Abstract
Enzyme electrophoresis and rRNA sequencing were used to analyze relationships of Bradyrhizobium sp. nodule bacteria from four papilionoid legumes (Clitoria javitensis, Erythrina costaricensis, Rhynchosia pyramidalis, and Desmodium axillare) growing on Barro Colorado Island (BCI), Panama. Bacteria with identical multilocus allele profiles were commonly found in association with two or more legume genera. Among the 16 multilocus genotypes (electrophoretic types [ETs]) detected, six ETs formed a closely related cluster that included isolates from all four legume taxa. Bacteria from two other BCI legumes (Platypodium and Machaerium) sampled in a previous study were also identical to certain ETs in this group. Isolates from different legume genera that had the same ET had identical nucleotide sequences for both a 5′ portion of the 23S rRNA and the nearly full-length 16S rRNA genes. These results suggest that Bradyrhizobium genotypes with low host specificity may be prevalent in this tropical forest. Parsimony analysis of 16S rRNA sequence variation indicated that most isolates were related to Bradyrhizobium japonicum USDA 110, although one ET sampled from C. javitensis had a 16S rRNA gene highly similar to that of Bradyrhizobium elkanii USDA 76. However, this isolate displayed a mosaic structure within the 5′ 23S rRNA region: one 84-bp segment was identical to that of BCI isolate Pe1-3 (a close relative of B. japonicum USDA 110, based on 16S rRNA data), while an adjacent 288-bp segment matched that of B. elkanii USDA 76. This mosaic structure is one of the first observations suggesting recombination in nature between Bradyrhizobium isolates related to B. japonicum versus B. elkanii.
Although lateral gene transfer is a major process affecting genome structure in many prokaryotes (15, 18, 28), the frequency of transfer for ribosome genes remains uncertain. Because ribosome gene sequences have been so widely used to infer phylogenetic relationships (12, 29, 39), it is important to analyze how often such genes have been affected by lateral transfer events. In the presence of horizontal transfer, the true genealogical history will vary among different genes or different portions of the same gene (8, 43). As a result, a phylogenetic tree for one gene may not reflect organismal relationships for the remainder of the genome. Recently, a number of analyses have provided evidence for transfer events involving entire ribosome genes (41) or portions of genes (9, 36). However, the number of bacterial groups analyzed remains limited, so it is important to study more species from a diversity of habitats in order to understand the prevalence of ribosome gene transfer in nature.
In this study, I analyzed ribosome genes from bradyrhizobia associated with four genera of papilionoid legumes growing on Barro Colorado Island (BCI), Panama, a biological preserve covered by moist tropical forest (11). The four plants had diverse growth forms, but all came from two related tribes in the legume subfamily Papilionoideae. Within the tribe Phaseoleae, nodule bacteria were isolated from the tree Erythrina costaricensis (subtribe Erythrininae) and the lianas Clitoria javitensis (subtribe Clitoriinae) and Rhynchosia pyramidalis (subtribe Cajaninae). The fourth species sampled was the herbaceous vine Desmodium axillare, which is traditionally placed in a separate legume tribe from the other genera (tribe Desmodieae). However, phylogenetic analyses of chloroplast DNA sequences have indicated that Desmodium has a closer relationship with certain Phaseoleae subtribes (such as Erythrininae) than these have with other subtribes in the Phaseoleae (4).
Bacterial isolates were characterized by starch gel electrophoresis (25) and by 16S rRNA sequencing. In addition, sequence data were obtained for a region in the 5′ end of the 23S rRNA gene which contains an intervening sequence (IVS) that is cleaved during RNA processing (16, 26). The IVS region is highly polymorphic among nodule bacteria and therefore provides more characters than 16S rRNA for analyzing closely related bacterial strains. However, an analysis of the 23S rRNA region revealed an apparent case of lateral gene transfer among the BCI isolates. While this creates difficulties for inferring phylogenetic relationships, it also provides a new perspective about evolutionary mechanisms affecting symbiotic bacteria in this habitat.
MATERIALS AND METHODS
Isolate sampling.
Nodule bacteria were collected from three individuals each of C. javitensis and E. costaricensis, four of R. pyramidalis, and six plants of D. axillare (Table 1). Isolates were named by an abbreviation of the host's name followed by a number designating an individual plant. Nodules were sampled from small juvenile plants by carefully excavating around the base of the stem until roots with attached nodules were located. All nodules observed were collected. In most cases, two or more isolates from separate nodules on each individual plant were obtained (Table 1); these were designated by a dash and a number following the plant number. All sampled plants were <2.2 km apart on the east side of BCI.
TABLE 1.
Origin of bacterial strains from BCI, Panama, used in this study
| Host | No. of nodules sampled | ET(s) |
|---|---|---|
| Clitoria javitensis 1 | 2 | ET1, ET6 |
| Clitoria javitensis 2 | 3 | ET1 |
| Clitoria javitensis 3 | 3 | ET16 |
| Desmodium axillare 1 | 2 | ET3, ET5 |
| Desmodium axillare 2 | 3 | ET7, ET13, ET14 |
| Desmodium axillare 3 | 4 | ET4, ET6, ET15 |
| Desmodium axillare 4 | 3 | ET6 |
| Desmodium axillare 5 | 2 | ET8, ET12 |
| Desmodium axillare 6 | 1 | ET2 |
| Erythrina costaricensis 1 | 1 | ET3 |
| Erythrina costaricensis 2 | 1 | ET3 |
| Erythrina costaricensis 3 | 4 | ET1, ET2, ET3 |
| Rhynchosia pyramidalis 1 | 1 | ET6 |
| Rhynchosia pyramidalis 2 | 3 | ET3, ET9 |
| Rhynchosia pyramidalis 3 | 2 | ET10, ET11 |
| Rhynchosia pyramidalis 4 | 1 | ET2 |
Nodules were washed and then stored in vials with calcium sulfate desiccant. Within 1 week, nodules were rehydrated in 0.04 M sodium phosphate buffer (pH 7.0) and then surface disinfected in 3.2% sodium hypochlorite. One isolate was purified from each nodule as described (27). All isolates grew slowly on yeast-mannitol (YM) agar plates (colonies not visible before 4 days), suggesting that they were members of the genus Bradyrhizobium.
Enzyme electrophoresis.
Bacterial isolates were grown in YM broth (34), and enzymes were obtained from sonicated cells (27). Isolates were characterized by starch-gel electrophoresis at the following 11 enzyme loci as described (21): alcohol dehydrogenase, alanine dehydrogenase, butyrate esterase, glucose-6-phosphate dehydrogenase, β-hydroxybutyrate dehydrogenase, isocitrate dehydrogenase, malic enzyme, malate dehydrogenase, phosphoglucose isomerase, 6-phosphogluconate dehydrogenase, and shikimate dehydrogenase. Each isolate was characterized by its allelic profile for the 11 enzymes, and each unique multilocus genotype was designated an electrophoretic type (ET). On each gel, two standards representing ETs from the BCI legumes Platypodium elegans and Machaerium milleflorum analyzed in a prior study (21) were included for comparison. Pairwise genetic distances between ETs were estimated by the proportion of enzyme loci at which allelic differences occurred. ETs were then clustered by the neighbor-joining method (22).
DNA amplification and sequencing.
DNA was purified from bacterial cells by the protocol of Wilson (38). PCR used 25-μl reaction mixtures containing 10 mM Tris buffer with 0.1% Triton X-100, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 0.5 μM each primer, 0.5 μl of genomic DNA, and 0.5 U of Taq polymerase. Tubes were incubated for 90 s at 94°C and then subjected to 33 cycles of 94°C (20 s), 58°C (20 s), and 72°C (1 min), with a final extension of 4 min at 72°C. One bacterial isolate was chosen at random from each of the four legume hosts and sequenced on both strands for a nearly full-length portion of 16S rRNA (1,410 bp) and a 5′ portion of 23S rRNA (472 to 496 bp) as described (19), using an Applied Biosystems model 310 automated sequencer with protocols recommended by the manufacturer. The 23S rRNA region was also sequenced in Bradyrhizobium elkanii USDA 76 (kindly provided by L. D. Kuykendall) to provide a reference for the Panama strains.
Phylogenetic analyses.
Sequences were first aligned using ClustalW (32), and trees were constructed by maximum parsimony using the PAUP software (version 4.0b1, from D. L. Swofford, Smithsonian Institution, Washington, D.C.). To determine the degree of statistical support for branches in the phylogeny (10), 1,000 bootstrap replicates of the data were analyzed. The 16S rRNA data were compared to the following reference sequences (strains without formal names are identified by host legume genus in parentheses): Bradyrhizobium japonicum USDA 6 (GenBank U69638) (2) and USDA 110 (Z35330); B. elkanii USDA 76 (U35000) and USDA 94 (D13429); B. liaoningense (AJ250813); strain DSM 30140 (Lupinus) (X87273) and LMG 12187 (Aeschynomene) (AJ133779) (17); three Bradyrhizobium strains from Australia (14): 5040B (Bossiaea) (Z94811), 5680G (Bossiaea) (Z94804), and 5111P (Daviesia) (Z94805); LMG 9514 (Lonchocarpus) (X70405) (7); jwc91-2 (Amphicarpaea) (AF178437) and ApB16 (Apios) (AF178434) (19); SH283012 (Amorpha) (AF041446) (35); and four isolates from the BCI legumes P. elegans, M. milleflorum, and Tachigali versicolor (AF15936 to AF15938 and AF216780) (20, 21). Azorhizobium caulinodans (X67221) was used as the outgroup, because 16S rRNA phylogenies place it at a basal position relative to the other taxa (42).
Nodulation and nitrogenase activity.
No seeds of the four host legume species were available to use for inoculation studies. I thus used plants of Vigna unguiculata and Macroptilium atropurpureum, which are known to be nodulated by a broad range of tropical bradyrhizobia (21, 31, 33). Fourteen isolates representing different genotypes revealed by isozyme and rRNA sequence analysis were used to inoculate four plants of each species as described (37). Seeds were surface disinfected with 50% ethanol and then germinated. Seedlings were planted in pots using a Bradyrhizobium-free mixture of sand, perlite, and potting soil and then inoculated with approximately 109 cells of a particular isolate grown in YM broth. Plants were grown in a greenhouse for 34 days with precautions to avoid bacterial contamination across inoculation treatments (37). Uninoculated control plants grown simultaneously in the same room were found to be completely free of nodules. Plants were fertilized weekly with a nitrogen-free nutrient solution (21). At harvest, nodule numbers were recorded, and each plant's root system was analyzed for acetylene reduction activity using a Hewlett-Packard 5890 series II gas chromatograph as described (27)
Nucleotide sequence accession numbers.
The four 16S rRNA sequences for isolates from C. javitensis, E. costaricensis, R. pyramidalis, and D. axillare have been placed in GenBank under accession numbers AF321212 to AF321215, respectively. Five partial 23S rRNA sequences for isolates from these plants have been assigned accession numbers AF321219 to AF321223, and a partial 23S rRNA sequence for B. elkanii USDA 76 has accession number AF321224.
RESULTS
Isozyme diversity.
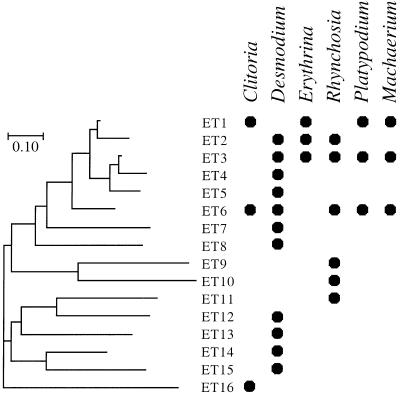
Among the 36 isolates analyzed, variation was detected at all 11 enzyme loci examined, with a mean of 5.8 alleles per locus (range, 2 to 10). A total of 16 distinct ETs were detected. Eleven ETs were represented by only a single isolate, while the remaining five ETs were recovered multiple times (Table 1). A neighbor-joining analysis grouped the ETs into several divergent lineages (Fig. 1). Separate nodules from a single host individual were often occupied by very different ETs. For example, the three nodules sampled from D. axillare plant 2 contained ET7, ET13, and ET14, which were as divergent from each other as from any three ETs in the entire sample (Fig. 1).
FIG. 1.
Neighbor-joining tree for relationships among 16 multilocus genotypes (ETs) of Bradyrhizobium spp. from the legumes. C. javitensis, D. axillare, E. costaricensis, and R. pyramidalis. Dots indicate the host plants for each ET. ETs that also occurred in association with the BCI legumes P. elegans and M. milleflorum (21) are shown. Scale bar for genetic distances along branches shows a 10% difference in the number of alleles shared between ETs.
In the neighbor-joining tree, ET1 to ET6 formed a relatively homogenous cluster (mean number of allelic differences among these six ETs was 1.9 out of 11 loci) that encompassed 24 of the 36 isolates analyzed. ETs in this group not only predominated on all four host legumes of this study but also were the most prevalent type of root nodule bacteria on two additional genera of BCI legumes (P. elegans and M. milleflorum, papilionoid tribe Dalbergieae) analyzed in a previous study (21). The three most abundant ETs in Fig. 1 (ET1, ET3, and ET6) had multilocus genotypes identical to those of three ETs sampled multiple times from P. elegans and M. milleflorum. The finding of identical ETs in nodules from four or more distantly related legume genera suggests that certain bacterial genotypes with low host specificity may be common in this environment.
16S rRNA variation.
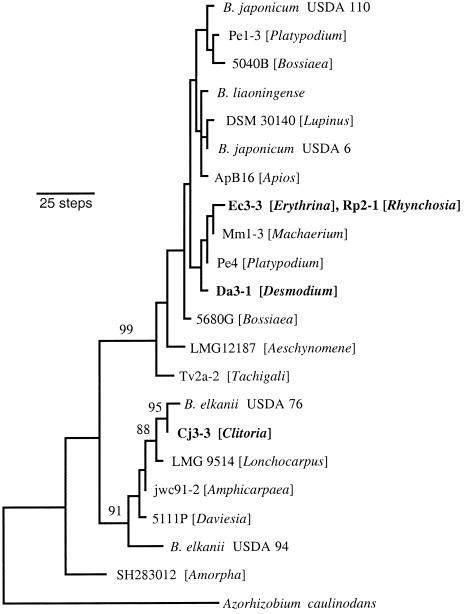
Nearly full-length sequences were obtained for isolates Ec3-3, Rp2-1 (both representing ET3), Da3-1 (ET15), and Cj3-3 (ET16). Ec3-3 and Rp2-1 had completely identical sequences and differed by 10 nucleotide substitutions from isolate Da3-1. Each of these three isolates was closely related to strains sampled from other BCI legumes. The 16S rRNA sequences of Ec3-3 and Rp2-1 differed by only four nucleotide substitutions from that of isolate Mm1-3, associated with the liana M. milleflorum (21). Da3-1 was most similar to isolate Pe4, sampled from the tree P. elegans (these two isolates differed by five substitutions). Isolate Cj3-3 was highly divergent, differing by at least 45 substitutions from all other BCI strains. A search of sequences in GenBank revealed that its closest known relative was the North American soybean symbiont B. elkanii USDA 76, which differed at only four nucleotide positions. A parsimony analysis indicated that all BCI isolates except Cj3-3 formed a well-defined clade (99% bootstrap support) related to B. japonicum (Fig. 2). Cj3-3 formed a clade together with B. elkanii USDA 76 in 95% of bootstrap resampling replicates. This pair clustered together with B. elkanii USDA 94 and three other isolates of diverse geographic origin (LMG9514 from Brazil, 5111P from Australia, and jwc91-2 from North America) in 91% of bootstrap replicates.
FIG. 2.
Phylogenetic relationships of four Bradyrhizobium isolates from Erythrina, Desmodium, Rhynchosia, and Clitoria (shown in bold) based on parsimony analysis of 16S rRNA gene sequences. Numbers above branches are bootstrap percentages (for clarity, only values >85% are shown).
23S rRNA variation.
To further clarify relationships, sequence data were obtained from a 5′ portion of 23S rRNA that is highly polymorphic among Bradyrhizobium strains (19, 21, 30). PCR amplification of DNA from isolates Ec3-3, Rp2-1, and Da3-1 yielded a 496-bp product (identical in size to that of B. japonicum USDA 110), while Cj3-3 yielded a 472-bp product (similar to the 468-bp product of B. elkanii USDA 76 [AF321224] and USDA 94 [AF081266]) (30). Isolates Ec3-3 and Rp2-1 had completely identical sequences (as in their 16S rRNA genes) and were also identical to strains from the BCI legumes P. elegans and M. milleflorum which had the same ET (AF159430 and AF159431) (21). The sequence from isolate Da3-1 did not match that of any other currently known isolates from BCI or elsewhere, but was most similar to isolates Ec3-3 and Rp2-1, from which it differed by five nucleotide substitutions.
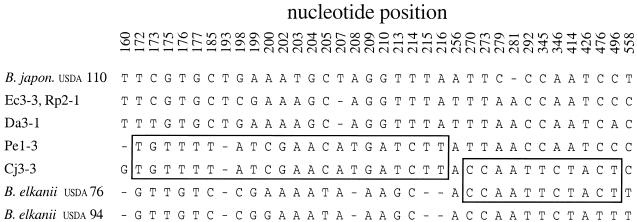
Although 16S rRNA data suggested a close relationship of isolate Cj3-3 to B. elkanii USDA 76 (Fig. 2), these isolates differed at 3.8% of nucleotide positions in this portion of the 23S rRNA gene. However, inspection of aligned 23S rRNA sequences indicated that nearly all of these differences were clustered in one small region (Fig. 3). Cj3-3 and B. elkanii USDA 76 had identical sequences for 288 bp after nucleotide position 270 (numbering based on the B. japonicum USDA 110 gene [Z35330]), but these two isolates differed at 65% of nongap polymorphic sites in the preceding 84 positions. In this portion of the 23S rRNA gene, Cj3-3 was instead identical to isolate Pe1-3 (Fig. 3), which was quite distantly related in the 16S rRNA tree (Fig. 2). A runs test (23), as implemented in modified form (http://www.math.wustl.edu/∼sawyer), revealed the existence of statistically significant mosaic structure in this region. For example, Cj3-3 and B. elkanii USDA 76 show highly significant sequence congruence in the 3′ end of this segment (P = 0.0081 based on 10,000 permutations), while in the 5′ end of 23S rRNA, Cj3-3 instead matched isolate Pe1-3 (P = 0.0096). The site likelihood test of Grassly and Holmes (13) also identified the segment from sites 172 to 216 (Fig. 3) as being significantly anomalous (P < 0.0005) relative to the maximum-likelihood phylogeny for the entire region. These results imply that different segments within the Cj3-3 23S rRNA gene were derived from two divergent ancestors.
FIG. 3.
Polymorphic sites in a 5′ portion of the 23S rRNA gene in eight Bradyrhizobium isolates. Sites are numbered relative to the B. japonicum USDA 110 sequence (Z35330). For isolate Cj3-3, two blocks of contiguous nucleotides identical to either Pe1-3 (AF159433) or B. elkanii USDA 76 (AF321224) are highlighted. All polymorphisms in this portion of 23S rRNA are shown except for two insertion-deletion differences where Pe1-3, Cj3-3, and B. elkanii USDA 76 and USDA 94 had gaps corresponding to invariant nucleotides at positions 161 to 171 and 217 to 229 in the other isolates.
To help rule out the possibility that the Cj3-3 23S rRNA sequence might be some sort of PCR artifact, this region was sequenced in the two other isolates having the same ET (Cj3-1 and Cj3-2). Both isolates were completely identical in sequence to Cj3-3. This indicates that mosaic structure (Fig. 3) was a general property of the ET16 clonal group. Because mosaic structure due to horizontal gene transfer creates incongruent phylogenies for different portions of a gene, no overall phylogenetic analysis of the 23S rRNA data was performed.
The size of the recombined segment in these isolates cannot be precisely determined due to a scarcity of polymorphisms around the 3′ end. The segment spans a minimum of 45 bp (positions 172 to 216), yet cannot extend past site 269 (Fig. 3), which would yield a maximum length of 97 bp. Since there are no informative polymorphisms between sites 216 and 270 (Fig. 3), the data do not permit any more detailed estimate within this range of values.
Analysis of rRNA secondary structure for the putative recombined region indicated that it occupied the tip of a long helix that is highly variable in length among different genera of proteobacteria (helix 9 [16]). Using MFOLD (44), the predicted base-pairing within this structure was compared for Cj3-3 and for isolates Pe1-3 and B. elkanii USDA 76 (which resemble strains that may have contributed different portions of Cj3-3's mosaic gene [Fig. 3]). These three isolates showed very similar degrees of base pairing. The number of paired/unpaired nucleotides in this helix was 82/23, 82/22, and 80/21 for Cj3-3, Pe1-3, and B. elkanii USDA 76, respectively. Moreover, the overall thermodynamic stability of Cj3-3's helix was marginally better than that of Pe1-3 or B. elkanii USDA 76 (ΔG = −53.4 versus −52.5 and −52.9 for the three isolates, respectively). These results indicate that the recombination event that created the mosaic 23S rRNA gene of Cj3-3 had a minimal effect on its secondary structure.
Nodulation and nitrogenase activity.
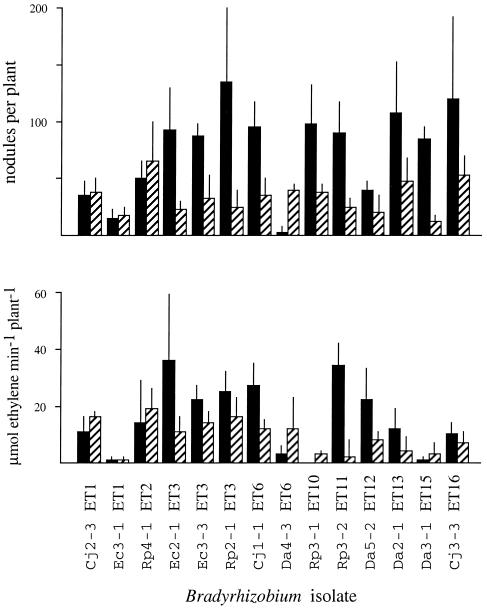
All Bradyrhizobium isolates tested formed nodules on both host legumes (Fig. 4), although the mean number of nodules developing per plant ranged from 5 (with Da4-3) to 134 (Rp2-1) on V. unguiculata. Likewise, substantial acetylene reduction activity was evident in most combinations of plants and bacteria (Fig. 4). The range of values for acetylene reduction activity was very similar to that in a previous study of BCI bradyrhizobia (21). For nodules of isolate Rp3-1 developing on V. unguiculata, there was no measurable ethylene production even though this isolate formed almost 100 nodules per plant. A few other isolates had low acetylene reduction activity with one or both hosts (e.g., Ec3-1 and Da3-1). In the few cases where multiple isolates within the same ET were analyzed, there was often wide disparity in symbiotic performance. For example, among the two isolates having ET1, one was a moderately effective symbiont on both host legumes (Cj2-3), while the other (Ec3-1) showed negligible acetylene reduction activity on both types of plants. Thus, isozyme phenotypes may be a poor predictor of symbiotic behavior.
FIG. 4.
Nodulation (top) and acetylene reduction activity (bottom) of Vigna unguiculata (solid bars) and Macroptilium atropurpureum (hatched bars) inoculated with Bradyrhizobium isolates from four genera of BCI legumes. Data are means + 1 standard deviation.
DISCUSSION
The main finding of the isozyme survey was that Bradyrhizobium genotypes with broad host range appear to be prevalent in this environment. For example, most bacterial ETs represented by >1 isolate occurred in association with several of the legume genera sampled in this study (Table 1, Fig. 1). Sharing of bacterial genotypes by these plants may not be surprising in view of the fact that the hosts are from two related legume tribes (Phaseoleae and Desmodieae). However, the most common bacterial ETs recorded from these four legume genera were identical to multilocus genotypes that also predominated on two additional BCI genera (Platypodium and Machaerium) from a different tribe (Dalbergieae) that is not a close relative of the Phaseoleae and Desmodieae (5).
The inference that ETs shared by multiple legumes represent strains with broad potential host range is supported by two other observations. First, analysis of 16S and 23S rRNA genes indicated that isolates with the same ET from different hosts invariably had identical sequences. This suggests that such bacteria are members of a single clone or at least have a very close phylogenetic relationship. Second, the inoculation experiment demonstrated that all of the isolates had potential host ranges that extended to other legume species as well (Fig. 4). Therefore, a lack of specificity toward various BCI legume genera would not be surprising.
To date, bacteria have only been sampled from a handful of the more than 35 BCI legume genera that form root nodules. Thus, for some of these bacterial genotypes, the actual range of legumes utilized may be much more extensive than is known at present. Bradyrhizobium isolates from one taxonomically divergent BCI legume (T. versicolor, subfamily Caesalpinioideae) had no genotypes in common with isolates from the papilionoid tribes Phaseoleae, Desmodieae, and Dalbergieae (20). This suggests that some legumes in this community may harbor relatively distinctive symbiont populations. Nevertheless, it is important to more fully survey all BCI legume species in order to better understand patterns of host legume utilization by bacteria in this tropical forest environment.
Obtaining more precise information about host specificity is important for accurate predictions concerning the global biodiversity of nodule bacteria. Among intensively studied domesticated legumes, it is common for a single host species to have several disparate species of nodule bacteria associated with it (1, 24, 35). The high diversity of isozyme lineages associated with most individual BCI legume species (Fig. 1) is not inconsistent with this finding and might lead to the inference that the total species diversity of symbiotic bacteria will greatly exceed that of their host plants. However, most of the isolates from this site belonged to a small number of apparently unspecialized ETs that each occurred on four or more host genera. Thus, it is not possible to estimate overall bacterial diversity by simply counting the number of lineages per host and then multiplying this by the total number of legume species. To the extent that communities of nodule bacteria in the tropics are dominated by genotypes with low host specificity, as is apparently the case in other regions (14, 19), then the overall species diversity of nodule bacteria may prove to be relatively restricted. This issue can only be resolved by further studies that include sampling from all legume hosts within a single environment, together with surveys of particular host taxa across their geographic ranges.
All but one of the BCI Bradyrhizobium strains have proved to be relatives of B. japonicum, based on rRNA sequence data (Fig. 2) (20, 21). The sole exception, isolate Cj3-3, had a 16S rRNA sequence highly similar to that of B. elkanii USDA 76. Isolates of Bradyrhizobium with 16S rRNA related to B. elkanii have also been recovered from a variety of woody legumes in the Amazon basin (7). However, the 5′ portion of the 23S rRNA gene in isolate Cj3-3 appeared to have a mosaic structure, with one portion being identical to that in B. elkanii USDA 76 and an adjacent segment matching that of isolate Pe1-3 (from the BCI tree P. elegans). Based on 16S rRNA, isolate Pe1-3 is a close relative of B. japonicum USDA 110, differing at only 7 nucleotides out of 1,410 bp analyzed (21). The mosaic structure of isolate Cj3-3's 23S rRNA gene is most readily explained by postulating an event of horizontal gene transfer. The main alternative possibility is that the similarity of isolates Cj3-3 and Pe1-3 represents nothing more than a chance outcome of independent evolution. However, based on Sawyer's (23) test, it is very unlikely that these isolates independently acquired identical nucleotides at >20 consecutive polymorphic sites in a highly variable portion of the 23S rRNA gene (Fig. 3) while showing little similarity for other portions of 23S rRNA (or for their 16S rRNA genes). Thus, the most parsimonious interpretation is that one of these isolates acquired a portion of its 23S rRNA gene by horizontal transfer from a strain related to the other isolate.
The mechanism of gene transfer remains unclear. The inferred size of the transferred region (Fig. 3) falls near the lower end of the size range of intragenic recombination events estimated for other bacteria (6, 43). However, there is no reason to doubt that ecological opportunities for transfer exist in this environment. Cj3-3 and Pe1-3 came from different host legumes, from sites 1.1 km apart on BCI. However, both of these legume taxa may grow in the same microenvironment <1 m apart (personal observations), and it should also be noted that even within a single host individual, separate nodules are often occupied by very divergent bacterial genotypes (Fig. 1) (21). Thus, there appears to be fine-scale cooccurrence of different Bradyrhizobium strains (a prerequisite for any of the known mechanisms of gene transfer).
The 23S rRNA IVS region has no clear function (26), so there may be little fitness benefit (or penalty) associated with transfer of portions of this region between different bacterial genotypes. The frequency of such transfers affects the evolution of neutral divergence among lineages (3), but it is uncertain whether such events have a major impact on bacterial adaptation. Other similar cases have been reported, where part or all of 16S rRNA genes have apparently been transferred (9, 36, 41). Nevertheless, the current finding of horizontal transfer in a relatively small-scale 23S rRNA sequencing survey raises the question of whether divergent Bradyrhizobium strains may be affecting each other's evolution through exchange of various other genes as well. To date, few studies of Bradyrhizobium spp. have analyzed multiple gene loci in a set of isolates from the same local environment. As a result, there are few comparable data for judging how often divergent lineages of Bradyrhizobium (such as B. japonicum versus B. elkanii) may participate in gene transfer in the environment. In future research to characterize Bradyrhizobium diversity in natural communities, it will be important to focus on obtaining sequence data from several loci in order to resolve this issue.
Lateral transfer is also significant because it complicates phylogenetic inference. In the presence of lateral transfer, the true genealogical history will vary among different genes or different portions of the same gene (8, 43). Thus, an unbiased picture of evolutionary relationships is best obtained by comparative analysis of phylogenetic patterns across multiple loci. This approach should be a priority for future work on the biodiversity of Bradyrhizobium in natural communities.
ACKNOWLEDGMENTS
I am grateful to J. Pfeil for assistance with sequencing, to S. Vyas for help with inoculation experiments, and to L. D. Kuykendall for providing bacterial isolates. I also thank the Smithsonian Tropical Research Institute and the Republic of Panama's Instituto de Recursos Nacionales Renovables for permission to collect on BCI.
I thank the Harpur College Dean of Arts and Sciences for financial support.
REFERENCES
- 1.Amarger N, Macheret V, Laguerre G. Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris nodules. Int J Syst Bacteriol. 1997;47:996–1006. doi: 10.1099/00207713-47-4-996. [DOI] [PubMed] [Google Scholar]
- 2.Barrera L L, Trujillo M E, Goodfellow M, Garcia F J, Hernandez-Lucas I, Davila G, van Berkum P, Martinez-Romero E. Biodiversity of bradyrhizobia nodulating Lupinus spp. Int J Syst Bacteriol. 1997;47:1086–1091. doi: 10.1099/00207713-47-4-1086. [DOI] [PubMed] [Google Scholar]
- 3.Cohan F M. Does recombination constrain neutral divergence among bacterial taxa? Evolution. 1995;49:164–175. doi: 10.1111/j.1558-5646.1995.tb05968.x. [DOI] [PubMed] [Google Scholar]
- 4.Doyle J J, Doyle J L. Chloroplast DNA phylogeny of the Papilionoid legume Tribe Phaseoleae. Syst Bot. 1993;18:309–327. [Google Scholar]
- 5.Doyle J J, Doyle J L, Ballenger J A, Dickson E E, Kajita T, Ohashi H. A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. Am J Bot. 1997;84:541–554. [PubMed] [Google Scholar]
- 6.DuBose R F, Dykhuizen D E, Hartl D L. Genetic exchange among natural isolates of bacteria: recombination within the phoA gene of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:7036–7040. doi: 10.1073/pnas.85.18.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupuy N, Willems A, Pot B, Dewettinck D, Vandenbruaene I, Maestrojuan G, Dreyfus B, Kersters K, Collins M D, Gillis M. Phenotypic and genotypic characterization of bradyrhizobia nodulating the leguminous tree Acacia albida. Int J Syst Bacteriol. 1994;44:461–473. doi: 10.1099/00207713-44-3-461. [DOI] [PubMed] [Google Scholar]
- 8.Dykhuizen D E, Green L. Recombination in Escherichia coli and the definition of biological species. J Bacteriol. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eardly B D, Wang F-S, van Berkum P. Corresponding 16S rRNA gene segments in Rhizobiaceae and Aeromonas yield discordant phylogenies. Plant Soil. 1996;186:69–74. [Google Scholar]
- 10.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Foster R B, Brokaw N V L. Structure and history of the vegetation on Barro Colorado Island. In: Leigh E G, Rand A S, Windsor D M, editors. The ecology of a tropical forest: seasonal rhythms and long-term changes. 2nd ed. Washington, D.C.: Smithsonian Institution; 1996. pp. 67–81. [Google Scholar]
- 12.Graham P H, Sadowsky M J, Keyser H H, Barnet Y M, Bradley R S, Cooper J E, de Ley D J, Jarvis B D W, Roslycky E B, Strijdom B W, Young J P W. Proposed minimum standards for the description of new genera and species of root- and stem-nodulating bacteria. Int J Syst Bacteriol. 1991;41:582–587. [Google Scholar]
- 13.Grassly N C, Holmes E C. A likelihood method for the detection of selection and recombination using nucleotide sequences. Mol Biol Evol. 1997;14:239–247. doi: 10.1093/oxfordjournals.molbev.a025760. [DOI] [PubMed] [Google Scholar]
- 14.Lafay B, Burdon J J. Molecular diversity of rhizobia occurring on native shrubby legumes in southeastern Australia. Appl Environ Microbiol. 1998;64:3989–3997. doi: 10.1128/aem.64.10.3989-3997.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence J G, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig W, Rossello-Mora R, Aznar R, Klugbauer S, Spring S, Reetz K, Beimfohr C, Brockmann E, Kirchof G, Dorn S, Bachleitner M, Klugbauer N, Springer N, Lane D, Nietupsky R, Weizenegger M, Schleifer K-H. Comparative sequence analysis of 23S rRNA from Proteobacteria. Syst Appl Microbiol. 1995;18:164–188. [Google Scholar]
- 17.Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B, Gillis M, de Lajudie P, Masson-Boivin C. Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulating species and form a separate 16S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol. 1999;65:3084–3094. doi: 10.1128/aem.65.7.3084-3094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson K E, Clayton R A, Gill S R, et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 19.Parker M A. Relationships of bradyrhizobia from the legumes Apios americana and Desmodium glutinosum. Appl Environ Microbiol. 1999;65:4914–4920. doi: 10.1128/aem.65.11.4914-4920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker M A. Divergent Bradyrhizobium symbionts on Tachigali versicolor from Barro Colorado Island, Panama. Syst Appl Microbiol. 2000;23:585–590. doi: 10.1016/S0723-2020(00)80034-X. [DOI] [PubMed] [Google Scholar]
- 21.Parker M A, Lunk A. Relationships of bradyrhizobia from Platypodium and Machaerium (Papilionoideae tribe Dalbergieae) on Barro Colorado Island, Panama. Int J Syst Evol Microbiol. 2000;50:1179–1186. doi: 10.1099/00207713-50-3-1179. [DOI] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- 24.Segovia L, Young J P W, Martinez-Romero E. Reclassification of American Rhizobium leguminosarum biovar phaseoli type I strains as Rhizobium etli sp. nov. Int J Syst Bacteriol. 1993;43:374–377. doi: 10.1099/00207713-43-2-374. [DOI] [PubMed] [Google Scholar]
- 25.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selenska-Pobell S, Evguenieva-Hackenberg E. Fragmentation of the large-subunit rRNA in the family Rhizobiaceae. J Bacteriol. 1995;177:6993–6998. doi: 10.1128/jb.177.23.6993-6998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spoerke J M, Wilkinson H H, Parker M A. Nonrandom genotypic associations in a legume-Bradyrhizobium mutualism. Evolution. 1996;50:146–154. doi: 10.1111/j.1558-5646.1996.tb04481.x. [DOI] [PubMed] [Google Scholar]
- 28.Spratt B G, Maiden M C J. Bacterial population genetics, evolution and epidemiology. Phil Trans R Soc Lond B. 1999;354:701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 30.Sterner J P, Parker M A. Diversity and relationships of bradyrhizobia from Amphicarpaea bracteata based on partial nod and ribosomal sequences. Syst Appl Microbiol. 1999;22:387–392. doi: 10.1016/S0723-2020(99)80047-2. [DOI] [PubMed] [Google Scholar]
- 31.Thies J E, Bohlool B B, Singleton P W. Subgroups of the cowpea miscellany: symbiotic specificity within Bradyrhizobium spp. for Vigna unguiculata, Phaseolus lunatis, Arachis hypogaea, and Macroptilium atropurpureum. Appl Environ Microbiol. 1991;57:1540–1545. doi: 10.1128/aem.57.5.1540-1545.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J D, Higgans D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment thorugh sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turk D, Keyser H H. Rhizobia that nodulate tree legumes: specificity of the host for nodulation and effectiveness. Can J Microbiol. 1992;38:451–460. [Google Scholar]
- 34.Vincent J M. A manual for the practical study of the root-nodule bacteria. Oxford, U.K: Blackwell; 1970. [Google Scholar]
- 35.Wang E T, van Berkum P, Sui X H, Beyene D, Chen W X, Martinez-Romero E. Diversity of rhizobia associated with Amorpha fruticosa isolated from Chinese soils and description of Mesorhizobium amorphae sp. nov. Int J Syst Bacteriol. 1999;49:51–65. doi: 10.1099/00207713-49-1-51. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Zhang Z. Comparative sequence analyses reveal frequent occurrence of short segments containing an abnormally high number of non-random base variations in bacterial rRNA genes. Microbiology. 2000;146:2845–2854. doi: 10.1099/00221287-146-11-2845. [DOI] [PubMed] [Google Scholar]
- 37.Wilkinson H H, Spoerke J M, Parker M A. Divergence in symbiotic compatibility in a legume-Bradyrhizobium mutualism. Evolution. 1996;50:1470–1477. doi: 10.1111/j.1558-5646.1996.tb03920.x. [DOI] [PubMed] [Google Scholar]
- 38.Wilson J K. Preparation of genomic DNA from bacteria. In: Ausubel F M, editor. Current protocols in molecular biology. New York, N.Y: John Wiley; 1994. pp. 2.4.1–2.4.5. [DOI] [PubMed] [Google Scholar]
- 39.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L M, Ge C, Cui Z, Li J, Fan H. Bradyrhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int J Syst Bacteriol. 1995;45:706–711. doi: 10.1099/00207713-45-4-706. [DOI] [PubMed] [Google Scholar]
- 41.Yap W H, Zhang Z, Wang Y. Distinct types of rRNA operons exist in the genome of the acctinomycete Thermomonospora chromogena and evidence for horozontal transfer of an entire rRNA operon. J Bacteriol. 1999;181:5201–5209. doi: 10.1128/jb.181.17.5201-5209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young J P W, Haukka K E. Diversity and phylogeny of rhizobia. New Phytol. 1996;133:87–94. [Google Scholar]
- 43.Zhou J, Bowler L D, Spratt B G. Interspecies recombination, and phylogenetic distortions, within the glutamine synthetase and shikimate dehydrogenase genes of Neisseria meningitidis and commensal Neisseria species. Mol Microbiol. 1997;23:799–812. doi: 10.1046/j.1365-2958.1997.2681633.x. [DOI] [PubMed] [Google Scholar]
- 44.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. NATO ASI Series. Amsterdam, The Netherlands: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]