Abstract
Chaperones and other quality control machinery guard proteins from inappropriate aggregation, which is a hallmark of neurodegenerative diseases. However, how the systems that regulate the “foldedness” of the proteome remain buffered under stress conditions and in different cellular compartments remains incompletely understood. In this study, we applied a FRET-based strategy to explore how well quality control machinery protects against the misfolding and aggregation of “bait” biosensor proteins, made from the prokaryotic ribonuclease barnase, in the nucleus and cytosol of human embryonic kidney 293T cells. We found that those barnase biosensors were prone to misfolding, were less engaged by quality control machinery, and more prone to inappropriate aggregation in the nucleus as compared with the cytosol, and that these effects could be regulated by chaperone Hsp70-related machinery. Furthermore, aggregation of mutant huntingtin exon 1 protein (Httex1) in the cytosol appeared to outcompete and thus prevented the engagement of quality control machinery with the biosensor in the cytosol. This effect correlated with reduced levels of DNAJB1 and HSPA1A chaperones in the cell outside those sequestered to the aggregates, particularly in the nucleus. Unexpectedly, we found Httex1 aggregation also increased the apparent engagement of the barnase biosensor with quality control machinery in the nucleus suggesting an independent implementation of “holdase” activity of chaperones other than DNAJB1 and HSPA1A. Collectively, these results suggest that proteostasis stress can trigger a rebalancing of chaperone abundance in different subcellular compartments through a dynamic network involving different chaperone–client interactions.
Keywords: Hsp70, Hsp40, chaperone DnaJ, chaperone DnaK, protein quality control, proteostasis, protein aggregation, protein folding, FRET, flow cytometry
Abbreviations: BSA, bovine serum albumin; HBSS, Hank’s balanced salt solution; HEK293T, human embryonic kidney 293T cell line; NES, nuclear export sequence; NLS, nuclear localization sequence; ROI, region of interest; TBS, Tris-buffered saline
Protein homeostasis involves a network of quality control systems for overseeing proteins through translation, folding, delivery to the correct cellular location, and turnover at appropriate times. When protein homeostasis becomes unbalanced, proteins become prone to misfolding leading to their mislocalization and accumulation as aggregates (1, 2). The imbalance of protein homeostasis is hypothesized to underlie the inappropriate protein misfolding and aggregation that arise in the brain of patients with common neurodegenerative diseases including Alzheimer’s, Parkinson’s, Huntington’s, and motor neuron diseases (3, 4). Tools that can measure proteostasis imbalance therefore offer capacity to explore the mechanisms involved.
Previously, we developed a method that enabled a measure of the effectiveness of the quality control system in maintaining protein homeostasis (5). The method involved the use of a biosensor that comprised a model protein that engages with protein quality control machinery such as chaperones. The biosensor reported on amount of the model protein bound to quality control proteins (which we hereon call holdase activity) and on the ability of quality control proteins to repress inappropriate protein aggregation of the model protein. The model protein was a catalytically inactive variant of the prokaryotic ribonuclease barnase. Unfolded barnase is both permissive to aggregation and able to bind to Hsp70 and Hsp40 family chaperones (5). Under equilibrium conditions, barnase is either folded or unfolded, and the distribution of the two species follows a two-state unfolding mechanism in vitro. The proportion of unfolded barnase relative to folded barnase can be predictably tuned by mutation (6). Hence, a panel of barnase variants enable different biosensors tuned to different ratios of folded versus unfolded proteins. When the biosensor is expressed in cells, the proportion of folded proteins can be measured by FRET through N- and C-terminal fusions to fluorescent protein donors and acceptors (Fig. 1A). We previously showed that the abundance of unfolded barnase is increased in cells relative to that predicted by analysis of purified proteins because of quality control machinery forming complexes with the unfolded-like conformations of the biosensor and partitioning it from the equilibrium of folding (5). Therefore, alterations in quality control levels influence the total abundance of unfolded-like barnase, which we can detect by FRET, and therefore determine changes in the overall quality control supply available as a measure of proteostasis capacity.
Figure 1.
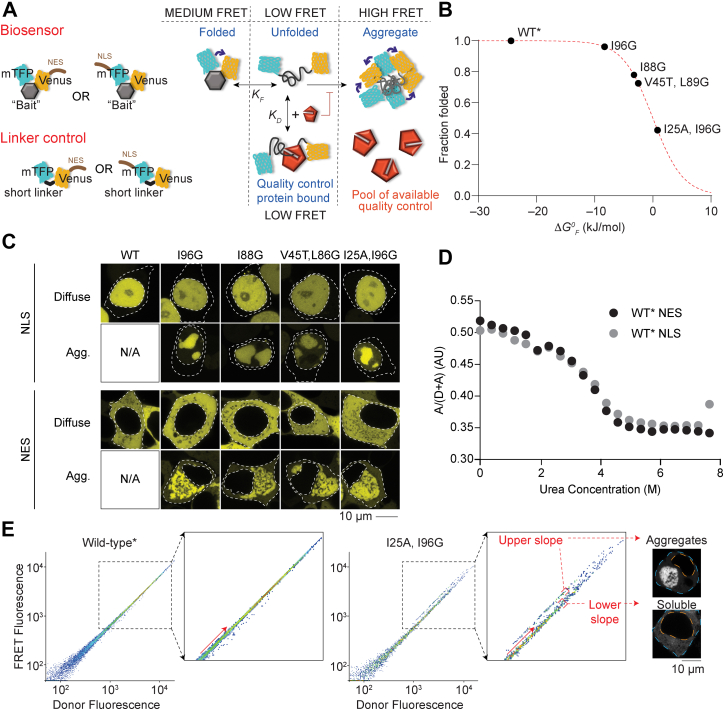
Targeting the barnase biosensor into the cytosol and nucleus.A, schematic of how the biosensor works (diagram adapted from Ref. (5)). The barnase protein is used as the “bait” for chaperones and flanked with fluorescence proteins for FRET measurements. A nuclear localization sequence (NLS) or nuclear export sequence (NES) is appended to the construct. B, shown is the relationship between mutations in barnase, the effect on standard free energy of folding (ΔG°F at 20 °C), and predicted fraction folded for the various biosensor variants used in the study. WT barnase is marked with ∗ to denote it contains the catalytic inactivation mutation H102A. This mutation is present in all constructs in the study. C, confocal images of HEK293T cells transiently transfected with either nuclear- or cytosol-targeting biosensor variants. The nucleus and cell edges are indicated by dashed lines. The localization of the biosensor is shown by Venus fluorescent protein fluorescence (yellow). Representative cells with only diffuse or aggregated proteins are shown. D, urea denaturation curves of WT∗ barnase biosensor variants as measured in cell lysates by FRET. E, flow cytometry strategy for monitoring foldedness and aggregation. Here, the donor and acceptor fluorescence of cells were measured by channels (FRET and donor fluorescence were gated by the PE (575/25) and V500 (525/50) filters, respectively, with the 405 nm laser). The inset highlights the changes that arise for cells bifurcated into “upper” and “lower” slope populations (division shown with red arrow). Representative cells collected from gates corresponding to the upper and lower slope populations imaged by confocal microscopy (grayscale). The orange dashed line denotes the nucleus boundary and the cyan dashed line the cell boundary. HEK293T, human embryonic kidney 293T cell line.
The amount of biosensor aggregation can also provide a measure of overall chaperone activity that acts on the biosensor. Our prior work devised a strategy that quantitatively measured aggregation as a complementary approach to foldedness (5). Here, we applied our biosensor system to examine how proteostasis balance is affected specifically within the nucleus and cytosol. We examine these local proteostasis changes that result from different triggers of stress either globally to the cell or locally within the cytoplasm or nucleus.
Results
Generation of nuclear-targeted biosensors and validation of folding stabilities
The biosensor system comprises a suite of constructs whereby the barnase moiety has been mutated to display different standard free energies of folding (ΔG0F), which define the thermodynamic equilibrium of folding Kf (Fig. 1B). These constructs contained a nuclear export sequence (NES), which leads to them being restricted to a cytosolic localization (5). To direct the constructs to the nucleus, we removed the NES and fused a nuclear localization sequence (NLS) from the SV40 protein to the N terminus (see Fig. 1A and Table 1 for sequences used). The biosensor containing WT∗ barnase (which is marked with ∗ to denote it contains the catalytic inactivation mutation H102A that is used in all our constructs), as well as four mutants, was efficiently targeted into the nucleus (Fig. 1C).
Table 1.
Sequence overview of the barnase biosensor constructsa
| Biosensor variant layout |
|---|
| pTriEx4 barnase WTa + NES |
| MAHHHHHHGSGEQKLISEEDLGSGSGSGGHHRVDFKTIYRAKKAVKLPDYHFVDHRIEILNHDKDYNKVTVYESAVARNSTDGMDELYKGASGGMVSKGEETTMGVIKPDMKIKLKMEGNVNGHAFVIEGEGEGKPYDGTNTINLEVKEGAPLPFSYDILTTAFAYGNRAFTKYPDDIPNYFKQSFPEGYSWERTMTFEDKGIVKVKSDISMEEDSFIYEIHLKGENFPPNGPVMQKKTTGWDASTERMYVRDGVLKGDVKHKLLLEGSGAQVINTFDGVADYLQTYHKLPDNYITKSEAQALGWVASKGNLADVAPGKSIGGDIFSNREGKLPGKSGRTWREADINYTSGFRNSDRILYSSDWLIYKTTDAYQTFTKIRAAAMDGGVQLADHYQQNTPIGDGPVLLPDNHYLSYQSALSKDPNEKRDHMVLLEFVTAAGITLGMDELYKGGSGGMVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKLICTTGKLPVPWPTLVTTLGYGLMCFARYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYITADKQKNGIKANFKIRHNIEGTDILQKKLEELELDE |
| pTriEx4 barnase WTa + NLS |
| MCGGGPKKKRKVEDPGGSGSGGHHRVDFKTIYRAKKAVKLPDYHFVDHRIEILNHDKDYNKVTVYESAVARNSTDGMDELYKGASGGMVSKGEETTMGVIKPDMKIKLKMEGNVNGHAFVIEGEGEGKPYDGTNTINLEVKEGAPLPFSYDILTTAFAYGNRAFTKYPDDIPNYFKQSFPEGYSWERTMTFEDKGIVKVKSDISMEEDSFIYEIHLKGENFPPNGPVMQKKTTGWDASTERMYVRDGVLKGDVKHKLLLEGSGAQVINTFDGVADYLQTYHKLPDNYITKSEAQALGWVASKGNLADVAPGKSIGGDIFSNREGKLPGKSGRTWREADINYTSGFRNSDRILYSSDWLIYKTTDAYQTFTKIRAAAMDGGVQLADHYQQNTPIGDGPVLLPDNHYLSYQSALSKDPNEKRDHMVLLEFVTAAGITLGMDELYKGGSGGMVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKLICTTGKLPVPWPTLVTTLGYGLMCFARYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYITADKQKNGIKANFKIRHNIEGTKDEL |
Sequences are coded as: mTFP1 fluorescent protein (cp 175);WT (WT)∗ barnase; Venus fluorescent protein (cp 173).
Because the NLS and NES could themselves affect the folding equilibrium, we measured their effect on folding by a urea denaturation curve, which showed no noticeable difference (Fig. 1D). Hence, we concluded that the biosensors with NLS and NES are amenable to directly measure and compare protein homeostasis balances between the cytosol and nucleus.
The strategy to monitor both the abundances of unfolded barnase and aggregation behavior involved a flow cytometry protocol we previously developed (5). In essence, cells expressing the biosensor bifurcate into two distinct FRET populations when cells are gated on FRET fluorescence versus donor fluorescence (Dataset S1). The two populations describe cells with different expression levels of barnase and a corresponding FRET signal that is linearly proportional to the donor fluorescence signal (which is proportional to the abundance of the barnase). We describe these two populations as the lower slope and upper slope populations (Fig. 1E). The lower slope population contains cells that display a lower FRET signal relative to a particular concentration of barnase. We previously had found by microscopy that these cells only contained soluble barnase without visible biosensor aggregates (5) (i.e., a mixture of medium and low FRET states; Fig. 1, A and E). The upper slope population contains cells with a higher FRET signal relative to a particular concentration of barnase, and we showed previously that these cells were enriched with visibly aggregated biosensor (5). The higher FRET signal relates to the aggregates directing a high level of intermolecular FRET (5). Because the two populations are linearly distributed, we can deduce that the gradient of the linear regression through the populations provides information on the conformational states that barnase adopts in the cells. This assumption holds true for the lower slope population whereby the FRET signal arises from intramolecular interactions and reflects the combination of unfolded and folded states of barnase in equilibrium (i.e., the average signal of low and medium FRET states from Fig. 1A). We refer hereon to the gradients derived from the linear regression of the lower slope population as lower slope values. However, for the upper slope population, the intermolecular nature of the FRET confounds a similar interpretation for enabling levels of aggregate to be estimated. Instead of slope values as a measure of aggregation, we use membership of the upper slope population as a binary indicator to whether an aggregate is present or absent (more details are explained later as to how we use that information quantitatively). We had previously shown that elements of quality control machinery including Hsp70 and Hsp40 proteins HSPA1A and DNAJB1, respectively, can bind to unfolded biosensor and hold it in an unfolded-like state that has a low FRET signal (5) as summarized conceptually in Figure 1A. This binding creates a pool of chaperone-bound biosensor that is partitioned from the equilibrium of folding. Greater partitioning leads to lower FRET signals, which can thus be used as a measure of the capacity of the quality control system to engage with the biosensor.
Next, we examined the effectiveness of protein quality control systems to interact with the biosensor in the nucleus versus the cytosol. For this, we examined four previously characterized mutants of barnase that have variable ΔG0F values, which correlates to different proportions of folded to unfolded barnase at equilibrium (Fig. 2A). The partly destabilized barnase variants (I88G and V45T, L89G) had higher lower slope values in the nucleus compared with the cytosol, which suggested that these two mutants were more folded in the nucleus (two-way ANOVA of the control samples provided I88G and V45T, L89G with p < 0.001 for both nucleus versus cytosol). The corresponding differences were minimal or negligible for WT∗ barnase and the most destabilized mutant I25A, I96G (and not significant by two-way ANOVA, with p > 0.05). We note that the lack of changes for WT∗ and the I25A, I96G may arise through these variants being close to the extreme ends of the dynamic range of the biosensor readouts. One explanation for the higher apparent foldedness in the nucleus for the partially destabilized barnase mutants is that there is less supply of chaperones in the nucleus that can otherwise bind to the unfolded states of these variants and hold them partitioned from the equilibrium of folding. Another possibility is that differences in the FRET arise from nonspecific effects arising from the cellular localization.
Figure 2.
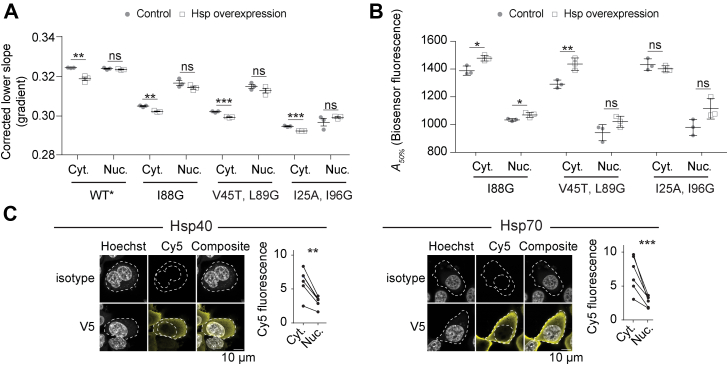
Cytosolic Hsp70 and Hsp40 activity buffer against protein misfolding and aggregation.A, lower slope values derived from flow cytometry analysis of HEK293T cells cotransfected with the biosensors, DNAJB1 and HSPA1A or control (a nonfluorescent derivative of GFP (Y66L Emerald (12)) for 48 h. Data points indicate biological replicates, and bars indicate means ± SD. Student’s t test results are shown (2-tailed; control versus overexpression); ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns ≥ 0.05. B, aggregation analysis (A50%) using the same treatments and conditions as for A. C, shown are immunofluorescence micrographs of HEK293T cells transiently transfected with either V5-tagged Hsp40 or Hsp70 proteins (DNAJB1 and HSPA1A, respectively). The nucleus is stained with Hoechst 33342 and chaperone with Cy5-labeled anti-V5 antibody (or isotype control for specificity). Graphs indicate quantitation with paired Student’s t test results shown (2-tailed, paired); ∗∗∗p < 0.001, ∗∗p < 0.01. Data points represent immunofluorescence intensity in single cells (paired by mean). HEK293T, human embryonic kidney 293T cell line; ns, not significant.
To test for whether cellular localization may explain the results, we examined a biosensor variant whereby the barnase moiety has been excised, which we call the linker variant. The linker therefore contains mTFP1 flanked directly by Venus as a tandem fluorescent protein heterodimer, and the FRET signal should not report on changes in foldedness (5). The linker target showed higher lower slope values in the cytosol compared with the nucleus (1.76%; Fig. S1, A and B), which therefore suggested that the localization affects the inherent FRET independently to foldedness. However, these effects were generally opposite in direction to the changes for the biosensor variants (which ranged from 1.66% to 0.78%; Fig. S1C), suggesting that the differences in foldedness were larger than distinct off-target effects arising from the different intracellular locations.
To examine whether the pool of chaperones that can bind to the biosensor is lower in the nucleus than the cytosol, we first assessed the aggregation propensity of the barnase biosensor using a previously devised method of determining the concentration of barnase at which 50% of the cells contain aggregates (A50%) (5). A50% values are derived from plots of the proportions of cells partitioning in the upper slope population for a given expression level of barnase in cells. This is achieved by binning flow cytometry based on expression level and then deriving the proportion of cells that are in the upper slope (Fig.S2 and Dataset S1). Consistent with prior findings (5), WT∗ barnase did not aggregate meaning that there was no upper slope population to derive an A50% value from. The less stable barnase variants (i.e., those with higher ΔG0F values) did form upper slope populations, and hence, these variants were amenable to derivation of A50% values (Fig. S2). The mutants that were more destabilized had lower A50% values, indicating their heightened propensity to form aggregates (Fig. 2B). For all the barnase variants that aggregated, the A50% values were lower in the nucleus compared with the cytosol (two-way ANOVA comparing the cytoplasmic versus nucleus control samples gave p < 0.0001 for all three mutants; Fig. 2B). These results indicated that barnase is inherently more aggregation prone in the nucleus than the cytosol and therefore strengthens the conclusion that there are less quality control proteins in the nucleus that are able to bind to and stabilize barnase and prevent aggregation.
Hsp70 and Hsp40 chaperone systems more robustly mitigate unfolded proteins from aggregating in the cytosol than the nucleus
Hsp70 isoforms HSPA1B and HSPA8 were previously found in immunoprecipitation experiments as major chaperone interactors to the destabilized barnase mutants (5). The related Hsp70 family member HSPA1A was also previously found to modulate both the amount of unfolded-like barnase and the amount of barnase aggregation, suggesting it could bind to and stabilize an unfolded-like conformation of barnase (5). While these Hsp70 isoforms are reported to be highly abundant in the cytosol (7), it was unclear as to how modulating their supply might propagate changes in the nucleus or cytosol.
To investigate the effect of upregulating the chaperones, we cotransfected HSPA1A and a specific Hsp40 cofactor DNAJB1 (8) with the barnase biosensors and analyzed the cells after 48 h culture. The transfected HSPA1A and DNAJB1 showed a mostly cytosolic enrichment (Fig. 2C). Transfected HSPA1A was present at the same level as the endogenous HSPA1A (i.e., total levels were twofold normal), whereas transfected Hsp40 was present at ∼48-fold higher concentration than normal (Fig. S3). The cotransfected chaperones significantly reduced the biosensor lower slope values in the cytosol (Fig. 2A), consistent with a greater abundance of chaperone bound to unfolded barnase. The treatments also increased the A50% values indicating that the chaperones effectively suppressed inappropriate aggregation (Fig. 2B). We also monitored the effect of chaperone overexpression on the total level of the I88G barnase aggregates versus WT∗ barnase with a pelleting assay, which showed that the chaperones suppressed aggregation of the cytosolic barnase, although in this context appeared to slightly increase aggregation for nuclear barnase (Fig. S4). The reason for the opposite readouts in the two assays regarding nuclear aggregation of the I88G is unclear but may relate to challenges in efficiently lysing the nucleus in the pelleting assay and the context of a small effect. Nonetheless, the key point from these experiments is that overexpression preferentially deepens the pool of chaperone supply in the cytosol, which likely is explained by the transfected chaperone being mostly restricted to the cytosol.
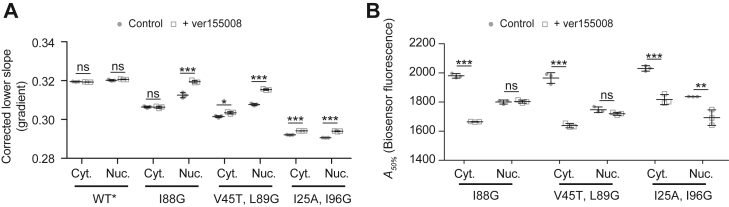
To further probe the role of Hsp70 activity, we inhibited Hsp70 in cells (without overexpressed chaperones) using the small-molecule inhibitor VER-155008, which competitively binds to the ATP-binding pocket of Hsp70 family proteins and impairs substrate binding (9). VER-155008 significantly reduced the viability or growth rate of the cells after treatment, which is consistent with its role in suppressing the proteostasis systems (Fig. S5). VER-155008 increased the lower slope values in the nucleus but not the cytosol (Fig. 3A; and note that the flow cytometry assay only includes live cells in the analysis). This result suggested that while Hsp40 and Hsp70 proteins are more effective at binding barnase in the cytosol, there was higher redundancy and flexibility in the cytosol to absorb a reduced Hsp70 activity than in the nucleus. This raises the possibility that the proteostasis more broadly may be more vulnerable to collapse in the nucleus upon stresses to proteostasis systems. However, the increased sensitivity to proteostasis imbalance in the nucleus was not seen in terms of aggregation. Indeed, aggregation of the biosensor was far more disproportionately enhanced in the cytosol than the nucleus (Fig. 3B). These findings suggested that the correlation of holdase activity and aggregation can be decoupled when specific elements of the proteostasis network are impaired. This effect may arise through redundant holdase activity in the cytosol from non-Hsp70 chaperones that are overall less effective at preventing aggregation than Hsp70.
Figure 3.
Effect of Hsp70 inhibition on the biosensor readouts.A, lower slope values derived by flow cytometry analysis of HEK293T cells after transfection with the biosensors for 18 h and a further treatment with 20 μM Hsp70 inhibitor VER-155008 for 18 h (versus vehicle control). Data points indicate biological replicates, and bars indicate means ± SD. Student’s t test results are shown (2-tailed; control versus overexpression); ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns ≥ 0.05. B, aggregation analysis (A50%) using the same treatments and conditions as for A. HEK293T, human embryonic kidney 293T cell line; ns, not significant.
Aggregation of mutant Htt exon 1 in the cytosol propagates proteostasis imbalances in the cytosol and nucleus
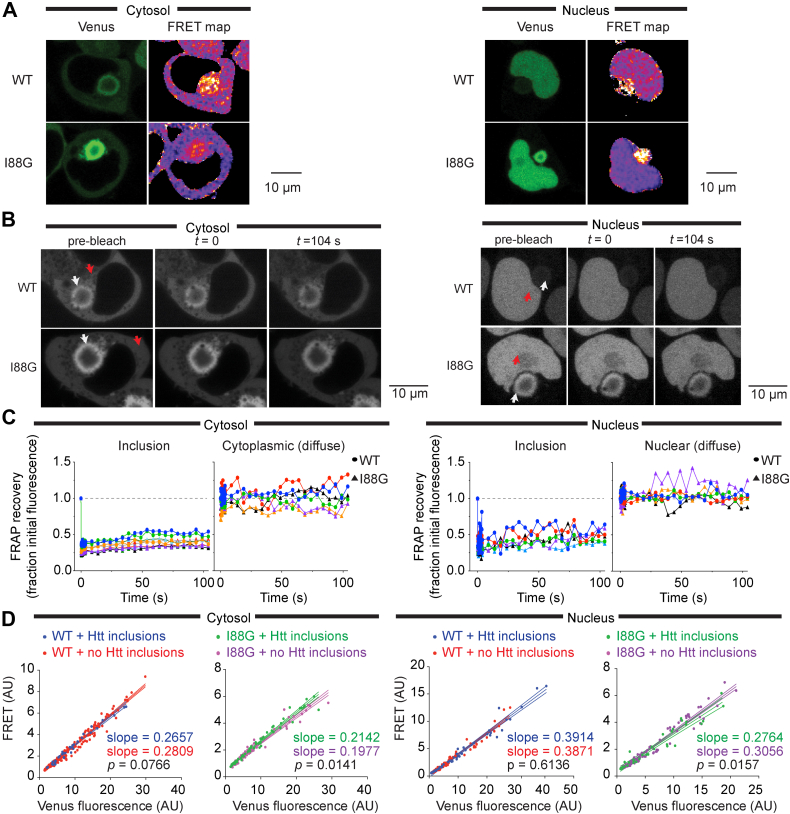
Next, we investigated the quality control supply in the nucleus and cytosol in the context of disease-related protein misfolding and aggregation. For this, we coexpressed the biosensors with mutant Huntington exon 1 fragment containing 97 glutamines in the polyglutamine repeat sequence (Httex197Q) fused to GFP, which forms cytosolic perinuclear inclusions in human embryonic kidney 293T (HEK293T) cells (10). Mutant Httex1 fragments with polyglutamine sequences longer than 36 glutamines accumulate into inclusion bodies in neurons of Huntington's disease patients and have been implicated to direct a maladaptation in protein quality control (11). Because the fluorescence of GFP interferes with our capacity to monitor FRET, we used a variant of GFP that is nonfluorescent as characterized previously (12) and also added a 6-amino acid tetracysteine motif for post hoc detection by ReAsH biarsenical dye binding (13). Live cells were examined for inclusions, which were detectable as spherical pearl-like structures under transmission imaging of confocal microscopy, imaged for FRET, and then post hoc analyzed by ReAsH staining to validate the inclusion structure. Because we needed to fix the cells after imaging, which reduces ReAsH staining, we were not able to ascertain cells containing only diffuse cytosolic Httex197Q. Using this approach, we observed the barnase biosensor as enriched at the periphery of the inclusions suggesting a degree of coaggregation or corecruitment to the inclusions (Fig. 4A). This was both true for the WT∗ barnase biosensor, which does not aggregate by its own volition, and for the nucleus-targeted biosensors suggesting that the biosensors were kinetically trapped on the surface of the Httex1 inclusion. This colocalization was directed to a partial extent by the barnase moiety, since the mTFP1 and Venus-alone constructs were much less associated at the inclusion surface (Fig. S6). To assess whether the biosensor was self-aggregated at the molecular scale, we determined their FRET using a ratiometric analysis of the fluorescence (Fig. 4A). Indeed, the biosensor enriched at the inclusion periphery appeared to have higher FRET than when more distal from the inclusions in either the nucleus or the cytosol. We further assessed the aggregation state using fluorescence recovery after photobleaching (Fig. 4B). A small section of the biosensor was targeted for bleaching on the periphery of the Httex1 inclusion. Both nucleus- and cytosol-targeted biosensors showed little to no recovery after bleaching, indicating the protein was in an immobile state on the seconds timescale (Fig. 4C). By contrast, diffuse biosensor instantly recovered within the first seconds upon recovery, which is consistent with its unaggregated status.
Figure 4.
Huntingtin exon 1 aggregation in the cytosol manifests proteostasis imbalances in the nucleus and cytoplasm.A, confocal micrographs showing the values proportional to the fluorescence ratio of acceptor/donor (Venus/mTFP) of the biosensors coexpressed with mutant Httex197Q fused to a nonfluorescent mutant of GFP. Selected cells are those with Httex1 inclusions (marked with red arrows), identified post hoc as described in the Experimental procedures section. Nuclear-targeted biosensors are on the right and cytosolic-targeted biosensors are on the left (same format for each panel). The scale of the FRET map is color coded from blue to magenta to yellow corresponding to lowest to highest FRET. B, fluorescence recovery after photobleaching (FRAP) of biosensor at the periphery of the Httex197Q inclusion. Arrows indicate region of bleaching (white arrow for Httex1 inclusions and red for diffuse [nuclear or cytoplasmic] barnase). C, quantitation of the data in B, tracking the recovery of fluorescence in the bleached zone. Each color depicts the time course of an individual cell. Data are normalized to the starting fluorescence value (shown as a dashed line). D, confocal microscopy FRET fluorescence values within cells distal to the inclusion. FRET fluorescence was measured by exciting at 458 nm (mTFP1 excitation) and collecting the emission at 510 to 560 nm (Venus emission). Each dot represents the average fluorescence derived from a single cell value. Solid lines show line of best fit from a linear regression with dashed lines showing 95% confidence intervals. p Value was determined by two-tailed t test.
Because the aggregation of the biosensor at the periphery of the inclusion is a confounding factor in whole-cell fluorescence analysis by our flow cytometry methods, we instead measured FRET by microscopy targeting small subregions of the cells away from the inclusions. This analysis revealed that the presence of Httex197Q inclusions had no bearing on the FRET of the WT∗ barnase biosensor outside that associated with the inclusion periphery (Fig. 4D). By contrast, the I88G barnase biosensor showed significantly changed FRET values in regions outside the Httex197Q inclusions compared with cells without inclusions at all. In the case of the I88G barnase biosensor targeted to the cytosol, the FRET was increased, which suggested a reduced overall holdase activity of chaperones in the cytosol arising from Httex197Q aggregation. However, in the case of the I88G barnase biosensor targeted to the nucleus, the FRET was decreased. This result is consistent with an elevated holdase activity in the nucleus when Httex197Q aggregates form.
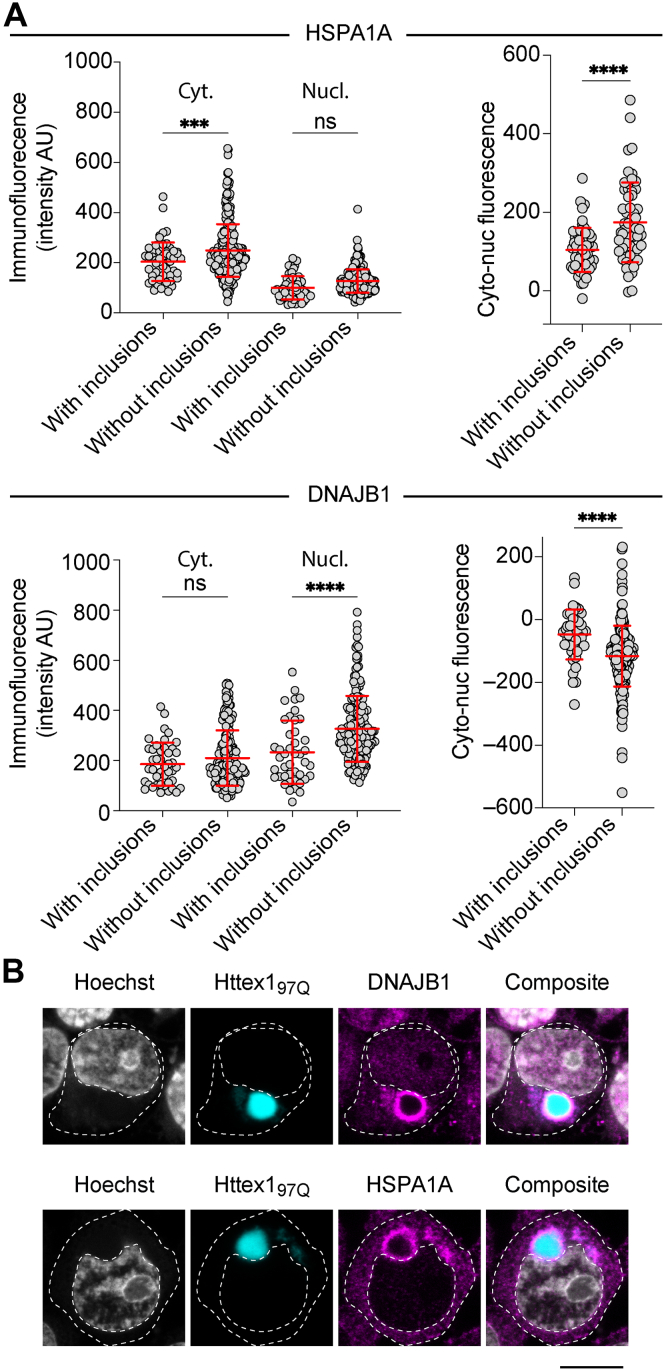
This result therefore suggests that when protein aggregation occurs in the cytosol that cells can move a subset of quality control machinery (holdases) from the cytosol into the nucleus as part of a global coordinated stress response. To test whether this effect arises through increased nuclear accumulation of DNAJB1 and HSPA1A in cells with Httex1 inclusions, we performed immunofluorescence to measure endogenous DNAJB1 and HSPA1A levels. When Httex197Q was coexpressed, both DNAJB1 and HSPA1A were also recruited to the surface of the inclusions suggesting a level of sequestration (Fig. 5A). The levels of endogenous DNAJB1 in the nucleus were decreased, and the levels of Hsp70 in the cytosol away from the inclusion were decreased (Fig. 5). These data collectively suggested that the sequestration of the chaperones by the Httex1 inclusion reduces the wider supply of these chaperones in both the nucleus and the cytosol. As such, these data are not consistent with an increased holdase activity arising from there being more DNAJB1 and HSPA1A transported into the nucleus. One explanation to account for this result is that other chaperone systems are deployed to bind to unfolded biosensor when Httex1 inclusions form, or that there is altered activity of other proteostatic functions such as degradation.
Figure 5.
Httex197Q-induced relocalization of the cellular reserves of DNAJB1 and HSPA1A.A, quantitation of the immunofluorescence levels in the nucleus or cytosol of cells with versus without inclusions of Httex197Q. Data points represent either average immunostaining in regions of cells (excluding the areas close to the inclusions) or differences of immunostaining intensity in individual cells between the nucleus and the cytosol. Differences (means and SD shown of cells from one slide) assessed by t test with significance defined as p < 0.05 (and ∗∗∗∗ for p <0.0001, ∗∗∗ for p < 0.001). B, immunostaining of endogenous DNAJB1 and HSPA1A in cells expressing the Httex197Q. The scale bar indicates 10 μm.
Discussion
Our studies show that the balance of resources required to manage proteostasis is different in the cytosol relative to the nucleus. We find evidence for there being a lower supply of Hsp40 and Hsp70-based chaperone capacity in the nucleus that is able to bind to the unfolded barnase and prevent its aggregation. When we supplemented the cells with additional Hsp70 and Hsp40 protein by their overexpression (HSPA1A and DNAJB1), we increased holdase activity in the cytosol and lowered the aggregation potential consistent with these chaperones exerting a critical activity to bind unfolded proteins and prevent their aggregation. When we pharmacologically inhibited the Hsp70 chaperone system, we observed a disproportionate impact on aggregation in the cytosol, concordant with the cytosol being more dependent on Hsp40 and Hsp70 chaperone–based networks to prevent protein aggregation. We further found that the presence of Httex1 inclusions could sequester total cellular levels of DNAJB1 and HSPA1A, which was detrimental to barnase aggregation in the cytosol, but led unexpectedly to an apparent increase in holdase activity in the nucleus through seemingly distinct quality control mechanisms (discussed later in more detail).
Overall, these results are consistent with the greater requirement of the Hsp70 chaperone system to engage with unfolded or aggregation-prone proteins in the cytosol. This finding is consistent with the high abundance of these chaperones in the cytosol (7), which is not surprising given that most proteins are synthesized in the cytosol or endoplasmic reticulum. Our findings are in agreement with prior work suggesting that the nucleus was more sensitive to heat stress compared with a variety of other organelles using luciferase targeted to different organelles (14). In that study, HSPA1A overexpression could also improve the refolding of denatured luciferase in the cytosol and the nucleus when targeted to those compartments. A heat shock treatment, which upregulated Hsp70 levels, could also induce the preconditioning of cells to protect from luciferase heat denaturation in both the cytosol and the nucleus (14).
Our findings with mutant Httex197Q indicate that the aggregation in the cytosol can manifest dysfunction in quality control capacity in both the cytosol and nucleus. Consistent with prior findings that protein aggregation can sequester quality control resources away from “housekeeping” activities and lead to metastably folded proteins aggregating (15), we found that the pool of resources binding to unfolded barnase biosensor decreased in the cytosol. Prior studies have found that Hsp70 and Hsp40 proteins are recruited into inclusions formed by Httex197Q and similar proteins with expanded polyglutamine sequences (16, 17, 18, 19). One function for this recruitment may be disaggregation, in light of recent findings showing the Hsp70-based chaperone machinery can dissociate amyloid fibrils (20). More unexpected however was the finding that there was an increase in unfolded barnase in the nucleus, which suggests that chaperones are redirected into the nucleus under stress to form a complex with the unfolded biosensor. Hsp70 is known to translocate from the cytosol into the nucleus upon heat shock (21, 22, 23), suggesting there is a dynamic capacity for quality control machinery activity in the nucleus under times of stress. This translocation is regulated by the Hikeshi nuclear import carrier, which is crucial for cells to recover from heat shock stress (24). DNAJB1 can also deliver misfolded protein into the nucleus for degradation (25). However, our data suggest that the chaperones that are recruited to this role for the barnase biosensor are independent to DNAJB1 or HSPA1A since these become depleted in the nucleus when Httex1 forms cytosolic inclusions. Candidate systems that might be engaged under such situations include the small heat shock protein family, which are known to buffer proteins from aggregating under acute proteome stress and which can relocalize to the nucleus after heat shock (26, 27). An alternative possibility is that the increased unfolded barnase in the nucleus is caused by changes in other proteostastic functions. We performed investigations to look at degradation of the biosensors in the presence of translation inhibitor cycloheximide, but the generally slow rate of degradation and variable nature of the degradation rates across different cells meant our findings were inconclusive (not shown).
The other curious result from our study was the recruitment of WT∗ barnase biosensor to the Httex197Q inclusion. We have never observed the WT∗ biosensor to aggregate when expressed on its own suggesting that the inclusion provides a mechanism to recruit this protein to the surface. We also saw a small amount of mTFP1 recruit to the inclusion but little Venus, when these proteins were expressed alone, suggesting that the phenomenon may extend to other proteins more generally. One interesting possibility is that a small fraction of proteins that remain in complex with chaperones can be recruited to the surface of the inclusion by Hsp70-based triage mechanisms that more generally handle misfolded proteins in the cell. The different extent of WT∗ biosensor foldedness in the nucleus compared with the cytoplasm supports the conclusion that some of the WT∗ barnase is partitioned from the equilibrium of folding in an unfolded-like state. Discrete bodies containing misfolded protein including the JUNQ, aggresome, and Q-bodies have been proposed as cellular depots for processing protein aggregates and are enriched with different Hsp70 and Hsp40 proteins (28, 29). In addition, Hsp70 has been proposed to engage with the surface of protein aggregates to act as a disaggregase (20). Hence, the capture of WT∗ biosensor may be indicative of a wider network of chaperone–client interactions, protein aggregate bodies in the cell, and a broader interconnected quality control network. And thus chaperones may have a broader function as a kind of lubricant constantly interfacing with unfolded proteins and aggregating proteins.
Experimental procedures
Expression constructs
The cytosolic FRET barnase biosensor library expressed in the pTriEx4 vector was prepared as previously described (5). In brief, the barnase moiety was flanked by circularly permuted mTFP1 cp175 and Venus cp173 fluorescent proteins. Nuclear localized FRET barnase was generated by the addition of an N-terminal SV40 NLS to the original cytosolic barnase using a synthesized gene cassette containing the relevant localization sequences (GeneArt [Thermo Fisher Scientific]) and standard restriction endonuclease–based ligation methods. For generation of individual mutants of targeting biosensor, the WT∗ barnase biosensor kernel was replaced by the barnase mutant of choice. This was achieved by double digestion of both the desired barnase mutant and nuclear targeting construct plasmids at BamHI and KpnI restriction sites. The tetracysteine-tagged Httex1 construct containing a tetracysteine tag at the C terminus of the Httex1 (TC1 (13)), and a nonfluorescent mutant of Emerald fluorescent protein (Em), Y66L (12), was generated in-house to yield a plasmid named Httex197QTC1-Em Y66L in the pT-Rex vector (Invitrogen). The pT-Rex Em Y66L construct alone was also generated in-house as described previously (12). V5-tagged chaperone proteins were overexpressed from pcDNA5/FRT/TO V5 DNAJB1 and pcDNA5/FRT/TO V5 HSPA1A provided as gifts from Harm Kampinga (30) via Addgene.
Cell culture
HEK293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (w/v) fetal calf serum and 1 mM glutamine in a 37 °C humidified incubator with 5% v/v atmospheric CO2. Cells were seeded in poly-l-lysine–coated plates. For microscopy experiments, cells were plated at 3 × 105 cells/ml in an 8-well μ-slide (Ibidi). For flow cytometry experiments, cells were seeded at 1.1 × 105 cells/ml in a 48-well plate. Cells were transiently transfected with Lipofectamine 3000 reagent as per manufacturer’s instructions (Life Technologies, Thermo Fisher Scientific). For Barnase and Httex1 cotransfections, the transfection was done in a way to decouple the expression of the two plasmids.
HSP70 was inhibited with 20 μM VER-155008 (catalog no.: SML0271; Sigma–Aldrich) in culture media for 18 h.
Microscopy
Cells were imaged on a TCS SP5 confocal microscope or LCS SP8 confocal microscope (Leica Biosystems). For anti-V5 immunofluorescence, cells were fixed in 4% w/v paraformaldehyde for 15 min at room temperature, washed with PBS, and permeabilized in 0.5% v/v Triton X-100 in PBS (Sigma–Aldrich) for 30 min. After incubation in blocking solution (5% w/v bovine serum albumin [BSA] in 0.3% v/v Triton X-100 in PBS), cells were incubated with anti-V5 (1:250 dilution in 1% w/v BSA in 0.3% v/v Triton X-100 in PBS) (catalog no.: ab27671; Abcam) overnight at 4 °C. Cells were then washed in 1% w/v BSA in 0.3% v/v Triton X-100 in PBS before being stained with anti-mouse cy5 (1:500 dilution in PBS) for 30 min at room temperature. Prior to confocal imaging, cells were stained with Hoechst 33342 (Thermo Fisher Scientific).
For anti-Hsp40 and Hsp70 immunofluorescence, cells were fixed with prechilled methanol (−20 °C) for 7 min at 4 °C and washed extensively with PBS, ensuring that the samples did not dry out in between wash steps. Cells were blocked in 5% w/v BSA in PBS for 30 min and stained with either mouse anti-Hsp40 antibody (Abcam; catalog no.: ab78437; 1:200 dilution) or mouse anti-Hsp70 antibody (Abcam; catalog no.: ab5439; 1:50 dilution) for 2 h at room temperature. Cells were then washed with PBS and incubated with antimouse Cy5 (1:500 dilution in PBS) for 1 h. Cells were washed with PBS, and the nucleus was stained with Hoechst 33342 before imaging.
For ReAsH staining, 24 h post-transfection, cells were washed twice with Hank’s balanced salt solution (HBSS) and stained with 1 μM ReAsH and 10 μM 1,2-ethanedithiol in HBSS for 30 min at 37 °C. Cells were then washed with 250 μM BAL for 15 min at 37 °C followed by a final wash in HBSS.
Image analysis
Confocal images were analyzed using custom analysis scripts for FIJI (31) and Python (version 3.6.7; Python Software Foundation), available alongside example datasets at doi.org/10.5281/zenodo.5661911.
In the case of immunofluorescence measurements, whole cells, nuclei and, where necessary, Httex1 inclusions, were initially identified using the machine learning package CellPose (32). Segmentation was performed on the Cy5-labeled anti-V5 antibody (633 nm excitation, 650–750 nm emission), Hoechst (405 nm excitation, 410–450 nm emission), and enhanced GFP (488 m excitation, 490–520 nm emission) channels to identify the whole cell, nuclei, and inclusion regions of interest (ROIs), respectively. The accuracy of each round of segmentation was manually inspected before per-pixel information for each ROI collected, and then the nuclei and inclusion ROIs were removed from the whole-cell ROI to yield the cytosolic population. Finally, the fluorescence intensity for the nucleus and cytosol was determined from the mean of all pixels in each compartment.
To quantify fluorescence recovery after photobleaching, ROIs for individual bleach spots were defined via automatic Otsu thresholding of the first bleaching frame. Identical ROIs were then manually placed for the adjacent (nonbleached) and background regions. Where necessary, ROI positions were manually adjusted across time points to account for cellular drift. The mean intensity was calculated for each ROI, and both bleached and nonbleached ROIs were then corrected against the corresponding background ROI for each time point, generating Bcorr and NBcorr, respectively. The ratio of Bcorr/NBcorr at each time point was finally normalized to the prebleach ratio of Bcorr/NBcorr to yield the relative recovery.
In the case of FRET measurements, whole cells, nuclei, and Httex1 inclusions were initially identified using CellPose (32) as described previously. In this case, segmentation was performed on the acceptor channel (488 nm excitation, 510–560 nm emission), computationally inverted acceptor channel, and ReAsH channel (561 nm excitation, 610–680 nm emission) for whole cells, nuclei, and inclusions, respectively. After manual inspection to ensure the accuracy of each round of segmentation, per-pixel intensity values for each ROI were collected. In the case of cytosolic barnase variants, both the nuclei and aggregate features (which included both Httex1 inclusions and the surrounding accumulated barnase) were excluded from the whole cell to yield the diffuse barnase population. In the case of nuclear barnase variants, aggregate features (which included both Httex1 inclusions and the surrounding accumulated barnase) within the nuclei ROI were similarly excluded to yield the diffuse barnase population. Finally, the relative FRET for each ROI was calculated as the mean pixel intensity in the FRET channel (458 nm excitation, 510–560 nm emission).
Ratiometric FRET maps were created using Fiji via a pixelwise division of the FRET channel by the mTFP channel. The resultant image was then multiplied by a binary cell mask derived from manual thresholding of the Venus channel (whereby the threshold was set to capture the maximal number of cells above the threshold) to remove the background pixels.
Flow cytometry
After 24 h (drug treatments) or 48 h (cotransfections) post-transfection, cells were washed and harvested by gentle pipetting in PBS. Cells were analyzed via flow cytometry as described previously (5). In short, 150 μl of cell suspension was analyzed at a flow rate of 3 μl/s on a BD LSRFortessa Cell Analyzer (BD Biosciences). Acceptor (Venus) fluorescence was collected with the 488 nm laser and FITC (530/30) filter. Acceptor sensitized emission (FRET) and donor (mTFP1) fluorescence were collected with the 405 nm laser with PE (575/25) and V500 (525/50) filters, respectively. All flow cytometry data were processed with FlowJo (version 10; Tree Star, Inc) to exclude cell debris, cell aggregates, and untransfected cells. The Venus channel was compensated to remove bleed through from mTFP1 and FRET channels using donor only. Data were analyzed in MATLAB (version 9; MathWorks). The gating strategy and associated data analysis protocols are detailed previously (33).
Urea denaturation curves
Urea denaturation curves were measured on cell lysates expressing the biosensors in 96-well format. In essence, 80 μl of samples were prepared containing 0 M to 8 M urea in PBS. Lysates were prepared from cells 24 h after transfection (WT∗ with NES and NLS tags) by pipetting in 20 mM Tris (pH 8.0), 2 mM MgCl2, 1% v/v Triton X-100, 1× EDTA-free protease inhibitor (Roche), 150 mM NaCl, 20 U ml–1 benzonase, and 1 mM PMSF. Aggregates and cell debris were pelleted by centrifugation at 16,000g for 10 min at 4 °C. About 5 μl supernatant was added to each urea concentration. As the measurements were ratiometric and both fluorophores were on the same molecule, samples were not matched for protein concentration. Fluorescence readings (430 nm excitation, 492 nm emission and 532 nm emission) were measured at 23 °C using a Clariostar microplate reader (BMG Labtech) every 15 min for 4 h. Relative FRET efficiencies (calculated as acceptor fluorescence/[donor fluorescence + acceptor fluorescence]) were averaged across readings and fit to a two-state unfolding model as described previously (5).
Western blot
HEK293T cells were transfected with DNAJB1 or HSPA1A and harvested 48 h post-transfection. Cells were pelleted (200g, 6 min) and resuspended in radioimmunoprecipitation assay lysis buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% v/v Triton X-100, 0.5% v/v sodium deoxycholate, 0.1% v/v SDS, 250 U benzonase, and supplemented with cOmplete EDTA-free Protease Inhibitor Cocktail pills [Roche]) and incubated on ice for 30 min. Lysate was matched for total protein by Bicinchoninic Acid kit (Thermo Fisher Scientific; catalog no.: 23225), and specific amounts of protein were loaded on to a TGX Stain-Free FastCast Acrylamide gel (Bio-Rad; catalog no.: 1610185) and transferred to the blotting membrane with an iBlot2 gel transfer device (Thermo Fisher Scientific; catalog no.: IB21001) and a polyvinylidene difluoride iBlot2 transfer stack (Thermo Fisher Scientific; catalog no.: IB24001). The membrane was blocked with 5% w/v BSA in Tris-buffered saline (TBS) for 1 h at room temperature. Anti-Hsp40 (Abcam; catalog no.: ab69402) and anti-Hsp70 (Abcam; catalog no.: ab194360) were diluted 1:10,000 and 1:4000, respectively, in 5% w/v BSA in TBS containing 0.1% v/v Tween-20 and incubated on the membrane for 1 h at room temperature. The membranes were then incubated in anti-rabbit secondary (Invitrogen; catalog no.: 65-6120; 1:20,000 dilution) or antimouse secondary (Thermo Fisher Scientific; catalog no.: 31430; 1:20,000 dilution) in TBS containing 0.1% v/v Tween-20 for 1 h at room temperature. Horseradish peroxidase was detected by enhanced chemiluminescence (Bio-Rad; catalog no.: 17050S) using the ChemiDoc MP Imaging system (Bio-Rad). For analysis, the band intensities were measured and background intensities were subtracted using the ImageLabs software (Bio-Rad). For each replicate, band intensity was normalized to the untransfected sample, and the normalized values were plotted and linear regression calculated. Fold overexpression was calculated as ([band intensitychaperone – band intensityuntransfected]/band intensityuntransfected).
Aggregation pelleting assay
At 48 h post-transfection, cells were harvested by lifting from the plate and pelleting (200g, 6 min). The pellets were resuspended in M-PER (Thermo Fisher Scientific; catalog no.: 78501) and incubated with shaking for 10 min. The resultant lysates were adjusted with M-PER so that they were all the same concentration of total protein by Bicinchoninic Acid kit (Thermo Fisher Scientific; catalog no.: 23225) and then spun at 16,000g for 30 min to pellet aggregates. The supernatant was transferred to a new tube, and the pellet was resuspended in M-PER to match the supernatant volume. The supernatant and pellet suspensions were transferred to black 384-well plate for measurement of Venus fluorescence (513 nm excitation, 570/60 nm emission) using a Clariostar microplate reader (BMG Labtech).
CellTitre-Glo assay
At 24 h post-transfection, HEK293T cells were detached from the plate by gentle pipetting in PBS. The cell suspensions, and a media-only control, were transferred to a white 96-well plate and equilibrated at room temperature for 30 min. Reconsitintued CellTitre-Glo reagent (Promega; catalog no.: G7570) was added to each well and incubated on an orbital shaker for 2 min followed by incubation at room temperature for 10 min. Luminesence was measured using a Clariostar microplate reader (BMG Labtech) according to the CellTitre-Glo manufacturer’s instructions.
Data availability
All data are provided within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
C. B. R., A. R. O., and D. M. H. conceptualization; D. C. and D. M. H. methodology; D. C. software; A. R. O. validation; C. B. R., A. R. O., D. C., and D. M. H. formal analysis; C. B. R., A. R. O., C. A. G., C. M., and N. S. M. investigation; D. M. H. resources; D. C. data curation; C. B. R. and D. M. H. writing–original draft; A. R. O., D. C., C. A. G., C. M., N. S. M., S. E., A. D., G. M., and D. M. H. writing–review & editing; C. B. R., A. R. O., C. A. G., N. S. M., and D. M. H. visualization; D. M. H. supervision; D. M. H. project administration; S. E., A. D., G. M., and D. M. H. funding acquisition.
Funding and additional information
The work was funded by a Human Frontier Science Program Research Grant (grant no.: RGP0022/2017) to S. E., A. D., G. M., and D. M. H.
Edited by Ursula Jakob
Supporting information
References
- 1.Schneider K., Bertolotti A. Surviving protein quality control catastrophes--from cells to organisms. J. Cell Sci. 2015;128:3861–3869. doi: 10.1242/jcs.173047. [DOI] [PubMed] [Google Scholar]
- 2.Knowles T.P., Vendruscolo M., Dobson C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 3.Bertram L., Tanzi R.E. The genetic epidemiology of neurodegenerative disease. J. Clin. Invest. 2005;115:1449–1457. doi: 10.1172/JCI24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hipp M.S., Park S.H., Hartl F.U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Wood R.J., Ormsby A.R., Radwan M., Cox D., Sharma A., Vopel T., et al. A biosensor-based framework to measure latent proteostasis capacity. Nat. Commun. 2018;9:287. doi: 10.1038/s41467-017-02562-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serrano L., Kellis J.T., Jr., Cann P., Matouschek A., Fersht A.R. The folding of an enzyme. II. Substructure of barnase and the contribution of different interactions to protein stability. J. Mol. Biol. 1992;224:783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- 7.Finka A., Goloubinoff P. Proteomic data from human cell cultures refine mechanisms of chaperone-mediated protein homeostasis. Cell Stress Chaperones. 2013;18:591–605. doi: 10.1007/s12192-013-0413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kampinga H.H., Craig E.A. The HSP70 chaperone machinery: j proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlecht R., Scholz S.R., Dahmen H., Wegener A., Sirrenberg C., Musil D., et al. Functional analysis of Hsp70 inhibitors. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramdzan Y.M., Trubetskov M.M., Ormsby A.R., Newcombe E.A., Sui X., Tobin M.J., et al. Huntingtin inclusions trigger cellular quiescence, deactivate apoptosis, and lead to delayed necrosis. Cell Rep. 2017;19:919–927. doi: 10.1016/j.celrep.2017.04.029. [DOI] [PubMed] [Google Scholar]
- 11.Cox D., Raeburn C., Sui X., Hatters D.M. Protein aggregation in cell biology: an aggregomics perspective of health and disease. Semin. Cell Dev. Biol. 2020;99:40–54. doi: 10.1016/j.semcdb.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Olshina M.A., Angley L.M., Ramdzan Y.M., Tang J., Bailey M.F., Hill A.F., et al. Tracking mutant huntingtin aggregation kinetics in cells reveals three major populations that include an invariant oligomer pool. J. Biol. Chem. 2010;285:21807–21816. doi: 10.1074/jbc.M109.084434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramdzan Y.M., Nisbet R.M., Miller J., Finkbeiner S., Hill A.F., Hatters D.M. Conformation sensors that distinguish monomeric proteins from oligomers in live cells. Chem. Biol. 2010;17:371–379. doi: 10.1016/j.chembiol.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hageman J., Vos M.J., van Waarde M.A.W.H., Kampinga H.H. Comparison of intra-organellar chaperone capacity for dealing with stress-induced protein unfolding. J. Biol. Chem. 2007;282:34334–34345. doi: 10.1074/jbc.M703876200. [DOI] [PubMed] [Google Scholar]
- 15.Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 16.Mitsui K., Nakayama H., Akagi T., Nekooki M., Ohtawa K., Takio K., et al. Purification of polyglutamine aggregates and identification of elongation factor-1alpha and heat shock protein 84 as aggregate-interacting proteins. J. Neurosci. 2002;22:9267–9277. doi: 10.1523/JNEUROSCI.22-21-09267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay D.G., Sathasivam K., Tobaben S., Stahl B., Marber M., Mestril R., et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum. Mol. Genet. 2004;13:1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 18.Seidel K., Meister M., Dugbartey G.J., Zijlstra M.P., Vinet J., Brunt E.R., et al. Cellular protein quality control and the evolution of aggregates in spinocerebellar ataxia type 3 (SCA3) Neuropathol. Appl. Neurobiol. 2012;38:548–558. doi: 10.1111/j.1365-2990.2011.01220.x. [DOI] [PubMed] [Google Scholar]
- 19.Radwan M., Lilley J.D., Ang C.S., Reid G.E., Hatters D.M. Immiscible inclusion bodies formed by polyglutamine and poly(glycine-alanine) are enriched with distinct proteomes but converge in proteins that are risk factors for disease and involved in protein degradation. PLoS One. 2020;15 doi: 10.1371/journal.pone.0233247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nillegoda N.B., Kirstein J., Szlachcic A., Berynskyy M., Stank A., Stengel F., et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015;524:247–251. doi: 10.1038/nature14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelham H.R. Hsp70 accelerates the recovery of nucleolar morphology after heat shock. EMBO J. 1984;3:3095–3100. doi: 10.1002/j.1460-2075.1984.tb02264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch W.J., Feramisco J.R. Nuclear and nucleolar localization of the 72,000-dalton heat shock protein in heat-shocked mammalian cells. J. Biol. Chem. 1984;259:4501–4513. [PubMed] [Google Scholar]
- 23.Velazquez J.M., Lindquist S. hsp70: nuclear concentration during environmental stress and cytoplasmic storage during recovery. Cell. 1984;36:655–662. doi: 10.1016/0092-8674(84)90345-3. [DOI] [PubMed] [Google Scholar]
- 24.Kose S., Furuta M., Imamoto N. Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell. 2012;149:578–589. doi: 10.1016/j.cell.2012.02.058. [DOI] [PubMed] [Google Scholar]
- 25.Park S.H., Kukushkin Y., Gupta R., Chen T., Konagai A., Hipp M.S., et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154:134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Geum D., Son G.H., Kim K. Phosphorylation-dependent cellular localization and thermoprotective role of heat shock protein 25 in hippocampal progenitor cells. J. Biol. Chem. 2002;277:19913–19921. doi: 10.1074/jbc.M104396200. [DOI] [PubMed] [Google Scholar]
- 27.Webster J.M., Darling A.L., Uversky V.N., Blair L.J. Small heat shock proteins, big impact on protein aggregation in neurodegenerative disease. Front. Pharmacol. 2019;10:1047. doi: 10.3389/fphar.2019.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaganovich D., Kopito R., Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sontag E.M., Vonk W.I.M., Frydman J. Sorting out the trash: the spatial nature of eukaryotic protein quality control. Curr. Opin. Cell Biol. 2014;26:139–146. doi: 10.1016/j.ceb.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hageman J., Kampinga H.H. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones. 2009;14:1–21. doi: 10.1007/s12192-008-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stringer C., Wang T., Michaelos M., Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Met. 2021;18:100–106. doi: 10.1038/s41592-020-01018-x. [DOI] [PubMed] [Google Scholar]
- 33.Hatters D., Wood R., Raeburn C., Ormsby A., Oliveberg M., Dickson A., et al. Measuring proteostasis capacity using transiently transfected bait proteins by flow cytometry. Protoc. Exchange. 2018 doi: 10.1038/protex.2017.156. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided within the article.