Abstract
Detached Arabidopsis thaliana leaves can regenerate adventitious roots, providing a platform for studying de novo root regeneration (DNRR). However, the comprehensive transcriptional framework of DNRR remains elusive. Here, we provide a high-resolution landscape of transcriptome reprogramming from wound response to root organogenesis in DNRR and show key factors involved in DNRR. Time-lapse RNA sequencing (RNA-seq) of the entire leaf within 12 h of leaf detachment revealed rapid activation of jasmonate, ethylene, and reactive oxygen species (ROS) pathways in response to wounding. Genetic analyses confirmed that ethylene and ROS may serve as wound signals to promote DNRR. Next, time-lapse RNA-seq within 5 d of leaf detachment revealed the activation of genes involved in organogenesis, wound-induced regeneration, and resource allocation in the wounded region of detached leaves during adventitious rooting. Genetic studies showed that BLADE-ON-PETIOLE1/2, which control aboveground organs, PLETHORA3/5/7, which control root organogenesis, and ETHYLENE RESPONSE FACTOR115, which controls wound-induced regeneration, are involved in DNRR. Furthermore, single-cell RNA-seq data revealed gene expression patterns in the wounded region of detached leaves during adventitious rooting. Overall, our study not only provides transcriptome tools but also reveals key factors involved in DNRR from detached Arabidopsis leaves.
Key words: single-cell RNA-seq, time-lapse RNA-seq, de novo root regeneration, plant regeneration, wounding, Arabidopsis thaliana
This study provides a high-resolution transcriptome atlas that profiles wound response and root organogenesis during de novo root regeneration from detached Arabidopsis leaves. ROS and ethylene serve as the wound signals to promote regeneration, and BOP1/2, PLT3/5/7, and ERF115 are key genes that regulate root organogenesis.
Introduction
After wounding, many plant organs are able to regenerate adventitious roots in a process known as de novo root regeneration (DNRR), which is widely used in many agricultural techniques such as cutting propagation (De Klerk et al., 1999; Bellini et al., 2014; Liu et al., 2018; Perez-Garcia and Moreno-Risueno, 2018; Sang et al., 2018; Xu, 2018; Druege et al., 2019; Ikeuchi et al., 2019; Mironova and Xu, 2019).
We have developed a simple method for studying DNRR from detached leaves (leaf explants) of Arabidopsis thaliana on hormone-free medium (Chen et al., 2014; Liu et al., 2014). Based on this method, the DNRR framework has been proposed at the physiological, genetic, and molecular levels (Xu, 2018). Upon detachment, leaf explants sense many early signals, including wound signals, stress signals, environmental signals, and the developmental status of the leaf. Guided by these early signals, auxin is produced and transported into regeneration-competent cells (such as those in the vascular procambium and some vascular parenchyma cells) within the wounded region to initiate cell fate transition and root organogenesis.
Jasmonate (JA) has been identified as one of the wound signals that promote DNRR from leaf explants (Zhang et al., 2019b). JA accumulates rapidly after leaf detachment. The JA-mediated wound-signaling pathway directly activates the expression of the AP2/EREBP (APETALA2/ethylene-responsive element binding protein) transcription factor family gene ETHYLENE RESPONSE FACTOR109 (ERF109) at about 10 min after leaf detachment, and ERF109 then promotes DNRR by upregulating genes involved in auxin biosynthesis (Sun et al., 2009; Cai et al., 2014; Zhang et al., 2019b). In addition, another AP2/EREBP family gene, ABSCISIC ACID REPRESSOR1 (ABR1), participates in the wound-signaling pathway to promote DNRR and, together with ERF109, functions in the microRNA165-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL)-mediated leaf age pathway (Ye et al., 2020). However, our understanding of the early signals in DNRR is still limited. It is largely unclear whether and how other wound-induced hormones and chemicals, such as ethylene and reactive oxygen species (ROS), together with stress and environmental signals, function in DNRR from detached leaves.
Auxin is the pivotal cell-fate-controlling hormone that links early signals to cell fate transition during DNRR (Thimann and Went, 1934; Zimmerman and Wilcoxon, 1935; Hitchcock and Zimmerman, 1936; Ahkami et al., 2013; Della Rovere et al., 2013; Liu et al., 2014; Pacurar et al., 2014; Chen et al., 2016a; Sun et al., 2016; Bustillo-Avendaño et al., 2018; Xu, 2018; Druege et al., 2019; Pan et al., 2019). When auxin is polarly transported into regeneration-competent cells, the auxin signaling pathway directly activates the expression of the transcription factor gene WUSCHEL-RELATED HOMEOBOX11 (WOX11) for fate transition from regeneration-competent cells to root founder cells (Liu et al., 2014). WOX11 then directly activates expression of the transcription factor genes LATERAL ORGAN BOUNDARIES DOMAIN16 (LBD16) and WOX5 for the cell fate transition from root founder cells to root primordium with cell division (Hu and Xu, 2016; Sheng et al., 2017). The AP2/EREBP family genes PLETHORA1 and 2 (PLT1/2) and PLT3/5/7 (Aida et al., 2004; Prasad et al., 2011) and the cell cycle genes CYCB1;1/1;2 (Nowack et al., 2012; Scofield et al., 2014) are also required for DNRR from leaf explants (Bustillo-Avendaño et al., 2018; Shanmukhan et al., 2021). The transcription factor genes NAC1 (petunia NAM and Arabidopsis ATAF1, ATAF2, and CUC2) and NAC1-RELATED1 (NAR1) promote cell wall metabolism at the wounded site of leaf explants to facilitate root primordium emergence via upregulation of KDEL-TAILED CYS ENDOPEPTIDASE1/2 (CEP1/2) to degrade EXTENSIN (EXT) (Chen et al., 2016b). However, the transcriptome framework for cell fate transition in DNRR remains largely unclear.
Recently, single-cell RNA sequencing (RNA-seq) technology was used to analyze the Arabidopsis root apical meristem (RAM) (Zhang et al., 2019a; Denyer et al., 2019; Jean-Baptiste et al., 2019; Ryu et al., 2019; Shulse et al., 2019), and it has since been widely used for many plant tissues and organs (Seyfferth et al., 2021). In this study, single-cell RNA-seq and time-lapse RNA-seq analyses reveal the high-resolution transcriptome framework of DNRR from leaf explants.
Results
Strategy for RNA-seq analyses of DNRR
We conducted time-lapse and single-cell RNA-seq experiments to characterize the high-resolution transcriptome framework in DNRR using our previously established system for adventitious rooting from detached Arabidopsis leaves (Chen et al., 2014) (Figure 1A–1D). The first pair of rosette leaves was cut, and the detached leaves were cultured on B5 medium without added hormones. Usually, one to three adventitious roots regenerated from the wounded region per leaf explant, and the root tip became visible after 6 d on B5 medium (Figure 1A). Each time-lapse RNA-seq experiment comprised two biological replicates and was quality controlled (Supplemental Figure 1A).
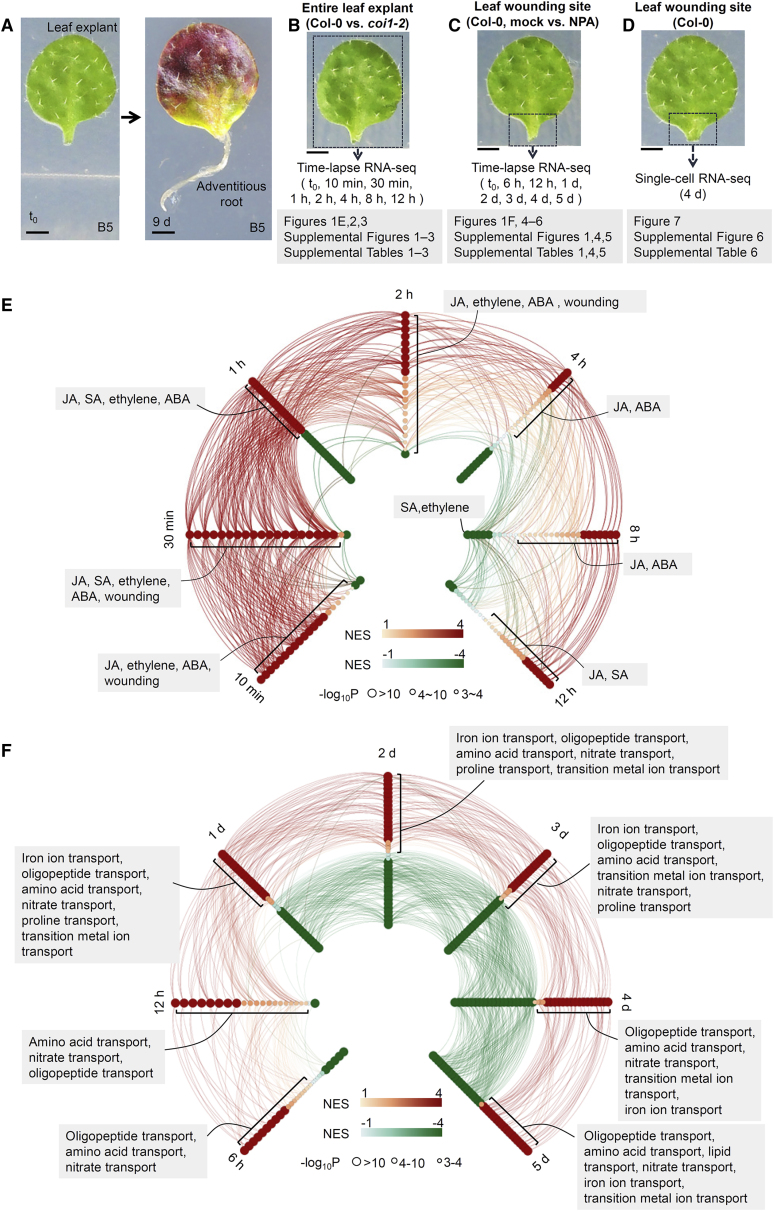
Figure 1.
Overview of RNA-seq experiments in this study.
(A) Adventitious rooting from Arabidopsis leaf explant.
(B) Time-lapse RNA-seq analysis of whole leaf explants (boxed region) of Col-0 and coi1-2 at t0, 10 min, 30 min, 1 h, 2 h, 4 h, 8 h, and 12 h after detachment. Two biological replicates were analyzed.
(C) Time-lapse RNA-seq analysis of wounded region of Col-0 leaf explant (boxed region) cultured on B5 medium without (mock) or with 5 μM NPA from t0, 6 h, 12 h, 1 d, 2 d, 3 d, 4 d, and 5 d after detachment. Because NPA was dissolved in dimethyl sulfoxide (DMSO), the mock control was treated with the same amount of DMSO in B5 medium. Two biological replicates were analyzed.
(D) Single-cell RNA-seq analysis of the wounded region of Col-0 leaf explant (boxed region) at 4 d after detachment. The data were from one biological replicate.
(E and F) Gene set enrichment analysis (GSEA) of GO terms enriched in up- or downregulated genes at each time point compared with t0. The time-lapse RNA-seq data for Col-0 (E, the entire detached leaf) or mock (F, the wounded region of detached leaves) were used for GSEA. Each dot represents the gene set of a GO term. Red dots represent gene sets enriched in upregulated genes (NES > 1), and green dots represent gene sets enriched in downregulated genes (NES < −1). GO terms with high gene overlap are connected by lines (enrichment of GO term pairwise comparison >5 and count of overlapped genes >20). GO terms related to JA, SA, ethylene, ABA, and wounding are listed in (E) as examples. GO terms related to transport are listed in (F) as examples. See Supplemental Table 1 for the full list of GO terms.
Scale bar, 1 mm in (A–D).
First, to study transcriptional regulation by early signals, we performed time-lapse RNA-seq analysis using whole leaf explants at time 0 (t0), 10 min, 30 min, 1 h, 2 h, 4 h, 8 h, and 12 h after detachment (Figure 1B). Usually, early signals such as wound and stress signals function in many cells, including leaf margin cells, mesophyll cells, some vascular cells, and cells at the wound site (Chen et al., 2016a; Zhang et al., 2019b). Wild-type Columbia-0 (Col-0) and the JA receptor mutant coronatine insensitive 1-2 (coi1-2) (Xu et al., 2002) were used to analyze JA-mediated and non-JA-mediated signaling pathways. The overview of gene expression patterns (Supplemental Table 1), the gene ontology (GO) enrichment of up- and downregulated genes (Figure 1E; Supplemental Table 1), and the hub genes in the co-expression network (Zhang and Horvath, 2005) (Supplemental Figure 1B; Supplemental Table 1) indicated transcriptome reprogramming in response to early signals. To focus on regeneration, genes with undetectable or very low transcript levels (TPM < 2) at t0 but significantly increased transcript levels (log2[fold change] > 2 and false discovery rate [FDR] <0.05) at any time point after leaf detachment compared with that at t0 were selected as candidate genes for further analysis (Figures 2 and 3; Supplemental Figures 2 and 3; Supplemental Tables 2 and 3). We considered that these candidate genes might be early wound-signaling response genes that were minimally involved in normal leaf development before detachment and were activated for regeneration after detachment.
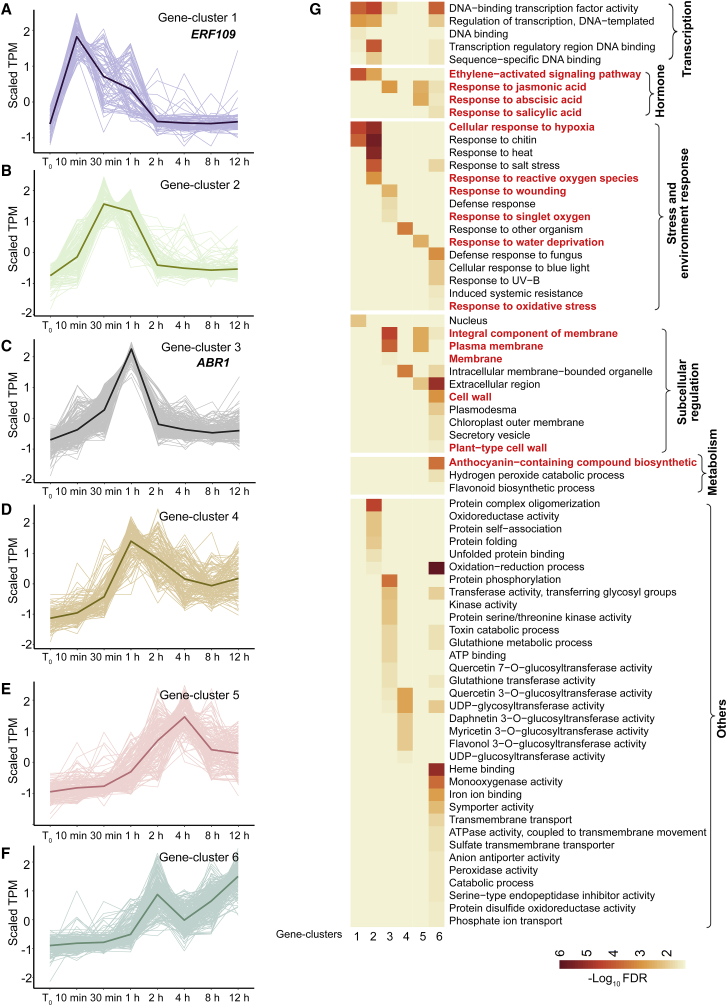
Figure 2.
Time-lapse RNA-seq analysis of candidate genes activated in response to early signals.
(A–F) Candidate genes activated in Col-0 leaf explants within 12 h after detachment, as determined from RNA-seq data (TPM < 2 at t0; log2[fold change] > 2 and FDR < 0.05 at any time point compared with that at t0). On the basis of expression patterns, candidate genes were grouped into six gene clusters (1–6). Lines indicate average values.
(G) GO analysis of gene clusters 1 to 6. GO terms mentioned in the results are labeled in red.
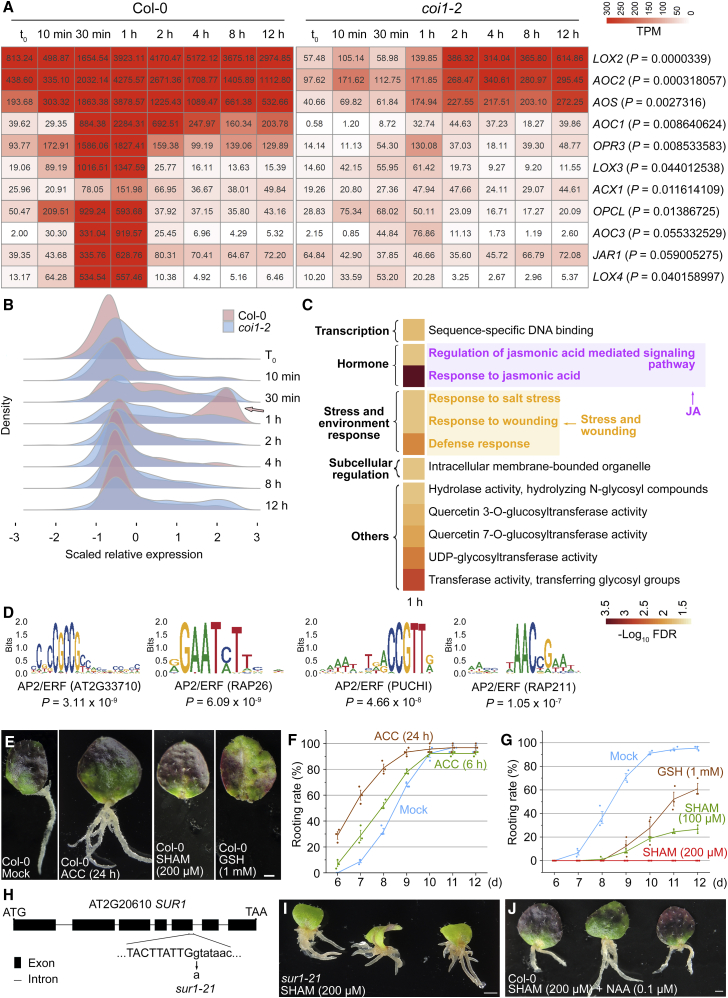
Figure 3.
Analysis of JA, ethylene, and ROS as wound signals in DNRR.
(A) Upregulated genes in the JA biosynthesis pathway, as identified from RNA-seq data of Col-0 and coi1-2 leaf explants from t0 to 12 h after detachment. Numbers in each box are TPM values. P values were calculated by performing a paired t-test between TPM values of Col-0 and coi1-2. One replicate was regarded as one sample.
(B) Distribution of genes with scaled relative expression at each time point from RNA-seq data of Col-0 and coi1-2. Vertical lines indicate gene count density; horizontal lines indicate transcript levels at each time point. We independently scaled the expression levels at each time point for Col-0 and coi1-2. Arrow shows a group of genes highly upregulated in Col-0 but not in coi1-2 at 1 h after leaf detachment.
(C) GO analysis of genes highly upregulated in Col-0 but not in coi1-2 at 1 h after leaf detachment. Upregulation of these genes may be dependent on the COI1-mediated JA signaling pathway. See Supplemental Table 3 for the full list of GO terms.
(D) Analysis of promoter cis elements that may be targeted by the JA pathway. Genes listed in Supplemental Table 3 were used for cis element analysis. See Supplemental Table 3 for the full list of cis element analysis.
(E–G) Phenotype (E) and statistical (F and G) analysis of DNRR after ethylene or ROS inhibitor treatment. For ethylene treatment, Col-0 leaf explants were cultured on B5 medium with 4 μM of the ethylene biosynthesis precursor 1-aminocyclopropane-1-carboxylic acid (ACC) for 6 or 24 h after detachment, then moved to ACC-free B5 medium for further culture. For the ROS inhibitor treatment, Col-0 leaf explants were cultured on B5 medium with 100 μM SHAM, 200 μM SHAM, or 1 mM GSH. Note that treatment with 200 μM SHAM could completely block rooting from leaf explants. Mock served as the control without chemical treatment. The rooting ratio (percentage of rooting leaves among all leaves tested at each time point) was tested (F and G). Error bars show the SEM of three biological replicates (n = 30 leaf explants per replicate), and individual values are indicated by dots (F and G).
(H) Map-based cloning showing mutations in the SUR1 gene. The mutation occurred in the first nucleotide of the fifth intron (G to A), resulting in a stop codon. Capital letters indicate exon nucleotides, and lowercase letters indicate intron nucleotides.
(I) Phenotype analysis of DNRR in wild-type Col-0 and sur1-21 after treatment with 200 μM SHAM. We tested more than 30 leaf explants of sur1-21, and all of them formed adventitious roots at 15 d after leaf detachment.
(J) Treatment with 0.1 μM of the synthetic auxin 1-naphtalene acetic acid (NAA) could partially rescue the rooting defect caused by 200 μM SHAM treatment at 15 d. Note that 82 out of 90 leaf explants formed adventitious roots upon co-treatment with SHAM and NAA.
Scale bars, 1 mm in (E, I, and J).
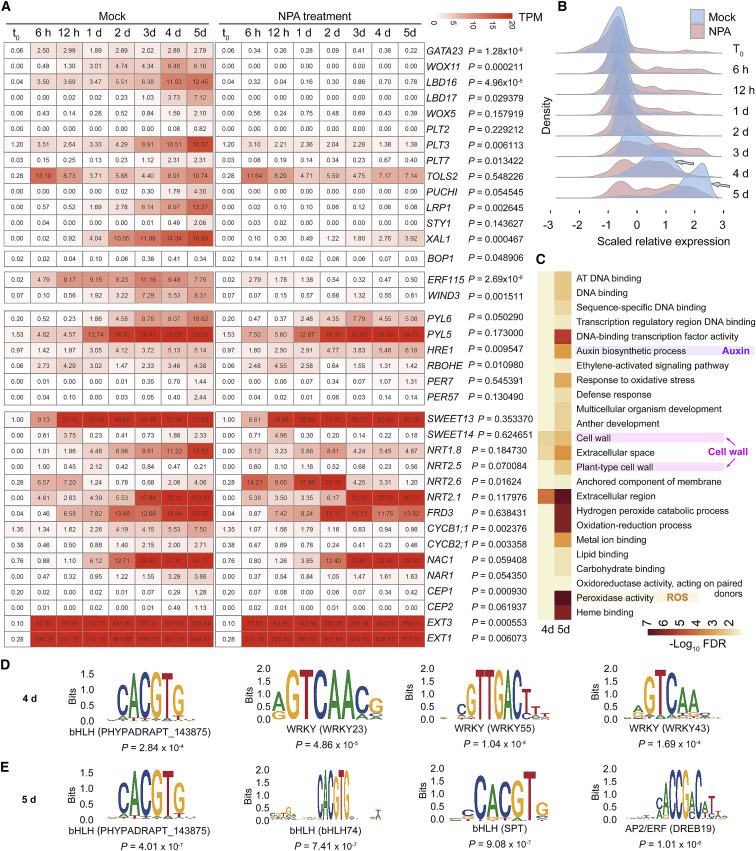
Second, to study transcriptome reprogramming in adventitious root organogenesis, we performed time-lapse RNA-seq analysis of the wounded region of leaf explants at t0, 6 h, 12 h, 1 d, 2 d, 3 d, 4 d, and 5 d after detachment (Figure 1C). Regeneration-competent cells are located in the vasculature of the wounded region, and they usually undergo a fate transition to form the root primordium within 5 d of detachment (Liu et al., 2014; Hu and Xu, 2016; Sheng et al., 2017; Xu, 2018; Bustillo-Avendaño et al., 2018). Besides regeneration-competent cells, many other cells in the wounded region may also be important for providing the cellular environment for root organogenesis. In this experiment, leaf explants were cultured on B5 medium (mock) or B5 medium containing the auxin polar transport inhibitor naphthylphthalamic acid (NPA) to reveal auxin-mediated and non-auxin-mediated gene expression profiles. Treatment with NPA can block auxin accumulation in regeneration-competent cells, resulting in a loss of root organogenesis (Liu et al., 2014). The overview of gene expression patterns (Supplemental Table 1), the GO enrichment of up- and downregulated genes (Figure 1F; Supplemental Table 1), and the hub genes in the co-expression network (Zhang and Horvath, 2005) (Supplemental Figure 1C and 1D; Supplemental Table 1) showed transcriptome reprogramming during adventitious root organogenesis. To focus on regeneration, candidate genes with undetectable or very low transcript levels (TPM < 2) at t0 and increased transcript levels after leaf detachment (log2[fold change] > 2 and FDR < 0.05) at any time point after t0 (Figures 4, 5, and 6; Supplemental Figures 4 and 5; Supplemental Tables 4 and 5) were selected for further analysis. These genes might be minimally involved in normal leaf development before detachment and activated in the wounded region of leaf explants in DNRR.
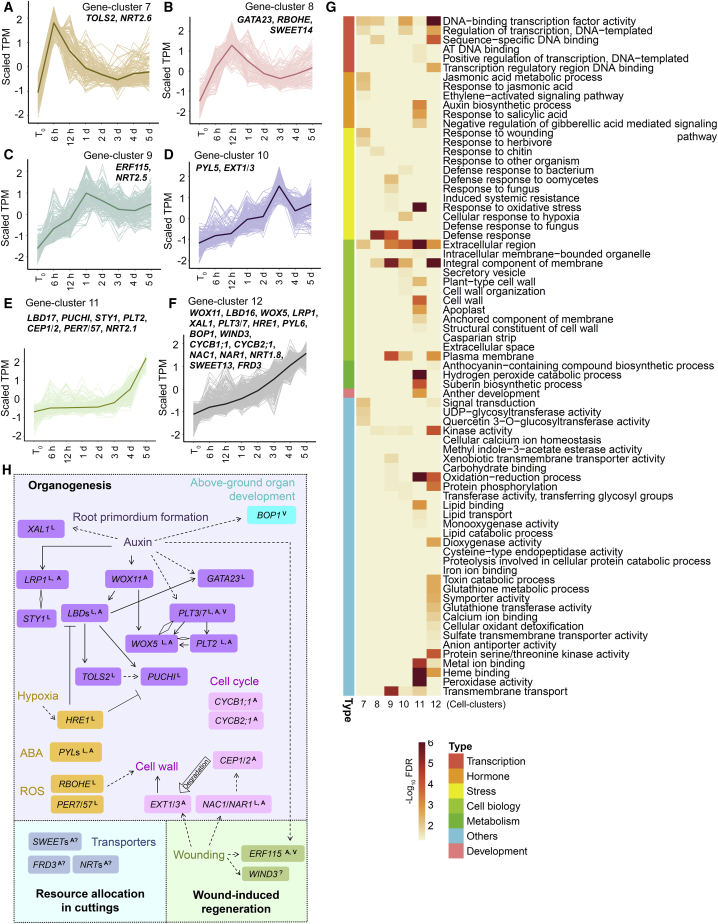
Figure 4.
Time-lapse RNA-seq analysis of candidate genes in adventitious root organogenesis.
(A–F) Candidate genes activated in the wounded region of Col-0 leaf explants (the mock without NPA treatment) cultured on B5 medium within 5 d after detachment, as determined from RNA-seq data (TPM < 2 at t0; log2[fold change] > 2 and FDR < 0.05 at any time point compared with that at t0). Candidate genes were grouped into six gene clusters (7–12) on the basis of changes in transcription patterns. Lines indicate average values.
(G) GO analysis of gene clusters 7 to 12. See Supplemental Table 4 for the full list of GO terms.
(H) Selected candidate genes identified in gene clusters 7 to 12. Genetic and molecular regulations are shown based on published data and this study. LValidated in lateral rooting by previous studies; Avalidated in adventitious rooting from hypocotyls and/or DNRR by previous studies; A?predicted or indicated in adventitious rooting in cuttings by previous studies; ?involved in regeneration but not yet validated in DNRR; Vvalidated in DNRR in this study. Rhombus indicates protein interactions.
Figure 5.
Auxin control of gene activation.
(A) Transcript profiles of selected candidate genes in gene clusters 7 to 12 as determined from RNA-seq data from the wounded region of Col-0 leaf explants cultured on B5 medium (mock) or B5 medium with 5 μM NPA. Numbers in each box are TPM values. P values were calculated by performing a paired t-test between TPM values of Col-0 and coi1-2. One replicate was regarded as one sample. Note that HRE1 showed a higher expression level in the NPA treatment than in the mock control. Although the expression levels of WOX5 and PLT2 were relatively low in the RNA-seq data, the regulation of these two genes by auxin in DNRR was shown in previous studies (Hu and Xu, 2016; Bustillo-Avendaño et al., 2018).
(B) Distribution of genes with scaled relative expression at each time point from time-lapse RNA-seq data from the wounded regions of Col-0 leaf explants cultured on B5 medium (mock) or B5 medium with 5 μM NPA. Vertical lines indicate gene count density, and horizontal lines indicate transcript levels at each time point. We independently scaled the expression levels at each time point for mock and NPA treatment. Arrows show groups of genes that were highly upregulated in mock but not in NPA treatment at 4 and 5 d after leaf detachment.
(C) GO analysis of genes activated in mock but not in NPA treatment at 4 and 5 d after leaf detachment. Upregulation of these genes may be dependent on the auxin signaling pathway. See Supplemental Table 5 for the full list of GO terms.
(D and E) Analysis of promoter cis elements that may be targeted by the auxin pathway at 4 d (D) and 5 d (E) after leaf detachment. See Supplemental Table 5 for the full list of cis element analyses.
Figure 6.
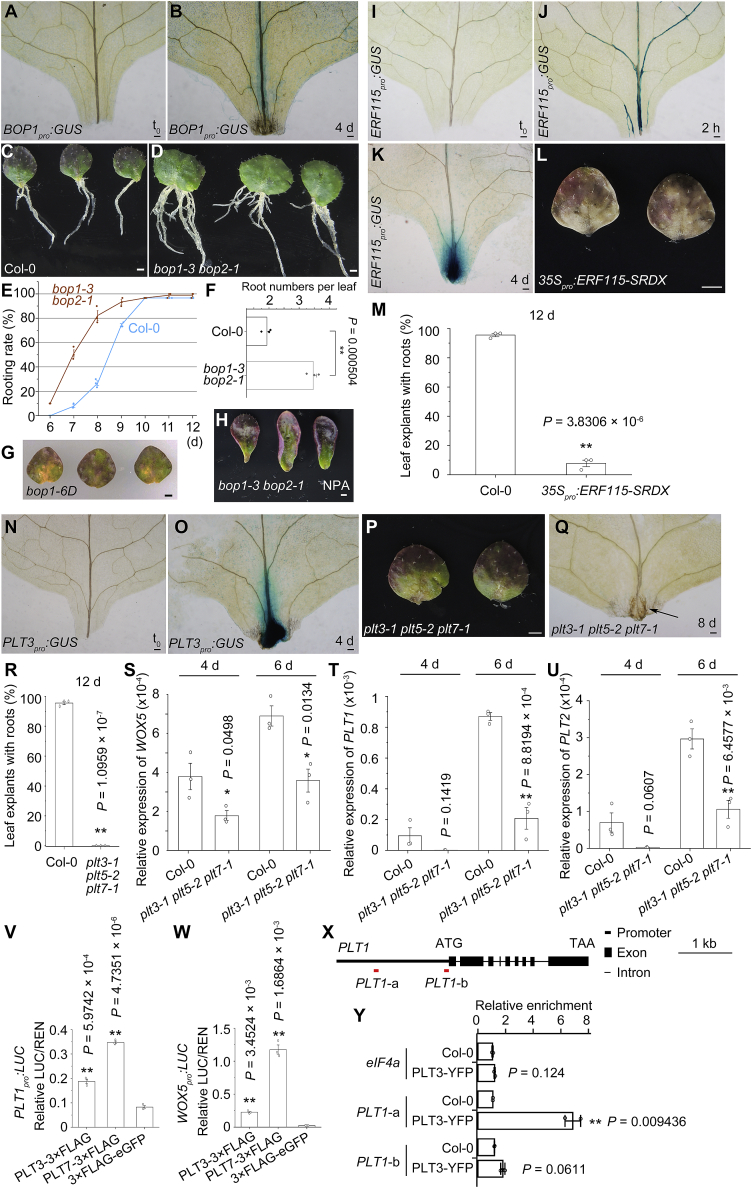
BOPs, ERF115, and PLTs in DNRR.
(A and B) GUS staining of BOP1pro:GUS leaf explants at t0(A) and 4 d (B) cultured on B5 medium.
(C–F) Phenotype (C and D) and statistical (E and F) analysis of DNRR from wild-type Col-0 and bop1-3 bop2-1 leaf explants. Note that rooting rate (E) and root numbers per leaf explant at 15 d (C, D, and F) are higher in bop1-3 bop2-1 than in Col-0. Error bars show the SEM of three biological replicates (n = 30 in each replicate), and individual values are indicated by dots (E and F). ∗∗P < 0.01 in two-sided Student's t-test compared with Col-0 control (F).
(G) Phenotype analysis of bop1-6D. We tested 51 leaf explants from bop1-6D heterozygous or homozygous plants, and only two of them produced adventitious roots at 15 d.
(H) Phenotype analysis of bop1-3 bop2-1 leaf explants cultured on B5 medium with 5 μM NPA treatment. We tested 90 leaf explants, and none of them produced adventitious roots at 15 d.
(I–K) GUS staining of ERF115pro:GUS leaf explants at t0(I), 2 h (J), and 4 d (K) cultured on B5 medium.
(L) Phenotype analysis of DNRR from 35Spro:ERF115-SRDX leaf explants. See (C) for Col-0 control.
(M) Percentages of Col-0 and 35Spro:ERF115-SRDX leaf explants that regenerated roots by 12 d on B5 medium. Error bars show the SEM of three biological replicates (n = 30 in each replicate), and individual values are indicated by dots. ∗∗P < 0.01 in two-sided Student's t-test compared with Col-0 control.
(N and O) GUS staining of PLT3pro:GUS leaf explants at t0(N) and 4 d (O) cultured on B5 medium.
(P) Phenotype analysis of DNRR from plt3-1 plt5-2 plt7-1 leaf explants. See (C) for Col-0 control.
(Q) DIC observation of the wounded region of plt3-1 plt5-2 plt7-1 leaf explants. Arrow indicates the primordium-like structure. We observed 60 leaf explants from plt3-1 plt5-2 plt7-1, and all of them showed the primordium-like structure.
(R) Percentages of Col-0 and plt3-1 plt5-2 plt7-1 leaf explants that regenerated roots by 12 d on B5 medium. Error bars show the SEM of three biological replicates (n = 30 in each replicate), and individual values are indicated by dots. ∗∗P < 0.01 in two-sided Student's t-test compared with Col-0 control.
(S–U) qRT-PCR analysis of WOX5(S), PLT1(T), and PLT2(U) in the wounded region of leaf explants from Col-0 and plt3-1 plt5-2 plt7-1 at 4 and 6 d after leaf detachment. Error bars show the SEM of three biological replicates, and each biological replicate was analyzed with three technical replicates. ∗P < 0.05 and ∗∗P < 0.01 in two-sided Student's t-test compared with Col-0 control.
(V and W) Relative ratio of firefly LUC to Renilla luciferase (REN) activity in Arabidopsis protoplasts co-transformed with PLT1pro:LUC(V) or WOX5pro:LUC(W) with 35Spro:PLT3-3×FLAG or 35Spro:PLT7-3×FLAG. UBQ10pro:3×FLAG-eGFP served as the control. Error bars show the SEM of three biological replicates, and each biological replicate was analyzed with three technical replicates. ∗∗P < 0.01 in two-sided Student's t-test compared with control.
(X) Schematic of PLT1 gene structure. Red horizontal lines show positions of PCR fragments in ChIP analysis in (Y).
(Y) ChIP analysis showing relative enrichment of PLT3-YPF in the promoter of PLT1. eIF4a served as a negative control. Error bars show the SEM of two biological replicates, and each biological replicate was analyzed with three technical replicates. ∗∗P < 0.01 in two-sided Student's t-test compared with control.
Scale bars, 100 μm (A, B, I–K, N, O, and Q), 1 mm (C, D, L, G, H, and P).
Third, we performed single-cell RNA-seq to study the transcriptome profile in the wounded region of leaf explants during adventitious root organogenesis at the single-cell level (Figure 1D). The wounded regions of leaf explants at 4 d after detachment were collected for single-cell extraction and RNA-seq analysis (Figure 7; Supplemental Figure 6; Supplemental Table 6). A mixture of cells in the wounded region, including those that underwent cell fate transition at different stages, could be observed at this time point. This is because rooting efficiency and speed were not precisely synchronized among different leaf explants, and leaf explants therefore harbored cells at different stages of fate transition. In addition, because each leaf explant could successively produce one to three adventitious roots, it could contain cells at different stages of fate transition. An estimated 7225 cells were obtained, with mean values of 57 768 reads per cell and 2321 genes per cell.
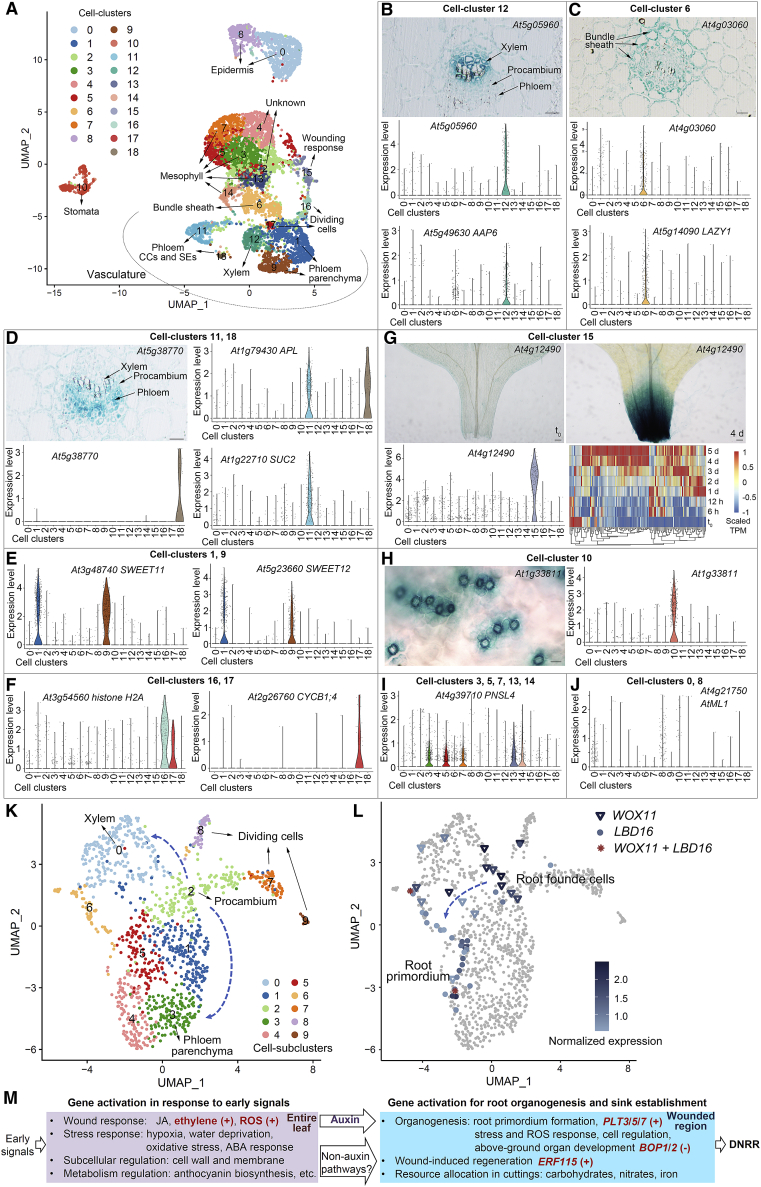
Figure 7.
Single-cell atlas in the wounded region of detached leaves.
(A) UMAP plot of 7225 cells within the wounded region of detached Col-0 leaves at 4 d. Cells were grouped into cell clusters 0 to 18. CC, companion cells; SE, sieve elements.
(B–J) Identification of each cell cluster based on promoter-fused GUS marker lines and published marker genes. Violin plots indicate the expression patterns of marker genes, and each dot indicates a single cell. In cell cluster 15 (G), the marker genes (pct.2 < 0.25) were analyzed in time-lapse RNA-seq data, and most showed upregulation after wounding.
(K) UMAP plot showing cell subcluster analysis of cell clusters 1, 12, 16, and 17 in (A). Cells were grouped into cell subclusters 0–9.
(L) UMAP plot showing WOX11 and LBD16 expression patterns. Blue triangles indicate cells expressing WOX11, blue dots indicate cells expressing LBD16, and red asterisks indicate cells co-expressing WOX11 and LBD16.
(M) Framework of wound-induced root organogenesis during DNRR from detached leaves. After leaf detachment, many genes are activated progressively in response to early signals in the entire leaf and during root organogenesis in the wounded region. Auxin may be the key hormone that links early signals to the fate transition of regeneration-competent cells in the wounded region. Roles of ethylene, ROS, PLT3/5/7, BOP1/2, and ERF115 were validated in this study. Plus sign (+) indicates involvement in the promotion of DNRR; minus sign (−) indicates involvement in the inhibition of DNRR.
Scale bars, 20 μm (B–D and H), 100 μm (G).
The RNA-seq data obtained in these analyses can be accessed using the online tool (http://xulinlab.cemps.ac.cn/), and gene IDs can be used to search for gene expression patterns.
Time-lapse RNA-seq analysis of transcriptional response to early signals
The candidate genes activated by early signals in whole detached leaves of Col-0 from t0 to 12 h (Figure 1B) could be grouped into six gene clusters based on their expression patterns (gene clusters 1 to 6; Figure 2A–2F; Supplemental Table 2). Gene clusters 1 (Figure 2A) and 2 (Figure 2B) were activated from 10 min to 1 h of detachment and were downregulated afterward. Gene clusters 3 and 4 were gradually activated and showed peak expression at 1 h after detachment, followed by sharp downregulation (gene cluster 3; Figure 2C) or gradual downregulation (gene cluster 4; Figure 2D). Gene cluster 5 was gradually activated after leaf detachment, showed peak expression at 2–8 h, and was downregulated afterward (Figure 2E). Gene cluster 6 was continuously upregulated after 1 h with various patterns (Figure 2F). ERF109 and ABR1, which are involved in the wound response in DNRR (Zhang et al., 2019b; Ye et al., 2020), were in gene clusters 1 and 3, respectively, suggesting that these two ERF family genes function at different times to promote DNRR (Figure 2A and 2C). We validated selected gene expression patterns in gene clusters 1–6 by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) (Supplemental Figure 2).
GO analyses were conducted to analyze the genes in gene clusters 1 to 6 (Figure 2G; Supplemental Table 2). Genes in the JA, ethylene, and ROS pathways were immediately activated after leaf detachment, indicating that, in addition to JA, ethylene and ROS may also serve as wound signals in the rapid response to leaf detachment during DNRR (see below for genetic validation analyses of ethylene and ROS) (Druege et al., 2016, 2019; Xu, 2018). Leaves experience multiple stress conditions after detachment, and genes that are responsive to such stress conditions (e.g., hypoxia, water deprivation, and oxidative stress) and are involved in the abscisic acid (ABA) and salicylic acid (SA) pathways may be rapidly activated and affect the efficiency of adventitious rooting (Mandadi et al., 2009; Guénin et al., 2011; Kotula et al., 2015; Chen et al., 2016b; Dawood et al., 2016). In addition, many membrane-related and cell-wall-related pathways were activated, indicating that adjustment of membrane and cell wall status is probably required in response to wounding, stress, and environmental changes (Guénin et al., 2011; Chen et al., 2016b). Gene cluster 6 included genes in some metabolic pathways, including anthocyanin biosynthesis (Figure 2G). For example, the upregulation of anthocyanin biosynthetic genes may explain why the leaf explants turned purple during DNRR on B5 medium (Figure 1A).
Taken together, our data indicate that genes involved in wound-signaling pathways, stress response, subcellular regulation, and metabolism pathways may be upregulated during DNRR.
Role of JA-mediated wound-signaling pathway in DNRR
We compared transcriptomes between whole leaf explants of Col-0 and coi1-2 from t0 to 12 h to analyze the role of JA in the wound response (Figure 1B).
We first examined genes in the JA biosynthesis pathway. A group of JA biosynthesis genes were quickly upregulated within 10–30 min of detachment, and their expression levels were lower in coi1-2 than in Col-0 (Figure 3A), indicating that positive feedback is involved in the regulation of JA biosynthesis and signaling pathways. qRT-PCR analysis confirmed that upregulation of JASMONATE RESISTANT 1 (JAR1) and ALLENE OXIDE CYCLASE 1 (AOC1) after wounding was dependent on COI1 (Supplemental Figure 3).
We next analyzed all six gene clusters at different time points after leaf detachment. We found a peak at 1 h showing enrichment of activated genes in Col-0 but not in coi1-2 (Figure 3B, arrow), suggesting that these genes were upregulated through the JA signaling pathway. GO analysis of these genes showed that, in addition to the JA pathway, many stress- and wounding-response pathways were also affected by JA signaling at 1 h after detachment (Figure 3C; Supplemental Table 3). Analysis of promoter cis elements (Supplemental Table 3) indicated that AP2/ERF family transcription factors may be involved in the JA-mediated activation of these genes (Figure 3D).
Therefore, JA may act as a key wound-response hormone in the regulation of many pathways in addition to the promotion of auxin production during DNRR (Fattorini et al., 2009, 2018; Lischweski et al., 2015; Zhang et al., 2019b; Lakehal et al., 2019; Park et al., 2019).
Ethylene and ROS act as wound signals to promote DNRR
To verify the role of ethylene in the regulation of DNRR, we carried out phenotype analysis. Compared with leaf explants in the mock control, leaf explants treated with ethylene for a short period (6 h or 24 h) after detachment exhibited enhanced rooting ability (Figure 3E and 3F). This is consistent with many previous observations that a wound-induced early ethylene pulse promotes adventitious root formation in cuttings of diverse plant species, probably via crosstalk with auxin (Riov and Yang, 1989; Clark et al., 1999; De Klerk and Hanecakova, 2008; da Costa et al., 2013; Veloccia et al., 2016).
Next, we tested the role of ROS in DNRR. Previous studies have indicated that ROS can be produced quickly upon wounding (Orozco-Cardenas and Ryan, 1999; Prasad et al., 2017, 2020). Treatment with the ROS inhibitor salicylhydroxamic acid (SHAM) or glutathione (GSH) reduced the rooting ability of leaf explants (Figure 3E and 3G). These results support a role for ROS in the regulation of adventitious root formation in cuttings (Falasca et al., 2004; Liao et al., 2009; Li and Xue, 2010; Bai et al., 2012; Lin et al., 2014; Takáč et al., 2016; Huang et al., 2019).
To further analyze the role of ROS in DNRR from leaf explants, we performed ethyl methane sulfonate (EMS) mutagenesis-based genetic screening to identify suppressor mutants that can rescue the rooting defect caused by SHAM treatment. Map-based cloning of one of the candidate suppressor mutants revealed a mutation in the SUPERROOT 1 (SUR1) gene (Figure 3H), leading to over-accumulation of auxin in the mutant (Boerjan et al., 1995; Mikkelsen et al., 2004). The suppressor sur1-21 mutant exhibited rescued rooting ability under SHAM treatment (Figure 3I). This was confirmed by the addition of auxin to the medium, which partially rescued the rooting defect in Col-0 leaf explants caused by SHAM treatment (Figure 3J). Therefore, ROS may be involved in DNRR via crosstalk with auxin.
Together, these data suggest that ethylene and ROS may act as wound signals, in addition to JA, to promote DNRR after wounding.
Time-lapse RNA-seq analysis of transcriptome reprogramming during adventitious root organogenesis
The candidate genes activated in the wounded region of leaf explants of the mock control (Col-0 without NPA treatment) from t0 to 5 d (Figure 1C) could be grouped into six gene clusters (gene clusters 7 to 12; Figure 4A–4F; Supplemental Table 4) based on their expression patterns. Gene clusters 7 (Figure 4A) and 8 (Figure 4B) were activated with peak transcript levels from 6 to 12 h and then downregulated afterward. Gene clusters 9 (Figure 4C) and 10 (Figure 4D) were gradually activated and showed peak transcript levels from 1 to 3 d. Gene cluster 11 (Figure 4E) showed low transcript levels before 3 d and was upregulated after 3 d. Gene cluster 12 (Figure 4F) was continuously upregulated throughout these time-lapse analyses. GO analysis of gene clusters 7 to 12 showed that multiple pathways were activated, including those related to transcription, hormones, the cell wall, stress response, development, and transport (Figure 4G; Supplemental Table 4). We validated the expression patterns of some candidate genes in gene clusters 7–12 by qRT-PCR (Supplemental Figure 4A–4G). We further analyzed selected candidate genes from the six gene clusters (Figure 4A–4F; Supplemental Figure 4H; Supplemental Table 4) based on their published biological functions in organogenesis, wound-induced regeneration, and resource allocation in cuttings (Figure 4H), as DNRR in this study was a process of wound-induced root organogenesis from leaf cuttings.
Many genes involved in organogenesis were identified, including those involved in root primordium formation, stress and ROS response, and cell regulation (Figure 4H). WOX11 is directly activated by the auxin signaling pathway in adventitious root founder cells to promote DNRR, but it is not expressed in lateral root founder cells (Liu et al., 2014; Sheng et al., 2017). GATA23 is a lateral root founder cell marker (De Rybel et al., 2010). LBDs (Okushima et al., 2007; Sheng et al., 2017; Liu et al., 2018), WOX5 (Sarkar et al., 2007; Hu and Xu, 2016), and PLTs are involved in the formation of both lateral and adventitious root primordia (Hofhuis et al., 2013; Du and Scheres, 2017; Bustillo-Avendaño et al., 2018; Shimotohno et al., 2018; Shanmukhan et al., 2021), and WOX5 can directly interact with PLTs (Burkart et al., 2021; Zhai and Xu, 2021). The SHORT INTERNODES/STYLISH (SHI/STY) family gene LATERAL ROOT PRIMORDIUM1 (LRP1) is directly activated by the auxin signaling pathway and is expressed at all stages of root primordium initiation during both lateral and adventitious rooting. It promotes root organogenesis via the regulation of auxin biosynthesis and interaction with its family member STY1 (Smith and Fedoroff, 1995; Kuusk et al., 2006; Singh et al., 2020). The peptide gene TARGET OF LBD SIXTEEN 2 (TOLS2) and the AP2/EREBP gene PUCHI are involved in positioning and patterning of the lateral root primordium (Hirota et al., 2007; Kang et al., 2013; Toyokura et al., 2019; Trinh et al., 2019). Auxin-induced XAANTAL1 (XAL1) may be involved in lateral root meristem cell proliferation (Tapia-López et al., 2008). PYRABACTIN RESISTANCE1-LIKEs (PYLs), which encode receptors for the stress hormone ABA, are involved in both lateral rooting (Zhao et al., 2014; Xing et al., 2016; Belda-Palazon et al., 2018; Li et al., 2020a) and adventitious rooting (Zeng et al., 2021). The ERF-VII gene HYPOXIA RESPONSIVE1 (HRE1) mediates hypoxic responses in the lateral root primordium via the repression of LBDs and PUCHI (Abbas et al., 2015; Shukla et al., 2019). PEROXIDASE7 (PER7) and PER57 are involved in lateral root primordium development to maintain ROS homeostasis (Manzano et al., 2014). The RESPIRATORY BURST OXIDASE HOMOLOG (RBOH) gene RBOHE functions in the ROS production pathway and may facilitate cell wall remodeling of overlying tissues to allow outgrowth of the lateral root primordium (Orman-Ligeza et al., 2016). The cell cycle genes CYCB1;1 and CYCB2;1 have been shown by genetic analyses to participate in DNRR from leaf explants (Bustillo-Avendaño et al., 2018). NAC1, NAR1, CEP1/2, and EXT1/3 are involved in a pathway that promotes DNRR from leaf explants via the regulation of cell wall metabolism (Chen et al., 2016b), and NAC1 is also involved in lateral rooting (Xie et al., 2000). Furthermore, activation of the BTB/POZ domain gene BLADE-ON-PETIOLE1 (BOP1) (Ha et al., 2003, 2004; Hepworth et al., 2005) indicated that pathways controlling aboveground organs may be recruited to facilitate DNRR (see genetic validation of BOP1 and PLT3/7 in DNRR, below).
Genes involved in wound-induced regeneration were identified (Figure 4H). We observed activation of the AP2/EREBP genes WIND3 and ERF115 during DNRR. Both genes are upregulated by wounding and are involved in wound-induced tissue repair and regeneration of plant organs (Iwase et al., 2011; Heyman et al., 2013, 2016; Ikeuchi et al., 2015; Bustillo-Avendaño et al., 2018; Rymen et al., 2019; Zhou et al., 2019; Lakehal et al., 2020). In addition, ERF115 was shown to be involved in adventitious rooting from hypocotyls (Lakehal et al., 2020). Therefore, wound-induced tissue repair programs may be involved in DNRR (see genetic validation of ERF115 in DNRR, below).
Studies in cuttings provided the idea that there is strict nutritional and metabolic control of resource allocation during adventitious root regeneration (Druege et al., 2019), involving the establishment of a carbohydrate sink at the stem base of cuttings (Ahkami et al., 2009, 2013; Klopotek et al., 2016), mobilization and translocation of N within cuttings (Svenson and Davies, 1995; Dong et al., 2004; Zerche et al., 2016), and iron accumulation at the stem base of cuttings (Svenson and Davies, 1995; Hilo et al., 2017). In the time-lapse RNA-seq data, we identified many genes involved in resource allocation (Figure 4H), including the sucrose transport genes SWEET13 and 14 (Chen et al., 2012; Kanno et al., 2016; Julius et al., 2017; Zhang and Turgeon, 2018), the nitrate transport genes NRT1.8 and NRT2.1/2.5/2.6 (Cerezo et al., 2001; Filleur et al., 2001; Li et al., 2010; Camañes et al., 2012; Dechorgnat et al., 2012; Kiba et al., 2012; Kechid et al., 2013; Lezhneva et al., 2014; Zhang et al., 2014; Wang et al., 2018), and the MULTIDRUG AND TOXIN EFFLUX (MATE) family gene FERRIC REDUCTASE DEFECTIVE 3 (FRD3) for iron homeostasis (Delhaize, 1996; Rogers and Guerinot, 2002). This result was also supported by the overview of the GO analysis, which showed that many genes involved in resource transport were upregulated during DNRR (Figure 1F).
Taken together, our results show that multiple genes involved in organogenesis, wound-induced regeneration, and resource allocation in cuttings were activated in the wounded region of leaf explants during DNRR. Our observations also suggest that, although adventitious rooting involves many unique mechanisms (especially early signal response and root founder cell establishment) that do not function in lateral rooting (Bellini et al., 2014; Verstraeten et al., 2014; Sheng et al., 2017), the two types of rooting still share many common molecular pathways for root organogenesis (e.g., GATA23, LBDs, PLTs, LRP1, TOLS2, PUCHI, HRE1, PERs, RBHOE, and NAC1).
Auxin control of gene expression in DNRR
To analyze auxin regulation in DNRR, we compared gene transcript levels in the wounded region of leaf explants between the mock control and the NPA treatment from t0 to 5 d (Figure 1C).
The activation of many of the selected candidate genes (Figure 4H) was completely blocked or partially impaired by NPA treatment (Figure 5A), indicating that auxin may be a master regulator of DNRR. However, auxin barely affected the activation of some genes, such as TOSL2, PYLs, PERs, SWEETs, NRTs, and NACs (Figure 5A), indicating that they may be activated by non-auxin pathways. qRT-PCR analysis confirmed that upregulation of PLT3, WOX11, LBD16, and ERF115 was dependent on auxin, whereas upregulation of SWEET13 and PYL5 was not affected by NPA treatment (Supplemental Figure 5A–5F). β-glucuronidase (GUS) marker lines confirmed that upregulation of TOLS2 and PYL5 after leaf detachment was independent of auxin (Supplemental Figure 5G–5L). Therefore, non-auxin pathways may also be required to assist in the DNRR process.
We next analyzed gene clusters 7 to 12 at different time points after leaf detachment and found marked differences in gene transcript levels at 4 and 5 d after detachment between the mock and NPA treatments (Figure 5B, arrows), suggesting that auxin-mediated gene activation occurred mainly during this time period. GO analyses confirmed that, in addition to auxin-related pathways, auxin also regulated developmental, cell wall-related, and ROS-related pathways (Figure 5C; Supplemental Table 5). Promoter cis element analysis (Supplemental Table 5) indicated that bHLH, WRKY, and AP2/ERF family transcription factors may be involved in auxin-mediated activation of these genes (Figure 5D and 5E).
BOPs, ERF115, and PLTs are involved in DNRR
BOP1 and BOP2 are two redundant homologous genes that control leaf and floral patterning and restrict organogenesis in Arabidopsis (Ha et al., 2003, 2004, 2007; Hepworth et al., 2005; McKim et al., 2008; Xu et al., 2010; Khan et al., 2012; Ding et al., 2015). However, the roles of BOP genes in root development and regeneration are poorly understood. BOP1pro:GUS leaf explants showed that BOP1 expression was upregulated after leaf detachment (Figure 6A and 6B). The loss-of-function bop1-3 bop2-1 double mutant (Ha et al., 2004; Hepworth et al., 2005; Norberg et al., 2005) showed an accelerated rooting rate from leaf explants and produced more adventitious roots per leaf explant compared with the wild-type Col-0 (Figure 6C–6F). The gain-of-function bop1-6D mutant (Norberg et al., 2005) showed defective adventitious rooting from detached leaves (Figure 6G). In addition, promotion of rooting in the bop1-3 bop2-1 double-mutant background was dependent on auxin, as NPA treatment led to a loss of root regeneration from detached bop1-3 bop2-1 leaf explants (Figure 6H). Taken together, these results show that BOP genes function as negative regulators to restrict adventitious root organogenesis.
ERF115 is critically involved in root tip repair in response to wounding (Heyman et al., 2013, 2016; Zhou et al., 2019). ERF115pro:GUS leaf explants showed that ERF115 expression was not detected at t0, was quickly upregulated in the vasculature within 2 h of leaf detachment, and was highly expressed during adventitious root primordium formation at 4 d (Figure 6I–6K). Detached leaves could barely produce adventitious roots in the line overexpressing ERF115 fused with the repression domain SRDX (35Spro:ERF115-SRDX) (Hiratsu et al., 2003; Heyman et al., 2013), and the leaves were severely stressed (Figure 6L and 6M).
PLT3, PLT5, and PLT7 are redundant genes that function in lateral root organogenesis and phyllotaxis (Prasad et al., 2011; Du and Scheres, 2017). A recent study indicated that PLT3/5/7 are also required for DNRR from leaf explants (Shanmukhan et al., 2021). PLT3pro:GUS leaf explants showed that PLT3 expression was not detected at t0 and was upregulated during adventitious root primordium formation at 4 d after leaf detachment (Figure 6N and 6O). We confirmed that adventitious roots did not develop normally in the plt3-1 plt5-2 plt7-1 triple mutant during DNRR (Figure 6P–6R). Some primordium-like structures were observed at the wounding site of plt3-1 plt5-2 plt7-1 leaf explants, but these structures failed to form normal adventitious root tips (Figure 6Q). qRT-PCR analysis confirmed that the expression levels of WOX5, PLT1, and PLT2, which are known to participate in cell fate transition during DNRR (Hu and Xu, 2016; Bustillo-Avendaño et al., 2018), were downregulated in the wounded region of leaf explants from plt3-1 plt5-2 plt7-1 compared with Col-0 (Figure 6S–6U) (Kareem et al., 2015; Du and Scheres, 2017; Shimotohno et al., 2018; Shanmukhan et al., 2021). Dual-luciferase assays showed that PLT3 and PLT7 could activate the expression of WOX5 and PLT1 in Arabidopsis protoplasts (Figure 6V and 6W), and a chromatin immunoprecipitation (ChIP) assay indicated that PLT3 could bind directly to the PLT1 promoter (Figure 6X and 6Y). Therefore, PLT3/5/7 may directly promote the expression of WOX5 and PLT1/2 during the cell fate transition in adventitious root organogenesis.
Taken together, our results confirmed that BOP1/2, which control aboveground organs, ERF115, which controls wound-induced regeneration, and PLT3/5/7, which control root organogenesis, are upregulated and participate in adventitious root organogenesis during DNRR from leaf explants.
Single-cell RNA-seq analysis of the wounded region of leaf explants
Based on single-cell data from the wounded region of Col-0 leaves at 4 d after detachment (Figure 1D), we identified 19 cell clusters using the uniform manifold approximation and projection (UMAP) algorithm (Becht et al., 2019) (Figure 7A; Supplemental Table 6).
We annotated the cell clusters based on published marker genes and our promoter-fusion GUS reporter lines (Figure 7A–7J; Supplemental Table 6). Cell cluster 12 was closely related to xylem cells, validated by At5g05960pro:GUS and the marker genes AMINO ACID PERMEASE 6 (AAP6) (Okumoto et al., 2002) and XYLEM NAC DOMAIN 1 (XND1) (Zhao et al., 2005) (Figure 7B; Supplemental Table 6). Cell cluster 6 represented bundle sheath cells, validated by At4g03060pro:GUS and the marker gene LAZY1 (Taniguchi et al., 2017) (Figure 7C; Supplemental Table 6). Cell clusters 11, 18, 1, and 9 were related to phloem cells, validated by At5g38770pro:GUS, the phloem companion cell and sieve element marker ALTERED PHLOEM DEVELOPMENT (APL) (Bonke et al., 2003), the phloem companion cell marker SUCROSE-PROTON SYMPORTER 2 (SUC2) (Truernit and Sauer, 1995; Chen et al., 2012), and the phloem parenchyma markers SWEET11 and 12 (Chen et al., 2012) (Figure 7D and 7E; Supplemental Table 6). Cell clusters 16 and 17 may represent dividing cells because we observed many histone genes and CYCLIN genes enriched in these two clusters (Figure 7F; Supplemental Table 6). Cell cluster 15 appears to exhibit wounding-related gene expression (Figure 7G; Supplemental Table 6). Cell cluster 10 represents stomata, as shown by At1g33811pro:GUS (Figure 7H; Supplemental Table 6). Cell clusters 3, 5, 7, 13, and 14 may represent mesophyll cells, as these cells expressed many genes involved in photosynthesis, such as PNSL4 (Figure 7I; Supplemental Table 6). Cell clusters 0 and 8 may be related to epidermal cells, as shown by A. thaliana MERISTEM L1 LAYER (AtML1) (Lu et al., 1996; Sessions et al., 1999) (Figure 7J; Supplemental Table 6). Overall, the cell cluster analysis reflected the original tissues present in the detached leaves.
Single-cell RNA-seq analysis of adventitious root organogenesis
We have previously shown that the adventitious root is initiated from regeneration-competent cells in the vasculature, and WOX11 and LBD16 are marker genes of the adventitious root founder cells and the root primordium, respectively (Liu et al., 2014; Hu et al., 2017; Sheng et al., 2017). Analysis of WOX11 and LBD16 expression patterns in the vasculature and dividing cells showed that the two marker genes were highly enriched in cell clusters 1, 12, 16, and 17 (Supplemental Figure 6A). Therefore, these four cell clusters were further analyzed, and 10 cell subclusters were identified (Figure 7K; Supplemental Figure 6B; Supplemental Table 6).
Cell subcluster 2 was related to the procambium in the original leaf explants and expressed procambium marker genes, e.g., the AP2-like transcription factor gene AINTEGUMENTA (ANT) (Elliott et al., 1996; Klucher et al., 1996; Randall et al., 2015; Smetana et al., 2019) and the receptor-like kinase gene PHLOEM INTERCALATED WITH XYLEM (PXY) (Fisher and Turner, 2007; Hirakawa et al., 2008; Etchells and Turner, 2010; Smetana et al., 2019) (Figure 7K; Supplemental Figure 6C–6E; Supplemental Table 6). Cell subcluster 0 could be related to xylem cells in the original leaf explants and was enriched in the xylem cell marker genes At5g05960 (Figure 7B), AAP6 (Okumoto et al., 2002), and XND1 (Zhao et al., 2005) (Figure 7K; Supplemental Figure 6G; Supplemental Table 6). The class III homeodomain-leucine zipper (HD-Zip III) family gene ATHB8, which was expressed in xylem parenchyma cells and procambium xylem-side daughter cells (Smetana et al., 2019) in the wounded region of leaf explants at 4 d after detachment, was primarily enriched in cell subclusters 0 and 8 (Figure 7K; Supplemental Figure 6F and 6H; Supplemental Table 6). Cell subcluster 3 could be related to phloem parenchyma cells in the original leaf explants, and it expressed the marker gene SWEET11 (Chen et al., 2012) (Figure 7K; Supplemental Figure 6G; Supplemental Table 6). Cell subclusters 7, 8, and 9 could be related to dividing cells because many histone and CYCLIN genes were enriched in these subclusters (Figure 7K; Supplemental Figure 6G; Supplemental Table 6).
The root founder cell marker WOX11 was expressed in some cells of cell subclusters 0, 2, 6, and 8 (Figure 7L). The expression of WOX11 in cell subcluster 2 is consistent with the finding of a previous study that root founder cells could be formed from the procambium (Liu et al., 2014). Cells expressing the root primordium marker LBD16 were located primarily in a region within cell subclusters 4, 5, and 6 (Figure 7L). ERF115 was highly enriched in cell subcluster 6 (Supplemental Table 6).
Taken together, results from the cell subcluster analysis provided information on gene expression patterns in tissues from the wounded region of the original leaf explants as well as cells involved in adventitious root organogenesis. Further studies will be needed to explore and identify more cell-type-specific markers to improve the accuracy of data interpretation in the future.
Discussion
Based on time-lapse and single-cell RNA-seq data, we constructed a high-resolution transcriptome framework for DNRR from leaf explants of Arabidopsis (summarized in Figure 7M). After leaf detachment, JA, ethylene, and ROS may act as wound signals to promote DNRR, and it is possible that the promotion of auxin production may be one of the events that occur downstream of these wound signals. Stress response pathways, subcellular regulation pathways, and metabolic pathways are also upregulated in response to early signals. Although the ABA pathway is known to participate in adventitious rooting (Mandadi et al., 2009; Dawood et al., 2016; Druege et al., 2016, 2019; Xu, 2018), the roles of many of these early signaling pathways in DNRR are largely unclear. When auxin is transported and accumulated in the vasculature near the wounded region of leaf explants, many genes are activated in the wounded region. In addition to genes that control organogenesis, genes involved in wound-induced regeneration and resource allocation may also be important for ensuring adventitious rooting and sink establishment in the wounded region of leaf explants.
Interestingly, some pathways, such as the ethylene, ROS, ABA, and hypoxia pathways, were found to act in both the response to early signals and in adventitious root organogenesis. Although an early ethylene pulse acts as a wound signal to promote adventitious rooting in cuttings, ethylene may have an inhibitory role in adventitious root development in the later stages of adventitious rooting (De Klerk and Hanecakova, 2008; Druege et al., 2014). A recent study showed that constant ethylene treatment could inhibit DNRR from leaf explants (Li et al., 2020b). The ROS and ABA pathways were both identified in response to wound signals and stress signals, and genes involved in ROS and ABA pathways were also activated during adventitious root organogenesis. It has been reported that ROS have multiple roles in stem cell control in roots and shoots (Tsukagoshi et al., 2010; Zeng et al., 2017; Yang et al., 2018), indicating that they probably also act as a stem-cell-controlling factor during the cell fate transition in DNRR. Therefore, we can speculate that ethylene, ROS, ABA, hypoxia, and probably many other pathways may serve dual roles in the regulation of DNRR, both in response to early signals and in adventitious root organogenesis. This is similar to the finding that JA plays dual roles in adventitious rooting (Pan et al., 2021; Zhang et al., 2021).
The genes and pathways described here are selected examples that may be involved in DNRR, but many other genes and pathways have not been shown. In addition, it is possible that the auxin pathway and non-auxin pathways act together to regulate adventitious root organogenesis (Figure 7M). Furthermore, it is important to note that gene expression changes do not happen only in regeneration-competent cells. Many other cells in the wounded region may also undergo transcriptional changes to make the cellular environment conducive to root organogenesis. We believe that transcriptome reprogramming during DNRR from leaf explants is far more complex than the preliminary framework shown here. We anticipate that further exploration of the database will reveal many previously unknown or ambiguous factors and pathways and will provide a better understanding of how adventitious roots are regenerated upon wounding.
Methods
Plant materials and culture conditions
Arabidopsis Col-0 was the wild type in this study. Arabidopsis seeds were germinated and grown on 1/2 Murashige and Skoog (MS) basal medium with 1% (w/v) sucrose at 22°C under a 16-h light/8-h dark photoperiod for 12 d. The first pair of rosette leaves from 12-d-old seedlings was detached and cultured on sucrose-free B5 medium at 22°C under 24-h light conditions. The bop1-3 bop2-1 double mutant, the bop1-6D mutant, the plt3-1 plt5-2 plt7-1 triple mutant, the 35Spro:ERF115-SRDX line, the ERF115pro:GUS marker line, the PLT3pro:GUS marker line, and the PLT3pro:PLT3-YFP marker line have been described previously (Ha et al., 2004; Hepworth et al., 2005; Norberg et al., 2005; Prasad et al., 2011; Heyman et al., 2013; Du and Scheres, 2017). For the construction of other GUS marker lines, the promoters were PCR amplified and inserted into pBI101. The constructs were then introduced into Col-0 via Agrobacterium tumefaciens-mediated transformation. Two independent transgenic lines were tested at the T2 generation and showed similar results.
Dual-luciferase assay, qRT-PCR, ChIP, and GUS staining
For the dual-luciferase assay, cDNAs encoding PLT3-3×FLAG and PLT7-3×FLAG were cloned into the pMD19T vector under the control of the 35S promoter to generate 35Spro:PLT3-3×FLAG and 35Spro:PLT7-3×FLAG, respectively. Construction of UBQ10pro:3×FLAG-eGFP was described previously (Zhai and Xu, 2021). The 4.5-kb and 5.7-kb promoters of WOX5 and PLT1 were cloned into the pAB287 vector to generate WOX5pro:LUC and PLT1pro:LUC, respectively. The dual-luciferase assay was carried out in Arabidopsis protoplasts (Wu et al., 2009; Zhai and Xu, 2021) using the Dual-Luciferase Reporter Assay System (Promega). UBQ10pro:Rluc was used as the normalization control.
qRT-PCR, ChIP, and GUS staining were performed as previously described (He et al., 2012; Liu et al., 2014; Hu and Xu, 2016). Relative expression levels of genes were normalized against that of ACTIN in qRT-PCR. For the ChIP assay, detached leaves cultured on B5 medium with 0.9 μM indole-3-acetic acid (IAA) for 4 d under dark conditions from PLT3pro:PLT3-YFP and Col-0 (negative control) were used. Relative enrichment levels were normalized to that of ACTIN in ChIP. Primers used in molecular cloning, ChIP, and qRT-PCR are listed in Supplemental Table 7.
EMS mutagenesis and map-based cloning
To generate suppressor mutants that could rescue the rooting defect caused by 200 μM SHAM treatment, >4000 Col-0 seeds were mutagenized with EMS (0.2% w/v). Suppressors in the M2 generation were identified as those that formed roots under 200 μM SHAM treatment. One of the suppressors, designated squid1, was selected for further analysis. The squid1 locus was mapped by analyzing the F2 population from a cross between squid1 and Landsberg erecta (Ler) plants. The mutation was mapped to the SUR1 locus, and we renamed the squid1 mutant sur1-21. Mutation of SUR1 in sur1-21 was confirmed by testing the published sur1-1 allele (Sugawara et al., 2009), which showed the same DNRR phenotype as sur1-21.
Analysis of time-lapse RNA-seq data
For time-lapse RNA-seq, entire tissues from ∼30 leaf explants or the wounded regions from ∼40 leaf explants were collected and pooled as single samples for RNA extraction. Library construction and deep sequencing were performed using the Illumina HiSeq 3000 platform (Genergy Biotechnology, Shanghai, China). The paired-end RNA-seq reads were mapped to the A. thaliana genome (TAIR10) using STAR v2.7.2b (Dobin et al., 2013) (see Supplemental Table 7 for mapping efficiency). Gene transcript abundance was determined using RSEM v1.3.2 (Li and Dewey, 2011). Fold change and FDR were calculated using EBSeq (Leng et al., 2013). For an overview of the time-lapse RNA-seq data (Figure 1E and 1F; Supplemental Table 1), differentially expressed genes at each time point compared with t0 were identified using the following combined criteria: log2[fold change] > −1 and FDR < 0.05 for upregulated genes, and log2[fold change] < −1 and FDR < 0.05 for downregulated genes. Activation of candidate genes (Figures 2 and 4; Supplemental Figures 2 and 4; Supplemental Tables 2 and 4) was identified based on the following combined criteria: TPM < 2 at t0, log2[fold change] > −2 and FDR < 0.05. One of the biological replicates of Col-0 leaf explants from t0 to 12 h for early signals was from a previously published study (GEO: GSE120418; samples GSM3400572, GSM3400574, GSM3400576, GSM3400578, GSM3400580, GSM3400582, GSM3400584, and GSM3400586) (Zhang et al., 2019b), which was performed under the same growth and culture conditions described in this study. The mock and NPA treatments used the same t0 control.
For cluster analysis (Figures 2 and 4), the activated candidate genes were subjected to unsupervised clustering by the fuzzy c-means algorithm as implemented in the Mfuzz package (Kumar and Futschik, 2007).
GSEA, GO term analysis, and motif enrichment
For gene set enrichment analysis (GSEA) (Figure 1E and 1F; Supplemental Table 1), up- and downregulated genes at each time point were ordered by log2[fold change] as a ranked list of genes. Biological process GO terms were used for enrichment analysis by GSEAPreranked of GSEA v4.1.0 (Subramanian et al., 2005). GO terms with normalized P value <0.05 were considered to be significantly enriched in upregulated (NES > 1) or downregulated (NES < −1) genes. The significantly enriched GO terms were visualized using Gephi v0.9.2 (Bastian et al., 2009).
The GO term (Figures 2G, 3C, 4G, and 5C; Supplemental Tables 2, 3, 4, and 5) and motif enrichment (Figures 3D, 5D, and 5E; Supplemental Tables 3 and 5) analyses were performed using the online platform Plant Regulomics (Ran et al., 2020). A modified Fisher's exact test defined as an Expression Analysis Systematic Explorer (EASE) score (Huang et al., 2007) was used to test whether the input gene list significantly overlapped with the gene sets curated in the database.
Construction of the co-expression network
We constructed two co-expression networks using the R package WGCNA v1.69 (Zhang and Horvath, 2005). The first co-expression network was built using the time-lapse RNA-seq data from whole leaf explants of Col-0 and coi1-2 from t0 to 12 h (Supplemental Table 1), and the second co-expression network was built using the time-lapse RNA-seq data from the wounded region of leaf explants of mock and NPA treatments from t0 to 5 d (Supplemental Table 1). The two biological replicates of each set of RNA-seq data were treated as two samples. Genes with TPM >1 in at least six samples were used for the gene expression matrix to build the co-expression networks. The signedKME function was used to calculate the correlation (membership) between the expression patterns of each gene and the module eigengene. The top 20% of genes ranked by membership from high to low were defined as the hub genes in a module.
JA-, auxin-, and root-development-related co-expression sub-networks (Supplemental Figure 1B–1D) were built using JA-, auxin-, and root-development-related genes identified from associated GO terms and published data (Supplemental Table 7). Gene pairs with toplogical overlap matrix (TOM) similarity value >0.2 were considered to be co-expressed genes and were output for visualization. The JA-related co-expression sub-network was built using the time-lapse RNA-seq data of Col-0 and coi1-2, and the auxin- and root-development-related co-expression sub-networks were built using the time-lapse RNA-seq data from the mock and NPA treatments. The classes of co-expression sub-networks were determined by the function modularity of Gephi, and the co-expression sub-networks were visualized with Gephi (Blondel et al., 2008).
Single-cell RNA-seq data analysis
Tissue dissociation and preparation of single-cell suspensions were performed as described previously (Zhang et al., 2019a; Denyer et al., 2019). In brief, the detached Arabidopsis leaf base regions (0.3–0.5 cm in length from petiole) were collected and digested for 2 h at room temperature in digestion buffer (1.5% cellulase R10, 1.2% macerozyme R10, 0.4 M mannitol, 3 mM β-mercaptoethanol, 10 mM CaCl2, and 0.3% BSA). The protoplast solution was strained through a 40-μm filter and then through a 30-μm filter. The filtered protoplast solution was centrifuged at 500 g for 3 min, washed twice with wash buffer (8% mannitol), and resuspended in 50 μl of 8% mannitol and 0.04% BSA. Cell viability (about 94%) was confirmed by trypan blue staining.
Single-cell suspensions (around 500 cells/μl) were loaded onto 10× Chromium to capture approximately 8000 single cells following the manufacturer’s instructions for the 10× Genomics Chromium Single-Cell 3′ kit (V3). cDNA amplification, library construction, and sequencing on the Illumina NovaSeq 6000 platform (paired-end multiplexing run, 150 bp) were performed by LC-Bio Technology (HangZhou, China).
The single-cell RNA-seq raw data were mapped to the A. thaliana genome (TAIR10) using Cell Ranger v3.1.0 (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/3.1/what-is-cell-ranger). The gene expression matrix from Cell Ranger was used as input for the R package Seurat v3.0.2 (Butler et al., 2018) for cell clustering and dimensionality reduction analysis. Genes expressed in fewer than three cells and cells expressing more than 6500 genes and less than 650 genes were removed from analyses. The gene transcript level was log-transformed using the Seurat function NormalizeData. Highly variable genes were identified by FindVariableFeatures with the parameter “selection.method = mvp”. The mvp method was used to calculate the average expression and standard deviation of each gene, and genes were divided into 20 bins based on their average expression. Z scores were calculated for the standard deviation of genes within each bin. Genes with Z score >1 were identified as highly variable genes. After scaling the gene-cell expression matrix of highly variable genes, the first 20 PCs were used as input to identify cell clusters with a graph-based approach. We used UMAP (Becht et al., 2019) to visualize dimensionality-reduced cells. Cells with normalized expression of a gene >0.5 were considered to be a subset of cells expressing that gene. The Seurat function FindAllMarkers was used to identify the enriched genes in the cell clusters, and genes with pct.2 >0.25 were removed (Supplemental Table 6).
Funding
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB27030103), the National Natural Science Foundation of China (32000175/31770285/32070397), the Youth Innovation Promotion Association CAS (2014241), and the Chinese Academy of Sciences. A portion of this research was supported by a Global Research Collaboration Grant from the Offices of Research and Global Engagement to L.Y. from the University of Georgia and National Science Foundation under Grant NO. IOS2039313 to L.Y.
Author contributions
L.X. designed the research. X.F. performed time-lapse RNA-seq. W.L., X.F., F.G., and H.B. performed single-cell RNA-seq. W.L., X.F., L.C., J.Y., T.Z., and L.S. analyzed ROS, sur1-21, and ethylene in DNRR. W.L., S.T., M.S.I., and L.Y. analyzed BOPs in DNRR. W.L. analyzed ERF115 in DNRR. W.L. and N.Z. analyzed PLTs in DNRR. W.L. and Z.Y. performed qRT-PCR and GUS staining. Yuyun Z. and Yijing Z. performed RNA-seq bioinformatics analysis. All authors analyzed and discussed the data. L.X. wrote the manuscript.
Acknowledgments
We thank the ABRC, B. Scheres, D. Xie, L. De Veylder, W. Zhou, and O. Nilsson for Arabidopsis seeds used in this work. We thank V. Willemsen, J. Xu, and J.M. Pérez-Pérez for helpful discussion. We thank G. Zhang for technical assistance in ethylene research, H. Wang for GUS line analysis, and J. He for exploring online tools. No conflict of interest is declared.
Published: February 25, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Yijing Zhang, Email: zhangyijing@fudan.edu.cn.
Li Yang, Email: li.yang1@uga.edu.
Lin Xu, Email: xulin@cemps.ac.cn.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the accession numbers listed in Supplemental Table 7. Scripts used in this study are listed in Supplemental Data 1. The RNA-seq data obtained in this study have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and assigned the identifier accession numbers GSE147301 (one of the biological replicates of time-lapse RNA-seq for early signals), GSE147296 (two biological replicates of time-lapse RNA-seq for root organogenesis), and GSE147289 (single-cell RNA-seq) and in the Genome Sequence Archive (https://ngdc.cncb.ac.cn/gsa/) under the accession number CRA006050.
Supplemental information
References
- Abbas M., Berckhan S., Rooney D.J., Gibbs D.J., Vicente Conde J., Sousa Correia C., Bassel G.W., Marín-de la Rosa N., León J., Alabadí D., et al. Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr. Biol. 2015;25:1483–1488. doi: 10.1016/j.cub.2015.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahkami A.H., Lischewski S., Haensch K.-T., Porfirova S., Hofmann J., Rolletschek H., Melzer M., Franken P., Hause B., Druege U., et al. Molecular physiology of adventitious root formation in Petunia hybrida cuttings: involvement of wound response and primary metabolism. New Phytol. 2009;181:613–625. doi: 10.1111/j.1469-8137.2008.02704.x. [DOI] [PubMed] [Google Scholar]
- Ahkami A.H., Melzer M., Ghaffari M.R., Pollmann S., Ghorbani Javid M., Shahinnia F., Hajirezaei M.R., Druege U. Distribution of indole-3-acetic acid in Petunia hybrida shoot tip cuttings and relationship between auxin transport, carbohydrate metabolism and adventitious root formation. Planta. 2013;238:499–517. doi: 10.1007/s00425-013-1907-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.-S., Amasino R., Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Bai X., Todd C.D., Desikan R., Yang Y., Hu X. N-3-Oxo-Decanoyl-l-Homoserine-Lactone activates auxin-induced adventitious root formation via hydrogen peroxide- and nitric oxide-dependent cyclic GMP signaling in mung bean. Plant Physiol. 2012;158:725–736. doi: 10.1104/pp.111.185769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M., Heymann S., Jacomy M. Int. AAAI Conf. Weblogs Soc. Media. 2009. Gephi: an open source software for exploring and manipulating networks. [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Belda-Palazon B., Gonzalez-Garcia M.-P., Lozano-Juste J., Coego A., Antoni R., Julian J., Peirats-Llobet M., Rodriguez L., Berbel A., Dietrich D., et al. PYL8 mediates ABA perception in the root through non-cell-autonomous and ligand-stabilization-based mechanisms. Proc. Natl. Acad. Sci. U S A. 2018;115:E11857–E11863. doi: 10.1073/pnas.1815410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini C., Pacurar D.I., Perrone I. Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 2014;65:639–666. doi: 10.1146/annurev-arplant-050213-035645. [DOI] [PubMed] [Google Scholar]
- Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. Theor. Exp. 2008;2008:P10008. [Google Scholar]
- Boerjan W., Cervera M.T., Delarue M., Beeckman T., Dewitte W., Bellini C., Caboche M., Van Onckelen H., Van Montagu M., Inzé D. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonke M., Thitamadee S., Mähönen A.P., Hauser M.-T., Helariutta Y. APL regulates vascular tissue identity in Arabidopsis. Nature. 2003;426:181–186. doi: 10.1038/nature02100. [DOI] [PubMed] [Google Scholar]
- Burkart R.C., Strotmann V.I., Kirschner G.K., et al. PLETHORA-WOX5 interaction and subnuclear localisation control Arabidopsis root stem cell maintenance. bioRxiv. 2021 doi: 10.1101/818187. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustillo-Avendaño E., Ibáñez S., Sanz O., Barros J.A.S., Gude I., Perianez-Rodriguez J., Micol J.L., del Pozo J.C., Moreno-Risueno M.A., Pérez-Pérez J.M., et al. Regulation of hormonal control, cell reprogramming, and patterning during de novo root organogenesis. Plant Physiol. 2018;176:1709–1727. doi: 10.1104/pp.17.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X.-T., Xu P., Zhao P.-X., Liu R., Yu L.-H., Xiang C.-B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014;5:5833. doi: 10.1038/ncomms6833. [DOI] [PubMed] [Google Scholar]
- Camañes G., Pastor V., Cerezo M., García-Andrade J., Vicedo B., García-Agustín P., Flors V. A deletion in NRT2.1 attenuates Pseudomonas syringae -induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol. 2012;158:1054–1066. doi: 10.1104/pp.111.184424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo M., Tillard P., Filleur S., Muños S., Daniel-Vedele F., Gojon A. Major alterations of the regulation of root NO 3 − uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in arabidopsis. Plant Physiol. 2001;127:262–271. doi: 10.1104/pp.127.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-Q., Qu X.-Q., Hou B.-H., Sosso D., Osorio S., Fernie A.R., Frommer W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 2012;335:207–211. doi: 10.1126/science.1213351. [DOI] [PubMed] [Google Scholar]
- Chen X., Qu Y., Sheng L., Liu J., Huang H., Xu L. A simple method suitable to study de novo root organogenesis. Front. Plant Sci. 2014;5:1–6. doi: 10.3389/fpls.2014.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Tong J., Xiao L., Ruan Y., Liu J., Zeng M., Huang H., Wang J.W., Xu L. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016;67:4273–4284. doi: 10.1093/jxb/erw213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Cheng J., Chen L., Zhang G., Huang H., Zhang Y., Xu L. Auxin-independent NAC pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiol. 2016;170:2136–2145. doi: 10.1104/pp.15.01733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D.G., Gubrium E.K., Barrett J.E., Nell T.A., Klee H.J. Root formation in ethylene-insensitive plants. Plant Physiol. 1999;121:53–60. doi: 10.1104/pp.121.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa C.T., de Almeida M.R., Ruedell C.M., Schwambach J., Maraschin F.S., Fett-Neto A.G. When stress and development go hand in hand: main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013;4:133. doi: 10.3389/fpls.2013.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood T., Yang X., Visser E.J.W., te Beek T.A.H., Kensche P.R., Cristescu S.M., Lee S., Floková K., Nguyen D., Mariani C., et al. A Co-opted hormonal cascade activates dormant adventitious root primordia upon flooding in solanum dulcamara. Plant Physiol. 2016;170:2351–2364. doi: 10.1104/pp.15.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Klerk G.-J., Hanecakova J. Ethylene and rooting of mung bean cuttings. The role of auxin induced ethylene synthesis and phase-dependent effects. Plant Growth Regul. 2008;56:203–209. [Google Scholar]
- De Klerk G.J., Van Der Krieken W., De Jong J.C. The formation of adventitious roots: new concepts, new possibilities. Vitr. Cell. Dev. Biol. - Plant. 1999;35:189–199. [Google Scholar]
- De Rybel B., Vassileva V., Parizot B., Demeulenaere M., Grunewald W., Audenaert D., Van Campenhout J., Overvoorde P., Jansen L., Vanneste S., et al. A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 2010;20:1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Dechorgnat J., Patrit O., Krapp A., Fagard M., Daniel-Vedele F. Characterization of the Nrt2.6 gene in Arabidopsis thaliana: a link with plant response to biotic and abiotic stress. PLoS One. 2012;7:e42491. doi: 10.1371/journal.pone.0042491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E. A metal-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 1996;111:849–855. doi: 10.1104/pp.111.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Rovere F., Fattorini L., D’Angeli S., Veloccia A., Falasca G., Altamura M.M. Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of arabidopsis. Ann. Bot. 2013;112:1395–1407. doi: 10.1093/aob/mct215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer T., Ma X., Klesen S., Scacchi E., Nieselt K., Timmermans M.C.P. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell. 2019;48:840–852.e5. doi: 10.1016/j.devcel.2019.02.022. [DOI] [PubMed] [Google Scholar]
- Ding L., Yan S., Jiang L., Zhao W., Ning K., Zhao J., Liu X., Zhang J., Wang Q., Zhang X. HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in arabidopsis. PLoS Genet. 2015;11:e1005479. doi: 10.1371/journal.pgen.1005479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Cheng L., Scagel C.F., Fuchigami L.H. Nitrogen mobilization, nitrogen uptake and growth of cuttings obtained from poplar stock plants grown in different N regimes and sprayed with urea in autumn. Tree Physiol. 2004;24:355–359. doi: 10.1093/treephys/24.3.355. [DOI] [PubMed] [Google Scholar]
- Druege U., Franken P., Lischewski S., Ahkami A.H., Zerche S., Hause B., Hajirezaei M.R. Transcriptomic analysis reveals ethylene as stimulator and auxin as regulator of adventitious root formation in petunia cuttings. Front. Plant Sci. 2014;5:494. doi: 10.3389/fpls.2014.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druege U., Franken P., Hajirezaei M.R. Plant hormone homeostasis, signaling, and function during adventitious root formation in cuttings. Front. Plant Sci. 2016;7:381. doi: 10.3389/fpls.2016.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druege U., Hilo A., Pérez-Pérez J.M., Klopotek Y., Acosta M., Shahinnia F., Zerche S., Franken P., Hajirezaei M.R. Molecular and physiological control of adventitious rooting in cuttings: phytohormone action meets resource allocation. Ann. Bot. 2019;123:929–949. doi: 10.1093/aob/mcy234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Scheres B. PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth. Proc. Natl. Acad. Sci. U S A. 2017;114:11709–11714. doi: 10.1073/pnas.1714410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R.C., Betzner A.S., Huttner E., Oakes M.P., Tucker W.Q., Gerentes D., Perez P., Smyth D.R. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells J.P., Turner S.R. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development. 2010;137:767–774. doi: 10.1242/dev.044941. [DOI] [PubMed] [Google Scholar]
- Falasca G., Zaghi D., Possenti M., Altamura M.M. Adventitious root formation in Arabidopsis thaliana thin cell layers. Plant Cell Rep. 2004;23:17–25. doi: 10.1007/s00299-004-0801-3. [DOI] [PubMed] [Google Scholar]
- Fattorini L., Falasca G., Kevers C., Mainero Rocca L., Zadra C., Altamura M.M. Adventitious rooting is enhanced by methyl jasmonate in tobacco thin cell layers. Planta. 2009;231:155–168. doi: 10.1007/s00425-009-1035-y. [DOI] [PubMed] [Google Scholar]
- Fattorini L., Hause B., Gutierrez L., Veloccia A., Della Rovere F., Piacentini D., Falasca G., Altamura M.M. Jasmonate promotes auxin-induced adventitious rooting in dark-grown Arabidopsis thaliana seedlings and stem thin cell layers by a cross-talk with ethylene signalling and a modulation of xylogenesis. BMC Plant Biol. 2018;18:182. doi: 10.1186/s12870-018-1392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filleur S., Dorbe M.F., Cerezo M., Orsel M., Granier F., Gojon A., Daniel-Vedele F. An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 2001;489:220–224. doi: 10.1016/s0014-5793(01)02096-8. [DOI] [PubMed] [Google Scholar]
- Fisher K., Turner S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 2007;17:1061–1066. doi: 10.1016/j.cub.2007.05.049. [DOI] [PubMed] [Google Scholar]
- Guénin S., Mareck A., Rayon C., Lamour R., Assoumou Ndong Y., Domon J.-M., Sénéchal F., Fournet F., Jamet E., Canut H., et al. Identification of pectin methylesterase 3 as a basic pectin methylesterase isoform involved in adventitious rooting in Arabidopsis thaliana. New Phytol. 2011;192:114–126. doi: 10.1111/j.1469-8137.2011.03797.x. [DOI] [PubMed] [Google Scholar]
- Ha C.M., Kim G.-T., Kim B.C., Jun J.H., Soh M.S., Ueno Y., Machida Y., Tsukaya H., Nam H.G. The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development. 2003;130:161–172. doi: 10.1242/dev.00196. [DOI] [PubMed] [Google Scholar]
- Ha C.M., Jun J.H., Nam H.G., Fletcher J.C. BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1361–1370. doi: 10.1093/pcp/pch201. [DOI] [PubMed] [Google Scholar]
- Ha C.M., Jun J.H., Nam H.G., Fletcher J.C. BLADE-ON-PETIOLE1 and 2 control arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell. 2007;19:1809–1825. doi: 10.1105/tpc.107.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Chen X., Huang H., Xu L. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 2012;8:e1002911. doi: 10.1371/journal.pgen.1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S.R., Zhang Y., McKim S., Li X., Haughn G.W. BLADE-ON-PETIOLE–Dependent signaling controls leaf and floral patterning in arabidopsis. Plant Cell. 2005;17:1434–1448. doi: 10.1105/tpc.104.030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J., Cools T., Vandenbussche F., Heyndrickx K.S., Van Leene J., Vercauteren I., Vanderauwera S., Vandepoele K., De Jaeger G., Van Der Straeten D., et al. ERF115 controls root quiescent center cell division and stem cell replenishment. Science. 2013;342:860–863. doi: 10.1126/science.1240667. [DOI] [PubMed] [Google Scholar]
- Heyman J., Cools T., Canher B., Shavialenka S., Traas J., Vercauteren I., Van den Daele H., Persiau G., De Jaeger G., Sugimoto K., et al. The heterodimeric transcription factor complex ERF115–PAT1 grants regeneration competence. Nat. Plants. 2016;2:16165. doi: 10.1038/nplants.2016.165. [DOI] [PubMed] [Google Scholar]
- Hilo A., Shahinnia F., Druege U., Franken P., Melzer M., Rutten T., von Wirén N., Hajirezaei M.-R.R. A specific role of iron in promoting meristematic cell division during adventitious root formation. J. Exp. Bot. 2017;68:4233–4247. doi: 10.1093/jxb/erx248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y., Fukuda H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. U S A. 2008;105:15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Hirota A., Kato T., Fukaki H., Aida M., Tasaka M. The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of arabidopsis. Plant Cell. 2007;19:2156–2168. doi: 10.1105/tpc.107.050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock A.E., Zimmerman P.W. Effect of the use of growth substances on the rooting response of cuttings. Contrib. Boyce Thompson Inst. 1936;8:63–79. [Google Scholar]
- Hofhuis H., Laskowski M., Du Y., Prasad K., Grigg S., Pinon V., Scheres B. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three plethora transcription factors. Curr. Biol. 2013;23:956–962. doi: 10.1016/j.cub.2013.04.048. [DOI] [PubMed] [Google Scholar]
- Hu X., Xu L. Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 2016;172:2363–2373. doi: 10.1104/pp.16.01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Zhang G., Liu W., Shi J., Wang H., Qi M., Li J., Qin P., Ruan Y., Huang H., et al. Divergent regeneration-competent cells adopt a common mechanism for callus initiation in angiosperms. Regeneration. 2017;4:132–139. doi: 10.1002/reg2.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A., Wang Y., Liu Y., Wang G., She X. Reactive oxygen species regulate auxin levels to mediate adventitious root induction in Arabidopsis hypocotyl cuttings. J. Integr. Plant Biol. 2019;62:912–926. doi: 10.1111/jipb.12870. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M., Iwase A., Rymen B., Harashima H., Shibata M., Ohnuma M., Breuer C., Morao A.K., de Lucas M., De Veylder L., et al. PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat. Plants. 2015;1:15089. doi: 10.1038/nplants.2015.89. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M., Favero D.S., Sakamoto Y., Iwase A., Coleman D., Rymen B., Sugimoto K. Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 2019;70:377–406. doi: 10.1146/annurev-arplant-050718-100434. [DOI] [PubMed] [Google Scholar]
- Iwase A., Mitsuda N., Koyama T., Hiratsu K., Kojima M., Arai T., Inoue Y., Seki M., Sakakibara H., Sugimoto K., et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 2011;21:508–514. doi: 10.1016/j.cub.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Jean-Baptiste K., McFaline-Figueroa J.L., Alexandre C.M., Dorrity M.W., Saunders L., Bubb K.L., Trapnell C., Fields S., Queitsch C., Cuperus J.T. Dynamics of gene expression in single root cells of Arabidopsis thaliana. Plant Cell. 2019;31:993–1011. doi: 10.1105/tpc.18.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius B.T., Leach K.A., Tran T.M., Mertz R.A., Braun D.M. Sugar transporters in plants: new insights and discoveries. Plant Cell Physiol. 2017;58:1442–1460. doi: 10.1093/pcp/pcx090. [DOI] [PubMed] [Google Scholar]
- Kang N.Y., Lee H.W., Kim J. The AP2/EREBP gene PUCHI Co-acts with LBD16/ASL18 and LBD18/ASL20 downstream of ARF7 and ARF19 to regulate lateral root development in arabidopsis. Plant Cell Physiol. 2013;54:1326–1334. doi: 10.1093/pcp/pct081. [DOI] [PubMed] [Google Scholar]
- Kanno Y., Oikawa T., Chiba Y., Ishimaru Y., Shimizu T., Sano N., Koshiba T., Kamiya Y., Ueda M., Seo M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016;7:13245. doi: 10.1038/ncomms13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareem A., Durgaprasad K., Sugimoto K., Du Y., Pulianmackal A.J., Trivedi Z.B., Abhayadev P.V., Pinon V., Meyerowitz E.M., Scheres B., et al. PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 2015;25:1017–1030. doi: 10.1016/j.cub.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechid M., Desbrosses G., Rokhsi W., Varoquaux F., Djekoun A., Touraine B. The NRT2.5 and NRT2.6 genes are involved in growth promotion of Arabidopsis by the plant growth-promoting rhizobacterium (PGPR) strain Phyllobacterium brassicacearum STM196. New Phytol. 2013;198:514–524. doi: 10.1111/nph.12158. [DOI] [PubMed] [Google Scholar]
- Khan M., Xu M., Murmu J., Tabb P., Liu Y., Storey K., McKim S.M., Douglas C.J., Hepworth S.R. Antagonistic interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE regulates arabidopsis inflorescence architecture. Plant Physiol. 2012;158:946–960. doi: 10.1104/pp.111.188573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Feria-Bourrellier A.-B., Lafouge F., Lezhneva L., Boutet-Mercey S., Orsel M., Bréhaut V., Miller A., Daniel-Vedele F., Sakakibara H., et al. The arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell. 2012;24:245–258. doi: 10.1105/tpc.111.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopotek Y., Franken P., Klaering H.-P., Fischer K., Hause B., Hajirezaei M.-R., Druege U. A higher sink competitiveness of the rooting zone and invertases are involved in dark stimulation of adventitious root formation in Petunia hybrida cuttings. Plant Sci. 2016;243:10–22. doi: 10.1016/j.plantsci.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Klucher K.M., Chow H., Reiser L., Fischer R.L. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotula L., Clode P.L., Striker G.G., Pedersen O., Läuchli A., Shabala S., Colmer T.D. Oxygen deficiency and salinity affect cell-specific ion concentrations in adventitious roots of barley ( Hordeum vulgare ) New Phytol. 2015;208:1114–1125. doi: 10.1111/nph.13535. [DOI] [PubMed] [Google Scholar]
- Kumar L., Futschik M.E. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusk S., Sohlberg J.J., Magnus Eklund D., Sundberg E. Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J. 2006;47:99–111. doi: 10.1111/j.1365-313X.2006.02774.x. [DOI] [PubMed] [Google Scholar]
- Lakehal A., Dob A., Novák O., Bellini C. A DAO1-mediated circuit controls auxin and jasmonate crosstalk robustness during adventitious root initiation in Arabidopsis. Int. J. Mol. Sci. 2019;20:4428. doi: 10.3390/ijms20184428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakehal A., Dob A., Rahneshan Z., Novák O., Escamez S., Alallaq S., Strnad M., Tuominen H., Bellini C. ETHYLENE RESPONSE FACTOR 115 integrates jasmonate and cytokinin signaling machineries to repress adventitious rooting in Arabidopsis. New Phytol. 2020;228:1611–1626. doi: 10.1111/nph.16794. [DOI] [PubMed] [Google Scholar]
- Leng N., Dawson J.A., Thomson J.A., Ruotti V., Rissman A.I., Smits B.M.G., Haag J.D., Gould M.N., Stewart R.M., Kendziorski C. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva L., Kiba T., Feria-Bourrellier A.B., Lafouge F., Boutet-Mercey S., Zoufan P., Sakakibara H., Daniel-Vedele F., Krapp A. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 2014;80:230–241. doi: 10.1111/tpj.12626. [DOI] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-W., Xue L. The interaction between H2O2 and NO, Ca2+, cGMP, and MAPKs during adventitious rooting in mung bean seedlings. Vitr. Cell. Dev. Biol. - Plant. 2010;46:142–148. [Google Scholar]
- Li J.-Y., Fu Y.-L., Pike S.M., Bao J., Tian W., Zhang Y., Chen C.-Z., Zhang Y., Li H.-M., Huang J., et al. The arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell. 2010;22:1633–1646. doi: 10.1105/tpc.110.075242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang Y., Tan S., Li Z., Yuan Z., Glanc M., Domjan D., Wang K., Xuan W., Guo Y., et al. Root growth adaptation is mediated by PYLs ABA receptor-PP2A protein phosphatase complex. Adv. Sci. 2020;7:1901455. doi: 10.1002/advs.201901455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yao L., Sun L., Zhu Z. ETHYLENE INSENSITIVE 3 suppresses plants de novo root regeneration from leaf explants and mediates age-regulated regeneration decline. Development. 2020;147:dev179457. doi: 10.1242/dev.179457. [DOI] [PubMed] [Google Scholar]
- Liao W., Xiao H., Zhang M. Role and relationship of nitric oxide and hydrogen peroxide in adventitious root development of marigold. Acta Physiol. Plant. 2009;31:1279–1289. [Google Scholar]
- Lin Y., Zhang W., Qi F., Cui W., Xie Y., Shen W. Hydrogen-rich water regulates cucumber adventitious root development in a heme oxygenase-1/carbon monoxide-dependent manner. J. Plant Physiol. 2014;171:1–8. doi: 10.1016/j.jplph.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Lischweski S., Muchow A., Guthörl D., Hause B. Jasmonates act positively in adventitious root formation in petunia cuttings. BMC Plant Biol. 2015;15:229. doi: 10.1186/s12870-015-0615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sheng L., Xu Y., Li J., Yang Z., Huang H., Xu L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell. 2014;26:1081–1093. doi: 10.1105/tpc.114.122887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Yu J., Ge Y., Qin P., Xu L. Pivotal role of LBD16 in root and root-like organ initiation. Cell. Mol. Life Sci. 2018;75:3329–3338. doi: 10.1007/s00018-018-2861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Porat R., Nadeau J.A., O’Neill S.D. Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell. 1996;8:2155–2168. doi: 10.1105/tpc.8.12.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi K.K., Misra A., Ren S., Mcknight T.D. BT2, a BTB protein, mediates multiple responses to nutrients, stresses, and hormones in Arabidopsis. Plant Physiol. 2009;150:1930–1939. doi: 10.1104/pp.109.139220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano C., Pallero-Baena M., Casimiro I., De Rybel B., Orman-Ligeza B., Van Isterdael G., Beeckman T., Draye X., Casero P., del Pozo J.C. The emerging role of reactive oxygen species signaling during lateral root development. Plant Physiol. 2014;165:1105–1119. doi: 10.1104/pp.114.238873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim S.M., Stenvik G.-E., Butenko M.A., Kristiansen W., Cho S.K., Hepworth S.R., Aalen R.B., Haughn G.W. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development. 2008;135:1537–1546. doi: 10.1242/dev.012807. [DOI] [PubMed] [Google Scholar]
- Mikkelsen M.D., Naur P., Halkier B.A. Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004;37:770–777. doi: 10.1111/j.1365-313x.2004.02002.x. [DOI] [PubMed] [Google Scholar]
- Mironova V., Xu J. A single-cell view of tissue regeneration in plants. Curr. Opin. Plant Biol. 2019;52:149–154. doi: 10.1016/j.pbi.2019.09.003. [DOI] [PubMed] [Google Scholar]
- Norberg M., Holmlund M., Nilsson O. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development. 2005;132:2203–2213. doi: 10.1242/dev.01815. [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Harashima H., Dissmeyer N., Zhao X., Bouyer D., Weimer A.K., De Winter F., Yang F., Schnittger A. Genetic framework of cyclin-dependent kinase function in arabidopsis. Dev. Cell. 2012;22:1030–1040. doi: 10.1016/j.devcel.2012.02.015. [DOI] [PubMed] [Google Scholar]
- Okumoto S., Schmidt R., Tegeder M., Fischer W.N., Rentsch D., Frommer W.B., Koch W. High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of arabidopsis. J. Biol. Chem. 2002;277:45338–45346. doi: 10.1074/jbc.M207730200. [DOI] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman-Ligeza B., Parizot B., de Rycke R., Fernandez A., Himschoot E., Van Breusegem F., Bennett M.J., Périlleux C., Beeckman T., Draye X. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development. 2016;143:3328–3339. doi: 10.1242/dev.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cardenas M., Ryan C.A. Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. U S A. 1999;96:6553–6557. doi: 10.1073/pnas.96.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacurar D.I., Perrone I., Bellini C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plant. 2014;151:83–96. doi: 10.1111/ppl.12171. [DOI] [PubMed] [Google Scholar]
- Pan J., Zhao F., Zhang G., Pan Y., Sun L., Bao N., Qin P., Chen L., Yu J., Zhang Y., et al. Control of de novo root regeneration efficiency by developmental status of Arabidopsis leaf explants. J. Genet. Genom. 2019;46:133–140. doi: 10.1016/j.jgg.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Pan X., Yang Z., Xu L. Dual roles of jasmonate in adventitious rooting. J. Exp. Bot. 2021;72:6808–6810. doi: 10.1093/jxb/erab378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park O.-S., Bae S.H., Kim S.-G., Seo P.J. JA-pretreated hypocotyl explants potentiate de novo shoot regeneration in Arabidopsis. Plant Signal. Behav. 2019;14:1618180. doi: 10.1080/15592324.2019.1618180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Garcia P., Moreno-Risueno M.A. Stem cells and plant regeneration. Dev. Biol. 2018;442:3–12. doi: 10.1016/j.ydbio.2018.06.021. [DOI] [PubMed] [Google Scholar]
- Prasad K., Grigg S.P., Barkoulas M., Yadav R.K., Sanchez-Perez G.F., Pinon V., Blilou I., Hofhuis H., Dhonukshe P., Galinha C., et al. Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr. Biol. 2011;21:1123–1128. doi: 10.1016/j.cub.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Prasad A., Sedlářová M., Kale R.S., Pospíšil P. Lipoxygenase in singlet oxygen generation as a response to wounding: in vivo imaging in Arabidopsis thaliana. Sci. Rep. 2017;7:9831. doi: 10.1038/s41598-017-09758-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A., Sedlářová M., Balukova A., Rác M., Pospíšil P. Reactive oxygen species as a response to wounding: in vivo imaging in Arabidopsis thaliana. Front. Plant Sci. 2020;10:1660. doi: 10.3389/fpls.2019.01660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran X., Zhao F., Wang Y., Liu J., Zhuang Y., Ye L., Qi M., Cheng J., Zhang Y. Plant Regulomics: a data-driven interface for retrieving upstream regulators from plant multi-omics data. Plant J. 2020;101:237–248. doi: 10.1111/tpj.14526. [DOI] [PubMed] [Google Scholar]
- Randall R.S., Miyashima S., Blomster T., Zhang J., Elo A., Karlberg A., Immanen J., Nieminen K., Lee J.-Y., Kakimoto T., et al. AINTEGUMENTA and the D-type cyclin CYCD3;1 regulate root secondary growth and respond to cytokinins. Biol. Open. 2015;4:1229–1236. doi: 10.1242/bio.013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riov J., Yang S.F. Ethylene and auxin-ethylene interaction in adventitious root formation in mung bean (Vigna radiata) cuttings. J. Plant Growth Regul. 1989;8:131–141. [Google Scholar]
- Rogers E.E., Guerinot M. Lou. FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell. 2002;14:1787–1799. doi: 10.1105/tpc.001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B., Kawamura A., Lambolez A., Inagaki S., Takebayashi A., Iwase A., Sakamoto Y., Sako K., Favero D.S., Ikeuchi M., et al. Histone acetylation orchestrates wound-induced transcriptional activation and cellular reprogramming in Arabidopsis. Commun. Biol. 2019;2:404. doi: 10.1038/s42003-019-0646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu K.H., Huang L., Kang H.M., Schiefelbein J. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 2019;179:1444–1456. doi: 10.1104/pp.18.01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y.L., Cheng Z.J., Zhang X.S. Plant stem cells and de novo organogenesis. New Phytol. 2018;218:1334–1339. doi: 10.1111/nph.15106. [DOI] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Scofield S., Jones A., Murray J.A.H. The plant cell cycle in context. J. Exp. Bot. 2014;65:2557–2562. doi: 10.1093/jxb/eru188. [DOI] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M.F. The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 1999;20:259–263. doi: 10.1046/j.1365-313x.1999.00594.x. [DOI] [PubMed] [Google Scholar]
- Seyfferth C., Renema J., Wendrich J.R., Eekhout T., Seurinck R., Vandamme N., Blob B., Saeys Y., Helariutta Y., Birnbaum K.D., et al. Advances and opportunities in single-cell transcriptomics for plant research. Annu. Rev. Plant Biol. 2021;72:847–866. doi: 10.1146/annurev-arplant-081720-010120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmukhan A.P., Mathew M.M., Aiyaz M., Varaparambathu V., Kareem A., Radhakrishnan D., Prasad K. Regulation of touch-stimulated de novo root regeneration from Arabidopsis leaves. Plant Physiol. 2021;187:52–58. doi: 10.1093/plphys/kiab286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng L., Hu X., Du Y., Zhang G., Huang H., Scheres B., Xu L. Non-canonical WOX11-mediated root branching contributes to plasticity in Arabidopsis root system architecture. Development. 2017;144:3126–3133. doi: 10.1242/dev.152132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno A., Heidstra R., Blilou I., Scheres B. Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes Dev. 2018;32:1085–1100. doi: 10.1101/gad.314096.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla V., Lombardi L., Iacopino S., Pencik A., Novak O., Perata P., Giuntoli B., Licausi F. Endogenous hypoxia in lateral root primordia controls root architecture by antagonizing auxin signaling in arabidopsis. Mol. Plant. 2019;12:538–551. doi: 10.1016/j.molp.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Shulse C.N., Cole B.J., Ciobanu D., Lin J., Yoshinaga Y., Gouran M., Turco G.M., Zhu Y., O’Malley R.C., Brady S.M., et al. High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 2019;27:2241–2247.e4. doi: 10.1016/j.celrep.2019.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Yadav S., Singh A., Mahima M., Singh A., Gautam V., Sarkar A.K. Auxin signaling modulates LATERAL ROOT PRIMORDIUM 1 (LRP1) expression during lateral root development in Arabidopsis. Plant J. 2020;101:87–100. doi: 10.1111/tpj.14520. [DOI] [PubMed] [Google Scholar]
- Smetana O., Mäkilä R., Lyu M., Amiryousefi A., Sánchez Rodríguez F., Wu M.-F., Solé-Gil A., Leal Gavarrón M., Siligato R., Miyashima S., et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature. 2019;565:485–489. doi: 10.1038/s41586-018-0837-0. [DOI] [PubMed] [Google Scholar]
- Smith D.L., Fedoroff N.V. LRP1, a gene expressed in lateral and adventitious root primordia of arabidopsis. Plant Cell. 1995;7:735–745. doi: 10.1105/tpc.7.6.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S., Hishiyama S., Jikumaru Y., Hanada A., Nishimura T., Koshiba T., Zhao Y., Kamiya Y., Kasahara H. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2009;106:5430–5435. doi: 10.1073/pnas.0811226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Xu Y., Ye S., Jiang H., Chen Q., Liu F., Zhou W., Chen R., Li X., Tietz O., et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell. 2009;21:1495–1511. doi: 10.1105/tpc.108.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Chen L., Liu J., Zhang X., Yang Z., Liu W., Xu L. TAA family contributes to auxin production during de novo regeneration of adventitious roots from Arabidopsis leaf explants. Sci. Bull. 2016;61:1728–1731. [Google Scholar]
- Svenson S.E., Davies F.T. Change in tissue mineral elemental concentration during root initiation and development of poinsettia cuttings. HortScience. 1995;30:617–619. [Google Scholar]
- Takáč T., Obert B., Rolčík J., Šamaj J. Improvement of adventitious root formation in flax using hydrogen peroxide. N. Biotechnol. 2016;33:728–734. doi: 10.1016/j.nbt.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Taniguchi M., Furutani M., Nishimura T., Nakamura M., Fushita T., Iijima K., Baba K., Tanaka H., Toyota M., Tasaka M., et al. The arabidopsis LAZY1 family plays a key role in gravity signaling within statocytes and in branch angle control of roots and shoots. Plant Cell. 2017;29:1984–1999. doi: 10.1105/tpc.16.00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-López R., García-Ponce B., Dubrovsky J.G., Garay-Arroyo A., Pérez-Ruíz R.V., Kim S.-H., Acevedo F., Pelaz S., Alvarez-Buylla E.R. An AGAMOUS -related MADS-box gene, XAL1 ( AGL12 ), regulates root meristem cell proliferation and flowering transition in arabidopsis. Plant Physiol. 2008;146:1182–1192. doi: 10.1104/pp.107.108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann K.V., Went F.W. On the chemical nature of the root forming hormone. Proc. K. Ned. Akad. Wet. Ser. C Biol. Med. Sci. 1934;37:456–459. [Google Scholar]
- Toyokura K., Goh T., Shinohara H., Shinoda A., Kondo Y., Okamoto Y., Uehara T., Fujimoto K., Okushima Y., Ikeyama Y., et al. Lateral inhibition by a peptide hormone-receptor cascade during arabidopsis lateral root founder cell formation. Dev. Cell. 2019;48:64–75.e5. doi: 10.1016/j.devcel.2018.11.031. [DOI] [PubMed] [Google Scholar]
- Trinh D.-C., Lavenus J., Goh T., Boutté Y., Drogue Q., Vaissayre V., Tellier F., Lucas M., Voß U., Gantet P., et al. PUCHI regulates very long chain fatty acid biosynthesis during lateral root and callus formation. Proc. Natl. Acad. Sci. U S A. 2019;116:14325–14330. doi: 10.1073/pnas.1906300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Sauer N. The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta. 1995;196:564–570. doi: 10.1007/BF00203657. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H., Busch W., Benfey P.N. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Veloccia A., Fattorini L., Della Rovere F., Sofo A., D’Angeli S., Betti C., Falasca G., Altamura M.M. Ethylene and auxin interaction in the control of adventitious rooting in Arabidopsis thaliana. J. Exp. Bot. 2016;67:6445–6458. doi: 10.1093/jxb/erw415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten I., Schotte S., Geelen D. Hypocotyl adventitious root organogenesis differs from lateral root development. Front. Plant Sci. 2014;5:1–13. doi: 10.3389/fpls.2014.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-Y., Cheng Y.-H., Chen K.-E., Tsay Y.-F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018;69:85–122. doi: 10.1146/annurev-arplant-042817-040056. [DOI] [PubMed] [Google Scholar]
- Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S. Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods. 2009;5:16. doi: 10.1186/1746-4811-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Frugis G., Colgan D., Chua N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Zhao Y., Gao J., Xiang C., Zhu J.-K. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci. Rep. 2016;6:27177. doi: 10.1038/srep27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. De novo root regeneration from leaf explants: wounding, auxin, and cell fate transition. Curr. Opin. Plant Biol. 2018;41:39–45. doi: 10.1016/j.pbi.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. The SCF COI1 ubiquitin-ligase complexes are required for jasmonate response in arabidopsis. Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hu T., McKim S.M., Murmu J., Haughn G.W., Hepworth S.R. Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J. 2010;63:974–989. doi: 10.1111/j.1365-313X.2010.04299.x. [DOI] [PubMed] [Google Scholar]
- Yang S., Yu Q., Zhang Y., Jia Y., Wan S., Kong X., Ding Z. ROS: the fine-tuner of plant stem cell fate. Trends Plant Sci. 2018;23:850–853. doi: 10.1016/j.tplants.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Ye B.-B., Shang G.-D., Pan Y., Xu Z.-G., Zhou C.-M., Mao Y.-B., Bao N., Sun L., Xu T., Wang J.-W. AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell. 2020;32:226–241. doi: 10.1105/tpc.19.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Dong Z., Wu H., Tian Z., Zhao Z. Redox regulation of plant stem cell fate. EMBO J. 2017;36:e201695955. doi: 10.15252/embj.201695955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Verstraeten I., Trinh H.K., Heugebaert T., Stevens C.V., Garcia-Maquilon I., Rodriguez P.L., Vanneste S., Geelen D. Arabidopsis hypocotyl adventitious root formation is suppressed by ABA signaling. Genes (Basel) 2021;12:1141. doi: 10.3390/genes12081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerche S., Haensch K.-T., Druege U., Hajirezaei M.-R. Nitrogen remobilisation facilitates adventitious root formation on reversible dark-induced carbohydrate depletion in Petunia hybrida. BMC Plant Biol. 2016;16:219. doi: 10.1186/s12870-016-0901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai N., Xu L. Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nat. Plants. 2021;7:1453–1460. doi: 10.1038/s41477-021-01015-8. [DOI] [PubMed] [Google Scholar]
- Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zhang C., Turgeon R. Mechanisms of phloem loading. Curr. Opin. Plant Biol. 2018;43:71–75. doi: 10.1016/j.pbi.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Zhang G.-B., Yi H.-Y., Gong J.-M. The arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell. 2014;26:3984–3998. doi: 10.1105/tpc.114.129296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.-Q., Xu Z.-G., Shang G.-D., Wang J.-W. A single-cell RNA sequencing profiles the developmental landscape of arabidopsis root. Mol. Plant. 2019;12:648–660. doi: 10.1016/j.molp.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Zhang G., Zhao F., Chen L., Pan Y., Sun L., Bao N., Zhang T., Cui C.-X., Qiu Z., Zhang Y., et al. Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants. 2019;5:491–497. doi: 10.1038/s41477-019-0408-x. [DOI] [PubMed] [Google Scholar]
- Zhang G., Liu W., Gu Z., Wu S., E Y., Zhou W., Lin J., Xu L. Roles of the wound hormone jasmonate in plant regeneration. J. Exp. Bot. 2021:erab508. doi: 10.1093/jxb/erab508. [DOI] [PubMed] [Google Scholar]
- Zhao C., Craig J.C., Petzold H.E., Dickerman A.W., Beers E.P. The xylem and phloem transcriptomes from secondary tissues of the arabidopsis root-hypocotyl. Plant Physiol. 2005;138:803–818. doi: 10.1104/pp.105.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Xing L., Wang X., Hou Y.-J., Gao J., Wang P., Duan C.-G., Zhu X., Zhu J.-K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 2014;7:ra53. doi: 10.1126/scisignal.2005051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Lozano-Torres J.L., Blilou I., Zhang X., Zhai Q., Smant G., Li C., Scheres B. A jasmonate signaling network activates root stem cells and promotes regeneration. Cell. 2019;177:942–956.e14. doi: 10.1016/j.cell.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Zimmerman P.W., Wilcoxon F. Several chemical growth substances which cause initiation of roots and other responses in plants. Contrib. Boyce Thompson Inst. 1935;7:209–229. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.