Graphical abstract
The ICTV proposed AAI threshold of 50% is widely used for Iridoviridae taxonomy that is used as the genus boundary. However, we found a more accurate cut-off value for better class members in this family, which is supported by comparative genomic analysis of 179 iridovirids. Furthermore, nine genes were selected from eighteen strict core genes as hallmark genes for preliminary viral identification in a more convenient and economical manner.

Keywords: Iridoviridae, Taxonomy, Core genes, Synteny analysis, Codon usage, Phylogenetics
Abstract
Members of the family Iridoviridae (iridovirids) are globally distributed and trigger adverse economic and ecological impacts on aquaculture and wildlife. Iridovirids taxonomy has previously been studied based on a limited number of genomes, but this is not suitable for the current and future virological studies as more iridovirids are emerging. In our study, 57 representative iridovirids genomes were selected from a total of 179 whole genomes available on NCBI. Then 18 core genes were screened out for members of the family Iridoviridae. Average amino acid sequence identity (AAI) analysis indicated that a cut-off value of 70% is more suitable for the current iridovirids genome database than ICTV-defined 50% threshold to better clarify viral genus boundaries. In addition, more subgroups were divided at genus level with the AAI threshold of 70%. This observation was further confirmed by genomic synteny analysis, codon usage preference analysis, genome GC content and length analysis, and phylogenic analysis. According to the pairwise comparison analysis of core genes, 9 hallmark genes were screened out to conduct preliminary identification and investigation at the genus level of iridovirids in a more convenient and economical manner.
1. Introduction
Members of the Iridoviridae family (designated iridovirids) are a diverse collection of large DNA viruses (approximately 120–300 nm in diameter) with linear, double-stranded circularly permutated and terminally redundant DNA genomes enclosed within an icosahedral capsid [1]. The family is currently devided into two subfamilies: Alphairidovirinae and Betairidovirinae. The former comprised three genera: Ranavirus, whose members mainly infects fish, amphibians, and reptiles; Lymphocystivirus and Megalocytivirus, whose members target only on bony fish. Another subfamily contains four genera (Iridovirus, Chloriridovirus, Decapodiridovirus, and Daphniairidovirus), which mainly infect invertebrates such as crustaceans and insects [2].
In the past two decades, reports of iridovirids infections have markedly increased, which reflects the fact that viruses of this family, once viewed as obscure viruses with little economic or ecological impact, are now known to be widespread in nature with significant impact on modern aquaculture and wildlife [3]. For example, the annual production of freshwater bass in China exceeds 620,000 tons (China Fishery Statistics Yearbook, 2021), and this large-scale aquaculture industry has been severely affected by Santee-Cooper ranavirus infection, especially in seasons with higher temperature, causing considerable economic losses [4]. Some ranaviruses have been linked to declining amphibian populations and represent a range of emerging infectious diseases that may even lead to population extinctions [5], [6]. Moreover, human activities can accelerate the spread of certain iridovirids, as seen in the case of tiger salamander (Ambystoma tigrinum) die-offs throughout western North America [7].
Nowadays, sequence comparisons using both pairwise sequence similarities and phylogenetic relationships have become one of the primary sets of characters used to define and differentiate virus taxa [8]. With the identification of shrimp hemocyte iridescent virus (SHIV) [9] and Cherax quadricarinatus iridovirus (CQIV) [10], a new genus (Decapodiridovirus) was established, followed by the seventh genus (Daphniairidovirus) within the family Iridoviridae that contains a single species (Daphniairidovirus tvaerminne, DIT) (ICTV proposal: 2020.018D). However, with the emergence of iridovirids and the advancement of modern sequencing technologies and bioinformatics, there are controversies regarding iridovirids taxonomy. Some researchers have proposed the construction of new genera to distinguish classified members of the family Iridoviridae [3], [11]. Therefore, we would like to verify the feasibility of the current ICTV proposed genus demarcation criteria used for iridovirids, which is that members of a given genus share less than 50% amino acid sequence identity (AAI) with members of other genera (file code: 2018.007D). Otherwise, we will find an more appropriate AAI threshold for members of the Iridoviridae taxonomy.
In this study, we re-annotated and systematically compared 179 Iridoviridae virus genomic nucleic acid sequences available in National Center for Biotechnology Information (NCBI) virus database. Eighteen core genes were redefined based on 57 representative genomes. Importantly, we proposed a new AAI cut-off value (70%) that is more suitable for current iridovirids genome databases and more conducive to understand the genera demarcation within the Iridoviridae family. This proposal was further confirmed by genomic-based synteny, phylogeny and codon usage preference analysis. In addition, 9 hallmark genes were selected for iridovirids identification and investigation at genus level.
2. Results
2.1. Data collection and remove replicate genomes
The finalized dataset comprises 196 iridovirids genomes, including 22 species among 7 genera, of which the taxonomy of 179 strains can be found in the ICTV Master Species List 2020 (Supplementary Table S1). Iridovirids genome size ranging from 100 to 288kbp, with GC contents ranging between 26% ∼ 55%. Pairwise comparisons of 196 iridovirids genomes were performed by CompareM v0.1.2 and the average amino-acid identity (AAI) values were calculated. Genomes with AAI values ≥ 99% were grouped into a cluster which are considered to be replicate genomes. As shown in Fig. 1A, 35 clusters and 22 singleton viral genomes were generated from 196 iridovirids genomes. Phylogenetic analysis of whole genome sequences also confirmed the similar evolutionary distances of viruses in the same group of AAI analysis (Fig. 1B). Eventually, a total of 57 representative genomes (the most studied genome from each of clusters), were screened out for later analysis (Supplementary Table S2).
Fig. 1.
(A) The AAI network built using the genomes of 196 Iridoviridae viruses. The edge represents AAI ≥ 99% between two nodes, and each node and color represent one genome and a cluster, respectively. (B) The viral proteomic tree (ViPTree) based on whole genome sequences. Different colored branches and outermost circles indicate different clusters. Branch length indicates evolutionary distance.
2.2. Determination of iridovirids strict core genes
The Prokka v1.14.6 package was used to re-annotate 57 representative iridovirids genomes, generated 6922 coding sequences (CDS) in total. Pairwise comparisons of all CDS were performed using BLASTp with an E-value threshold of 1e-5, which eventually generated 485 orthogroups. Conserved genes of top 28 orthogroups and Eaton previously identified core genes are shown in Fig. 2 [5]. For selection of eligible core genes, orthogroups that contain subgroups due to paralogous genes, or possess less than 57 iridovirids genomes, should be excluded. For instance, orthogroup #1 contains gene cg7, cg18, and cg19; only 48 of 57 representative iridovirids genomes encode gene that belong to orthogroup #26 (Table 1). Finally, eighteen core genes qualified (Supplementary file_1).
Fig. 2.
The BLASTp network of top 28 orthogroups. Each node represents one amino acid sequence. The edge represents percentage of identical matches >0 between two nodes (E-value threshold of 1e-5). Core genes defined by Eaton are colored.
Table 1.
Conserved genes and core genes of iridovirids.
| Orthogroups | Number of nodes | Number of iridoviridsa | Core genes defined by Eatonb | Gene name | Qualified core genec |
|---|---|---|---|---|---|
| #1 | 946 | 57 | cg7,cg18,cg19 | Putative tyrosin kinase, Serine-threonine protein kinase | no |
| #2 | 134 | 34 | NA | Hypothetical protein | no |
| #3 | 96 | 57 | cg14 | Ribonuclease III | no |
| #4 | 60 | 57 | cg10 | Myristilated membrane protein | yes |
| #5 | 60 | 57 | cg3 | Putative NTPase I | yes |
| #6 | 60 | 57 | cg2 | DNA-dep RNA pol-II Largest subunit | yes |
| #7 | 58 | 57 | cg9 | Unknown | yes |
| #8 | 58 | 57 | cg17 | Putative XPPG-RAD2-type nuclease | yes |
| #9 | 58 | 57 | cg6 | D5 family NTPase involved in DNA replication | yes |
| #10 | 58 | 57 | cg12 | DNA-dep RNA pol-II second largest subunit | yes |
| #11 | 57 | 57 | new_cg2 | Unknown | yes |
| #12 | 57 | 57 | cg8 | NIF-NLI interacting factor | yes |
| #13 | 57 | 57 | new_cg4 | Deoxynucleoside kinase | yes |
| #14 | 57 | 57 | new_cg6 | Immediate early protein ICP-46 | yes |
| #15 | 57 | 57 | cg1 | Putative replication factor and/or DNA binding-packing | yes |
| #16 | 57 | 57 | cg4 | ATPase-like protein | yes |
| #17 | 57 | 57 | cg5 | Helicase family | yes |
| #18 | 57 | 57 | cg11 | DNA pol Family B exonuclease | yes |
| #19 | 57 | 57 | cg16 | Major capsid protein | yes |
| #20 | 57 | 57 | new_cg5 | Erv1/Alr family | yes |
| #21 | 57 | 57 | new_cg7 | Hypothetical protein | yes |
| #22 | 56 | 56 | new_cg3 | Transcription elongation factor TFIIS | no |
| #23 | 56 | 56 | NA | Hypothetical protein | no |
| #24 | 55 | 53 | NA | Hypothetical protein | no |
| #25 | 55 | 55 | cg13 | Ribonucleotide reductase small subunit | no |
| #26 | 48 | 48 | cg15 | Proliferating cell nuclear antigen | no |
| #27 | 46 | 46 | NA | Hypothetical protein | no |
| #28 | 47 | 47 | NA | Hypothetical protein | no |
a: This value represents the number of viral genome that encoded corresponding genes. Value = 57 indicate strict core genes (genes present in all strains), values < 57 indicate soft core genes (genes present in partial strains).
b: NA means not defined by Eaton.
c: Eligibility of core gene needs to meet two conditions, namely (1) being a strict core gene and (2) no more than three paralogous genes in an orthogroup (number of iridoviridsa ≤ 60).
2.3. Whole genome AAI analysis
According to the ICTV proposed genus demarcation criteria of Iridoviridae viruses, we obtained the AAI network for 57 iridovirids genomes based on a cut-off value of 50%, generating seven genera (Fig. 3A1). However, average amino acid identity analysis of 57 representative viral genomes showed that an AAI threshold of 50% is prone to include some dispersed CDS into one group, whereas a threshold of around 70% is able to concentrate similar proteins into one group (Fig. 3B). With the AAI cut-off value at 70%, members in Iridoviridae were divided into fourteen subgroups (Fig. 3A2). Ranavirus and Chloriridovirus are split into three subgroups; Megalocytivirus, Lymphocystivirus, and Iridovirus were divided into two subgroups, separately. Therefore, we propose a new AAI threshold of genus boundary for the family Iridoviridae, and verify this proposal in the remainder of this study.
Fig. 3.
(A) AAI network of 57 iridovirids genomes (A1: cut-off ≥ 50%, A2: cut-off ≥ 70%). Each node represents one genome. Nodes connected by lines indicate that the AAI value of connected nodes is ≥ 50% or 70%. (B) Violin plot of overall identity analysis of 6922 CDS encoded by 57 iridovirids genomes. Each point represents an identity value.
2.4. Synteny analysis
The amino acid sequences and nucleotide sequences of 18 core genes encoded by 57 representative iridovirids were compared pairwise and identity value were calculated (Fig. 4A). A threshold of 75% was revealed to be suitable for classifying iridovirids at single gene level using amino acid sequences. Whereas the division boundary was not clear by analyzing nucleic acid sequences. Subsequently, the full landscape of protein sequences linear relationship of representative iridovirids was assessed, with an identity threshold set at 75% (Fig. 4B). It showed that the genera Ranavirus and Chloriridovirus had three subgroups each, and the genera Megalocytivirus and Lymphocystivirus were divided into two subgroups, separately.
Fig. 4.
(A) Violin plots of the percentage of identical matches of amino acid sequences and nucleic acid sequences of core genes. Each point represents the percentage of identical matches between the two aligned sequences (Left). Points have been removed for clarity of observation (Right). (B) Synteny analysis of representative iridovirids amino acid sequences (identity threshold at 75%). Each corresponding block represents the collinear comparison of two viruses. If there were no collinear amino acid sequence at a 75% identity between two viruses, the block would be blank.
2.5. Phylogenetic analysis
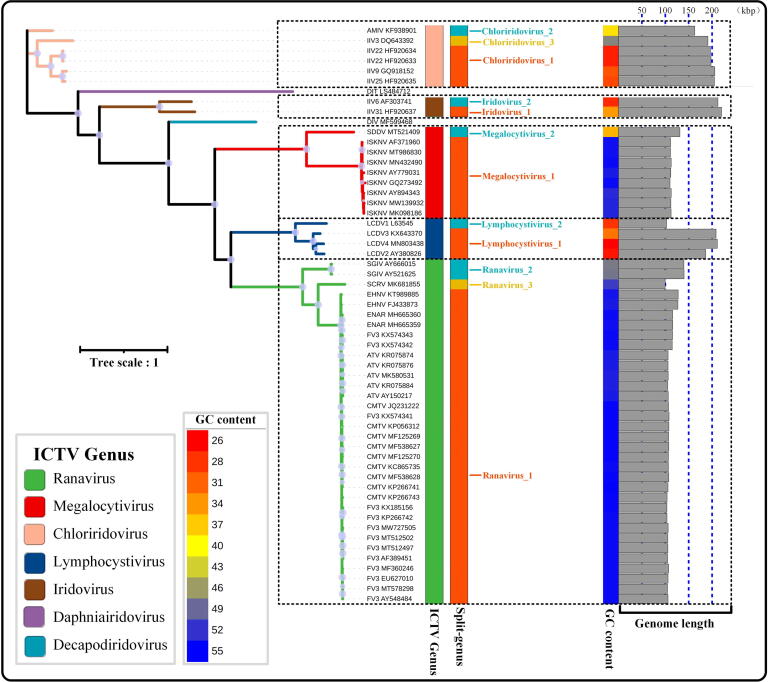
The IQ-TREE program and the iTol web server was used to preformed a maximum likelihood-based phylogenetic analysis of concatenated core genes of 57 representative iridovirids (Fig. 5). More than seven major clades come out in the phylogenetic tree. ICTV genus Chloriridovirus and Ranavirus are each divided into three monophyletic clades; Iridivirus, Megalocytivirus, and Lymphocystivirus were each divided into two monophyletic clades. Megalocytivirus_2 and Ranavirus_2 subgroup contain only one genome and diverge significantly from other subgroups of the respective ICTV genus, as indicated by the relatively long branch lengths at this node. Meanwhile, genome GC content and size statistics also provided strong evidence of differences among members in all monophyletic clades (Fig. S1, Fig. 5). For example, Lymphocystivirus 1 and Lymphocystivirus 2 have an average genome size of 200 and 105kbp, respectively; Megalocytivirus 1 and Megalocytivirus 2 have a GC content of 55% and 36%, respectively (Fig. S1).
Figure 5.
Phylogenic tree of iridovirids. Maximum likelihood analysis based on concatenated core genes of representative iridovirids (best-fit model according to BIC: Q.yeast + R6). The tree was rooted on midpoint. The first column of colored branches and bars represents ICTV classified iridovirids genera. The second column of colored bars represents genera or subgroup classified in this study (AAI identity cut-off ≥ 75%). The third column of colored bars represents a heat map of the GC content of the viral genome (Fig. S1). The grey bars in the last column represent viral genome size (Fig. S1). Branch length indicates evolutionary distance. The size of the point on the branch represents the bootstrap value >75.
2.6. Codon usage bias analysis
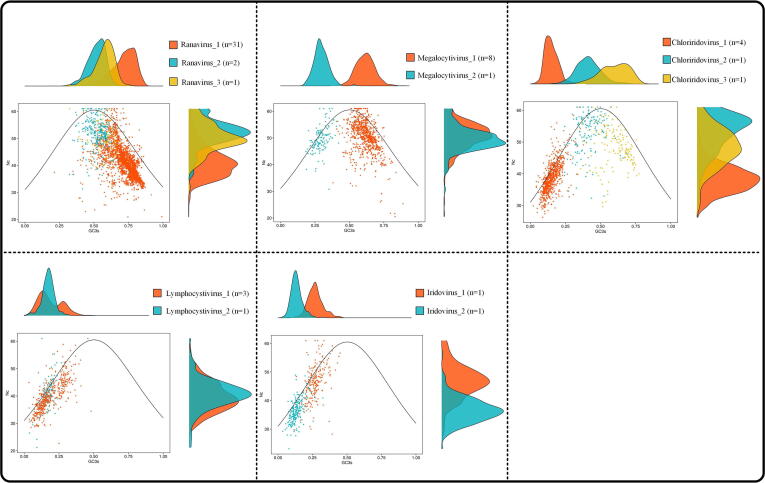
Codon usage bias (CUB) is the mechanism of unequal usage of synonymous codons in mature mRNA molecules, and a distinctive property of viral genome and very specific even for a species [12], [13], [14]. Correspondence analysis (CoA) based on the relative synonymous codon usage (RSCU) matrix was able to minimize the effect of amino acid composition and reduce the dimensionality of datasets to obtain awareness of multiple variables (Fig. 6). An effective number of codons (ENC) plot can clarify the relationship between the ENC and GC content at the third codon position (GC3), enabling assessment of the effects of natural selection and mutational pressure on viral genome evolution (Fig. 7) [12]. In both CoA and ENC plot analysis, the CUB properties of Megalocytivirus and Iridovirus were clearly divided into two subgroups; Chloriridoviru and Ranavirus were divided into three subgroups. Furthermore, ENC-GC3 plot indicate that the codon usage bias of iridovirids is mainly shaped by mutational pressure, and Ranavirus_3 is the subgroup that most affected by natural selection (Fig. 7).
Fig. 6.
Correspondence analysis of Ranavirus, Megalocytivirus, Chloriridovirus, Lymphocystivirus, and Iridovirus. Each dot represents the RSCU value of one gene. Density statistics for the two axes are shown above and to the right of the plot, respectively.
Fig. 7.
The relationship between the ENC values and GC3s. Each dot represents the ENC value (Y axes) and GC3 value (X axes) of one gene. The solid line indicates the expected curve of ENC and GC3 only in the absence of natural selection. Points on or close to the expected curve mean that the bias is caused by mutation pressure, while points below the curve indicate the presence of other influential factors such as natural selection. Density statistics for the two axes are shown above and to the right of the plot, respectively.
2.7. Iridovirids hallmark gene identification
Complete genomes or concatenated core gene sequences are commonly used for virus taxonomic studies, but single gene-based taxonomy is easier and convenient to conduct. In order to clarify hallmark genes of members in the family Iridoviridae, pairwise comparisions of each core gene of 57 iridovirids were performed using BLAST (Supplementary file_2, Supplementary file_3 filtered the data for pident ≤ 75%). The criteria for selecting hallmark gene is that the similarity of all amino acid sequences within a group of viruses is ≥ 75%, but their similarity with viruses from other groups is <75%. Finally, nine core genes were selected as iridovirids hallmark genes that are able to identify unknown iridovirids at the genus level (Table 2).
Table 2.
Selection of iridovirids hallmark genes.
| Core genes | Length (Nucleic acid/Amino acid) |
Qualified hallmark proteins | ||
|---|---|---|---|---|
| min_length | avg_len | max_len | ||
| cg1 | 723/240 | 841.7/279.6 | 1203/400 | Yes |
| cg2 | 2352/783 | 3759.2/1252.1 | 4134/1377 | Yes |
| cg3 | 2607/868 | 2853.7/950.2 | 3516/1171 | Yes |
| cg4 | 720/239 | 858.1/285 | 972/323 | No |
| cg5 | 495/164 | 743.4/246.8 | 1395/464 | Yes |
| cg6 | 2145/714 | 2837.2/944.7 | 3060/1019 | No |
| cg8 | 531/176 | 604.1/200.4 | 642/213 | Yes |
| cg9 | 1215/404 | 3364.7/1120.6 | 4152/1383 | No |
| cg10 | 1365/454 | 1531.4/509.5 | 1608/535 | Yes |
| cg11 | 2799/932 | 3130.6/1042.5 | 4773/1590 | Yes |
| cg12 | 1395/464 | 3309.3/1102.1 | 3597/1198 | No |
| cg16 | 1362/453 | 1387.7/461.6 | 1455/484 | No |
| cg17 | 606/201 | 1052.6/349.9 | 1248/415 | No |
| newcg2 | 369/122 | 881.2/292.7 | 1083/360 | Yes |
| newcg4 | 567/188 | 585.4/194.1 | 639/212 | Yes |
| newcg5 | 336/111 | 437.7/144.9 | 714/237 | No |
| newcg6 | 1011/336 | 1189.6/395.5 | 1902/633 | No |
| newcg7 | 402/133 | 482.9/160 | 594/197 | No |
3. Discussion
Characters consisting of any viral property or feature can be used to distinguish one virus from another, including genomic characterization, viral capsid structure, gene expression program, host range, and pathogenicity [8]. The genus demarcation criteria for Iridoviridae viruses proposed by ICTV is that members of a given genus share less than 50% amino acid sequence identity with members of other genera. Furthermore, additional criteria, such as phylogenetic analysis to clearly distinguish one genus from others, principal host species, presence of a DNA methytransferase, and characteristic pathology, can also distinguish genera within the family (file code: 2018.007D). Previously, methods used to classify members of the Iridoviridae included molecular analysis of restriction endonuclease (REN) profiles, mcp amplicons sequencing, DNA hybridization, terminal redundancies, and DNA-DNA homologies [15], [16]. However, with rapid expansion of viral genome databases, these advances have led the ICTV to present a consensus statement suggesting a shift from “traditional” taxonomy toward a genome-centered, and perhaps one day largely automated, viral taxonomy [17], [18].
Whole-genome average amino acid identity (AAI) is calculated based on protein-coding genes between a pair of genomes as determined by whole-genome pairwise sequence comparisons using the BLAST algorithm, which have been widely applied for microbial taxonomy [19]. Rohwer and Edwards successfully grouped phages into taxa by AAI analysis and highlighted genetic markers useful for monitoring phage biodiversity [20]. Furthermore, AAI analysis is also important for revealing bacterial genetic relatedness, whether at a single gene level (for instance, 16S rRNA and 23S rRNA) or at the whole-genome level [19].
Due to the controversy taxonomy on some members in the Iridoviridae family, we analyzed 179 iridovirids genomes available at NCBI. The AAI cut-off value (50%) for iridovirids genus demarcation proposed by ICTV included some dispersed genomes into the same group (Fig. 3B and Fig. 4A). In our study, an AAI cut-off value of 70% was found to be more suitable for iridovirids classification based on existing sequenced genomes, indicating that the Iridoviridae family should be divided into more genera, or subgroup at least. Further, synteny analysis, concatenated strict core gene phylogenetic analysis, genome codon usage preference, GC content and length statistics all supported our classification proposal.
It should be noted that we are not the first to call for an update of the taxonomy of members in the Iridoviridae family. The genus Ranavirus is the most researched and contains most of the iridovirids discovered so far. One of our previous studies of Santee-cooper ranavirus showed that Asian isolates are quite different from European and American isolates based on mcp phylogeny [4]. Genomic dot plot analysis in this study showed collinearity between the genomes of GIV and SGIV, but they possessed few regions of collinearity with other ranaviruses. In addition, GIV/SGIV lack the DNA methyltransferase gene that seen in other ranaviruses, which as a result, may need to be considered as a new genus, or recognized as a distinct species in the genus Ranavirus [3]. In our study, SGIV have the farthest evolutionary distance from the other two subgroups (Fig. 5). This is consistent with previous studies that the codon usage bias and genomic length of GIV and SGIV were different as compared to other members of Ranavirus [21]. Previous phylogenic analysis showed that scale drop disease virus (SDDV) clusters with megalocytiviruses, but form a separate branch within this genus [11]. Furthermore, the major infection symptoms of members of the same Iridoviridae genus are different. For instance, a symptom of SDDV infection in seabream is severe scale loss [22], whereas infection with infectious spleen and kidney necrosis virus (ISKNV) mainly observes diffuse necrosis in the haematopoietic tissues [23].
To date, phylogenetic analysis based on viral genomes or the 26 core genes identified by Eaton is the most commonly used method to elucidate evolutionary relationships among iridovirids, as seen in the genus or species renewal ICTV proposal for Iridoviridae in recent years [5]. However, the prerequisite is that the viral genomes should have been sequenced or the sequences of whole core genes are available. Previously, the major capsid protein (mcp) was thought to be reliable for the evolutionary analysis of iridovirids [10], [24], [25]. However, we found that the mcp gene is not accurate enough to allocate viruses at the genus level which is not recommended for future research (Supplementary file_2, Supplementary file_3). Instead, the identification of nine hallmark genes in this study provides an easy-to-use framework for virologists to accurately group viruses and form the basis of genus-level taxonomy in the future.
4. Methods
4.1. Genomic data and annotation
All Iridoviridae virus genomics listed in the National Center for Biotechnology Information (NCBI) Virus database (https://www. ncbi.nlm.nih.gov/labs/virus/) (as of December 2021) were collected. Genomes were re-annotated by using Prokka v1.14.6 package uniformly with the same parameters (settings: --kingdom Viruses, remaining settings: default) [26].
4.2. Repetitive genomes filtration
The program CompareM v0.1.2 (https://github.com/dparks1134/CompareM) was used to pairwise align collected genomes and calculate the AAI values of extracted CDS. The AAI value of 99% was set as a threshold to group similar viral genomics and then the generated network diagram matrix file was visualized by Cytoscape v3.8.2 [27]. Meanwhile, genomic phylogenic analysis was performed to examine the reliability of AAI analysis. All the genomic nucleic acid sequences were merged into a single file and subsequently submitted to ViPTreeGen (v.1.1.2) to construct a phylogenic tree [28]. From each group, select the most studied genome as the representative virus for later analysis. The GC content and genome size were calculated and visualized by seqkit v0.16.1 and the ggplot2 package in R [29].
4.3. Evaluation of core genes
After re-annotated collected iridovirids, each genome has a greater consensus among their annotated CDS. All protein sequences generated by Prokka annotation were merged into a single file (all.fa) using the “cat” command of Linux. Then, the all.fa file were submitted to BLAST (2.11.0+) for calculating the percentage of identical matches (makeblastdb -in all.fa -dbtype prot -out index/all -parse_seqids; blastp -query all.fa -db index/all -out all_blast.out -evalue 1e-5 -num_threads 8 -outfmt 6). After grouping conserved homologous genes by using Cytoscape, core genes of iridovirids were screened out by filtering groups including paralogous genes or genes that were not shared by all 57 representative genomes.
4.4. AAI analysis
The program CompareM v0.1.2 was used to calculate average amino acid identity (AAI) of representative Iridoviridae genomes. The AAI value of 50% (according to the ICTV proposal) and 70% (generated in this study) were separately set as threshold to group iridovirids genome, then visualize generated matrix file by using Cytoscape v3.8.2.
4.5. Synteny analysis of core genes
Synteny analysis serves as an alternative method to determine viral taxonomy and evolutionary relationships. BLAST v2.11.0+ (E-value threshold of 1e-5) and MCScanX were performed to determine synteny of concatenated core genes of representative iridovirids genes (Table 3). Firstly, annotated amino acid sequence files of representative iridovirids were merged into a dataset, using the “makeblast” command of BLAST. Secondly, the merged sequence file iridovirus.fa was aligned by using “blastp” command of BLAST. Then, comparison results were filtered according to the identity threshold of 75%. Finally, both the annotation information file (gff format) and the aligned file were imported into MCScanX to generate synteny images.
Table 3.
The detailed steps of synteny analysis.
| Step | Codes |
|---|---|
| Step 1: Create database | makeblastdb -in iridovirus.fa -dbtype prot -out index/all -parse_seqids |
| Step 2: BLAST | blastp -query iridovirus.fa -db index/all -out out.blast -evalue 1e-5 -num_threads 8 -outfmt 6 |
| Step 3: Filtration | cat out.blast | awk ‘{ if ($3 > 75) print $0}’ > iridovirus.blast (identity threshold set as 75%) |
| Step 4: MCScanX | ./MCScanX input_file/iridovirus |
| Step 5: Visualization | java dot_plotter -g iridovirus.gff –s iridovirus.collinearity -c dot.ctl -o dot.PNG |
4.6. Phylogenic analysis
The maximum likelihood phylogenetic tree (ML-Tree) was constructed based on core genes of representative iridovirids. The MAFFT software was used to pairwise align sequences using the default setting [30]. The aligned core genes were concatenated by using PhyloSuite [31]. ML-Trees were then constructed by using IQ-TREE v1.6.12 [32]. Finally, iTol was used to annotate the phylogenetic trees [33].
4.7. Indicators for codon performance
In this study, correspondence Analysis (CoA) on RSCU and ENC-Plot Analysis were performed to evaluate viral codon usage preference as previously described [12]. In brief, each viral coding region was represented as 59-dimensional vector corresponding to RSCU value of each synonymous codon (excluding AUG, UGG, and stop codons) calculated by CodonW program. The effective number of codons (ENC) ranging from 20 (only one specific codon is recruited for each amino acid) to 61 (the recruitment percentage for all synonymous codons is equal) were also calculated. The expected ENC value corresponding to GC3 was calculated as previously described [12]. All data was finally visualized by R ggplot2 package.
CRediT authorship contribution statement
Ruoxuan Zhao: Conceptualization, Data curation, Software, Writing – original draft, Writing – review & editing. Congwei Gu: Conceptualization, Data curation, Writing – review & editing. Xiaoxia Zou: Conceptualization, Data curation, Writing – review & editing. Mingde Zhao: Software, Writing – review & editing. Wudian Xiao: Software, Writing – review & editing. Manli He: Methodology, Writing – review & editing. Lvqin He: Methodology, Writing – review & editing. Qian Yang: Data curation, Writing – review & editing. Yi Geng: Conceptualization, Writing – review & editing. Zehui Yu: Conceptualization, Methodology, Software, Writing – original draft, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This work was supported by The Science and Technology Strategic Cooperation Programs of Sichuan University and Luzhou Municipal People's Government, and Suining First People's Hospital – Southwest Medical University Strategic Cooperation Project (2021SNXNYD03).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.06.049.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
References
- 1.Chinchar V.G., Hick P., Ince I.A., Jancovich J.K., Marschang R., Qin Q., et al. ICTV virus taxonomy profile: Iridoviridae. J Gen Virol. 2017 May 1;98(5):890–891. doi: 10.1099/jgv.0.000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chinchar V.G., Duffus A.L.J. Molecular and Ecological Studies of a Virus Family (Iridoviridae) Infecting Invertebrates and Ectothermic Vertebrates. Viruses. 2019;11(6) doi: 10.3390/v11060538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray M.J., Chinchar V.G. Ranaviruses: lethal pathogens of ectothermic vertebrates. Springer. Nature. 2015 [Google Scholar]
- 4.Zhao R., Geng Y., Qin Z., Wang K., Ouyang P., Chen D., et al. A new ranavirus of the Santee-Cooper group invades largemouth bass (Micropterus salmoides) culture in southwest China. Aquaculture. 2020;526 [Google Scholar]
- 5.Eaton H.E., Metcalf J., Penny E., Tcherepanov V., Upton C., Brunetti C.R. Comparative genomic analysis of the family Iridoviridae: re-annotating and defining the core set of iridovirus genes. Virol J. 2007;4:11. doi: 10.1186/1743-422X-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teacher A.G.F., Cunningham A.A., Garner T.W.J. Assessing the long-term impact of Ranavirus infection in wild common frog populations. Anim Conserv. 2010;13(5):514–522. [Google Scholar]
- 7.Jancovich J.K., Davidson E.W., Parameswaran N., Mao J., Chinchar V.G., Collins J.P., et al. Evidence for emergence of an amphibian iridoviral disease because of human-enhanced spread. Mol Ecol. 2005;14(1):213–224. doi: 10.1111/j.1365-294X.2004.02387.x. [DOI] [PubMed] [Google Scholar]
- 8.Lefkowitz E.J., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acids Res. 2018;46(D1):D708. doi: 10.1093/nar/gkx932. D717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu L., Chen M.M., Wan X.Y., Li C., Zhang Q.L., Wang R.Y., et al. Characterization of a new member of Iridoviridae, Shrimp hemocyte iridescent virus (SHIV), found in white leg shrimp (Litopenaeus vannamei) Sci Rep. 2017;7(1):11834. doi: 10.1038/s41598-017-10738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L., Wang T., Li F., Yang F. Isolation and preliminary characterization of a new pathogenic iridovirus from redclaw crayfish Cherax quadricarinatus. Dis Aquat Organ. 2016;120(1):17–26. doi: 10.3354/dao03007. [DOI] [PubMed] [Google Scholar]
- 11.de Groof A., Guelen L., Deijs M., van der Wal Y., Miyata M., Ng K.S., et al. A Novel Virus Causes Scale Drop Disease in Lates calcarifer. PLoS Pathog. 2015;11(8):e1005074. doi: 10.1371/journal.ppat.1005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh N.K., Tyagi A., Kaur R., Verma R., Gupta P.K. Characterization of codon usage pattern and influencing factors in Japanese encephalitis virus. Virus Res. 2016;221:58–65. doi: 10.1016/j.virusres.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Williams T., Cory J.S. Proposals for a new classification of iridescent viruses. J Gen Virol. 1994;75(6):1291–1301. doi: 10.1099/0022-1317-75-6-1291. [DOI] [PubMed] [Google Scholar]
- 14.Webby R.J., Kalmakoff J. Comparison of the major capsid protein genes, terminal redundancies, and DNA–DNA homologies of two New Zealand iridoviruses. Virus Res. 1999 Feb 1;59(2):179–189. doi: 10.1016/s0168-1702(98)00135-x. [DOI] [PubMed] [Google Scholar]
- 15.Ince I.A., Ozcan O., Ilter-Akulke A.Z., Scully E.D., Ozgen A. Invertebrate Iridoviruses: A Glance over the Last Decade. Viruses. 2018;10(4) doi: 10.3390/v10040161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson C.C., Chimetto L., Edwards R.A., Swings J., Stackebrandt E., Thompson F.L. Microbial genomic taxonomy. BMC Genomics. 2013 Dec;14(1):1–8. doi: 10.1186/1471-2164-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohwer F., Edwards R. The Phage Proteomic Tree: a genome-based taxonomy for phage. J Bacteriol. 2002;184(16):4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng Z., Wang J., Zhang W., Geng Y., Zhao M., Gu C., et al. The Insights of Genomic Synteny and Codon Usage Preference on Genera Demarcation of Iridoviridae Family. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.657887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Y, Li Y, Fu W, Su H, Zhang L, Huang C, et al. Scale Drop Disease Virus Associated Yellowfin Seabream (Acanthopagrus latus) Ascites Diseases, Zhuhai, Guangdong, Southern China: The First Description. Viruses. 2021;13(8). [DOI] [PMC free article] [PubMed]
- 20.He J.G., Zeng K., Weng S.P., Chan S.M. Experimental transmission, pathogenicity and physical–chemical properties of infectious spleen and kidney necrosis virus (ISKNV) Aquaculture. 2002 Jan 21;204(1–2):11–24. [Google Scholar]
- 21.Geng Y., Wang K.Y., Zhou Z.Y., Li C.W., Wang J., He M., et al. First report of a ranavirus associated with morbidity and mortality in farmed Chinese giant salamanders (Andrias davidianus) J Comp Pathol. 2011;145(1):95–102. doi: 10.1016/j.jcpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Wei J., Huang Y., Zhu W., Li C., Huang X., Qin Q. Isolation and identification of Singapore grouper iridovirus Hainan strain (SGIV-HN) in China. Arch Virol. 2019;164(7):1869–1872. doi: 10.1007/s00705-019-04268-z. [DOI] [PubMed] [Google Scholar]
- 23.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 24.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimura Y., Yoshida T., Kuronishi M., Uehara H., Ogata H., Goto S. ViPTree: the viral proteomic tree server. Bioinformatics. 2017;33(15):2379–2380. doi: 10.1093/bioinformatics/btx157. [DOI] [PubMed] [Google Scholar]
- 26.Shen W., Le S., Li Y., Hu F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE. 2016;11(10):e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K., Asimenos G., Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 28.Zhang D., Gao F., Jakovlic I., Zou H., Zhang J., Li W.X., et al. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 2020;20(1):348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293. doi: 10.1093/nar/gkab301. W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.