Abstract
Herbicides are vital for modern agriculture, but their utility is threatened by genetic or metabolic resistance in weeds, as well as regulatory barriers. Of the known herbicide modes of action, 7,8-dihydropterin synthase (DHPS), which is involved in folate biosynthesis, is targeted by just one commercial herbicide, asulam. A mimic of the substrate para-aminobenzoic acid, asulam is chemically similar to sulfonamide antibiotics, and although it is still in widespread use, asulam has faced regulatory scrutiny. With an entire mode of action represented by just one commercial agrochemical, we sought to improve the understanding of its plant target. Here we solve a 2.3 Å resolution crystal structure for Arabidopsis thaliana DHPS that is conjoined to 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase (HPPK), and we reveal a strong structural conservation with bacterial counterparts at the sulfonamide-binding pocket of DHPS. We demonstrate that asulam and the antibiotic sulfamethoxazole have herbicidal as well as antibacterial activity, and we explore the structural basis of their potency by modeling these compounds in mitochondrial HPPK/DHPS. Our findings suggest limited opportunity for the rational design of plant selectivity from asulam and indicate that pharmacokinetic or delivery differences between plants and microbes might be the best ways to safeguard this mode of action.
Key words: structural biology, folate metabolism, mode of action
This study presents a 2.3 Å crystal structure for the bifunctional enzyme HPPK/DHPS, the protein target of the herbicide asulam. The binding site for asulam in Arabidopsis thaliana HPPK/DHPS is highly conserved relative to its microbial counterparts, suggesting that the best way to improve the plant specificity of asulam-like DHPS inhibitors may be through different pharmacokinetic or delivery approaches.
Introduction
Asulam is the only commercial herbicide that targets the folate biosynthesis pathway. Although humans obtain folate from their diet, plants and microorganisms synthesize tetrahydrofolic acid (THF) using 6-hydroxymethylpterin, glutamic acid, and para-aminobenzoic acid (p-ABA) as precursors (Hossain et al., 2004). THF and its derivatives methyl-THF, methenyl-THF, and formyl-THF are involved in one-carbon transfer reactions and are part of the methylation cycle and the biosynthesis of DNA and amino acids (Hanson and Gregory, 2002, 2011). In plants, THF is synthesized by the sequential activities of five mitochondrial enzymes, 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase (HPPK), 7,8-dihydropterin synthase (DHPS), dihydrofolate synthetase (DHFS), dihydrofolate reductase (DHFR), and folylpolyglutamate synthetase (FPGS) (Hanson and Gregory, 2011). HPPK initiates tetrahydrofolate biosynthesis by ATP-dependent pyrophosphorylation of 6-hydroxymethyl-7,8-dihydropterin (6-HMDP; Figure 1A). The resulting product is combined with p-ABA by DHPS to yield 7,8-dihydropteroate (7,8-DHP; Figure 1A). To produce THF, 7,8-dihydropteroate undergoes consecutive conjugations with glutamic acid followed by reduction of the pteridine core (Cossins and Chen, 1997).
Figure 1.
HPPK/DHPS is a herbicide target for sulfonamides that mimic DHPS substrate p-ABA.
(A) Schematic of HPPK/DHPS mechanism. HPPK catalyzes ATP-mediated pyrophosphorylation of 6-hydroxymethyl-7,8-dihydropterin (6-HMDP) to yield DHPP. DHPS then adds para-aminobenzoic acid (p-ABA) to DHPP, forming 7,8-dihydropteroate (7,8-DHP).
(B) DHPS substrate p-ABA and its antibiotic and herbicidal mimics sulfanilamide and asulam.
Although several of these steps in folate biosynthesis are targeted by antibacterials, antimalarials, or human chemotherapies, asulam is the only herbicide targeting folate biosynthesis in plants. Asulam is a sulfonamide like the antibacterial sulfa drug sulfanilamide (Figure 1B). By mimicking the DHPS substrate p-ABA, asulam and other sulfa drugs competitively inhibit enzyme activity by forming inactive adducts with 6-HMDP that impede downstream steps in folate synthesis (Roland et al., 1979; Chakraborty et al., 2013). Although they are popular for treating bacterial infections and malaria (Fernández-Villa et al., 2019), resistance to these drugs is seen with mutations to residues capping the DHPS active site, making sulfa drugs that protrude beyond the DHPS surface more susceptible to resistance (Yun et al., 2012; Pornthanakasem et al., 2016; Griffith et al., 2018). Despite the similar structure of sulfa drugs and asulam, no equivalent resistance mutations in plant DHPS have been reported to date according to the International Herbicide-Resistant Weed Database (www.weedscience.org).
Although most bacteria have separate genes for HPPK and DHPS, these proteins are expressed from a single gene in plants to give a conjoined, bifunctional enzyme in which the N-terminal HPPK is connected by a short linker region to DHPS (Prabhu et al., 1997; Rébeillé et al., 1997). Like plants, protozoans including Toxoplasma gondii (Pashley et al., 1997) and Plasmodium spp. (Triglia and Cowman, 1994), also have bifunctional HPPK/DHPS. Some bacteria are unusual in having bifunctional HPPK/DHPS (e.g., Francisella tularensis; Pemble et al., 2010) or a bifunctional dihydroneopterin aldolase (DHNA)-HPPK like that of Streptococcus pneumoniae, in which DHNA catalyzes the formation of the HPPK substrate (Arnaud et al., 2006). Saccharomyces cerevisiae expresses a trifunctional DHNA-HPPK/DHPS (Ulrich et al., 2004; Lawrence et al., 2005).
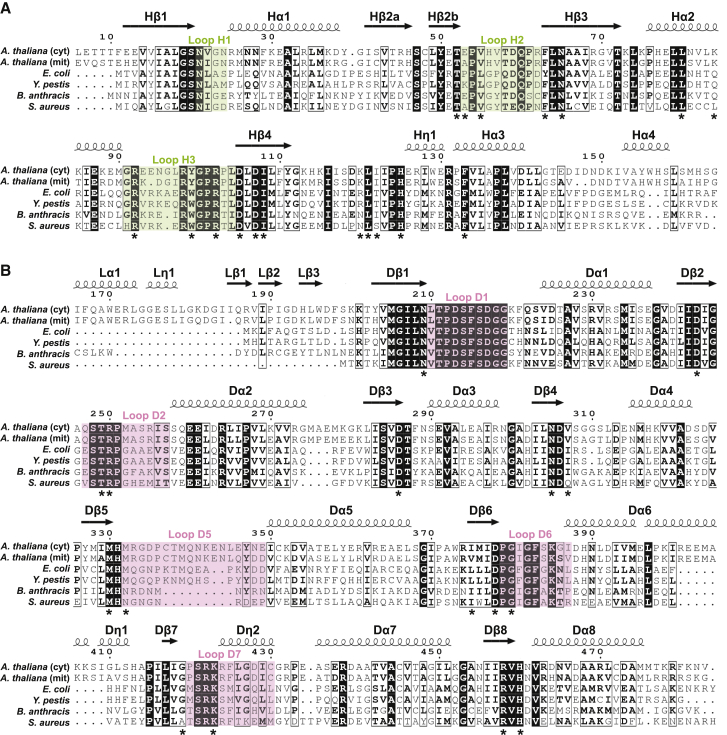
Although the HPPK/DHPS gene is widespread in plants, its protein product has been biochemically characterized only in Pisum sativum (Rébeillé et al., 1997) and Arabidopsis thaliana (Prabhu et al., 1997; Storozhenko et al., 2007). Some plants, such as pea (Rébeillé et al., 1997) and wheat (McIntosh et al., 2008), have only a mitochondrial HPPK/DHPS gene, whereas A. thaliana has two 80% identical genes encoding a mitochondrial (mitHPPK/DHPS, At4g30000) and a cytosolic (cytHPPK/DHPS, At1g69190) form (Storozhenko et al., 2007). MitHPPK/DHPS is directly involved in folate biosynthesis and is essential, whereas the non-essential cytHPPK/DHPS has been shown to play a role in seed germination and stress response by an unknown mechanism that is apparently folate independent (Navarrete et al., 2012). Although non-essential, the A. thaliana cytHPPK/DHPS gene rescued a yeast mutant devoid of HPPK/DHPS, demonstrating in vivo that it has the catalytic capability of the mitochondrial enzyme and suggesting that it has a catalytic role in the cytosol (Storozhenko et al., 2007). Current mechanistic insights into HPPK and DHPS catalysis rely on crystal structures of these enzymes from bacteria, fungi, and protozoa. Whether expressed individually or as a part of multicomponent enzymes, HPPK and DHPS enzymes are structurally conserved across species, with a high sequence similarity of active site residues (Figure 2).
Figure 2.
The HPPK/DHPS active site residues are conserved.
(A and B) Alignment of A. thaliana cytosolic and mitochondrial forms of (A) HPPK and (B) DHPS domains with microbial sequences. Identical (white font in black boxes) and similar residues (bold black font) are noted. Catalytic loops H1–H3 in HPPK are highlighted in green, whereas DHPS catalytic loops D1–D7 are in pink. Residues hypothesized to participate in catalysis are marked with asterisks; β strands (marked as arrows), α helices, and 310 helices (marked as coils) are annotated by the symbols β, α, and η, respectively.
Given the similarity of asulam to antibacterial sulfonamide drugs, it is important to contrast the structures of the plant enzymes with those of bacterial homologs in order to develop novel DHPS inhibitors as herbicides. To better understand the interaction of DHPS and asulam, we solved the crystal structure for A. thaliana cytHPPK/DHPS at a resolution of 2.3 Å. We found that both components are similar to their microbial counterparts, with an overall structural root-mean-square deviation (RMSD) of 1.1–2.3 Å in Cα atoms. We used this structure to model A. thaliana mitHPPK/DHPS, which we were unable to express in a soluble form. We also compared the herbicidal and antibiotic activities of seven sulfonamides (including asulam) to determine which had cross-kingdom efficacy. Overall, the structural data presented here provide insight to help protect this mode of action and highlight HPPK as a new potential herbicide target for the class of folate biosynthesis inhibitors.
Results and discussion
The HPPK/DHPS active site is conserved across kingdoms
Comparing A. thaliana HPPK/DHPS with sequences of microbial species shows conservation of the catalytically relevant HPPK and DHPS regions (Figure 2, Supplemental Figure 1). The DHPS domains in S. cerevisiae and Plasmodium species contain long insertions (Supplemental Figure 1). By contrast, cytDHPS and mitDHPS are closer in length to bacterial DHPS enzymes and share 28%–43% identity (Supplemental Table 1). Secondary structure is conserved between plants and microbes, but the linker region varies in length for microbial species with a bifunctional HPPK/DHPS (Figure 2, Supplemental Figure 1).
Alignments of HPPK/DHPS sequences from throughout the plant kingdom show active site conservation with sequence identity of 48%–80% (Supplemental Figure 2, Supplemental Table 3). For agricultural relevance, we included weed species that are commonly treated with asulam, including the grasses Lolium multiflorum, Echinochloa crus-galii, Cyperus rotundus, and Poa annua (Supplemental Table 3). Collectively, the sequence alignments reveal that the HPPK/DHPS active sites are conserved not only within the plant kingdom but also across microbial species.
Crystal structure of A. thaliana cytosolic HPPK/DHPS
An N-terminally 6-His-tagged A. thaliana cytHPPK/DHPS expressed and purified from Escherichia coli (Supplemental Figure 3) was crystallized and its structure determined by molecular replacement in an unliganded form. Crystals of cytHPPK/DHPS diffracted to a resolution of 2.3 Å and belong to the space group C 2 2 21 with two protein molecules per asymmetric unit (Supplemental Table 3; Figure 3A and 3B). Each monomer consists of an N-terminal HPPK domain (residues 1–160) linked to the C-terminal DHPS domain (residues 203–483) by a structured linker region (residues 161–202). Inspection of the asymmetric unit revealed that the two monomers do not form a dimer. The interaction between the monomers occurs between HPPK catalytic loops H1, H2, and H3 (Figure 2, Figure 3B), which is in contrast to bifunctional HPPK/DHPS structures for Plasmodium falciparum and Plasmodium vivax, which interact at their DHPS domains (Chitnumsub et al., 2020; Lawrence et al., 2005; Yogavel et al., 2018). However, each cytHPPK/DHPS monomer within the asymmetric unit forms a homotypic dimer with the same monomer from a neighboring asymmetric unit (A:A′ and B:B′ rather than A:B). The dimerization interface is via the DHPS domain of each monomer (Figure 3C) through helices Dα6, Dη3, Dα7, and Dα8. Assignment of these interaction interfaces is supported by the PDBePISA server (Krissinel and Henrick, 2007), which predicts that the A:B interaction is not a probable interface (ΔiG p value 0.66), whereas the A:A′ (-x, y, -z+1/2; Figure 3C), or B:B′ (-x, y, -z-1/2; Supplemental Figure 4A) interfaces are strongly predicted as real (ΔiG p values <0.01). Furthermore, the A:A′ and B:B′ dimers are very similar (RMSD 1.2 Å for 423 pairs of Cα atoms), and the observed arrangement resembles previously determined HPPK/DHPS dimer structures (Supplemental Figure 4B) (Lawrence et al., 2005; Yogavel et al., 2018; Chitnumsub et al., 2020). In summary, the asymmetric unit contains two halves of two crystallographic dimers.
Figure 3.
A. thaliana cytHPPK/DHPS forms a crystallographic dimer.
(A) Simplified schematic of cytHPPK/DHPS.
(B) Monomers in the crystallographic asymmetric unit interact at their HPPK (green) domain, which is connected to DHPS (pink) by a structured linker (gray).
(C) Crystallographic dimer between chain A of the asymmetric unit and its symmetry mate A′ with an interface comprising Dα6, Dη3, Dα7, and Dα8.
The two molecules in the asymmetric unit differ mainly in their flexible loop regions (RMSD of 1.2 Å in Cα atoms). In addition, disordered loops account for one region missing electron density in HPPK (chain A, loop H3 residues 93–100) and four regions in DHPS (chain A, loop D1 residues 213–222, loop D2 residues 247–255, residues 309–312, loop D5 residues 333–348; chain B, loop D1 residues 213–221, loop D2 residues 247–258, residues 311–313, loop D5 residues 333–347). Because of the flexibility of the catalytic loops in HPPK and DHPS, they are often disordered (Babaoglu et al., 2004; Lawrence et al., 2005; Pemble et al., 2010), as reflected by missing loops within the structure.
Like its microbial homologs, cytHPPK adopts an αβα ferredoxin-like fold composed of a central, four-strand antiparallel β sheet (Hβ2-Hβ3-Hβ1-Hβ4) sandwiched between four α helices (Hα1-Hα2 on one face and Hη1-Hα3-Hα4 on the other face) (Figure 2, Figure 4A). In contrast to monofunctional HPPKs, the 42-residue linker of cytHPPK/DHPS begins with two successive helices (Lα1 and Lη1) that stabilize the HPPK domain, followed by a short three-residue β strand (Lβ1) that associates with Hβ3 in HPPK, and terminates in a β-hairpin cap (Lβ2-turn-Lβ3) that closes over the N-terminal end of the TIM barrel in DHPS (Figure 2, Figure 3B). The cytDHPS structure has a typical TIM-barrel-like fold (Babaoglu et al., 2004; Lawrence et al., 2005) composed of an eight-stranded β barrel surrounded by eight α helices (Figures 2 and 5A). The β sheet appearance of residues Ile303 to Asp306 in what should be Dβ4 is not defined in the chain B structure, probably owing to the disordered loop connecting this segment to Dα4.
Figure 4.
cytHPPK has a conserved active site flanked by catalytic loops in different conformations.
(A) Superposition of chain A and chain B cytHPPK domains, with catalytic loops 1 to 3 colored yellow (chain A) or orange (chain B). Disordered regions in loop H3 (chain A) are indicated by a dashed line.
(B) HPPK substrates 6-HMDP and the non-hydrolysable ATP-mimic diphosphomethylphosphonic acid adenosyl ester (AMPcPP) from the F. tularensis structure (PDB: 3MCO) are superposed in cytHPPK, with catalytically relevant residues shown as sticks. Ligand atoms are shown in purple (carbon), blue (nitrogen), red (oxygen), orange (phosphorus), and green (magnesium).
(C) Residues that differ between mitHPPK and cytHPPK are highlighted (blue sticks), showing general active site conservation between homologs, with a few labeled exceptions.
Figure 5.
cytDHPS is structurally conserved with microbial counterparts.
(A) Superposition of cytDHPS domains from chain A and chain B shows that both monomers adopt similar conformations. Catalytic loops 1, 2, 5, 6, and 7 are highlighted in dark pink, with regions of disorder indicated by dashed lines.
(B) DHPS substrates DHP+, PPi, and p-ABA from Y. pestis (PDB: 3TYZ) were superposed to show catalytically relevant residues (sticks). Ligand atoms are shown in purple (carbon), blue (nitrogen), red (oxygen), orange (phosphorus), and green (magnesium).
(C) Residues non-identical to mitDHPS are highlighted in blue, revealing active site conservation apart from Glu346 (in disordered loop D5, not observed) and Gly386 (labeled).
In P. falciparum HPPK/DHPS, a patch of positively charged residues adorn the surface between the HPPK and DHPS active sites and are hypothesized to facilitate channeling of the negatively charged DHPP from one site to the other (Supplemental Figure 5) (Chitnumsub et al., 2020). In some microbial parasites, another bifunctional folate biosynthesis enzyme is hypothesized to channel the substrate by a path of positively charged surface residues connecting the active sites of the two domains (Anderson, 2017). Here, for cytHPPK/DHPS and a model of mitHPPK/DHPS generated using the cytHPPK/DHPS crystal structure as a template using iTASSER (Yang et al., 2015a), we found that the domains were connected by a largely electronegative stretch of residues (Supplemental Figure 5), making the hypothesis of substrate channeling less probable. It is worth noting that P. falciparum and P. vivax have multiple long insertions within their HPPK and DHPS domains that alter surface architecture (Supplemental Figure 1) (Chitnumsub et al., 2020; Yogavel et al., 2018). By contrast, the A. thaliana enzymes are closer in length and sequence identity to bacterial monofunctional enzymes (Figure 2; Supplemental Table 2). Enzyme kinetics of P. sativum HPPK/DHPS showed that catalysis of 6-HMDP by HPPK is rate limiting for the production of 7,8-DHP by the bifunctional enzyme (Mouillon et al., 2002). In the absence of p-ABA, the product of HPPK, DHPP accumulated in the medium, but it was rapidly turned over upon the addition of p-ABA (Mouillon et al., 2002). The authors reasoned that substrate channeling seemed less likely given the propensity of DHPP to freely equilibrate with the external media and that its rapid clearance by DHPS was due to high specific activity (Mouillon et al., 2002). Taken together with our structural data, this suggests that substrate channeling may not play a role in HPPK/DHPS activity in A. thaliana.
The cytHPPK catalytic loops adopt different conformations between monomers
Superposition of individual monomers within the cytHPPK/DHPS asymmetric unit revealed differences in the catalytic loops (Figure 4A). HPPK catalysis requires participation from residues in loops H1, H2, and H3 (Figures 2 and 4A) that delineate the enzyme active site. In cytHPPK, loop H1 adopts similar conformations in each chain. In chain B, loop H2 projects outward relative to chain A, and loop H3 projects toward the active site, showing clear electron density for key catalytic residues Arg92 and Arg102. Chain A loop H2 is angled slightly inward, and eight residues in chain A loop H3 are disordered, with Arg92 removed from the active site and Arg102 positioned within the active site (Figure 4A).
To examine active site architecture, cytHPPK chain B was used, as clear electron density was observed for the entire domain. Enzyme substrates 6-HMDP and the non-hydrolysable ATP mimic AMPcPP were superposed onto the cytHPPK site based on structural similarity to the F. tularensis enzyme (RMSD of 1.7 Å in Cα atoms of HPPK domains only) (Figure 4B) (Pemble et al., 2010). The binding site for AMPcPP is situated closer to the surface, where residues Leu80, Lys84, Ile108, Leu121, Ile122, and His125 could stabilize the pocket around the adenosine ring of ATP. The phosphate groups are proposed to be further stabilized by electrostatic interactions with residues Lys84, Arg92, Arg102, His125, and Arg131 (Pemble et al., 2010). As the cytHPPK-binding site is unliganded, Lys84 and His125 are moved slightly away from the active site. The essential Mg+2 cofactors are coordinated by two conserved aspartate residues, Asp105 and Asp107; however, in cytHPPK, Asp105 is pointed away from Asp107 in the absence of Mg+2 atoms. In FtHPPK, this Mg+2 atom can be seen to coordinate the α- and β-phosphates of AMPcPP, and the second Mg+2 atom coordinates the β- and γ-phosphate and orients the 6-hydroxymethyl group of 6-HMDP toward the pyrophosphate moiety of AMPcPP for transfer (Pemble et al., 2010). At the base of the pterin-binding pocket, Phe63 and Phe133 are clearly positioned in a highly conserved π-π stacking interaction where they could flank the pterin ring of 6-HMDP. Hydrogen-bonding interactions between Thr52 and the conserved Asn65 tether 6-HMDP to the base of the pocket by its nitrogen face, and Val55 and Tyr99 complete the binding pocket.
HPPK catalysis is proposed to involve six steps, beginning with Mg+2-ATP binding to HPPK (Blaszczyk et al., 2000). Then, outward movement of loop H3 (by ∼20 Å) triggers key arginine residues (Arg92 and Arg102 in cytHPPK) to complete the active site. Next, 6-HMDP binds, and loops H1 and H3 close around it, with a tryptophan residue (Tyr99 in cytHPPK) moving in to seal the active site closed. After the reaction is completed, loop H3 moves back outward to release the product DHPP.
Based on the currently known reaction trajectory of HPPK catalysis (Blaszczyk et al., 2000), it would appear that chain A in unliganded cytHPPK adopts a conformation either poised to accept 6-HMDP or to release the product DHPP. Chain B, on the other hand, reflects a conformation facilitating catalysis within the active site, as loop H3 is moved inward, with both Arg92 and Arg102 facing the substrate-binding pockets and Tyr99 partially sealing the active site.
To investigate how sequence differences in the mitochondrial HPPK/DHPS of A. thaliana might influence HPPK catalysis, we mapped the different residues onto the cytHPPK structure (highlighted in blue, Figure 4C). Within catalytically relevant loop regions, cytHPPK differs from mitHPPK at six amino acid residues. In cytHPPK, loop H1 residue Val22 (Ile97 in mitHPPK) and loop H3 residue Leu97 (Ile171 in mitHPPK) are similar in size and hydrophobicity. Interestingly, where cytHPPK has Glu53 in loop H2, there is an Ala128 in mitHPPK. Whether a larger, negatively charged residue affects the ability of loop H2 to stabilize the active site pocket in HPPK is unclear. Although most microbial HPPK enzymes have a charged residue at this site, E. coli HPPK has a proline and Staphylococcus aureus an alanine, suggesting that glutamate at this position may not adversely affect enzyme catalysis. In loop H3, where mitHPPK contains two oppositely charged residues (Lys168-Asp169), cytHPPK contains two negatively charged residues followed by a polar uncharged residue (Glu93-Glu94-Asn95). Considering the important role of loop H3 in mediating product binding and release, it is possible that these differences in net charge and the presence of an additional charged residue in cytHPPK alter its interactions with substrate/product sufficiently to affect its activity. Aside from these residues in the catalytic loops, variations between the mitHPPK and cytHPPK largely decorate surfaces away from the active site.
The cytDHPS active site is structurally conserved
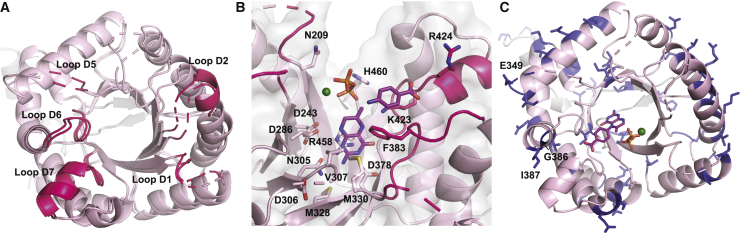
In contrast to the differences between HPPK monomers, superposition of the DHPS crystallographic chains A and B revealed only subtle differences (Figure 5A). Loops D1, D2, D4, and D5 were disordered in both crystallographic monomers, and they are often disordered in DHPS crystal structures. Among the loops connecting individual β sheets to α helices in the TIM barrel, loops D1, D2, D5, D6, and D7 have been implicated in mediating catalysis or stabilizing the active site in microbial DHPS (Babaoglu et al., 2004; Yun et al., 2012).
To understand the A. thaliana DHPS structure in the context of its bound substrates, we superposed DHP+, pyrophosphate, and p-ABA into the cytDHPS active site (Figure 5B) based on a crystal structure of the substrate-bound DHPS from Yersinia pestis (RMSD of 1.2 Å in Cα atoms) (Yun et al., 2012). When crystals of Y. pestis DHPS were soaked in DHPP and p-ABA, the pyrophosphoryl group from DHPP was cleaved and remained in the active site (Yun et al., 2012). Comparison with microbial DHPS shows these two binding pockets in cytDHPS: a pterin-binding site at the opening of the β-barrel, and a p-ABA-binding site closer to the surface formed by loops D1, D2, and D7.
The pterin-binding pocket contains two aspartate residues (Asp286 and Asp378) proposed to stabilize the resonance forms of the pterin ring during pyrophosphoryl cleavage, with the latter residue essential for DHPP binding (Yun et al., 2012). In addition to Asp286 and Asp378, Asn305 and Lys423 would form a part of the hydrogen bond network stabilizing the pterin ring (Yun et al., 2012). In the unliganded cytDHPS, Arg458 overlaps the pterin-binding site; in a substrate-bound structure, this arginine would probably engage in π-stacking interactions parallel to the pterin ring such that it could also interact with the pyrophosphate moiety. The superposed pyrophosphate group sits in an adjacent cleft composed of Ser214, Ser216, and Asp217 (from loop D1), Ser248 and Thr249 (from loop D2), and Arg458 and His460, which form the anion-binding pocket (Yun et al., 2012). At this site, a Mg+2 ion would form part of a coordinated network between the pyrophosphate oxygen atoms, Asn209, and water molecules. This Mg+2 ion is proposed to assist pyrophosphate release from the DHPS active site, as well as stabilize the substructure formed by catalytic loops D1 and D2 (Yun et al., 2012; Bourne, 2014).
Key interacting residues in the p-ABA-binding pocket include Phe215 (loop D1), Pro251 (loop D2), Phe383 (loop D6), and Lys423 (loop D7). In YpDHPS, the p-ABA carboxylate group hydrogen bonds to a neighboring serine residue in loop D7 (Arg424 in cytDHPS) and is stabilized by the helix dipole of Dα7 (Yun et al., 2012). In cytDHPS, the involvement of functionally critical loops D1 and D2 in p-ABA binding cannot be seen owing to loop disorder; however, Phe383, Lys423, and Arg424 can be seen to adopt orientations similar to those of their counterparts in YpDHPS.
Drawing from a series of bacterial DHPS structures crystallized with various substrate/products, a mechanism was proposed for 7,8-DHP synthesis (Babaoglu et al., 2004; Yun et al., 2012). The reaction begins with DHPP binding, which stabilizes mobile catalytic loops, including loops D1 and D2, into forming the p-ABA-binding pocket. Pyrophosphate cleavage from DHPP occurs first by an SN1 reaction, with both molecules retained in their respective binding pockets. Mg+2 facilitates departure of the pyrophosphate from the active site, allowing p-ABA to be covalently attached to the unphosphorylated pterin to yield 7,8-DHP (reaction scheme in Figure 1).
It is proposed that, in the absence of pterin substrate, its binding site is occupied by an arginine residue in loop D2 (Arg250 in cytDHPS) where the guanidinium group of arginine coordinated with water molecules mimic the pterin substrate. This conformation was observed in native Bacillus anthracis DHPS, where loop D2 also blocked access to the p-ABA-binding site (Babaoglu et al., 2004). It is hypothesized that DHPP entry displaces the Arg-containing loop D2, which now forms the p-ABA-binding pocket. Following product formation, this loop is suggested to swing back in to displace product from the active site. This is predicted to be a mechanism to prevent p-ABA binding prior to pterin binding, as this would occlude the pterin-binding site and hinder catalysis (Babaoglu et al., 2004). This might explain our inability to crystallize DHPS with asulam alone. Considering that cytDHPS is unliganded, it could be expected that loop D2 would be moved in toward the pterin-binding pocket. However, several crystal structures of microbial DHPS enzymes also display disorder in loop D2 in their native and ligand-bound forms (Babaoglu et al., 2004; Morgan et al., 2011; Yun et al., 2012). This could be due to the sheer flexibility of the longer catalytic loops and the associated difficulty of capturing a crystal structure of DHPS in a catalytically relevant conformation.
The DHPS active site and the base of the pterin-binding pocket in particular are almost completely conserved across species. As with A. thaliana cytHPPK, we mapped onto the cytosolic DHPS structure those sites at which amino acid sequences differed from the mitochondrial enzyme (Figure 5C). Within the cytDHPS active site, there is a Glu346 (Gln417 in mitDHPS) and a Glu349 (Asp420 in mitDHPS) in loop D5 and a Gly386 (Ser457 in mitDHPS) and an Ile387 (Val458 in mitDHPS) in loop D6. All other variable residues were strictly either on the outside of the enzyme surface or facing away from the substrate-binding sites, hinting at the structural basis for why cytHPPK/DHPS can fully complement the function of its mitochondrial counterpart in S. cerevisiae (Storozhenko et al., 2007).
Sulfonamides demonstrate cross-kingdom activity against A. thaliana and E. coli
Based on the sequence and structural similarity of A. thaliana cytDHPS to microbial DHPS, sulfonamides could be expected to exhibit similar in vivo inhibition of plants and microbes. Accordingly, we contrasted the herbicidal activity of selected sulfonamides with varying R groups against their antibacterial activity (Figure 6). Our results show that asulam is indeed the most potent herbicide among tested sulfonamides, displaying A. thaliana growth inhibition from 0.25 mM onward after pre-emergence treatment (Figure 6). Sulfacetamide and sulfamethoxazole, both commercially used antibacterials, also demonstrate reasonable potency, although their herbicidal activity is approximately four- to eight-fold weaker than that of asulam (Figure 6). Conversely, although sulfamethoxazole strongly inhibits E. coli growth (minimum inhibitory concentration (MIC) of 0.031 mM), inhibition by asulam and sulfacetamide is weaker at 0.125 mM and 0.25 mM, respectively (Figure 6C). Compared with sulfamethoxazole, sulfadoxine demonstrated weaker herbicidal and antibiotic activities (Figure 6). Among the sulfonamides tested, sulfamethoxazole was the strongest inhibitor of the gram-positive Bacillus subtilis (MIC of 0.25 mM), whereas asulam and sulfacetamide were not effective within the tested range. It is also interesting to note that altering an R group from a ketone (as in sulfacetamide) to an ester (as in asulam) enhanced the herbicidal and antibacterial effect of this class of DHPS inhibitors (Figure 6). Although the uptake and transport of sulfonamides may vary between plants and microbes, as a class of inhibitors, they consistently demonstrate cross-kingdom potency.
Figure 6.
Asulam displays the strongest herbicidal activity against A. thaliana among a range of sulfonamides.
(A and B) Pre- and post-emergence herbicidal activities are shown for (A) sulfonamides (sulfanilamide, 1; sulfacetamide, 2; asulam, 3; sulfamethoxazole, 4; sulfadiazine, 5; sulfamethazine, 6; and sulfadoxine, 7) and (B) the herbicide controls glyphosate and oryzalin.
(C) Minimum inhibitory concentration (mM) for DHPS inhibitors.
Asulam has similar physicochemical properties to sulfonamide antibiotics
To investigate whether the lack of herbicidal activity by some sulfonamides could be a result of their physicochemical properties, we compared them with those of 359 commercial herbicides (Supplemental Figure 6) (Sukhoverkov et al., 2021). The sulfonamides we tested largely cluster together within the expected range of commercial herbicides (Supplemental Figure 6). Overall, the cluster analysis shows that sulfonamides, including asulam, have similar physicochemical properties that probably underpin their cross-kingdom activity and are unlikely to be the basis for the individual herbicidal potencies of asulam, sulfacetamide, and sulfamethoxazole.
A model of mitDHPS and asulam shows the difficulty in developing plant-specific inhibitors
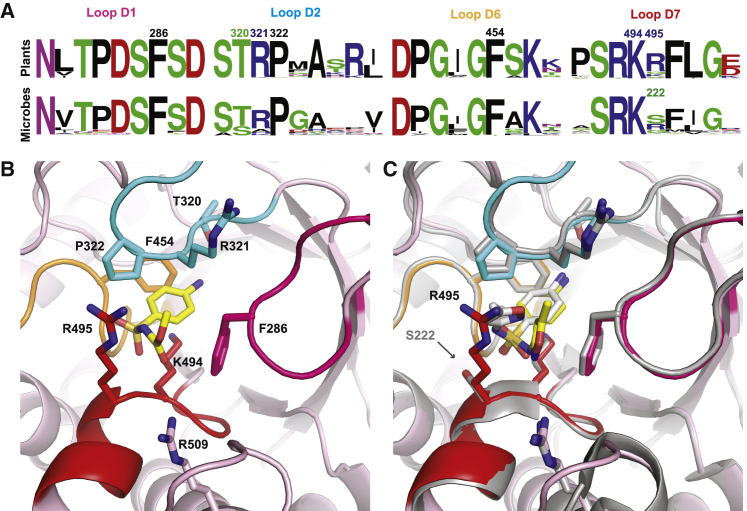
To study the conservation of DHPS residues comprising the p-ABA/sulfonamide-binding pocket, we examined key regions of catalytic loops D1, D2, D6, and D7 from 40 microbial species (Supplemental Table 2) and 40 plant species (Supplemental Table 3) using the WebLogo server (Crooks et al., 2004). These sequence logos illustrate which residues are conserved across plants and microbes (Figure 7A). To visualize the positioning of these residues relative to asulam, we created a mitDHPS homology model with SWISS-MODEL (Waterhouse et al., 2018) based on a Y. pestis DHPS holoenzyme crystal structure in which the enzyme was captured in a catalytically active conformation (Yun et al., 2012). Using AutoDock Vina (Trott and Olson, 2010), the sulfonamides asulam and sulfamethoxazole were docked into the p-ABA-binding pocket, resulting in poses comparable to related structures and predicted binding affinities of −5.7 and −6.5 kcal mol−1, respectively (Figure 7B and 7C).
Figure 7.
The sulfonamide-binding pocket is conserved between plants and microbes.
(A) Relative abundance of amino acid residues within subsections of DHPS catalytic loops D1, D2, D6, and D7 that form the p-ABA-binding pocket. Contact residues that interact with p-ABA or sulfonamides are labeled with mitDHPS numbering. Arg509 falls within Dα7, adjacent to loop D7.
(B) mitDHPS model with asulam docked in the p-ABA-binding pocket within range of catalytic loop residues (colored as in A). Ligand atoms are shown in yellow (carbon), blue (nitrogen), and red (oxygen).
(C) mitDHPS:sulfamethoxazole model superposed with a sulfamethoxazole-bound Y. pestis DHPS crystal structure (shown in gray; PDB: 3TZF) showing conservation of contact residues except for Ser222 in YpDHPS. Carbon atoms in sulfamethoxazole are shown in yellow (docked molecule) or gray (YpDHPS-bound crystal structure).
The p-ABA-binding pocket is formed by loops D1, D2, D6, D7, and the N-terminal region of Dα7. Key residues contributing to stabilizing p-ABA or sulfonamides at this site in mitDHPS would be Phe286 (loop D1); Thr320, Arg321, and Pro322 (loop D2); Gly453 and Phe454 (loop D6); and Lys494 and Arg495 (loop D7) (Figure 7A and 7B). Arg509 on Dα7 is within 3.5 Å of the docked sulfonamides and would be expected to assist Arg495 in stabilizing the carboxylate moiety (or SO2 group in sulfonamides) by inference from the YpDHPS structure bound to p-ABA or sulfamethoxazole (Yun et al., 2012). The catalytic loops in the mitDHPS model presented in Figure 7 adopt a closed conformation as in the DHPS holoenzyme (Yogavel et al., 2018; Yun et al., 2012), whereas an unliganded E. coli DHPS crystal structure captures loop D1 in an open conformation (Supplemental Figure 7) (Achari et al., 1997).
Superposition of the sulfamethoxazole-bound YpDHPS crystal structure to the mitDHPS:sulfamethoxazole model reveals absolute conservation of key residues except for Ser221 in YpDHPS at the site of Arg495 (Figure 7C). Although many plants have an arginine residue at this position, serine residues are also observed (Figure 7A), as in the weed L. multiflorum (Supplemental Figure 2). In contrast to the large, positively charged Arg495, a serine residue at this position (as observed in Y. pestis and E. coli; see Figure 7C and Supplemental Figure 7) can potentially accommodate larger substituents and may explain our observation that sulfadiazine was not herbicidal but was more antibacterial than asulam (Figure 6). This position is a frequent site of mutation in microbial DHPS that results in sulfonamide resistance, which predominantly arises from mutations within loops D1, D2, and D7 (Yun et al., 2012; Yogavel et al., 2018; Chitnumsub et al., 2020). It has been noted that sulfa drugs with substituents extending beyond the van der Waals surface of DHPS are more likely to select for resistance mutations, such as sulfadoxine, whose bulky substituent moves catalytic loop D2 into a partially open conformation less able to stabilize inhibitor binding (Yun et al., 2012; Chitnumsub et al., 2020). Accordingly, lead compounds mimicking p-ABA must be designed to stay within the overall surface of DHPS, and the role of loop D2 in mediating drug affinity as well as resistance must be considered. Interestingly, in the sulfamethoxazole-bound YpDHPS structure, the R substituent still permits loop D2 to adopt a closed conformation covering the active site. Our findings that sulfamethoxazole, sulfacetamide, and asulam demonstrated potent herbicidal and antibiotic activity could be a result of the similar size of their R groups, which are smaller than those of sulfadoxine and sulfamethazine (Figure 6) and less likely to disrupt the stabilizing influence of loop D2. Failed attempts at docking sulfadoxine and sulfamethazine to mitDHPS may also be due to an inability to accommodate large conformational changes in loops D2 and D1 that may occur in the vicinity of their bulky R groups.
Exploring alternative HPPK/DHPS inhibitors as herbicides
Microbial drug discovery efforts have targeted the p-ABA- and/or pterin-binding site of DHPS (Hammoudeh et al., 2014; Dennis et al., 2018) with the pterin-like molecules pyrimido[4,5-c]pyridazines (Zhao et al., 2012) and derivatives of 5-nitroisocytosine (Babaoglu et al., 2004). Pterin-based DHPS inhibitors, although potent enzyme inhibitors in vitro (Zhao et al., 2012; Dennis et al., 2018), are often poor antibacterials in vivo (Lever et al., 1985, 1986). To determine whether pterin-based inhibitors have potential as herbicides, we synthesized a series of guanine-based inhibitors that mimic the pterin substrate of both HPPK and DHPS. Although compounds like these have promise as leads against microbial enzymes (Hevener et al., 2010; Dennis et al., 2014, 2018; Shaw et al., 2014), we found them to have low solubility, which could be a contributing factor to their complete lack of herbicidal and antibacterial activity (Supplemental Figure 8; Supplemental Table 4). However, analysis of the physicochemical properties of these pterins revealed that they are within the expected range of commercial herbicides (Supplemental Figure 9). Pterin-mimicking inhibitors are less likely to give rise to resistance owing to the absolute conservation of residues at the base of the DHPS active site, and screening a library of suitably substituted pterin mimics might therefore uncover lead compounds that provide plant specificity and might be optimized using the structural data presented here.
Another avenue for herbicide design could be to exploit the opportunities presented by two substrate-binding sites in each of HPPK and DHPS. Herbicides that occupy both substrate sites simultaneously in each individual enzyme could be developed. Indeed, bisubstrate pterin-sulfa conjugate inhibitors targeting DHPS have been shown to have antibacterial activity and bind to Y. pestis DHPS by occupying both the pterin and p-ABA pockets (Zhao et al., 2016). They could be a viable solution for developing potent inhibitors that are less prone to resistance, as the absolute conservation of the pterin-binding pocket in DHPS is less likely to tolerate resistance mutations, even if resistance is more frequent at the sulfonamide-binding pocket. Success with HPPK inhibitors, including bisubstrate HPPK inhibitors that occupy both the pterin pocket and the ATP pocket, has been achieved mostly in vitro and has yet to reveal promising in vivo antibacterial candidates (Shi et al., 2012; Chhabra et al., 2013; Dennis et al., 2014).
In summary, our crystal structure of the bifunctional A. thaliana cytHPPK/DHPS shows that plant enzymes are structurally conserved compared with their microbial counterparts and provides a structural context for the herbicidal activity of asulam and the sulfonamide antibiotics sulfacetamide and sulfamethoxazole. Although studies have investigated the off-target and toxicity effects of asulam, such as its inhibition of mammalian sepiapterin reductase (Yang et al., 2015b), its cross-kingdom activity against soil microbiota is less well studied (European Food Safety Authority et al., 2021). Sulfonamide antibiotics are used in the livestock industry and have been reported to have phytotoxic effects (Liu et al., 2009; Piotrowicz-Cieślak et al., 2010; Cheong et al., 2020). We demonstrated the herbicidal potency of sulfamethoxazole against A. thaliana, and a previous study showed that it was also herbicidal against duckweed (Lemna gibba) (Brain et al., 2008). The conservation of key sulfonamide-interacting residues within the DHPS active site suggests that achieving plant specificity for a p-ABA-mimicking inhibitor may not be trivial. Exploiting differences in pharmacokinetic or delivery mechanisms between plants and microbes might be an effective way to develop greater plant specificity. Although asulam resistance in weeds has not yet been reported, the combined inhibition of HPPK and DHPS could provide a means to preempt resistance.
Materials and methods
Sequence alignments and analysis
HPPK/DHPS sequences were retrieved from NCBI Protein BLAST using A. thaliana mitHPPK/DHPS as the query. Transcriptomes of weed species commonly targeted by asulam in the field were assembled using CLC Genomics Workbench v20.0.3 (Qiagen Aarhus A/S) from data in the NCBI Sequence Read Archive: L. multiflorum (accession SRR1648407) (Czaban et al., 2015), E. crus-galli (SRR8633067) (Fang et al., 2019), C. rotundus (SRR12887711) (Ji et al., 2021), and P. annua (SRR1633980) (Chen et al., 2016). Protein sequences of HPPK-DHPS from these species were obtained using tBLASTn by searching the transcriptomes against the A. thaliana mitHPPK/DHPS protein sequence. Sequences were aligned using Clustal Omega (Sievers et al., 2011) and rendered using the ESPript 3.0 server (https://espript.ibcp.fr) (Robert and Gouet, 2014). Logos of selected sequences within catalytically relevant regions across 40 microbe and plant species (details in Supplemental Tables 2 and 3) were generated using the WebLogo server (Crooks et al., 2004).
Expression and purification of cytHPPK/DHPS
E. coli (Shuffle Express; NEB) containing the plasmid pREP4 (Qiagen) was transformed with an N-terminally His-tagged fusion protein of cytHPPK/DHPS (At1g69190) in the vector pQE30 (Qiagen). Overnight cultures grown at 30°C in LB broth supplemented with 35 μg/ml kanamycin and 100 μg/ml ampicillin were sub-cultured to an optical density at 600 nm (OD600) of 0.6–0.7 and induced overnight at 16°C with 100 μM isopropyl β-D-1-thiogalactopyranoside. The bacterial culture was centrifuged at 4000 g for 15 min at 4°C (Beckman), and the cell pellet was resuspended in 30–40 ml of lysis buffer (50 mM HEPES [pH 8.0], 500 mM sodium chloride, 10 mM imidazole, 10 mM β-mercaptoethanol, 5% glycerol, and lysozyme to a final concentration of 1 mg/ml). After a 30-min incubation on ice, the cell suspension was sonicated on ice for 6 min (40% amplitude, 3-s impulse, 9-s relapse) and centrifuged twice at 11 000 g for 20 min at 4°C (Beckman) to pellet cell debris. The cell extract was clarified by filtering through 0.4-μm and 0.22-μm filters prior to loading onto ∼4 ml of Ni-NTA resin (Profinity IMAC resin, Bio-Rad). To allow the His-tag protein to bind the resin, it was incubated overnight at 4°C on a rolling shaker. To remove contaminant proteins, the resin was loaded into polypropylene gravity columns (Bio-Rad) and washed sequentially with 50 ml of ice-cold lysis buffer and 50 ml of washing buffer containing 50 mM HEPES (pH 8.0), 500 mM sodium chloride, 30 mM imidazole, 10 mM β-mercaptoethanol, and 5% glycerol. CytHPPK/DHPS was eluted from the columns using 10 ml of ice-cold elution buffer containing 50 mM HEPES (pH 8.0), 500 mM sodium chloride, 250 mM imidazole, 10 mM β-mercaptoethanol, and 5% glycerol. The eluate was centrifuged at 11 000 g at 4°C to remove aggregated protein, and the supernatant was concentrated by centrifugation on Amicon Ultra filters (MWCO 10 kDa) to a final volume of 0.2–0.3 ml. To remove excessive imidazole, the protein sample was diluted with ∼30 ml of buffer containing 50 mM HEPES (pH 8.0), 500 mM sodium chloride, 10 mM β-mercaptoethanol, and 5% glycerol and concentrated again to a final volume of 0.6–0.8 ml. To remove aggregates and the remaining traces of imidazole, the protein sample was purified on an S200 size-exclusion column (HiLoad 16/600 Superdex 200 pg, GE) using 50 mM HEPES (pH 8.0), 500 mM sodium chloride, 10 mM β-mercaptoethanol, and 5% glycerol. The cytHPPK/DHPS peak was collected and concentrated by centrifugation on 10 kDa MWCO Amicon Ultra filters (Millipore) to a final volume of 0.2–0.3 ml and kept on ice before reductive methylation was performed. A similar expression and purification approach was attempted for mitHPPK/DHPS (without its 68-residue N-terminal mitochondrial transit peptide), but we were unable to obtain soluble protein.
Reductive methylation of cytHPPK/DHPS
We were unable to crystallize purified cytHPPK/DHPS until its surface lysine residues were reductively methylated according to the protocol described by Tan et al. (2014). Purified cytHPPK/DHPS diluted to 16 mg/ml in 50 mM HEPES (pH 8.0), 500 mM sodium chloride, 10 mM β-mercaptoethanol, and 5% glycerol was mixed gently with 20 μl of 1 M dimethylamine-borane complex (Sigma) per 1 ml of protein solution. Immediately after addition of dimethylamine-borane complex, 40 μl of 1 M formaldehyde per 1 ml of protein solution was added to the reaction mixture and mixed gently. The solution was incubated at 4°C for 2 h, and the procedure was repeated. After 2 h of incubation, an additional 10 μl of 1 M dimethylamine-borane complex per 1 ml of protein solution was added to the reaction mixture, which was incubated at 4°C for 12–14 h. To quench the reaction, 80 μl of 1 M glycine and 6 μl of 1 M dithiothreitol were added to the reaction mixture, and the solution was left on ice for 2 h. To remove unreacted dimethylamine-borane complex, formaldehyde, and high-molecular-weight protein aggregates, the reaction mixture was purified by size-exclusion chromatography on an S200 size-exclusion column (HiLoad 16/600 Superdex 200 pg, GE) and eluted in 20 mM HEPES (pH 8.0), 250 mM sodium chloride, and 2 mM dithiothreitol. The cytHPPK/DHPS peak was collected and concentrated by centrifugation on 10 kDa MWCO Amicon Ultra filters (Millipore). To remove excess salt, the sample was diluted with 20 mM HEPES (pH 8.0), 125 mM sodium chloride, and 2 mM dithiothreitol, then concentrated to ∼10–20 mg/ml for use in crystallization trials.
Crystallization and structure determination of A. thaliana cytHPPK/DHPS
Crystals of reductively methylated cytHPPK/DHPS were grown at 20°C using the sitting drop vapor-diffusion method by mixing reservoir buffer (30%–40% PEG 3350 and 0.2 M sodium fluoride) with protein solution (13–20 mg/ml) in a 1:3 or 1:4 ratio. Prior to screening, crystals were cryo-protected in 25% glycerol, 30%–40% PEG 3350, and 0.2 M sodium fluoride and flash-cooled in liquid nitrogen. X-ray data were collected using the MX2 beamline at the Australian Synchrotron, part of Australia's Nuclear Science and Technology Organisation (ANSTO) (Aragao et al., 2018). The X-ray data were indexed and integrated using iMosflm (Battye et al., 2011), space group assigned using Pointless, and scaled using Aimless (CCP4 package) (Winn et al., 2011). The cytHPPK/DHPS structure was determined by molecular replacement using PHASER (from within the PHENIX package) (Adams et al., 2010), and an ensemble model was generated in Chainsaw (CCP4 package) (Winn et al., 2011) using S. cerevisiae HPPK/DHPS (PDB: 2BMB). The molecular replacement solution was subsequently rebuilt using PHENIX.AUTOBUILD (Terwilliger et al., 2008) and refined in Refmac5 (CCP4 package) (Winn et al., 2011) including TLS and Jelly body options. The data showed some evidence of anisotropy; however, attempts to correct for this did not improve refinement statistics or the quality of electron density, so a conservative isotropic resolution cutoff of 2.3 Å was chosen for refinement. Data processing and refinement statistics are presented in Supplemental Table 1.
Chemical synthesis of pterin-mimic inhibitors
Compounds 8–12 were synthesized in yields ranging from 9%–19% as previously described (Dennis et al., 2014, 2016, 2018; Yun et al., 2014) and purified by C18 chromatography (0%–100% acetonitrile/0.1% trifluoracetic acid in water).
MIC assay
MICs were performed in triplicate using the broth microdilution method (Clinical and Laboratory Standards Institute) (CLSI, 2021) in a 96-well plate with serial dilutions of sulfonamides (AK Scientific), pterin-mimicking inhibitors (synthesis described above), glyphosate (Sigma-Aldrich), and oryzalin (Sigma-Aldrich) in 100 μl of cation-adjusted Mueller–Hinton broth. To account for the dimethyl sulfoxide used to dissolve HPPK/DHPS inhibitors, an equivalent quantity was added to the Mueller–Hinton broth to a final 0.6%–2% concentration. Wells were inoculated with 100 μl of ∼105 cells of E. coli or B. subtilis grown to an OD600 of ∼0.5, with the final inhibitor concentration ranging from 0.04 to 2 mM. MICs were determined by visualizing wells with no growth after incubation of the cells for 20 h at 37°C in a shaker incubator.
Sulfonamide treatments on soil-grown A. thaliana
Approximately 60 A. thaliana Col-0 seeds were sown in pots (63 × 63 × 59 mm) of Seedling Substrate Plus + soil (Bord na Móna Horticulture, Newbridge, Ireland) consisting of Irish peat. Soil was pre-wetted to saturation before sowing and then watered throughout the experiment to maintain adequately moist conditions. No fertilizer was added to the soil. Seeds were cold-treated for 3 days in the dark at 4°C to synchronize germination and then grown in a chamber at 23°C with 55% relative humidity and a 16 h light/8 h dark photoperiod. Sulfonamides (AK Scientific), glyphosate (Sigma-Aldrich), and oryzalin (Sigma-Aldrich) were initially dissolved in DMSO at 20 mg/ml and further diluted in water prior to treatments. The surfactant Brushwet (SST Dandenong, Australia) was added to a final concentration of 0.2 ml/l. The active ingredients glyphosate and oryzalin of commercial herbicides were used as positive controls. Each compound was tested on seeds (pre-emergence) or seedlings (post emergence) by directly spraying the plants in each pot with 0.5 ml of 0.125, 0.25, 0.5, 1, 2, or 4 mM solutions, which are equivalent to ∼27–1562 g/ha a.i. The soil density in each pot was 0.29 g/cm3, based on dry soil. The total volume of herbicide solution per gram of soil was 10 μl/g for pre-emergence and 20 μl/g for post emergence because a single 0.5 ml application was used pre-emergence, whereas two applications of 0.5 ml were used post emergence. The day that the trays were moved into their first long day was considered to be day 0, on which the single pre-emergence treatments were given. Post-emergence treatments were carried out twice post germination, at days 4 and 8, and seedlings were grown for 15 days before photographs were taken. The herbicidal effect of each compound was assessed visually.
Funding
K.V.S. was supported by the Australian Research Training Program scholarship. This work and G.V. were supported by Australian Research Council grant DP190101048 to J.S.M., K.A.S., and J.H., who was also supported by an ARC Discovery Early Career Researcher Award (grant no. DE180101445).
Author contributions
J.S.M., G.V., and K.V.S. conceived the study. K.V.S. performed recombinant protein purification and crystallization. G.V. determined the crystal structure with J.H. and C.S.B. providing input. G.V., J.H., J.S.M., and K.V.S. performed plant and microbial sensitivity testing. K.J.B. and K.A.S. performed chemical syntheses of mercaptoguanine inhibitors. M.F.F. assembled transcriptomes of weed species and determined their HPPK/DHPS sequences. G.V. and J.S.M. wrote the manuscript with input from all authors.
Acknowledgments
This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector. No conflict of interest is declared.
Published: April 9, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information can be found online at Plant Communications Online.
Accession numbers
The atomic factors and structure factors have been deposited in the Protein Data Bank under accession code 7MPY.
Supplemental information
References
- Achari A., Somers D.O., Champness J.N., Bryant P.K., Rosemond J., Stammers D.K. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 1997;4:490–497. doi: 10.1038/nsb0697-490. [DOI] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Section D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.S. Understanding the molecular mechanism of substrate channeling and domain communication in protozoal bifunctional TS-DHFR. Protein Eng. Des. Sel. 2017;30:253–261. doi: 10.1093/protein/gzx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragao D., Aishima J., Cherukuvada H., Clarken R., Clift M., Cowieson N.P., Ericsson D.J., Gee C.L., Macedo S., Mudie N., et al. MX2: a high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 2018;25:885–891. doi: 10.1107/S1600577518003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud G., Colin L., Jeremy P.D. Crystal structure of the bifunctional dihydroneopterin aldolase/6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Streptococcus pneumoniae. J. Mol. Biol. 2006;360:644–653. doi: 10.1016/j.jmb.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Babaoglu K., Qi J., Lee R.E., White S.W. Crystal structure of 7,8-dihydropteroate synthase from Bacillus anthracis: mechanism and novel inhibitor design. Structure. 2004;12:1705–1717. doi: 10.1016/j.str.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Battye T.G.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.W. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczyk J., Shi G., Yan H., Ji X. Catalytic center assembly of HPPK as revealed by the crystal structure of a ternary complex at 1.25 Å resolution. Structure. 2000;8:1049–1058. doi: 10.1016/s0969-2126(00)00502-5. [DOI] [PubMed] [Google Scholar]
- Bourne C.R. Utility of the biosynthetic folate pathway for targets in antimicrobial discovery. Antibiotics (Basel) 2014;3:1–28. doi: 10.3390/antibiotics3010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brain R.A., Ramirez A.J., Fulton B.A., Chambliss C.K., Brooks B.W. Herbicidal effects of sulfamethoxazole in Lemna gibba: using p-aminobenzoic acid as a biomarker of effect. Environ. Sci. Technol. 2008;42:8965–8970. doi: 10.1021/es801611a. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Gruber T., Barry C.E., Boshoff H.I., Rhee K.Y. Para-aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science. 2013;339:88–91. doi: 10.1126/science.1228980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., McElroy J.S., Dane F., Goertzen L.R. Transcriptome assembly and comparison of an allotetraploid weed species, annual bluegrass, with its two diploid progenitor species, Poa supina Schrad and Poa infirma Kunth. Plant Genome. 2016;9:1–11. doi: 10.3835/plantgenome2015.06.0050. [DOI] [PubMed] [Google Scholar]
- Cheong M.S., Seo K.H., Chohra H., Yoon Y.E., Choe H., Kantharaj V., Lee Y.B. Influence of sulfonamide contamination derived from veterinary antibiotics on plant growth and development. Antibiotics (Basel) 2020;9:456. doi: 10.3390/antibiotics9080456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra S., Barlow N., Dolezal O., Hattarki M.K., Newman J., Peat T.S., Graham B., Swarbrick J.D. Exploring the chemical space around 8-mercaptoguanine as a route to new inhibitors of the folate biosynthesis enzyme HPPK. PLoS One. 2013;8:e59535. doi: 10.1371/journal.pone.0059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnumsub P., Jaruwat A., Talawanich Y., Noytanom K., Liwnaree B., Poen S., Yuthavong Y. The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance. FEBS J. 2020;287:3273–3297. doi: 10.1111/febs.15196. [DOI] [PubMed] [Google Scholar]
- CLSI . Clinical and Laboratory Standards Institute; 2021. Performance Standards for Antimicrobial Susceptibility Testing, 31st Edition M100-S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossins E.A., Chen L. Folates and one-carbon metabolism in plants and fungi. Phytochemistry. 1997;45:437–452. doi: 10.1016/s0031-9422(96)00833-3. [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaban A., Sharma S., Byrne S.L., Spannagl M., Mayer K.F., Asp T. Comparative transcriptome analysis within the Lolium/Festuca species complex reveals high sequence conservation. BMC Genomics. 2015;16:1–13. doi: 10.1186/s12864-015-1447-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M.L., Pitcher N.P., Lee M.D., DeBono A.J., Wang Z.C., Harjani J.R., Rahmani R., Cleary B., Peat T.S., Baell J.B., et al. Structural basis for the selective binding of inhibitors to 6-Hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Staphylococcus aureus and Escherichia coli. J. Med. Chem. 2016;59:5248–5263. doi: 10.1021/acs.jmedchem.6b00002. [DOI] [PubMed] [Google Scholar]
- Dennis M.L., Chhabra S., Wang Z.C., Debono A., Dolezal O., Newman J., Pitcher N.P., Rahmani R., Cleary B., Barlow N., et al. Structure-based design and development of functionalized Mercaptoguanine derivatives as inhibitors of the folate biosynthesis pathway enzyme 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Staphylococcus aureus. J. Med. Chem. 2014;57:9612–9626. doi: 10.1021/jm501417f. [DOI] [PubMed] [Google Scholar]
- Dennis M.L., Lee M.D., Harjani J.R., Ahmed M., DeBono A.J., Pitcher N.P., Wang Z.C., Chhabra S., Barlow N., Rahmani R., et al. 8-mercaptoguanine derivatives as inhibitors of dihydropteroate synthase. Chemistry. 2018;24:1922–1930. doi: 10.1002/chem.201704730. [DOI] [PubMed] [Google Scholar]
- European Food Safety Authority. Alvarez F., Arena M., Auteri D., Borroto J., Brancato A., Carrasco Cabrera L., Castoldi A.F., Chiusolo A., Colagiorgi A., et al. Updated peer review of the pesticide risk assessment of the active substance asulam (variant evaluated asulam-sodium) EFSA J. 2021;19:e06921. doi: 10.2903/j.efsa.2021.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J.P., Zhang Y.H., Liu T.T., Yan B.J., Li J., Dong L.Y. Target-site and metabolic resistance mechanisms to penoxsulam in barnyardgrass (Echinochloa crus-galli (L.) P. Beauv) J. Agric. Food Chem. 2019;67:8085–8095. doi: 10.1021/acs.jafc.9b01641. [DOI] [PubMed] [Google Scholar]
- Fernández-Villa D., Aguilar M.R., Rojo L. Folic acid antagonists: antimicrobial and immunomodulating mechanisms and applications. Int. J. Mol. Sci. 2019;20:4996. doi: 10.3390/ijms20204996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith E.C., Wallace M.J., Wu Y., Kumar G., Gajewski S., Jackson P., Phelps G.A., Zheng Z., Rock C.O., Lee R.E., et al. The structural and functional basis for recurring sulfa drug resistance mutations in Staphylococcus aureus dihydropteroate synthase. Front Microbiol. 2018;9:1369. doi: 10.3389/fmicb.2018.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoudeh D.I., Daté M., Yun M.K., Zhang W., Boyd V.A., Viacava Follis A., Griffith E., Lee R.E., Bashford D., White S.W. Identification and characterization of an allosteric inhibitory site on dihydropteroate synthase. ACS Chem. Biol. 2014;9:1294–1302. doi: 10.1021/cb500038g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson A.D., Gregory J.F. Synthesis and turnover of folates in plants. Curr. Opin. Plant Biol. 2002;5:244–249. doi: 10.1016/s1369-5266(02)00249-2. [DOI] [PubMed] [Google Scholar]
- Hanson A.D., Gregory J.F. Folate biosynthesis, turnover, and transport in plants. Annu. Rev. Plant Biol. 2011;62:105–125. doi: 10.1146/annurev-arplant-042110-103819. [DOI] [PubMed] [Google Scholar]
- Hevener K.E., Yun M.-K., Qi J., Kerr I.D., Babaoglu K., Hurdle J.G., Balakrishna K., White S.W., Lee R.E. Structural studies of pterin-based inhibitors of dihydropteroate synthase. J. Med. Chem. 2010;53:166–177. doi: 10.1021/jm900861d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain T., Rosenberg I., Selhub J., Kishore G., Beachy R., Schubert K. Enhancement of folates in plants through metabolic engineering. Proc. Natl. Acad. Sci. USA. 2004;101:5158–5163. doi: 10.1073/pnas.0401342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Liu D., Yang Z. High oil accumulation in tuber of yellow nutsedge compared to purple nutsedge is associated with more abundant expression of genes involved in fatty acid synthesis and triacylglycerol storage. Biotechnol. Biofuels. 2021;14:1–24. doi: 10.1186/s13068-021-01909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lawrence M.C., Iliades P., Fernley R.T., Berglez J., Pilling P.A., Macreadie I.G. The three-dimensional structure of the bifunctional 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase/dihydropteroate synthase of Saccharomyces cerevisiae. J. Mol. Biol. 2005;348:655–670. doi: 10.1016/j.jmb.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Lever O.W., Bell L.N., McGuire H.M., Ferone R. Monocyclic pteridine analogues. Inhibition of Escherichia coli dihydropteroate synthase by 6-amino-5-nitrosoisocytosines. J. Med. Chem. 1985;28:1870–1874. doi: 10.1021/jm00150a019. [DOI] [PubMed] [Google Scholar]
- Lever O.W., Bell L.N., Hyman C., McGuire H.M., Ferone R. Inhibitors of dihydropteroate synthase: substituent effects in the side-chain aromatic ring of 6-[[3-(aryloxy)propyl]amino]-5-nitrosoisocytosines and synthesis and inhibitory potency of bridged 5-nitrosoisocytosine-p-aminobenzoic acid analogues. J. Med. Chem. 1986;29:665–670. doi: 10.1021/jm00155a014. [DOI] [PubMed] [Google Scholar]
- Liu F., Ying G.G., Tao R., Zhao J.L., Yang J.F., Zhao L.F. Effects of six selected antibiotics on plant growth and soil microbial and enzymatic activities. Environ. Pollut. 2009;157:1636–1642. doi: 10.1016/j.envpol.2008.12.021. [DOI] [PubMed] [Google Scholar]
- McIntosh S.R., Brushett D., Henry R.J. GTP cyclohydrolase 1 expression and folate accumulation in the developing wheat seed. J. Cereal Sci. 2008;48:503–512. [Google Scholar]
- Morgan R.E., Batot G.O., Dement J.M., Rao V.A., Eadsforth T.C., Hunter W.N. Crystal structures of Burkholderia cenocepacia dihydropteroate synthase in the apo-form and complexed with the product 7,8-dihydropteroate. BMC Struct. Biol. 2011;11:21. doi: 10.1186/1472-6807-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillon J.-M., Ravanel S., Douce R., Rébeillé F. Folate synthesis in higher-plant mitochondria: coupling between the dihydropterin pyrophosphokinase and the dihydropteroate synthase activities. Biochem. J. 2002;363:313–319. doi: 10.1042/0264-6021:3630313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete O., Van Daele J., Stove C., Lambert W., Van Der Straeten D., Storozhenko S. A folate independent role for cytosolic HPPK/DHPS upon stress in Arabidopsis thaliana. Phytochemistry. 2012;73:23–33. doi: 10.1016/j.phytochem.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Pashley T.V., Volpe F., Pudney M., Hyde J.E., Sims P.F., Delves C.J. Isolation and molecular characterization of the bifunctional hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase gene from Toxoplasma gondii. Mol. Biochem. Parasitol. 1997;86:37–47. [PubMed] [Google Scholar]
- Pemble C.W., Mehta P.K., Mehra S., Li Z., Nourse A., Lee R.E., White S.W. Crystal structure of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase dihydropteroate synthase bifunctional enzyme from Francisella tularensis. PLoS One. 2010;5:e14165. doi: 10.1371/journal.pone.0014165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowicz-Cieślak A.I., Adomas B., Nałecz-Jawecki G., Michalczyk D.J. Phytotoxicity of sulfamethazine soil pollutant to six legume plant species. J. Toxicol. Environ. Health A. 2010;73:1220–1229. doi: 10.1080/15287394.2010.492006. [DOI] [PubMed] [Google Scholar]
- Pornthanakasem W., Riangrungroj P., Chitnumsub P., Ittarat W., Kongkasuriyachai D., Uthaipibull C., Yuthavong Y., Leartsakulpanich U. Role of Plasmodium vivax dihydropteroate synthase polymorphisms in sulfa drug resistance. Antimicrob. Agents Chemother. 2016;60:4453–4463. doi: 10.1128/AAC.01835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu V., Lui H., King J. Arabidopsis dihydropteroate synthase: general properties and inhibition by reaction product and sulfonamides. Phytochemistry. 1997;45:23–27. doi: 10.1016/s0031-9422(96)00793-5. [DOI] [PubMed] [Google Scholar]
- Rébeillé F., Macherel D., Mouillon J.-M., Garin J., Douce R. Folate biosynthesis in higher plants: purification and molecular cloning of a bifunctional 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase/7,8-dihydropteroate synthase localized in mitochondria. EMBO J. 1997;16:947–957. doi: 10.1093/emboj/16.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland S., Ferone R., Harvey R.J., Styles V.L., Morrison R.W. The characteristics and significance of sulfonamides as substrates for Escherichia coli dihydropteroate synthase. J. Biol. Chem. 1979;254:10337–10345. [PubMed] [Google Scholar]
- Shaw G.X., Li Y., Shi G., Wu Y., Cherry S., Needle D., Zhang D., Tropea J.E., Waugh D.S., Yan H., et al. Structural enzymology and inhibition of the bi-functional folate pathway enzyme HPPK–DHPS from the biowarfare agent Francisella tularensis. FEBS J. 2014;281:4123–4137. doi: 10.1111/febs.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G., Shaw G., Liang Y.-H., Subburaman P., Li Y., Wu Y., Yan H., Ji X. Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: new design with improved properties. Bioorg. Med. Chem. 2012;20:47–57. doi: 10.1016/j.bmc.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhenko S., Navarrete O., Ravanel S., De Brouwer V., Chaerle P., Zhang G.-F., Bastien O., Lambert W., Rébeillé F., Van Der Straeten D. Cytosolic hydroxymethyldihydropterin pyrophosphokinase/dihydropteroate synthase from Arabidopsis thaliana: a specific role in early development and stress response. J. Biol. Chem. 2007;282:10749–10761. doi: 10.1074/jbc.M701158200. [DOI] [PubMed] [Google Scholar]
- Sukhoverkov K.V., Corral M.G., Leroux J., Haywood J., Johnen P., Newton T., Stubbs K.A., Mylne J.S. Improved herbicide discovery using physico-chemical rules refined by antimalarial library screening. RSC Adv. 2021;11:8459–8467. doi: 10.1039/d1ra00914a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K., Kim Y., Hatzos-Skintges C., Chang C., Cuff M., Chhor G., Osipiuk J., Michalska K., Nocek B., An H., et al. Salvage of failed protein targets by reductive alkylation. Methods Mol. Biol. 2014;1140:189–200. doi: 10.1007/978-1-4939-0354-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.-W., Read R.J., Adams P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T., Cowman A.F. Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 1994;91:7149–7153. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich G., Gabriele J.K., Christoph H., Adelbert B., Jörn K., Dietmar B., Johannes H.H. Characterization of the Saccharomyces cerevisiae Fol1 protein: starvation for C1 carrier induces pseudohyphal growth. Mol. Biol. Cell. 2004;15:3811–3828. doi: 10.1091/mbc.E03-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Jan Y.H., Mishin V., Richardson J.R., Hossain M.M., Heindel N.D., Heck D.E., Laskin D.L., Laskin J.D. Sulfa drugs inhibit sepiapterin reduction and chemical redox cycling by sepiapterin reductase. J. Pharmacol. Exp. Ther. 2015;352:529–540. doi: 10.1124/jpet.114.221572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogavel M., Nettleship J.E., Sharma A., Harlos K., Jamwal A., Chaturvedi R., Sharma M., Jain V., Chhibber-Goel J., Sharma A. Structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase-dihydropteroate synthase from Plasmodium vivax sheds light on drug resistance. J. Biol. Chem. 2018;293:14962–14972. doi: 10.1074/jbc.RA118.004558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M.-K., Hoagland D., Kumar G., Waddell M.B., Rock C.O., Lee R.E., White S.W. The identification, analysis and structure-based development of novel inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase. Bioorg. Med. Chem. 2014;22:2157–2165. doi: 10.1016/j.bmc.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M.-K., Wu Y., Li Z., Zhao Y., Waddell M.B., Ferreira A.M., Lee R.E., Bashford D., White S.W. Catalysis and sulfa drug resistance in dihydropteroate synthase: crystal structures reveal the catalytic mechanism of DHPS and the structural basis of sulfa drug action and resistance. Science. 2012;335:1110–1114. doi: 10.1126/science.1214641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Hammoudeh D., Yun M.-K., Qi J., White S.W., Lee R.E. Structure-based design of novel pyrimido[4,5-c]pyridazine derivatives as dihydropteroate synthase inhibitors with increased affinity. ChemMedChem. 2012;7:861–870. doi: 10.1002/cmdc.201200049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Shadrick W.R., Wallace M.J., Wu Y., Griffith E.C., Qi J., Yun M.-K., White S.W., Lee R.E. Pterin-sulfa conjugates as dihydropteroate synthase inhibitors and antibacterial agents. Bioorg. Med. Chem. Lett. 2016;26:3950–3954. doi: 10.1016/j.bmcl.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.