Abstract
It is widely accepted that activating the transcription factor NRF2 will blast the physiological anti-inflammatory mechanisms, which will help combat pathologic inflammation. Much effort is being put in inhibiting the main NRF2 repressor, KEAP1, with either electrophilic small molecules or disrupters of the KEAP1/NRF2 interaction. However, targeting β-TrCP, the non-canonical repressor of NRF2, has not been considered yet. After in silico screening of ∼1 million compounds, we now describe a novel small molecule, PHAR, that selectively inhibits the interaction between β-TrCP and the phosphodegron in transcription factor NRF2. PHAR upregulates NRF2-target genes such as Hmox1, Nqo1, Gclc, Gclm and Aox1, in a KEAP1-independent, but β-TrCP dependent manner, breaks the β-TrCP/NRF2 interaction in the cell nucleus, and inhibits the β-TrCP-mediated in vitro ubiquitination of NRF2. PHAR attenuates hydrogen peroxide induced oxidative stress and, in lipopolysaccharide-treated macrophages, it downregulates the expression of inflammatory genes Il1b, Il6, Cox2, Nos2. In mice, PHAR selectively targets the liver and greatly attenuates LPS-induced liver inflammation as indicated by a reduction in the gene expression of the inflammatory cytokines Il1b, TNf, and Il6, and in F4/80-stained liver resident macrophages. Thus, PHAR offers a still unexplored alternative to current NRF2 activators by acting as a β-TrCP/NRF2 interaction inhibitor that may have a therapeutic value against undesirable inflammation.
Keywords: NRF2, β-TrCP, Protein-protein interaction inhibitor, Inflammation, Liver
Graphical abstract
Highlights
-
•
We report the first disrupter of the β-TrCP/NRF2 interaction.
-
•
This protein/protein interaction inhibitor activates NRF2 in a KEAP1-independent but β-TrCP-dependent manner.
-
•
This molecule protects against hydrogen peroxide-induced oxidative stress.
-
•
This molecule targets NRF2 specifically in liver, but not in brain or kidney.
-
•
This molecule attenuates liposaccharide-induced acute liver inflammation.
1. Introduction
Transcription factor NRF2 (Nuclear factor (erythroid-derived 2)-like 2) is a cap'n’collar (CNC) basic-region leucine zipper (bZIP) transcription factor that is a master regulator of cellular homeostasis [1]. It controls both the basal and inducible expression of more than 250 genes that contain a specific enhancer, termed the Antioxidant Response Element (ARE, 5′-TGACNNNGC-3′), located in their regulatory regions [2]. These genes encode a spectrum of cytoprotective proteins that protect against multiple stressors by participating in physiological processes such as detoxification, biotransformation, antioxidant reactions, inflammation, and intermediary metabolism [3]. One of the best-characterized roles of NRF2 is the attenuation of exacerbated inflammation [4,5]. NRF2 attenuates inflammatory responses directly by activating the expression of the scavenger receptors MARCO and CD36, and the anti-inflammatory cytokine IL-17D, or by inhibiting the expression of the pro-inflammatory cytokines IL-1β and IL-6 [6,7]. Moreover, NRF2 attenuates inflammation indirectly by inducing the anti-inflammatory and antioxidant enzymes, such as heme oxygenase-1 (HO1) [6,8], and by preventing oxidative stress-induced up-regulation of NF-κB, the master regulator of inflammation [9]. Pharmacological activation of NRF2 represents an underappreciated alternative, or addition to the anti-inflammatory drugs used to treat inflammatory conditions, which is based on the fact that activation of NRF2 bolsters endogenous anti-inflammatory programs.
NRF2 is principally controlled through regulation of its protein stability by the ubiquitin-proteasome system, which is achieved through the presence of two degradation sequences (degrons) within its Neh2 and Neh6 domains, that under non-stressed conditions, confer on this CNC-bZIP transcription factor a half-life of ∼30 min, depending on the cell type [[10], [11], [12]]. The best-characterized of these is the E3 ubiquitin ligase adapter KEAP1 (Kelch-like-ECH-associated protein 1) [13,14]. Briefly, under homeostatic conditions, a KEAP1 homodimer binds one molecule of NRF2 at two amino acid motifs in the transcription factor that exhibit low (DLG) and high (ETGE) affinity, and hence presents NRF2 for ubiquitination by the CUL3/RBX1(Cullin-3/RING-box protein 1) complex, leading to subsequent degradation by the proteasome [15]. KEAP1 is a redox and electrophile sensor that upon modification of critical cysteines loses its ability to repress NRF2 [6]. Under these conditions, newly synthesized NRF2 translocates to the nucleus, heterodimerizes with small MAF protein, and transactivates its target genes. Alternatively, activation of SQSTM1-mediated autophagy directs KEAP1 for degradation, thus relieving NRF2 from repression by KEAP1 [16]. Pharmacological inhibition of KEAP1 is a field of intensive research [17,18]. The most successful case is dimethyl fumarate (DMF), which was approved by the FDA and EMA as a KEAP1 inhibitor for the treatment of psoriasis and multiple sclerosis [[19], [20], [21]]. However, DMF, like many other electrophilic compounds, lacks specificity for KEAP1, because these molecules modify cysteines of other proteins that can lead to many cellular changes [22]. To overcome this problem, a new class of NRF2 inducers has emerged that prevent coupling between NRF2 and KEAP1 and are called Protein-Protein Interaction (PPI) inhibitors for NRF2/KEAP1 [22].
An alternative mechanism of regulation of NRF2 stability is its ubiquitination driven by the E3 ligase adapter β-TrCP. We described a phosphorylation-dependent degradation motif (phosphodegron) in the Neh6 domain of NRF2 that is phosphorylated by glycogen synthase kinase 3 (GSK-3), which is then recognized by the E3 ligase adapter β-TrCP (beta-transducin repeat-containing protein). Acting in concert, GSK-3β and β-TrCP target NRF2 for degradation by a CUL1/RBX1 complex [10,12,23]. We also identified another GSK-3-independent phosphodegron that is also recognized by β-TrCP in the Neh6 domain which suggests β-TrCP, like KEAP1, binds NRF2 in a two-site manner [24].
To date, exploitation of the β-TrCP-NRF2 pathway in development of pharmacological NRF2 activators has not been explored. Here, we report the activation of NRF2 by the first PPI inhibitor of β-TrCP-NRF2 to be developed, which we term PHAR. Our study shows that inhibition of β-TrCP-NRF2 association by PHAR significantly antagonized lipopolysaccharide (LPS)-stimulated inflammation in macrophages as well as acute liver inflammation in the mouse.
2. Material and methods
Cell culture and reagents. MCF-7 c32 ARE-Luc, human HEK293T cells, and mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS, HyClone, CH30160.03) with 80 μg/ml gentamicin (Normon Laboratories). Raw264.7 macrophage cells were cultured in Roswell Park Memorial Institute medium (RPMI1640) (Sigma-Aldrich, R6504) with 10% fetal bovine serum (FBS) and 80 μg/ml gentamicin. MCF-7 c32 ARE-luc cells were kindly provided by Professor C. Roland Wolf (University of Dundee, UK). Keap1+/+ and Keap1−/− MEFs were kindly provided by Professor Ken Itoh (Center for Advanced Medical Research, Hirosaki University Graduate School of Medicine, Hirosaki, Japan). Nrf2+/+ and Nrf2−/− MEFs were obtained from respective wild type and Nrf2-knockout C57BL/6 mice kindly provided by Professor Masayuki Yamamoto (Department of Medical Biochemistry, Tohoku University Graduate School of Medicine, Sendai, Japan). Other reagents, including 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium(MTT), Lipopolysaccharide from Escherichia coli O111:B4 (LPS) and H2O2, were purchased from Sigma-Aldrich. Other reagents were obtained as follows: R, S-sulforaphane (SFN, LKT Laboratories, Inc. ID S8044), LY294002 (TOCRIS). Pevonedistat (MLN-4924) was purchased from MedKoo Bioscience (201924). Pevonedistat, LY294002, and PHAR were dissolved in dimethyl sulfoxide (DMSO). The final concentration of DMSO in cell culture was less than 0.2%.
Animals and treatments. All experimental procedures were performed according to the Guide for the Care and Use of Laboratory Animals and had been previously approved by the Autonomous Community of Madrid (PROEX 105/18). For HPLC analysis of organ exposure to PHAR, 2–4-month-old C57BL/6 mice were used for each experimental condition (n = 5). Mice were randomly divided into three groups that received an intraperitoneal (i.p.) injection of either vehicle (Tween-80: PBS; 1:13), PHAR (50 mg/kg body weight dissolved in the vehicle) or SFN (50 mg/kg body weight dissolved in Saline). After 2 h, livers were removed and used for biochemical analyses and HPLC. For chronic treatments, mice were randomly divided into two groups that received a daily i.p. injection of either vehicle or PHAR. After 5 days, the liver, brain, and kidney were removed for biochemical analyses. For induction of acute liver inflammation, 2–4-month-old mice were randomly divided into four groups (n = 5) that received i.p. either vehicle (groups 1 and 3) or PHAR (groups 2 and 4) for 5 days. Then, mice received i.p. either saline (control mice, groups 1 and 2) or 10 mg/kg body weight LPS. Mice were sacrificed after 4 h, and livers were collected for biochemical analyses.
Chemical Libraries. The 3D structures of all the tested compounds belong to ZINC natural products, and the National Center for Biotechnology Information (NCBI) PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The SDS files were converted to MOL2 format using Marvin Suite 6.0 tools, The MOL2 files were then converted to PDBQT files using Open Babel 2.4.1 suite, as described previously [[25], [26], [27]].
Protein Structures. Crystallographic data for human β-TrCP1-Skp1-β-catenin (PDB code: 1P22), β-TrCP2- Skp1-β-catenin (PDB code: 6WNX), and KEAP1 (PDB code: 4IQK) were obtained from the Protein Data Bank in the PDB format. The specific edition of protein structures was made using PyMol 2.0 software (PyMOL Molecular Graphics System, v2.3.3 Schrödinger, LLC, at http://www.pymol.org/). For each model, water molecules, ions, or inhibitors were removed.
ADMET prediction. Molecular descriptors, such as the topological polar surface area (TPSA), molecular weight (MW), the estimated logarithm (base 10) of the solubility measured in molarity (cLogS), the estimated logarithm (base 10) of the partition coefficient between n-octanol and water (cLogP), the number of hydrogen bond acceptors (H-acceptors), the number of hydrogen bond donors (H-donors), number of rotatable bonds, and the violations of Lipinski's rule of five (Ro5 violations) were calculated using DataWarrior v5.0.0 software (Allschwil, Switzerland) [[25], [26], [27]]. The in-silico absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of all compounds were estimated using AdmetSAR web application [28] and the DataWarrior v5.0 [29].
Molecular Docking Simulation. Molecular docking simulations were carried out using YASARA structure v19.12.14 software, executing the AutoDock 4 algorithm with AMBER99 as a force field [[25], [26], [27]]. A total of 100 flexible docking runs were set and clustered (7 Å) around the ligand-binding domain cavity, i.e., two complexed compounds belonged to different clusters if the ligand root-mean-square deviation of their atomic positions was greater than 7 Å around certain hot spot conformation [30]. The YASARA software calculated the Gibbs free energy variation (ΔG, Kcal mol−1), with more positive energy values indicating stronger binding [31]. To calculate this parameter, Autodock Vina uses a force field scoring function that considers the strength of electrostatic interactions, hydrogen bonding between all the atoms of the two binding partners, in the complex, intermolecular van der Waals forces, and solvation and entropy contributions [32]. The ligand-protein interactions were detected with the protein–ligand interaction profiler (PLIP) algorithm [33]. Molecular docking simulations were carried out in the known binding sites of NRF2 for β-TrCP (WD40 domain) [10] and for KEAP1 (KELCH domain) [34]. For each region, the grid dimensions were 23 × 23 × 23. The points were centered to the co-crystallized ligands to ensure the coverage of the binding site of the structure. The appropriate pH 7.4 protonation state of the β-TrCP or KEAP1 side chains was created using the YASARA structure v19.12.14 software [35]. YASARA software generated a file containing the molecular coordinates of different poses of the conformer docked to the binding site in the protein, as well the Gibbs free energy variation (ΔG, kcal/mol) for each pose. Compounds with ΔG ≤ 9.5 kcal/mol were chosen as potential inhibitors. YASARA structure v19.12.14 software was set up on a Linux cluster at https://www.ccc.uam.es/(Center for Scientific Computing CCC-UAM). All the figures were prepared using PyMol 2.3.3 software.
Molecular dynamics (MD) simulations. YASARA structure v19.12.14 (Vienna, Austria) was employed to carry out all the MD simulations with AMBER14 as a force field as described previously [[36], [37], [38]]. The simulation cell was allowed to include 20 Å surrounding the protein that was filled with water at a density of 0.997 g/ml. Initial energy minimization was carried out under relaxed constraints using the steepest descent minimization. Simulations were performed in water under constant pressure and constant temperature (25 °C) conditions. To mimic a physiological environment, counter ions were added to neutralize the system (Na+ or Cl− were added as a replacement for water to give a final NaCl concentration of 0.9% and pH maintained at 7.4). Hydrogen atoms were added to the protein structure at the appropriate ionizable groups according to both the calculated pKa and the simulation pH (i.e., a hydrogen atom was added if the computed pKa was higher than the pH). The pKa was computed for each residue according to the Ewald method [39]. All simulation steps were run using a pre-installed macro (md_run.mcr) within the YASARA suite. Data were collected every 100 ps. The molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) calculations were used to determine the alchemical binding free energy of drug candidates against β-TrCP or KEAP1 using the YASARA macro md_analyzebindenergy.mcr, as previously described [[36], [37], [38]]. When a compound remains bound to β-TrCP during the entire time of the MD simulation, the measurement of its trajectory should not exceed a RMSD value of 10 Å. Compounds with an RMSD value greater than 10 Å (i.e. binding is not stable over 200 ns) were discarded. Additionally, the value of MM|PBSA solvation binding energy [40,41] was considered in this filtering process. Values lower than 10 kcal/mol for the last 50 ns of the MD simulation were also discarded.
Rosetta Modeling of Binding Affinity. The program InterfaceAnalyzer from Rosetta Package [42] was used to predict the binding affinity of the phospho-peptides by docking with human β-TrCP1 and β-TrCP2. The peptide of the first site of NRF2 bound with β-TrCP1was modelled with the program MODELLER [43], using as template the conformation of the peptide of human β-Catenin in the structure of 1P22 from PDB. Phospho-serines 335 and 338 were built by adding two phosphate groups using the program Chimera [44]. The phosphorylated peptide bound with β-TrCP2 was similarly modelled using the structure of β-TrCP2 from 6WNX in PDB. Both models were optimized using 5 cycles of optimization with the program “relax” of Rosetta Package [45]. The same approach was repeated for the peptide of the second site of NRF2, with phospho-serines 342 and 345. The other peptides (hEpoR, hGli2, hMcl-1, hFGD1, hIFNAR1, hIkBa, hPRLR, hSNAIL3, hYAP) were modelled with MODELLER. Due to the different amino-acid lengths between phospho-serines, the conformations of hEpoR, hGli2 and hMcl-1 were modelled using the conformation of the modelled peptide of NRF2 in the first site, while the conformation of β-Catenin was used to model the rest. The models of the peptides bound with β-TrCP1 and β-TrCP2 were obtained by superimposition with their respective templates in the structures of PDB encoded by 1P22 (for β-TrCP1) and 6WNX (for β-TrCP2). The structures of both phospho-serines of all peptides were built with Chimera by adding the phosphate groups in the structures with their interacting partners (β-TrCP1 and β-TrCP2) and they were all optimized with 5 cycles of the program “relax” of the package of Rosetta. Then, all the structures were further optimized, repeating 100 times of 15 cycles of optimization. The optimization protocol produced two collections of 100 conformations of each phospho-peptide, one with peptides bound with β-TrCP1 and another with β-TrCP2. Binding affinities were calculated for all the modelled structures using a docking approach with the program InterfaceAnalyzer of Rosetta. The average of each collection is used as a prediction of the ΔΔG and the standard deviation shows the margins of the prediction (Table 1). The distributions can be used to identify similar binding affinities between phospho-peptides.
Table 1.
Prediction of ΔG of docking of phosphorylated peptides from several β-TrCP substrates including the two reported sites in NRF2 [12,24,51,88]. β-TrCP1 (PDB code IP22) and to β-TrCP2 (PDB code 6WNX).
| β-TrCP1 ΔG(kcal/mol) | β-TrCP2 ΔG(kcal/mol) | |
|---|---|---|
| NRF2 (first site) | −14.08 ± 1.97 | −11.76 ± 2.93 |
| 333-PESTAEFNDSDpSGIpSLNTSPSVASPEHSVE-362 | ||
| NRF2 (second site; reported to bind β-TrCP as well) | −17.29 ± 2.71 | −17.52 ± 2.26 |
| 365-SYGDTLLGLSDSEVEELDpSAPGpSVKQNGPK-394 | ||
| β-CATENIN | −22.74 ± 3.11 | −21.61 ± 3.31 |
| 24-VSHWQQQSYLDpSGIHpSGATTTAPSLSGKGN-53 | ||
| hEpoR | −17.21 ± 2.71 | −15.31 ± 3.80 |
| 449-PPHLKYLYLVVSDpSGIpSTDYSSGDSQGAQG-478 | ||
| hFGD1 | −19.20 ± 2.67 | −23.57 ± 1.91 |
| 267-LAPGPRDGEKVPNRDpSGIDpSISSPSNSEET-296 | ||
| hGli2 | −12.67 ± 2.02 | −13.81 ± 3.21 |
| 667-SEPSPLGSAPNNDpSGVEMPGTGPGSLGDLT-696 | ||
| hIFNAR1 | −21.12 ± 2.83 | −19.00 ± 2.00 |
| 269-DHKKYSSQTSQDpSGNYpSNEDESESKTSEEV-298 | ||
| hIkBa | −22.56 ± 2.60 | −22.66 ± 2.03 |
| 19-GLKKERLLDDRHDpSGLDpSMKDEEYEQMVKE-48 | ||
| hMCL-1 | −12.35 ± 5.15 | −10.62 ± 4.19 |
| 145-ELVGESGNNTSTDGpSLPpSTPPPAEEEEDEL-174 | ||
| hPRLR | −18.30 ± 2.21 | −18.49 ± 2.34 |
| 336-QGMKPTYLDPDTDpSGRGpSCDSPSLLSEKCE-365 | ||
| hSNAIL3 | −21.65 ± 2.58 | −23.91 ± 2.31 |
| 83-PRVAELTSLSDEDpSGKGpSQPPSPPSPAPSS-112 | ||
| hSECURIN | −19.08 ± 2.60 | −22.57 ± 2.50 |
| 94-TEKTVKAKSSVPASDDAYPEIEKFFPFNPL-123 | ||
| hYAP | −12.44 ± 2.30 | −8.28 ± 2.27 |
| 390-LNSGTYHSRDESTDpSGLpSMSSYSVPRTPDD-419 |
Plasmids. The expression vectors pcDNA3.1/V5HisB-mNrf2, pcDNA3.1/V5His-mNrf2ΔETGE, pcDNA3.1/V5His-mNrf2ΔETGE-6S6A, pcDNA3.1/HismKEAP1-HA have been described previously [46]. A plasmid encoding pcDNA3-Flag-β-TrCP1 was kindly provided by Dr. Tomoki Chiba (Department of Molecular Biology, University of Tsukuba, Japan).
Luciferase Assays. MCF-7c32 ARE-LUC [47] cells were seeded on 24-well plates (75,000 cells per well) and incubated with PHAR for 16 h. Then, cells were lysed and assayed with a luciferase assay system (Promega) according to the manufacturer's instructions. Relative light units were measured in a GloMax 96 microplate luminometer with dual injectors (Promega). Each sample was measured from at least triplicate samples.
Cell viability assessed by MTT reduction. In live cells but not in dead ones, the tetrazolium ring of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) can be reduced by active dehydrogenases to produce a formazan precipitate. At the end of the experiments, cells were washed three times with phosphate-buffered saline (PBS) followed by the addition of MTT (0.125 mg/ml) and incubation for 1 h at 37 °C. Thereafter, the media was removed and DMSO was added to each well to dissolve the formazan precipitate for 30 min, thereby determining the relative number of alive cells. An aliquot (100 μl) of the supernatants were analyzed in 96-well multiwell plates at 550 nm in a VERSAmax microplate reader (Molecular Devices).
Immunoblotting. This method was performed essentially as described in Ref. [48]. Briefly, cells were homogenized in lysis buffer (50 mM TRIS pH 7.6, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1% SDS), denatured at 95 °C for 15 min, sonicated, and pre-cleared by centrifugation. Twenty μg of protein lysate were resolved in SDS-PAGE, transferred to Immobilon-P (Millipore) membranes and proteins of interest were detected with the following primary antibodies: NRF2 (homemade, validated in Ref. [12]) HO1 (homemade, validated in Ref. [49]), AKT (610860, BD Transduction Laboratories), β-Catenin (610153, BD Transduction laboratories), phospho-AKT (S473) (4058L, Cell Signaling Technology), GSK3β (sc-7291, Santa Cruz Biotechnology), phospho-GSK3β (S9) (9336L, cell signaling technology), KEAP1 (80475, Cell Signaling technology), GAPDH (CB1001, Merck Millipore), LAMINB (sc-6217, Santa Cruz Biotechnology), β-actin (sc-1616, Santa Cruz Biotechnology), Vinculin (E1E9V, Cell Signaling Technology), COX2 (sc-1747, Santa Cruz Biotechnology), NOS2 (sc-650, Santa Cruz Biotechnology), pre-IL1β (AF-401-NA, RD Systems), p65 (PC138, Calbiochem), Ubiquitin (Chemicon, Millipore). Proper peroxidase-conjugated secondary antibodies were used for detection by enhanced chemiluminescence (GE Healthcare). Phosphospecific antibodies against the critical phosphoserines 335, 338, 342, and 347 were generated in sheep and were used to probe in vitro phosphorylated recombinant NRF2.
Analysis of mRNA levels. Total RNA extraction, reverse transcription, and quantitative polymerase chain reaction (qRT-PCR) were done as detailed in Ref. [50]. Primer sequences are shown in Supplementary Table 1. To ensure that equal amounts of cDNA were added to the PCR reaction, Actb, Tbp, and Gapdh housekeeping genes were amplified. Data analyses were based on the ΔΔCT method, with normalization of the raw data by the geometric mean of the housekeeping genes Actb, Gapdh and Tbp (Applied Biosystems). All PCRs amplifications were performed from at least triplicate samples.
In vitro kinase assays.In vitro phosphorylation was performed using bacterially expressed EYFP-tagged His-mNrf2ΔNeh2 as substrate, isolated using the ProBond purification system (Invitrogen). GSK-3β kinase was purchased from Merck Millipore. For in vitro phosphorylation studies, the substrate (0.5 μg protein) was incubated with 5 ng of active recombinant GSK-3β in 10 μl of reaction buffer (10 mM MgCl2, 10 mM ATP in 40 mM morpholinepropanesulfonic acid (MOPS), pH 7.0, and 1 mM EDTA) for 30 min at 30 °C with continuous shaking.
In vitro ubiquitination assay. Purified recombinant proteins for β-TrCP-dependent ubiquitination were kindly provided by N. W. Pierce and R. J. Deshaies (Howard Hughes Medical Institute, Division of Biology, Pasadena, CA). Ubiquitination reactions were carried out as described previously [12] and contained ATP (2 mM), ubiquitin (30 μM), E1 (1 μM), Cdc34b (5 μM), SCF- β TrCP (450 nM), and non-phosphorylated or phosphorylated NRF2 (20 ng protein) in ubiquitination buffer (30 mM Tris [pH 7.6], 5 mM MgCl2, 2 mM dithiothreitol [DTT], 100 mM NaCl). Prior to the initiating the NRF2 ubiquitination reactions, E1, Cdc34b, and ubiquitin were pre-incubated together for 2 min to allow E2 thioester formation. NRF2 ubiquitination reaction mixtures were allowed to proceed for 1 h at 25 °C, before being quenched with SDS-PAGE buffer (62.5 mM Tris-HCl [pH 6.8], 12.5% [vol/vol] glycerol, 2% [wt/vol] SDS, 0.06% [wt/vol] bromophenol blue, 0.04% [vol/vol] 2-mercaptoethanol). The reaction products were resolved by SDS-PAGE, followed by transfer to Immobilon-P membranes. Ubiquitinated proteins were detected with an anti-ubiquitin antibody (Chemicon, Millipore).
Lentiviral and retroviral vector production and infection. Pseudotyped lentiviral vectors were produced in HEK293T cells transiently co-transfected with 10 μg of the corresponding lentiviral vector pWXL, 6 μg of the packaging plasmid pSPAX2 (12260, Addgene), and 6 μg of the VSV-G envelope protein plasmid pMD2G (12259, Addgene) using Lipofectamine Plus reagent according to the manufacturer's instructions (Invitrogen). Retrovirus supernatant was prepared by transfection of phoenix-Ampho cells (Garry Nolan, Baxter Laboratory in Genetic Pharmacology, Department of Microbiology and Immunology, Stanford University, 450 Serra Mall) with 5 μg of each plasmid using Lipofectamine Plus. Lentiviral vector shRNA control (shCTRL) (1864, Addgene), shβ-TrCP1 (NM_009771 TRCN-0000012807), shβ-TrCP2 (NM_134015 TRCN-0000231303) were purchased from Sigma-Aldrich (MISSION shRNA). Cells were infected in the presence of 4 μg/ml polybrene (Sigma-Aldrich) and selected with 1 μg/ml puromycin (Sigma-Aldrich) for 5 days.
Proximity ligation assay (PLA). The proximity of β-TrCP or KEAP1 to NRF2 was assessed using the Duolink® PLA In Situ Orange Starter Kit (Mouse/Rabbit) (Sigma-Aldrich, St. Louis, MO, USA). Transient transfections of HEK293T cells were performed with the expression vectors pcDNA3.1/V5His-mNrf2, pcDNA3.1/V5His-mNrf2ΔETGE, pcDNA3.1/V5His-mNrf2ΔETGE−6S6A, pcDNA3-Flag-β-TrCP1 and pcDNA3.1/HAHis-KEAP1. HEK293T cells were seeded in 24-well plates (75,000 cells per well), cultured for 16 h, and transfected using Transfectin reagent. After 24 h, cells were treated with PHAR or vehicle for 6 h. Then, cells were fixed in 4% paraformaldehyde (Merck) for 10 min and washed with PBS. A blocking solution provided with the kit was added and slides were incubated in a humidified chamber at 37 °C for 60 min. Slides were incubated with anti-HA (1:200, 16B12, Covance), anti-FLAG (1:200, F3167, Sigma-Aldrich) or anti-NRF2 (1:150, D1Z9C, Cell signaling) overnight at 4 °C. Subsequently, slides were washed twice for 5 min in 1 × wash buffer A at room temperature, followed by incubation with Duolink PLA PLUS and MINUS probes diluted 1:5 in Duolink antibody diluent at 37 °C for 1 h. Then, 5 × Duolink Ligation buffer was diluted in high-purity water at a 1:5 ratio and slides were washed. Slides were incubated in ligation buffer for 30 min at 37 °C. All subsequent steps were performed in the dark. The 5 × amplification buffer was diluted 1:5 in high-purity water. Slides were washed as above. DNA polymerase was added to the diluted amplification buffer (1:80) and slides were incubated at 37 °C for 100 min. To define the localization of target proteins, the appropriate secondary antibodies coupled to Alexa Fluor 488 or 647 (1:500) (Life Technologies-Molecular Probes, Grand Island, NY, USA) was added for 120 min at 37 °C. Slides were washed twice for 10 min with 1 × wash buffer B, with one final wash for 1 min in 0.01 × wash buffer B. Coverslips were mounted on slides using Duolink In Situ Mounting Medium with DAPI and sealed. Fluorescent images were captured using the LSM710 spectral microscope confocal (Zeiss). Quantification of puncta per cells was done with Image J.
Determination of reactive oxygen species by Flow Cytometry. Intracellular reactive oxygen species (ROS) were detected in a FACSCanto II (BD Biosciences) flow cytometer with hydroethidine (HE) (ThermoFisher Scientific), which upon oxidation emits orange fluorescence (BP 575/24 nm). Cells were incubated for 1 h at 37 °C with 2 μM HE and then detached from the plate, washed once with cold PBS, and analyzed immediately.
High-performance liquid chromatography. The mouse liver samples (∼80 mg) were treated with 0.5 ml methanol, ground in a potter, and centrifuged. The supernatant collected and filtered through a 0.45 μm PTFE microfilter (Fisherbrand). The samples were first analyzed by HPLC-UV. The eluted peaks were further analyzed by HPLC-MS. As a standard, PHAR was prepared in DMEM and analyzed by HPLC-MS.
Histological analysis. Livers were fixed for 24 h in PBS-buffered 4% paraformaldehyde and then submerged in 70% ethanol. Next, they were embedded in paraffin and cut into 5 μm thick sections and stained with hematoxylin and eosin (H&E). For immunohistochemical analyses, sections were deparaffinized and rehydrated in water, and antigen retrieval was carried out by incubation with citrate buffer pH 6.0 at 50 °C for 30 min. Endogenous peroxidase and nonspecific antibody reactivity were blocked by treatment with 3% H2O2 at room temperature for 10 min. The sections were then incubated for 16 h at 4 °C with the corresponding peroxidase-conjugated primary antibody (diluted in PBS containing 1% normal goat serum) F4/80 (1:150, Serotec) and developed with 3,3′-diaminobenzidine (DAB). The sections were counterstained with hematoxylin, dehydrated in ethanol, then in xylol, and then mounted in DePex (Thermo Fisher Scientific). Negative controls with goat normal serum replacing the primary antibody were used. Densitometric quantification was done using macros of the ImageJ software.
Statistical analyses. Unless otherwise indicated, all experiments were performed at least 3 times and all data presented in the graphs are the mean of at least 3 independent samples. Data are presented as mean ± S.D. (standard deviation). Statistical differences between groups were assessed using GraphPad Prism 8 software by the unpaired Student's t-test. One and two-way analyses of variance with post-Bonferroni's test were used for multiple comparisons. Statistically significant differences are indicated in the figures (*** indicate p values < 0.001, ** <0,01 and * <0,05).
3. Results
3.1. In silico search for candidate inhibitors of the β-TrCP-NRF2 interaction
A chemical library of 954,861 compounds from SuperNatural II and ZINC Natural Products was analyzed using sequential filters: 1) molecular docking to the 1P22 crystal of β-TrCP with a Gibbs free energy variation of more than 9.5 kcal/mol (473 compounds selected); 2) prediction of a suitable absorption, distribution, metabolism, excretion, and toxicity (ADMET) profile, based on the analysis by DataWarrior and ADMETsar software (87 compounds selected); 3) analysis of molecular dynamics (MD) simulation over 200 ns (30 compounds selected). The selected compounds represent 0.00032% of the initial library. In this study, we will present the experimental results obtained with the compound ZINC70705153, that we have designated PHAR for simplicity, as a candidate inhibitor of the NRF2-β-TrCP interaction.
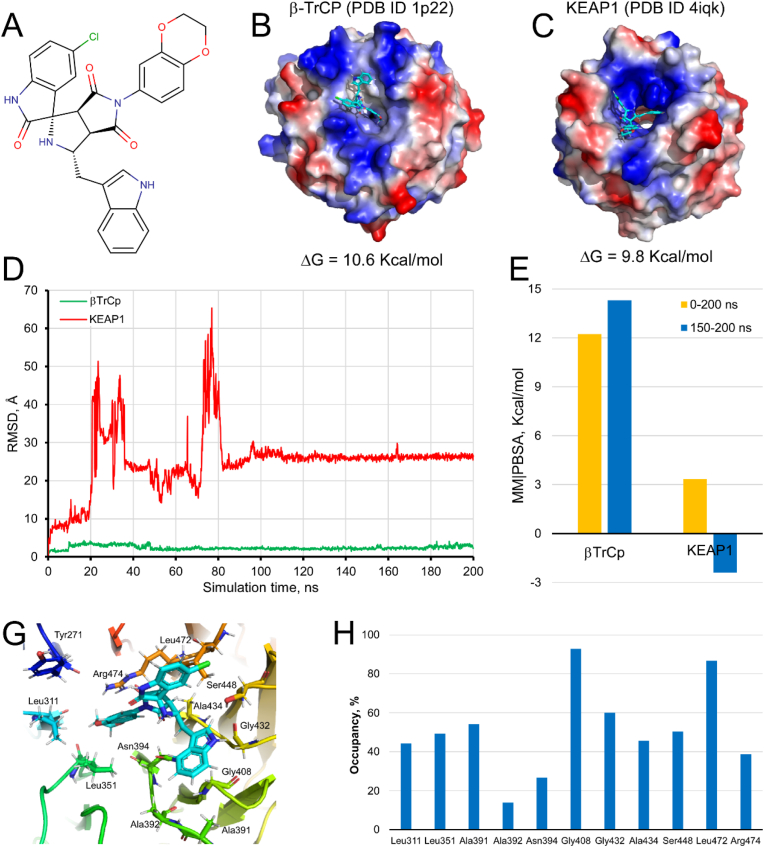
PHAR is (1S,3R,3aR,6aS)-5′-chloro-5-(2,3-dihydro-1,4-benzodioxin-6-yl)-1-(1H-indol-3-ylmethyl)spiro[1,2,3a,6a-tetrahydropyrrolo [3,4-c]pyrrole-3,3′-1H-indole]-2′,4,6-trione (Fig. 1A). Fig. 1B and C shows PHAR docked on the interaction surface of β-TrCP (ΔG = 10.6 kcal/mol) and KEAP1 (ΔG = 9.8 kcal/mol), respectively. Fig. 1D and E shows the trajectory of PHAR docked β-TrCP or KEAP1 protein domains throughout 200 ns of MD simulation and the solvation binding energy (MM|PBSA) for their ligand-protein interaction, respectively. While the compound remains bound at the NRF2-β-TrCP interface region (RMSD less than 3 Å), binding to NRF2-KEAP1 interface is lost within 20 ns of MD simulation (Fig. 1D). Therefore, in silico predictions indicate that PHAR interacts selectively with β-TrCP. For the PHAR-β-TrCP complex, the MM|PBSA values (12.224 and 14.299 kcal/mol for the last 50 ns or the complete MD simulation, respectively) reflect stable binding to NRF2-β-TrCP interface (Fig. 1E). Finally, the analysis of the “fingerprint” that PHAR generates on the amino acids of the NRF2-β-TrCP interface region indicates that mostly hydrophobic interactions are established between the compound and the protein. The hydrogen bonding interactions with Tyr271, Ser448, and Arg474 represent the exception (Fig. 1G). The amino acids Leu311, Leu351, Ala391, Gly408, Gly432, Ala434, Ser448, and Leu472 show an occupancy time forming hydrophobic interaction with PHAR greater than 40% (Fig. 1H).
Fig. 1.
Molecular docking and dynamics simulations-based selection of compound PHAR as an inhibitor candidate of the β-TrCP-NRF2 interaction. A, shows the structure of the PHAR compound. B–C, PHAR docked on the interaction surface of β-TrCP and KEAP1, respectively. Both proteins represent their electrostatic surface potential within PyMol 2.3. D, trajectory of the PHAR compound bound to β-TrCP (green trace) or KEAK1 (red trace) throughout 200 ns MD simulation. E, calculated MM|PBSA free energy values of the PHAR-βTrCP and PHAR-KEAP1 complexes. YASARA-calculated MM|PBSA provides positive values when the predicted binding is strong and stable whereas negative values indicate unstable or no binding. G, spatial location of the amino acids of β-TrCP that maintain hydrophobic or electrostatic interactions with the PHAR compound. H, the occupation time of the amino acids of β-TrCP that establish interactions with the compound PHAR throughout the 200 ns of MD simulation. Only amino acids with an occupancy time greater than 10% have been represented. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. PHAR activates NRF2 transcriptional activity
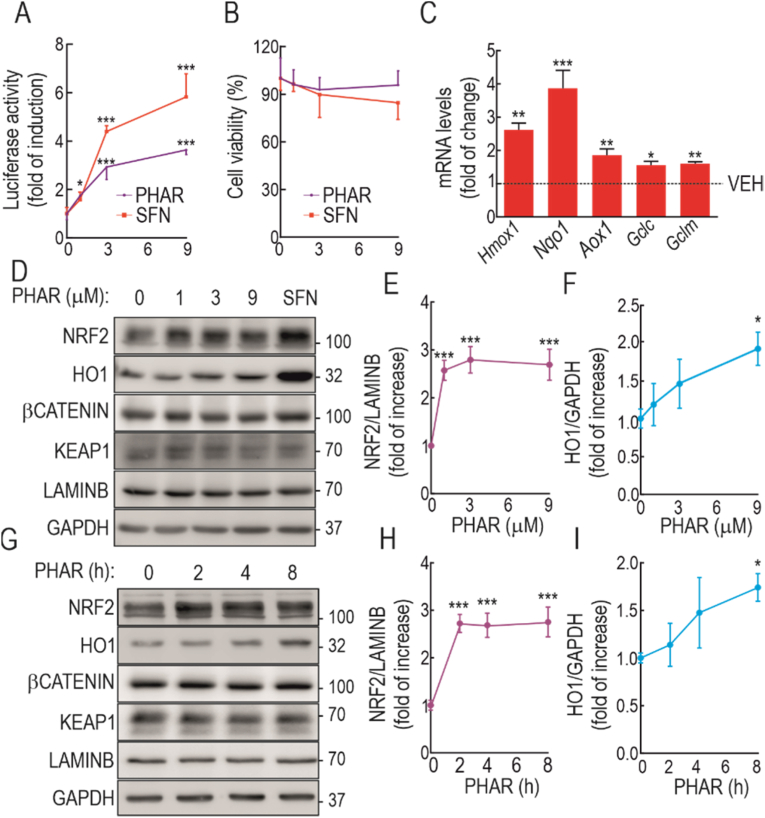
The ability of PHAR to activate NRF2 was first examined in the reporter MCF-7 c32 ARE-LUC cell line, which carries 8 tandem repeats of the ARE sequence driving the expression of firefly luciferase, and is well documented to be under the control of NRF2 [47]. Because GSK-3β is active in the absence of cell signaling, but is inhibited by growth factors, cells were serum-depleted (<0.5%) for 16 h to activate the GSK-3/β-TrCP-mediated NRF2 degradative pathway, and then were treated with PHAR (1, 3, and 9 μM, 16 h). Since at present no disrupter of the NRF2/β-TrCP interaction has been reported, we used the KEAP1 inhibitor SFN as positive control. As shown in Fig. 2A, PHAR produced a ∼3-fold increase in the expression of the ARE-driven reporter gene which was about half the effect observed with SFN. This result is consistent with the notion that KEAP1 is the main repressor of NRF2 and that SFN, by inhibiting KEAP1, elicits a strong activation of NRF2. The analysis of cell viability in the same cells, evaluated with an MTT assay, indicated that PHAR is not toxic at any of the concentrations used (Fig. 2B).
Fig. 2.
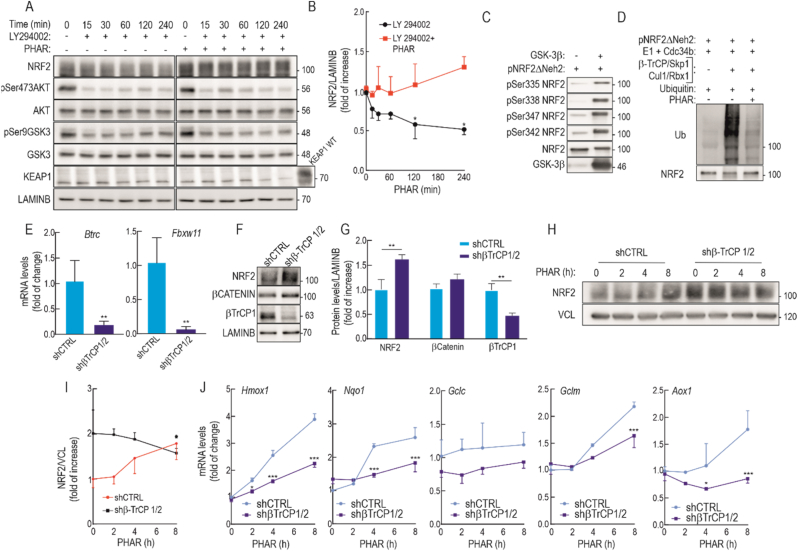
Evaluation of PHAR in ARE activation and cell viability. A, MCF-7 c32 ARE-LUC reporter cells were serum-depleted for 16 h and then subjected to the indicated PHAR concentrations or 10 μM SFN, as a positive control. 0.1% DMSO was used as vehicle. Luciferase activity was measured after 16 h of treatment. Data are mean ± S.D. (n = 4). *p < 0,5; ***p < 0.001 vs. vehicle according to a one-way ANOVA test. B, MTT assay was performed to assess cell viability of PHAR and SFN in serum-depleted cells. Data are mean ± S.D. (n = 4). C, serum-depleted MEFs were subjected to 10 μM PHAR for 8 h. Transcript levels of Hmox1, Nqo1, Aox1, Gclc, and Gclm were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp, and Actb levels. Data are mean ± S.D. (n = 4). *p < 0.05; **p < 0.01 ***p < 0.001 according to a Student's t-test. D, Serum-depleted MEFs subjected to the indicated PHAR concentrations or 10 μM SFN, as a positive control, for 16 h. Representative immunoblots of NRF2, HO1, β-CATENIN, KEAP1, LAMINB, and GAPDH as a loading control from. E-F, densitometric quantification of NRF2 and HO1 protein levels from representative immunoblots of D expressed as a ratio LAMINB and GAPDH, respectively. Data are mean ± S.D. (n = 4). *p < 0.05; ***p < 0.001 vs. vehicle according to a one-way ANOVA test. G, representative immunoblots of NRF2, HO1, β-CATENIN, KEAP1, LAMINB, and GAPDH as a loading control from serum-depleted MEFs were submitted to 10 μM PHAR for the indicated times. H–I, densitometric quantification of NRF2 and HO1 protein levels from representative immunoblots of G expressed as a ratio of LAMINB, and GAPDH, respectively. Data are mean ± S.D. (n = 4). *p < 0.05; ***p < 0.001 vs. vehicle according to a one-way ANOVA test.
Then, we evaluated the effect of PHAR on activation of the endogenous NRF2-target genes using wildtype MEFs that had been serum-depleted for 16 h to have a significant activation of the GSK-3/β-TrCP axis. As shown in Fig. 2C, transcript levels of the ARE-genes Hmox1, Nqo1, Aox1, Gclc, and Gclm were significantly increased by PHAR (10 μM, 8 h). In dose-response experiments (Fig. 2D–F) from 1 to 9 μM, PHAR increased NRF2 protein levels, albeit less than 10 μM SFN. The increase in NRF2 correlated with a slight increase in the expression of the bona fide gene target heme oxygenase-1 (HO1), with 9 μM PHAR. In time-course experiments up to 8 h (Fig. 2G–I), 10 μM PHAR also induced the accumulation of NRF2 from 2 h of treatment, which correlated with the increased HO1 levels after 8 h of treatment.
Of note, β-Catenin, a bona fide target of GSK-3β/β-TrCP was not affected by PHAR (Fig. 2D and G) suggesting that this small molecule might not compete efficiently with the binding to β-TrCP of other substrates. To further explore this hypothesis, we did an in silico binding analysis of the phosphopeptides derived from NRF2, β-Catenin and other β-TrCP substrates to the WD40 propeller of β-TrCP1 and of β-TrCP2 using the Rossetta software. In the case of NRF2, we simulated the binding of both sites within the Neh6 domain that have been reported to bind β-TrCP. As shown in Table 1, the predicted ΔG of binding for phospho-β-Catenin is about twice lower than for the two phospho-NRF2 peptides, suggesting that β-TrCP exhibits much higher affinity for phospho-β-Catenin than for phospho-NRF2. This is consistent with our hypothesis that PHAR is selective for inhibition of the NRF2/β-TrCP interaction as it probably cannot displace the β-Catenin/β-TrCP interaction. Moreover, the two phospho-NRF2 peptides bind β-TrCP with lower affinity than most other β-TrCP substrates tested (Table 1). Because the NRF2/β-TrCP interaction is among the weakest, our results suggest that PHAR might disrupt the interaction β-TrCP only with a subset of substrates that, like phospho-NRF2, would exhibit low affinity for β-TrCP binding.
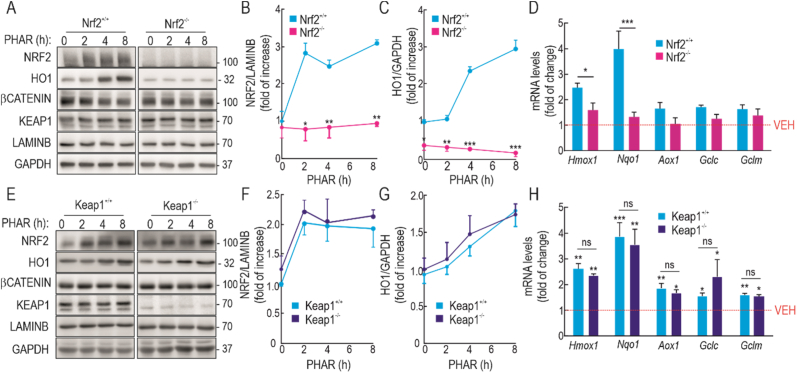
Taken together, these results suggest that PHAR, increases NRF2 protein levels and leads to activation of ARE-genes, but to determine that the effect of PHAR is channeled through NRF2, HO1 levels were examined in MEFs from NRF2 wild-type (Nrf2+/+) and knock-out (Nrf2−/−) mice from the same littermates that had been incubated with 10 μM PHAR (2, 4 and 8 h). In MEFs from Nrf2+/+ mice, PHAR produced a significant increase in HO1 that was not observed in MEFs from Nrf2−/− mice (Fig. 3A–C). Moreover, in Nrf2−/− MEFs, PHAR did not significantly induce the expression of two robust NRF2 target genes Hmox1, and Nqo1, or other weaker targets, such as Aox1, Gclc and Gclm (Fig. 3D), though it should be noted that the 8 h PHAR treatment interval may be suboptimal for some of these NRF2-target genes. Therefore, in conclusion, PHAR treatment activates NRF2, and this is responsible for the increase in the expression of its target genes.
Fig. 3.
PHAR induces ARE-genes in an NRF2-dependent but KEAP1-independent manner. A, representative immunoblots of NRF2, HO1, β-CATENIN, KEAP1, LAMINB and GAPDH as a loading control from serum-depleted MEFs from wild type (Nrf2+/+) and NRF2-knockout (Nrf2−/−) mice subjected to 10 μM PHAR for the indicated times. B–C, densitometric quantification of NRF2 and HO1 protein levels from representative immunoblots from A, expressed as a ratio of LAMINB and GAPDH, respectively. Data are mean ± S.D. (n = 3). *p < 0,5; **p < 0,01; ***p < 0.001 vs vehicle of Nrf2+/+ according to a two-way ANOVA test. D, mRNA levels of several ARE-genes after 8 h of incubation with 10 μM PHAR were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp, and Actb levels. Data are mean ± S.D. (n = 4). *p < 0,5; ***p < 0.001 vs Nrf2+/+ according to a two-way ANOVA test. E representative immunoblots of NRF2, HO1, β-CATENIN, KEAP1, LAMINB, and GAPDH as a loading control from serum-depleted MEFs from wild type (Keap1+/+) and Keap1-knockout (Keap1−/−) mice subjected to 10 μM PHAR for the indicated times. F-G, densitometric quantification of NRF2 and HO1 protein levels from representative immunoblots from E, expressed as a ratio of LAMINB, and GAPDH, respectively. Data are mean ± S.D. (n = 3). H, mRNA levels of ARE-genes after 8 h of incubation with 10 μM PHAR were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp and Actb levels. Data are mean ± S.D. (n = 4). *p < 0.05; **p < 0.01; ***p < 0.001 vs Keap1+/+ according to a two-way ANOVA test; ns means non-significant differences between Keap1+/+ and Keap1−/−.
3.3. Activation of NRF2 by PHAR is KEAP1-independent
From the point of view of its biopharmaceutical utility, it is necessary to determine if PHAR might act via inhibition of KEAP1, or whether it indeed represents a novel approach for selective inhibition of the β-TrCP/NRF2 interaction. We addressed this question by comparing the effects of PHAR on NRF2-target gene expression in MEFs from wild type (Keap1+/+) and Keap1-knockout (Keap1−/−) mice (Fig. 3E–G). A time course revealed that the increase in NRF2 and HO1 protein levels was similar in both cell lines treated with 10 μM PHAR. Consistently, the transcript levels of several NRF2-target genes were also activated by PHAR to a similar extent in Keap1−/− MEFs (Fig. 3H).
By contrast with most NRF2 activators, PHAR is not an electrophile or oxidative stressor, and is therefore not expected to alter cell signaling by triggering redox alterations. Nevertheless, we considered it important to determine if PHAR might alter the PI3K/AKT/GSK-3 axis as this pathway participates in β-TrCP-mediated regulation of NRF2 [10,23,51]. We therefore first inhibited the PI3K/AKT signaling pathway with the PI3K inhibitor, LY294002, thereby activating GSK-3β which would result in phosphorylation of NRF2 in its Neh6 domain and lead to its degradation by interaction with β-TrCP [23]. Therefore, if PHAR disrupts the β-TrCP/NRF2 interaction, NRF2 levels will remain stable regardless of activation and phosphorylation by GSK-3β. This hypothesis was tested in MEFs from Keap1−/− mice to still avoid a possible alternative mechanism mediated by KEAP1. These cells were pre-treated with 10 μM PHAR or vehicle for 30 min and then treated with 20 μM LY294002 for the indicated times. Mainly after 120 and 240 min, LY294002 caused a decrease in pSer473-AKT (inhibition) and pS9-GSK-3β (activation) that correlated with the expected reduction in NRF2 protein levels (Fig. 4A). By contrast, pre-treatment with PHAR prevented the reduction in NRF2 protein levels stimulated by activation of GSK-3β elicited by LY294002 (Fig. 4A–B). This protection was also observed after 4 and 8 h of LY294002 treatment (Fig. S1 of supplemental material). Conversely, GSK-3 inhibition with SB216763, increased NRF2 protein levels as expected from our previous results [52] like PHAR did, but when cells were submitted together to both SB216763 and PHAR, NRF2 protein levels were increased to a similar extent as with any of them alone (Fig. S2 of supplemental material). These results suggest that both drugs target the same pathway, SB216763 by inhibiting GSK-3 activity towards NRF2 phosphorylation and PHAR by inhibiting binding of GSK-3-phosphorylated NRF2 to β-TrCP.
Fig. 4.
PHAR increases NRF2 protein levels in a β-TrCP dependent-manner. Serum-depleted Keap1−/− MEFs were subjected to the vehicle (0.1% DMSO) or 10 μM of PHAR for 30 min. Then, cells were treated with 20 μM LY294002 for the indicated times. A, representative immunoblots of NRF2, pSer473AKT, AKT, pSer9GSK3β, GSK3, KEAP1, and LAMINB as a loading control. B, densitometric quantification of NRF2 protein levels from representative immunoblots from A, expressed as a ratio of GAPDH. Data are mean ± S.D. (n = 3). *p < 0.05 vs LY2940002 at point 0, according to a two-way ANOVA test. C, In vitro kinase assay of NRF2 by GSK-3β. Recombinant pNRF2ΔNeh2, lacking the KEAP1 binding domain Neh2 [89], was submitted to an in vitro kinase assay in the absence or presence of recombinant GSK-3β. Representative immunoblots of pSer335NRF2, pSer338NRF2, pSer347NRF2, pSer342NRF2, NRF2, and GSK3β from in vitro kinase assay. D, In vitro ubiquitylation assay of NRF2 by the β-TrCP complex. Phospho-NRF2ΔNeh2 (20 ng) was incubated at 25 °C for 1 h with purified ubiquitin, E1/cdc34b, β-TrCP/Skp1, and Cul1/Rbx1 as indicated in the presence or absence of 1 μM PHAR. E, knockdown of Btrc (encoding β-TrCP1) and Fbxw11 (encoding β-TrCP2). Keap1−/− MEFs were transduced with control lentivirus encoding shCTRL or a combination of two lentiviruses encoding sh-Btrc and sh-Fbxw11. After 5 days, cells were serum-depleted for 16 h and then subjected to 10 μM PHAR for the indicated times. Transcript levels of Btrc and Fbxw11 were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp, and Actb. Data are mean ± S.D. (n = 4). *p < 0.05; **p < 0.01; ***p < 0.001 vs. shCTRL according to a Student's t-test. F, representative immunoblots of NRF2, β-CATENIN, β-TrCP1, and LAMINB as a loading control. G, densitometric quantification of NRF2, β-CATENIN, and β-TrCP1 protein levels from representative immunoblots from F, expressed as a ratio of LAMINB. Data are mean ± S.D. (n = 4). **p < 0.01 vs. shCTRL according to a Student's t-test. H, representative immunoblots of NRF2 and Vinculin (VCL) as a loading control, from control (shCTRL) and β-TrCP knocked-down (shβ-TrCP1/2) MEFs that were submitted to 10 μM PHAR for the indicated times. I, densitometric quantification of NRF2 protein levels from representative immunoblots from H, expressed as a ratio of NRF2/VCL. Data are mean ± S.D. (n = 4). *p < 0,05 vs. shCTRL at point 0 according to a one-way ANOVA test. J, expression of five NRF2-regulated genes in shCTRL vs. shβ-TrCP1/2 MEFs. Cells were submitted to 10 μM PHAR for the indicated times and transcript levels were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp, and Actb levels. Data are mean ± S.D. (n = 4). *p < 0.05; ***p < 0.001 vs. shCTRLat point 0 according to a two-way ANOVA test.
3.4. PHAR increases NRF2 protein levels in a β-TrCP dependent-manner
To determine if PHAR inhibits β-TrCP-mediated ubiquitination of NRF2, we performed an in vitro ubiquitination assay using a recombinant EYFP-NRF2ΔNeh2 protein chimera which lacks the domain of NRF2 that binds KEAP1. Because β-TrCP recognizes phosphorylated NRF2, we first phosphorylated this protein with recombinant GSK-3β in an in vitro kinase assay (Fig. 4C). The use of phospho-specific antibodies raised against specific phospho-serines within the Neh6 phopho-degron demonstrated that EYFP-NRF2ΔNeh2 was phosphorylated by GSK-3β in the critical serines (335, 338, 347 and 342) that participate in docking to β-TrCP. Then, phospho-EYFP-NRF2ΔNeh2 (p-NRF2ΔNeh2) was used as the substrate in an in vitro ubiquitination assay in the presence of recombinant β-TrCP1 and the Cul1/RBX1 complex (Fig. 4D), following the protocol reported in Ref. [52], where it had been shown that in vitro ubiquitination of NRF2 by β-TrCP is highly dependent on its phosphorylation in the Neh6 domain by GSK-3. p-NRF2ΔNeh2 was ubiquitinated in the presence of the complete β-TrCP-E3 ligase complex, as expected, but importantly 1 μM PHAR significantly reduced the levels of polyubiquitinated NRF2.
In additional experiments, we knocked down the expression of β-TrCP1, β-TrCP2, or both isoforms in MEFs from Keap1−/− mice. Cells were infected with silencing lentiviral vectors followed by 5 days of puromycin selection before they were treated with 10 μM PHAR. The silencing achieved was >80% for β-TrCP1 (Btrc) and >90% for β-TrCP2 (Fbxw11) coding genes (Fig. 4E), which was sufficient to modestly increase the levels of the bona fide β-TrCP target β-Catenin and also NRF2 (Fig. 4F–G). In shCTRL infected cells, PHAR increased NRF2 levels starting at 4 h of treatment and was most obvious at 8 h (Fig. 4H–I). By contrast, β-TrCP1/2 knockdown cells exhibited high NRF2 levels that were not further increased by PHAR. The PHAR-dependent expression of five NRF2-regulated genes, Hmox1, Nqo1, Gclc, Gclm, and Aox1, was also impaired in shβ-TrCP1/2 MEFs (Fig. 4J). Interestingly, the reduction of NRF2 levels in LY294002-treated cells was lost in cells silenced for β-TrCP (Supplemental Fig. S3).
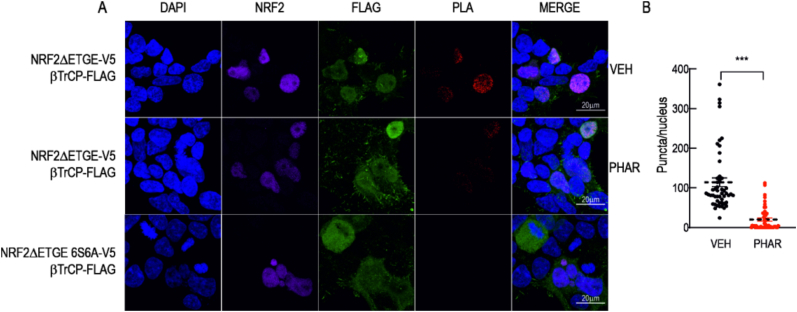
To further investigate the physical association between β-TrCP and NRF2 and the possible disruption by PHAR, we employed a proximity ligation assay (see Material and methods). HEK293T cells were transfected with expression vectors for NRF2ΔETGE-V5, and Flag-β-TrCP1. After 24 h transfection, cells were treated with vehicle or 10 μM PHAR, 20 μM LY294002 (to activate the GSK-3β/β-TrCP signaling pathway) and 1 μM pevonedistat for 6 h. In the latter case, pevonedistat (MLN-4924) is a NEDD8-activating enzyme inhibitor that prevents neddylation of Cullin-RING E3 ligases (CRLs), leading to the stabilization of substrates including phosphorylated NRF2 [53]. With this treatment, the degradation of NRF2 by β-TrCP is avoided and we can therefore analyze the interaction (Supplemental Fig. S4). We observed red fluorescent puncta in the nuclei of cells treated with vehicle, indicating proximity of less than 40 nm between Flag-β-TrCP1 and NRF2ΔETGE-V5. These fluorescent puncta were significantly reduced when cells were incubated with 10 μM PHAR for 6 h (Fig. 5A and B). We validated the proximity ligation assay using a specific negative control: NRF2ΔETGE−6S6A-V5. In this construct, the 6 serines of NRF2 that upon phosphorylation participate in β-TrCP binding, had been mutated into alanines [12]. In this case, we obtained a total absence of fluorescent puncta, confirming that these serines are necessary for the recognition of NRF2 by β-TrCP. Moreover, PHAR did not affect the interaction between NRF2 and KEAP1 in either the cytoplasm or the nucleus (Supplemental Fig. S3), confirming that PHAR specifically prevents the interaction between NRF2 and β-TrCP but not between NRF2 and KEAP1. Collectively, these results indicate that PHAR disrupts the β-TrCP/NRF2 interaction.
Fig. 5.
Proximity ligation assay (PLA) for β-TrCP and NRF2. HEK293T cells were transfected with expression vectors for NRF2ΔETGE-V5, or β-TrCP insensitive NRF2ΔETGE-6S6A-V5 mutant. After 24 h, cells were incubated in the presence vehicle (VEH) or PHAR (10 μM) for 6 h. Cells were subjected to the PLA assay using rabbit anti-FLAG and mouse anti-V5. Upon PLA reaction, red puncta represent colocalization of β-TrCP and NRF2. A, representative fluorescence images of DAPI, NRF2, FLAG, PLA results and MERGE. B, Quantification of puncta per nucleus of a total of 50 nuclei per condition. Calibration bar = 20 μm ***p < 0.001 vs. vehicle according to a Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. PHAR attenuates hydrogen peroxide-induced oxidative stress
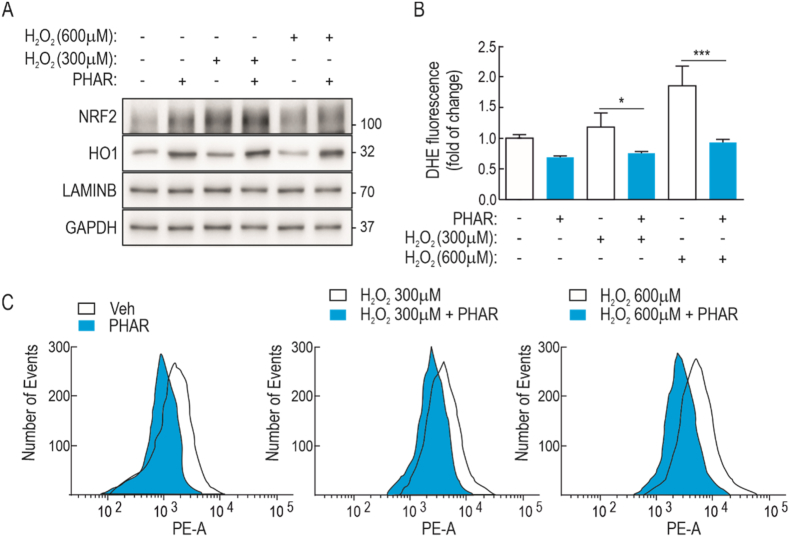
Because NRF2 is a master regulator of redox homeostasis, we examined whether PHAR might exert protection against a strong oxidant insult, such as H2O2. Serum-depleted MEFs were incubated with PHAR (10 μM, 16 h) and then treated with 300 or 600 μM H2O2 for 3 h. As expected, PHAR increased the levels of NRF2 and the target gene product HO1 (Fig. 6A). Hydrogen peroxide also increased NRF2 levels after 3 h incubation, although this time frame was not long enough to significantly induce HO1. The oxidant environment was analyzed by flow cytometry in parallel cell cultures incubated with 2 μM dihydroethidine (DHE) for 60 min. As shown in Fig. 6B–C, pre-incubation with PHAR significantly attenuated DHE staining in response to both H2O2 concentrations. These results indicate that PHAR protects against redox dysregulation.
Fig. 6.
PHAR attenuates redox dysregulation induced by hydrogen peroxide. A, representative immunoblots of NRF2, HO1, LAMINB, and GAPDH as a loading control from serum-depleted MEFs that were pre-treated with 10 μM PHAR for 16 h and then submitted to hydrogen peroxide as indicated for 3 h. B–C, Flow cytometry analysis of hydrogen peroxide-induced intracellular ROS production in 2 μM hydroethydine (HE) stained cells. A representative sample of 10,000 cells is shown for each condition. Data are mean ± S.D. (n = 3). *p < 0.05; ***p < 0.001 vs. PHAR according to a Student's t-test.
3.6. PHAR attenuates LPS-induced inflammation in macrophages
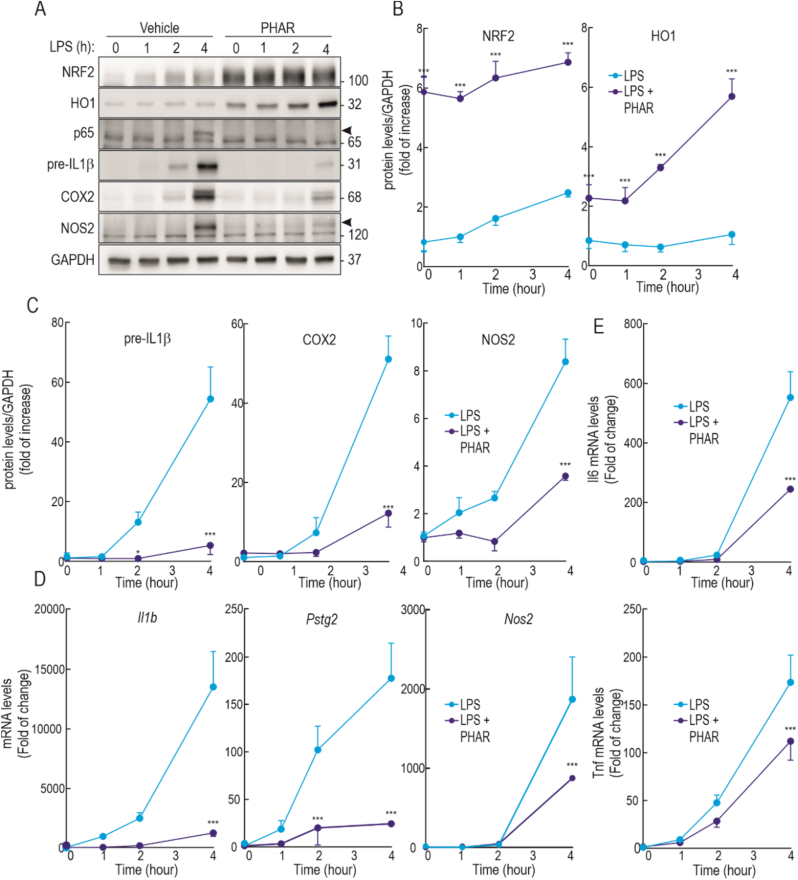
Considering the well-established role of NRF2 in the resolution of inflammation [6], we tested if PHAR might oppose the inflammatory response elicited by LPS in the Raw264.7 mouse macrophage cell line. These cells were pre-treated with PHAR (10 μM, 8 h) or vehicle (DMSO) and then subjected to LPS (100 ng/ml; 1, 2, and 4 h). As expected, LPS induced an inflammatory response in vehicle-treated cells that was greatest after 4 h, as determined by the increase in the levels of p65-NFκB, pre-IL1β, COX2, and NOS2. By contrast, PHAR led to the expected increase of NRF2 and HO1 protein levels, but also to a significant attenuation of the LPS-induced inflammatory markers (Fig. 7A–C). Consistently, the induction of mRNA levels of inflammatory markers Il1b, Cox2, Nos2, Il6 and Tnf was also attenuated in PHAR-treated cells (Fig. 7E). To further determine if the anti-inflammatory effect of PHAR was driven by NRF2, we used peritoneal macrophages isolated from wild type and Nrf2-knockout mice (Fig. S6 of supplemental material). We found that PHAR attenuates the LPS-induced expression of several inflammatory markers, including COX2 and NOS2 proteins, and Pstg2, Nos2, Il6 and Tnf transcripts, in wild type macrophages but not in Nrf2-knockout macrophages. Taken together, we conclude that besides inducing antioxidant genes, PHAR exerts anti-inflammatory effects, mechanism of which is the activation of NRF2 through inhibition of its interaction with β-TrCP.
Fig. 7.
PHAR decreases inflammatory response in Raw264.7 cells stimulated with LPS. Low serum-depleted Raw264.7 cells were pre-treated to 10 μM of PHAR for 8 h. Then, cells were treated to 100 ng/ml of LPS for the indicated times. A, representative immunoblots of NRF2, HO1, p65, pre-IL1β, COX2, NOS2, and GAPDH as a loading control. The black arrowhead marks p65 and NOS2 specific bands. B–C, densitometric analysis of NRF2, pre-IL1b, COX2, and NOS2 protein levels from representative immunoblot from (A), expressed as a ratio of protein levels/GAPDH. Data are mean ± S.D. (n = 3). *p < 0.05; ***p < 0,001 vs vehicle or LPS according to a two-way ANOVA test. D-E, mRNA levels of Il1b, Ptgs2, Nos2, Il6, and Tnf were determined by qRT-PCR and normalized by Gapdh, Tbp, and Actb average. Data are mean ± S.D. (n = 4). *p < 0.05; ***p < 0.001 vs LPS according to a two-way ANOVA test.
3.7. PHAR increases NRF2 levels in liver
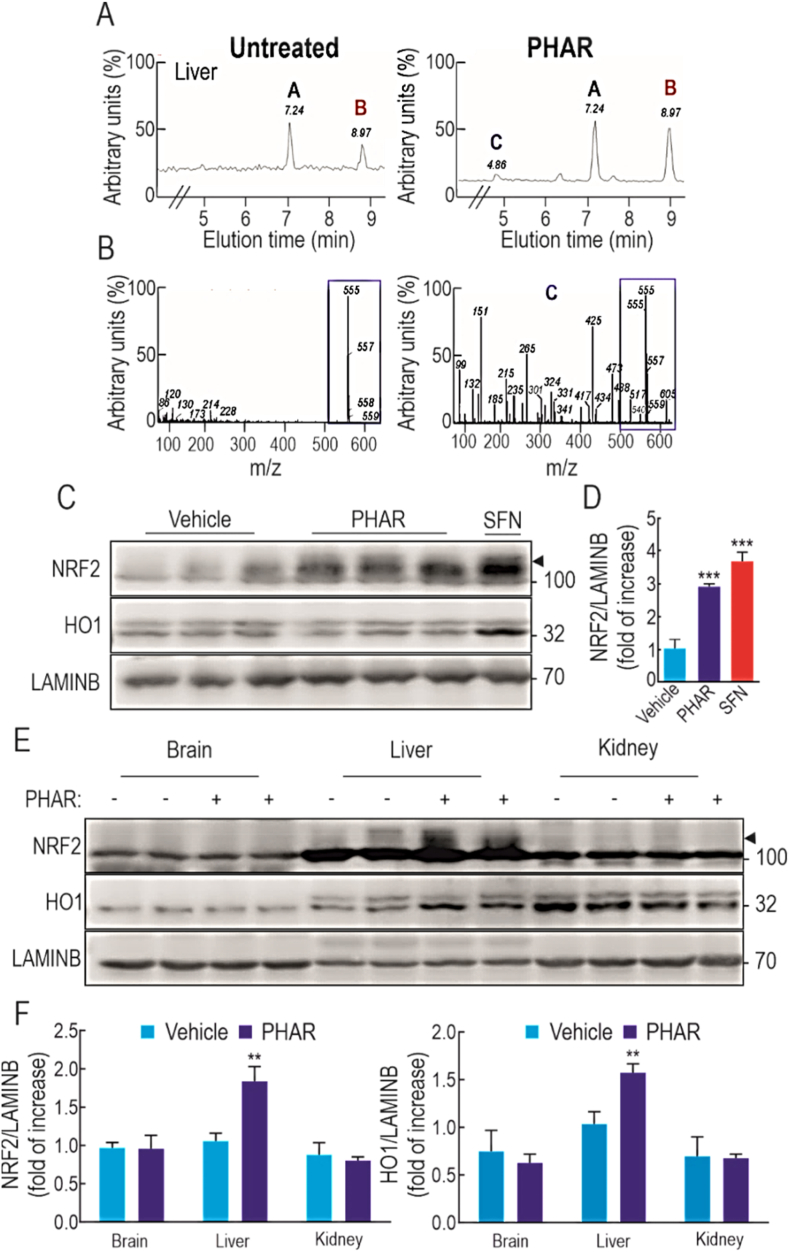
In a preliminary pharmacokinetics analysis, we examined the distribution of PHAR in C57BL/6 mice following a single intraperitoneal (i.p.) injection of 50 mg/kg body weight of the PPI inhibitor (vehicle was Tween-80 + PBS, 1:13). We did not detect exposure to PHAR in the brain or kidney (data not shown). However, in the liver the comparison of the HPLC-UV profile of vehicle-vs. PHAR-treated mice demonstrated two common nonspecific peaks at approximately 7 and 9 min of elution time (Fig. 8A, letters A and B) plus one peak only in PHAR-treated mice at 4 min of elution time (Fig. 8A, letter C). The HPLC-MS analysis of this peak revealed the existence of a compound with the molecular weight of 555 Da consistent with PHAR (Fig. 8B, right graph). This compound was identical to the standard obtained for PHAR prepared in DMEM (Fig. 8B, left graph). These results indicate that PHAR can be detected in liver 2 h after i.p. administration.
Fig. 8.
PHAR activates NRF2 in the liver. In A-D, C57Bl/6 male mice received one intraperitoneal (i.p.) injection of 50 mg/kg PHAR and livers were analyzed after 2 h. A, analysis by HPLC-UV of liver extracts comparing vehicle (Tween-80 + PBS, 1:13) vs. PHAR treated mice (Tween-80 + PBS, 1:13). B, left graph, analysis by HPLC-MS of PHAR standard elution in DMEM. B, right graph, analysis by HPLC-MS of peak C detected in A in liver of PHAR-treated mice. Note the identification of a 555 da molecule in both graphs surrounded by a blue rectangle that corresponds to PHAR molecular weight. C, representative immunoblots of NRF2, HO1 and LAMINB as a loading control from liver extracts of vehicle and PHAR treated mice and from a positive control mouse treated i.p. with 50 mg/kg SFN. D, densitometric quantification of NRF2 levels from D expressed as a ratio of NRF2/LAMINB. Data are mean ± S.D. (n = 5). ***p < 0.001 vs. vehicle according to a One-way ANOVA test. E-F, mice were treated daily with vehicle (Tween-80 + PBS, 1:13) or 50 mg/kg PHAR by i.p. injection for 5 days. F, representative immunoblots showing the levels of NRF2, HO1, and LAMINB as a loading control from brain, liver and kidney extracts. G, densitometric quantification of NRF2 and HO1 protein levels from representative liver immunoblots showed in E, expressed as a ratio of protein/LAMINB. Data are mean ± S.D. (n = 5). **p < 0.01 vs. vehicle according to a Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Regarding NRF2 activation, hepatic NRF2 protein levels were significantly increased 2 h after administration of PHAR, which to a lesser extent resembled the accumulation of NRF2 in SFN-treated mice (50 mg/kg, 2 h) (Fig. 8C–D). Contrary to sulforaphane, the accumulation of NRF2 was not accompanied by an increase in HO1 within this time frame. The difference between SFN and PHAR is consistent with a stronger response of NRF2 activation upon KEAP1 inhibition. For this reason, the mice were treated i.p. daily for 5 consecutive days with vehicle or 50 mg/kg PHAR. On the fifth day, 2 h after the administration of the last dose of PHAR, a significant increase in both NRF2 and HO1 protein levels were observed in the liver but not in the brain and kidney (Fig. 8E–F), indicating that the liver is the main target of PHAR.
3.8. PHAR attenuates acute liver inflammation in response to LPS
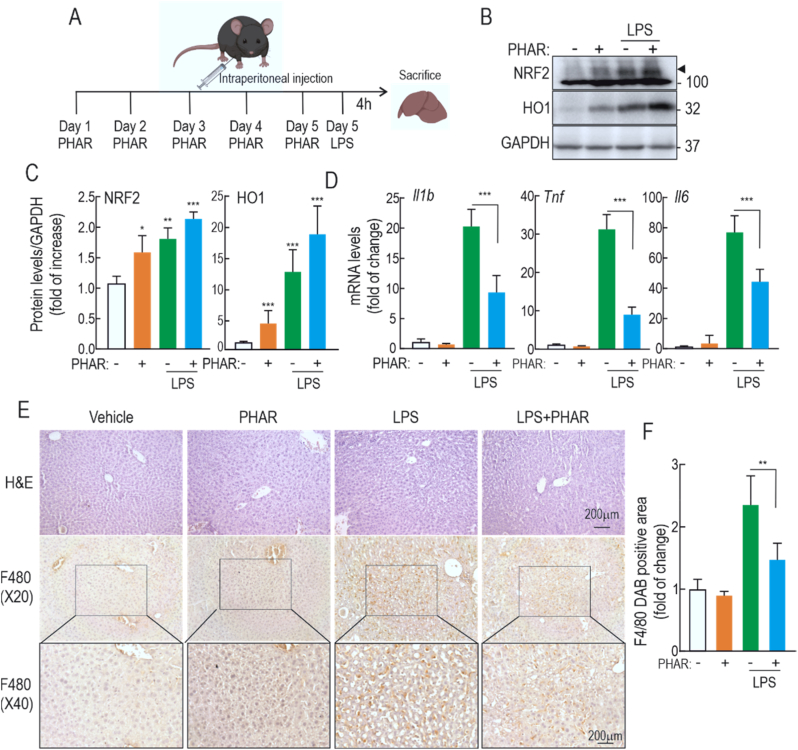
Lastly, we evaluated whether PHAR could prevent LPS-mediated inflammation in vivo. Mice were treated i.p daily with vehicle or 50 mg/kg PHAR for 5 consecutive days. Two hours after the last treatment, mice were given 10 mg/kg of LPS by i.p. injection and 4 h later they were sacrificed to allow analyses of hepatic inflammatory responses (Fig. 9A). As expected, protein immunoblot of liver lysates revealed an increase in the levels of NRF2 protein and its target gene product HO1 in response to PHAR and LPS (Fig. 9B–C). The mRNA levels of three pro-inflammatory cytokines, Il1β, Il6, and Tnf, revealed the expected induction by LPS in vehicle-treated mice. However, this induction was significantly attenuated in PHAR-treated mice (Fig. 9D). Examination of H&E-stained liver sections revealed that the hepatic structure is not obviously altered by administration of PHAR (Fig. 9E). Immunohistochemical staining with anti-F4/80 of hepatic macrophages (Kupffer cells) revealed that LPS significantly increases staining of this cell type, as expected, and that this increase is greatly diminished in PHAR-treated mice (Fig. 9F). Therefore, these results confirm that PHAR favors an inflammation-protective environment in mouse liver in response to acute inflammation.
Fig. 9.
PHAR alleviates inflammatory response in mice treated with LPS. A, C57Bl/6 male mice were treated i.p with vehicle (Tween-80 + PBS, 1:13, groups 1 and 3) or 50 mg/kg PHAR (groups 2 and 4) for 5 days. Two hours after the last administration, mice received vehicle or 10 mg/kg LPS and were sacrificed after 4 h for liver analysis. B, representative immunoblots in liver extracts of NRF2, HO1, and GAPDH as a loading control. C, densitometric quantification of the NRF2 and HO1 protein levels from representative immunoblots shown in A, expressed as a ratio of protein/GAPDH. Data are mean ± S.D (n = 6). *p < 0,05; **p < 0.01 and ***p < 0.001 vs. vehicle according to a two-way ANOVA test. D, mRNA levels of Il1b, Tnf, and Il6 were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp, and Actb levels. Data are mean ± S.D (n = 6). ***p < 0.001 vs. LPS according to a two-way ANOVA test. E, paraffin-embedded liver section stained with H&E and immunohistochemistry for F4/80. F, Quantification of DAB-staining positive area of F4/80. Data are mean ± S.D. (n = 6). **p < 0.01 vs LPS according to a two-way ANOVA test.
4. Discussion
A plethora of NRF2 activators have been described [54,55]. Most of them are electrophiles that inhibit the ability of KEAP1 to repress NRF2 through thiol modification of any one of several reactive cysteines in this Cul3/RBX1 substrate adaptor [18]. However, electrophiles react with sulfhydryl groups of many other proteins and are therefore not selective NRF2 activators [18,56,57]. A novel strategy is being considered, based on the generation of small molecules that dock at the interface between KEAP1 and NRF2, thus preventing KEAP1-mediated ubiquitination and subsequent degradation of NRF2 [58]. The drug development pipelines along this strategy are just emerging and it is not clear if these drugs will also inhibit other substrates that are regulated by KEAP1. Moreover, somatic mutations in the interphase of KEAP1/NRF2 have been reported in a large variety of human tumours [[59], [60], [61]]. Whilst there is no evidence that such mutations initiate cancer, it is possible that prolonged and potent inhibition of KEAP1 might promote tumorigenesis once cancer has been initiated.
By contrast with the KEAP1/NRF2 interaction, disruption of the β-TrCP/NRF2 interaction is a completely unexplored pathway. Focusing on the β-TrCP/NRF2 interaction as a drug target is attractive for at least three reasons: 1) the β-TrCP/NRF2 interaction is weaker than those between β-TrCP and other substrates (Table 1), thus enabling the development of protein/protein interaction inhibitors that might displace NRF2 but not several other β-TrCP substrates; 2) the activation of NRF2 following β-TrCP inhibition is weaker than that resulting from KEAP1 inhibition [23], thus remaining close to homeostatic variations; 3) somatic mutations in the interface of interaction between NRF2 and β-TrCP have not been reported, suggesting that this pathway is safer than the KEAP1 pathway for cancer risk.
Compared to electrophilic inhibitors of KEAP1 such as SFN, PHAR was found to activate NRF2 to a lesser extent in most cell types tested. This was expected because KEAP1 is the main negative controller of NRF2 stability in most cell types investigated to date. Nevertheless, PHAR activation of NRF2-target gene expression took place in KEAP1-depleted cells further demonstrating the KEAP1-independent mechanism of NRF2 activation. It is interesting that we found the capacity of PHAR to activate NRF2 different in the cell lines of study, MCF-7, HEK293T, MEFs, RAW264.7, and primary peritoneal macrophages. We attribute this variation to the extent of GSK-3β activity in each for these cell lines that occurs in the absence of serum, because this is a crucial requisite for the formation of the phosphodegron in NRF2 by GSK-3. Autocrine stimulation has been reported in tumour cell lines and continuous passage of cells in culture also favours spontaneous transformation. Therefore, the different sensitivities to PHAR observed in the cell lines studied are probably connected with GSK-3-related signaling events. For this reason, it was important to determine if PHAR could inhibit β-TrCP-mediated degradation of NRF2 within the context of strong GSK-3 activity. Under active signaling conditions, the PI3K/AKT pathway leads to phosphorylation of GSK3-α at Ser-21 and GSK3-β at Ser-9. This phosphorylation takes place at the N-terminal pseudosubstrate and results in GSK-3 inhibition. Therefore, in the presence of the PI3K inhibitor LY294002 we could maintain GSK-3α/β in the non-phosphorylated, active form that leads to NRF2 phosphorylation and subsequent β-TrCP-mediated degradation. Under these conditions, NRF2 levels decreased as expected (Fig. 4A, Fig. S1 and [62,63]), but, importantly, PHAR fully prevented NRF2 degradation and even slightly increased NRF2 levels. Moreover, pharmacological inhibition of GSK-3 increased NRF2 protein levels, also as expected (Fig. S2, and [62]), but rendered cells unresponsive to PHAR, further suggesting that PHAR targets the β-TrCP/NRF2 interaction when NRF2 is phosphorylated by GSK-3.
GSK-3 shows a preference for phosphorylation of Ser/Thr amino acids close to Ser/Thr residues that have previously been phosphorylated by other kinases, which are called priming kinases. In the case of NRF2, we have speculated previously that Ser347 and Ser342 might be phosphorylated by a priming kinase(s) and so might provide a recognition sites for GSK-3, which in turn enables GSK-3 to phosphorylate Ser338 and Ser335 [10]. However, in the in vitro kinase assay that employed bacterially expressed EYFP-NRF2ΔNeh2 protein, GSK-3 phosphorylated Ser347, Ser342, Ser338 and Ser335, as determined using phospho-specific antibodies, without the cooperation of any other kinase. A possible interpretation of this finding is that GSK-3 can also phosphorylate Ser/Thr amino acids in proximity to prolines, i.e., it can act as a proline-directed kinase. However, that said, these are in vitro experiments and they do not preclude the involvement of priming kinases under in vivo conditions, particularly putative priming kinases that are activated by inflammatory stimuli. Due to the forced in vitro phosphorylation conditions, we were able to obtain a highly phosphorylated NRF2 for further ubiquitination analysis.
The in vitro β-TrCP ubiquitination assay on phospho-NRF2 indicates that PHAR is an inhibitor of ubiquitination of NRF2 but this does not necessarily prove that PHAR is a protein/protein interaction inhibitor. As such, PHAR should dissociate the physical interaction between β-TrCP and NRF2. We tested this hypothesis in cells submitted to LY294002 to have active GSK-3 and to the NEDD8 inhibitor MLN-4924. Unlike the widely used MG132 proteasome inhibitor that stabilizes NRF2 after ubiquitination, MLN-4924 stabilizes NRF2 prior to the enzymatic reaction that leads to ubiquitination, and, therefore, should maintain the association between NRF2 and the E3 ligase adapter, in our case β-TrCP. We found that indeed, NRF2 and β-TrCP associate as their distance is less than 40 nm, which is the maximal distance that gives a positive signal in the proximity ligation assay. In a former report [24], we speculated that the β-TrCP/NRF2 interaction should take place in the nucleus because other components of the ubiquitylation machinery, including CDC34, SKP1, and CUL1, have been identified within the nucleus [64]. However, now we can provide experimental evidence (see Fig. 5), that indeed the association between β-TrCP and NRF2 occurs at least in the nucleus. Importantly, PHAR disrupted this interaction, demonstrating that it behaves as a β-TrCP/NRF2 interaction inhibitor. Validation of this method was undertaken by visualizing the interaction between ectopically expressed HA-tagged KEAP1 and wild-type NRF2-V5 in HEK293T cells, using as negative control the NRF2ΔETGE-V5 mutant that does not bind KEAP1 (Supplemental Fig. S5). We found that KEAP1 and NRF2 are tightly associated within both the nucleus and the cytoplasm as would be expected from reports showing that these two proteins are located in both compartments [65]. Importantly, PHAR did not have a significant effect on the HA-KEAP1/NRF2-V5 association, once again demonstrating that PHAR is not a disrupter of this interaction.
Upon i.p. administration of PHAR, there was an apparently selective accumulation of the PPI inhibitor in the liver, relative to kidney and brain. Although more through pharmacokinetics analysis is necessary, these findings suggest that the liver may behave as a trap for PHAR. One possible explanation is that this hydrophobic molecule might be transported selectively to the liver in lipoproteins. In fact, we have found that over 90% of PHAR is bound to plasma proteins (to be reported elsewhere). The hypothesis that lipoproteins could behave as nanocarriers to deliver hydrophobic compounds to the liver has been enunciated several times [66]. Lipoproteins, are the major carriers of lipids in the systemic circulation, and therefore may transport lipophilic drugs such as PHAR. This association may limit PHAR uptake into most cell types. However, because lipoproteins will be finally recycled by receptor-mediated uptake in the liver, we speculate that this mechanism explains the selective exposure and activation of NRF2 in this organ. An alternative explanation has been suggested for a PPI inhibitor of KEAP1/NRF2, which was also active in liver [67]. The authors suggested that the selectivity for this organ could be due relatively low bioavailability, therefore restricting its activity to the liver. In any case, despite of the fact that NRF2 is a ubiquitous protein, our finding demonstrates the possibility of targeted organ activation.
Nonsteroidal anti-inflammatory drugs (NSAIDs) are the main strategy used for the treatment of inflammatory disorders. However, continuous uptake of NSAIDs have many side effects, including gastrointestinal adverse reactions [68,69], hepatotoxicity [70,71], arterial hypertension [72,73], renal damage [74,75], and heart failure [68,76]. For this reason, it is desirable to find new alternatives to treat inflammatory diseases. An innovative strategy to combat inflammation is the activation of transcription factor NRF2. A potential advantage of targeting NRF2 over classical NSAIDs that target just one pro-inflammatory enzyme, i.e. COX2, is that NRF2 enhances the endogenous anti-inflammatory signature, by either antagonizing the effect NFκB, the master regulator of inflammation [[77], [78], [79], [80]], or by directly regulating the expression of pro-inflammatory or anti-inflammatory genes. NRF2 enhances antioxidant defense system and increases HO1 expression and GSH levels. This antioxidant response inhibits ROS mediated activation of NF-κB [81,82]. In addition, NRF2 directly inhibits the expression of pro-inflammatory genes encoding IL-6 and IL-1β [7]. Other studies have described that NRF2 also up-regulates the expression of some immunomodulatory genes such as platelet glycoprotein 4 (CD36), interleukin 17D (IL17D), and leukotriene B4 dehydrogenase (LTB4DH) [83].
Macrophages play crucial roles in inflammation through their ability to produce pro-inflammatory mediators [84,85]. Lipopolysaccharide (LPS) represents a bacterial endotoxin that triggers inflammation by activating NFκB in macrophages and is widely used in experiments. Several studies have reported that disruption of the gene encoding NRF2 increases the mortality of mice in response to LPS-induced septic shock in mice [86,87]. Therefore, we analyzed if NRF2 activation by PHAR might exert anti-inflammatory effects. In cultured macrophages, we found the expression of LPS-induced pro-inflammatory markers (pre-IL1β, COX-2, and NOS2 protein, as well as mRNA for Tnf, Il6 and Il1b in mRNA) were drastically attenuated by previous activation of NRF2 by PHAR. The role of NRF2 in this mechanism was further demonstrated by comparing the LPS-induced inflammatory response in peritoneal macrophages obtained from wild type vs. Nrf2-knockout mice. In NRF2-deficient macrophages PHAR could hardly reduce the expression of specific pro-inflammatory markers. However, it is interesting to note that, although not statistically significant, there was a very mild tendency for PHAR to slightly attenuate the expression of these markers in NRF2-deficient macrophages. If PHAR might exert some NRF2-independent downregulation of inflammation, one possible explanation might be that PHAR could be partially disrupting the interaction between β-TrCP and inflammatory signals. Nevertheless, the dependency of NRF2 for PHAR-mediated attenuation of inflammation was the most conclusive result. Importantly, a similar anti-inflammatory effect was also observed in the liver of mice submitted to acute inflammation induced by LPS. The livers of these mice exhibited significant attenuation of LPS-induced levels of pro-inflammatory markers and reduced activation of the liver resident macrophages (Kupffer cells). Our results report for the first time a disrupter of the interaction between NRF2 and β-TrCP that may have a clinical value in attenuation of liver inflammation. Future studies will be directed towards a detailed characterization and optimization of its absorption, distribution, metabolism, excretion, and toxicity (ADMET) profile.
Author contributions
Conceptualization and methodology RFG, AIR and AC; investigation, formal analysis, visualization and validation, RFG and AIR; writing—original draft preparation RFG and AC; writing—review and editing JAE, JDH, BO, MR, AIR, RFG and AC; supervision, project administration, resources and funding acquisition, AC.
Funding
This research was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) (grants PID2019-110061RB-I00, PID2021-122650OB-I00, and PDC2021-121421-I00) and The Autonomous Community of Madrid (grant B2017/BMD-3827). RFG enjoyed FPI contract of MINECO (FPI-2017). The Spanish Ministry of Economy and Competitiveness (MINECO, Project RTI2018-096724-B-C21) and the Generalitat Valenciana (PROMETEO/2021/059) supported the work in the Encinar laboratory.
Conflicts of interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Declaration of competing interest
The authors of this study declare no conflict of interest.
Acknowledgments
This article is based upon work from COST Action CA20121, supported by COST (European Cooperation in Science and Technology) (www.cost.eu) (https://benbedphar.org/about-benbedphar/). We are grateful to the Centro de Computación Científica (CCC-UAM) for letting us take advantage of the computer cluster Cibeles (https://www.ccc.uam.es/) for providing computing facilities. We thank Professor Calum Sutherland for expert advice and for helping raise NRF2 phospho-specific antibodies. PHAR has been submitted for international patent (application number PCT/EP2022/050657; priority date of January 13, 2022) for use as PPI inhibitor of beta-TrCP/NRF2 and use un therapy of liver disease.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102396.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Taguchi K., Motohashi H., Yamamoto M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Gene Cell. 2011;16:123–140. doi: 10.1111/J.1365-2443.2010.01473.X. [DOI] [PubMed] [Google Scholar]
- 2.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2006;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 3.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/J.TIBS.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Ren J., Li L., Wang Y., Zhai J., Chen G., Hu K. Gambogic acid induces heme oxygenase-1 through Nrf2 signaling pathway and inhibits NF-κB and MAPK activation to reduce inflammation in LPS-activated RAW264.7 cells. Biomed. Pharma. 2019;109:555–562. doi: 10.1016/j.biopha.2018.10.112. [DOI] [PubMed] [Google Scholar]
- 5.Kim J., Cha Y.N., Surh Y.J. A protective role of nuclear factor-erythroid 2-related factor-2 (Nrf2) in inflammatory disorders. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2010;690:12–23. doi: 10.1016/j.mrfmmm.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Cuadrado A., Manda G., Hassan A., Alcaraz J., Barbas C., Daiber A., Ghezzi P., León R., López M.G., Oliva B., Pajares M., Rojo A.I., Robledinos-Antón N., Valverde A.M., Guney E., Schmidt H.H.H.W., Victor N.R. Transcription factor NRF2 as a therapeutic target for chronic diseases: a systems medicine approach. Pharmacol. Rev. 2018;70:348–383. doi: 10.1124/pr.117.014753. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., Yamamoto M. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7 doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su W.C., Liu X., Macias A.A., Baron R.M., Perrella M.A. Heme oxygenase-1-derived carbon monoxide enhances the host defense response to microbial sepsis in mice. J. Clin. Invest. 2008;118:239–247. doi: 10.1172/JCI32730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellezza I., Giambanco I., Minelli A., Donato R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:721–733. doi: 10.1016/j.bbamcr.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Rada P., Rojo A.I., Evrard-Todeschi N., Innamorato N.G., Cotte A., Jaworski T., Tobon-Velasco J.C., Devijver H., Garcia-Mayoral M.F., Van Leuven F., Hayes J.D., Bertho G., Cuadrado A. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/-TrCP Axis. Mol. Cell Biol. 2012;32:3486–3499. doi: 10.1128/mcb.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004;279:31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 12.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell Biol. 2011;31:1121–1133. doi: 10.1128/mcb.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki T., Motohashi H., Yamamoto M. Toward clinical application of the Keap1-Nrf2 pathway. Trends Pharmacol. Sci. 2013;34:340–346. doi: 10.1016/j.tips.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Ma Q. Role of Nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong K.I., Padmanabhan B., Kobayashi A., Shang C., Hirotsu Y., Yokoyama S., Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as Hinge and Latch in oxidative stress response. Mol. Cell Biol. 2007;27:7511–7521. doi: 10.1128/mcb.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichimura Y., Waguri S., shin Sou Y., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., Hoshii T., Hirao A., Takagi K., Mizushima T., Motohashi H., Lee M.S., Yoshimori T., Tanaka K., Yamamoto M., Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Magesh S., Chen Y., Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med. Res. Rev. 2012;32:687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robledinos-Antón N., Fernández-Ginés R., Manda G., Cuadrado A. Activators and inhibitors of NRF2: a review of their potential for clinical development. Oxid. Med. Cell. Longev. 2019 doi: 10.1155/2019/9372182. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Höxtermann S., Nüchel C., Altmeyer P. Fumaric acid esters suppress peripheral CD4- and CD8-positive lymphocytes in psoriasis. Dermatology. 1998;196:223–230. doi: 10.1159/000017903. [DOI] [PubMed] [Google Scholar]
- 20.Xu Z., Zhang F., Sun F., Gu K., Dong S., He D. Dimethyl fumarate for multiple sclerosis. Cochrane Database Syst. Rev. 2015:CD011076. doi: 10.1002/14651858.CD011076.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schimrigk S., Brune N., Hellwig K., Lukas C., Bellenberg B., Rieks M., Hoffmann V., Pöhlau D., Przuntek H. Oral fumaric acid esters for the treatment of active multiple sclerosis: an open-label, baseline-controlled pilot study. Eur. J. Neurol. 2006;13:604–610. doi: 10.1111/j.1468-1331.2006.01292.x. [DOI] [PubMed] [Google Scholar]
- 22.Richardson B.G., Jain A.D., Speltz T.E., Moore T.W. Non-electrophilic modulators of the canonical Keap1/Nrf2 pathway. Bioorg. Med. Chem. Lett. 2015;25:2261–2268. doi: 10.1016/j.bmcl.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuadrado A. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/β-TrCP. Free Radic. Biol. Med. 2015;88:147–157. doi: 10.1016/j.freeradbiomed.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 24.Chowdhry S., Zhang Y., Mcmahon M., Sutherland C., Cuadrado A., Hayes J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antonio Encinar J., Fernández-Ballester G., Galiano-Ibarra V., Micol V. Drug Design, Development and Therapy Dovepress in silico approach for the discovery of new PParγ modulators among plant-derived polyphenols. Drug Des. Dev. Ther. 2015;9:5877–5895. doi: 10.2147/DDDT.S93449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galiano V., Garcia-Valtanen P., Micol V., Antonio Encinar J. Drug Design, Development and Therapy Dovepress looking for inhibitors of the dengue virus ns5 rna-dependent rna-polymerase using a molecular docking approach. Drug Des. Dev. Ther. 2016;10:10–3163. doi: 10.2147/DDDT.S117369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz-Torres V., Losada-Echeberría M., Herranz-López M., Barrajón-Catalán E., Galiano V., Micol V., Encinar J.A. New mammalian target of Rapamycin (mTOR) modulators derived from natural product databases and marine extracts by using molecular docking techniques. Mar. Drugs. 2018;16:385. doi: 10.3390/md16100385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng F., Li W., Zhou Y., Shen J., Wu Z., Liu G., Lee P.W., Tang Y. AdmetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012;52:3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- 29.Sander T., Freyss J., Von Korff M., Rufener C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015;55:460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- 30.Cuyàs E., Verdura S., Micol V., Joven J., Bosch-Barrera J., Encinar J.A., Menendez J.A. Revisiting silibinin as a novobiocin-like Hsp90 C-terminal inhibitor: computational modeling and experimental validation. Food Chem. Toxicol. 2019;132 doi: 10.1016/j.fct.2019.110645. [DOI] [PubMed] [Google Scholar]
- 31.Rubio-Camacho M., Encinar J.A., José Martínez-Tomé M., Esquembre R., Reyes Mateo C. The interaction of temozolomide with blood components suggests the potential use of human serum albumin as a biomimetic carrier for the drug. Biomolecules. 2020;10:1015. doi: 10.3390/biom10071015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43:443–447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canning P., Sorrell F.J., Bullock A.N. Structural basis of Keap1 interactions with Nrf2. Free Radic. Biol. Med. 2015;88:101–107. doi: 10.1016/j.freeradbiomed.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krieger E., Vriend G., Kelso J. YASARA View-molecular graphics for all devices-from smartphones to workstations. Bioinf. Appl. 2014;30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Encinar J.A., Menendez J.A. Potential drugs targeting early innate immune evasion of SARS-coronavirus 2 via 2’-O-methylation of viral RNA. Viruses. 2020;12:525. doi: 10.3390/v12050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marroqui L., Martinez-Pinna J., Castellano-Muñoz M., dos Santos R.S., Medina-Gali R.M., Soriano S., Quesada I., Gustafsson J.A., Encinar J.A., Nadal A. Bisphenol-S and Bisphenol-F alter mouse pancreatic β-cell ion channel expression and activity and insulin release through an estrogen receptor ERβ mediated pathway. Chemosphere. 2021;265 doi: 10.1016/j.chemosphere.2020.129051. [DOI] [PubMed] [Google Scholar]
- 38.Verdura S., Cuyàs E., Cortada E., Brunet J., Lopez-Bonet E., Martin-Castillo B., Bosch-Barrera J., Encinar J.A., Menendez J.A., Encinar A. Resveratrol targets PD-L1 glycosylation and dimerization to enhance antitumor T-cell immunity. Aging (Albany NY) 2020;12:8–34. doi: 10.18632/aging.102646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krieger E., Nielsen J.E., Spronk C.A.E.M., Vriend G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006;25:481–486. doi: 10.1016/j.jmgm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Genheden S., Ryde U. Expert Opinion on Drug Discovery the MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expet Opin. Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C., Nguyen P.H., Pham K., Huynh D., Le T.-B.N., Wang H., Ren P., Luo R. Calculating protein-ligand binding affinities with MMPBSA: method and error analysis. J. Comput. Chem. 2016;37:2436–2446. doi: 10.1002/jcc.24467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stranges P.B., Kuhlman B. A comparison of successful and failed protein interface designs highlights the challenges of designing buried hydrogen bonds. Protein Sci. 2012;22(1):74–82. doi: 10.1002/pro.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr, Protoc. Bioinform. 2016;54:5–6. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z., Lasker K., Schneidman-Duhovny D., Webb B., Huang C.C., Pettersen E.F., Goddard T.D., Meng E.C., Sali A., Ferrin T.E. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J. Struct. Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conway P., Tyka M.D., Dimaio F., David E. Konerding, Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2013;23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rada P., Rojo A.I., Offergeld A., Feng G.J., Velasco-Martín J.P., González-Sancho J.M., Valverde Á.M., Dale T., Regadera J., Cuadrado A. WNT-3A regulates an Axin1/NRF2 complex that regulates antioxidant metabolism in hepatocytes. Antioxidants Redox Signal. 2015;22:555–571. doi: 10.1089/ars.2014.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X.J., Hayes J.D., Wolf C.R. Generation of a stable Antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of Nrf2 by cancer chemotherapeutic agents. Cancer Res. 2006;66:10983–10994. doi: 10.1158/0008-5472.CAN-06-2298. [DOI] [PubMed] [Google Scholar]
- 48.Pajares M., Jiménez-Moreno N., García-Yagüe Á.J., Escoll M., de Ceballos M.L., Van Leuven F., Rábano A., Yamamoto M., Rojo A.I., Cuadrado A., Jim enez-Moreno N., Garc ıa-Yag A.J., ıa de Ceballos M.L., Abano A.R. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12:1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojo A.I., Rada P., Mendiola M., Ortega-Molina A., Wojdyla K., Rogowska-Wrzesinska A., Hardisson D., Serrano M., Cuadrado A. The PTEN/NRF2 axis promotes human carcinogenesis. Antioxidants Redox Signal. 2014;21:2498–2514. doi: 10.1089/ars.2014.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rojo A.I., Innamorato N.G., Mart In-Moreno A.M., De Ceballos I.L., Yamamoto M., Cuadrado A. Nrf2 regulates microglial dynamics and Neuroinflammation in experimental Parkinson's disease. Glia. 2010;8:588–598. doi: 10.1002/glia.20947. [DOI] [PubMed] [Google Scholar]
- 51.Rojo A.I., Medina-Campos O.N., Rada P., Zúñiga-Toalá A., López-Gazcón A., Espada S., Pedraza-Chaverri J., Cuadrado A. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: role of glycogen synthase kinase-3. Free Radic. Biol. Med. 2012;52:473–487. doi: 10.1016/j.freeradbiomed.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 52.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell Biol. 2011;31:1121–1133. doi: 10.1128/mcb.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Best S., Lam V., Liu T., Bruss N., Adam Kittai, Danilova O.V., Murray S., Berger A., Pennock N.D., Lind E.F., Danilov A.V. Immunomodulatory effects of pevonedistat, a NEDD8-activating enzyme inhibitor, in chronic lymphocytic leukemia-derived T cells, Leukemia. 2021. 35. 156–168. [DOI] [PMC free article] [PubMed]
- 54.Boo Y.C. Natural Nrf2 modulators for skin protection. Antioxidants. 2020;9:1–31. doi: 10.3390/antiox9090812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W., Dinkova-Kostova A.T. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 56.Liu P., Tian W., Tao S., Tillotson J., Wijeratne E.M.K., Gunatilaka A.A.L., Zhang D.D., Chapman E. Non-covalent NRF2 activation confers greater cellular protection than covalent activation. Cell Chem. Biol. 2019;26:1427–1435. doi: 10.1016/j.chembiol.2019.07.011. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.L S., Satoh T. Recent Advances in Understanding NRF2 as a Druggable Target: Development of Pro-electrophilic and Non-covalent NRF2 Activators to Overcome Systemic Side Effects of Electrophilic Drugs like Dimethyl Fumarate. F1000Res. 2017;vol. 6:2138. doi: 10.12688/f1000research.12111.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tran K.T., Pallesen J.S., Solbak S.M., Narayanan D., Baig A., Zang J., Aguayo-Orozco A., Carmona R.M.C., Garcia A.D., Bach A. A comparative assessment study of known small-molecule Keap1-Nrf2 protein-protein interaction inhibitors: chemical synthesis, binding properties, and cellular activity. J. Med. Chem. 2019;62:8028–8052. doi: 10.1021/acs.jmedchem.9b00723. [DOI] [PubMed] [Google Scholar]
- 59.Padmanabhan B., Tong K.I., Ohta T., Nakamura Y., Scharlock M., Ohtsuji M., Il Kang M., Kobayashi A., Yokoyama S., Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 60.Shibata T., Ohta T., Tong K.I., Kokubu A., Odogawa R., Tsuta K., Asamura H., Yamamoto M., Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taguchi K., Yamamoto M. The KEAP1-NRF2 system as a molecular target of cancer treatment. Cancers. 2020;13:46. doi: 10.3390/cancers13010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L., Chen Y., Sternberg P., Cai J. Essential roles of the PI3 kinase/akt pathway in regulating Nrf2-dependent antioxidant functions in the RPE. Invest. Ophthalmol. Vis. Sci. 2008;49:1671–1678. doi: 10.1167/iovs.07-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin D., Rojo A.I., Salinas M., Diaz R., Gallardo G., Alam J., Ruiz De Galarreta C.M., Cuadrado A. Regulation of heme oxygenase-1 expression through the phosphatidylinositol 3-kinase/akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J. Biol. Chem. 2004;279:8919–8929. doi: 10.1074/jbc.M309660200. [DOI] [PubMed] [Google Scholar]
- 64.Pons J., Evrard-Todeschi N., Bertho G., Gharbi-Benarous J., Tanchou V., Benarous R., Girault J.P. Transfer-NMR and docking studies identify the binding of the peptide derived from activating transcription factor 4 to protein ubiquitin ligase β-TrCP. Competition STD-NMR with β-catenin. Biochemistry. 2008;47:14–29. doi: 10.1021/bi7014212. [DOI] [PubMed] [Google Scholar]
- 65.Baird L., Swift S., Llères D., Dinkova-Kostova A.T. Monitoring Keap1-Nrf2 interactions in single live cells. Biotechnol. Adv. 2014;32:1133–1144. doi: 10.1016/j.biotechadv.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jigar D.R.B., Patel P. The effect of oral lipids and circulating lipoproteins on the metabolism of drugs. Expet Opin. Drug Metabol. Toxicol. 2009;5:1385. doi: 10.1517/17425250903176439. Abstract. [DOI] [PubMed] [Google Scholar]
- 67.Dayalan Naidu S., Suzuki T., Dikovskaya D., Knatko E.V., Higgins M., Sato M., Novak M., Villegas J.A., Moore T.W., Yamamoto M., Dinkova-Kostova A.T. The isoquinoline PRL-295 increases the thermostability of Keap1 and disrupts its interaction with Nrf2. iScience. 2022;25 doi: 10.1016/j.isci.2021.103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Al-Saeed A. Gastrointestinal and cardiovascular risk of nonsteroidal anti-inflammatory drugs. Oman Med. J. 2011;26:385–391. doi: 10.5001/omj.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sostres C., Gargallo C.J., Arroyo M.T., Lanas A. Adverse effects of non-steroidal anti-inflammatory drugs (NSAIDs, aspirin and coxibs) on upper gastrointestinal tract. Best Pract. Res. Clin. Gastroenterol. 2010;24:121–132. doi: 10.1016/j.bpg.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 70.Meunier L., Larrey D. Recent advances in hepatotoxicity of non steroidal anti-inflammatory drugs. Ann. Hepatol. 2018;17:187–191. doi: 10.5604/01.3001.0010.8633. [DOI] [PubMed] [Google Scholar]
- 71.Rubenstein J.H., Laine L. Systematic review: the hepatotoxicity of non-steroidal anti-inflammatory drugs. Aliment. Pharmacol. Ther. 2004;20:373–380. doi: 10.1111/j.1365-2036.2004.02092.x. [DOI] [PubMed] [Google Scholar]
- 72.Zheng L., Du X. Non-steroidal anti-inflammatory drugs and hypertension. Cell Biochem. Biophys. 2014;69:209–211. doi: 10.1007/s12013-013-9791-5. [DOI] [PubMed] [Google Scholar]
- 73.Bavry A.A., Khaliq A., Gong Y., Handberg E.M., Cooper-Dehoff R.M., Pepine C.J. Harmful effects of NSAIDs among patients with hypertension and coronary artery disease. Am. J. Med. 2011;124:614–620. doi: 10.1016/j.amjmed.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nawaz F.A., Larsen C.P., Troxell M.L. Membranous nephropathy and nonsteroidal anti-inflammatory agents. Am. J. Kidney Dis. 2013;62:1012–1017. doi: 10.1053/j.ajkd.2013.03.045. [DOI] [PubMed] [Google Scholar]
- 75.Mérida E., Praga M. NSAIDs and nephrotic syndrome. Clin. J. Am. Soc. Nephrol. 2019;14:1280–1282. doi: 10.2215/CJN.08090719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghosh R., Alajbegovic A., Gomes A.V. NSAIDs and cardiovascular diseases: role of reactive oxygen species. Oxid. Med. Cell. Longev. 2015 doi: 10.1155/2015/536962. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X., Qin, Jiang Hao, Li Yan, Chen Wenfei, Li Hanmei, Peng Ke, Zhang Zhirong, Sun Targeting NF-kB signaling with polymeric hybrid micelles that co-deliver siRNA and dexamethasone for arthritis therapy. Biomaterials. 2017;122:10–22. doi: 10.1016/j.biomaterials.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 78.Christian F., Smith E.L., Carmody R.J. Cells the regulation of NF-κB subunits by phosphorylation. Cells. 2016;5:12. doi: 10.3390/cells5010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu T., Zhang L., Joo D., Sun S.-C. NF-κB signaling in inflammation. Signal Transduct. Targeted Ther. 2017;2 doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun S.C., Ley S.C. New insights into NF-κB regulation and function. Trends Immunol. 2008;29:469–478. doi: 10.1016/j.it.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bellezza I., Tucci A., Galli F., Grottelli S., Mierla A.L., Pilolli F., Minelli A. Inhibition of NF-κB nuclear translocation via HO-1 activation underlies α-tocopheryl succinate toxicity. J. Nutr. Biochem. 2012;23:1583–1591. doi: 10.1016/j.jnutbio.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 82.Ganesh Yerra V., Negi G., Sharma S.S., Kumar A. Potential therapeutic effects of the simultaneous targeting of the Nrf2 and NF-κB pathways in diabetic neuropathy. Redox Biol. 2013;1:394–397. doi: 10.1016/j.redox.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dick R.A., Kwak M.K., Sutter T.R., Kensler T.W. Antioxidative function and substrate specificity of NAD(P)H-dependent alkenal/one oxidoreductase. A new role for leukotriene B4 12-hydroxydehydrogenase/15-oxoprostaglandin 13-reductase. J. Biol. Chem. 2001;276:40803–40810. doi: 10.1074/jbc.M105487200. [DOI] [PubMed] [Google Scholar]
- 84.Rani M., Nicholson S.E., Zhang Q., Schwacha M.G. Damage-associated molecular patterns (DAMPs) released after burn are associated with inflammation and monocyte activation. Burns. 2017;43:297–303. doi: 10.1016/j.burns.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Funakoshi-Tago M., Nonaka Y., Tago K., Takeda M., Ishihara Y., Sakai A., Matsutaka M., Kobata K., Tamura H. pyrocatechol, a component of coffee, suppresses LPS-induced inflammatory responses by inhibiting NF-κB and activating Nrf2. Sci. Rep. 2020;10:2584. doi: 10.1038/s41598-020-59380-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rangasamy T., Guo J., Mitzner W.A., Roman J., Singh A., Fryer A.D., Yamamoto M., Kensler T.W., Tuder R.M., Georas S.N., Biswal S. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J. Exp. Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thimmulappa R.K., Lee H., Rangasamy T., Reddy S.P., Yamamoto M., Kensler T.W., Biswal S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Invest. 2006;116 doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang B., Li Y., Beachy P.A. Evidence for the direct involvement of TrCP in Gli3 protein processing. Proc. Natl. Acad. Sci. U. S. A. 2006;3:33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rada P., Rojo A.I., Evrard-Todeschi N., Innamorato N.G., Cotte A., Jaworski T., Tobon-Velasco J.C., Devijver H., Garcia-Mayoral M.F., Van Leuven F., Hayes J.D., Bertho G., Cuadrado A. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/-TrCP Axis. Mol. Cell Biol. 2012;32:3486–3499. doi: 10.1128/mcb.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.