Abstract
The prognostic significance of hypoxia markers, hypoxia-inducible factor-1α (HIF-1α), hypoxia-inducible factor-2α (HIF-2α), and carbonic anhydrase IX (CAIX), was investigated in estrogen receptor (ER)-positive breast cancer patients. Immunohistochemistry determined the expression of makers in two independent ductal ER-positive cohorts (Training set, n=373 and Validation set, n=285) and was related to clinicopathological parameters and disease-free survival (DFS). In the training cohort, nuclear HIF-1α (1) was independently associated with poorer DFS in luminal A tumors [hazard ratio (HR) = 0.53 95% confidence interval (CI): 0.30–0.94, p=0.030]. In the validation cohort, both HIF-1α (1) and CAIX were independently associated with decreased DFS in the entire cohort (HR = 1.85 95% CI: 1.10–3.11, p=0.019; HR = 1.74 95% CI: 1.08–2.82, p=0.023), in luminal A disease (HR = 1.98 95% CI: 1.02–3.83, p=0.042), and in luminal B disease (HR = 2.75 95% CI: 1.66–4.55, p<0.001), respectively. Taken together, elevated cytoplasmic HIF-1α (1) expression was an independent prognostic factor in luminal A disease, whereas CAIX was an independent prognostic factor in luminal B disease. Further work in large tissue cohorts is required.
Keywords: breast cancer, carbonic anhydrase IX, hypoxia-inducible factor-1α, hypoxia-inducible factor-2α, survival, tumor hypoxia
Introduction
Breast cancer is the most commonly diagnosed cancer in women and the second leading cause of cancer mortality among women in more developed countries.1,2 Although the improvements in early detection and treatment strategies have resulted in decreased mortality, more reliable prognostic and predictive markers of breast carcinoma are required. 3 Breast cancer is a heterogeneous disease, with a high degree of diversity between and within tumors as well as among cancer-bearing patients. 4 According to gene expression, carcinomas have been classified into luminal A, luminal B, Her-2-enriched, and triple-negative disease. These subtypes have a strong prognostic impact and represent the basis for therapy. 5 , 6
Estrogen receptor (ER)-positive breast cancer represents approximately 70% of all breast cancers. 7 Endocrine therapy is critical to the success of controlling hormone-positive breast cancers, including tumors bearing the ER for early-stage and metastatic breast disease. 8 – 10 However, approximately 30% of patients either present with initial resistance called intrinsic (de novo resistance) or with resistance that arises during treatment (acquired resistance). 11
Tumor microenvironment influences the behavior of cancer cells including characteristics of aggressiveness, such as invasiveness, metastasis, angiogenesis, and therapy resistance. 12 A number of factors have been implicated in hormone resistance mechanism including altered ER-binding and crosstalk with other pathways, for example, the PI3K–AKT–mTOR pathway. 11 Hypoxia, a condition commonly occurring in solid tumors, is a major driver of invasiveness and metastasis in breast cancer, and it is associated with chemotherapy and radiotherapy resistance. 13 , 14
At the molecular level, the adaptation of tumor cells to hypoxic stress is regulated mainly by hypoxia-inducible factors (HIFs), which are transcription factors that accumulate in response to decreased cellular oxygen levels. 15 , 16 The main mediator of signaling these poorly oxygenated areas is hypoxia-inducible factor-1α (HIF-1α), which is stabilized at low oxygen levels activating the expression of several hypoxia response genes, including erythropoiesis, angiogenesis, glucose metabolism, and pH regulation. 17 , 18 Hypoxia-associated enzyme carbonic anhydrase IX (CAIX) is a direct transcriptional target of HIF-1α and is one of the most commonly upregulated genes in response to hypoxia. 19 , 20 CAIX, a transmembrane glycoprotein, plays a major role in maintaining the pH gradient between tumor cells and their extracellular space by reversible hydration of carbonic dioxide to carbonic acid (CO2+ H2O = HCO3– + H+). 21 Therefore, CAIX might increase metastatic potential by allowing aggressive cancer cells to survive the hostile environment imposed by hypoxia and may further function to potentiate extracellular acidosis, facilitating growth and invasion of the surviving tumor cells. 22
HIFs have been demonstrated to be involved in the resistance mechanism. 23 – 25 Significantly, the response to tamoxifen is decreased in hypoxic breast cancer cells compared with cells grown under normoxic environments. 26 , 27 The possibility of using HIFs and CAIX as prognostic and predictive markers in breast cancer has already been discussed by several authors. 15 ,28–32 The prognostic role of these proteins was dependent on cellular distribution and luminal subtypes. However, the interplay between hypoxia, HIFs and CAIX, activity and their impact on survival has not been clarified in ER-positive breast cancer cells. Therefore, the aim of this study was to examine the prognostic value of HIF-1α (1), HIF-1α (2), HIF-2α, and CAIX in ER-positive invasive breast cancer.
Materials and Methods
Patient TMA
Tissue microarrays were previously constructed from formalin-fixed paraffin-embedded tissue (FFPE) blocks from breast cancer patients in triplicate tissue microarrays (TMAs). Cores of 0.6 mm from archival paraffin blocks of each tumor were placed in separate TMA blocks. Sections of 2.5 μm thickness from each TMA block were placed on silanized glass slides.
Exclusion criteria included the following: (1) cases with ER-negative status, (2) cases with non-ductal carcinoma, (3) cases with HIF-1α (2) expression not available, (4) cases whose TMAs’ tumor samples were not sufficiently representative for evaluation of protein expression, and (5) cases in which clinicopathological data of interest could not be properly collected from the review of medical records.
ER-positive Cohort 1
In all, 570 female patients with ER-positive invasive breast cancer operated on at Royal Infirmary, Western Infirmary, and Stobhill Hospital, Glasgow, in the period from 1995 to 1998 were included in this study. Selection criteria of specimens having ductal histological subtype (n=476) and HIF-1α (2) expression available for analysis (n=373) were applied, resulting in the exclusion of 197 patients (Fig. 1A). Clinicopathological data including age, histological tumor type, grade, tumor size, lymph node status, and adjuvant treatment (hormonal therapy and chemotherapy) were retrieved from the routine reports. The molecular subtypes were defined as follows: Luminal A: ER-positive and/or progesterone receptor (PR)-positive, Her-2-negative, low proliferative index (≤15%); Luminal B: hormone receptor–positive, Her-2-positive, high proliferative index (>15%).
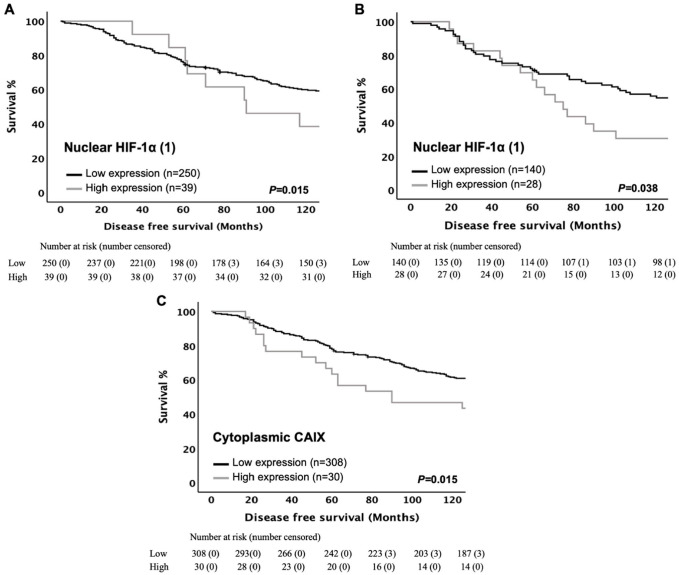
Figure 1.
Consort diagram of patient inclusion in the study. (A) ER-positive cohort 1 and (B) ER-positive cohort 2. Patients with missing cores or insufficient tumor for analysis were excluded. Abbreviations: ER, estrogen receptor; TMA, tissue microarray; HIF-1α, hypoxia-inducible factor-1α; IHC, immunohistochemistry; CAIX, carbonic anhydrase IX.
ER-positive Cohort 2
In all, 392 female patients with ER-positive invasive breast cancer operated at Western Infirmary, Victoria Hospital, and Stobhill Hospital, Glasgow, between 1980 and 1999 (Fig. 1B) were included in the study. Selection criteria of specimens having ductal histological subtype (n=314) and HIF-1α (2) expression available for analysis (n=285) were applied, resulting in the exclusion of 107 patients. The clinicopathological data available on the database included patient’s age, histological tumor type, tumor size, tumor grade, involved lymph node status, PR status, Her-2 status, and Ki67 proliferation index. All patients in the study were treated with the adjuvant tamoxifen.
Disease-free survival (DFS) was used as the primary end point defined as survival in months from the date of surgery until recurrence or all-cause mortality. All tumor samples were collected following approval by the Research Ethics Committee of North Glasgow University Hospitals (NHS GG&C REC reference: 16/WS/0207).
Immunohistochemistry (IHC)
Immunohistochemical expression of HIF-1α (1), HIF-1α (2), HIF-2α, and CAIX was carried out using a previously constructed TMA. HIF-1α (1) and HIF-1α (2) were from different suppliers. Two HIF-1α antibodies were tested. The analysis was performed with HIF-1α (2) and was used to select the cohort; however, data from HIF-1α (1) were more robust, so the results from that are presented in the article. Sections were dewaxed in Histoclear (National Diagnostics; CA) and then rehydrated through a decreasing gradient of alcohols. Heat-induced antigen retrieval was carried out under pressure in a microwave in either Tris-EDTA buffer at pH 9 for anti-HIF-1α (1), HIF-1α (2), and HIF-2α or citrate buffer at pH 6 for anti-CAIX for 14 min at 96C. Endogenous peroxidase activity was blocked using 3% hydrogen peroxide (H2O2) for 30 min. Prior to incubating in primary antibody, nonspecific antibody binding was blocked using either 1.5% horse serum (Vector Laboratories, USA) for anti-HIF-1α (1), anti-HIF-1α (2), and anti-HIF-2α or 10% casein (Vector Laboratories; Newark, CA) for anti-CAIX for 60 min at room temperature. TMAs were stained with anti-HIF-1α (1) antibody (clone monoclonal antibody HIF-1α 67, NB 100-105; Novus Biologicals, Abingdon, UK) at a dilution of 1:150, anti-HIF-1α (2) antibody (clone polyclonal antibody HIF-1α, NB 100-449; Novus Biologicals) at a dilution of 1:400, and anti-HIF-2α antibody (clone polyclonal antibody HIF-2α, NB 100-122; Novus Biologicals) at a dilution of 1:1000. For CAIX immunohistochemical detection, a monoclonal anti-CAIX antibody (Bioscience, Slovakia) at a dilution of 1:500 was used, followed by an overnight incubation at 4C for all proteins. TMAs were incubated for 30 min in ImmPRESS reagent (Vector Laboratories) and visualized with DAB chromogen substrate (Vector Laboratories). Samples were then counterstained in Haeamtoxylin Gill III (Leica Microsystems, Milton Keynes, UK cat. No. 3801540E) before being dehydrated in increasing alcohol gradients and Histoclear. Cover slips were applied using Pertex as mounting agent (Cellpath, Newton, UK cat no. SEA-0100-00A).
Slide Staining and Scanning
Stained TMAs were scanned using Hamamatsu NanoZoomer Digital Slide Scanner (Hamamatsu Photonics K.K.; Shizuoka, Japan) and visualized in NDP.serve3 image viewer platform system. Slides were visualized at 20× magnification. The expression of protein levels was assessed at each cellular compartment separately, nuclear and cytoplasmic for HIF-1α (1), HIF-1α (2), and HIF-2α and membranous and cytoplasmic for CAIX. No negative control antibody was used in the breast cancer TMA to rule out nonspecific staining (Supplementary Figs. S1–S3).
Scoring for Hypoxic Markers
Weighted Histoscore
Scoring was performed by a single observer (S.A.K.S.) blinded to the clinical data. Tumor cell expression of HIF-1α (2), HIF-2α, and CAIX was assessed using the weighted Histoscore method. 33 , 34 The weighted Histoscore was calculated as follows: (% of unstained tumor cells × 0) + (% of weakly stained tumor cells × 1) + (% of moderately stained tumor cells × 2) + (% of strongly stained tumor cells × 3) to give a range from 0 to 300. All three cores were scored separately, and an average score was taken. S.A.K.S. subsequently scored all slides for analysis.
QuPath Scoring
HIF-1α (1) was scored using QuPath digital pathology software v0.2.3 (QuPath; Edinburgh, UK). In brief, after using the TMA Dearrayer function to create a TMA grid with cores in their correct positions, stain vectors were estimated during preprocessing by the visual stain editor available in QuPath to increase staining quality. Then, cells were detected using a watershed cell detection method, and annotations were made to allow QuPath to recognize different tissue types, which are tumor and stroma. Then, a random trees classifier was trained using more than 40 features such as perimeter, area, and optical density. Three intensity thresholds were used to represent negative, weak, moderate, and strong staining, and after the classifier was built, the auto-update feature was used to revalidate the classifier’s accuracy in real-time. The classifier was then saved and applied to all TMA slides that were subjected to QuPath analysis.
To ensure reproducibility of scoring, 10% of cores for each marker was co-scored by a second observer (J.E.) blinded to the previous observer score as well as clinicopathological information. Reliability analysis was performed with SPSS software to ensure consistency and objectivity between the main scorer and the co-scorers giving an interclass correlation coefficient (ICCC) for all markers. Values above 0.75 are indicative of good reliability. 35 Scatter plots and Bland–Altman plots were constructed to visualize the correlation between scores. Manual scores for HIF-2α, and CAIX and QuPath scores for HIF-1α (1) were used for optimal thresholding and thus all subsequent analyses.
Statistical Analysis
To set threshold values for categorizing the expression of each protein into two groups, “low” and “high,” log-rank statistics were performed in R Studio (R Studio; Boston, MA) using survminer, survival, tidyverse, and maxstat packages. Survminer package in R studio was used to determine the optimal threshold for different antibodies based on overall survival (OS). The same threshold value was also prognostic for DFS. The threshold values from cohort 1 were applied for analysis in cohort 2. IBM SPSS software version 27 (SPSS Inc.; Chicago, IL) was used for statistical analysis. Kaplan–Meier log-rank curves were used to identify the associations between protein expression and DFS within the entire cohort and subtype group. Univariate Cox regression survival analysis with 95% confidence intervals (CIs) was used to calculate hazard ratio (HR) and 95% CI. Multivariable Cox regression survival analysis was carried out using a backward conditional elimination model. Chi-square testing was also used to determine the association between markers. Statistical significance was set to p<0.05.
Results
ER-positive Cohort 1
Supplementary Table S1 shows the clinicopathological characteristics of selected patients. The majority of patients (73%) were aged above 50 years, had small tumors ≤20 mm (62%), grade II (53%) or grade III (24%), and negative lymph node (59%). Of the 373 patients, 252 (68%) had PR-positive and 329 (89%) had Her-2-negative tumors. In all, 234 (65%) patients had luminal A disease and 127 (35%) had luminal B disease. Furthermore, 281 (88%) patients received tamoxifen and 110 (30%) received chemotherapy. Sixty-seven patients (19%) experienced recurrence. Of these patients, 15 (4%) had local recurrence, 49 (14%) had distant recurrence, and 3 patients had both.
Of the 373 patients, in 73 (20%) there was a tissue core missing and so HIF-1α (1) staining could not be carried out, and they were excluded from the analysis. Therefore, expression of HIF-1α (1) was assessed in 300 patients. HIF-1α (1) was clearly expressed in the nuclei and cytoplasm of tumor cells. Representative images of cytoplasmic and nuclear HIF-1α (1) staining and scoring, which was performed using QuPath digital image analysis software, are shown in Supplementary Fig. S1A. Cytoplasmic HIF-1α (1) scores ranged from 0 to 167.75, with a mean score of 93.41, and nuclear scores ranged from 0 to 205.55, with a mean score of 109.4. A histogram was plotted to visualize the range of scores and data were relatively normally distributed (Supplementary Fig. S1B). A correlation coefficient of 0.842 and 0.898 for cytoplasmic and nuclear, respectively, was obtained between QuPath and manual scores, and data were visualized in the form of a scatter plot and a Bland–Altman plot (Supplementary Fig. S1C). The optimal threshold was 104 for cytoplasmic and 156 for nuclear staining (Supplementary Fig. S1D).
Kaplan–Meier curves were plotted to determine any association between HIF-1α (1) expression and DFS in the full cohort. No significant association of cytoplasmic HIF-1α (1) expression with DFS (log-rank, p=0.114) was found. In contrast, statistically significant poorer DFS (log-rank, p=0.015) was observed with high level of nuclear HIF-1α (1) expression (Fig. 2A). Based on text life table analysis, the 10-year DFS was 53% for low nuclear HIF-1α (1) expression versus 10% for high protein expression (p=0.096). However, when entered into multivariate analysis, nuclear HIF-1α (1) was not significantly independently associated with reduced DFS when combined with age, tumor size, lymph node status, Ki67, molecular subtype, lymphatic vessel invasion, tumor necrosis, Klintrup–Mäkinen grade, tumor stroma percentage, tumor budding, and adjuvant radiotherapy (HR = 0.66 95% CI: 0.27–1.64, p=0.373) (Supplementary Table S2).
Figure 2.
Hypoxic marker expression in ER-positive cohort 1 is significantly associated with survival. Kaplan–Meier curves showing associations between disease-free survival and (A) nuclear HIF-1α (1) in the entire cohort, (B) nuclear HIF-1α (1) in luminal A disease, and (C) cytoplasmic CAIX in the entire cohort. Log-rank test was used. Abbreviations: ER, estrogen receptor; HIF-1α, hypoxia-inducible factor-1α; CAIX, carbonic anhydrase IX.
Chi-square analysis was performed to determine whether the expression of nuclear HIF-1α (1) was associated with any clinicopathological characteristic of the patients. Expression of nuclear HIF-1α (1) was inversely associated with blood vessel invasion (p=0.028) and tumor necrosis (p=0.017), and trends toward significance with patient’s age (p=0.053) and luminal A molecular subtype (p=0.083) as shown in Supplementary Table S3.
To determine whether HIF-1α (1) expression was associated with clinical outcome in ER-positive patients, the cohort was subdivided into luminal A and luminal B tumors. Kaplan–Meier survival curves for cytoplasmic and nuclear expression of HIF-1α (1) were plotted. Patients with high immunostaining for nuclear HIF-1α (1) had significantly poorer DFS (log-rank, p=0.038) in patients with luminal A but not with luminal B disease (log-rank, p=0.720) (Fig. 2B). According to text life table analysis, the 10-year DFS was 46% for low nuclear HIF-1α (1) expression versus 23% for high protein expression (p=0.090). Multivariate analysis suggested that nuclear expression of HIF-1α (1) was an independent prognostic marker for poorer DFS when combined with tumor size, lymph node status, and tumor stroma percentage in luminal A tumor (HR = 0.53 95% CI: 0.30–0.94, p=0.030) (Table 1).
Table 1.
Univariate and Multivariate Analyses for Disease-free Survival of Nuclear HIF-1α (1) and Clinicopathological Characteristics in ER-positive Cohort 1, Luminal A Tumors (n=175).
| Clinicopathological Characteristics | Luminal A Disease | |||
|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| Age (≤50/>50years) | 2.07 (1.17–3.65) | 0.013* | 1.48 (0.77–2.82) | 0.240 |
| Tumor size (≤2/2.1–5/>5 cm) | 1.73 (1.26–2.38) | 0.001* | 1.87 (1.24–2.81) | 0.003* |
| Grade (I/II/III) | 1.21 (0.89–1.66) | 0.229 | — | — |
| Involved lymph node (negative/positive) | 2.09 (1.38–3.19) | 0.001* | 1.75 (1.04–2.95) | 0.037* |
| PR status (negative/positive) | 0.95 (0.61–1.48) | 0.810 | — | — |
| Lymphatic vessel invasion (no/yes) | 1.46 (0.82–2.61) | 0.199 | — | — |
| Blood vessel invasion (no/yes) | 1.50 (0.68–3.34) | 0.315 | — | — |
| Tumor necrosis (low/high) | 1.03 (0.66–1.61) | 0.885 | — | — |
| Klintrup–Mäkinen grade (low/high) | 0.64 (0.45–0.91) | 0.013* | 0.64 (0.41–1.01) | 0.055 |
| CD68+ (low/moderate/high) | 0.94 (0.63–1.39) | 0.743 | — | — |
| CD8+ (low/moderate/high) | 1.07 (0.73–1.58) | 0.713 | — | — |
| CD138+ (low/moderate/high) | 1.04 (0.74–1.46) | 0.820 | — | — |
| Tumor stroma percentage (low/high) | 2.06 (1.35–3.13) | 0.001* | 2.18 (1.32–3.61) | 0.002* |
| Tumor budding (low/high) | 1.39 (0.92–2.12) | 0.122 | — | — |
| Adjuvant endocrine therapy (no/yes/ATAC trial) | 1.43 (0.73–2.84) | 0.300 | — | — |
| Adjuvant chemotherapy (no/yes) | 0.92 (0.58–1.47) | 0.725 | — | — |
| Adjuvant radiotherapy (no/yes) | 0.68 (0.45–1.04) | 0.078 | — | — |
| Nuclear HIF-1α (1) (low/high) | 0.56 (0.32–0.98) | 0.038* | 0.53 (0.30–0.94) | 0.030* |
Abbreviations: HIF-1α (1), hypoxia-inducible factor-1α (1); ER, estrogen receptor; HR, hazard ratio; CI, confidence interval; PR, progesterone receptor; ATAC, Arimidex, Tamoxifen, Alone or in Combination.
Statistically significant p value <0.05.
Of the 373 patients, 37 (10%) had a tissue core missing and so HIF-2α staining could not be carried out, and they were excluded from the analysis. Therefore, expression of HIF-2α was assessed in 336 patients. Nuclear and cytoplasmic staining was detected in tumor cells with HIF-2α expression. Representative profiles of immunostainings of cytoplasmic and nuclear HIF-2α with examples of weak and strong staining can be seen in Supplementary Fig. S2A. Histograms showing the distribution of histoscores for cytoplasmic HIF-2α ranged from 0 to 247.5 with a mean score of 143.67 and for nuclear ranged from 0 to 300 with a mean score of 155.67, and data were relatively normally distributed as shown in histogram plot (Supplementary Fig. S2B). An ICCC value of 0.884 and 0.867 for cytoplasmic and nuclear, respectively, was obtained between both observer scores, and data were visualized in the form of a scatter plot and a Bland–Altman plot (Supplementary Fig. S2C). A density and scatter plot were constructed to visualize the threshold of 113 for cytoplasmic and 173 for nuclear HIF-2α (Supplementary Fig. S2D).
Kaplan–Meier survival curves for cytoplasmic and nuclear expression of HIF-2α were plotted. The log-rank test was used to compare low and high protein expression in tumor cells. No association or trend between cytoplasmic HIF-2α expression and DFS (log-rank, p=0.881) was observed. In contrast, high nuclear HIF-2α expression showed a trend toward prolonged DFS (log-rank, p=0.086). Because there was no significant association with survival, HIF-2α would not be investigated further.
The expression of CAIX in tumor cell was examined in 353 of 373 patients. Twenty (5%) patients were excluded from the analysis due to a tissue core missing. Cytoplasmic and membranous staining was detected in tumor cells with CAIX expression. Representative profiles of immunostainings can be seen in Supplementary Fig. S3A. Weighted histoscores for cytoplasmic expression ranged from 0 to 217.5 with a mean score of 5.53 and for membranous scores ranged from 0 to 260 with a mean score of 6.72. A histogram was plotted to visualize the range of scores, and data showed a positively skewed pattern (Supplementary Fig. S3B). There was good correlation between observers with an ICCC score of 0.986 and 0.987 for cytoplasmic and membranous expression, respectively. Validation was visualized by plotting a scatter plot and a Bland–Altman plot (Supplementary Fig. S3C). This method yielded an optimum threshold score of 18 and 30 for cytoplasmic and membranous expression (Supplementary Fig. S3D).
Kaplan–Meier survival curves for cytoplasmic and membranous expression of CAIX were plotted. As shown in Fig. 2C, the expression of cytoplasmic CAIX was associated with poorer DFS (log-rank, p=0.015), whereas membranous CAIX expression failed to reach statistical significance with DFS (log-rank, p=0.262). In multivariate logistic regression analysis, cytoplasmic CAIX was not significant independently associated with worse DFS in full cohort when combined with age, tumor size, lymph node status, Ki67, molecular subtype, lymphatic vessel invasion, tumor necrosis, Klintrup–Mäkinen grade, tumor stroma percentage, tumor budding, and adjuvant radiotherapy (HR = 1.04 95% CI: 0.46–2.35, p=0.926) (Table 2).
Table 2.
Univariate and Multivariate Analyses for Disease-free Survival of Cytoplasmic CAIX and Clinicopathological Characteristics in the Entire ER-positive Cohort 1 (n=373).
| Clinicopathological Characteristics | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| Age (≤50/>50years) | 1.73 (1.16–2.57) | 0.007* | 2.56 (1.34–4.88) | 0.004* |
| Tumor size (≤2/2.1–5/>5 cm) | 1.59 (1.24–2.03) | <0.001* | 1.01 (0.67–1.52) | 0.973 |
| Grade (I/II/III) | 1.22 (0.97–1.53) | 0.089 | — | — |
| Involved lymph node (negative/positive) | 2.29 (1.67–3.15) | <0.001* | 2.29 (1.40–3.77) | 0.001* |
| PR status (negative/positive) | 0.89 (0.64–1.23) | 0.477 | — | — |
| Her-2 status (negative/positive) | 1.52 (0.97–2.38) | 0.070 | — | — |
| Ki67 index (low/high) | 1.68 (1.22–2.32) | 0.002* | 3.11 (1.92–5.04) | <0.001* |
| Molecular subtype (luminal A/luminal B) | 1.63 (1.19–2.24) | 0.003* | 0.90 (0.22–3.81) | 0.891 |
| Lymphatic vessel invasion (no/yes) | 1.98 (1.32–2.96) | 0.001* | 1.69 (1.05–2.74) | 0.033* |
| Blood vessel invasion (no/yes) | 1.69 (0.98–2.95) | 0.059 | — | — |
| Tumor necrosis (low/high) | 1.48 (1.09–2.03) | 0.014* | 1.78 (1.09–2.92) | 0.022* |
| Klintrup–Mäkinen grade (low/high) | 0.78 (0.62–0.98) | 0.036* | 0.24 (0.15–0.41) | <0.001* |
| CD68+ (low/moderate/high) | 1.07 (0.80–1.42) | 0.654 | — | — |
| CD8+ (low/moderate/high) | 0.77 (0.59–1.02) | 0.066 | — | — |
| CD138+ (low/moderate/high) | 1.17 (0.92–1.48) | 0.200 | — | — |
| Tumor stroma percentage (low/high) | 1.84 (1.34–2.54) | <0.001* | 1.61 (1.01–2.57) | 0.047* |
| Tumor budding (low/high) | 1.46 (1.06–1.99) | 0.019* | 1.43 (0.89–2.27) | 0.133 |
| Adjuvant endocrine therapy (no/yes/ATAC trial) | 1.12 (0.69–1.82) | 0.658 | — | — |
| Adjuvant chemotherapy (no/yes) | 0.99 (0.71–1.39) | 0.985 | — | — |
| Adjuvant radiotherapy (no/yes) | 0.71 (0.52–0.98) | 0.035* | 0.67 (0.41–1.10) | 0.116 |
| Cytoplasmic CAIX (low/high) | 1.81 (1.12–2.92) | 0.018* | 1.04 (0.46–2.35) | 0.926 |
Abbreviations: CAIX, carbonic anhydrase IX; ER, estrogen receptor; HR, hazard ratio; CI, confidence interval; PR, progesterone receptor; ATAC, Arimidex, Tamoxifen, Alone or in Combination.
Statistically significant p value <0.05.
Chi-square analysis was performed to determine whether the expression of cytoplasmic CAIX was associated with any clinicopathological characteristic of the patients in ER-positive cohort 1 as shown in Supplementary Table S4. Overexpression of cytoplasmic CAIX was significantly associated with tumor size (p=0.045), lymph node positivity (p=0.007), high proliferative index (p=0.004), luminal B subtypes (p=0.04), high tumor necrosis (p=0.01), and low CD8+ infiltrate (p=0.03). No other associations were observed, but high levels of CAIX had an association of borderline significance with Her-2 negativity (p=0.071) and no adjuvant chemotherapy received (p=0.059).
To determine whether cytoplasmic CAIX expression was associated with clinical outcome in specific ER-positive subtype, the cohort was subdivided into luminal A and luminal B tumors. Patients with high cytoplasmic CAIX had a trend toward an association with poorer DFS (log-rank, p=0.078) in luminal B disease but not in luminal A tumor (log-rank, p=0.464).
Chi-square analysis was used to examine the possible associations between hypoxic markers. CAIX expression was not significantly associated with HIF-1α (1) expression. However, there was a significant association between nuclear and cytoplasmic HIF-1α (1) and cytoplasmic and membranous CAIX (both p<0.001) (Table 3).
Table 3.
Association Between Hypoxic Markers.
| Hypoxic Markers | ER-positive Cohort 1 | ER-positive Cohort 2 | ||||
|---|---|---|---|---|---|---|
| Nuclear HIF-1α (1) | Cytoplasmic CAIX | Membranous CAIX | Nuclear HIF-1α (1) | Cytoplasmic CAIX | Membranous CAIX | |
| Cytoplasmic HIF-1α (1) | <0.001* | 0.304 | 0.916 | <0.001* | 0.366 | 0.035* |
| Nuclear HIF-1α (1) | — | 0.411 | 0.622 | — | 0.432 | 0.071 |
| Cytoplasmic CAIX | — | <0.001* | — | — | <0.001* | |
Abbreviations: ER, estrogen receptor; HIF-1α, hypoxia inducible factor-1α; CAIX, carbonic anhydrase IX.
Statistically significant p value <0.05.
ER-positive Cohort 2
A total of 285 patients who presented with ER-positive early-stage invasive ductal carcinoma were included in the study. Supplementary Table S5 shows the clinicopathological characteristics of patients. The majority of patients (82%) were aged above 50 years, had tumors size ≤20 (48%), and had grade II carcinoma (51%). There were 137 patients with axillary lymph node involvement (52%). A total of 168 patients (61%) had PR-positive tumors and 263 patients (93%) had Her-2 negative tumors. In all, 169 (71%) patients had luminal A disease and 69 (29%) had luminal B disease. Only 71 (25%) patients received chemotherapy and 86 (30%) patients received radiotherapy. Two hundred two patients (71%) had no recurrences, and 82 patients (29%) experienced recurrences. Of these patients, 7 (3%) had bilateral recurrence, 76 cancer-associated deaths and 60 non-cancer deaths.
The expression of HIF-1α (1) was analyzed in 217 patients from ER-positive cohort 2 (n=285). In 68 (24%) patients, there was a tissue core missing and so HIF-1α (1) staining could not be carried out, and they were excluded from the analysis. Kaplan–Meier curves were plotted to visualize association with the expression of HIF-1α (1) and DFS. When patients were split into two groups based on low and high HIF-1α (1), there was statistically significant poorer DFS (log-rank, p=0.032) with high level of cytoplasmic HIF-1α (1) (Fig. 3A). In addition, high nuclear HIF-1α (1) expression was associated with DFS (log-rank, p=0.009) (Fig. 3B). Based on text life table analysis, the 10-year DFS of patients with high nuclear HIF-1α (1) expression was 13% vs 26% with low HIF-1α (1) expression (p=0.220). However, when entered into multivariate Cox regression analysis and comparing them directly against DFS, nuclear but not cytoplasmic HIF-1α (1) retained an independent prognostic value when combined with tumor size, lymph node status, NPI (Nottingham prognostic index), Ki67, and molecular subtype (HR = 1.85 95% CI: 1.10–3.11, p=0.019) (Table 4).
Figure 3.
Hypoxic marker expression in ER-positive cohort 2 is significantly associated with survival. Kaplan–Meier curves showing associations between disease-free survival and (A) cytoplasmic HIF-1α (1) in the entire cohort, (B) nuclear HIF-1α (1) in the entire cohort, (C) nuclear HIF-1α (1) in luminal A disease, (D) cytoplasmic CAIX in the entire cohort, and (E) cytoplasmic CAIX in luminal B disease. Log-rank test was used. Abbreviations: ER, estrogen receptor; HIF-1α, hypoxia inducible factor-1α; CAIX, carbonic anhydrase IX.
Table 4.
Univariate and Multivariate Analyses for Disease-free Survival of HIF-1α (1) and Clinicopathological Characteristics in the Entire ER-positive Cohort 2 (n=285).
| Clinicopathological Characteristics | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| Age (<50/>50) | 1.57 (0.93–2.65) | 0.089 | — | — |
| Size (≤20/21–50/>50 mm) | 1.65 (1.24–2.21) | 0.001* | 2.11 (1.36–3.28) | 0.001* |
| Grade (I/II/III) | 1.25 (0.99–1.59) | 0.066 | — | — |
| Lymph node status (negative/positive) | 1.69 (1.19–2.40) | 0.004* | 1.16 (0.64–2.12) | 0.622 |
| NPI (<3.5/3.5–5.5/>5.5) | 1.63 (1.27–2.09) | <0.001* | 1.25 (0.84–1.86) | 0.273 |
| PR (negative/positive) | 0.78 (0.56–1.09) | 0.152 | — | — |
| Her-2 (negative/positive) | 0.86 (0.42–1.76) | 0.682 | — | — |
| Ki67 (proliferation index) (low/high) | 1.83 (1.24–2.70) | 0.002* | 1.14 (0.26–4.93) | 0.860 |
| Molecular subtype (luminal A/luminal B) | 1.69 (1.16–2.46) | 0.007* | 1.99 (1.23–3.24) | 0.005* |
| Chemotherapy (no/yes) | 0.99 (0.65–1.51) | 0.964 | — | — |
| Radiotherapy (no/yes) | 0.78 (0.54–1.13) | 0.189 | — | — |
| Cytoplasmic HIF-1α (1) (low/high) | 1.54 (1.04–2.29) | 0.032* | 0.89 (0.43–1.87) | 0.774 |
| Nuclear HIF-1α (1) (low/high) | 1.75 (1.14–2.68) | 0.006* | 1.85 (1.10–3.11) | 0.019* |
Abbreviations: HIF-1α, hypoxia inducible factor-1α; ER, estrogen receptor; HR, hazard ratio; CI, confidence interval; PR, progesterone receptor; NPI, Nottingham prognostic index.
Statistically significant p value <0.05.
Chi-square analysis was performed to determine whether the expression of nuclear HIF-1α (1) was associated with any clinicopathological characteristic of ER-positive patients as shown in Supplementary Table S6. The expression of nuclear HIF-1α (1) was associated with increasing patient’s age (p=0.004), tumor size (p=0.017), and positive PR status (p=0.033).
To determine whether the nuclear expression of HIF-1α (1) was associated with clinical outcome in specific ER-positive subtype, the cohort was subdivided into luminal A and luminal B tumors. High nuclear HIF-1α (1) was significantly associated with poorer DFS in luminal A disease (log-rank, p=0.013) (Fig. 3C), but not in luminal B disease (log-rank, p=0.587). In multivariate logistic regression analysis, nuclear expression of HIF-1α (1) was an independent prognostic marker for DFS when combined with NPI in luminal A tumor (HR = 1.98 95% CI: 1.02–3.83, p=0.042) (Table 5).
Table 5.
Univariate and Multivariate Analyses for Disease-free Survival of Nuclear HIF-1α (1) and Clinicopathological Characteristics in ER-positive Cohort 2, Luminal A Tumors (n=169).
| Clinicopathological Characteristics | Luminal A | |||
|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| Age (<50/>50) | 1.97 (0.91–4.27) | 0.087 | — | — |
| Size (≤20/21–50/>50 mm) | 1.34 (0.90–1.99) | 0.151 | — | — |
| Grade (I/II/III) | 1.07 (0.76–1.49) | 0.714 | — | — |
| Lymph node status (negative/positive) | 1.43 (0.92–2.23) | 0.117 | — | — |
| NPI (<3.5/ 3.5-5.5/>5.5) | 1.42 (1.02–1.99) | 0.040* | 1.64 (1.07–2.51) | 0.023* |
| PR status (negative/positive) | 0.92 (0.59–1.43) | 0.705 | — | — |
| Chemotherapy (no/yes) | 0.94 (0.49–1.79) | 0.853 | — | — |
| Radiotherapy (no/yes) | 0.64 (0.38–1.06) | 0.085 | — | — |
| Nuclear HIF-1α (1) (low/high) | 2.04 (1.15–3.62) | 0.013* | 1.98 (1.02–3.83) | 0.042* |
Abbreviations: HIF-1α, hypoxia inducible factor-1α; ER, estrogen receptor; HR, hazard ratio; CI, confidence interval; PR, progesterone receptor; NPI, Nottingham prognostic index.
Statistically significant p value <0.05.
Of the 285 patients, 31 (11%) had a tissue core missing and so CAIX staining could not be carried out, and they were excluded from the analysis. Therefore, the expression of CAIX was assessed in 254 patients. To determine whether CAIX expression was significantly associated with clinical outcome, Kaplan–Meier survival curves for cytoplasmic and membranous expression of CAIX were plotted and low and high expression were compared using the log-rank test. Univariate survival analysis for the entire group of patients showed association between cytoplasmic CAIX and DFS (log-rank, p=0.008) as shown in Fig. 3D. In contrast, no correlation was found with membranous CAIX and poorer DFS (log-rank, p=0.612). Based on text life table analysis, the 10-year DFS of patients with high cytoplasmic CAIX expression compared with patients with low expression was significant (p=0.012). When entered into multivariate analysis, cytoplasmic CAIX was an independent prognostic marker for DFS when combined with tumor size, lymph node status, NPI, Ki67, and molecular subtype (HR = 1.74 95% CI: 1.08–2.82, p=0.023) (Table 6).
Table 6.
Univariate and Multivariate Analyses for Disease-free Survival of Cytoplasmic CAIX and Clinicopathological Characteristics in the Entire ER-positive Cohort 2 (n=285).
| Clinicopathological Characteristics | Cytoplasmic CAIX | |||
|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| Age (<50/>50) (283) | 1.57 (0.93–2.65) | 0.089 | — | — |
| Size (≤20/21–50/>50 mm) | 1.65 (1.24–2.21) | 0.001* | 1.84 (1.25–2.69) | 0.002* |
| Grade (I/II/III) (275) | 1.25 (0.99–1.59) | 0.066 | — | — |
| Lymph node status (negative/positive) | 1.69 (1.19–2.40) | 0.004* | 1.40 (0.91–2.15) | 0.123 |
| NPI (<3.5/3.5–5.5/>5.5) | 1.63 (1.27–2.09) | <0.001* | 1.07 (0.69–1.64) | 0.759 |
| PR (negative/positive) | 0.78 (0.56–1.09) | 0.152 | — | — |
| Her-2 (negative/positive) | 0.86 (0.42–1.76) | 0.682 | — | — |
| Ki67 (proliferation index) (low/high) | 1.83 (1.24–2.70) | 0.002* | 1.65 (1.06–2.57) | 0.026* |
| Molecular subtype (luminal A/luminal B) | 1.69 (1.16–2.46) | 0.007* | 1.03 (0.24–4.39) | 0.974 |
| Chemotherapy (no/yes) | 0.99 (0.65–1.51) | 0.964 | — | — |
| Radiotherapy (no/yes) | 0.78 (0.54–1.13) | 0.189 | — | — |
| Cytoplasmic CAIX (low/high) | 1.64 (1.14–2.37) | 0.008* | 1.74 (1.08–2.82) | 0.023* |
Abbreviations: CAIX, carbonic anhydrase IX; ER, estrogen receptor; HR, hazard ratio; CI, confidence interval; PR, progesterone receptor; NPI, Nottingham prognostic index.
Statistically significant p-value < 0.05.
Chi-square analysis was performed to determine the relationship between cytoplasmic CAIX and various clinicopathological features, as shown in Supplementary Table S7. Overexpression of cytoplasmic CAIX demonstrated only correlation with patient’s age (p= 0.027).
To determine whether CAIX expression was associated with clinical outcome in specific ER-positive subtype, the cohort was subdivided into luminal A and luminal B tumors. High levels of cytoplasmic CAIX were correlated strongly with shortened DFS (log-rank, p=0.001) in patients with luminal B but not with luminal A disease (log-rank, p=0.814) as shown in Fig. 3E. Multivariate logistic regression analysis suggested that cytoplasmic expression of CAIX was an independent prognostic marker for DFS (HR = 2.75 95% CI: 1.66–4.55, p<0.001) (Table 7).
Table 7.
Univariate and Multivariate Analyses for Disease-free Survival of Cytoplasmic CAIX and Clinicopathological Characteristics in ER-positive Cohort 2, Luminal B Tumors (n=69).
| Clinicopathological Characteristics | Luminal B | |||
|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| Age (<50/>50) | 1.73 (0.67–4.46) | 0.259 | — | — |
| Size (≤20/21–50/>50 mm) | 2.38 (1.34–4.21) | 0.003* | 2.26 (1.26–4.06) | 0.006* |
| Grade (I/II/III) | 0.95 (0.55–1.63) | 0.847 | — | — |
| Lymph node status (negative/positive) | 2.25 (0.97–5.19) | 0.058 | — | — |
| NPI (<3.5/3.5–5.5/>5.5) | 1.63 (0.90–2.95) | 0.106 | — | — |
| PR status (negative/positive) | 0.73 (0.39–1.37) | 0.324 | — | — |
| Her-2 status (negative/positive) | 0.49 (0.22–1.08) | 0.078 | — | — |
| Ki67 (proliferation index) (low/high) | 1.42 (0.50–4.03) | 0.507 | — | — |
| Chemotherapy (no/yes) | 0.67 (0.34–1.33) | 0.256 | — | — |
| Radiotherapy (no/yes) | 1.01 (0.50–2.02) | 0.984 | — | — |
| Cytoplasmic CAIX (low/high) | 2.23 (1.38–3.61) | 0.001* | 2.75 (1.66–4.55) | <0.001* |
Abbreviations: CAIX, carbonic anhydrase IX; ER, estrogen receptor; HR, hazard ratio; CI, confidence interval; PR, progesterone receptor; NPI, Nottingham prognostic index.
Statistically significant p value <0.05.
Chi-square analysis was used to examine possible association between markers. There was an association between cytoplasmic HIF-1α (1) and membranous CAIX expression (p=0.035). Also, there was a significant association between cytoplasmic and nuclear HIF-1α (1) (p<0.001), and cytoplasmic and membranous CAIX (p<0.001) (Table 3).
Discussion
To our knowledge, no previous study has carried out a comprehensive analysis of hypoxic marker expression in patients with ER-positive ductal breast carcinoma. This study was carried out in two cohorts of ER-positive ductal tumors. The prognostic role of HIF-1α (1), HIF-2α, and CAIX tumor cell expression in different molecular subtypes (luminal A and luminal B) was examined.
In this study, there were differences in survival associated with nuclear HIF-1α (1) and cytoplasmic CAIX expression seen between luminal A and luminal B patients. High nuclear HIF-1α (1) expression was independently associated with DFS in luminal A subtypes in the two prospective cohort studies. Although this study did not report consistent significance with high cytoplasmic CAIX expression in the tumor cells in ER-positive cohort 1, cytoplasmic CAIX expression was a consistent independent prognosticator in the entire cohort 2 and in luminal B disease. These differences in the clinical outcomes between luminal A and B types might reflect difference in the biology between luminal A and luminal B. However, it may also reflect that luminal B breast cancer subtypes are associated with greater tumor aggressiveness and with significantly worse prognosis than the luminal A subtypes. 36 , 37 Also, luminal B subtypes have high expression of Ki67 (high proliferation rate). 38 Discrepancies in survival between the two cohorts are also probably linked to diverse type of treatment, the lack of power of statistical studies, and limited sample number particularly when the population is divided into too small subgroups. Also, because the samples were derived from different services, differences in the techniques of fixing and preserving the material should be considered, which could contribute to the reduction in antigenicity, decline in the sensitivity of the IHC reaction, and, of course, lower detection of protein expression. 39 , 40
The observation that nuclear HIF-1α (1) is associated with poor survival may be explained by its role in inducing treatment resistance. Previous results link HIF-1α to a worse outcome with tamoxifen resistance in breast cancer patients. 41 A previous study of 187 patients reported an association between HIF-1α expression and poorer DFS in ER-positive but not ER-negative patients. 28 This larger study confirms the prognostic significance of HIF-1α and through using validation cohort shows the independent prognostic value of HIF-1α (1). These findings suggest that the nuclear expression of HIF-1α (1) may be a hallmark of malignancy and is associated with the progression of ER-positive breast carcinoma. However, further molecular and mechanistic investigations are needed to fully elucidate the role of HIF-1α protein in ER-positive tumors.
In this study, although CAIX expression was elevated in approximately 28% of ER-positive breast cancer patients in cohort 2, this percentage was less and associated with poor prognosis of luminal B breast cancers. These results are consistent with previous studies. Ivanova et al. 42 evaluated breast cancer samples of more than 3000 breast cancer patients and showed that a high CAIX level was significantly associated with lower OS in luminal B but not in luminal A. Also, Generali et al. 43 reported that, in women with breast cancer treated with epirubicin and tamoxifen, CAIX expression was associated with lower DFS and OS.
It has been considered that as HIF-1α is only active when located in the nucleus, cytoplasmic staining is of little importance. 27 However, it should be noted that the HIF protein is synthesized and degraded in the cytoplasm. Moreover, it has been reported that there is an HIF-1α variant that is stable even in normoxia and does not translocate to the nucleus under hypoxic conditions. 44 Therefore, it is relevant to examine nuclear and cytoplasmatic HIF-1α as well as cytoplasmatic and membranous CAIX separately.
Although membranous CAIX expression in our patients was significantly associated with cytoplasmic expression of HIF-1α (1), there was no significant association between nuclear HIF-1α (1) and cytoplasmic CAIX. This absence of an association is consistent with other reports 45 – 47 and may reflect that rather than being regulated by hypoxia, HIF-1α expression may be modified by other factors. These factors include alterations in tumor suppressor genes and oncogenes. 48 – 50 HIF-1α may also lose its transcriptional ability such that CAIX induction does not happen despite high expression of HIF-1α. 51 Moreover, CAIX expression may be correlated with HIF-1α expression in tumors where HIF-1α expression is perinecrotic, but not in tumors in which HIF-1α expression is diffuse throughout the tumor. 46 , 52 Also, it may be that the difference in tissue half-lives of HIF-1α (degraded in minutes) 14 and CAIX (degraded in 2–3 days) 53 , 54 accounts for the present results. CAIX expression may also be increased in the absence of HIF-1α by high cell density via the PI3K pathway, 55 and increased expression of CAIX in the absence of hypoxia may also occur with hypomethylation of the CAIX gene promoter. 56 Nevertheless, there was a consistent association between cytoplasmic and nuclear HIF-1α (1), and between cytoplasmic and membranous CAIX indicating reliable methodology.
CAIX is functionally involved in diverse aspects of cancer progression and development. CAIX is important for hypoxic tumor cell survival by regulating acidification of the external tumor microenvironment, allowing cancer cells to adapt and metastasize to other tissues. 57 Recent meta-analyses showed that HIF-1α 58 and CAIX overexpression are predictive of poor prognosis in breast cancer patients. Recently, diagnostic and therapeutic agents targeting HIF-1α and CAIX have been developed. 59 Hypoxia-associated biomarker profiling in advanced breast cancer may provide additional information for staging, clinical decision, and prognosis and potentially have an important part in the development of personalized therapeutic drugs. Indeed, HIF-1α targeting is considered as a novel therapeutic modality for management of breast cancer patients and improving their prognosis, which could be used in combination with currently used therapies. In fact, many small molecules have been reported as HIF-1α inhibitors. 60 Knockdown of HIF-1α expression has been reported to cause complete inhibition of hypoxic induction in breast cancer stem cells. 61 Also, CAIX is under consideration, by both academic and pharmaceutical entities, as a potential target for intervention in breast carcinoma. 62 Assessment of CAIX in tumors before or during therapy may represent a more powerful prognostic and predictive biomarker as well as important targets for breast cancer especially in luminal B, which warrants further investigation.
The main limitation of this study was the relatively small number of patient samples analyzed, limiting the power of the present analysis, and so further confirmation of the present results is required. Compared with CAIX protein which is relatively stable, 21 the HIF proteins undergo a rapid degradation, 45 and this may have impacted the results obtained. This study is one of the few studies that have described the relationship with survival for markers of hypoxia in ER-positive breast cancer. The results showed that nuclear HIF-1α (1) was an independent prognostic factor for DFS in the entire cohort and in luminal A disease. Also, the current study demonstrated that cytoplasmic CAIX was an independent prognosticator for both DFS in the whole cohort and in the patient subpopulation of luminal B disease. This finding suggests that HIF-1α (1) and CAIX are biomarkers with potentially important therapeutic implications, which may help clinician to refine the treatment plan including therapeutic options of luminal B patients.
Supplemental Material
Supplemental material, sj-docx-1-jhc-10.1369_00221554221110280 for The Relationship Between the Tumor Cell Expression of Hypoxic Markers and Survival in Patients With ER-positive Invasive Ductal Breast Cancer by Suad A.K. Shamis, Jean Quinn, Elizabeth E.A. Mallon, Joanne Edwards and Donald C. McMillan in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-docx-2-jhc-10.1369_00221554221110280 for The Relationship Between the Tumor Cell Expression of Hypoxic Markers and Survival in Patients With ER-positive Invasive Ductal Breast Cancer by Suad A.K. Shamis, Jean Quinn, Elizabeth E.A. Mallon, Joanne Edwards and Donald C. McMillan in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-docx-3-jhc-10.1369_00221554221110280 for The Relationship Between the Tumor Cell Expression of Hypoxic Markers and Survival in Patients With ER-positive Invasive Ductal Breast Cancer by Suad A.K. Shamis, Jean Quinn, Elizabeth E.A. Mallon, Joanne Edwards and Donald C. McMillan in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-docx-4-jhc-10.1369_00221554221110280 for The Relationship Between the Tumor Cell Expression of Hypoxic Markers and Survival in Patients With ER-positive Invasive Ductal Breast Cancer by Suad A.K. Shamis, Jean Quinn, Elizabeth E.A. Mallon, Joanne Edwards and Donald C. McMillan in Journal of Histochemistry & Cytochemistry
Acknowledgments
The authors acknowledge the support of NHS Research Scotland (NRS) (NHS GGC Biorepository) for providing access to the clinical specimens and Glasgow Tissue Research Facility for TMA construction.
Footnotes
Author Contributions: JE and DCM conceived and planned the study. SAKS and JQ carried out the experiment. SAKS scored TMAs, analyzed data, and wrote the manuscript. JE aided in double-scoring TMAs. JE and DCM supervised the findings of this work and aided in editing the manuscript. EEAM is a consultant pathologist advising on all pathology and TMA construction. All authors discussed the results, reviewed the manuscript, and contributed to the final version for submission and publication.
Availability of Data and Materials: http://researchdata.gla.ac.uk/784/
Competing interest: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by Libyan government and Libyan Cultural Affairs Bureau, grant number AA274-518-54090.
Ethics Approval: This study was approved by the Research Ethics Committee of the West Glasgow University Hospitals NHS Trust (NHS GG&C REC reference: 16/WS/0207).
ORCID iDs: Jean Quinn  https://orcid.org/0000-0002-8267-0993
https://orcid.org/0000-0002-8267-0993
Joanne Edwards  https://orcid.org/0000-0002-7192-6906
https://orcid.org/0000-0002-7192-6906
Donald C. McMillan  https://orcid.org/0000-0003-4260-5334
https://orcid.org/0000-0003-4260-5334
Contributor Information
Suad A.K. Shamis, Academic Unit of Surgery, School of Medicine, University of Glasgow, Glasgow, United Kingdom; Unit of Molecular Pathology, Wolfson Wohl Cancer Research Centre, Institute of Cancer Sciences, University of Glasgow, Glasgow, United Kingdom.
Jean Quinn, Unit of Molecular Pathology, Wolfson Wohl Cancer Research Centre, Institute of Cancer Sciences, University of Glasgow, Glasgow, United Kingdom.
Elizabeth E.A. Mallon, Department of Pathology, Queen Elizabeth University Hospital, Glasgow, United Kingdom
Joanne Edwards, Unit of Molecular Pathology, Wolfson Wohl Cancer Research Centre, Institute of Cancer Sciences, University of Glasgow, Glasgow, United Kingdom.
Donald C. McMillan, Academic Unit of Surgery, School of Medicine, University of Glasgow, Glasgow, United Kingdom
Literature Cited
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 3. Marić P, Ozretić P, Levanat S, Oresković S, Antunac K, Beketić-Oresković L. Tumor markers in breast cancer–evaluation of their clinical usefulness. Coll Antropol. 2011;35(1):241–7. [PubMed] [Google Scholar]
- 4. Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011;121(10):3786–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J, Shi B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5(10):2929–43. [PMC free article] [PubMed] [Google Scholar]
- 6. Paquet ER, Hallett MT. Absolute assignment of breast cancer intrinsic molecular subtype. J Natl Cancer Inst. 2015;107(1):357. [DOI] [PubMed] [Google Scholar]
- 7. Castoria G, Migliaccio A, Giovannelli P, Auricchio F. Cell proliferation regulated by estradiol receptor: therapeutic implications. Steroids. 2010;75(8–9):524–7. [DOI] [PubMed] [Google Scholar]
- 8. Lumachi F, Luisetto G, Basso SM, Basso U, Brunello A, Camozzi V. Endocrine therapy of breast cancer. Curr Med Chem. 2011;18(4):513–22. [DOI] [PubMed] [Google Scholar]
- 9. Sainsbury R. The development of endocrine therapy for women with breast cancer. Cancer Treat Rev. 2013;39(5):507–17. [DOI] [PubMed] [Google Scholar]
- 10. Goldhirsch A, Colleoni M, Gelber R. Endocrine therapy of breast cancer. Ann Oncol. 2002;13:61–8. [DOI] [PubMed] [Google Scholar]
- 11. Liu CY, Wu CY, Petrossian K, Huang TT, Tseng LM, Chen S. Treatment for the endocrine resistant breast cancer: current options and future perspectives. J Steroid Biochem Mol Biol. 2017;172:166–75. [DOI] [PubMed] [Google Scholar]
- 12. Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu Y, Gong Z, Zhang S, Zhou J, Cao K, Li X, Xiong W, Li G, Zeng Z, Guo C. Role of tumor microenvironment in tumorigenesis. J Cancer. 2017;8(5):761–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11(6):393–410. [DOI] [PubMed] [Google Scholar]
- 14. Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14(6):430–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schito L, Rey S. Hypoxic pathobiology of breast cancer metastasis. Biochim Biophys Acta Rev Cancer. 2017;1868(1):239–45. [DOI] [PubMed] [Google Scholar]
- 16. Wolff M, Kosyna FK, Dunst J, Jelkmann W, Depping R. Impact of hypoxia inducible factors on estrogen receptor expression in breast cancer cells. Arch Biochem Biophys. 2017;613:23–30. [DOI] [PubMed] [Google Scholar]
- 17. Luo W, Wang Y. Epigenetic regulators: multifunctional proteins modulating hypoxia-inducible factor-α protein stability and activity. Cell Mol Life Sci. 2018;75(6):1043–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Masoud GN, Li W. HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5(5):378–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60(24):7075–83. [PubMed] [Google Scholar]
- 20. Potter C, Harris AL. Hypoxia inducible carbonic anhydrase IX, marker of tumour: hypoxia, survival pathway and therapy target. Cell Cycle. 2004;3(2):159–62. [PubMed] [Google Scholar]
- 21. Svastová E, Hulíková A, Rafajová M, Zat’ovicová M, Gibadulinová A, Casini A, Cecchi A, Scozzafava A, Supuran CT, Pastorek J, Pastoreková S. Hypoxia activates the capacity of tumor-associated carbonic anhydrase IX to acidify extracellular pH. FEBS Lett. 2004;577(3):439–45. [DOI] [PubMed] [Google Scholar]
- 22. Parks SK, Chiche J, Pouyssegur J. PH control mechanisms of tumor survival and growth. J Cell Physiol. 2011;226(2):299–308. [DOI] [PubMed] [Google Scholar]
- 23. Jia X, Hong Q, Lei L, Li D, Li J, Mo M, Wang Y, Shao Z, Shen Z, Cheng J, Liu G. Basal and therapy-driven hypoxia-inducible factor-1α confers resistance to endocrine therapy in estrogen receptor-positive breast cancer. Oncotarget. 2015;6(11):8648–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang J, AlTahan A, Jones DT, Buffa FM, Bridges E, Interiano RB, Qu C, Vogt N, Li JL, Baban D, Ragoussis J, Nicholson R, Davidoff AM, Harris AL. Estrogen receptor-α directly regulates the hypoxia-inducible factor 1 pathway associated with antiestrogen response in breast cancer. Proc Natl Acad Sci U S A. 2015;112(49):15172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alam MW, Persson CU, Reinbothe S, Kazi JU, Rönnstrand L, Wigerup C, Ditzel HJ, Lykkesfeldt AE, Påhlman S, Jögi A. HIF2α contributes to antiestrogen resistance via positive bilateral crosstalk with EGFR in breast cancer cells. Oncotarget. 2016;7(10):11238–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kurebayashi J, Otsuki T, Moriya T, Sonoo H. Hypoxia reduces hormone responsiveness of human breast cancer cells. Jpn J Cancer Res. 2001;92(10):1093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kronblad A, Jirström K, Rydén L, Nordenskjöld B, Landberg G. Hypoxia inducible factor-1α is a prognostic marker in premenopausal patients with intermediate to highly differentiated breast cancer but not a predictive marker for tamoxifen response. Int J Cancer. 2006;118(10):2609–16. [DOI] [PubMed] [Google Scholar]
- 28. Generali D, Berruti A, Brizzi MP, Campo L, Bonardi S, Wigfield S, Bersiga A, Allevi G, Milani M, Aguggini S, Gandolfi V, Dogliotti L, Bottini A, Harris AL, Fox SB. Hypoxia-inducible factor-1α expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin Cancer Res. 2006;12(15):4562–8. [DOI] [PubMed] [Google Scholar]
- 29. Yamamoto Y, Ibusuki M, Okumura Y, Kawasoe T, Kai K, Iyama K, Iwase H. Hypoxia-inducible factor 1α is closely linked to an aggressive phenotype in breast cancer. Breast Cancer Res Treat. 2008;110(3):465–75. [DOI] [PubMed] [Google Scholar]
- 30. Helczynska K, Larsson AM, Holmquist Mengelbier L, Bridges E, Fredlund E, Borgquist S, Landberg G, Påhlman S, Jirström K. Hypoxia-inducible factor-2α correlates to distant recurrence and poor outcome in invasive breast cancer. Cancer Res. 2008;68(22):9212–20. [DOI] [PubMed] [Google Scholar]
- 31. Betof AS, Rabbani ZN, Hardee ME, Kim SJ, Broadwater G, Bentley RC, Snyder SA, Vujaskovic Z, Oosterwijk E, Harris LN, Horton JK. Carbonic anhydrase IX is a predictive marker of doxorubicin resistance in early-stage breast cancer independent of HER2 and TOP2A amplification. Br J Cancer. 2012;106(5):916–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brennan DJ, Jirstrom K, Kronblad A, Millikan RC, Landberg G, Duffy MJ, Rydén L, Gallagher WM, O’Brien SL. CA IX is an independent prognostic marker in premenopausal breast cancer patients with one to three positive lymph nodes and a putative marker of radiation resistance. Clin Cancer Res. 2006;12(21):6421–31. [DOI] [PubMed] [Google Scholar]
- 33. Kirkegaard T, Edwards J, Tovey S, McGlynn LM, Krishna SN, Mukherjee R, Tam L, Munro AF, Dunne B, Bartlett JM. Observer variation in immunohistochemical analysis of protein expression, time for a change? Histopathology. 2006;48(7):787–94. [DOI] [PubMed] [Google Scholar]
- 34. Wu L, Yi B, Wei S, Rao D, He Y, Naik G, Bae S, Liu XM, Yang WH, Sonpavde G, Liu R, Wang L. Loss of FOXP3 and TSC1 accelerates prostate cancer progression through synergistic transcriptional and posttranslational regulation of c-MYC. Cancer Res. 2019;79(7):1413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15(2):155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Creighton CJ. The molecular profile of luminal B breast cancer. Biologics. 2012;6:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ades F, Zardavas D, Bozovic-Spasojevic I, Pugliano L, Fumagalli D, de Azambuja E, Viale G, Sotiriou C, Piccart M. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J Clin Oncol. 2014;32(25):2794–803. [DOI] [PubMed] [Google Scholar]
- 38. Zhang MH, Man HT, Zhao XD, Dong N, Ma SL. Estrogen receptor-positive breast cancer molecular signatures and therapeutic potentials. Biomed Rep. 2014;2(1):41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pinder SE, Brown JP, Gillett C, Purdie CA, Speirs V, Thompson AM, Shaaban AM. NCRI Breast Clinical Studies Group. The manufacture and assessment of tissue microarrays: suggestions and criteria for analysis, with breast cancer as an example. J Clin Pathol. 2013;66(3):169–77. [DOI] [PubMed] [Google Scholar]
- 40. O’Hurley G, Sjöstedt E, Rahman A, Li B, Kampf C, Pontén F, Gallagher WM, Lindskog C. Garbage in, garbage out: a critical evaluation of strategies used for validation of immunohistochemical biomarkers. Mol Oncol. 2014;8(4):783–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jögi A, Ehinger A, Hartman L, Alkner S. Expression of HIF-1α is related to a poor prognosis and tamoxifen resistance in contralateral breast cancer. PLoS ONE. 2019;14(12):e0226150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ivanova L, Zandberga E, Siliņa K, Kalniņa Z, Ābols A, Endzeliņš E, Vendina I, Romanchikova N, Hegmane A, Trapencieris P, Eglītis J, Linē A. Prognostic relevance of carbonic anhydrase IX expression is distinct in various subtypes of breast cancer and its silencing suppresses self-renewal capacity of breast cancer cells. Cancer Chemother Pharmacol. 2015;75(2):235–46. [DOI] [PubMed] [Google Scholar]
- 43. Generali D, Fox SB, Berruti A, Brizzi MP, Campo L, Bonardi S, Wigfield SM, Bruzzi P, Bersiga A, Allevi G, Milani M, Aguggini S, Dogliotti L, Bottini A, Harris AL. Role of carbonic anhydrase IX expression in prediction of the efficacy and outcome of primary epirubicin/tamoxifen therapy for breast cancer. Endocr Relat Cancer. 2006;13(3):921–30. [DOI] [PubMed] [Google Scholar]
- 44. Chun YS, Choi E, Kim TY, Kim MS, Park JW. A dominant-negative isoform lacking exons 11 and 12 of the human hypoxia-inducible factor-1α gene. Biochemical J. 2002;362(1):71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tomes L, Emberley E, Niu Y, Troup S, Pastorek J, Strange K, Harris A, Watson PH. Necrosis and hypoxia in invasive breast carcinoma. Breast Cancer Res Treat. 2003;81(1):61–9. [DOI] [PubMed] [Google Scholar]
- 46. Sobhanifar S, Aquino-Parsons C, Stanbridge EJ, Olive P. Reduced expression of hypoxia-inducible factor-1α in perinecrotic regions of solid tumors. Cancer Res. 2005;65(16):7259–66. [DOI] [PubMed] [Google Scholar]
- 47. Kuijper A, van der Groep P, van der Wall E, van Diest PJ. Expression of hypoxia-inducible factor 1 alpha and its downstream targets in fibroepithelial tumors of the breast. Breast Cancer Res. 2005;7(5):R808–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, Simons JW, Semenza GL. Modulation of hypoxia-inducible factor 1α expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60(6):1541–5. [PubMed] [Google Scholar]
- 49. Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1α. Gen Dev. 2000;14(1):34–44. [PMC free article] [PubMed] [Google Scholar]
- 50. Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–75. [DOI] [PubMed] [Google Scholar]
- 51. Vleugel MM, Shvarts D, van der Wall E, van Diest PJ. p300 and p53 levels determine activation of HIF-1 downstream targets in invasive breast cancer. Human Pathol. 2006;37(8):1085–92. [DOI] [PubMed] [Google Scholar]
- 52. Vleugel MM, Greijer AE, Shvarts A, van der Groep P, van Berkel M, Aarbodem Y, van Tinteren H, Harris AL, van Diest PJ, van der Wall E. Differential prognostic impact of hypoxia induced and diffuse HIF-1α expression in invasive breast cancer. J Clin Pathol. 2005;58(2):172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Turner KJ, Crew JP, Wykoff CC, Watson PH, Poulsom R, Pastorek J, Ratcliffe PJ, Cranston D, Harris AL. The hypoxia-inducible genes VEGF and CA9 are differentially regulated in superficial vs invasive bladder cancer. Br J Cancer. 2002;86(8):1276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rafajová M, Zatovicová M, Kettmann R, Pastorek J, Pastoreková S. Induction by hypoxia combined with low glucose or low bicarbonate and high posttranslational stability upon reoxygenation contribute to carbonic anhydrase IX expression in cancer cells. Int J Oncol. 2004;24(4):995–1004. [PubMed] [Google Scholar]
- 55. Kopacek J, Barathova M, Dequiedt F, Sepelakova J, Kettmann R, Pastorek J, Pastorekova S. MAPK pathway contributes to density-and hypoxia-induced expression of the tumor-associated carbonic anhydrase IX. Biochim Biophys Acta. 2005;1729(1):41–9. [DOI] [PubMed] [Google Scholar]
- 56. Nakamura J, Kitajima Y, Kai K, Hashiguchi K, Hiraki M, Noshiro H, Miyazaki K. Expression of hypoxic marker CA IX is regulated by site-specific DNA methylation and is associated with the histology of gastric cancer. Am J Pathol. 2011;178(2):515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Semenza GL. Molecular mechanisms mediating metastasis of hypoxic breast cancer cells. Trends Mol Med. 2012;18(9):534–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shamis SA, McMillan DC, Edwards J. The relationship between hypoxia-inducible factor 1α (HIF-1α) and patient survival in breast cancer: systematic review and meta-analysis. Crit Rev Oncol Hematol. 2021;159:103231. [DOI] [PubMed] [Google Scholar]
- 59. Ward C, Langdon SP, Mullen P, Harris AL, Harrison DJ, Supuran CT, Kunkler IH. New strategies for targeting the hypoxic tumour microenvironment in breast cancer. Cancer Treat Rev. 2013;39(2):171–9. [DOI] [PubMed] [Google Scholar]
- 60. Wigerup C, Påhlman S, Bexell D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol Ther. 2016;164:152–69. [DOI] [PubMed] [Google Scholar]
- 61. Conley SJ, Gheordunescu E, Kakarala P, Newman B, Korkaya H, Heath AN, Clouthier SG, Wicha MS. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci U S A. 2012;109(8):2784–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Supuran CT, Winum JY. Designing carbonic anhydrase inhibitors for the treatment of breast cancer. Expert Opin Drug Discov. 2015;10(6):591–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-jhc-10.1369_00221554221110280 for The Relationship Between the Tumor Cell Expression of Hypoxic Markers and Survival in Patients With ER-positive Invasive Ductal Breast Cancer by Suad A.K. Shamis, Jean Quinn, Elizabeth E.A. Mallon, Joanne Edwards and Donald C. McMillan in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-docx-2-jhc-10.1369_00221554221110280 for The Relationship Between the Tumor Cell Expression of Hypoxic Markers and Survival in Patients With ER-positive Invasive Ductal Breast Cancer by Suad A.K. Shamis, Jean Quinn, Elizabeth E.A. Mallon, Joanne Edwards and Donald C. McMillan in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-docx-3-jhc-10.1369_00221554221110280 for The Relationship Between the Tumor Cell Expression of Hypoxic Markers and Survival in Patients With ER-positive Invasive Ductal Breast Cancer by Suad A.K. Shamis, Jean Quinn, Elizabeth E.A. Mallon, Joanne Edwards and Donald C. McMillan in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-docx-4-jhc-10.1369_00221554221110280 for The Relationship Between the Tumor Cell Expression of Hypoxic Markers and Survival in Patients With ER-positive Invasive Ductal Breast Cancer by Suad A.K. Shamis, Jean Quinn, Elizabeth E.A. Mallon, Joanne Edwards and Donald C. McMillan in Journal of Histochemistry & Cytochemistry