Abstract
Post-translational modifications (PTMs) are closely linked to numerous diseases, playing a significant role in regulating protein structures, activities, and functions. Therefore, the identification of PTMs is crucial for understanding the mechanisms of cell biology and diseases therapy. Compared to traditional machine learning methods, the deep learning approaches for PTM prediction provide accurate and rapid screening, guiding the downstream wet experiments to leverage the screened information for focused studies. In this paper, we reviewed the recent works in deep learning to identify phosphorylation, acetylation, ubiquitination, and other PTM types. In addition, we summarized PTM databases and discussed future directions with critical insights.
Abbreviations: AAindex, Amino acid index; ATP, Adenosine triphosphate; AUC, Area under curve; Ac, Acetylation; BE, Binary encoding; BLOSUM, Blocks substitution matrix; Bi-LSTM, Bidirectional LSTM; CKSAAP, Composition of k-spaced amino acid Pairs; CNN, Convolutional neural network; CNNOH, CNN with the one-hot encoding; CNNWE, CNN with the word-embedding encoding; CNNrgb, CNN red green blue; CV, Cross-validation; DC-CNN, Densely connected convolutional neural network; DL, Deep learning; DNNs, Deep neural networks; EBGW, Encoding based on grouped weight; EGAAC, Enhanced grouped amino acids content; E. coli, Escherichia coli; IG, Information gain; K, Lysine; KNN, k nearest neighbor; LASSO, Least absolute shrinkage and selection operator; LSTM, Long short-term memory; LSTMWE, LSTM with the word-embedding encoding; MDCAN, Multilane dense convolutional attention network; MDC, Modular densely connected convolutional networks; ML, Machine learning; MLP, Multilayer perceptron; MMI, Multivariate mutual information; M.musculus, Mus musculus; NMBroto, Normalized Moreau-Broto autocorrelation; P, Proline; PSP, PhosphoSitePlus; PSSM, Position-specific scoring matrix; PTM, Post-translational modifications; Ph, Phosphorylation; PseAAC, Pseudo-amino acid composition; R, Arginine; RF, Random forest; RNN, Recurrent neural network; ROC, Receiver operating characteristic; SE, Squeeze and excitation; SEV, Split to Equal Validation; S, Serine; ST, Source and target; SUMO, Small ubiquitin-like modifier; SVM, Support vector machines; S.cerevisiae, Saccharomyces cerevisiae; S. typhimurium, Salmonella typhimurium; T, Threonine; Ub, Ubiquitination; Y, Tyrosine; ZSL, Zero-shot learning
Keywords: Post-translational modification, Machine learning, Deep learning, Prediction, Mass spectrometry
1. Introduction
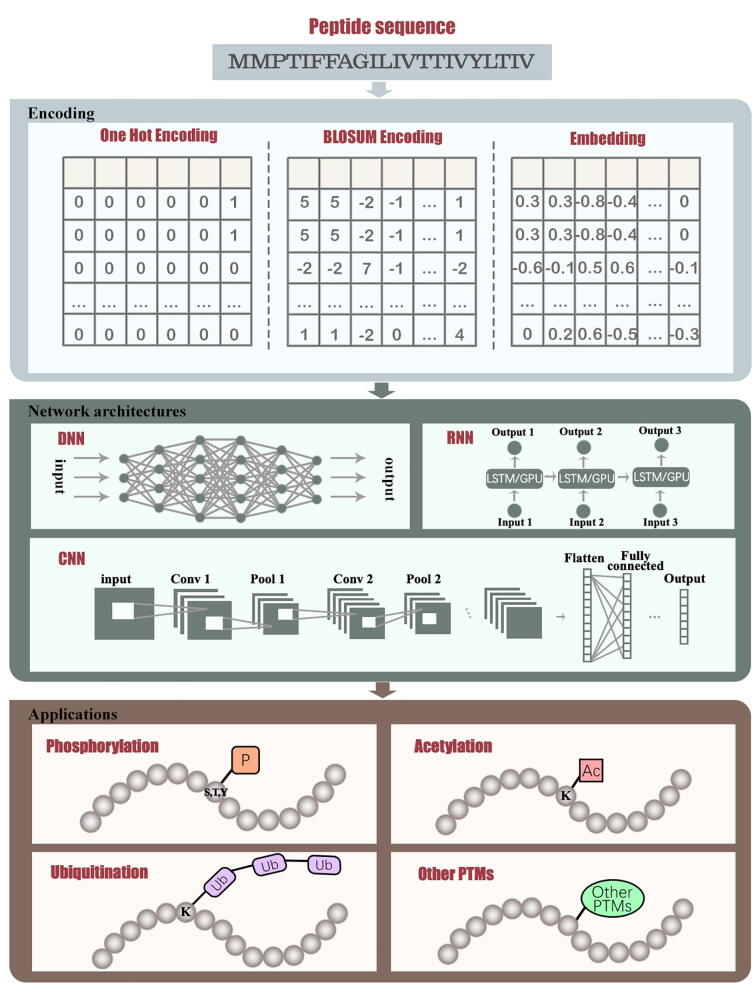
Post-translational modifications (PTMs) generally refer to the addition of functional groups (e.g., phosphates, acetates, small proteins, lipids, carbohydrates, etc.) to amino acids during translation [1]. After PTM, amino acids' chemical properties or structures will be changed, leading to functional changes. To date, over 600 different types of PTMs have been discovered in different proteins [2], [3]. It is known that phosphorylation, acetylation, and ubiquitination are the extensively studied PTMs, as quantified with the dbPTM [4] database. PTMs are critical in maintaining protein structures [5], functions [6], metabolic regulation [7], cellular signaling [8], and proteomic diversity [9], whereby our understanding of PTMs are essential to downstream consequences such as diseases. For example, S-nitrosylation is a promising therapeutic target for cancers and neurodegenerative diseases [10], [11], [12]; methyl glutamine is associated with the host defence mechanism against microorganisms [13], [14]. Different experimental techniques have been developed to reveal the mechanisms underlying PTMs, including chromatin immunoprecipitation (ChIP) [15], western blotting (WB) [16], mass spectrometry (MS) [17], [18], and isotope labeling [19]. In the recent decade, MS-based proteomic techniques [20] play a major role in PTM identification, which yield solid data with actual evidence [21]. In addition, computational methods can also explore and predict new modification sites by building a model from those data. In the last few years, machine learning has grown to be a cost-effective and labor-efficient method for the prediction of various PTM sites [22], [23], [24], [25], [26], [27], [28]. Specifically, deep learning is an advanced machine learning method that is capable of automatically exploring PTM patterns and capturing high-level abstraction (Fig. 1 [29]). Therefore, it is an appropriate solution to improve the efficiency of PTM sites' prediction with growing interest in recent years (Fig. 2). A lot of published works focused on adopting deep learning to predict PTM sites for phosphorylation [30], acetylation [31], ubiquitination [32], and many other types of modifications [33], [34]. One of the most famous tools is MusiteDeep [30], developed by Wang and Zeng, which leveraged convolutional neural network (CNN) and 2D attention mechanism for phosphorylation sites prediction. DeepPhos [35], which is created by Luo et al., is an efficient phosphorylation sites predictor to identify not only general but also kinase-specific sites. Moreover, Wu et al. [36] and Fu et al. [37] developed deep learning-based methods to predict acetylation and putative ubiquitination sites with promising results.
Fig. 1.
Overview of deep learning approaches for PTM prediction. [29].
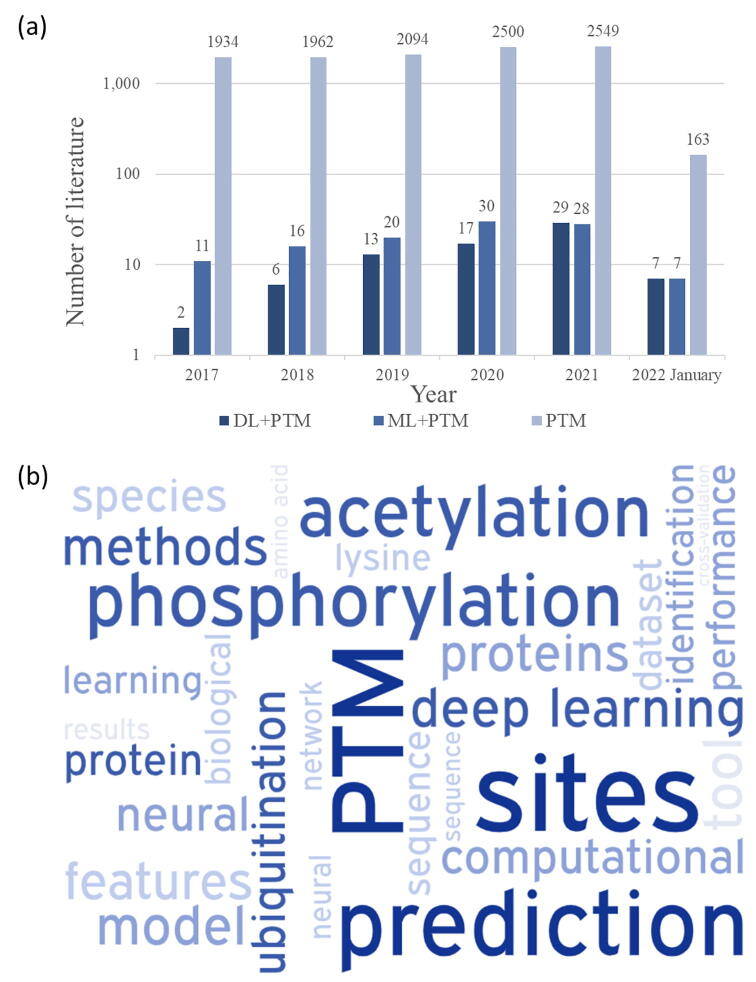
Fig. 2.
The statistics of published literature on machine/deep learning-based PTM prediction. (a) Number of articles published in different peer-reviewed journals. Note that the year 2022 only includes publications up to January 2022. Abbreviations: DL = deep learning, ML = machine learning, PTM = post-translational modification. (b) Word cloud based on the collective concordance ranking with the size of terms proportional to their frequency in the above articles.
In this mini-review, we summarized and discussed the most recent (2020–2022) progress made in the prediction of PTMs using deep learning-based methods with a particular emphasis on protein phosphorylation, acetylation, and ubiquitination sites. Moreover, we presented frequently used databases for deep learning-based PTM prediction, along with future directions in the computational identification of PTMs.
2. PTM databases
Available PTM datasets can mainly be retrieved from two sources: databases with various types of data and scientific literature data. The obtained data can be used to train a model for PTM prediction. Table 1 summarizes the leading databases with different data types based on recent literature [38], [39], [40], [41], [42], [43].
Table 1.
Summary of PTM databases harbored.
| Database | Development Year | Number of PTM Sites Deposited | Database Link | Annotation | Reference |
|---|---|---|---|---|---|
| UniProt | 2005 | Varies according to the keyword search | https://www.uniprot.org | Multiple-type PTM sites for multi-species | [38] |
| PLMD | 2017 | 284,780 | https://plmd.biocuckoo.org/ | Protein lysine modification sites for multi-species | [43] |
| PhosphoSitePlus | 2012 | 598,976 | https://www.phosphosite.org/ | Multiple-type PTM sites for multi-species | [40] |
| Phospho.ELM | 2010 | 42,914 | https://phospho.elm.eu.org/ | Phosphorylation sites for Eukaryotic | [39] |
| mUbiSida | 2014 | 110,976 | https://reprod.njmu.edu.cn/mUbiSiDa | Uniquitination sites mainly for Human and Mouse | [41] |
| DEPOD | 2015 | 1,215 | https://www.depod.org | Dephosphorylation interactions | [42] |
2.1. UniProt
UniProt [38] is one of the most comprehensive databases with PTM annotations; it contains annotations for a wide variety of PTMs. UniProt data is of high quality and was recognized as an ELIXIR Core Data Resource in 2017 [44]. The database received the CoreTrustSeal certification in 2020. It has four components customized for different uses: UniParc, UniProtKB, UniRef, and UniMES. Notably, the UniProtKB database has become the gateway to protein functional information. Over the last two years, UniProtKB's sequences have grown to about 190 million [45], despite efforts in sequence redundancy removal at the proteome level. According to the survey, we found that most of the literature collect datasets from UniProtKB as their benchmark datasets. The latest version of the UniProt database can be accessed by visiting https://www.uniprot.org/.
2.2. PLMD
There are 20 types of protein lysine modifications across 176 species in PLMD [43]. The PLMD database was constructed from the CPLA and CPLM databases with manual curations. It contains 284,780 protein lysine modification sites in 53,501 proteins, including 111,253 acetylation sites and 121,742 ubiquitination sites. To the best of our knowledge, it is the largest available database of protein acetylation, along with the largest database of protein ubiquitination sites, which has never been reported in any other ubiquitination sites prediction research. There is a free and open-source version of PLMD 3.0 at https://plmd.biocuckoo.org, which is implemented in PHP and MySQL.
2.3. PhosphoSitePlus
PhosphoSitePlus (PSP) [40] offers comprehensive data information for studying PTMs, such as phosphorylation, SUMOylation, ubiquitination, and others. Manually collected and organized data are curated to constitute this database, which primarily contains human and mouse protein data. At the time of writing, it has harbored 598,976 nonredundant modified sites, including 294,425 phosphorylation sites. The PSP database is versatile, offering a variety of information about the modification sites. PSP is a free database that can be accessed through https://www.phosphosite.org.
3. Phosphorylation site prediction
Phosphorylation is one the most frequently investigated PTM, referring to the transfer of phosphate groups (PO4) from adenosine triphosphate (ATP) sites to amino acid chains via the catalysis of various kinases [46]. Typically, phosphorylation of proteins occurs at serine (S), threonine (T), or tyrosine (Y) [47]. Approximately 13,000 human proteins can be phosphorylated, and 230,000 phosphorylation sites in human proteome were reported [48]. In the past decades, phosphorylation studies have gained widespread popularity due to their significance in characterizing signaling pathways [49], [50] and cellular processes, such as cell growth [51], cell division [52], and apoptosis [53]. With the development of high-throughput MS-based technology, a single proteomic experiment can detect large-scale phosphorylation. Therefore, various databases have been built to collect annotated phosphorylation sites [38], [39], [40]. The application of these databases in recent years has been enabled through the extensive development of computational methods for phosphorylation sites identification [22], [54], [55], [56], [57], [58]. In machine learning, we can formulate the phosphorylation site prediction problem as two classification tasks. The first task is the general site prediction, which aims to determine whether a given site can be modified. The second task is the kinase-specific prediction, which determines whether a site can be modified by a particular kinase [29]. In particular, the recent development of deep learning could speed up the progress of phosphorylation site prediction. A well-known deep learning-based predictor, MusiteDeep [30], incorporates one-hot encoding and CNN with attention layers and performs better than previous feature-based models. Another phosphorylation site prediction method, DeepPhos [35], exploits densely connected convolutional neural network (DC-CNN) blocks for predictions. The results of DeepPhos outperform MusiteDeep in not only general sites but also kinase-specific sites predictions. Recently, a single unified multi-label classification model, EMBER [58], was released. Unlike the previous deep learning methods, MusiteDeep and DeepPhos, which perform single-label classification, EMBER was designed to predict phosphorylation events for multiple kinases. In this tool, the input sequence is fifteen amino acids in length, of which the eighth site is to be predicted. The sequence is encoded using both one-hot encoding and embedding generated from a siamese neural network. After encoding, both sequences are fed into their corresponding identical CNNs. In the top layer, the two feature vectors are concatenated, followed by fully connected layers. Finally, the output is a vector of length eight, where each value represents the probability that a family of kinases will phosphorylate an input site. In addition, different tools are also proposed to predict protein-specific phosphorylation sites. In 2020, Chen et al. developed PROSPECT [56] which is a method for phosphorylation site prediction occur on histidine using deep learning. Three specific classifiers are set up in PROSPECT for histidine phosphorylation site prediction based on one-of-K, EGAAC, and CKSAAGP encodings [35], [59]. The classifier for one-of-K encoding is built with a multi-layer attention-based CNN; and the classifier for EGAAC encoding employs a multi-layer CNN. In the case of CKSAAGP encoding, the random forest (RF) algorithm is used to train the classifier. After that, an online web server of PROSPECT is developed. In the same year, Wang et al. also presented a web server named MusiteDeep based on their deep-learning models implemented in 2017. The server is capable of providing real-time prediction and batch submission for large-scale protein sequences, as listed in Table 4. Conclusively, we compare the performance of recent deep learning-based phosphorylation predictors in Table 2.
Table 4.
Summary of recently deep learning tools associated with PTM sites prediction.
| Tool name | PTM type | Species | Core network model |
Evaluation strategy |
Benchmark dataset size (modification sites) | Web server/ source code | Published year | Reference |
|---|---|---|---|---|---|---|---|---|
| MusiteDeep | Multiple | Human | CNN | 5-fold CV | 997,687 | https://www.musite.net | 2017/2020 | [30] |
| PROSPECT | Phosphorylation | Escherichia coli | CNN | 10-fold CV and independent test | 1,664 | *prospect.erc.monash.edu/ | 2020 | [56] |
| DeepKinZero | Phosphorylation | Human | ZSL | holdout | 12,901 | *https://github.com/Tastanlab/DeepKinZero | 2020 | [60] |
| PhosTransfer | Phosphorylation | – | CNN | holdout | 43,785 | https://github.com/yxu132/PhosTransfer | 2020 | [61] |
| GPS-PBS | Phosphorylation | Multiple | seven-layer DNNs | 10-fold CV | 4,458 | – | 2020 | [62] |
| DeepPPSite | Phosphorylation | Mammals and Arabidopsis thaliana | LSTM | 10-fold CV | 41,436 | github.com/saeed344/DeepPPSite | 2021 | [57] |
| DeepIPs | Phosphorylation | Human | CNN + LSTM | 5-fold CV | 10.978 |
https://lin-group.cn/server/DeepIPs https://github.com/linDing-group/DeepIPs |
2021 | [63] |
| PhosIDN | Phosphorylation | Human | Multi-layer DNNs | holdout | more than 160,000 | https://github.com/ustchangyuanyang/PhosIDN | 2021 | [64] |
| EMBER | Phosphorylation | Multiple | CNN + RNN | 5-fold CV | 8,389 | https://github.com/gomezlab/EMBER | 2022 | [58] |
| DNNAce | Acetylation | Multiple | DNN | 10-fold CV and independent test | 96,372 | https://github.com/QUSTAIBBDRC/DNNAce/ | 2020 | [78] |
| Deep-PLA | Acetylation | Human and Nonhuman |
DNN | 5- and 10-fold CV | 1,331 | https://deeppla.cancerbio.info | 2020 | [79] |
| MDC-Kace | Acetylation | Multiple | MDC | 10-fold CV and independent test | 11,583 | https://github.com/lianglianggg/MDC-Kace | 2020 | [80] |
| DeepTL-Ubi | Ubiquitination | Multiple | CNN | holdout | 94,518 | github.com/USTC-HIlab/DeepTL-Ubi | 2020 | [106] |
| Wang et al.’s work | Ubiquitination | Multiple | CNN | 10-fold CV | 121,742 | *https://github.com/wang-hong-fei/DL-plantubsites-prediction | 2020 | [105] |
| UbiComb | Ubiquitination | Multiple | LSTM | 10-fold CV | 121,742 | https://nsclbio.jbnu.ac.kr/tools/UbiComb | 2021 | [107] |
| SSMFN | Methylation | Human and Mouse | CNN + LSTM | holdout | 6,754 | *https://github.com/bharuno/SSMFNMethylation-Analysis | 2021 | [110] |
| Malebary et al.’s work | Methylation | Human | CNN | 10-fold CV and jackknife | 2000 | https://github.com/s2018 https://doi.org/1080001/WebServer.git |
2022 | [14] |
| RecSNO | S-Nitrosylation | – | BiLSTM | 5-fold CV | 4,762 | https://nsclbio.jbnu.ac.kr/tools/RecSNO/. | 2021 | [111] |
| MDCAN-Lys | Succinylation | Human | MDCAN | 10-fold CV and independent test | 77,418 | – | 2021 | [112] |
| LSTMCNNsucc | Succinylation | Multiple | LSTM + CNN | holdout | 18,593 | https://8.129.111.5/ | 2021 | [113] |
| DeepMal | Malonylation | Multiple | CNN + DNN | 10-fold CV and independent test | 17,288 | https://github.com/QUST-AIBBDRC/DeepMal/ | 2020 | [114] |
| K_net | Malonylation | Human and Mice | CNN | 10-fold CV and SEV | 85,204 | – | 2020 | [115] |
| DeepCSO | S-Sulphenylation | Homo sapiens and Arabidopsis thaliana | LSTMWE | 10-fold CV | 10,354 | *https://www.bioinfogo.org/DeepCSO. | 2020 | [116] |
| DeepSSPred | S-Sulphenylation | Homo Sapiens | 2D-CNN | jackknife | 7,756 | *https://github.com/zaheerkhancs/DeepSSPred | 2021 | [117] |
| pKcr | Crotonylation | Papaya | CNN | 10-fold CV and independent test | 58,769 | *https://www.bioinfogo.org/pkcr. | 2020 | [119] |
| Deep-Kcr | Crotonylation | Human | CNN | 10-fold CV | 19,928 | https://lin-group.cn/server/Deep-Kcr | 2020 | [120] |
| DeepKcrot | Crotonylation | Multiple | CNNWE | 10-fold CV and independent test | 10,702/1,265/2,044/5,995 | *https://www.bioinfogo.org/deepkcrot. | 2021 | [121] |
| nhKcr | Crotonylation | Human | CNNrgb | 10-fold CV and independent test | 180,312 | https://nhKcr.erc.monash.edu/ | 2021 | [118] |
| DeepKhib | 2-Hydroxyisobutyrylation | Multiple | CNNOH | 10-fold CV and independent test | 18,946/15,444/12,756/19,330/2,098 | *https://www.bioinfogo.org/DeepKhib. | 2020 | [122] |
| DeepGlut | Glutarylation | Prokaryotes and Eukaryote | CNN | 10-fold CV | 4,572 | *https://github.com/urmisen/DeepGlut. | 2020 | [123] |
| NPalmitoylDeep-PseAAC | N-Palmitoylation | Human | DNN | holdout | 4,364 | https://mega.nz/#F!s9cSiQIa!1jXO0NmgrhxUqOexmYuouA | 2021 | [124] |
| DTL-DephosSite | Dephosphorylation | Human | Bi-LSTM | 5-fold CV and independent test | 4,956 | https://github.com/dukkakc/DTLDephos | 2021 | [127] |
| PreCar_Deep | Carbonylation | Human and other Mammals | CNN + BiLSTM | 10-fold CV and independent test | 5,003 | https://github.com/QUST-SHULI/PreCar_Deep/ | 2021 | [125] |
| He et al.'s work | SUMOylation Ubiquitylation | – | CNN + DNN | 10-fold CV | 280,731 | https://github.com/lijingyimm/MultiUbiSUMO | 2021 | [126] |
Note: *, Link is not working at the time of writing. Multiple, more than three species or PTM types. -, data not available.
Table 2.
Comparison of deep learning-based phosphorylation sites predictors.
| Tool name | Framework | Encoding strategy | Window size | Average AUC | Reference |
|---|---|---|---|---|---|
| MusiteDeep | Keras/TensorFlow | One-hot | 33 | 0.880 | [30] |
| PROSPECT | PyTorch | One-hot, EGAAC, CKSAAGP | 27 | 0.770 | [56] |
| DeepKinZero | TensorFlow | Word embedding | 15 | – | [60] |
| PhosTransfer | TensorFlow | Word embedding | – | 0.898 | [61] |
| GPS-PBS | Keras/TensorFlow | BLOSUM62 | 21 | 0.832 | [62] |
| DeepPPSite | Keras/TensorFlow | BE, EBGW, CKSAAP, PSPM, IPCP | 21 | 0.872 | [57] |
| DeepIPs | Keras/TensorFlow | Word embedding | 15 | 0.909 | [63] |
| PhosIDN | Keras/TensorFlow | One-hot, PPI embedding | 21 | 0.939 | [64] |
| EMBER | PyTorch | One-hot | 15 | 0.928 | [58] |
Note: -, data not available. AUC: Area under the Curve of ROC.
4. Acetylation site prediction
Acetylation is a very common PTM that describes the modification of the acetyl group to amino acid residues. About 63% of mitochondrial proteins can be acetylated at their lysine residues [65]. During the protein acetylation process, the positive charge in lysine residues is neutralized, leading to the regulation of cell lifespan [66], DNA binding [67], the interactions between proteins [68], and the interactions between proteins and membranes [69]. In contrast, dysregulation of lysine acetylation is associated with several diseases, including cancers [70], cardiovascular diseases [71], Parkinson's diseases [72], and neurodegenerative disorders [73]. Thus, the identification of acetylation sites may benefit the understanding of its molecular mechanism and further experimental design. Proteomic and high-throughput MS-based techniques have identified massive acetylation sites. For example, Choudhary et al. detected 3,600 lysine acetylation sites on 1,750 proteins from a human cell line. [74]; Lundby et al. quantified 15,474 lysine acetylation sites on 4,541 proteins from 16 rat tissues [75]. Several public databases have been developed to facilitate the collection and maintenance of acetylation sites information [38], [43]. Therefore, to predict acetylation sites, many computational methods have been proposed [76], [77], [36]. Among them, deep learning methods are increasingly popular in bioinformatics, which also show encouraging results of acetylation sites identification [78], [79], [80]. For example, Wu et al. [36] presented an MLP architecture, DeepAcet, as an acetylation site prediction model. Feature embedding were performed with six methods (One-hot, IG, CKSAAP, PSSM, AAindex, and BLOSUM62); multilayer perceptron (MLP) is then applied to extract features. After adopting 10-fold cross-validation method [81] paired model evaluation on a separate test site, accuracies were reported to be 0.8495 and 0.8487, respectively. Yu et al. also developed a deep neural networks (DNN) based model called DNNAce for acetylation sites prediction [78]. First, they applied eight different encoding methods to extract information from multiple amino acid residues and then fused the encoded feature vectors to create a high-level feature representation. These encodings methods are BE, PseAAC, AAindex, NMBroto, EBGW, MMI, BLOSUM62, and KNN. Next, they employ LASSO to screen the optimal feature subsets to improve the model performance. As a final stage, nine prokaryotic acetylation site datasets are adopted to evaluate the performance and compared to state-of-the-art models such as AdaBoost, Naive Bayes, XGBoost, KNN, RF, SVM, CNN, and LSTM. An evaluation of DNNAce was conducted by comparing its results with ProAcePred [82]. The performance of DNNAce on the remaining eight species was significantly lower than that of ProAcePred except for S. typhimurium species. However, DNNAce outperforms ProAcePred for the other seven species during independent evaluation. Therefore, the advantages of DNNAce are trivial because there is performance discrepancy in training and independent testing. In contrast to deepAcet and DNNAce, which only consider the amino acid sequences and their physicochemical properties, MDC-Kace [80] pays attention to both sequence information and protein structural properties to predict acetylation sites. In MDC-Kace, modular densely connected convolutional networks (MDC), which consist of three independent modules (sequence, physicochemical and structure), is employed to extract features of lysine acetylation sites. In the next step, squeeze and excitation (SE) layer [83] is utilized to weight importance of features to build representation more accurately. Finally, the fused advanced feature is fed into a softmax layer for classification to predict acetylation sites efficiently. The authors compared MDC-Kace with state-of-the-art models (MusiteDeep [30], CapsNet [34], DeepAcet [36], PSKAcePred [84], EnsemblePail [85], GPS-PAIL2.0 [86] and ProAcePred [82]) to evaluate its performance. Three species (human, M. musculus, E. coli) datasets have been evaluated by10-fold cross-validation and independent testing. The results indicate that MDC-Kace has a similar performance as existing acetylation sites predictors.
5. Ubiquitination site prediction
Ubiquitination represents an enzymatic PTM on cellular protein by ubiquitin conjugation [87]. Multiple important cellular processes are related to ubiquitination, including protein degradation [88], cell division [89], and protein stability [90], [91]. Ubiquitination serves as a fundamental component of the ubiquitin–proteasome system, mediating more than 80% of protein degradation in eukaryotes [92]. Moreover, aberrant ubiquitination is highly related to the progression of aging [93] and many diseases; for example, the dysregulation of ubiquitin–proteasome system may contribute to the occurrence of neurodegenerative conditions [94] and inflammatory bowel diseases [95]. Therefore, the identification of ubiquitination sites is an essential step in exploring various ubiquitination-involved mechanisms. In order to identify the ubiquitination sites in proteins, a myriad of experimental [96], [97], [98] and computational methods [99], [100], [101] have been developed. In recent years, with the continuous growth in high-throughput experimental data [102], [103], [104], deep learning [105], [106], [107] has been increasingly applied to the prediction of ubiquitination. Fu et al. proposed a deep learning predictor, DeepUbi [37], based on CNN. In this tool, four feature encoding schemes are utilized for feature construction. Under 10-fold cross-validation, DeepUbi is able to achieve an AUC of 0.90, with the accuracy, sensitivity, and specificity being all over 0.85. Compared with DeepUbi, which is trained for general ubiquitination site prediction, DeepTL-Ubi [106] is a species-specific sites predictor which consists of three connected modules: a deep feature extractor, a source label classifier, and a target label classifier. Firstly, a densely connected convolutional neural network (DCCNN) is applied as the deep feature extractor, which is composed of six layers. Features of both source species and target species are extracted simultaneously by the deep feature extractor, mapping samples into a joint feature space. Secondly, the two parallel classifiers are employed to classify source species and target species at the same time. Thirdly, ST (source and target) loss assists the extractor in transferring knowledge from source species to target species by learning relevant features. Finally, as the performance optimization step, the classification loss is minimized to train the two classifiers. DeepTL-Ubi outperforms several existing tools, including Ubisite [108], Ubiprober [24], and MUscADEL [109], as shown in Table 3.
Table 3.
AUC values on different ubiquitination prediction tools. [106].
| AUC |
Species |
||||||
|---|---|---|---|---|---|---|---|
| H.sapiens | M.musculus | R.norvegicus | S.cerevisiae | T.gondii | A.nidulans | ||
| Tools | DeepTL-Ubi | 0.753 | 0.789 | 0.720 | 0.772 | 0.824 | 0.814 |
| Ubisite | 0.598 | 0.625 | 0.561 | 0.548 | 0.607 | 0.611 | |
| Ubiprober | 0.624 | 0.661 | 0.644 | 0.600 | 0.630 | 0.638 | |
| MUscADEL | 0.656 | 0.693 | 0.659 | 0.664 | 0.715 | 0.681 | |
6. Other PTMs
In addition to those discussed, deep learning can also be applied for other PTMs’ predictions, including methylation [110], S-nitrosylation [111], succinylation [112], [113], malonylation [114], [115], S-sulphenylation [116], [117], crotonylation [118], [119], [120], [121], 2- hydroxyisobutyrylation [122], glutarylation [123], N-palmitoylation [124] carbonylation [125], and SUMOylation [126]. In particular, crotonylation prediction has demonstrated highly accurate results based on deep-learning methods. Moreover, 2- hydroxyisobutyrylation, as a novel type of PTM, was predicted by deep learning method for the first time in 2020.
Along with predicting conventional PTMs associated with functional group addition, deep learning-based methods have also been applied to predict niche-type PTMs; for instance, Chaudhari et al. developed a transfer learning-based predictor (DTL-DephosSite) for dephosphorylation site prediction [127]. To collect datasets of S, T, and Y dephosphorylation sites, they integrated the experimentally verified datasets from the literature and datasets from the DEPOD database. They then employ bidirectional long short-term memory (Bi-LSTM), which can predict the modification of the target amino acid according to the knowledge of residues from both directions. To the best of our knowledge, it is the first tool that can predict the general dephosphorylation sites for protein S/T residues and Y residues. On the other hand, a novel prediction model focusing on carbonylation, Precar_Deep [125], is recently reported. Carbonylation is an irreversible covalent PTM and is a measure of protein oxidative damage. In this model, CNN and Bi-LSTM are combined under a deep learning framework. The AUC values of the four datasets (K, T, P, and R) reach 0.981, 0.982, 0.987, and 0.976, respectively. The AUC values of the independent test set reach 0.945, 0.978, 0.965, and 0.983, respectively. In addition, there is also a novel small protein-addition type PTM site predictor based on deep learning in 2021. He et al. built an ensemble learning model that adopts CNN and DNN, followed by the output result containing four types of sites. [126]. This is the first tool that predicts both ubiquitylation and SUMOylation sites at the same time based on deep learning. PTM prediction tools mentioned in this section, as well as predictors of phosphorylation, acetylation, and ubiquitination, are tabulated in Table 4.
7. Summary and outlook
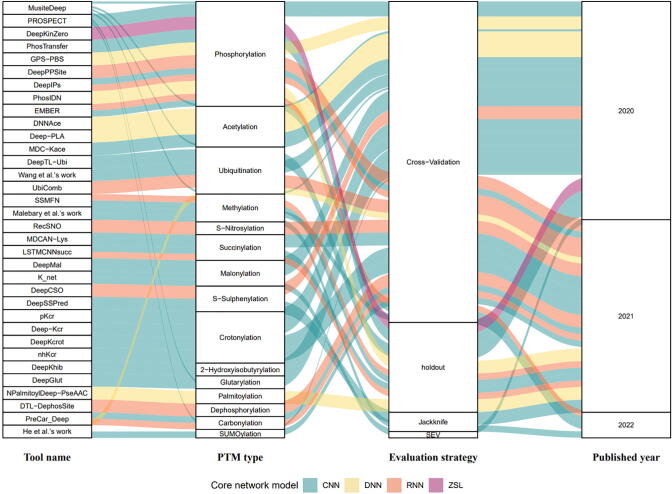
PTM identification is critical to a better understanding of molecular functions and diseases. Advanced MS-based technology has yielded an extensive list of identified PTMs, providing abundant data to support the development of downstream computational identification methods. Although the traditional machine learning methods can precisely predict the modified sites, deep learning features can be automatically deduced and optimally turned without encoding features ahead of time [29]. Thus, deep learning is highly effective in scientific fields with large and complex datasets. Researchers recently gradually shift their attention from traditional machine learning to deep learning for PTM site prediction (Fig. 2). Furthermore, with the growing number of PTM profiling datasets, deep learning models have been developed for not only phosphorylation, acetylation, and ubiquitination, but also many other PTM types. In this review, we summarized the recently (2020–2022) released deep learning tools and online web servers for protein PTM site prediction (Table 4). Among all these, CNN and cross-validation are the most widely used network model and evaluation strategy, respectively (Fig. 3).
Fig. 3.
Sankey diagram depicting the distribution of PTM types, core network models, evaluation strategies, and published years.
Although several deep learning methods have been built with high performance to predict PTM sites, there is still room for improvement. Most of the existing deep learning algorithms employed CNN, DNN, and LSTM classifiers. However, each classifier has its own advantages and disadvantages. Therefore, further research is required to evaluate more state-of-the-art frameworks such as attention and transformer-based models. On top of that, in many developed tools, although PTM sites are predicted based on certain characteristics, such as sequence information, physical properties, chemical properties, and protein structure properties, there are still other approaches that need to be explored, such as reduced amino acid compositions [128], [129], [130]. Additionally, most of web server links are not working, and few methods provide stand-alone versions. After testing all web servers, we found that they were difficult to operate.
By using deep learning based methods, PTM identification can be implemented in a non-invasive, efficient, and low-cost way. However, there is still a caveat before deep learning algorithms can directly diagnose diseases. Typical PTM prediction models lack sufficient interpretations due to the black-box nature of deep learning algorithms. Insufficient interpretability may not be an issue in many areas, but within healthcare, every misdiagnosis can pose a danger to a patient's health. Therefore, transparent and explainable models [131], [132], [133] will be needed, so that the technique can be applied in clinical practice.
CRediT authorship contribution statement
Lingkuan Meng: Writing, Conceptualization, Methodology, Visualization. Wai-Sum Chan: Methodology. Lei Huang: Methodology. Linjing Liu: Methodology. Xingjian Chen: Methodology. Weitong Zhang: Methodology. Fuzhou Wang: Methodology. Ke Cheng: Methodology. Hongyan Sun: Writing – review & editing, Supervision. Ka-Chun Wong: Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by the (1) National Natural Science Foundation of China (Grant No. 32170654, 32122003 and 32000464), (2) the Hong Kong Research Grants Council (No. 11102719, 11302320), (3) Health and Medical Research Fund, the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region [07181426], and (4) City University of Hong Kong (CityU 11202219, CityU 11203520, CityU 11203221).
Contributor Information
Hongyan Sun, Email: hongysun@cityu.edu.hk.
Ka-Chun Wong, Email: kc.w@cityu.edu.hk.
References
- 1.Walsh C.T., Garneau-Tsodikova S., Gatto G.J., Jr. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.https://www.uniprot.org/docs/ptmlist.
- 3.UniProt C., UniProt: a worldwide hub of protein knowledge, Nucleic Acids Res. 2019; 47: D506-D515. http://10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed]
- 4.Lee T-Y, Huang H-D, Hung J-H, Huang H-Y, Yang Y-S, Wang T-H. dbPTM: an information repository of protein post-translational modification, Nucleic Acids Res. 2006; 34: D622–D627. http://10.1093/nar/gkj083. [DOI] [PMC free article] [PubMed]
- 5.Craveur P., Narwani T.J., Rebehmed J., de Brevern A.G. Investigation of the impact of PTMs on the protein backbone conformation. Amino Acids. 2019;51:1065–1079. doi: 10.1007/s00726-019-02747-w. [DOI] [PubMed] [Google Scholar]
- 6.Lin H., Du J., Jiang H. Post-translational modifications to regulate protein function. Wiley Encycl Chem Biol. 2008 doi: 10.1002/9780470048672.wecb467. [DOI] [Google Scholar]
- 7.Humphrey S.J., James D.E., Mann M. Protein Phosphorylation: a major switch mechanism for metabolic regulation. Trends Endocrinol Metab. 2015;26:676–687. doi: 10.1016/j.tem.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Sharma K., D'Souza R.C., Tyanova S., Schaab C., Wisniewski J.R., Cox J., et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 9.Aebersold R., Agar J.N., Amster I.J., Baker M.S., Bertozzi C.R., Boja E.S., et al. How many human proteoforms are there? Nat Chem Biol. 2018;14:206–214. doi: 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura T., Lipton S.A. Protein S-nitrosylation as a therapeutic target for neurodegenerative diseases. TIP. 2016;37:73–84. doi: 10.1016/j.tips.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Lulu S., T. Ziv, P. Weisman-Shomer, M. Benhar, Nitrosothiol-trapping-based proteomic analysis of S-nitrosylation in human lung carcinoma cells, PLoS One. 2017; 12: e0169862. http://10.1371/journal.pone.0169862. [DOI] [PMC free article] [PubMed]
- 12.Huang G., Li J., Zhao C. computational prediction and analysis of associations between small molecules and binding-associated s-Nitrosylation sites. Molecules. 2018;23 doi: 10.3390/molecules23040954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wawro A.M., Gajera C.R., Baker S.A., Leśniak R.K., Fischer C.R., Saw N.L., et al. Enantiomers of 2-methylglutamate and 2-methylglutamine selectively impact mouse brain metabolism and behavior. Sci Rep. 2021;11:8138. doi: 10.1038/s41598-021-87569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malebary S.J., Alzahrani E., Khan Y.D. A comprehensive tool for accurate identification of methyl-Glutamine sites. J Mol Graph Model. 2022;110 doi: 10.1016/j.jmgm.2021.108074. [DOI] [PubMed] [Google Scholar]
- 15.Collas P. The current state of chromatin immunoprecipitation. Mol Biotechnol. 2010;45:87–100. doi: 10.1007/s12033-009-9239-8. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitas M.A., Sklenar A.R., Parthun M.R. Application of mass spectrometry to the identification and quantification of histone post-translational modifications. J Cell Biochem. 2004;92:691–700. doi: 10.1002/jcb.20106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witze E.S., Old W.M., Resing K.A., Ahn N.G. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 19.Kettenbach A.N., Rush J., Gerber S.A. Absolute quantification of protein and post-translational modification abundance with stable isotope-labeled synthetic peptides. Nat Protoc. 2011;6:175–186. doi: 10.1038/nprot.2010.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong A.T., Leprevost F.V., Avtonomov D.M., Mellacheruvu D., Nesvizhskii A.I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods. 2017;14:513–520. doi: 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang K.Y., Lee T.Y., Kao H.J., Ma C.T., Lee C.C., Lin T.H., et al. dbPTM in 2019: exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res. 2019;47:D298–D308. doi: 10.1093/nar/gky1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J., Thelen J.J., Dunker A.K., Xu D. Musite, a tool for global prediction of general and kinase-specific phosphorylation sites*. MCP. 2010;9:2586–2600. doi: 10.1074/mcp.M110.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Shao XJ, Wu LY, Deng NY, Chou KC. iSNO-AAPair: incorporating amino acid pairwise coupling into PseAAC for predicting cysteine S-nitrosylation sites in proteins, PeerJ. 2013; 1: e171. http://10.7717/peerj.171. [DOI] [PMC free article] [PubMed]
- 24.Chen X., Qiu J.-D., Shi S.-P., Suo S.-B., Huang S.-Y., Liang R.-P. Incorporating key position and amino acid residue features to identify general and species-specific Ubiquitin conjugation sites. Bioinformatics. 2013;29:1614–1622. doi: 10.1093/bioinformatics/btt196. [DOI] [PubMed] [Google Scholar]
- 25.Hou T, Zheng G, Zhang P, Jia J, Li J, Xie L, Wei C, Li Y. LAceP: lysine acetylation site prediction using logistic regression classifiers, PLoS One. 2014; 9: e89575. http://10.1371/journal.pone.0089575. [DOI] [PMC free article] [PubMed]
- 26.Liu Z., Xiao X., Yu D.-J., Jia J., Qiu W.-R., Chou K.-C. pRNAm-PC: Predicting N6-methyladenosine sites in RNA sequences via physical–chemical properties. Anal Biochem. 2016;497:60–67. doi: 10.1016/j.ab.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Qiu W.-R., Sun B.-Q., Xiao X., Xu Z.-C., Chou K.-C. iPTM-mLys: identifying multiple lysine PTM sites and their different types. Bioinformatics. 2016;32:3116–3123. doi: 10.1093/bioinformatics/btw380. [DOI] [PubMed] [Google Scholar]
- 28.Pupylation sites prediction with ensemble classification model, Int J Data Min Bioinformatics. 2017; 18: 91–104. http://10.1504/ijdmb.2017.086441
- 29.Wen B, Zeng WF, Liao Y, Shi Z, Savage SR, Jiang W, Zhang B. Deep learning in proteomics, Proteomics. 2020; 20: e1900335. http://10.1002/pmic.201900335. [DOI] [PMC free article] [PubMed]
- 30.Wang D., Zeng S., Xu C., Qiu W., Liang Y., Joshi T., et al. MusiteDeep: a deep-learning framework for general and kinase-specific phosphorylation site prediction. Bioinformatics. 2017;33:3909–3916. doi: 10.1093/bioinformatics/btx496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X., Li J., Wang R., He F., Yue L., Yin M. General and species-specific lysine acetylation site prediction using a bi-modal deep architecture. IEEE Access. 2018;6:63560–63569. doi: 10.1109/access.2018.2874882. [DOI] [Google Scholar]
- 32.He F., Wang R., Li J., Bao L., Xu D., Zhao X. Large-scale prediction of protein ubiquitination sites using a multimodal deep architecture. BMC Syst Biol. 2018;12:109. doi: 10.1186/s12918-018-0628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long H., Liao B., Xu X., Yang J. A hybrid deep learning model for predicting protein hydroxylation sites. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19092817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang D., Liang Y., Xu D. Capsule network for protein post-translational modification site prediction. Bioinformatics. 2018;35:2386–2394. doi: 10.1093/bioinformatics/bty977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo F., Wang M., Liu Y., Zhao X.M., Li A. DeepPhos: prediction of protein phosphorylation sites with deep learning. Bioinformatics. 2019;35:2766–2773. doi: 10.1093/bioinformatics/bty1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu M., Yang Y., Wang H., Xu Y. A deep learning method to more accurately recall known lysine acetylation sites. BMC Bioinform. 2019;20:49. doi: 10.1186/s12859-019-2632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu H., Yang Y., Wang X., Wang H., Xu Y. DeepUbi: a deep learning framework for prediction of ubiquitination sites in proteins. BMC Bioinform. 2019;20:86. doi: 10.1186/s12859-019-2677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bairoch A, Apweiler R, Wu CH, Barker WC, B. Boeckmann, S. Ferro, E. Gasteiger, H. Huang, R. Lopez, M. Magrane, M.J. Martin, D.A. Natale, C. O'Donovan, N. Redaschi, L.S. Yeh, The universal protein resource (UniProt), Nucleic Acids Res. 2005; 33: D154-9. http://10.1093/nar/gki070. [DOI] [PMC free article] [PubMed]
- 39.Dinkel H., C. Chica, A. Via, C.M. Gould, L.J. Jensen, T.J. Gibson, F. Diella, Phospho.ELM: a database of phosphorylation sites--update 2011, Nucleic Acids Res. 2011; 39: D261-7. http://10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed]
- 40.Hornbeck PV, Kornhauser JM, S. Tkachev, B. Zhang, E. Skrzypek, B. Murray, V. Latham, M. Sullivan, PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse, Nucleic Acids Res. 2012; 40: D261-D270. http://10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed]
- 41.Chen T, Zhou T, B. He, H. Yu, X. Guo, X. Song, J. Sha, mUbiSiDa: a comprehensive database for protein ubiquitination sites in mammals, PLoS One. 2014; 9: e85744. http://10.1371/journal.pone.0085744. [DOI] [PMC free article] [PubMed]
- 42.Duan G., Li X., Kohn M. The human DEPhOsphorylation database DEPOD: a 2015 update. Nucleic Acids Res. 2015;43:D531–D535. doi: 10.1093/nar/gku1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu H., Zhou J., Lin S., Deng W., Zhang Y., Xue Y. PLMD: An updated data resource of protein lysine modifications. JGG. 2017;44:243–250. doi: 10.1016/j.jgg.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Drysdale R, Cook CE, R. Petryszak, V. Baillie-Gerritsen, M. Barlow, E. Gasteiger, F. Gruhl, J. Haas, J. Lanfear, R. Lopez, N. Redaschi, H. Stockinger, D. Teixeira, A. Venkatesan, F. Elixir Core Data Resource, N. Blomberg, C. Durinx, J. McEntyre, The ELIXIR Core Data Resources: fundamental infrastructure for the life sciences, Bioinformatics 2020; 36: 2636-2642. http://10.1093/bioinformatics/btz959. [DOI] [PMC free article] [PubMed]
- 45.UniProt C., UniProt: the universal protein knowledgebase in 2021, Nucleic Acids Res. 2021; 49: D480-D489. http://10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed]
- 46.Johnson L.N. The regulation of protein phosphorylation. Biochem Soc Trans. 2009;37:627–641. doi: 10.1042/BST0370627. [DOI] [PubMed] [Google Scholar]
- 47.Potel C.M., Lin M.H., Heck A.J.R., Lemeer S. Widespread bacterial protein histidine phosphorylation revealed by mass spectrometry-based proteomics. Nat Methods. 2018;15:187–190. doi: 10.1038/nmeth.4580. [DOI] [PubMed] [Google Scholar]
- 48.Vlastaridis P., Kyriakidou P., Chaliotis A., Van de Peer Y., Oliver S.G., Amoutzias G.D. Estimating the total number of phosphoproteins and phosphorylation sites in eukaryotic proteomes. GigaScience. 2017;6:1–11. doi: 10.1093/gigascience/giw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun S.C. Non-canonical NF-kappaB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka Y, Chen Zhijian J. STING Specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway, Sci Signal. 2012; 5: ra20-ra20. http://10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed]
- 51.Wang H, Owens C, N. Chandra, M.R. Conaway, D.L. Brautigan, D. Theodorescu, Phosphorylation of RalB is important for bladder cancer cell growth and metastasis, Cancer Res. 2010; 70: 8760-8769. http://10.1158/0008-5472.CAN-10-0952. [DOI] [PMC free article] [PubMed]
- 52.Hans F., Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- 53.Wei Y, Sinha SC, Levine B. Dual Role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation, Autophagy. 2008; 4: 949-951. http://10.4161/auto.6788. [DOI] [PMC free article] [PubMed]
- 54.Trost B., Kusalik A. Computational prediction of eukaryotic phosphorylation sites. Bioinformatics. 2011;27:2927–2935. doi: 10.1093/bioinformatics/btr525. [DOI] [PubMed] [Google Scholar]
- 55.Dou Y., Yao B., Zhang C. PhosphoSVM: prediction of phosphorylation sites by integrating various protein sequence attributes with a support vector machine. Amino Acids. 2014;46:1459–1469. doi: 10.1007/s00726-014-1711-5. [DOI] [PubMed] [Google Scholar]
- 56.Chen Z., Zhao P., Li F., Leier A., Marquez-Lago T.T., Webb G.I., et al. PROSPECT: A web server for predicting protein histidine phosphorylation sites. J Bioinform Comput Biol. 2020;18:2050018. doi: 10.1142/S0219720020500183. [DOI] [PubMed] [Google Scholar]
- 57.Ahmed S., Kabir M., Arif M., Khan Z.U., Yu D.-J. DeepPPSite: A deep learning-based model for analysis and prediction of phosphorylation sites using efficient sequence information. Anal Biochem. 2021;612 doi: 10.1016/j.ab.2020.113955. [DOI] [PubMed] [Google Scholar]
- 58.Kirchoff KE, Gomez SM. EMBER: multi-label prediction of kinase-substrate phosphorylation events through deep learning, Bioinformatics. 2022; btac083. http://10.1093/bioinformatics/btac083. [DOI] [PMC free article] [PubMed]
- 59.Chen Z., Zhao P., Li F., Leier A., Marquez-Lago T.T., Wang Y., et al. iFeature: a Python package and web server for features extraction and selection from protein and peptide sequences. Bioinformatics. 2018;34:2499–2502. doi: 10.1093/bioinformatics/bty140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deznabi I., Arabaci B., Koyuturk M., Tastan O. DeepKinZero: zero-shot learning for predicting kinase-phosphosite associations involving understudied kinases. Bioinformatics. 2020;36:3652–3661. doi: 10.1093/bioinformatics/btaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu Y., Wilson C., Leier A., Marquez-Lago T.T., Whisstock J., Song J. In: Advances in Knowledge Discovery and Data Mining. Lauw H.W., Wong R.-C.W., Ntoulas A., Lim E.-P., Ng S.-K., Pan S.J., editors. Springer International Publishing; Cham: 2020. PhosTransfer: a deep transfer learning framework for kinase-specific phosphorylation site prediction in hierarchy; pp. 384–395. [Google Scholar]
- 62.Guo Y., Ning W., Jiang P., Lin S., Wang C., Tan X., et al. A deep learning framework to predict phosphorylation sites that specifically interact with phosphoprotein-binding domains. Cells. 2020;9 doi: 10.3390/cells9051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lv H., Dao F.Y., Zulfiqar H., Lin H. DeepIPs: comprehensive assessment and computational identification of phosphorylation sites of SARS-CoV-2 infection using a deep learning-based approach. Brief Bioinform. 2021;22 doi: 10.1093/bib/bbab244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang H., Wang M., Liu X., Zhao X.-M., Li A. PhosIDN: an integrated deep neural network for improving protein phosphorylation site prediction by combining sequence and protein–protein interaction information. Bioinformatics. 2021;37:4668–4676. doi: 10.1093/bioinformatics/btab551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baeza J., Smallegan M.J., Denu J.M. Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem Sci. 2016;41:231–244. doi: 10.1016/j.tibs.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dang W., Steffen K.K., Perry R., Dorsey J.A., Johnson F.B., Shilatifard A., et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sykes S.M., Mellert H.S., Holbert M.A., Li K., Marmorstein R., Lane W.S., et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shogren-Knaak M., Ishii H., Sun J.-M., Pazin Michael J., Davie James R., Peterson Craig L. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 69.Okada A.K., Teranishi K., Ambroso M.R., Isas J.M., Vazquez-Sarandeses E., Lee J.Y., et al. Lysine acetylation regulates the interaction between proteins and membranes. Nat Commun. 2021;12:6466. doi: 10.1038/s41467-021-26657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalvik T.V., Arnesen T. Protein N-terminal acetyltransferases in cancer. Oncogene. 2013;32:269–276. doi: 10.1038/onc.2012.82. [DOI] [PubMed] [Google Scholar]
- 71.Pons D., de Vries F.R., van den Elsen P.J., Heijmans B.T., Quax P.H.A., Jukema J.W. Epigenetic histone acetylation modifiers in vascular remodelling: new targets for therapy in cardiovascular disease. Eur Heart J. 2009;30:266–277. doi: 10.1093/eurheartj/ehn603. [DOI] [PubMed] [Google Scholar]
- 72.Toker L., Tran G.T., Sundaresan J., Tysnes O.-B., Alves G., Haugarvoll K., et al. Genome-wide histone acetylation analysis reveals altered transcriptional regulation in the Parkinson’s disease brain. Mol Neurodegener. 2021;16:31. doi: 10.1186/s13024-021-00450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saha R.N., Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13:539–550. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choudhary C., Kumar C., Gnad F., Nielsen Michael L., Rehman M., Walther Tobias C., et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 75.Lundby A., Lage K., Weinert B.T., Bekker-Jensen D.B., Secher A., Skovgaard T., et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012;2:419–431. doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen G., Cao M., Yu J., Guo X., Shi S. Prediction and functional analysis of prokaryote lysine acetylation site by incorporating six types of features into Chou's general PseAAC. J Theor Biol. 2019;461:92–101. doi: 10.1016/j.jtbi.2018.10.047. [DOI] [PubMed] [Google Scholar]
- 77.Ning Q., Yu M., Ji J., Ma Z., Zhao X. Analysis and prediction of human acetylation using a cascade classifier based on support vector machine. BMC Bioinform. 2019;20:346. doi: 10.1186/s12859-019-2938-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu B., Yu Z., Chen C., Ma A., Liu B., Tian B., et al. DNNAce: Prediction of prokaryote lysine acetylation sites through deep neural networks with multi-information fusion. Chemom Intell Lab. 2020;200 doi: 10.1016/j.chemolab.2020.103999. [DOI] [Google Scholar]
- 79.Yu K., Zhang Q., Liu Z., Du Y., Gao X., Zhao Q., et al. Deep learning based prediction of reversible HAT/HDAC-specific lysine acetylation. Brief Bioinform. 2020;21:1798–1805. doi: 10.1093/bib/bbz107. [DOI] [PubMed] [Google Scholar]
- 80.Wang H., Yan Z., Liu D., Zhao H., Zhao J. MDC-Kace: A model for predicting lysine acetylation sites based on modular densely connected convolutional networks. IEEE Access. 2020;8:214469–214480. doi: 10.1109/access.2020.3041044. [DOI] [Google Scholar]
- 81.Liu L., Petinrin O., Zhang W., Rahaman S., Tang Z., Wong K. Machine learning protocols in early cancer detection based on liquid biopsy: a survey. Life. 2021;11 doi: 10.3390/life11070638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen G., Cao M., Luo K., Wang L., Wen P., Shi S. ProAcePred: prokaryote lysine acetylation sites prediction based on elastic net feature optimization. Bioinformatics. 2018;34:3999–4006. doi: 10.1093/bioinformatics/bty444. [DOI] [PubMed] [Google Scholar]
- 83.Hu J, Shen L, Sun G. Squeeze-and-Excitation Networks, 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2018, pp. 7132–7141.
- 84.Suo S.-B., Qiu J.-D., Shi S.-P., Sun X.-Y., Huang S.-Y., Chen X., et al. Position-specific analysis and prediction for protein lysine acetylation based on multiple features. PLoS ONE. 2012;7:e49108. doi: 10.1371/journal.pone.0049108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Y., Wang X.-B., Ding J., Wu L.-Y., Deng N.-Y. Lysine acetylation sites prediction using an ensemble of support vector machine classifiers. J Theor Biol. 2010;264:130–135. doi: 10.1016/j.jtbi.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 86.Deng W., Wang C., Zhang Y., Xu Y., Zhang S., Liu Z., et al. GPS-PAIL: prediction of lysine acetyltransferase-specific modification sites from protein sequences. Sci Rep. 2016;6:39787. doi: 10.1038/srep39787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Glickman M.H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 88.Wilkinson K.D. Ubiquitination and deubiquitination: Targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 89.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 90.Li C., Xiao Z.-X. Regulation of p63 protein stability via ubiquitin-proteasome pathway. Biomed Res Int. 2014;2014 doi: 10.1155/2014/175721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hicke L., Schubert H.L., Hill C.P. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 92.Collins G.A., Goldberg A.L. The logic of the 26S proteasome. Cell. 2017;169:792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kevei É., Hoppe T. Ubiquitin sets the timer: impacts on aging and longevity. Nat Struct Mol Biol. 2014;21:290–292. doi: 10.1038/nsmb.2806. [DOI] [PubMed] [Google Scholar]
- 94.Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 95.Chen R., Pang X., Li L., Zeng Z., Chen M., Zhang S. Ubiquitin-specific proteases in inflammatory bowel disease-related signalling pathway regulation. Cell Death Dis. 2022;13:139. doi: 10.1038/s41419-022-04566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marotti L.A., Newitt R., Wang Y., Aebersold R., Dohlman H.G. Direct identification of a G protein ubiquitination site by mass spectrometry. Biochemistry. 2002;41:5067–5074. doi: 10.1021/bi015940q. [DOI] [PubMed] [Google Scholar]
- 97.Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., et al. A proteomics approach to understanding protein ubiquitination. Nature Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 98.Merbl Y., Kirschner Marc W. Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. PNAS. 2009;106:2543–2548. doi: 10.1073/pnas.0812892106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cai B., Jiang X. Computational methods for ubiquitination site prediction using physicochemical properties of protein sequences. BMC Bioinform. 2016;17:116. doi: 10.1186/s12859-016-0959-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen Z., Chen Y.-Z., Wang X.-F., Wang C., Yan R.-X., Zhang Z. Prediction of ubiquitination sites by using the composition of k-spaced amino acid pairs. PLoS ONE. 2011;6:e22930. doi: 10.1371/journal.pone.0022930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen J., Zhao J., Yang S., Chen Z., Zhang Z. Prediction of protein ubiquitination sites in arabidopsis thaliana. Curr Bioinform. 2019;14:614–620. doi: 10.2174/1574893614666190311141647. [DOI] [Google Scholar]
- 102.Chernorudskiy A.L., Garcia A., Eremin E.V., Shorina A.S., Kondratieva E.V., Gainullin M.R. UbiProt: a database of ubiquitylated proteins. BMC Bioinform. 2007;8:126. doi: 10.1186/1471-2105-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Du Y., Xu N., Lu M., Li T. hUbiquitome: a database of experimentally verified ubiquitination cascades in humans. Database. 2011;2011:bar055. doi: 10.1093/database/bar055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gao T, Liu Z, Y. Wang, H. Cheng, Q. Yang, A. Guo, J. Ren, Y. Xue, UUCD: a family-based database of ubiquitin and ubiquitin-like conjugation, Nucleic Acids Res. 2013; 41: D445-D451. http://10.1093/nar/gks1103. [DOI] [PMC free article] [PubMed]
- 105.Wang H., Wang Z., Li Z., Lee T.Y. Incorporating deep learning with word embedding to identify plant ubiquitylation sites. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.572195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Y., Li A., Zhao X.M., Wang M. DeepTL-Ubi: A novel deep transfer learning method for effectively predicting ubiquitination sites of multiple species. Methods. 2021;192:103–111. doi: 10.1016/j.ymeth.2020.08.003. [DOI] [PubMed] [Google Scholar]
- 107.Siraj A., Lim D.Y., Tayara H., Chong K.T. UbiComb: A hybrid deep learning model for predicting plant-specific protein ubiquitylation sites. Genes (Basel) 2021;12 doi: 10.3390/genes12050717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang C.-H., Su M.-G., Kao H.-J., Jhong J.-H., Weng S.-L., Lee T.-Y. UbiSite: incorporating two-layered machine learning method with substrate motifs to predict ubiquitin-conjugation site on lysines. BMC Syst Biol. 2016;10:S6. doi: 10.1186/s12918-015-0246-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen Z., Liu X., Li F., Li C., Marquez-Lago T., Leier A., et al. Large-scale comparative assessment of computational predictors for lysine post-translational modification sites. Brief Bioinform. 2019;20:2267–2290. doi: 10.1093/bib/bby089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lumbanraja F.R., Mahesworo B., Cenggoro T.W., Sudigyo D., Pardamean B. SSMFN: a fused spatial and sequential deep learning model for methylation site prediction. PeerJ Comput Sci. 2021;7:e683. doi: 10.7717/peerj-cs.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Siraj A., Chantsalnyam T., Tayara H., Chong K.T. RecSNO: prediction of protein S-nitrosylation sites using a recurrent neural network. IEEE Access. 2021;9:6674–6682. doi: 10.1109/ACCESS.2021.3049142. [DOI] [Google Scholar]
- 112.Wang H., Zhao H., Yan Z., Zhao J., Han J. MDCAN-Lys: A model for predicting succinylation sites based on multilane dense convolutional attention network. Biomolecules. 2021;11 doi: 10.3390/biom11060872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang G., Shen Q., Zhang G., Wang P., Yu Z.-G. LSTMCNNsucc: A Bidirectional LSTM and CNN-based deep learning method for predicting lysine succinylation sites. Biomed Res Int. 2021;2021:9923112. doi: 10.1155/2021/9923112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang M., Cui X., Li S., Yang X., Ma A., Zhang Y., et al. DeepMal: Accurate prediction of protein malonylation sites by deep neural networks. Chemom Intell Lab Syst. 2020;207 doi: 10.1016/j.chemolab.2020.104175. [DOI] [Google Scholar]
- 115.Sun J., Cao Y., Wang D., Bao W., Chen Y. K_net: lysine malonylation sites identification with neural network. IEEE Access. 2020;8:47304–47311. doi: 10.1109/ACCESS.2019.2961941. [DOI] [Google Scholar]
- 116.Lyu X., Li S., Jiang C., He N., Chen Z., Zou Y., et al. DeepCSO: A deep-learning network approach to predicting cysteine S-sulphenylation sites. Front Cell Dev Biol. 2020;8:1489. doi: 10.3389/fcell.2020.594587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Khan U.Z., Pi D. DeepSSPred: a deep learning based sulfenylation site predictor via a novel nsegmented optimize federated feature encoder. Protein Pept Lett. 2021;28:708–721. doi: 10.2174/0929866527666201202103411. [DOI] [PubMed] [Google Scholar]
- 118.Chen Y.-Z., Wang Z.-Z., Wang Y., Ying G., Chen Z., Song J. nhKcr: a new bioinformatics tool for predicting crotonylation sites on human nonhistone proteins based on deep learning. Brief Bioinform. 2021;22:bbab146. doi: 10.1093/bib/bbab146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao Y., He N., Chen Z., Li L. Identification of protein lysine crotonylation sites by a deep learning framework with convolutional neural networks. IEEE Access. 2020;8:14244–14252. doi: 10.1109/ACCESS.2020.2966592. [DOI] [Google Scholar]
- 120.Lv H., Dao F.-Y., Guan Z.-X., Yang H., Li Y.-W., Lin H. Deep-Kcr: accurate detection of lysine crotonylation sites using deep learning method. Brief Bioinform. 2021;22:bbaa255. doi: 10.1093/bib/bbaa255. [DOI] [PubMed] [Google Scholar]
- 121.Wei X., Sha Y., Zhao Y., He N., Li L. DeepKcrot: a deep-learning architecture for general and species-specific lysine crotonylation site prediction. IEEE Access. 2021;9:49504–49513. doi: 10.1109/ACCESS.2021.3068413. [DOI] [Google Scholar]
- 122.Zhang L., Zou Y., He N., Chen Y., Chen Z., Li L. DeepKhib: A deep-learning framework for lysine 2-hydroxyisobutyrylation sites prediction. Front Cell Dev Biol. 2020;8:897. doi: 10.3389/fcell.2020.580217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sen U., Hasan M.A.M. 2020 IEEE Region 10 Symposium (TENSYMP) 2020. DeepGlut: A deep learning framework for prediction of glutarylation sites in proteins; pp. 941–944. [Google Scholar]
- 124.Naseer S., Hussain W., Khan D.Y., Rasool N. NPalmitoylDeep-PseAAC: A predictor of N-palmitoylation sites in proteins using deep representations of proteins and PseAAC via modified 5-steps rule. Curr Bioinform. 2021;16:294–305. doi: 10.2174/1574893615999200605142828. [DOI] [Google Scholar]
- 125.Song L., Xu Y., Wang M., Leng Y. PreCar_Deep: A deep learning framework for prediction of protein carbonylation sites based on Borderline-SMOTE strategy. Chemom Intell Lab Syst. 2021;218 doi: 10.1016/j.chemolab.2021.104428. [DOI] [Google Scholar]
- 126.He F., Li J., Wang R., Zhao X., Han Y. an ensemble deep learning based predictor for simultaneously identifying protein ubiquitylation and SUMOylation sites. BMC Bioinform. 2021;22:519. doi: 10.1186/s12859-021-04445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chaudhari M., Thapa N., Ismail H., Chopade S., Caragea D., Kohn M., et al. DTL-DephosSite: Deep transfer learning based approach to predict dephosphorylation sites. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.662983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen W., Feng P., Liu T., Jin D. Recent advances in machine learning methods for predicting heat shock proteins. Curr Drug Metab. 2019;20:224–228. doi: 10.2174/1389200219666181031105916. [DOI] [PubMed] [Google Scholar]
- 129.Pan Y., Wang S., Zhang Q., Lu Q., Su D., Zuo Y., et al. Analysis and prediction of animal toxins by various Chou's pseudo components and reduced amino acid compositions. J Theor Biol. 2019;462:221–229. doi: 10.1016/j.jtbi.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 130.Zuo Y., Li Y., Chen Y., Li G., Yan Z., Yang L. PseKRAAC: a flexible web server for generating pseudo K-tuple reduced amino acids composition. Bioinformatics. 2017;33:122–124. doi: 10.1093/bioinformatics/btw564. [DOI] [PubMed] [Google Scholar]
- 131.Barredo A.A., Díaz-Rodríguez N., Del Ser J., Bennetot A., Tabik S., Barbado A., et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Information Fusion. 2020;58:82–115. doi: 10.1016/j.inffus.2019.12.012. [DOI] [Google Scholar]
- 132.Mann M., Kumar C., Zeng W.F., Strauss M.T. Artificial intelligence for proteomics and biomarker discovery. Cell Syst. 2021;12:759–770. doi: 10.1016/j.cels.2021.06.006. [DOI] [PubMed] [Google Scholar]
- 133.Adadi A., Berrada M. In: Embedded Systems and Artificial Intelligence. Bhateja V., Satapathy S.C., Satori H., editors. Springer Singapore; Singapore: 2020. Explainable AI for healthcare: from black box to interpretable models; pp. 327–337. [Google Scholar]