of transformant pssCsMnP1-4 were grown in minimal medium containing 5% maltose. The treatments varied with respect to the timing of hemin addition; 0.25 mg of hemin per ml was added with the inoculum (grey bars) or 24 h after inoculation (solid bars) or 0.5 mg of hemin per ml was added with the inoculum (bars with vertical lines) or 24 h after inoculation (open bars). Controls contained no hemin (stippled bars). Activity was measured at the times indicated.
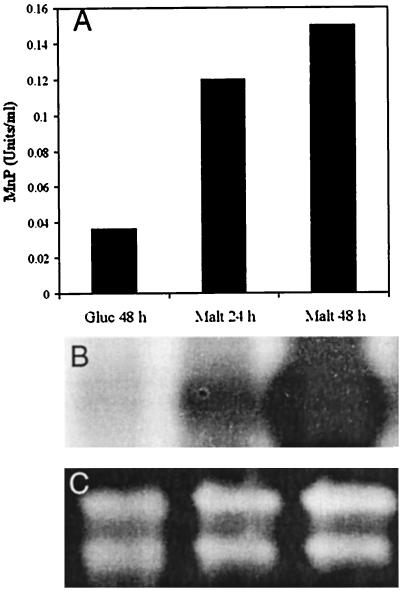
FIG. 3.
Production of Cs-rMnP1 in transformant pssCsMnP1-4. (A) Titers of MnP activity detected in extracellular medium containing maltose (Malt) or glucose (Gluc) as the carbon source. (B) Northern blot hybridization of total RNA from each culture. (C) Loading control ethidium bromide-stained formaldehyde gel, showing that the amounts of rRNA were approximately equal.
Enzyme purification.
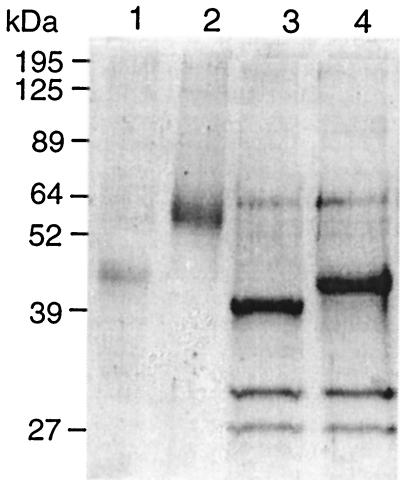
We purified both rMnPs with good yields, as determined by zymograms and SDS-PAGE (Fig. 4A). Zymograms of recombinant H4 (rH4) did not contain an activity band, suggesting that this enzyme is more labile than C. subvermispora recombinant MnP1 (Cs-rMnP1) under the conditions of this assay. When stained with Coomassie blue, the zymogram had no additional bands. Migration of both rMnPs in this gel was faster than migration in an SDS-PAGE gel electrophoresed under reducing conditions. In the latter case electrophoresis resulted in only one band for each enzyme, confirming that both rMnPs retained the molecular weights described. Indeed, Cs-rMnP1 migrated as a protein with a molecular mass of 62.7 kDa, the value obtained for the MnPs produced by C. subvermispora grown on wood (12), whereas rH4 had a molecular mass of 46.5 kDa (5) (Fig. 4B).
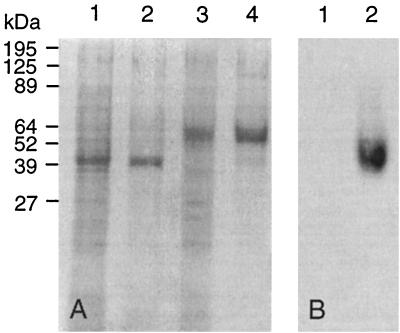
FIG. 4.
SDS-PAGE of rMnPs. (A) Extracellular fluids from 48-h cultures were concentrated 20-fold, and 0.05 U of MnP activity from transformant pTAAMnP1-3 (rH4) (lane 1) or pssCsMnP1-4 (Cs-rMnP1) (lane 3) was treated with β-mercaptoethanol, boiled, and applied to the gel. After Q-Sepharose chromatography, 0.05 U of rH4 and 0.05 U of Cs-rMnP1 were subjected to the same treatment and applied to the gel (lanes 2 and 4, respectively). (B) Zymogram obtained with o-dianisidine as the substrate, as described in Materials and Methods. Lane 1, 0.05 U of rH4; lane 2, 0.05 U of Cs-rMnP1.
Glycosylation.
When the rMnPs were treated with endoglycosidase F–N-glycosidase F for 30 h, the apparent molecular mass of Cs-rMnP1 decreased from 62.7 to 44.0 kDa and the molecular mass of rH4 decreased from 46.5 to 42 kDa (Fig. 5). The bands obtained with the glycosidase-treated sample were much sharper (Fig. 5, lanes 4 and 5) than those obtained with untreated rMnPs (lanes 2 and 3).
FIG. 5.
SDS-PAGE of endoglycosidse F–N-glycosidase F-treated rMnPs. The lanes contained 0.5 μg of untreated rH4 (lane 1), 1 μg of untreated Cs-rMnP1 (lane 2), 1 μg of N-glycosidase-treated rH4 (lane 3), and 1 μg of N-glycosidase-treated Cs-rMnP1 (lane 4). The two lowest bands in lanes 3 and 4 correspond to endoglycosidase F–N-glycosidase F.
pIs.
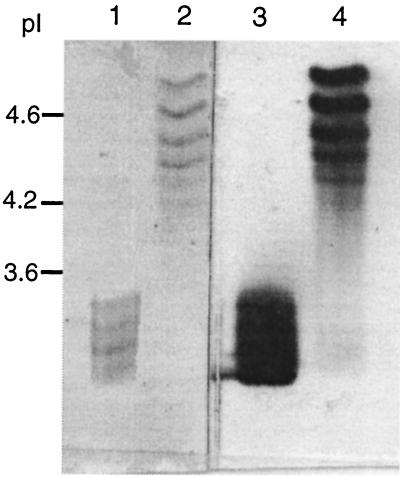
MnP isoenzymes have distinct pIs. P. chrysosporium isoenzyme H4 has a pI close to 4.6 (18). The precise pI of MnP1 from C. subvermispora is unknown, but isoenzymes produced in solid-state cultures have pIs ranging from 3.2 to 3.5 (13). IEF gels of both rMnPs contained several bands (Fig. 6). Cs-rMnP1 produced a diffuse four-band pattern that was the same as the pattern detected for MnPs extracted from cultures of C. subvermispora grown on wood chips (13), with pIs of 3.45, 3.39, 3.24, and 3.15. rH4 produced six bands with pIs of 4.83, 4.61, 4.41, 4.29, 4.17, and 4.06.
FIG. 6.
IEF of rMnPs. Portions (0.03 U) of Cs-rMnP1 (lanes 1 and 3) and rH4 (lanes 2 and 4) were loaded, and after the gel was electrophoresed, the proteins were either stained with Coomassie blue (lanes 1 and 2) or developed for activity with o-dianisidine (lanes 2 and 3).
Manganese dependence.
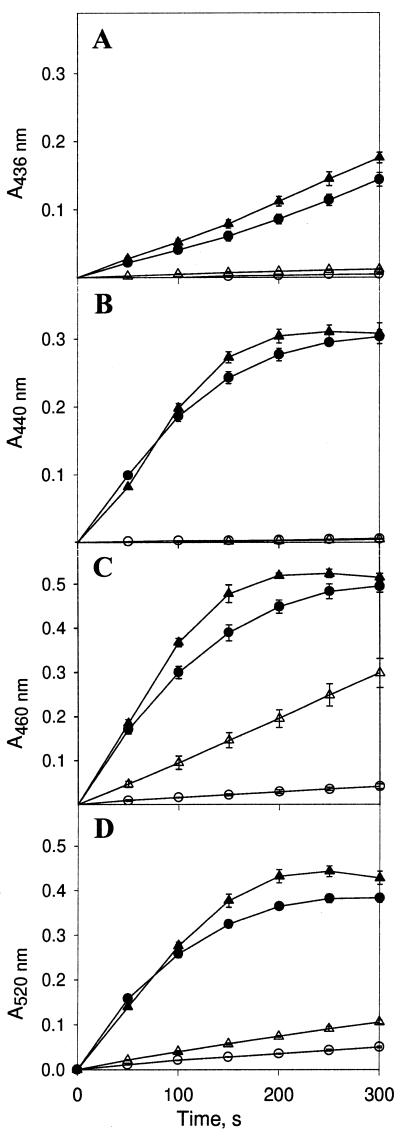
The two recombinant enzymes were compared with respect to their requirements for Mn(II) during oxidation of different substrates (Fig. 7). The same substrates were used previously to characterize several MnP isoenzymes isolated directly from C. subvermispora (24). rH4 required a higher concentration of Mn(II) than Cs-rMnP1. Among the various substrates tested, oxidation of guaicol (and vanillylcetone [data not shown]) had the highest demand for Mn(II) for both rMnPs, whereas oxidation of o-dianisidine and p-anisidine had the lowest demand for this metal.
FIG. 7.
Oxidation of aromatic compounds by rMnPs. One-tenth millimolar 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) (A), 0.1 mM guaiacol (B), 0.1 mM o-dianisidine (C), and 1 mM p-anisidine (D) were incubated with Cs-rMnP1 (triangles) or rH4 (circles) in the presence (solid symbols) or absence (open symbols) of 0.1 mM Mn(II). The incubation mixtures each contained 0.2 mM tartrate buffer (pH 5.0), 50 μM H2O2, and 0.02 U of rMnP. Reactions were monitored spectrophotometrically at the wavelengths indicated. Each point and error bar indicate the average and standard deviation based on three experiments. For points without error bars the standard deviation was less than 10%.
Determination of Km.
The Km values obtained with Cs-rMnP1 when o-dianisidine and p-anisidine were used as substrates were 0.586 and 0.106 mM, which are similar to the values observed in the same reactions with the MnPs derived from solid cultures of C. subvermispora (24). Km values of 1.59 and 0.058 mM were obtained with rH4 in the same reactions.
Activation by oxalate.
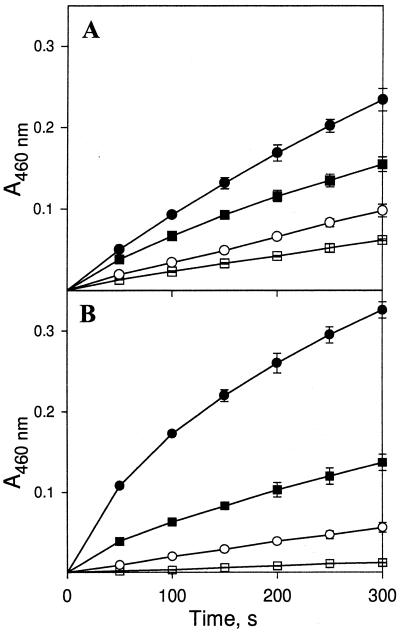
We tested the effect of oxalate on the oxidation of o-dianisidine by the rMnPs both in the presence and in the absence of externally added Mn(II). In these experiments, 100 mM sodium succinate (pH 5.0) replaced 100 mM sodium tartrate (pH 5.0) as the buffer. The rate of oxidation of o-dianisidine nearly doubled when 2 mM oxalate was included in the reaction mixtures either in the presence or in the absence of added Mn(II) (Fig. 8).
FIG. 8.
Effect of oxalate on oxidation of o-dianisidine by rMnPs. o-Dianisidine (0.1 mM) was incubated with 0.007 U of Cs-rMnp1 (A) or 0.02 U of rH4 (B) with or without oxalate either in the presence or in the absence of Mn(II). Symbols: □, oxalate absent and Mn(II) absent; ○, oxalate present and Mn(II) absent; ■ oxalate absent and Mn(II) present; ●, oxalate present and Mn(II) present. Each point and error bar indicate the average and standard deviation based on three experiments. For points without error bars the standard deviation was less than 10%.
DISCUSSION
Due to the difficulties encountered in purification of single MnP isoenzymes from C. subvermispora, heterologous expression in Aspergillus strains offers an attractive alternative for this purpose. Aspergillus spp. are well known for impressive protein secretion, and several studies have demonstrated efficient expression and secretion of heterologous peroxidases (19, 21; Andersen et al., European Patent Office).
Using a common laboratory strain of A. nidulans, we produced rMnP at levels that are three- to sevenfold higher than those obtained in the same host with MnP from Pleurotus eryngii (19). Concentrations of maltose less than 5% severely affected the yield of the recombinant enzymes. This result differs from the results obtained with A. oryzae; with this organism only 2% maltose is required in the medium. Higher levels of extracellular Cs-rMnP1 were obtained with its own signal peptide than with a signal peptide from the TAKA amylase gene, although the latter results in high yields of rH4. This finding was not unexpected considering the sequence heterogeneity in functional signal sequences (3, 25). In the case of Cs-rMnP1, four serine residues flank the cleavage site and may contribute to efficient secretion. Other structural characteristics of the hybrid protein, (e.g., glycosylation, folding, or proper hemin incorporation) also might influence secretion of active rMnP.
Most glycans in Cs-rMnP1 appear to be N linked, as deduced by digestion with endoglycosidse F–N-glycosidase F (Fig. 5). This treatment increased the migration of Cs-rMnP1 in the gel to the migration of a protein 18 kDa smaller than the recombinant enzyme, whereas the reduction in the molecular mass of rH4 after the same treatment was only 3 kDa. Even after digestion with N-glycosidase, however, molecular masses of both rMnPs were greater than the values predicted from their cDNA sequences (about 38 kDa for both enzymes). Assuming that digestion of N-glycans was complete, the difference in molecular mass could be attributed to the presence of O-linked glycans. This type of covalent posttranslational modification has been confirmed with isoenzyme H4 (14), which possesses 55 potential sites for O-glycosylation, as opposed to only 4 sites for N-glycosylation. In turn, MnP1 from C. subvermispora possesses 56 potential sites for O-glycosylation and seven potential sites for N-glycosylation. Nie et al. (14) showed that O-glycans play a role in the thermal stability of peroxidases from P. chrysosporium. O-Glycans might also have this role in H4, LiP2, or LiP8, since N-glycans represent only a small fraction of the total associated glycans. N-glycosylations also have been associated with thermal stability of yeast invertase (8, 27). For both rMnPs we think that both the content and the position of glycosylations are important for thermal stability (27).
When both rMnPs were subjected to IEF, several activity bands were observed. Partial proteolysis probably was not responsible for the IEF patterns because the bands in the SDS gels were sharp. On the other hand, differential glycosylation in filamentous fungi usually does not change the pIs of the proteins, since it does not normally involve participation of charged sugars (26, 30). Therefore, it is possible that the isoenzymes of the rMnPs which we obtained differ in the degree of covalent modification with a charged group, such as phosphate (10). Pease et al. (17) also observed more bands than expected during an IEF analysis of MnP1 from P. chrysosporium expressed in baculovirus.
Of the six activity bands obtained with rH4, only one had the expected pI, pI 4.6. The IEF pattern which we obtained for rH4 looks very similar to that described for nitrogen-limited cultures of P. chrysosporium (18). However, in that case the protein pattern detected with antibodies specific for H4 did not exactly coincide with the pattern developed for MnP activity. The pattern of posttranslational modification that takes place in Aspergillus may differ from the C. subvermispora or P. chrysosporium pattern. However, it is now clear that isoenzyme multiplicity can arise following expression of a single MnP gene, and despite the multiplicity of genes, the role of posttranslational processing may be significant.
Oxidation of o-dianisidine and p-anisidine by rH4 took place in reaction mixtures lacking externally added manganese. The fact that guaiacol was not oxidized at all under these conditions indicates that the manganese concentration in the reaction mixtures was negligible. On the other hand, chelation of Mn(III) by oxalate could explain the activation of both rMnPs by this organic acid. However, the same effect was observed in the absence of added manganese. The mechanism by which oxalate affects the reaction in the latter case remains unknown (24). Oxidation of o-dianisidine and p-anisidine by rH4 and activation of rH4 by oxalate, in both cases in the absence of added manganese, are previously unknown properties of this enzyme.
In this study we found that Aspergillus cells provide a very convenient expression system for MnPs from C. subvermispora. This finding is particularly important due to the high isoenzyme multiplicity observed in this fungus (12, 24). In addition, A. nidulans could secrete rMnPs with glycosylation similar to the glycosylation exhibited by native enzymes.
ACKNOWLEDGMENTS
This work was financed by grants 8990004 and 2000076 from FONDECYT-Chile and by U.S. Department of Energy grant DE-FG02-87ER13712. L.F.L is a Predoctoral Fellow supported by Fundacion Andes.
We thank Loreto Salas, Francisco Pizarro, and Marcela Avila for technical assistance.
REFERENCES
- 1.Ballance D J, Buxton F P, Turner G. Transformation of Aspergillus nidulans by the orotidine-5′-phosphate decarboxylase gene of Neurospora crassa. Biochem Biophys Res Commun. 1983;112:284–289. doi: 10.1016/0006-291x(83)91828-4. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Cullen D, Gray G L, Wilson L J, Hayenga K J, Lamsa M H, Rey M W, Norton S, Berka R M. Controlled expression and secretion of bovine chymosin in Aspergillus nidulans. Bio/Technology. 1987;5:369–375. [Google Scholar]
- 4.Cullen D. Recent advances on the molecular genetics of ligninolytic fungi. J Biotechnol. 1997;53:273–289. doi: 10.1016/s0168-1656(97)01684-2. [DOI] [PubMed] [Google Scholar]
- 5.Glenn J K, Gold M H. Purification and characterization of an extracellular Mn(II) dependent peroxidase from the lignin-degrading basidiomycete Phanerochaete chrysosporium. Arch Biochem Biophys. 1985;242:329–341. doi: 10.1016/0003-9861(85)90217-6. [DOI] [PubMed] [Google Scholar]
- 6.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 7.Karahanian E, Corsini G, Lobos S, Vicuña R. Structure and expression of a laccase gene from the ligninolytic basidiomycete Ceriporiopsis subvermispora. Biochim Biophys Acta. 1998;1443:65–74. doi: 10.1016/s0167-4781(98)00197-3. [DOI] [PubMed] [Google Scholar]
- 8.Kern G, Schulke N, Schmid F X, Jaenicke R. Stability, quaternary structure, and folding of internal, external, and core-glycosylated invertase from yeast. Protein Sci. 1992;1:120–131. doi: 10.1002/pro.5560010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kersten P J, Witek C, van den Wymelenberg A, Cullen D. Phanerochaete chrysosporium glyoxal oxidase is encoded by two allelic variants: structure, genomic organization, and heterologous expression of glx1 and glx2. J Bacteriol. 1995;177:6106–6110. doi: 10.1128/jb.177.21.6106-6110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuan I C, Tien M. Phosphorylation of lignin peroxidase from Phanerochaete chrysosporium. J Biol Chem. 1989;264:20350–20355. [PubMed] [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Lobos S, Larraín J, Salas L, Cullen D, Vicuña R. Isoenzymes of manganese-dependent peroxidase and laccase produced by the lignin-degrading basidiomycete Ceriporiopsis subvermispora. Microbiology. 1994;140:2691–2698. doi: 10.1099/00221287-140-10-2691. [DOI] [PubMed] [Google Scholar]
- 13.Lobos S, Larrondo L, Salas L, Karahanian E, Vicuña R. Cloning and molecular analysis of a cDNA and the Cs-mnp1 gene encoding a manganese peroxidase isoenzyme from the lignin-degrading basidiomycete Ceriporiopsis subvermispora. Gene. 1998;206:185–193. doi: 10.1016/s0378-1119(97)00583-0. [DOI] [PubMed] [Google Scholar]
- 14.Nie G, Reading N S, Aust S D. Relative stability of recombinant versus native peroxidases from Phanerochaete chrysosporium. Arch Biochem Biophys. 1999;365:328–334. doi: 10.1006/abbi.1999.1180. [DOI] [PubMed] [Google Scholar]
- 15.Oakley B R, Rinehart J E, Mitchell B L, Oakley C E, Carmona C, Gray G L, May G S. Cloning, mapping and molecular analysis of the pyrG (orotidine-5′-phosphate decarboxylase) gene of Aspergillus nidulans. Gene. 1987;61:385–399. doi: 10.1016/0378-1119(87)90201-0. [DOI] [PubMed] [Google Scholar]
- 16.Paszcynski A, Huynh V B, Crawford R L. Enzymatic activities of a extracellular, manganese-dependent peroxidase from Phanerochaete chrysosporium. FEMS Microbiol Lett. 1985;27:37–41. [Google Scholar]
- 17.Pease E, Aust S D, Tien M. Heterologous expression of active manganese peroxidase from Phanerochaete chrysosporium using the baculovirus expression system. Biochem Biophys Res Commun. 1991;179:897–903. doi: 10.1016/0006-291x(91)91903-p. [DOI] [PubMed] [Google Scholar]
- 18.Pease E, Tien M. Heterogeneity and regulation of manganese peroxidases from Phanerochaete chrysosporium. J Bacteriol. 1992;174:3532–3540. doi: 10.1128/jb.174.11.3532-3540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Dueñas F J, Martínez M J, Martínez A T. Heterologous expression of Pleurotus eryngii peroxidase confirms its ability to oxidize Mn2+ and different aromatic substrates. Appl Environ Microbiol. 1999;65:4705–4707. doi: 10.1128/aem.65.10.4705-4707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarath G, de la Motte R S, Wagner F W. Protease assay methods. In: Beynon R J, Bond J S, editors. Proteolytic enzymes: a practical approach. Oxford, United Kingdom: IRL Press; 1989. pp. 25–55. [Google Scholar]
- 21.Stewart P, Whitwam R E, Kersten P J, Cullen D, Tien M. Efficient expression of a Phanerochaete chrysosporium manganese peroxidase gene in Aspergillus oryzae. Appl Environ Microbiol. 1996;62:860–864. doi: 10.1128/aem.62.3.860-864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tani S, Kawaguchi T, Kato M, Kobayashi T, Tsukagoshi N. A novel nuclear factor, SREB, binds to a cis-acting element, SRE, required for inducible expression of the Aspergillus oryzae Taka-amylase A gene in A. nidulans. Mol Gen Genet. 2000;263:232–238. doi: 10.1007/s004380051164. [DOI] [PubMed] [Google Scholar]
- 23.Tello M, Corsini G, Larrondo L F, Salas L, Lobos S, Vicuña R. Characterization of three new manganese peroxidase genes from the ligninolytic basidiomycete Ceriporiopsis subvermispora. Biochim Biophys Acta. 2000;1490:137–144. doi: 10.1016/s0167-4781(99)00227-4. [DOI] [PubMed] [Google Scholar]
- 24.Urzúa U, Larrondo L F, Lobos S, Larraín J, Vicuña R. Oxidation reactions catalyzed by manganese peroxidase isoenzymes from Ceriporiopsis subvermispora. FEBS Lett. 1995;371:132–136. doi: 10.1016/0014-5793(95)00874-9. [DOI] [PubMed] [Google Scholar]
- 25.von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 26.Wallis G L F, Swift R J, Hemming F W, Trinci A P J, Peberdy J F. Glucoamylase overexpression and secretion in Aspergillus niger: analysis of glycosylation. Biochim Biophys Acta. 1999;1472:576–586. doi: 10.1016/s0304-4165(99)00188-9. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Eufemi M, Turano C, Giartosio A. Influence of the carbohydrate moiety on the stability of glycoproteins. Biochemistry. 1996;35:7299–7307. doi: 10.1021/bi9517704. [DOI] [PubMed] [Google Scholar]
- 28.Wariishi H, Akileswaran L, Gold M H. Manganese peroxidase from the basidiomycete Phanerochaete chrysosporium: spectral characterization of the oxidized states and the catalytic cycle. Biochemistry. 1988;27:5365–5370. doi: 10.1021/bi00414a061. [DOI] [PubMed] [Google Scholar]
- 29.Wariishi H, Dunford H B, MacDonald I D, Gold M H. Manganese peroxidase from the lignin-degrading basidiomycete Phanerochaete chrysosporium: transient-state kinetics and reaction mechanism. J Biol Chem. 1989;264:23688–23695. [PubMed] [Google Scholar]
- 30.Wyss M, Pasamontes L, Friedlein A, Rémy R, Tessier M, Kronenberger A, Middendorf A, Lehmann M, Schnoebelen L, Röthlisberger U, Kusznir E, Wahl G, Müller F, Lahm H W, Vogel K, van Loon A P G M. Biophysical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): molecular size, glycosylation pattern, and engineering of proteolytic resistance. Appl Environ Microbiol. 1999;65:359–366. doi: 10.1128/aem.65.2.359-366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]