Abstract
Plant mitochondrial genomes are a valuable source of genetic information for a better understanding of phylogenetic relationships. However, no mitochondrial genome of any species in Ebenaceae has been reported. In this study, we reported the first mitochondrial genome of an Ebenaceae model plant Diospyros oleifera. The mitogenome was 493,958 bp in length, contained 39 protein-coding genes, 27 transfer RNA genes, and 3 ribosomal RNA genes. The rps2 and rps11 genes were missing in the D. oleifera mt genome, while the rps10 gene was identified. The length of the repetitive sequence in the D. oleifera mt genome was 31 kb, accounting for 6.33%. A clear bias in RNA-editing sites were found in the D. oleifera mt genome. We also detected 28 chloroplast-derived fragments significantly associated with D. oleifera mt genes, indicating intracellular tRNA genes transferred frequently from chloroplasts to mitochondria in D. oleifera. Phylogenetic analysis based on the mt genomes of D. oleifera and 27 other taxa reflected the exact evolutionary and taxonomic status of D. oleifera. Ka/Ks analysis revealed that 95.16% of the protein-coding genes in the D. oleifera mt genome had undergone negative selections. But, the rearrangement of mitochondrial genes has been widely occur among D. oleifera and these observed species. These results will lay the foundation for identifying further evolutionary relationships within Ebenaceae.
Keywords: Diospyros oleifera, Mitochondrial genome, Phylogenetic analysis
Diospyros oleifera; Mitochondrial genome; Phylogenetic analysis.
1. Introduction
Mitochondria are the main organelle involved in energy metabolism in plants [1, 2]. They supply ATP via oxidative phosphorylation for metabolism, cell differentiation, apoptosis, cell growth, and cell division and are abundant in energy-consuming tissues involved in essential biological functions [1, 2, 3, 4]. Therefore, mitochondria play an important role in plant productivity and development [2, 5, 6]. According to endosymbiotic theory, plant mitochondria are believed to have descended from free-living bacteria-independent microorganisms, which explains the presence of their genomes [5, 7].
During evolution, the plant mitochondrial (mt) genome underwent dramatic changes in, for example, the gene order, genome structure, and migration of sequences from other organelles [5, 7, 8, 9]. Thus, plants have about 100–10,000 times larger and more structurally complex mitochondrial (mt) genomes than animals [10, 11, 12]. The mt genomes of plants demonstrate significant genome size variation, from 66 kb [13] to 11.3 Mb [14]; the number of protein-coding genes varies from 14 to 67 [15]; and the number of tRNA genes varies from 3 to 27 [9]. There are variations in mitochondrial genomes not only between plant species but also within the same species [9, 12, 16, 17], in stark contrast to the conserved structure of plant chloroplast genomes [16, 17, 18]. Thus, mt genomes have been used as a valuable source of genetic information and for investigation of essential cellular processes in many phylogenetic studies [18, 19, 20, 21].
While, these characteristics of plant mt genomes (bigger size, more structural complexity, and low conservation across species) make plant mitochondrial genome assembly difficult [1, 8, 10]. To date, more than 5000 plant chloroplast genomes have been sequenced, but only about 400 mt genome sequences are available (www.ncbi.nlm.nih.gov/genome/organelle/,11/11/2021). In addition, sequenced plants largely differ in their classification, and only three complete mitochondrial genomes of species from the order Ericales have been identified.
Diospyros L., from the Ebenaceae family, is a plant genus that includes over 500 species widely distributed across tropical and subtropical regions [22] and that is one of the largest angiosperm genera [23]. Among these species, Diospyros oleifera and Diospyros kaki have been cultivated as an important fruit crop in China, Korea, Japan for centuries, due to its edible fruit is rich in vitamins, sugars, nutrients and antioxidants vital for optimum health with various medicinal and chemical [24, 25]. Morphological, molecular, and genomic studies have shown that D. oleifera can be used as a model plant [24, 26]. Chloroplast genome sequencing has been performed in 15 species of Diospyros [26, 27], and nuclear genome sequencing has been performed in D. oleifera [23, 28] and Diospyros lotus [29, 30]. However, to date, no mt genome of any species in Ebenaceae has been reported.
Fortunately, advancements in long read sequencing, such as PacBio and Oxford Nanopore, have made organelle genome sequencing easier and faster. Therefore, in this study, we constructed the complete mt genome of D. oleifera based on PacBio and Illumina data, performed a phylogenetic analysis, and compared the complete mt genomes of D. oleifera and related genera. These results will help better understand the features of the D. oleifera mitochondrial genome and lay the foundation for identifying further evolutionary relationships within Ebenaceae.
2. Materials and methods
2.1. Samples and mitogenome sequencing
Due to the advancement of sequence technology, long reads, used for de novo assembly of organelle genomes without the need for organelle DNA isolation, could be easily generated from high throughput sequencing. The well-established methodology is quite efficient and well accepted in the scientific community [1, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42].
In this study, Mature leaves of D. oleifera (at latitude 34.27569 and longitude 107.75079) were used to isolate total DNA following the protocol for the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) and the SMRTbell Libraries protocol for PacBio data (Pacific Biosciences, Menlo Park, CA, USA). All these whole-genome Illumina HiSeq and PacBio sequencing data were deposited in the NCBI GenBank (accession no. PRJNA562043) and the Persimmon Genome Website (http://www.kakiwi.zju.edu.cn/cgi-bin/persimmon/about_genome.cgi). Sequencing reads of the mitochondria were filtered and extracted from these WGD sequencing data of D. oleifera. Raw data of second-generation sequencing were filtered using fastp version 0.20.0 software (https://github.com/OpenGene/fastp) [43]. The three-generation sequencing data of mitochondrial reads were error-corrected, trimmed, and de-novo-assembled using a Canu assembler (version 1.5) with default parameters [44]. Then, the contig sequence was obtained. The gene databases of plant mitochondria (the mitochondrial gene sequences of species published on the NCBI) were compared using blast v2.6 (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and contigs that matched with the mitochondrial gene as the seed sequence were selected. The original data were used to extend and circularize the contigs to obtain the ring-dominant structure (or secondary ring), and then, the assembly was polished using NextPolish 1.3.1 (https://github.com/Nextomics/NextPolish) [45]. The assembly results were calibrated using second- and third-generation data, and the parameters were set as rerun = 3 and -max_depth = 100. Then, the final assembly results were obtained.
2.2. Genome annotation
The assembled D. oleifera mt genome was annotated using the GeSeq tool [46]. To confirm the annotated results, the assembled D. oleifera mt genome was also BLAST-searched against protein-coding genes and ribosomal RNA (rRNA) genes of available plant mt genomes at the NCBI. Then, the sequence coordinates of the identified protein-coding genes (PCGs) were manually verified for start and stop codons. The annotations of transfer RNA (tRNA) genes were also confirmed by tRNAscan-SE [47]. ViennarNA-2.4.14 [48] was used to visualize the secondary structure of tRNA. The physical circular map was drawn using the Organellar Genome DRAW (OGDraw) v1.2 program [49]. The final annotated mt genome sequences of D. oleifera have been deposited in the NCBI GenBank (accession no. MW970112).
Strand asymmetry was calculated according to the formulas: AT-skew = [A − T]/[A + T] and GCskew = [G − C]/[G + C] [50]. The possible RNA-editing sites in the PCGs of D. oleifera were predicted using the online predictive RNA editor for plant mitochondrial genes (PREP-Mt) [51] suite of servers (http://prep.unl.edu/). The codon frequencies were calculated using the Codon Usage tool in the Sequence Manipulation Suite (bioinformatics.org/sms2/codon_usage.html) [52]. The relative synonymous codon usage (RSCU [53]) was calculated using the CAI Python package of Lee [54].
2.3. Analysis of repeated sequences
Three kinds of repeats (simple sequence, tandem, and dispersed) were detected in the D. oleifera mitochondrial genome. The MIcroSAtellite (MISA) identification tool Perl script was used to detect simple sequence repeats [55]. The repeats of mono-, di-, tri-, tetra-, penta-, and hexanucleotide bases with 12, 6, 4, 3, 3, and 3 repeat numbers, respectively, were identified. Tandem repeats (>6 bp repeat units) were detected using Tandem Repeats Finder v4.09 software (http://tandem.bu.edu/trf/trf.submit.options.html) [56] with default parameters (matching probability of 80 and indel probability of 10). Direct and inverted repeats were detected using the vmatch (v2.3.0) Perl script with the minimal repeat size set to 30 bp.
2.4. Chloroplast-to-mitochondrion-DNA transformation
The D. oleifera cp genome (NC_030787.1) was downloaded from the NCBI Organelle Genome Resources Database. The protein-coding and tRNA genes, which were transferred from chloroplasts to mitochondria, were identified using Blastn software with the following screening criteria: matching rate ≥70%, E-value ≤ 1e − 10, and length ≥30 bp.
2.5. Phylogenetic tree construction and Ka/Ks analysis
The D. oleifera and Twenty-seven other species with complete or nearly complete mitogenomes were used in phylogenetic analyses, representing twenty families. Two species from Conifers were used as outgroup. All species were listed in Table 1. The mt genomes were downloaded from the NCBI Organelle Genome Resources Database, and the conserved protein-coding genes (atp1, atp4, atp6, atp8, atp9, ccmB, ccmC, ccmFC, ccmFN, cob, cox1, cox2, cox3, matR, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, rpl5, rps12, rps13, rps3, and rps4) were extracted and aligned using MAFFT v7.402 [57] with default parameters. ModelTest-NG v0.1.3 was used to determine the best-fit model, and a maximum likelihood (ML) tree was generated using RAxMLv8.2.12 with the best-fit substitution model (GTRGAMMA) at 1000 bootstrap replicates [58].
Table 1.
GenBank accession numbers of mitochondrial genomes for species sampled in this study.
| Classification Status | Order | Familiy | Species | Length (bp) | Accession number | |
|---|---|---|---|---|---|---|
| Ingroup | Asterids | Apiales | Apiaceae | Daucus carota# | 281,132 | NC_017855 |
| Asterids | Aquifoliales | Aquifoliaceae | Ilex pubescens | 517,520 | NC_045078 | |
| Asterids | Asterales | Asteraceae | Chrysanthemum boreale | 211,002 | NC_039757 | |
| Asterids | Asterales | Asteraceae | Helianthus annuus | 300,945 | NC_023337 | |
| Asterids | Asterales | Asteraceae | Lactuca sativa | 363,324 | NC_042756 | |
| Asterids | Asterales | Asteraceae | Lactuca serriola | 363,328 | NC_042378 | |
| Asterids | Asterales | Campanulaceae | Codonopsis lanceolata | 403,704 | NC_037949 | |
| Asterids | Asterales | Campanulaceae | Platycodon grandiflorus | 1,249,593 | NC_035958 | |
| Asterids | Ericales | Ebenaceae | Diospyros oleifera∗# | 493,958 | MW970112 | |
| Asterids | Ericales | Ericaceae | Rhododendron simsii# | 802,707 | WJXA01000014 | |
| Asterids | Ericales | Ericaceae | Vaccinium macrocarpon# | 459,678 | NC_023338 | |
| Asterids | Gentianales | Rubiaceae | Scyphiphora hydrophyllacea | 354,155 | MT610041 | |
| Asterids | Lamiales | Lamiaceae | Salvia miltiorrhiza | 499,236 | NC_023209 | |
| Asterids | Lamiales | Lentibulariaceae | Utricularia reniformis | 857,234 | NC_034982 | |
| Asterids | Lamiales | Oleaceae | Olea europaea | 710,808 | MW262896 | |
| Asterids | Lamiales | Phrymaceae | Mimulus guttatus | 525,671 | NC_018041 | |
| Asterids | Solanales | Convolvulaceae | Ipomoea nil | 265,768 | NC_031158 | |
| Asterids | Solanales | Solanaceae | Capsicum annuum | 511,530 | KJ865410 | |
| Asterids | Solanales | Solanaceae | Nicotiana tabacum | 430,597 | NC_006581 | |
| Asterids | Solanales | Solanaceae | Solanum lycopersicum# | 446,257 | NC_035963 | |
| Commelinids | Poales | Poaceae | Oryza sativa | 637,692 | JF281153 | |
| Commelinids | Poales | Poaceae | Zea mays | 680,603 | DQ645539.1 | |
| Rosids | Brassicales | Brassicaceae | Arabidopsis thaliana | 367,808 | NC_037304 | |
| Rosids | Fabales | Fabaceae | Glycine max | 402,558 | NC_020455 | |
| Rosids | Rosales | Rosaceae | Malus domestica# | 396,947 | NC_018554 | |
| Rosids | Vitales | Vitaceae | Vitis vinifera | 773,279 | NC_012119 | |
| Outgroup | Conifers | Ginkgoales | Ginkgoaceae | Ginkgo biloba | 346,544 | KM672373 |
| Conifers | Pinales | Pinaceae | Pinus taeda | 1,191,054 | MF991879 | |
∗Represents the new mitogenome in this study. #Represents these species were used for mitogenome synteny and rearrangements through Mauve software.
The synonymous (Ks) and nonsynonymous (Ka) substitution rates of the protein-coding genes in the D. oleifera mt genome were analyzed using the 27 species. In this analysis, KaKs_Calculator (v2.0) [59] with the MLWL model was used to calculate Ka/Ks. Genome synteny and rearrangements among the using six representative species (Table 1) mitogenomes were analyzed using the progressive Mauve algorithm as implemented in Mauve ver. 2.4.0 software [60].
3. Results and discussion
3.1. Genomic features of the D. oleifera mt genome
The plant mitochondrial genome greatly varies in size, from 66 kb in Viscum scurruloideum [13] to 11.3 Mb in Silene conica [14]. We assembled the complete mt genome of D. oleifera in a single circular contig of 493,958 bp (GenBank accession number MW970112). The relatively medium size of the D. oleifera mt genome is similar to that of Vaccinium macrocarpon (459,678 bp) [3] and some asterids, such as Solanum lycopersicum (446,257 bp) [61], Salvia miltiorrhiza (499,236 bp) [62], and Capsicum annuum (511,530 bp) [63]; smaller than that of Rhododendron simsii (802,707 bp) [64] and Olea europaea (710,808 bp) [65]; and larger than that of Daucus carota (281,132 bp) [66] and Malus domestica (396,947 bp) [67].
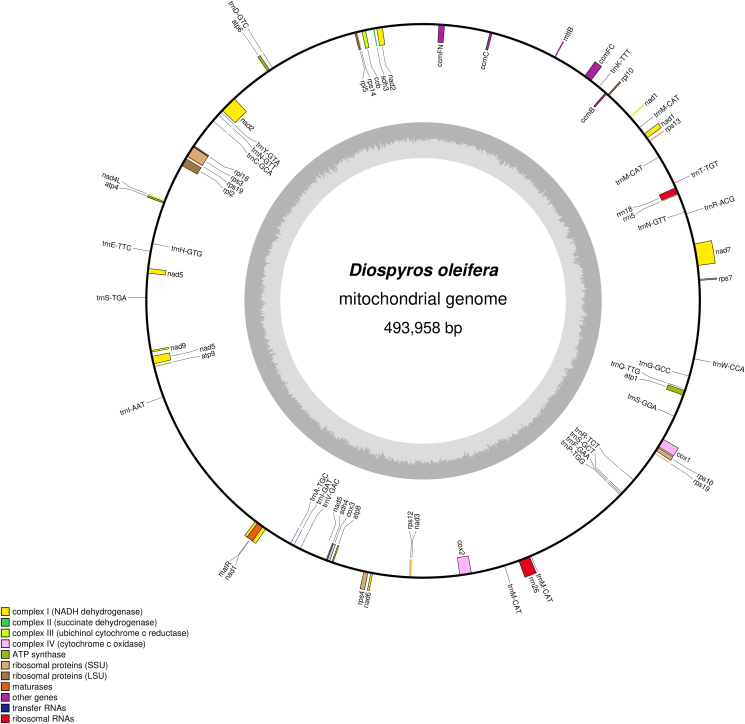
In the D. oleifera mt genome, 69 genes (39 protein-coding genes, 27 tRNA genes, and 3 rRNA genes (rrn5, rrn18, and rrn26)) were annotated. The functional categorization and physical locations of the annotated genes are shown in Figure 1. The 38 different proteins (rps19 has two copies) could be divided into 10 classes (Table 2): ATP synthase (five genes), cytochrome C biogenesis (four genes), ubiquinol cytochrome c reductase (one gene), cytochrome C oxidase (three genes), maturases (one gene), transport membrane protein (one gene), NADH dehydrogenase (nine genes), ribosomal proteins (LSU; four genes), ribosomal proteins (SSU; nine genes), and succinate dehydrogenase (two genes). ATG was used as the starting codon by almost all the protein-coding genes, and the four stop codons (TAA, TGA, TAG, and CGA) had utilization rates of 48.71%, 30.77%, 17.95%, and 2.57%, respectively.
Figure 1.
The circular map of D. oleifera mt genome. Gene map showing 69 annotated genes of different functional groups.
Table 2.
Gene profile and organization of the D. oleifera mt genome.
| Group of genes | Gene name | Length | Start codon | Stop codon | Amino acid |
|---|---|---|---|---|---|
| ATP synthase | atp1 | 1530 | ATG | TGA | 510 |
| atp4 | 579 | ATG | TAA | 193 | |
| atp6 | 807 | ATG | TGA | 269 | |
| atp8 | 480 | ATG | TAA | 160 | |
| atp9 | 285 | ATG | TAG | 95 | |
| Cytohrome c biogenesis | ccmB | 621 | ATG | TGA | 207 |
| ccmC | 753 | ATG | TGA | 251 | |
| ccmFC∗ | 1353 | ATG | TAA | 451 | |
| ccmFN | 1755 | ATG | TGA | 585 | |
| Ubichinol cytochrome c reductase | cob | 1182 | ATG | TGA | 394 |
| Cytochrome c oxidase | cox1∗ | 1584 | ACG (ATG) | TAA | 528 |
| cox2∗∗ | 780 | ATG | TAA | 260 | |
| cox3 | 798 | ATG | TGA | 266 | |
| Maturases | matR | 1968 | ATG | TAG | 656 |
| Transport membrance protein | mttB | 375 | ATG | TAG | 125 |
| NADH dehydrogenase | nad1∗∗∗∗ | 978 | ATG | TAA | 326 |
| nad2∗∗∗∗ | 1467 | ATG | TAA | 489 | |
| nad3 | 357 | ATG | TAA | 119 | |
| nad4∗∗∗ | 1488 | ATG | TGA | 496 | |
| nad4L | 273 | ATG | TAA | 91 | |
| nad5∗∗∗∗ | 2013 | ATG | TAA | 671 | |
| nad6 | 618 | ATG | TAA | 206 | |
| nad7∗∗∗∗ | 1185 | ATG | TAG | 395 | |
| nad9 | 588 | ATG | TAG | 196 | |
| Ribosomal proteins (LSU) | rpl10 | 489 | ATG | TAA | 163 |
| rpl16 | 435 | ND | TAA | 145 | |
| rpl2∗ | 1005 | ATG | TAA | 335 | |
| rpl5 | 564 | ATG | TAA | 188 | |
| Ribosomal proteins (SSU) | rps10∗ | 333 | ACG (ATG) | CGA | 111 |
| rps12 | 378 | ATG | TGA | 126 | |
| rps13 | 351 | ATG | TGA | 117 | |
| rps14 | 303 | ATG | TAG | 101 | |
| rps19(2) | (231,231) | ATG | TAA | 77 | |
| rps3∗ | 1752 | ATG | TAG | 584 | |
| rps4 | 1326 | ATG | TAA | 442 | |
| rps7 | 447 | ATG | TAA | 149 | |
| Succinate dehydrogenase | sdh3 | 306 | ATG | TGA | 102 |
| sdh4 | 432 | ATG | TGA | 144 | |
| Ribosomal RNAs | rrn18 | 1904 | |||
| rrn26 | 3373 | ||||
| rrn5 | 119 | ||||
| Transfer RNAs | trnA-TGC∗ | 67 | |||
| trnC-GCA | 71 | ||||
| trnD-GTC | 74 | ||||
| trnE-TTC | 72 | ||||
| trnF-GAA | 74 | ||||
| trnG-GCC | 72 | ||||
| trnH-GTG | 74 | ||||
| trnI-AAT | 69 | ||||
| trnI-GAT∗ | 74 | ||||
| trnK-TTT | 73 | ||||
| trnM-CAT(4) | (73,74,74,77) | ||||
| trnN-GTT(2) | (72,72) | ||||
| trnP-TGG | 75 | ||||
| trnQ-TTG | 72 | ||||
| trnR-ACG | 74 | ||||
| trnR-TCT∗ | 72 | ||||
| trnS-GCT | 88 | ||||
| trnS-GGA | 87 | ||||
| trnS-TGA | 87 | ||||
| trnT-TGT∗ | 75 | ||||
| trnV-GAC | 72 | ||||
| trnW-CCA | 74 | ||||
| trnY-GTA | 83 |
Previous studies have shown that rps10 is missing in the mt genomes of most plants, such as Arabidopsis thaliana, Brassica napus, and Beta vulgaris, and that its function is replaced by the nuclear gene [9]. However, the rps10 gene was found in the D. oleifera mt genome. The absence of rps2 and rps11 genes in the D. oleifera mt genome, consistent with R. simsii [64] and V. macrocarpon [3], supports Adams’ speculation that rps2 and rps11 genes were lost in the early evolution of eukaryotic plants [3]. Similar to Nicotiana tabacum [68] and M. luteus [4], the D. oleifera mt genome has no rps1 gene, whereas rps1 is present in the V. macrocarpon mt genome [3] and two copies of rps1 are present in the R. simsii mt genome [64].
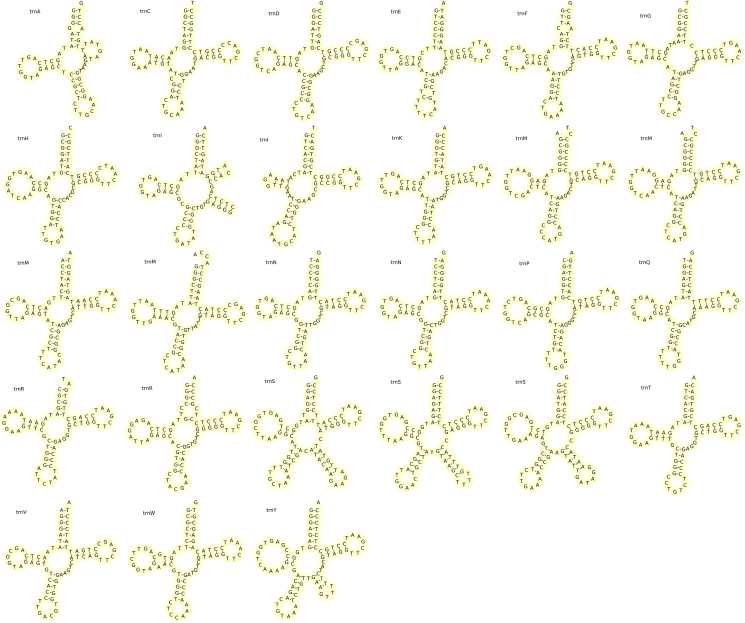
The persimmon mitochondria have 27 tRNAs (23 typical tRNA genes, one more trnN-GTT and three more trnM-CAT). The average length of these tRNAs is 67–88 bp, with a total length of 1479 bp. The number of tRNAs in the D. oleifera mt genome is more than that in other asterids, such as V. macrocarpon (18) [3], R. simsii (23) [64], M. luteus (24) [4], and N. tabacum (21) [68]. This may be because some tRNAs in the D. oleifera mt genome have multiple copies; for example, trnN-GTT has two copies and trnM-CAT has four copies. The secondary structures are shown in Figure 2. Following terms for Agris et al. [69, 70], secondary structures of most tRNAs were recovered as ordinal cloverleaf structures, which includes amino acid accepting stem (AAS), dihydrouridine stem and loop (DSL), anticodon stem and loop (ASL), thymidine stem and loop (TSL), furthermore, trnI-GAT, trnS-GCT, trnS-GGA, trnS-TGA, trnY-GTA were with an addition variable stem and loop (VSL). And, consist with many report [19, 21, 38, 39, 42], G-T (U) matches were also found in mostly tRNA secondary structures in the D. oleifera mt genome.
Figure 2.
Secondary structures of tRNAs of D. oleifera. Each region of tRNA is named as follows [69, 70]: Amino acid accepting stem, AAS (upper arm); dihydrouridine stem and loop, DSL (left arm); anticodon stem and loop, ASL (lower arm); thymidine stem and loop, TSL (right arm); variable stem and loop, VSL (between ASL and TSL).
The total gene length added up to 8% of the total mt genome length, with protein-coding regions comprising only 6.5% (32 kb) of the genome length. The gene content of D. oleifera is similar to that of the published mt genomes of asterids, especially Mimulus guttatus (7.4%) [4] and Helianthus annuus (8.5%) [71]. We found 54 genes with no introns, accounting for 78.26% of the total, consistent with the result conclusion that 63.2%–100% of mitochondrial genes in most plants have no introns [8, 9]. In addition, 30 introns were found in the other 15 D. oleifera mt genes; nad1, nad2, nad5, and nad7 had 4 introns; nad4, 3 introns; and cox2, 2 introns.
The nucleotide composition of the whole mt genome (Table 3) was found to be A (27.27%), T (27.03%), C (22.90%), and G (22.80%). The overall GC content was 45.7%, consistent with that of other asterids (V. macrocarpon 45.33% [3], D. carota 45.41% [66], Ilex pubescens 45.55% [35], Camellia sinensis 45.70% [33], and R. simsii 45.86% [64]). The GC skew was positive in CDS regions and negative in the mitochondrial genome. Strikingly, the GC content of the PCGs (43.11%) was lower than that of other CDS regions (tRNAs and rRNAs).
Table 3.
Composition and skewness of the D. oleifera mt genome.
| D.oleifera | Size (bp) | A% | T% | G% | C% | A + T% | G + C% | AT-skew | GC-skew |
|---|---|---|---|---|---|---|---|---|---|
| Mitogenome | 493958 | 27.27 | 27.03 | 22.8 | 22.9 | 54.3 | 45.7 | 0.004 | -0.002 |
| PCGs | 32400 | 26.59 | 30.3 | 21.9 | 21.21 | 56.89 | 43.11 | -0.065 | 0.016 |
| tRNAs | 2021 | 22.46 | 26.03 | 28.95 | 22.56 | 48.49 | 51.51 | -0.073 | 0.124 |
| rRNAs | 5396 | 25.8 | 22.07 | 29.23 | 22.91 | 47.87 | 52.13 | 0.078 | 0.121 |
3.2. Repeat sequences analysis
Simple sequence repeats (SSRs, or microsatellites) are DNA stretches consisting of short, tandem units of sequence repetitions 1–6 base pairs in length [72]. We identified 87 SSRs in the D. oleifera mt genome. The proportions of different repeat units are shown in Table 1. Tetranucleotide repeats were the most abundant SSR type, constituting 68.97% of all identified SSRs, and there were 7 SSRs in di-, tri-, and pentanucleotide repeats, accounting for 8.05% of all identified SSRs. There were only three mono- and hexanucleotide repeats in the D. oleifera mt genome. AAAG/CTTT motifs (16) were most recurrent motifs, representing 18.39% of all identified SSRs (Table S1).
Tandem repeats (satellite DNA) are core repeating units of 1–200 bases repeated several times in tandem [73]. As shown in Table 4, 12 tandem repeats 6 to 30 bp long were observed in the D. oleifera mt genome.
Table 4.
Distribution of perfect tandem repeats in the D. oleifera mt genome.
| NO. | Size | Copy | Repeat sequence | Percent Matches | Start | End |
|---|---|---|---|---|---|---|
| 1 | 30 | 1.9 | TACTACAATCCGTACGATAACTAGAATCCG | 82 | 123393 | 123450 |
| 2 | 18 | 2.2 | GCTTGATTCGGTGTAAAC | 90 | 143948 | 143987 |
| 3 | 20 | 2 | TTTGATTTCATCTTCATATAC | 90 | 176075 | 176115 |
| 4 | 14 | 2.8 | GGAGCTGACACCCT | 84 | 210479 | 210515 |
| 5 | 15 | 2.4 | AAATAAAAAAATAAA | 90 | 273479 | 273514 |
| 6 | 19 | 2.1 | AACAACCTATCTTGCGACA | 90 | 308468 | 308506 |
| 7 | 15 | 6.7 | ACAACCTATTATGCG | 70 | 308469 | 308572 |
| 8 | 18 | 2.1 | AATACTAATAGAATAGAA | 90 | 335217 | 335254 |
| 9 | 18 | 2.4 | CATAGTCGCGAGCTGTTT | 81 | 400200 | 400242 |
| 10 | 6 | 4.2 | AAAGAA | 100 | 409196 | 409220 |
| 11 | 18 | 5.2 | TATTGATGATAGTGACGA | 92 | 456597 | 456686 |
| 12 | 9 | 6.8 | ATTGATGAT | 73 | 456613 | 456673 |
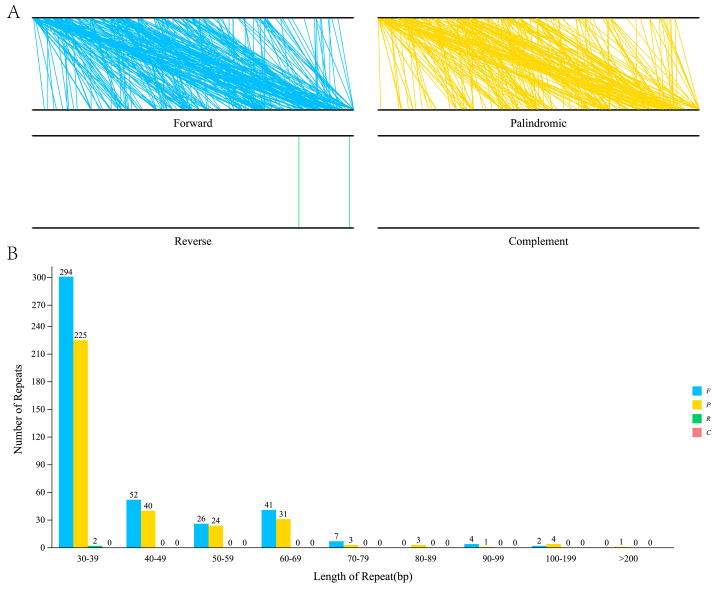
In addition, 760 non-tandem repeats, with 30 bp or more in length, were detected in the D. oleifera mt genome. Of the 760 non-tandem repeats, 426 were direct, 332 were palindromic, and 2 were reverse. The longest direct-type repeat was 115 bp long, while the longest inverted repeat was 331 bp long (Table S2). As shown in Figure 3, the 30–39 bp repeats were most abundant for both repeat types.
Figure 3.
The repeats in the D. oleifera mt genome. A: The synteny between the mt genome and its copy showing the direct repeats. B: The length distribution of reverse and inverted repeats in the D. oleifera mt genome. The number on the histograms represents the repeat number of designated lengths shown on the horizontal axi.
The repetitive sequence in the D. oleifera mt genome was 31 kb, accounting for 6.33% of the total mitochondria. This is considered a medium proportion of repeats, higher than that in Boea hygrometrica, (1.5%) and V. macrocarpon (3%) and lower than that in N. tabacum (13%) [68] and D. carota (16%) [66]. The different proportions of repeats may be because the mitochondria of B. hygrometrica, V. macrocarpon, and D. oleifera are mainly short repeating units, whereas those of tobacco and carrots are mainly longer repeating units [66].
3.3. The prediction of RNA editing
The number of RNA-editing sites varies in different species and is usually frequent in angiosperm and gymnosperm mitochondria. We predicted 515 RNA-editing sites within all the 38 protein-coding genes (Table 5) in the D. oleifera mt genome, which is more than those in A. thaliana (441) [5], Suaeda glauca (261) [73], Eucalyptus grandis (470) [74], and Citrullus lanatus (463) [75] and less than those in gymnosperms with larger mt genomes, such as Taxus cuspidata (974), Pinus taeda (1179), Cycas revoluta (1206), and Ginkgo biloba (1306) [32]. However, whether the number of RNA-editing sites is positively correlated with the size of the mt genome requires further research.
Table 5.
Prediction of RNA editing sites.
| Type | Effect | Number | Percentage (%) |
|---|---|---|---|
| Hydrophilic | CGT (R) => TGT (C) | 28 | 13.40 |
| CGC (R) => TGC (C) | 13 | ||
| CAT (H) => TAT (Y) | 20 | ||
| CAC (H) => TAC (Y) | 8 | ||
| Hydrophobic | GCT (A) => GTT (V) | 3 | 30.29 |
| GCG (A) => GTG (V) | 7 | ||
| GCC (A) => GTC (V) | 2 | ||
| CTT (L) => TTT (F) | 13 | ||
| CTC (L) => TTC (F) | 5 | ||
| CCT (P) => CTT (L) | 19 | ||
| CCG (P) => CTG (L) | 35 | ||
| CCC (P) => TTC (F) | 6 | ||
| CCC (P) => CTC (L) | 7 | ||
| CCA (P) => CTA (L) | 45 | ||
| CCT (P) => TTT (F) | 14 | ||
| Hydrophilic-hydrophobic | TCT (S) => TTT (F) | 44 | 47.57 |
| TCG (S) => TTG (L) | 49 | ||
| TCC (S) => TTC (F) | 29 | ||
| TCA (S) => TTA (L) | 78 | ||
| CGG (R) => TGG (W) | 30 | ||
| ACT (T) => ATT (I) | 4 | ||
| ACG (T) => ATG (M) | 6 | ||
| ACA (T) => ATA (I) | 5 | ||
| Hydrophilic-stop | CGA (R) => TGA (X) | 3 | 0.77 |
| CAA (Q) => TAA (X) | 1 | ||
| Hydrophobic-hydrophilic | CCT (P) => TCT (S) | 21 | 7.77 |
| CCC (P) => TCC (S) | 9 | ||
| CCA (P) => TCA (S) | 6 | ||
| CCG (P) => TCG (S) | 4 |
The selection of mitochondrial RNA-editing sites in D. oleifera shows a high degree of compositional bias. All RNA-editing sites are the C-T editing type, which is consistent with the fact that C-T is the most common editing type found in plant mt genomes [76, 77, 78]. In previous studies, almost half of the mitochondrial RNA editing occurred at the second codon position [73, 77]. The proportion of RNA-editing sites at the second codon position in the D. oleifera mt genome is also about 45.72% (235), slightly less than that at the first codon position (259; 50.39%). However, no editing site was found at the third position of triplet codons, consistent with the fact that RNA-editing sites are rare in plant mt genomes [73, 78].
Due to mitochondrial RNA editing, the D. oleifera mt genome has more RNA-editing sites but fewer editing types (Table 5). There were only 29 codon transfer types, corresponding to 14 amino acid transfer types, among the 515 RNA-editing sites. The types of transfer are comparable to those of most gymnosperms (30–40 codons; around 20 amino acids) [32, 76] but less than those of monocotyledonous and dicotyledonous plants (50–60 codons; around 30 amino acids) [74, 75, 78]. Among the 29 codon transfer types, TCA => TTA was the most common, with 78 sites. A leucine tendency after RNA editing, supported by the fact that 45.24% (233 sites) of the edits are converted to leucine, was found in the amino acids of predicted editing codons. After RNA editing, 43.59% of the amino acids remained hydrophobic. However, 47.57% of the amino acids were predicted to change from hydrophilic to hydrophobic, while 7.77% were predicted to change from hydrophobic to hydrophilic.
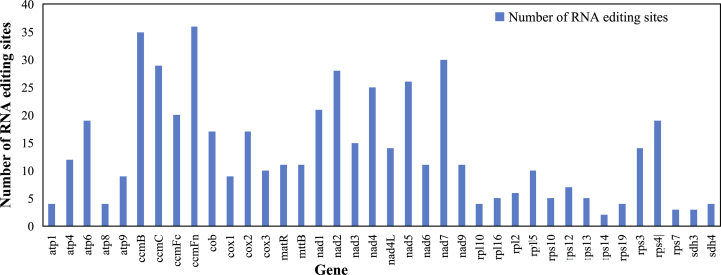
The number and type of RNA-editing sites differed among the mt genomes of D. oleifera and other species. Like with most angiosperms [73, 76], ribosomal proteins (except rps4) and ATPase subunits (except atp6) had a relatively small number of RNA-editing-derived substitutions (1–12 sites), while the transcripts of NADH dehydrogenase subunits and cytochrome c biogenesis genes were significantly edited (11–36 sites; Figure 4), and ccmFn and ccmB had the most RNA-editing sites predicted (36, 35).
Figure 4.
The distribution of RNA-editing sites in the D. oleifera mt protein-coding genes. The blue bars represent the number of RNA-editing sites of each gene.
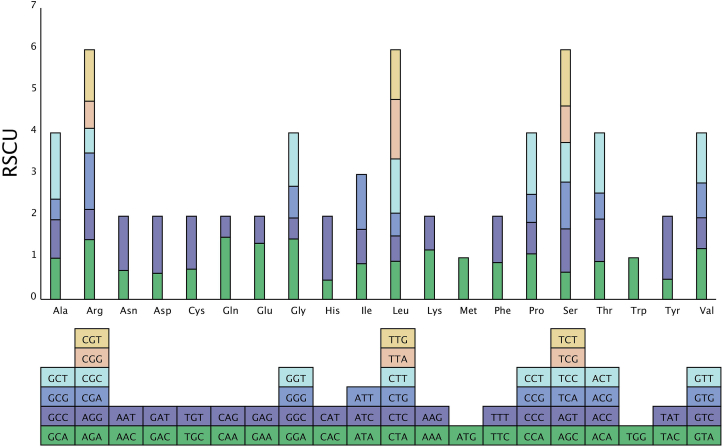
In D. oleifera, 10,611 amino acids were encoded. The most frequently used amino acids were Leu (10.25%), Ser (9.23%), and Arg (6.86%), and the least common amino acids were Trp (1.52%) and Met (2.65%) (Figure 4). The relative synonymous codon usage (RSCU) value for D. oleifera for the third codon position is shown in Figure 5. Consistent with most of the currently studied mitochondrial genomes [1, 73, 76], the use of both two- and four-fold degenerate codons was biased toward the use of codons abundant in A or T.
Figure 5.
Relative synonymous codon usage in the D. oleifera mt genome.
3.4. Chloroplast-derived mitogenomic sequences
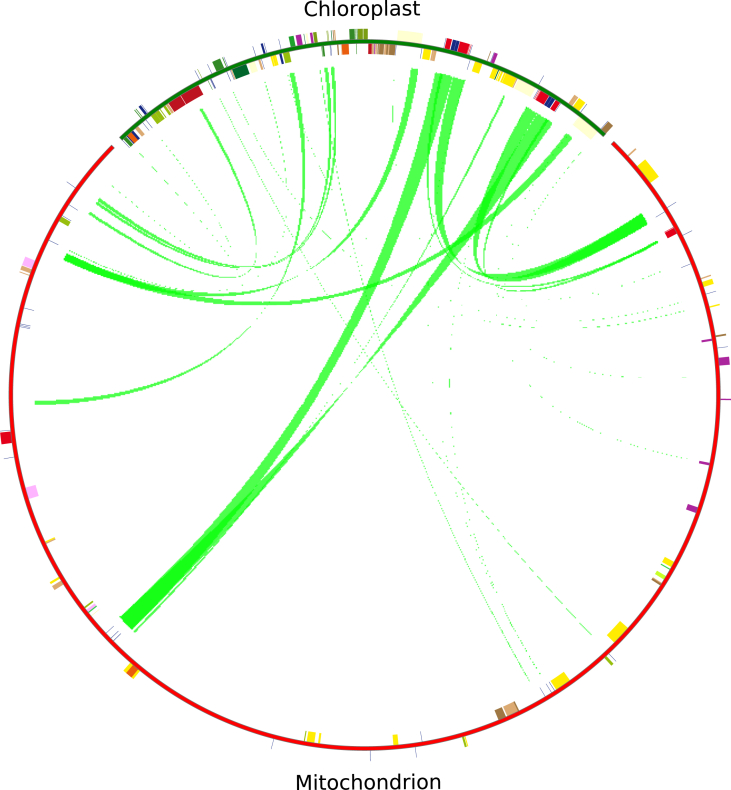
The transfer of DNA sequences among chloroplast and mt genomes has been frequently observed in the mt genomes of plants [79]. In many cases, the chloroplast DNA content in the mt genomes of most plants is 3%–6%, sometimes reaching up to about 10% [80]. The D. oleifera mt genome contained 28 chloroplast insertions, ranging in length from 32 to 5703 bp (Figure 6, Table 6), with a total length of 32.83 kb, accounting for 6.65% of the total length of the genome, which is greater than the mitochondrial genome lengths of Liriodendron tulipifera (3%) [31], and N. tabacum (2.5%) [68]; comparable to those of C. lanatus (6%) [75], E. grandis (6%) [74], and Oryza sativa (6.3%) [81]; and less than those of Vitis vinifera (8.8%) [36] and Cucurbita pepo (11.5%) [75].
Figure 6.
DNA and gene transfer between Chloroplast and Mitochondrial genomes in D. oleifera. The track shows complete genomes of cp and mt in green and red respectively.
Table 6.
Chloroplast insertions in the mitochondrial genome of D. oleifera.
| Chloroplast insertion | Start | End | Length | Chloroplast genes carried | Mitochondrial gene |
|---|---|---|---|---|---|
| 1 | 102039 | 107741 | 5703 | rps12-rrn16-rrn23-trnA-UGC-trnI-GAU-trnV-GAC | nad5/trnA-TGC/trnI-GAT/trnV-GAC/ |
| 2 | 137054 | 142756 | 5703 | rps12-rrn16-rrn23-trnA-UGC-trnI-GAU-trnVGAC | nad5/trnA-TGC/trnI-GAT/trnV-GAC |
| 3 | 132935 | 136943 | 4009 | rps12-rrn23-rrn4.5-rrn5-trnN-GUU-trnR-ACG | trnN-GTT/trnR-ACG/ |
| 4 | 107852 | 111860 | 4009 | rps12rrn23-rrn4.5-rrn5-trnN-GUU-trnR-ACG | trnN-GTT/trnR-ACG |
| 5 | 148183 | 150621 | 2439 | ycf15-ycf2 | ORF |
| 6 | 94174 | 96612 | 2439 | rps12-ycf15-ycf2 | ORF |
| 7 | 55481 | 56973 | 1493 | atpB-atpE | ORF |
| 8 | 66698 | 67774 | 1127 | psbE-psbF-psbJ-psbL | ORF |
| 9 | 24713 | 25651 | 939 | rpoB-rpoC1 | ORF |
| 10 | 68737 | 69666 | 939 | petG-petL-trnPUGG-trnWCCA | trnW-CCA |
| 11 | 103614 | 104477 | 888 | rps12-rrn16 | rrn18 |
| 12 | 140318 | 141181 | 888 | rps12-rrn16 | rrn18 |
| 13 | 124702 | 125419 | 719 | rps12-ndhA-ndhH | nad5 |
| 14 | 65137 | 65375 | 245 | petA | nad1 |
| 15 | 31892 | 32085 | 197 | trnDGUC | nad1/trnD-GTC |
| 16 | 47055 | 47223 | 171 | trnSGGA | trnS-GGA |
| 17 | 36672 | 36818 | 147 | psbC | nad1 |
| 18 | 1096 | 1190 | 96 | psbA | ORF |
| 19 | 9282 | 9370 | 92 | trnSGCU | trnS-GGA |
| 20 | 133140 | 133220 | 82 | rps12-trnN-GUU | nad1/trnN-GTT |
| 21 | 111575 | 111655 | 82 | rps12-trnN-GUU | nad1/trnN-GTT |
| 22 | 54808 | 54886 | 79 | TrnM-CAU | nad1/trnM-CAT |
| 23 | 155661 | 155735 | 77 | TrnI-CAU | nad1/ccmC/orf |
| 24 | 89060 | 89134 | 77 | rps12-trnI-CAU | nad1/ccmC/orf |
| 25 | 95837 | 95897 | 61 | rps12-ycf2 | nad1/orf |
| 26 | 148898 | 148958 | 61 | ycf2 | nad1/orf |
| 27 | 155661 | 155692 | 32 | trnICAU | ORF |
| 28 | 89103 | 89134 | 32 | rps12-trnICAU | ORF |
Among the transfer DNA sequences, some chloroplast protein-coding genes, such as atpB, atpE, rps12, rpoB, petA, psaA, and psbC, lost their integrity while migrating from the cp to the mitochondria, and only partial sequences of those cp-derived PCGs could be found in the D. oleifera mt genome (Table 6). In the D. oleifera mt genome, 11 chloroplast-derived tRNAs with a complete sequence were identified: trnA-UGC, trnD-GUC, trnI-GAU, trnM-CAU, trnN-GUU, trnP-UGG, trnR-ACG, trnS-GCU, trnS-GGA, trnV-GAC, and trnW-CCA. The different completeness levels of the transferred PCGs and tRNA genes showed that tRNA genes are much more conserved in the mt genome than PCGs, indicating that tRNA genes play an indispensable role in mitochondria. The transfer of these tRNAs can be traced back to the retention of an earlier horizontal gene transfer event. In accordance with the present results, cp-derived trnM-CAU first appeared in gymnosperms [82]; cp-derived trnD-GUC mainly appeared in dicotyledons, not in monocotyledons [76]; and cp-derived trnM-CAU and trnD-GUC were both found in the D. oleifera mt genome. However, the absence of cp-derived trnH-GTG, which is commonly found in angiosperms [3, 74, 76, 82], and the presence of cp-derived trnA-UGC, lost during early evolution of terrestrial plants [80, 83], indicate that special evolutionary events may be occurring during D. oleifera formation.
3.5. Phylogenetic, Ka/Ks and gene arrangement analysis
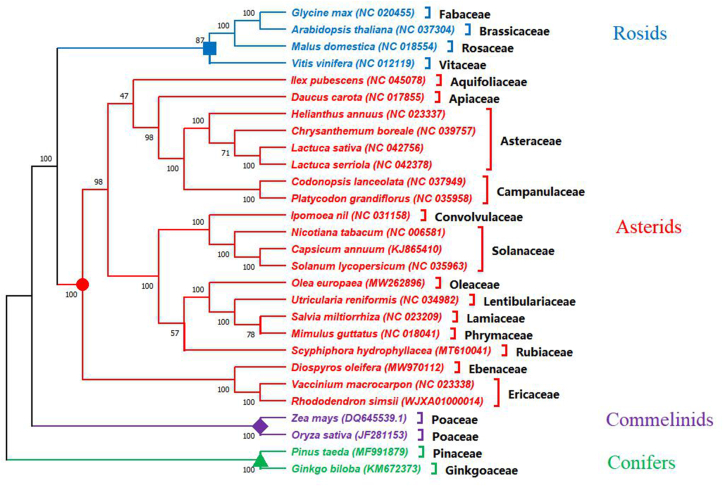
To detect the evolutionary status of the D. oleifera mt genome, a phylogenetic analysis was performed on D. oleifera, together with 27 other species: 23 eudicots (19 asterids and 4 rosids), 2 monocotyledons, and 2 gymnosperms (designated as outgroups). Phylogenetic relationships were analyzed using the concatenated dataset (28 PCGs: atp1, atp4, atp6, atp8, atp9, ccmB, ccmC, ccmFC, ccmFN, cob, cox1, cox2, cox3, matR, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, rpl5, rps12, rps13, rps3 and rps4) through ML phylogenetic analysis. The abbreviations and accession numbers of the mt genomes investigated in this study are listed in Table S1. As outgroups, the two gymnosperms were distinct from the other angiosperms. The phylogenetic tree (Figure 7) strongly supported the separation of asterids from rosids and the separation of eudicots from monocots. Moreover, the taxa from 20 families (Apiaceae, Aquifoliaceae, Asteraceae, Brassicaceae, Campanulaceae, Convolvulaceae, Ericaceae, Ebenaceae, Fabaceae, Ginkgoaceae, Lamiaceae, Lentibulariaceae, Oleaceae, Phrymaceae, Pinaceae, Poaceae, Rosaceae, Rubiaceae, Solanaceae, and Vitaceae) were well clustered. In addition, the monophyly of D. oleifera, which belongs to the single genus of Diospyros in the Ebenaceae family, was well supported based on mt genomes (Figure 7). Consistent with previous comparative genome studies [23, 28, 29], this study also found that the clade united V. macrocarpon and R. simsii and then formed a sister cluster with the Ebenaceae family with high confidence (bootstrap value of 100%). In general, the phylogenetic tree topology was in line with the evolutionary relationships among those species, indicating the consistency of traditional taxonomy with the molecular classification.
Figure 7.
The phylogenetic relationships of D. oleifera with other 27 plant species using the maximum likelihood (ML) analysis. The bootstrapping values are listed in each node. The number after the species name is the GenBank accession number. Colors indicate the groups that the specific species belongs.
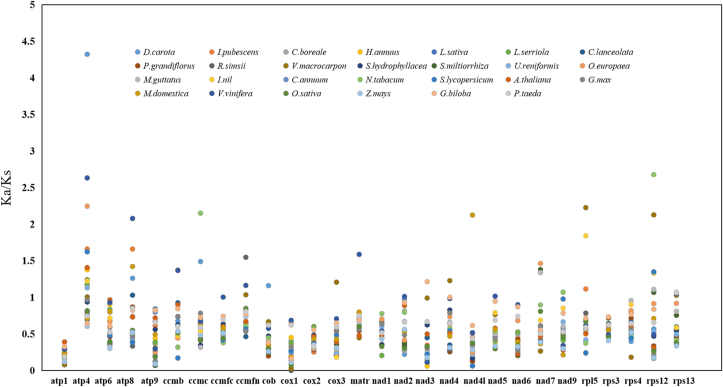
To evaluate selective pressures during the evolutionary dynamics of protein-coding genes among closely related species, the nonsynonymous (Ka) and synonymous (Ks) substitution ratio (Ka/Ks) was calculated. For the Ka/Ks calculation, 28 PCGs from the D. oleifera mt genome were compared with the mt genomes of 27 species.
As shown in Figure 8, for the gene-specific substitution rates, Ka/Ks ranged from 0.031 at the cox1 gene in V. macrocarpon to 4.321 at the atp4 gene in D. carota. In 58 cases (except Glycine max, O. sativa, Platycodon grandiflorus, Scyphiphora hydrophyllacea and Z. mays), the Ka/Ks values of D. oleifera gene-specific substitution rates were higher than 1, compared with 22 other species, suggesting positive selection during evolution. Among the 22 species, nine substitution genes with higher Ka/Ks values were found between the D. oleifera and V. vinifera mt genomes and six genes between the D. oleifera and V. macrocarpon mt genomes. The atp4 and atp8 genes exhibited the highest average rate (1.348 and 0.751) and 15 and 5 Ka/Ks values above 1, respectively, suggested to be the result of positive or relaxed selection [2]. However, most genes had undergone negative selection pressures during evolution, supported by the fact that the Ka/Ks values of 654 proteins, accounting for 91.59% of the proteins in D. oleifera, were less than 1 compared to the other plant species. The atp1 and cox1 genes have the smallest average Ka/Ks values (0.212 and 0.272), indicating strong purifying selection [34, 84]. These results show that mt genes are highly conserved during the evolutionary process in green plants.
Figure 8.
The Ka/Ks values of 28 protein-coding genes of D. oleifera versus 27 species.
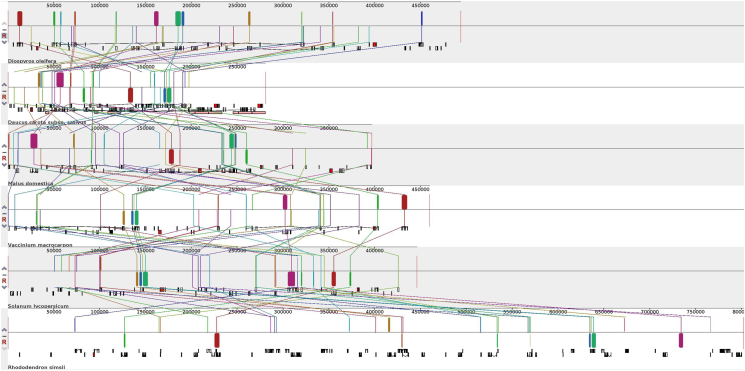
Because of no mt genome of any species in Ebenaceae has been reported, Synteny of entire mitochondrial genomes was only compared among four Asterids (including three Ericales, one Apiales, and one Solanales) and one Rosids species in this study to assess the degree of structural rearrangement between different lineages. Figure 9 and Figs.1 showed that the rearrangement of mitochondrial genes has been widely occur among these six species, which is accords with many mitogenome observations [20, 37, 38, 39, 41, 42]. When using D. oleifera as a reference genome, The dot-plot analyses showed sequences or synteny were seldom shared, and only short stretches of synteny among species (Figs.2). These Large rearrangement events have indicated differentiation within these six species mitogenome. Understandably, species that have close evolutionary relationships share more clusters [20, 41, 42], for example, In general, longer synteny sequences with higher similarity were found between D. oleifera and V. macrocarpon than that between D. oleifera and M. domestica.
Figure 9.
Synteny analysis of D. oleifera and other five species mitogenomes as generated with Mauve. The sizes and relative positions of the homologous fragments varied across mitogenomes.
Among genus Diospyros, The most well-known species is D. kaki, which has been cultivated as an important fruit crop, due to its edible fruit [25]. However, D. kaki are hexaploid (2n = 6× = 90) or nonaploid (2n = 9× = 135) and their origin, and polyploidization mechanisms are unclear, which has hampered genome sequencing and molecular breeding [23, 28]. Phylogenetic analyses based on the nuclear [23, 28] and chloroplast [26, 27] genome and mtDNA non-coding fragments [85] have indicated that D. oleifera is more closely related to D. kaki [24] and could be used as a model plant for studies of Diospyros [24, 26]. So, as the nuclear and mt genome of hexaploid cultivated persimmon both remains unpublished, the availability of the D. oleifera mt genome provides more alternative comparable reference for D. kaki than D. lotus does. In addition, our results will lay the foundation for identifying further evolutionary relationships within Ebenaceae. However, due to the lack of adequate representative mitogenomes, more Ebenaceae mitogenomes are needed to be sequenced to better resolve the phylogeny and evolutionary biology of Ebenaceae.
4. Conclusions
Here, we presented the first mitochondrial genome assembly and annotation of an Ebenaceae model plant Diospyros oleifera as well as the mitochondrial genome in the family Ebenaceae. The mitogenome was 493,958 bp in length, contained 39 protein-coding genes, 27 transfer RNA genes, and 3 ribosomal RNA genes. Comparative analysis of gene structure, codon usage, repeat regions and RNA-editing sites shows that rps2 and rps11 genes are missing, and a clear bias of RNA-editing sites is existing in the D. oleifera mt genome. In addition, the phenomenon that intracellular tRNA genes transferred frequently from chloroplasts to mitochondria was also observed in D. oleifera. Moreover, Phylogenetic analysis based on the mt genomes of D. oleifera and 27 other taxa indicates consistency in molecular and taxonomic classification. Furthermore, The Ka/Ks analysis based on code substitution revealed that most of the coding genes had undergone negative selections, indicating the conservation of mt genes during the evolution. These results will help in better understanding the features of the D. oleifera mitochondrial genome and lay the foundation for identifying further evolutionary relationships within Ebenaceae. However, due to the lack of adequate representative mitogenomes, more Ebenaceae mitogenomes are needed to be sequenced to better resolve the phylogeny and evolutionary biology of Ebenaceae.
Declarations
Author contribution statement
Yang Xu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yi Dong: Performed the experiments.
Wenqiang Cheng; Haidong Gao, Lei Liu and Lei Xu: Analyzed and interpreted the data.
Kaiyun Wu: Contributed reagents, materials, analysis tools or data.
Bangchu Gong: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Ph.D. Yang Xu was supported by National Key R & D Program of China [2018YFD1000606].
Ph.D. Yang Xu was supported by National Key R & D Program of China [2019YFD1000600].
Bangchu Gong was supported by Key Project for New Agricultural Cultivar Breeding in Zhejiang Province, China [2021C02066-10].
Data availability statement
Data associated with this study [The final annotated mt genome sequences of D. oleifera] has been deposited at NCBI GenBank under the accession number MW970112.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Analysis of conservative gene clusters between the D. oleifera mt genome and other plant mt genomes.
Dot-plot graphs indicating regions of synteny between mitochondrial genomes compared to D. oleifera as the reference.
Distribution to different repeat type classes in the D. oleifera mt genome.
Frequency of classified SSR motifs in the D. oleifera mt genome.
Repeats (≥30bp) in the D. oleifera mt genome.
References
- 1.Bi C., Lu N., Xu Y., He C., Lu Z. Characterization and analysis of the mitochondrial genome of common bean (Phaseolus vulgaris) by comparative genomic approaches. Int. J. Mol. Sci. 2020;21(11) doi: 10.3390/ijms21113778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozik A., Rowan B.A., Lavelle D., Berke L., Schranz M.E., Michelmore R.W., Christensen A.C. The alternative reality of plant mitochondrial DNA: one ring does not rule them all. PLoS Genet. 2019;15(8) doi: 10.1371/journal.pgen.1008373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fajardo D., Schlautman B., Steffan S., Polashock J., Vorsa N., Zalapa J. The American cranberry mitochondrial genome reveals the presence of selenocysteine (tRNA-Sec and SECIS) insertion machinery in land plants. Gene. 2014;536(2):336–343. doi: 10.1016/j.gene.2013.11.104. [DOI] [PubMed] [Google Scholar]
- 4.Vallejo-Marin M., Cooley A.M., Lee M.Y., Folmer M., McKain M.R., Puzey J.R. Strongly asymmetric hybridization barriers shape the origin of a new polyploid species and its hybrid ancestor. Am. J. Bot. 2016;103(7):1272–1288. doi: 10.3732/ajb.1500471. [DOI] [PubMed] [Google Scholar]
- 5.Greiner S., Bock R. Tuning a menage a trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. Bioessays. 2013;35(4):354–365. doi: 10.1002/bies.201200137. [DOI] [PubMed] [Google Scholar]
- 6.Ogihara Y., Yamazaki Y., Murai K., Kanno A., Terachi T., Shiina T., Miyashita N., Nasuda S., Nakamura C., Mori N., Takumi S., Murata M., Futo S., Tsunewaki K. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 2005;33(19):6235–6250. doi: 10.1093/nar/gki925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004;5(2):123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 8.Chevigny N., Schatz-Daas D., Lotfi F., Gualberto J.M. DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 2020;21(1) doi: 10.3390/ijms21010328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubo T., Newton K.J. Angiosperm mitochondrial genomes and mutations. Mitochondrion. 2008;8(1):5–14. doi: 10.1016/j.mito.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Best C., Mizrahi R., Ostersetzer-Biran O. Why so complex? The intricacy of genome structure and gene expression, associated with angiosperm mitochondria, may relate to the regulation of embryo quiescence or dormancy-intrinsic blocks to early plant life. Plants. 2020;9(5) doi: 10.3390/plants9050598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen A.C. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol. Evol. 2013;5(6):1079–1086. doi: 10.1093/gbe/evt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palumbo F., Vitulo N., Vannozzi A., Magon G., Barcaccia G. The mitochondrial genome assembly of fennel (Foeniculum vulgare) reveals two different atp6 gene sequences in cytoplasmic male sterile accessions. Int. J. Mol. Sci. 2020;21(13) doi: 10.3390/ijms21134664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skippington E., Barkman T.J., Rice D.W., Palmer J.D. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. U.S.A. 2015;112(27):E3515–E3524. doi: 10.1073/pnas.1504491112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sloan D.B., Alverson A.J., Chuckalovcak J.P., Wu M., McCauley D.E., Palmer J.D., Taylor D.R. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10(1) doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen G., Cuenca A., Moller I.M., Seberg O. Massive gene loss in mistletoe (Viscum, Viscaceae) mitochondria. Sci. Rep. 2015;5 doi: 10.1038/srep17588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr. Genet. 2004;46(3):123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 17.O'Conner S., Li L. Mitochondrial fostering: the mitochondrial genome may play a role in plant orphan gene evolution. Front. Plant Sci. 2020;11 doi: 10.3389/fpls.2020.600117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan L., Xu W., Zhang D., Li J. Comparative analysis of the mitochondrial genomes of flesh flies and their evolutionary implication. Int. J. Biol. Macromol. 2021;174:385–391. doi: 10.1016/j.ijbiomac.2021.01.188. [DOI] [PubMed] [Google Scholar]
- 19.Zhang R., Li J., Geng S., Yang J., Zhang X., An Y., Li C., Cui H., Li X., Wang Y. The first mitochondrial genome for Phaudidae (Lepidoptera) with phylogenetic analyses of Zygaenoidea. Int. J. Biol. Macromol. 2020;149:951–961. doi: 10.1016/j.ijbiomac.2020.01.307. [DOI] [PubMed] [Google Scholar]
- 20.Tyagi K., Kumar V., Poddar N., Prasad P., Tyagi I., Kundu S., Chandra K. The gene arrangement and phylogeny using mitochondrial genomes in spiders (Arachnida: araneae) Int. J. Biol. Macromol. 2020;146:488–496. doi: 10.1016/j.ijbiomac.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Sharma A., Siva C., Ali S., Sahoo P.K., Nath R., Laskar M.A., Sarma D. The complete mitochondrial genome of the medicinal fish, Cyprinion semiplotum: insight into its structural features and phylogenetic implications. Int. J. Biol. Macromol. 2020;164:939–948. doi: 10.1016/j.ijbiomac.2020.07.142. [DOI] [PubMed] [Google Scholar]
- 22.Duangjai S., Wallnofer B., Samuel R., Munzinger J., Chase M.W. Generic delimitation and relationships in Ebenaceae sensulato: evidence from six plastid DNA regions. Am. J. Bot. 2006;93(12):1808–1827. doi: 10.3732/ajb.93.12.1808. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Q.G., Xu Y., Yang Y., Guan C.F., Zhang Q.Y., Huang J.W., Grierson D., Chen K.S., Gong B.C., Yin X.R. The persimmon (Diospyros oleifera Cheng) genome provides new insights into the inheritance of astringency and ancestral evolution. Hortic. Res. 2019;6:138. doi: 10.1038/s41438-019-0227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanzaki S. The origin and cultivar development of Japanese Persimmon (Diospyros kaki Thunb.) J. Jpn. Soc. Food Sci. Technol. 2016;63:328–330. [Google Scholar]
- 25.Wang R.Z., Yang Y., Li G.C. Chinese persimmon germplasm resources. Acta Hortic. 1997;20(3):43–50. [Google Scholar]
- 26.Fu J., Liu H., Hu J., Liang Y., Liang J., Wuyun T., Tan X. Five complete chloroplast genome sequences from Diospyros: genome organization and comparative analysis. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0159566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W., Liu Y., Yang Y., Xie X., Lu Y., Yang Z., Jin X., Dong W., Suo Z. Interspecific chloroplast genome sequence diversity and genomic resources in Diospyros. BMC Plant Biol. 2018;18(1):210. doi: 10.1186/s12870-018-1421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suo Y., Sun P., Cheng H., Han W., Diao S., Li H., Mai Y., Zhao X., Li F., Fu J. A high-quality chromosomal genome assembly of Diospyros oleifera Cheng. GigaScience. 2020;9(1) doi: 10.1093/gigascience/giz164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akagi T., Shirasawa K., Nagasaki H., Hirakawa H., Tao R., Comai L., Henry I.M. The persimmon genome reveals clues to the evolution of a lineage-specific sex determination system in plants. PLoS Genet. 2020;16(2) doi: 10.1371/journal.pgen.1008566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao W., Yao G., Wang S., Zhou L., Hu G. Chromosome-level genomes of seeded and seedless date plum based on third-generation DNA sequencing and hi-c analysis. Forest. Res. 2021;1:9. [Google Scholar]
- 31.Dong S., Chen L., Liu Y., Wang Y., Zhang S., Yang L., Lang X., Zhang S. The draft mitochondrial genome of Magnolia biondii and mitochondrial phylogenomics of angiosperms. PLoS One. 2020;15(4) doi: 10.1371/journal.pone.0231020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kan S.L., Shen T.T., Gong P., Ran J.H., Wang X.Q. The complete mitochondrial genome of Taxus cuspidata (Taxaceae): eight protein-coding genes have transferred to the nuclear genome. BMC Evol. Biol. 2020;20(1):10. doi: 10.1186/s12862-020-1582-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawal H.C., Kumar P.M., Bera B., Singh N.K., Mondal T.K. Decoding and analysis of organelle genomes of Indian tea (Camellia assamica) for phylogenetic confirmation. Genomics. 2020;112(1):659–668. doi: 10.1016/j.ygeno.2019.04.018. [DOI] [PubMed] [Google Scholar]
- 34.Wang X., Cheng F., Rohlsen D., Bi C., Wang C., Xu Y., Wei S., Ye Q., Yin T., Ye N. Organellar genome assembly methods and comparative analysis of horticultural plants. Hortic. Res. 2018;5:3. doi: 10.1038/s41438-017-0002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Z., Hao Y., Xu Y. Characterization of the complete mitochondrial genome of lex pubescens. Mitochondrial. DNA. Resour. B. 2019;4(1):2003–2004. [Google Scholar]
- 36.Yin X., Gao Y., Song S., Hassani D., Lu J. Identification, characterization and functional analysis of grape (Vitis vinifera L.) mitochondrial transcription termination factor (mTERF) genes in responding to biotic stress and exogenous phytohormone. BMC Genom. 2021;22(1):136. doi: 10.1186/s12864-021-07446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren L., Zhang X., Li Y., Shang Y., Chen S., Wang S., Qu Y., Cai J., Guo Y. Comparative analysis of mitochondrial genomes among the subfamily Sarcophaginae (Diptera: sarcophagidae) and phylogenetic implications. Int. J. Biol. Macromol. 2020;161:214–222. doi: 10.1016/j.ijbiomac.2020.06.043. [DOI] [PubMed] [Google Scholar]
- 38.Xu X.-D., Guan J.-Y., Zhang Z.-Y., Cao Y.-R., Storey K.B., Yu D.-N., Zhang J.-Y. Novel tRNA gene rearrangements in the mitochondrial genomes of praying mantises (Mantodea: mantidae): translocation, duplication and pseudogenization. Int. J. Biol. Macromol. 2021;185:403–411. doi: 10.1016/j.ijbiomac.2021.06.096. [DOI] [PubMed] [Google Scholar]
- 39.Chen Z., Liu Y., Wu Y., Song F., Cai W., Li H. Novel tRNA gene rearrangements in the mitochondrial genome of Camarochiloides weiweii (Hemiptera: pachynomidae) Int. J. Biol. Macromol. 2020;165:1738–1744. doi: 10.1016/j.ijbiomac.2020.10.051. [DOI] [PubMed] [Google Scholar]
- 40.Feng Z., Wu Y., Yang C., Gu X., Wilson J.J., Li H., Cai W., Yang H., Song F. Evolution of tRNA gene rearrangement in the mitochondrial genome of ichneumonoid wasps (Hymenoptera: ichneumonoidea) Int. J. Biol. Macromol. 2020;164:540–547. doi: 10.1016/j.ijbiomac.2020.07.149. [DOI] [PubMed] [Google Scholar]
- 41.Wu Y., Yang H., Feng Z., Li B., Zhou W., Song F., Li H., Zhang L., Cai W. Novel gene rearrangement in the mitochondrial genome of Pachyneuron aphidis (Hymenoptera: pteromalidae) Int. J. Biol. Macromol. 2020;149:1207–1212. doi: 10.1016/j.ijbiomac.2020.01.308. [DOI] [PubMed] [Google Scholar]
- 42.Monnens M., Thijs S., Briscoe A.G., Clark M., Frost E.J., Littlewood D.T.J., Sewell M., Smeets K., Artois T., Vanhove M.P.M. The first mitochondrial genomes of endosymbiotic rhabdocoels illustrate evolutionary relaxation of atp8 and genome plasticity in flatworms. Int. J. Biol. Macromol. 2020;162:454–469. doi: 10.1016/j.ijbiomac.2020.06.025. [DOI] [PubMed] [Google Scholar]
- 43.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mondal T.K., Rawal H.C., Chowrasia S., Varshney D., Panda A.K., Mazumdar A., Kaur H., Gaikwad K., Sharma T.R., Singh N.K. Draft genome sequence of first monocot-halophytic species Oryza coarctata reveals stress-specific genes. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-31518-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J., Fan J., Sun Z., Liu S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2020;36(7):2253–2255. doi: 10.1093/bioinformatics/btz891. [DOI] [PubMed] [Google Scholar]
- 47.Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E.S., Fischer A., Bock R., Greiner S. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45(W1):W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan P.P., Lowe T.M. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019;1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lohse M., Drechsel O., Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52(5-6):267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 50.Perna N.T., Kocher T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995;41:353–358. doi: 10.1007/BF00186547. [DOI] [PubMed] [Google Scholar]
- 51.Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA package 2.0. Algorithm Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mower J.P. PREP-Mt: predictive RNA editor for plant mitochondrial genes. BMC Bioinf. 2005;6:96. doi: 10.1186/1471-2105-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28(6):1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 54.Lee Python implementation of codon adaptation index. J. Open. Source. Softw. 2018;30:905. [Google Scholar]
- 55.Thiel T., Michalek W., Varshney R.K., Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theor. Appl. Genet. 2003;106(3):411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 56.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Posada D., Crandall K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 59.Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Dev. Reprod. Biol. 2010;8(1):77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Causse M., Giovannoni J., Bouzayen M., Zouine M. In: Compendium of Plant Genomes. Causse M., Giovannoni J., Bouzayen M., Zouine M., editors. Springer; Berlin, Germany: 2016. The chloroplast and mitochondrial genomes of tomato; pp. 111–137. Chapter 7. [Google Scholar]
- 62.Chen H., Liu C. In: Compendium of Plant Genomes. Lu S., editor. Springer; Berlin, Germany: 2019. The chloroplast and mitochondrial genomes of Salvia miltiorrhiza; pp. 55–68. (2019) [Google Scholar]
- 63.Jo Y.D., Choi Y., Kim D.H., Kim B.D., Kang B.C. Extensive structural variations between mitochondrial genomes of CMS and normal peppers (Capsicum annuum L.) revealed by complete nucleotide sequencing. BMC Genom. 2014;15:561. doi: 10.1186/1471-2164-15-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu J., Luo H., Nie S., Zhang R.G., Mao J.F. The complete mitochondrial and plastid genomes of Rhododendron simsii, an important parent of widely cultivated azaleas. Mitochondrial DNA B Resour. 2021;6(3):1197–1199. doi: 10.1080/23802359.2021.1903352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nardi F., Carapelli A., Boore J.L., Roderick G.K., Dallai R., Frati F. Domestication of olive fly through a multi-regional host shift to cultivated olives: comparative dating using complete mitochondrial genomes. Mol. Phylogenet. Evol. 2010;57(2):678–686. doi: 10.1016/j.ympev.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 66.Iorizzo M., Senalik D., Szklarczyk M., Grzebelus D., Spooner D., Simon P. De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol. 2012;12:61. doi: 10.1186/1471-2229-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goremykin V.V., Lockhart P.J., Viola R., Velasco R. The mitochondrial genome of Malus domestica and the import-driven hypothesis of mitochondrial genome expansion in seed plants. Plant J. 2012;71(4):615–626. doi: 10.1111/j.1365-313X.2012.05014.x. [DOI] [PubMed] [Google Scholar]
- 68.Sugiyama Y., Watase Y., Nagase M., Makita N., Yagura S., Hirai A., Sugiura M. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol. Genet. Genom. 2005;272(6):603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 69.Agris P.F., Eruysal E.R., Narendran A., Väre V.Y.P., Vangaveti S., Ranganathan S.V. Celebrating wobble decoding: half a century and still much is new. RNA Biol. 2018;15:537–553. doi: 10.1080/15476286.2017.1356562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agris P.F., Vendeix F.A.P., Graham W.D. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 71.Grassa C.J., Ebert D.P., Kane N.C., Rieseberg L.H. Complete mitochondrial genome sequence of sunflower (Helianthus annuus L.) Genome Announc. 2016;4(5) doi: 10.1128/genomeA.00981-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Y., Cheng W., Xiong C., Jiang X., Wu K., Gong B. Genetic diversity and association analysis among germplasms of Diospyros kaki in Zhejiang Province based on SSR markers. Forest. 2021;12(4):422. [Google Scholar]
- 73.Cheng Y., He X., Priyadarshani S., Wang Y., Ye L., Shi C., Ye K., Zhou Q., Luo Z., Deng F., Cao L., Zheng P., Aslam M., Qin Y. Assembly and comparative analysis of the complete mitochondrial genome of Suaeda glauca. BMC Genom. 2021;22(1):167. doi: 10.1186/s12864-021-07490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pinard D., Myburg A.A., Mizrachi E. The plastid and mitochondrial genomes of Eucalyptus grandis. BMC Genom. 2019;20(1):132. doi: 10.1186/s12864-019-5444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alverson A.J., Wei X., Rice D.W., Stern D.B., Barry K., Palmer J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae) Mol. Biol. Evol. 2010;27(6):1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edera A.A., Sanchez-Puerta M.V. Computational detection of plant RNA editing events. Methods Mol. Biol. 2021;2181:13–34. doi: 10.1007/978-1-0716-0787-9_2. [DOI] [PubMed] [Google Scholar]
- 77.Robles P., Quesada V. Organelle genetics in plants. Int. J. Mol. Sci. 2021;22(4) doi: 10.3390/ijms22042104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verhage L. Targeted editing of the Arabidopsis mitochondrial genome. Plant J. 2020;104(6):1457–1458. doi: 10.1111/tpj.15097. [DOI] [PubMed] [Google Scholar]
- 79.Straub S.C., Cronn R.C., Edwards C., Fishbein M., Liston A. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (apocynaceae) Genome Biol. Evol. 2013;5(10):1872–1885. doi: 10.1093/gbe/evt140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adams K.L., Qiu Y.L., Stoutemyer M., Palmer J.D. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. U.S.A. 2002;99(15):9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Asaf S., Khan A.L., Khan A.R., Waqas M., Kang S.M., Khan M.A., Shahzad R., Seo C.W., Shin J.H., Lee I.J. Mitochondrial genome analysis of wild rice (Oryza minuta) and its comparison with other related species. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0152937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Richardson A.O., Palmer J.D. Horizontal gene transfer in plants. J. Exp. Bot. 2007;58(1):1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- 83.Bergthorsson U., Adams K.L., Thomason B., Palmer J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424(6945):197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- 84.Fay J.C., Wu C.I. Sequence divergence, functional constraint, and selection in protein evolution. Annu. Rev. Genom. Hum. Genet. 2003;4:213–235. doi: 10.1146/annurev.genom.4.020303.162528. [DOI] [PubMed] [Google Scholar]
- 85.Hu D.C., Luo Z.R. Polymorphisms of amplified mitochondrial DNA non-coding regions in Diospyros spp. Sci. Hortic. 2006;109(3):275–281. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of conservative gene clusters between the D. oleifera mt genome and other plant mt genomes.
Dot-plot graphs indicating regions of synteny between mitochondrial genomes compared to D. oleifera as the reference.
Distribution to different repeat type classes in the D. oleifera mt genome.
Frequency of classified SSR motifs in the D. oleifera mt genome.
Repeats (≥30bp) in the D. oleifera mt genome.
Data Availability Statement
Data associated with this study [The final annotated mt genome sequences of D. oleifera] has been deposited at NCBI GenBank under the accession number MW970112.