Abstract
Mammalian cells require activated folates to generate nucleotides for growth and division. The most abundant circulating folate species is 5-methyl tetrahydrofolate (5-methyl-THF), which is used to synthesize methionine from homocysteine via the cobalamin-dependent enzyme methionine synthase (MTR). Cobalamin deficiency traps folates as 5-methyl-THF. Here, we show using isotope tracing that methionine synthase is only a minor source of methionine in cell culture, tissues, or xenografted tumors. Instead, methionine synthase is required for cells to avoid folate trapping and assimilate 5-methyl-THF into other folate species. Under conditions of physiological extracellular folates, genetic MTR knockout in tumor cells leads to folate trapping, purine synthesis stalling, nucleotide depletion, and impaired growth in cell culture and as xenografts. These defects are rescued by free folate but not one-carbon unit supplementation. Thus, MTR plays a crucial role in liberating tetrahydrofolate for use in one-carbon metabolism.
Introduction
Dividing cells require activated one-carbon (1C) units to synthesize and package nucleic acids [1]. In mammals, tetrahydrofolate (THF) species carry the formyl- and methylene- 1C groups necessary for purine and thymidylate biosynthesis [2–4]. This essentiality is reflected in the efficacy of antifolate chemotherapeutic agents for treating malignancy, their associated toxicities in proliferating normal tissues, and alleviation of these effects by folate rescue agents (i.e. 5-formyl-THF, a.k.a. leucovorin) [5]. The 1C units of activated folates are locally sourced from serine, glycine, and other amino acids [6, 7], with the major 1C donor typically serine via the enzyme serine hydroxymethyltransferase (SHMT).
In contrast to the capacity to manipulate 1C units using THF, mammals lack the necessary machinery for de novo folate synthesis. Folate species are instead absorbed from food and converted in the gut and liver to 5-methyl tetrahydrofolate (5-methyl-THF), the dominant circulating folate species, before distributing to tissue folate pools [8, 9]. Deficiency in dietary folate causes neural tube defects during pregnancy [10].
Mammals also lack the ability to assimilate inorganic sulfur, and thus have a daily requirement for sulfur containing amino acids like methionine [11–13]. In addition to its role in protein synthesis, methionine serves as the primary methyl source for DNA, protein, and lipid methylation via S-adenosyl-methionine (SAM) [14]. Methyl donation by SAM ultimately yields the free thiol species homocysteine, which undergoes either degradation via the transsulfuration pathway to yield cysteine or remethylation to methionine [11]. SAM also acts as an allosteric regulator, inhibiting methylenetetrahydrofolate reductase (MTHFR), the enzyme producing 5-methyl-THF, and activating cystathionine β-synthase (CBS), the first enzyme of transsulfuration [15, 16]. Low SAM accordingly tends to preserve homocysteine and promote its conversion to methionine, allowing homocysteine to substitute for dietary methionine [17–19].
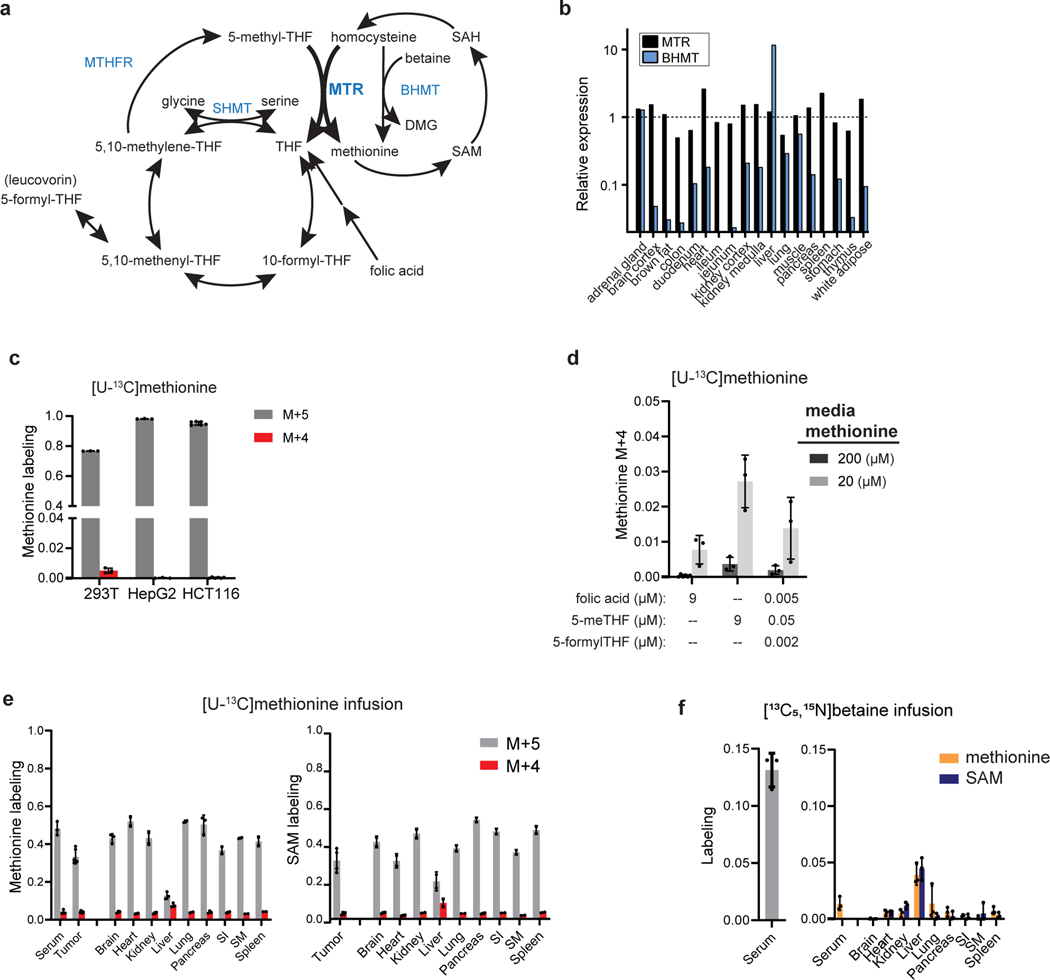
Methionine synthase (MTR) catalyzes the methyl transfer from 5-methyl-THF to homocysteine, yielding methionine and tetrahydrofolate (THF), linking folate and methionine cycling [11, 20] (Fig. 1A). MTR is one of two enzymes in humans known to utilize the cofactor vitamin B12 (cobalamin) [21]. Mutations in MTR leading to diminished enzyme activity result in methylcobalamin deficiency G (cblG) disorder, a condition characterized by hyperhomocysteinemia, hypomethioninemia and homocystinuria [22]. In development, MTR is essential, with its loss leading to embryonic death in mice [23]. MTR is universally expressed in tissues, while expression of betaine-homocysteine S-methyltransferase (BHMT), the other enzyme known to produce methionine via methyl transfer from betaine to homocysteine, is primarily expressed in the liver and, to a lesser degree, in the kidney ([15, 24–26]; Fig. 1B). Similarly, MTR is broadly expressed in human cancer cell lines [27] and tumors [28].
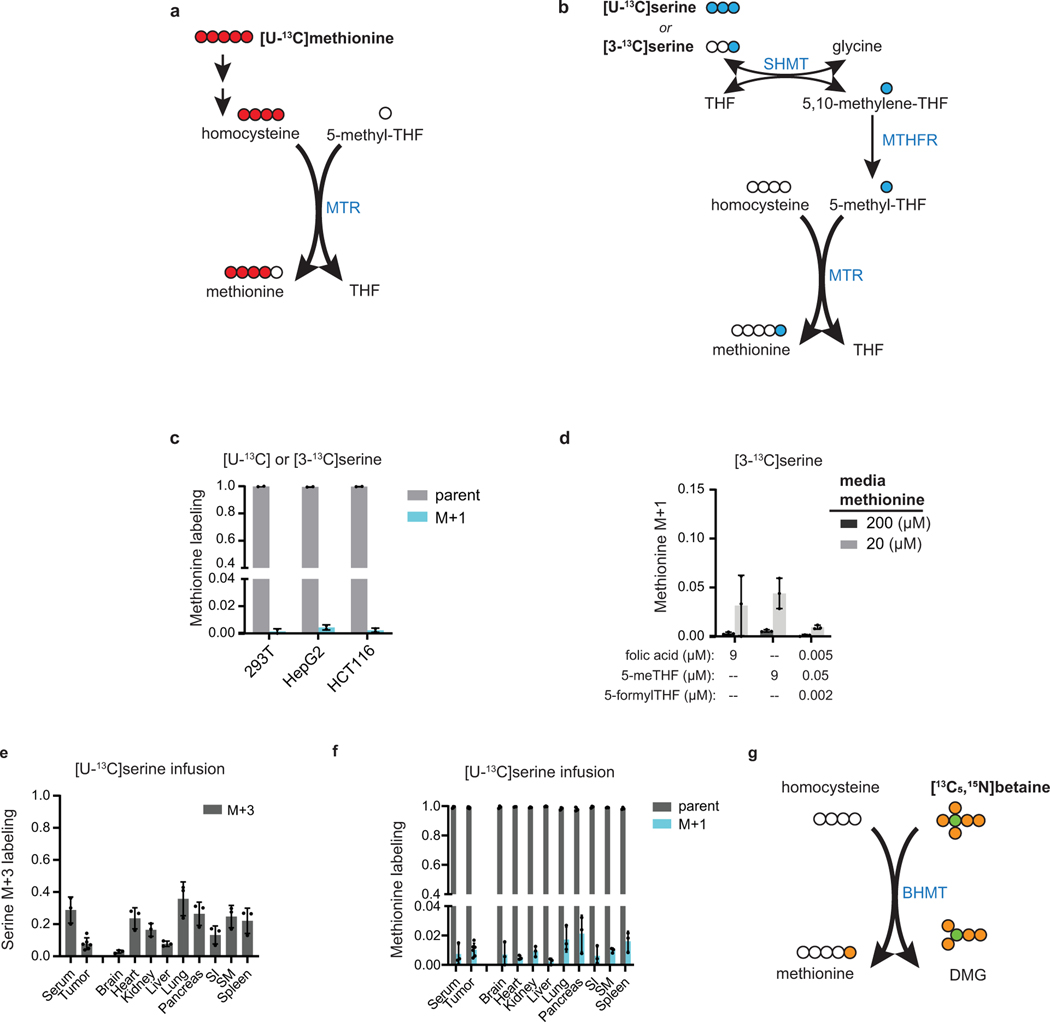
Figure 1. MTR is a minor source of methionine in cell lines, tissues and tumors.
(A) Schematic showing the activity of MTR at the intersection of the folate and methionine cycles. MTR = methionine synthase, MTHFR = methylenetetrahydrofolate reductase, BHMT = betaine-homocysteine S-methyltransferase, SHMT = serine hydroxymethyltransferase. (B) Relative protein levels of MTR and BHMT in mice from [24]. (C) Methionine labeling in cell lines after culturing for 4 h in media containing [U-13C]methionine (mean ± SD, n = 6 for HCT116 cells, n = 3 for 293T and HepG2). (D) Methionine M+4 fraction from 4 h [U-13C]methionine tracing in HCT116 cultured in media containing indicated methionine and folate concentrations (mean ± SD, n = 6 for 9 μM folic acid and 200 μM methionine, n = 3 for others). (E) Methionine (left) and S-adenosyl-methionine (SAM, right) labeling of PDAC tumors and normal tissues, and methionine labeling of serum in male C57BL/6 mice after [U-13C]methionine infusion for 2.5 h (mean ± SD, n = 3 mice for methionine labeling and n = 2 mice for SAM labeling; two technical replicates were included for each tumor). (F) Betaine labeling (M+6) in serum (left), and methionine and SAM labeling (M+1, right) in tissues of male C57BL/6 mice after [13C5,15N]betaine infusion for 4 h (mean ± SD, n = 3 mice).
The physiological role of MTR is evident from vitamin B12 deficiency. Classically, B12-deficient patients present with neurological and hematological manifestations, most notably a macrocytic, megaloblastic anemia, an indicator of impaired nucleotide synthesis. This phenotype similarly occurs in folate deficiency [29, 30]. Vitamin B12 restriction or chemical inactivation of vitamin B12 (and thus MTR) by nitrous oxide impairs MTR activity in vitro and in vivo [30–32]. Moreover, because methylene-THF reduction to 5-methyl-THF is nearly irreversible in cells (due to high cytosolic NADPH/NADP+) [33], MTR dysfunction leads to folate trapping as 5-methyl-THF [20, 30, 34–36].
Despite numerous studies over the last 70 years into folate and methionine metabolism, it is unclear how much methionine arises locally from remethylation in a given tissue. Similarly, the role of MTR in supporting cancer growth has not yet been robustly examined. Here, we combine modern genetic tools with isotope tracer studies to dissect the role of MTR in normal tissue and tumor biology. We show that, under standard dietary conditions, tissue and tumor methionine is overwhelmingly acquired from the systemic circulation, with MTR contributing only a minor fraction to local methionine pools. Conversely, MTR plays a major role in maintaining folate pools, with cells and xenografted tumors lacking MTR unable to grow when subjected to physiological folate conditions. Mechanistically, we show that MTR loss leads to folate trapping, purine synthesis stalling and nucleotide depletion, which can be mitigated with folate rescue agents.
Results
Methionine comes mainly from uptake rather than remethylation
Methionine cycling involves reduction of 5,10-methylene-THF to 5-methyl-THF mediated by MTHFR, followed by remethylation of homocysteine to methionine by MTR. Alternatively, homocysteine can be remethylated to methionine by betaine hydroxymethyltransferase (BHMT) (Fig. 1A). MTR is expressed throughout the body, while BHMT is preferentially expressed in liver (Fig. 1B).
To probe methionine cycling, we began with cell culture experiments where the media methionine component was substituted for [U-13C]methionine at the same concentration. We reasoned that untransformed methionine would retain all five labeled carbons (M+5). In contrast, methionine that had been recycled via homocysteine remethylation would lose one 13C carbon, retaining four labeled carbons (M+4; Fig. 1A, Extended Data Fig. 1A). The fraction of M+4 to M+5 methionine approximates the relative amount of cellular methionine arising via remethylation as opposed to uptake. All cell lines examined displayed a high fraction of M+5 and low fraction of M+4 methionine (<3%), consistent with intracellular methionine mainly arising from uptake (Fig. 1C). Similar experiments with either [U-13C] or [3-13C]serine tracing, looking for M+1 methionine coming from homocysteine remethylation via the SHMT-MTHFR-MTR pathway (Extended Data Fig. 1B), showed very low levels of enrichment (<1%) (Extended Data Fig. 1C). Remethylation was modestly greater in cells cultured with 5-methyl-THF rather than folic acid or a physiologic folate mixture as the folate source (Fig. 1D, Extended Data Fig. 1D) [37, 38]. Likely due to dilution of the same remethylation flux by a smaller total amount of extracellular methionine, decreasing the exogenous methionine concentrations to physiological serum levels increased fractional methionine from remethylation, but only to 3% of the total cellular methionine pool (Fig. 1D, Extended Data Fig. 1D). Thus, consistent with prior literature, methionine cycling is minimal in cell culture [3, 39].
To examine whether these observations hold in vivo, we infused [U-13C]methionine via jugular venous catheter into C57BL/6 mice bearing subcutaneous pancreatic ductal adenocarcinoma (PDAC) allograft tumors [40], achieving methionine enrichment of 30–60% in serum, tumors and most tissues (Fig. 1E). This extent of labeling is adequate for robust label detection while avoiding severe metabolic perturbation by the labeled methionine infusion. In all tissues except liver, as well as tumors, M+5 labeling was dominant, with M+4/M+5 ratio about 0.1, suggesting that approximately 10% of methionine arose from homocysteine remethylation (Fig. 1E). The fraction of M+4 was similar across serum, tumors, and non-liver tissues, and thus the observed M+4 labeling may be coming from circulating methionine rather than tissue- or tumor-intrinsic methionine cycling. In liver, M+5 labeling was markedly lower and M+4 higher, resulting in a M+4/M+5 ratio of ~0.6 and consistent with substantial methionine arising via homocysteine remethylation. However, infusion of labeled serine demonstrated limited folate-mediated methionine synthesis throughout the body, including the liver (Extended Data Fig. 1E–F). Thus, homocysteine remethylation in the liver must utilize a different methyl donor.
We next sought to assess the contribution of remethylated methionine from betaine, the alternative source of methionine in mammals. The BHMT reaction transfers a methyl group from the quaternary amine of betaine to homocysteine via a cobalamin-independent reaction mechanism [41]. Mice were infused with [13C5,15N]betaine tracer, which contains a 13C on each methyl group and thus will give rise to M+1 labeled methionine (Extended Data Fig. 1G). This infusion revealed preferential methionine synthesis from betaine in liver (Fig. 1F). SAM labeling largely aligned with that of methionine (Fig. 1E–F). HepG2, a hepatocellular carcinoma cell line, did not show comparable homocysteine remethylation activity to liver (Fig. 1C, Extended Data Fig. 1C), which can be explained by a splicing variant leading to loss of BHMT function in HepG2 [42]. Collectively, these studies support BHMT being a source of methionine and SAM in healthy liver while the MTHFR-MTR pathway operates more broadly throughout the body but, at least acutely, makes only a small contribution to overall methionine/SAM pools.
MTR is required for cell growth in physiological folates
The above studies raised questions about the physiological role of the MTHFR-MTR pathway, since it was not a dominant methionine producer anywhere in the body. As an alternative strategy to examine the role of MTR, we generated clonal MTR knockout cell lines (ΔMTR) using CRISPR-Cas9 gene editing in three cancer cell backgrounds: HCT-116 (colorectal carcinoma), 8988T (pancreatic ductal carcinoma), and HepG2 (hepatocellular carcinoma) (Fig. 2A). Each of these parental cell lines possesses reasonable expression of MTR (Extended Data Fig. 2A). In standard tissue culture media formulations containing folic acid, each of these MTR knockout lines grew reasonably well (with no growth rate impairment in the HCT116 background and some in the 8988T and HepG2 backgrounds) (Extended Data Fig. 2B). Expectedly, neither MTR knockout nor control lines grew in the complete absence of folates (Extended Data Fig. 2B). Control cells were, however, able to grow well in media where folic acid was replaced with 5-methyl-THF, the most abundant circulating folate species. In contrast, MTR knockout cells did not grow at all (Extended Data Fig. 2B). The defective growth of the MTR knockout cells was fully rescued through supplementation with 5-formyl-THF (leucovorin) (Extended Data Fig. 2B), a folate species that enters folate metabolism independently of MTR. Thus, MTR is required for cancer cell growth using 5-methyl-THF, the main circulating folate species, presumably because MTR is required to liberate THF from 5-methyl-THF.
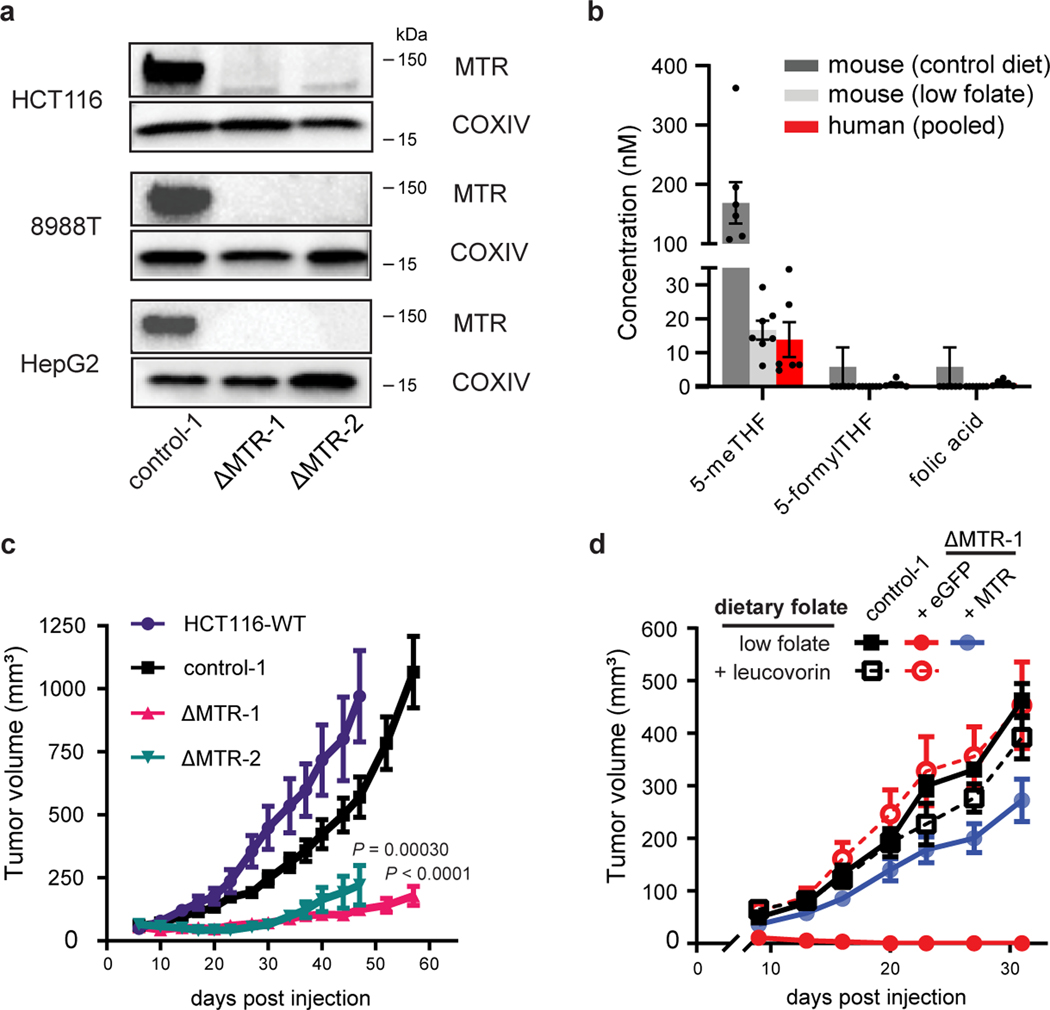
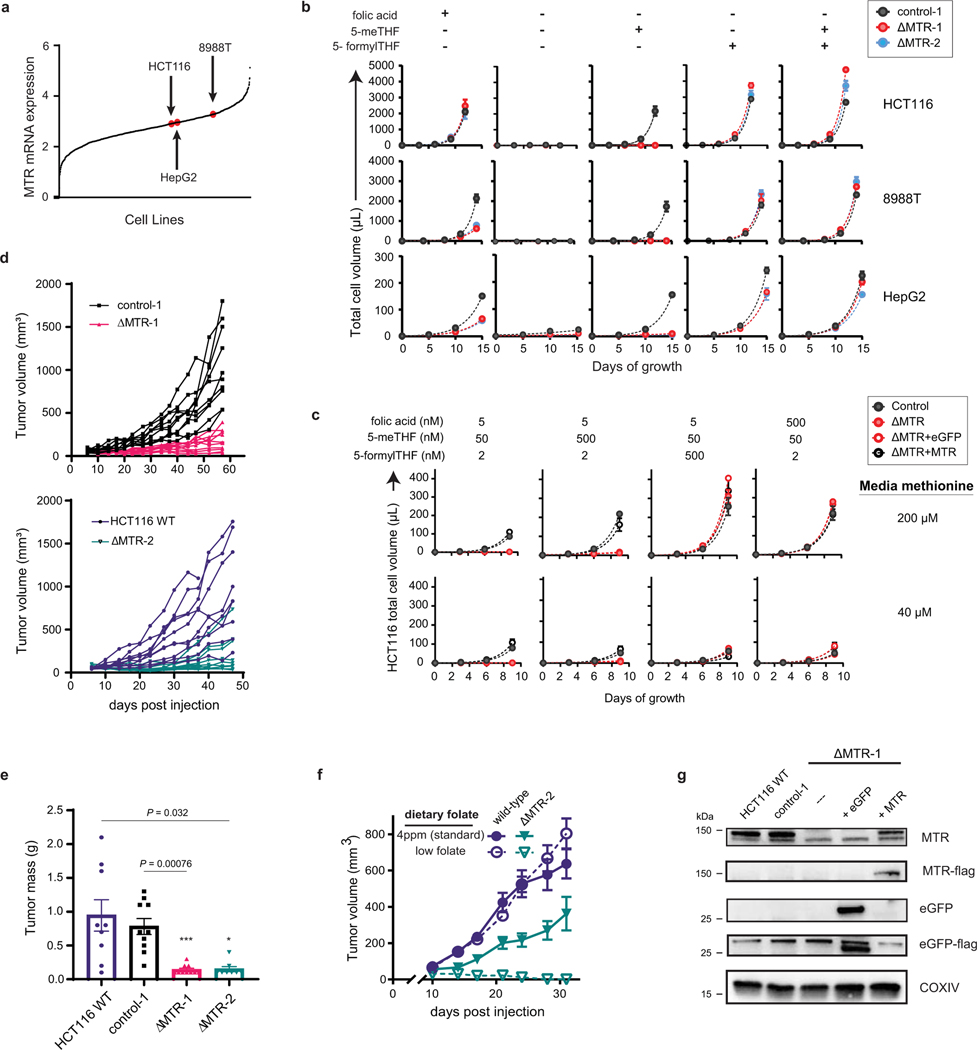
Figure 2. MTR is essential for cell and tumor growth under physiological folate conditions.
(A) Western blot analysis of MTR in HCT116, 8988T and HepG2 clonally isolated lines after treatment with lentiviral CRISPR-Cas9 targeting scrambled control (control-1) or MTR (ΔMTR-1 and ΔMTR-2). Results are representative of 3 biological replicates with similar results. (B) Folate concentrations in mouse and human serum measured by LC-MS (mean ± SEM, 6 technical replicates for pooled human serum, n = 7 male CD-1 nude mice on control diet with 4mg/kg folic acid and n = 7 male CD-1 nude mice on low folate diet). (C) Growth of subcutaneous HCT116 xenografts in female CD-1 nude mice on standard chow with 3 mg/kg folic acid (mean ± SEM, n = 10 mice, P values were determined by mixed-effects two-way ANOVA comparing tumors implanted on mice’s two flanks: wild-type (WT) and ΔMTR-2, control-1 and ΔMTR-1). (D) Growth of subcutaneous HCT116 xenografts in male CD-1 nude mice on a low folate diet with or without leucovorin supplement (mean ± SEM, for low folate diet: n = 7 mice for control-1, n = 8 mice for ΔMTR+eGFP, n = 15 mice for ΔMTR+MTR; low folate diet with leucovorin supplementation: n = 8 mice for each cell line).
In addition to 5-methyl-THF, other folate species are present in serum at much lower concentrations [37], which we confirmed through LC-MS analysis of murine and human sera (Fig. 2B). To investigate whether these alternative species could compensate for loss of MTR, we cultured MTR knockout cells in media containing a physiological folate mixture. Consistent with their low concentrations, physiological circulating levels of folic acid and 5-formyl-THF were insufficient to support MTR knockout cell growth, even though higher levels fully rescued knockout cell growth (Extended Data Fig. 2C).
To explore cell growth in vivo, we subcutaneously implanted HCT116 ΔMTR cells into the lateral flanks of nude mice. Tumors derived from MTR knockout cells demonstrated a growth deficiency (Fig. 2C, Extended Data Fig. 2D–E). Moreover, this effect was amplified when mice were fed a low folate diet (Fig. 2D), an intervention leading to closer alignment of serum folate pools with those of humans ([43], Fig. 2B), while wild-type tumor growth was unaltered (Extended Data Fig. 2F).
To further confirm this phenotype arises from MTR loss and subsequent THF deprivation, re-expression of MTR (ΔMTR + MTR), but not control (ΔMTR + eGFP), rescued in vitro and in vivo growth of ΔMTR cells (Fig. 2D, Extended Data Fig. 2C, 2G). Moreover, oral supplementation of leucovorin was sufficient to rescue in vivo ΔMTR tumor growth (Fig. 2D). Together, these findings implicate MTR as critical for folate access in the presence of a physiological mixture of circulating folates.
MTR loss depletes cells and tumors of reduced folate species
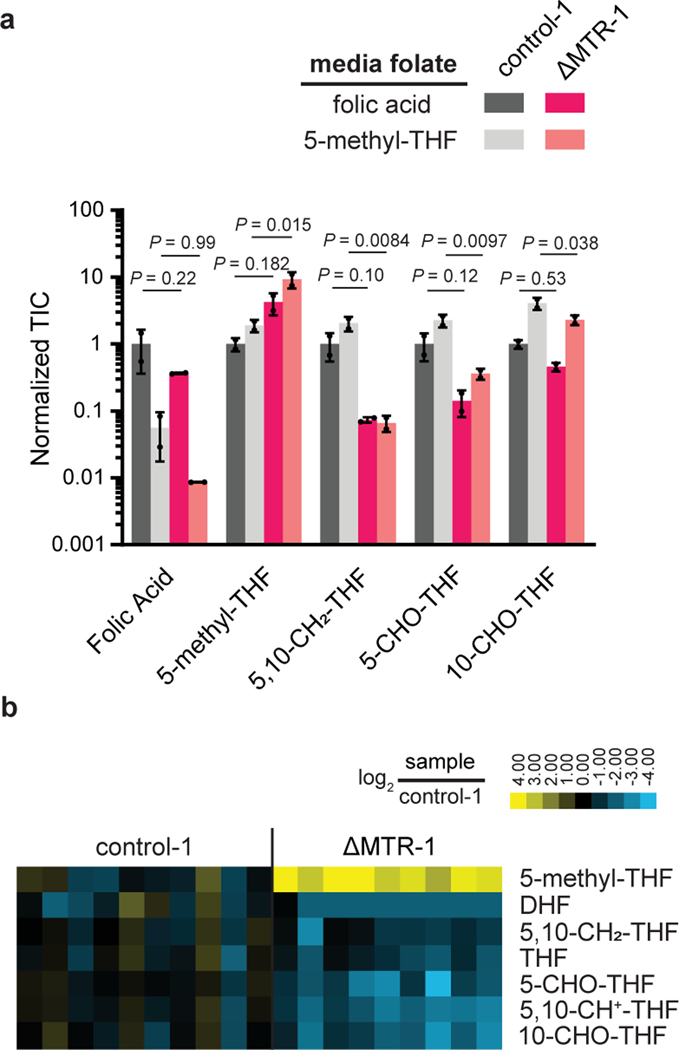
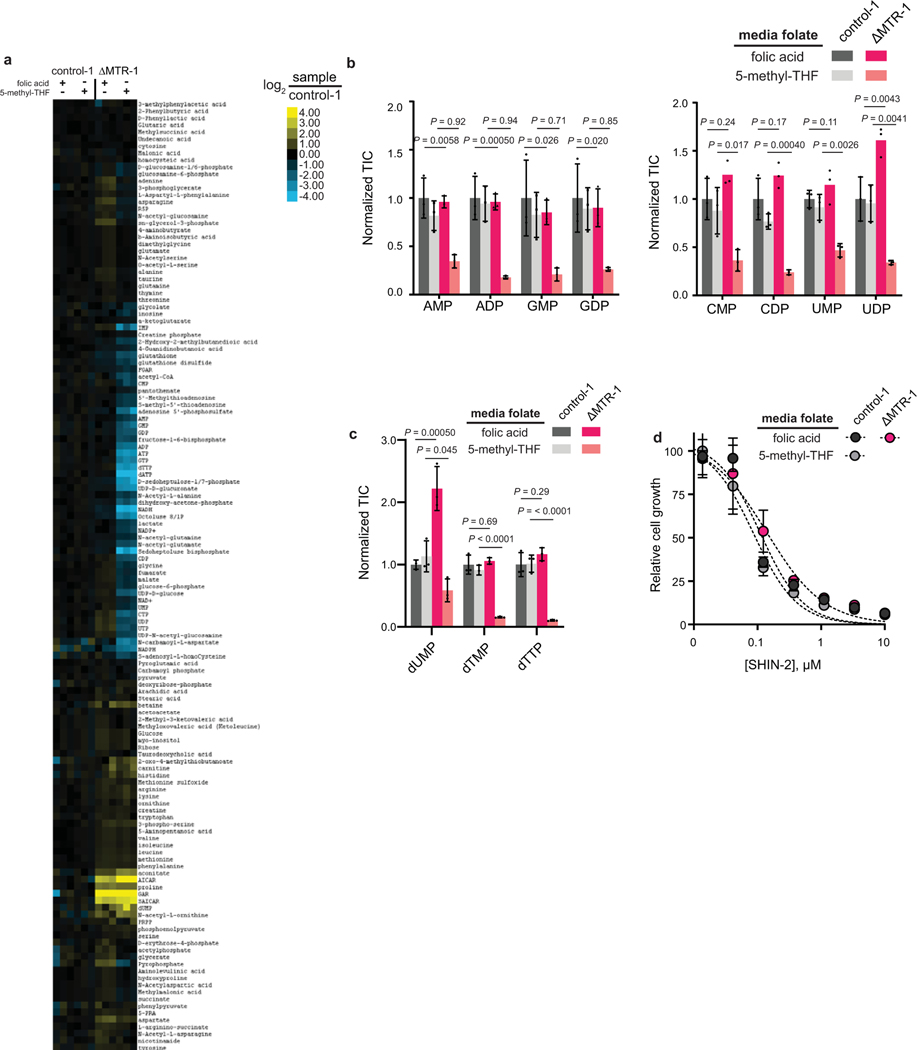
These above findings support the hypothesis that MTR is required to supply cancer cells with free THF when fed 5-methyl-THF, the most abundant physiologically relevant folate species. To evaluate this further, we measured cellular folate species in HCT116 cells cultured with either folic acid or 5-methyl-THF as the sole media folate source. Expectedly, in line with the concept of “methyl trapping”, MTR loss led to a significant buildup of 5-methyl-THF in both media conditions (Fig. 3A). Concurrently, most downstream folate species were substantially decreased. The most depleted species was 5,10-methylene-THF, which is required for thymidine biosynthesis. In contrast, 10-formyl-THF, which is used for purine synthesis, was not strongly depleted. This likely reflects inhibition of 10-formyl-THF consumption by purine pathway disruption due to overall folate species dysregulation. Interestingly, buildup of 5-methyl-THF and depletion of other folate species was triggered by MTR loss in cells growing in media containing folic acid, indicating the importance of MTR for folate pool homeostasis even when adequate free folate is available.
Figure 3. Loss of MTR results in the elevation of 5-methyl-THF and depletion of other folate species.
(A) Relative abundance of folate species in HCT116 control and MTR knockout cells in media with indicated folate sources. Intensities are normalized to the average of control-1 cells in folic acid (mean ± SD, n = 2, P values were determined by a one-way ANOVA comparing control to MTR knockout in the same medium followed by Dunnett’s post hoc analysis). (B) Relative abundance of folate species in individual HCT116 control and MTR knockout subcutaneous tumors (normalized to control tumor average). DHF = dihydrofolate, THF = tetrahydrofolate. TIC = total ion count.
In vivo, 5-methyl-THF was elevated ~12-fold in HCT116 ΔMTR-1 tumors when compared to control-1 tumors, while all other folate species were depleted by about 50–80% (Fig. 3B). This suggests that, although MTR produces THF and methionine equimolarly, it is more important for maintaining folate pools than for methionine pool, which is sensible given the lower abundance of folate species.
MTR activity maintains purine and pyrimidine biosynthesis
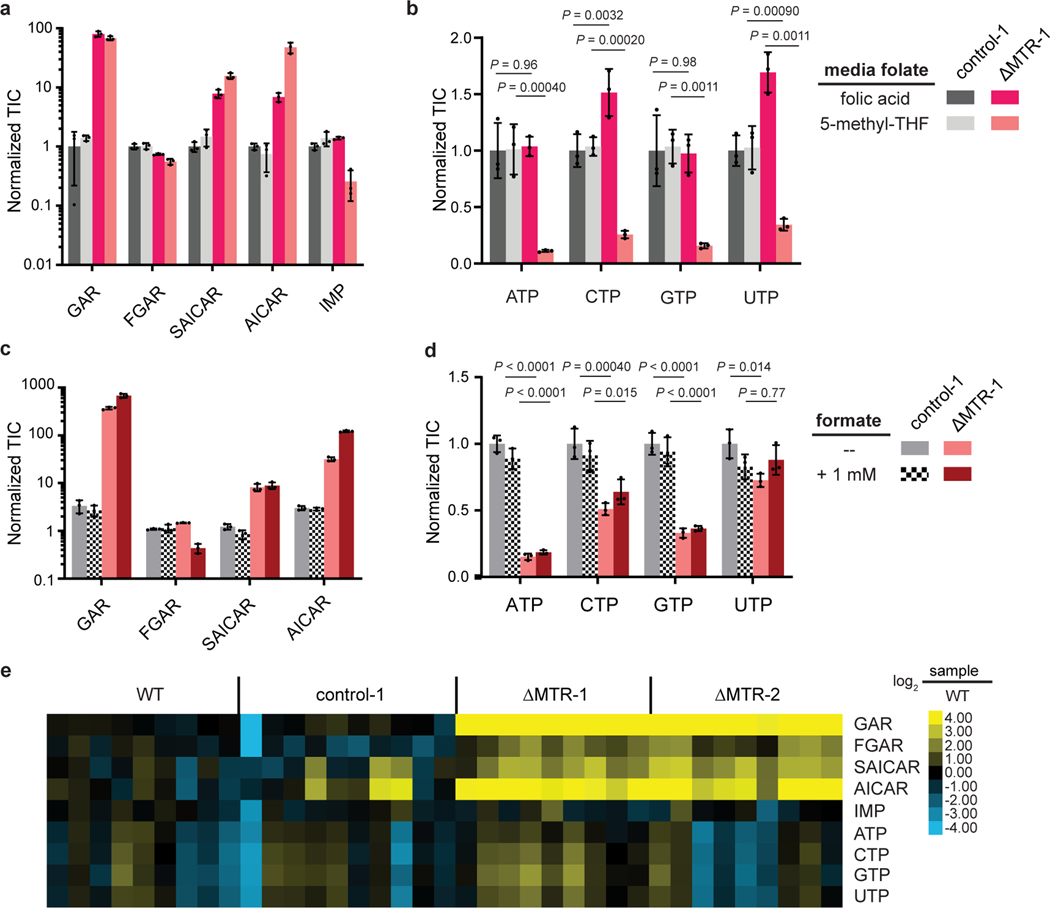
We next sought to investigate the downstream metabolic effects of MTR loss. To this end, HCT116 ΔMTR cells and controls were cultured in media containing either folic acid or 5-methyl-THF as the sole media folate. Metabolomics demonstrated global changes in metabolism due to MTR loss (Extended Data Fig. 3A). Most notably, we observed dramatic increases in several purine biosynthetic intermediate pools, including GAR, SAICAR and AICAR (Fig. 4A), and corresponding decreases in nucleotide species (Fig. 4B, Extended Data Fig. 3B). Nucleotide depletion was most substantial in media containing 5-methyl-THF as the sole folate source. Similarly, MTR loss in folic acid media led to a buildup of dUMP, consistent with impaired thymidine biosynthesis secondary to diminished 5,10-methylene-THF, though the thymidylate species dTMP and dTTP were unperturbed (Extended Data Fig. 3C). Interestingly, both dUMP and dTMP/dTTP were significantly decreased in 5-methyl-THF media, possibly reflecting the overall decrease in the precursor UMP, perhaps due to purine-pyrimidine crosstalk.
Figure 4. MTR supports purine biosynthesis by promoting folate availability.
Relative abundance of (A) purine biosynthetic intermediates and (B) nucleotides in HCT116 control and MTR knockout cells in media with indicated folate sources. Intensities are normalized to the average of control-1 cells in folic acid (mean ± SD, n = 3). Relative abundance of (C) purine biosynthetic intermediate and (D) nucleotides in control and MTR knockout cells grown in media with 5-methyl-THF, with and without the addition of 1 mM formate (mean ± SD, n = 3). (E) Relative abundance of purine biosynthetic intermediate and nucleotide triphosphates in individual HCT116 control and MTR knockout subcutaneous tumors (normalized to control tumor average). TIC = total ion count. For (B) and (D), P values were determined by a one-way ANOVA comparing control to MTR knockout in the same medium followed by Dunnett’s post hoc analysis.
To evaluate whether these changes were due to aberrant levels of folates, or deficiency of usable 1C units [3], we attempted to rescue these effects through supplementation with the 1C precursor formate. Consistent with this phenotype arising from folate dysregulation, formate did not rescue (Fig. 4C–Fig. 4D). Furthermore, MTR knockout cells showed equivalent sensitivity as wild-type to the SHMT1/2 inhibitor SHIN2 [44], which blocks loading of THF with 1C units from serine (Extended Data Fig. 3D). To investigate whether these metabolic changes occur in vivo, we examined nucleotide levels from the HCT116 xenografts (Extended Data Fig. 4A). Elevated levels of GAR (50-fold), FGAR (4-fold), SAICAR (4-fold), and AICAR (7-fold) were observed in the MTR knockouts relative to wild-type controls, mirroring the phenotype observed in cell culture (Fig. 4E). AICAR accumulation can lead to AMP-activated protein kinase (AMPK) activation and resulting cell cycle arrest [45]. While purine intermediates including AICAR were strongly elevated, IMP and nucleotides were not consistently depleted in tumors derived from ΔMTR knockouts (Fig. 4E, Extended Data Fig. 4A), suggesting that tumors in vivo can maintain nucleotide homeostasis despite impaired de novo synthesis, perhaps through import of circulating nucleosides or decreased nucleotide consumption for RNA and DNA synthesis. Overall, these results demonstrate that MTR is required for folate species and nucleotide homeostasis under physiological folate conditions.
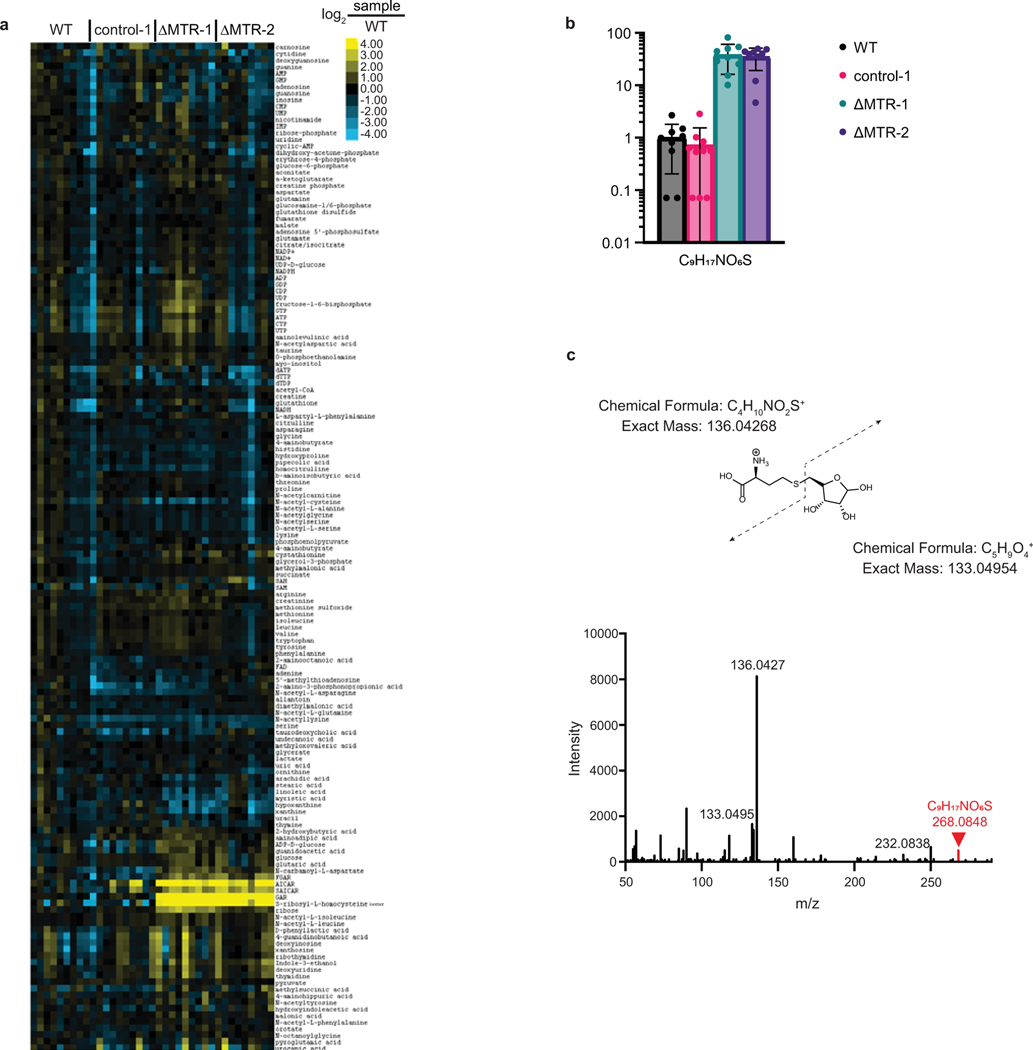
Untargeted metabolomic analysis also identified a consistently elevated (10- to 100-fold) unannotated metabolite (m/z of 267.0702, negative ion mode) in MTR knockout subcutaneous xenografts (Extended Data Fig. 4A–B). This peak’s exact mass and MS/MS spectra suggest that it is an isomer of the bacterial signaling metabolite S-ribosylhomocysteine (C9H17NO6S, Extended Data Fig. 4C). Thus, selectively in vivo, lack of methionine synthase activity, while not depleting methionine concentrations, leads to buildup of an S-ribosylhomocysteine isomer in tumors, suggesting yet-to-be-discovered enzyme(s) linking methionine cycle and nucleotide metabolites.
Discussion
Folate metabolism has long been recognized as a central player in supporting proliferation in tissues and tumors. However, given its complexity, many questions remain pertaining to folate species homeostasis and how dysregulation of folate levels perturbs other areas of cellular metabolism. In this work, we show that despite its name, methionine synthase (MTR) is a minor source of tissue and tumor methionine, but instead is acutely critical for maintaining folate pools when cells are exposed to physiological folate conditions. Furthermore, we find that MTR loss significantly impairs thymidine and purine biosynthesis, effects not rescued with 1C unit supplementation. The importance of MTR in supporting tumor growth is also supported by an accompanying paper by Sullivan et al., where A549 with MTR knockout was unable to form tumors in mice with humanized daily folate consumption [46].
The essentiality of MTR depends on the available extracellular folate species. In cell culture, the importance of MTR is lost when culturing cells using a standard (i.e. folic acid) media preparation. This reflects culture medium typically containing markedly supraphysiological levels of free folate, and lacking physiological levels of 5-methyl-THF. In mouse xenografts, dependence on MTR is augmented by reducing mouse serum folate levels to those in human. Murine “low-folate” diet, which more closely mimics human physiology, selectively ablated MTR knockout tumor growth without impairing that of the wild type. Altogether, these highlight the importance of carrying out folate-related experiments in physiologically accurate context, as has been previously recognized in antifolate drug development efforts [43]. For experiments in vitro, while there has been recent progress towards more physiological culture medium formulations [47, 48], these continue to employ vitamin levels, including folates, selected for convenience rather than physiological relevance. Future physiological medium should switch to 5-methyl-THF as the primary folate source, and more generally push towards physiological vitamin abundances.
The importance of MTR for growth using 5-methyl-THF as the folate source makes sense based on the pathway architecture. As expected for folate perturbations, intermediates accumulated in both the thymidine and purine pathways. But the tie between the observed changes in folate pools and nucleotide alterations was not obvious. Specifically, purine metabolism was dramatically perturbed by MTR loss, even though its direct folate substrate, 10-formyl-THF, was only modestly affected. Conversely, 5,10-methylene-THF was the most depleted folate species but thymidine levels were maintained. Why the stronger effects on purine metabolism? One possibility is that the structural similarity of different folate cofactors leads to enzyme inhibition by elevated species acting as competitive active site inhibitors. For example, elevated DHF arising from methotrexate treatment is known to inhibit multiple enzymes in folate and purine metabolism [49–51]. Similarly, 5-formyl-THF, and to a lesser extent 5-methyl-THF, have been shown to bind SHMT [52]. In the context of MTR loss, the most likely disrupter of purine synthesis is elevation of 5-methyl-THF. This possibility is supported by the observation that media supplemented with 5-methyl-THF exacerbates purine depletion (Fig. 4B, Extended Data Fig. 3B). Interestingly, recent studies in MTR-null fibroblasts demonstrated that buildup of 5-methyl-THF leads to impaired nuclear synthesis of thymidylate, but not purines [34]. However, these cells were cultured in media containing 5-formyl-THF, rather than 5-methyl-THF, bypassing the additional stress imposed by 5-methyl-THF media exposure. Though it remains unproven which enzymatic steps of purine synthesis are impacted by 5-methyl-THF, the excessive buildup of purine intermediates upstream of AICAR (Fig. 4A,C,E) suggests inhibition of AICAR transformylase, the enzyme catalyzing the conversion of AICAR to IMP.
Is there a therapeutic role for MTR inhibitors in the treatment of cancer? Given the substantial success of antifolate treatments in combatting malignancy and MTR’s role in maintaining tumor folate pools, targeting MTR, either with a single agent or in combination with other antifolates, merits investigation [53]. This is particularly true given the complete absence of MTR knockout xenograft growth in conditions mimicking human serum folate abundance.
Antifolate resistance presents a significant challenge for achieving remission [54, 55], and MTR inhibitors could potentially help in this regard by providing an orthogonal approach to depleting tumor folates and/or by promoting toxic 5-methyl-THF accumulation within tumor cells resistant to other antifolate agents. Accordingly, we expect that lower cellular folates arising from MTR inhibition in vivo are likely to sensitize tumors to antifolates. Side effects could potentially be mitigated by rescue therapy, perhaps with leucovorin, as is common for other anti-folates. To our knowledge, no clinically approved antifolate therapies are known to directly inhibit MTR, though some low potency, folate-like small molecule inhibitors have been reported [56–58]. We look forward to the advancement of such compounds into in vivo tools so that the therapeutic potential of MTR inhibition can be more rigorously evaluated.
Methods
Cell lines, culture conditions and reagents
HCT116 (CCL-247), 293T (CRL-3216), and HepG2 (HB-8065) were obtained from ATCC (Manassas, VA). 8988T cells (ACC 162) were obtained from DSMZ (Braunschweig, Germany). Cell lines were used as received. Unless otherwise stated, cells were maintained in DMEM (CellGro 10–017-CV, Corning) supplemented with 10% fetal bovine serum (FBS, F2442, Sigma-Aldrich). Cell lines were cultured in a 37 ⁰C incubator with 5% CO2 and routinely tested negative for mycoplasma contamination by PCR. Rowell Park Memorial Institute medium (RPMI, Gibco 11875093; Thermo Fisher) supplemented with 20% FBS (v/v) was used for single-cell plating MTR knockout cells. Folic acid (BP25195) and leucovorin (18–603-788) were obtained from Fisher Scientific; 5-methyl-THF was obtained from Sigma Aldrich (M0132). Isotopic tracers [U-13C]methionine (CLM-893-H), [U-13C]serine (CLM-1574-H), and [3-13C]serine (CLM-1572) were purchased from Cambridge Isotope Laboratories, while [13C5,15N]betaine (792322) was purchased from Sigma Aldrich. All tracers were stored in stocks of 10 mg/mL at −20 ⁰C. All primers were synthesized by IDT (Coralville, IA). Primers used for sequencing (FWD/REV): MTR-1 = (CCCCAAAGGACACAAGGCTAA/ ACTTGCATTTTCTCCCACCAC); MTR-2 = (AGAAAGCACAGCAGTCCTGAA/ TGGATGTGCCAGCTAGTCAG). Anti-MTR (25896–1-AP) and anti-COXIV (11242–1-AP) were obtained from Proteintech. Anti-GFP (2956T), anti-FLAG (2368S), and HRP-linked Anti-rabbit IgG (7074) were obtained from Cell Signaling. All other standard laboratory chemicals used to make media were obtained from Sigma-Aldrich.
Custom media preparation
For isotope tracing, metabolomics, and growth experiments, cells were cultured in DMEM formulated in house from their chemical components. Media were supplemented with 10% (v/v) dialyzed FBS (dFBS, F0392; Sigma-Aldrich). Standard DMEM contains folic acid (4 mg/L). For media composed of alternative folate species (5-methyl-THF, leucovorin, or both), total folate species concentration was 4 mg/L unless otherwise indicated. In addition, homocysteine (7.4 μM) and vitamin B12 (1 mg/L) were added to all custom media. For tracer media, the amino acid found in standard DMEM was replaced with an equal concentration of tracer amino acid. For formate rescue experiments, sodium formate (141–53-7, Sigma-Aldrich) was added to a final concentration of 1 mM. Final media pH was adjusted to ~7.4 prior to sterile filtration using a Stericup Sterile Vacuum filter (SCGPU02RE, MilliporeSigma).
SHMT inhibitor cell growth inhibition assays
HCT116 cells were cultured in DMEM with folic acid or 5-methyl-THF and supplemented with 10% dialyzed fetal bovine serum. For each cell line tested, cells were seeded in 96-well opaque-walled clear-bottomed plates at 3,000 cells per well in 100 μL growth media and incubated overnight. The following day, SHIN2 was dosed in triplicate as a three-fold serial dilution from 10 μM to 5 nM with DMSO concentration below 0.1%. Cells were incubated with the compound for 3 days. Growth was assayed before SHIN2 addition and at the end of the experiment by quantification of relative cell number using resazurin sodium salt (10 mg/L, 1 h incubation), and read as fluorescence intensity using a Synergy HT plate reader (BioTek).
Metabolite extraction
For extraction of metabolites from cultured cells, cell lines were plated in 6-well plates in regular DMEM supplemented with 10% FBS or in the described custom media and allowed to attach overnight. For isotope tracing and metabolomics experiments involving altered folate composition, cells were first cultured transiently in folate-free media (3 days for physiological folate condition, 1 day otherwise) to deplete their endogenous folate pool. Then the cells were cultured in the experimental condition (relevant custom medium) for 24 h, with cells reaching a final confluency of about 80%. For isotope tracing, medium was then replaced by tracer-containing medium followed by 4 h incubation. Then, the medium was rapidly removed by aspiration, cells were washed three times by 2 mL of ice-cold PBS, and metabolism was immediately quenched by 800 μL of ice-cold extraction buffer (40:40:20 acetonitrile:methanol:water, with 0.5% formic acid). After 1 min of incubation on ice, the extract was neutralized by the addition of NH4HCO3 (15% w/v in H2O, 8.8% of the volume of the extraction buffer). Samples were incubated at −20 °C for ~30 min, then scraped, transferred to Eppendorf tubes, and centrifuged (15 min, 21,000×g, 4 °C). The resulting supernatant was frozen on dry ice and kept at −80 °C until LC-MS analysis.
For extraction of metabolites from tissues, ~10 mg of snap-frozen tissue was stored at −80 °C, disrupted using a Cryomill and lysed in 1 mL of ice-cold 40:40:20 acetonitrile:methanol:water with 0.5% formic acid followed by neutralization with 8.8 % w/v ammonium bicarbonate. Solids were precipitated and supernatants were either utilized directly for LC-MS analysis (by HILIC LC-MS) or were dried and resuspended in water (for reverse phase LC-MS).
Folate species extraction
Folate species were isolated using a previously established protocol [59]. Briefly, cells in culture were seeded at 5×105 cells/well in triplicate in 6-well plates maintained for at least 2 doublings in their designated custom folate media. After aspirating media and washing three times with 2mL of ice-cold PBS, metabolism was quenched with ice-cold extraction buffer (50:50 H2O:MeOH, 25 mM sodium ascorbate, 25 mM NH4OAc, pH 7). For xenografts, tumors were weighed and 50 mg of tissue was disrupted via cryomilling. The tissue was then extracted in 1 mL ice-cold buffer (50:50 H2O:MeOH, 25 mM sodium ascorbate, 25 mM NH4OAc, pH 7). For serum, 15 μL (mouse) or 50 μL (human, H4522 Sigma) of serum was diluted in 400 μL of extraction buffer (20:80 H2O:MeOH, 25 mM sodium ascorbate, 25 mM NH4OAc, pH 7). Solids were precipitated by centrifuging at 21,000×g at 4 °C for 10 min. The resulting supernatant was dried under N2 flow and resuspended in 450 μL of resuspension buffer (50 mM K2HPO4, 30 mM ascorbic acid and 0.5% 2-mercaptoethanol, pH 7). 25 μL of rat serum (R9759, Sigma-Aldrich) that was pre-treated by activated charcoal (10% w/v for 2 h at 4 °C, and centrifuged at 21,000×g at 4°C for 10 min to collect supernatant) was added and samples were incubated at 37 °C for 2 hours to cleave polyglutamate tails on folate species. Sample pH was then adjusted to 4 with 3.5 μL of formic acid prior to loading on Bond Elut-PH SPE columns (Agilent, 14102005), washing with aqueous buffer (30 mM ascorbic acid, 25 mM NH4OAc, pH 4.0), and eluting with 50:50 H2O:MeOH (0.5% 2-mercaptoethanol, 25 mM NH4OAc, pH 7). Subsequent eluate was dried down under N2 flow, resuspended in HPLC-grade water, and centrifuged (21,000×g, 10 min, 4 ⁰C) to remove insolubles. The supernatant was then analyzed immediately or stored at −80°C until LC-MS analysis.
Generation of MTR knockout cell lines.
Generation of batch MTR knockout cells in each cell line was achieved using the lentiviral CRISPR–Cas9 vector system LentiCRISPR v2 (Addgene #52961). Briefly, RNA sequences targeting exon 15 and exon 28 of human MTR were designed using the crispr.mit.edu design tool. The guide RNA or scrambled control sequences (Supplementary Table 1) were subcloned into the lentiCRISPR v2 using the BsmBI restriction endonuclease (NEB R0580S). Virus was produced through PEI (MilliporeSigma, 408727) transfection of vectors and lentiviral packaging plasmids psPax2 and VSVG in 293T cells. Medium containing lentivirus was collected after two days and filtered through a PES filter (0.22 μm, MilliporeSigma). Cells were transfected with virus containing scrambled control or targeting MTR and Polybrene (8 μg/mL, Invitrogen). Cells were split after 48 h into standard RPMI-media (10% FBS) containing puromycin (2 μg/mL) and cultured for 3 days. The resulting batch knockout cells were suspended in RPMI-media (20% FBS) and plated at a dilution of ~1 cell per well in 96-well plates. After 10 days, single colonies were identified and passaged. MTR knockout of clonally isolated knockout cells was confirmed by Sanger sequencing and by western blotting.
Generation of MTR re-expression cell lines
An MTR cDNA clone with a C-terminal FLAG TAG in pcDNA3.1 was obtained from Genscript (Clone ID: OHu24754D, Accession No.: NM_000254.2). The CRISPR target sites were modified to achieve resistance using alternate codons (exchange wild type CTTTGTGCATCGACTCCTCCAATT with CCTTGTGTATTGATTCTTCAAACT and wild type CCACGTTTATTGGGACCCAGGTCT with CTACATTCATCGGGACACAAGTTT) using standard PCR based methods (NEB Phusion High-Fidelity Master Mix - E2621L) making use of the unique 5’ XbaI and the 3’ ApaI site for Gibson Assembly (NEBuilder® HiFi DNA Assembly Master Mix - E2621L). Due to the toxicity of the MTR plasmid in standard NEBstable and similar strains, the cloning and plasmid amplification were performed using EPI400 (CopyCutter, Lucigen - C400CH10). As a control eGFP with a FLAG TAG was also cloned into pcDNA3.1.
The two parental cell lines (HCT116 ΔMTR-1 and ΔMTR-2) were plated in 6 well plates with a density of about 8×105. Next day they were transfected using “X-tremeGENE HP DNA Transfection Reagent” (Sigma-Aldrich 6366244001) and 2 μg plasmid followed by 2-week selection with G418 (500 μg/mL). Expression of the proteins was confirmed by Western blot using anti-FLAG and anti-MTR antibodies. HCT116 ΔMTR-1 + eGFP and HCT116 ΔMTR-1 + MTR were used for experiments unless otherwise indicated.
Western blotting
5 ×106 cells were washed with 1 mL of PBS, and centrifuged (1,000×g, 3 min, room temperature) to collect the pellet. Cell pellet was lysed by 100 μL of RIPA lysis buffer (Millipore Sigma 20–188) with protease inhibitor (Roche 11697498001). Protein concentration was determined by BCA assay kit (Thermo Scientific 23225). Samples were mixed with one-third volume of Laemmli sample buffer (Bio-Rad 161–0747) which was supplemented with 10 vol% 2-mercaptoethanol. Such samples were directly loaded without boiling onto 8–16% precast polyacrylamide gels (Bio-Rad 4568104) with 20 ng of protein per lane. After electrophoresis, proteins were transferred onto 0.2 μm nitrocellulose membranes (Bio-Rad 1620112). The antibodies anti-MTR (Proteintech 25896–1-AP), anti-COXIV (Proteintech 11242–1-AP), anti-GFP (Cell Signaling 2956T), and anti-FLAG (Cell Signaling 2368S) were diluted 1:1000 in TBS-0.1% Tween 20, 5% non-fat milk. Membranes were incubated with diluted primary antibodies at 4 ⁰C overnight, washed five times with 10 mL of TBS-0.1% Tween 20, and incubated with HRP-linked anti-rabbit IgG (Cell Signaling 7074, 1:1000 dilution) at room temperature for 1 h. Blots were imaged by HRP reaction with ChemiDoc MP Imaging System (Bio-Rad) using Image Lab 6.0.1.
Cell growth
Media containing different folate species were formulated in house from their chemical components. Clonally isolated MTR knockout cells and their respective controls in HCT116, 8988T, and HepG2 backgrounds, as well as batch eGFP/MTR expressing cells in HCT116 ΔMTR background were first cultured in folate-free media to deplete intracellular folates (6 days for MTR-expressing and 4 days for MTR-knockout). Then they were plated (1 μL of cells per well) in 6-well plates in the indicated conditions. When approaching confluency (3–5 days depending on cell line), cells were passaged and re-plated at the same dilution (1 μL of cells per well). Growth was monitored at each point by packed cell volume (PCV). If total well PCV reached 0.5 μL or lower, the measurements were terminated for that condition.
Mouse handling
Animal studies were approved and conducted under the supervision of the Princeton University Institutional Animal Care and Use Committee (Protocol# 2032). 8-week-old C57BL/6 male mice, and 7-week-old male and female CD-1 nude mice were purchased from Charles River Laboratories. Mice were housed in a normal 12-h light (7 am to 7 pm)/12-h dark cycle room with temperature 20–26 ⁰C and humidity 40–60%. Food (PicoLab Rodent Diet 20 unless otherwise indicated) and water was provided ad libitum. Mice were allowed to adapt for at least five days before any experimental manipulation. For tumor-bearing mice, the maximum allowed tumor volume is 2000 mm3, which was not exceeded in experiments. Mice were euthanized when tumors became necrotic, or when they displayed signs of distress.
Mouse infusion studies
PDAC tumor tissues derived from LSL-KrasG12D/+; LSL-Trp53R172H/+; and Pdx-1-Cre (KPC) mice were used to generate a syngeneic model. Briefly, tumor tissues were minced, mixed with Matrigel (Corning 354234 in 1:1 ratio (v/v)) and injected (200 μL) into the subcutaneous left flank of male C57BL/6 mice. Tumors were allowed to grow until palpable, at which point jugular vein catheter placement surgery was performed. Mice were single-housed and allowed to recover for at least 5 days following surgery. On day of infusion, food was removed ~6 h prior to start of infusion. Isotope tracers in saline ([U-13C]methionine, 40 mM; [U-13C]serine, 50 mM) were infused via the catheter at the rate of 0.1 μL/g/min. After 2.5 h, serum was collected by tail snipping. Mice were euthanized via cervical dislocation and tissues (including tumor) were isolated immediately and snap-frozen in liquid nitrogen using a Wollenberger clamp to preserve metabolite levels. For betaine infusions [13C5,15N]betaine (3 mM, infusion rate 0.1 μL/g/min), pre-catheterized, non-tumor bearing male C57BL/6 mice from Charles River were utilized and infused for 4 h. All tissues were snap-frozen using a Wollenberger clamp and stored at −80 ⁰C prior to analysis.
Tumor xenograft growth with standard chow
HCT-116 wild-type, control-1, ΔMTR-1/ 2 cells were suspended in 1:1 PBS:Matrigel matrix (Corning 354234) at 1×107 cells/mL. Female CD-1 nude mice aged between 8 and 10 weeks were bilaterally injected in the rear flanks with 100 μL of cell suspension per flank. Wild-type and ΔMTR-2 were injected in two flanks of the same mouse, and so were control-1 and ΔMTR-1. After tumors became palpable, tumors were monitored twice per week using caliper measurements [volume=0.5×(length×width2)]. At the end of the study, mice were fasted for 8 h, and euthanized by cervical dislocation. Tumors were isolated, weighed and snap-frozen in liquid nitrogen using a Wollenberger clamp. Tumor samples were stored at −80 ⁰C prior to analysis.
Tumor xenograft growth with dietary folate manipulation
Male CD-1 nude mice aged between 8 and 10 weeks were fed on either control diet with 4 mg/kg folic acid (TD.160607, Envigo) or diet without added folic acid (TD.130431, Envigo) starting at least 5 days before tumor implantation. For rescue with leucovorin, mice were also provided water medicated with leucovorin calcium (100 mg/L, PHR1541, Sigma-Aldrich), with medicated water replenished twice a week. HCT-116 wild-type, control-1, ΔMTR-2, ΔMTR-1+eGFP, and ΔMTR-1+MTR cells were suspended in 1:1 PBS:Matrigel matrix (Corning 354234) at 1×107 cells/mL.100 μL of cell suspension were injected pair-wise in the two flanks of mice, with the following pairs: control-1 and ΔMTR-1+eGFP in leucovorin-supplemented mice, control-1 and ΔMTR-2 in mice on control diet and low folate diet, control-1 and ΔMTR-1+MTR in mice on low folate diet, ΔMTR-1+eGFP and ΔMTR-1+MTR in mice on low folate diet. After tumors became palpable, tumors were monitored twice per week using caliper measurements [volume=0.5×(length×width2)]. At the end of the study, mice were fasted for 8 h, and blood was collected by tail snipping directly into a collection tube without anticoagulant (MVCB-S-300, SAI Infusion Technologies). Blood was kept on ice for 1 h and centrifuged (10 min, 21,000×g, 4 °C) to collect serum. Serum was stored at −80 ⁰C until metabolite extraction.
LC-MS metabolite measurement
Metabolites except for folate species were analyzed by a quadrupole-orbitrap mass spectrometer (Q Exactive Plus, Thermo Fisher Scientific, Waltham, MA) coupled to hydrophilic interaction chromatography (HILIC), or a stand-alone orbitrap (Thermo-Fisher Exactive) coupled to reversed-phase ion-pairing chromatography. Folate species were analyzed by Q Exactive Plus coupled to reverse-phase chromatography.
HILIC separation was on a an Xbridge BEH Amide column (150 mm×2 mm, 2.5 μm particle size; Waters, Milford, MA) at 25 °C. Solvent A was 20 mM ammonium acetate and 20 mM ammonium hydroxide, in 95% H2O 5% acetonitrile, pH 9.4. Solvent B was acetonitrile. The gradient was 0 min, 90% B; 2 min, 90% B; 3 min, 75%; 7 min, 75% B; 8 min, 70%, 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 14 min, 25% B; 16 min, 0% B, 21 min, 0% B; 21 min, 90% B; 25 min, 90% B. The flow rate was 150 μL/min. MS full scans were in the range of 75–1000 m/z with a resolution of 140,000 (at m/z 200) and AGC was target 1×106 [60]. Betaine and SAM were measured in the positive ion mode, and other metabolites were measured in the negative ion mode.
Reversed-phase ion-pairing chromatography was on a Synergy Hydro-RP column (100 mm×2 mm, 2.5 μm particle size, Phenomenex, Torrance, CA) at room temperature. Solvent A was 10 mM tributylamine and 15 mM acetic acid in 97% H2O 3% methanol, pH 5. Solvent B was methanol. The gradient was 0 min, 0% B; 2.5 min, 0% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 15.5 min, 95% B; 18.5 min, 95% B; 19 min, 0% B; 25 min, 0% B. The flow rate was 200 μL/min. MS full scans were in the negative ion mode with a resolution 100,000 at 1 Hz and a scan range of 0–5 min, m/z 85–800; 5–6.7 min, m/z 100–800; 6.7–9 min, m/z 85–800; 9–16 min, m/z 110–1000; 16–24 min, m/z 220–1000. AGC target was 3×106 [61].
Folates were separated on a Poroshell 120 Bonus-RP column (150 mm×2.1 mm, 2.7 μm particle size, Agilent, Santa Clara, CA). Solvent A was 10 mM ammonium acetate in 99.9% H2O 0.1% acetic acid, pH 4. Solvent B was acetonitrile. The gradient was 4 min, 80% B; 10 min, 2% B; 6 min, 30% B; 11 min, 100% B; 15 min, 100% B; 16 min, 2% B; 20 min, 2% B, with a flow rate of 200 μL/min. For tumor and cell samples, MS scans were in negative ion mode with m/z range 350–1000, resolution of 35,000 and AGC target 1×106. For serum samples, MS scans were in positive ion mode with m/z range 430–480, resolution of 35,000 and AGC target 5×106.
MS/MS analysis of SRH isomer was performed on Q Exactive Plus at m/z 268.0848 in positive ion mode with MS2 resolution of 17,500 (at m/z 200), at the retention time of 10.6 min (HILIC method). MS2 spectra were collected using a high-energy collision disassociation (HCD) energy of 20 eV; other MS parameters include AGC target = 1×106, maximum IT = 500 ms and isolation window = 1.5 m/z.
Cell culture metabolite abundances were normalized by packed cell volume. Tissues metabolite abundances were normalized by mass. Isotopic labeling of metabolites arising from 13C-labeled nutrients were corrected for natural abundance using “Accucor” package in R [62]. Isotopic impurity was set to 0.1% based on manufacturer specifications for the labeled tracers. All LC-MS data was analyzed using the ElMaven software (v 0.2.4, Elucidata), with compounds identified based on exact mass and retention time matched to commercial standards. For metabolomics analysis, metabolites data were normalized to control condition and clustered using Cluster 3.0 software. Heatmaps were plotted using Java Treeview (1.1.6r4).
Statistics
Samples sizes are defined in each figure legend. Unless otherwise indicated, sample size (n) refers to independent biological replicates, whose results are presented as mean ± SD or mean ± SEM, as noted in figure legends. Statistical significance was calculated using two-sided Student’s t-test when comparing two groups, and one-way ANOVA followed by Dunnett’s post hoc analysis when comparing three or more groups. P < 0.05 was considered statistically significant. Student’s t-test was performed by Microsoft Excel 2107, and ANOVA was performed by GraphPad Prism 7.03.
Data availability
Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author upon request.
Extended Data
Extended Data Fig. 1. MTR is a minor source of methionine in vitro and in vivo.
(A) Schematic of methionine labeling from [U-13C]methionine. Red circles indicate 13C atoms. MTR = methionine synthase. (B) Schematic of methionine labeling from [U-13C] or [3–13C]serine. Blue circles indicate 13C atoms. MTHFR = methylenetetrahydrofolate reductase, SHMT = serine hydroxymethyltransferase. (C) Methionine labeling in cell lines after culturing for 4 h in media containing [U-13C]serine (for 293T) or [3–13C]serine (for HCT116 and HepG2) (mean ± SD, n = 2). (D) Methionine M+1 fraction from 4 h [3–13C]serine tracing in HCT116 cultured in media containing indicated methionine and folate concentrations (mean ± SD, n = 3 for each condition). Labeling of (E) serine and (F) methionine in serum, PDAC tumors, and normal tissues of male C57BL/6 mice after [U-13C]serine infusion for 2.5 h. (mean ± SD, n = 3 mice; two technical replicates were included for each tumor). (G) Schematic of methionine labeling from [13C5,15N]betaine. Orange circles indicate 13C atoms, green circles indicate 15N atoms. BHMT = betaine-homocysteine S-methyltransferase, DMG = dimethylglycine.
Extended Data Fig. 2. MTR is important for cell growth in physiological folates.
(A) Expression of MTR in the HCT116, 8988T, and HepG2 cell lines as reported in the Cancer Cell Line Encyclopedia[63]. (B) Cell growth curves in the media containing indicated folate sources (mean ± SD, n = 2). (C) Cell growth curves in media containing indicated folate and methionine concentrations (mean ± SD, n = 3). (D) Individual tumor volumes for HCT116 xenografts in female CD-1 nude mice (n = 10 mice). (E) Terminal tumor mass of HCT116 xenografts in female CD-1 nude mice (mean ± SEM, n = 10 mice). P values were determined by a two-sided paired Student’s t-test comparing ΔMTR-1 to wild-type, and ΔMTR-2 to CRISPR control (control-1). (F) Growth of subcutaneous HCT116 xenografts in male CD-1 nude mice on a standard folate (4ppm) or low folate diet (mean ± SEM, n = 10 mice). (G) Western blot analysis of MTR and eGFP in HCT116 wild-type (WT), CRISPR control-1 or ΔMTR-1 which was also engineered to express a vector containing either eGFP or MTR cDNA. Loading control (COXIV) was analyzed on a separate gel from parallel experiments. Results are representative of 2 independent biological replicates with similar results.
Extended Data Fig. 3. Loss of MTR disrupts nucleotide synthesis.
(A) Water-soluble metabolite levels from HCT116 control and MTR knockout cells cultured in indicated media conditions. Each box reflects one independent biological measurement, normalized to the average of control cells cultured in folic acid. (B) Relative nucleotide mono- and diphosphate abundances in HCT116 control and MTR knockout cells in indicated media. Intensities are normalized to the average of control-1 cells in folic acid (mean ± SD, n = 3). (C) Relative thymidylate species abundances in HCT116 control and knockout cells in indicated media. Intensities are normalized to the average of control-1 cells in folic acid (mean ± SD, n = 3). TIC = total ion count. (D) Cell growth dose response curves for HCT116 WT and MTR knockout cells treated with SHMT1/2 inhibitor SHIN2 under different folate conditions (mean ± SD, n = 5). For (B) and (C), P values were determined by a one-way ANOVA comparing control to MTR knockout in the same medium followed by Dunnett’s post hoc analysis.
Extended Data Fig. 4. Metabolomics of MTR knockout tumors.
(A) Water-soluble metabolites levels from individual HCT116 control and MTR knockout subcutaneous tumors (normalized to wild-type tumor average). (B) Relative abundance of an S-ribosylhomocysteine isomer in HCT116 control and MTR knockout subcutaneous tumors (mean ± SD, n = 10 tumors for control-1, and n = 9 tumors for each other group). (C) MS/MS spectrum of m/z 268.0848 peak in positive-ion mode. Fragmentation pattern suggests an S-ribosylhomocysteine (SRH) isomer. WT = wild-type.
Supplementary Material
Acknowledgments
We thank Wenyun Lu and other members of the Rabinowitz laboratory for helpful comments and suggestions. LentiCRISPR v2 was a gift from Feng Zhang (Addgene plasmid # 52961). This work was supported by NIH grants 1DP1DK113643 and R01 CA163591 to J.D.R.
Footnotes
Code availability
“Accucor” package for natural isotope correction is publicly available through GitHub (https://github.com/lparsons/accucor).
Competing Interests
J.D.R. is a paid advisor and stockholder in Kadmon Pharmaceuticals, L.E.A.F. Pharmaceuticals, and Rafael Pharmaceuticals; a paid consultant of Pfizer; a founder, director, and stockholder of Farber Partners and Serien Therapeutics. J.D.R. and J.M.G. are inventors of patents in the area of folate metabolism held by Princeton University. Other authors declare no competing interests.
References
- 1.Voet D, Voet JG, and Pratt CW, Fundamentals of biochemistry: life at the molecular level. 2016. [Google Scholar]
- 2.Tibbetts AS and Appling DR, Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr, 2010. 30: p. 57–81. [DOI] [PubMed] [Google Scholar]
- 3.Ducker GS, et al. , Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab, 2016. 23(6): p. 1140–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox JT and Stover PJ, Chapter 1 Folate-Mediated One-Carbon Metabolism, in Vitamins & Hormones. 2008, Academic Press. p. 1–44. [DOI] [PubMed] [Google Scholar]
- 5.Dudman NP, Slowiaczek P, and Tattersall MH, Methotrexate rescue by 5-methyltetrahydrofolate or 5-formyltetrahydrofolate in lymphoblast cell lines. Cancer Res, 1982. 42(2): p. 502–7. [PubMed] [Google Scholar]
- 6.Ducker GS and Rabinowitz JD, One-Carbon Metabolism in Health and Disease. Cell Metab, 2017. 25(1): p. 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labuschagne CF, et al. , Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep, 2014. 7(4): p. 1248–58. [DOI] [PubMed] [Google Scholar]
- 8.Wright AJ, et al. , Differential kinetic behavior and distribution for pteroylglutamic acid and reduced folates: a revised hypothesis of the primary site of PteGlu metabolism in humans. J Nutr, 2005. 135(3): p. 619–23. [DOI] [PubMed] [Google Scholar]
- 9.Patanwala I, et al. , Folic acid handling by the human gut: implications for food fortification and supplementation. Am J Clin Nutr, 2014. 100(2): p. 593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Običan SG, et al. , Folic acid in early pregnancy: a public health success story. The FASEB Journal, 2010. 24(11): p. 4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelstein JD, Methionine metabolism in mammals. J Nutr Biochem, 1990. 1(5): p. 228–37. [DOI] [PubMed] [Google Scholar]
- 12.Rose WC, Johnson JE, and Haines WJ, The amino acid requirements of man. 1. The role of valine and methionine. Journal of Biological Chemistry, 1950. 182: p. 541–556. [Google Scholar]
- 13.Sowers JE, Stockland WL, and Meade RJ, L-Methionine and L-Cystine Requirements of the Growing Rat. Journal of Animal Science, 1972. 35(4): p. 782–788. [DOI] [PubMed] [Google Scholar]
- 14.Lu SC, S-adenosylmethionine. The international journal of biochemistry & cell biology, 2000. 32(4): p. 391–395. [DOI] [PubMed] [Google Scholar]
- 15.Finkelstein JD, Metabolic regulatory properties of S-adenosylmethionine and S-adenosylhomocysteine. Clin Chem Lab Med, 2007. 45(12): p. 1694–9. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Y, et al. , Regulation of folate and methionine metabolism by multisite phosphorylation of human methylenetetrahydrofolate reductase. Scientific Reports, 2019. 9(1): p. 4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker DH and Czarnecki GL, Transmethylation of Homocysteine to Methionine: Efficiency in the Rat and Chick. The Journal of Nutrition, 1985. 115(10): p. 1291–1299. [DOI] [PubMed] [Google Scholar]
- 18.Bennett MA, UTILIZATION OF HOMOCYSTINE FOR GROWTH IN PRESENCE OF VITAMIN B12 AND FOLIC ACID. Journal of Biological Chemistry, 1950. 187(2): p. 751–756. [PubMed] [Google Scholar]
- 19.du Vigneaud V, Ressler C, and Rachele JR, The Biological Synthesis of “Labile Methyl Groups”. Science, 1950. 112(2906): p. 267–271. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee RV and Matthews RG, Cobalamin-dependent methionine synthase. The FASEB Journal, 1990. 4(5): p. 1450–1459. [DOI] [PubMed] [Google Scholar]
- 21.Watkins D. and Rosenblatt DS, Inborn errors of cobalamin absorption and metabolism. American Journal of Medical Genetics Part C: Seminars in Medical Genetics, 2011. 157(1): p. 33–44. [DOI] [PubMed] [Google Scholar]
- 22.Watkins D, et al. , Hyperhomocysteinemia due to methionine synthase deficiency, cblG: structure of the MTR gene, genotype diversity, and recognition of a common mutation, P1173L. American journal of human genetics, 2002. 71(1): p. 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson DA, et al. , Targeted disruption of the methionine synthase gene in mice. Mol Cell Biol, 2001. 21(4): p. 1058–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geiger T, et al. , Initial Quantitative Proteomic Map of 28 Mouse Tissues Using the SILAC Mouse. Molecular & Cellular Proteomics, 2013. 12(6): p. 1709–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlen M, et al. , Proteomics. Tissue-based map of the human proteome. Science, 2015. 347(6220): p. 1260419. [DOI] [PubMed] [Google Scholar]
- 26.Pajares MA and Pérez-Sala D, Betaine homocysteine S-methyltransferase: just a regulator of homocysteine metabolism? Cellular and Molecular Life Sciences CMLS, 2006. 63(23): p. 2792–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, et al. , Expression profiling of homocysteine junction enzymes in the NCI60 panel of human cancer cell lines. Cancer Res, 2005. 65(4): p. 1554–60. [DOI] [PubMed] [Google Scholar]
- 28.Uhlen M, et al. , A pathology atlas of the human cancer transcriptome. Science, 2017. 357(6352). [DOI] [PubMed] [Google Scholar]
- 29.Hoffbrand AV and Waters AH, Observations on the Biochemical Basis of Megaloblastic Anaemia. British Journal of Haematology, 1972. 23(s1): p. 109–118. [DOI] [PubMed] [Google Scholar]
- 30.Shane B. and Stokstad ER, Vitamin B12-folate interrelationships. Annual review of nutrition, 1985. 5(1): p. 115–141. [DOI] [PubMed] [Google Scholar]
- 31.Kondo H, et al. , Nitrous oxide has multiple deleterious effects on cobalamin metabolism and causes decreases in activities of both mammalian cobalamin-dependent enzymes in rats. The Journal of clinical investigation, 1981. 67(5): p. 1270–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horne DW and Briggs WT, Effect of Dietary and Nitrous Oxide-Induced Vitamin B-12 Deficiency on Uptake of 5-Methyltetrahydrofolate by Isolated Rat Hepatocytes. The Journal of Nutrition, 1980. 110(2): p. 223–230. [DOI] [PubMed] [Google Scholar]
- 33.Matthews RG and Drummond JT, Providing one-carbon units for biological methylations: mechanistic studies on serine hydroxymethyltransferase, methylenetetrahydrofolate reductase, and methyltetrahydrofolate-homocysteine methyltransferase. Chemical Reviews, 1990. 90(7): p. 1275–1290. [Google Scholar]
- 34.Palmer AM, et al. , Folate rescues vitamin B12 depletion-induced inhibition of nuclear thymidylate biosynthesis and genome instability. Proceedings of the National Academy of Sciences, 2017. 114(20): p. E4095–E4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herbert V. and Zalusky R, INTERRELATIONS OF VITAMIN B12 AND FOLIC ACID METABOLISM: FOLIC ACID CLEARANCE STUDIES. The Journal of Clinical Investigation, 1962. 41(6): p. 1263–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noronha J. On folic acid, vitamin B12, methionine and formiminoglutamic acid metabolism. in Second European Symposium on Vitamin B12 and Intrinsic Factor, 1962. 1962. [Google Scholar]
- 37.Fazili Z, Pfeiffer CM, and Zhang M, Comparison of Serum Folate Species Analyzed by LC-MS/MS with Total Folate Measured by Microbiologic Assay and Bio-Rad Radioassay. Clinical Chemistry, 2007. 53(4): p. 781–784. [DOI] [PubMed] [Google Scholar]
- 38.Fazili Z, et al. , A high-throughput LC-MS/MS method suitable for population biomonitoring measures five serum folate vitamers and one oxidation product. Analytical and bioanalytical chemistry, 2013. 405(13): p. 4549–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maddocks OD, et al. , Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol Cell, 2016. 61(2): p. 210–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L, et al. , Serine Catabolism Feeds NADH when Respiration Is Impaired. Cell Metab, 2020. 31(4): p. 809–821 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunden SL, et al. , Betaine-homocysteine methyltransferase expression in porcine and human tissues and chromosomal localization of the human gene. Arch Biochem Biophys, 1997. 345(1): p. 171–4. [DOI] [PubMed] [Google Scholar]
- 42.Pellanda H, et al. , A splicing variant leads to complete loss of function of betaine–homocysteine methyltransferase (BHMT) gene in hepatocellular carcinoma. The International Journal of Biochemistry & Cell Biology, 2012. 44(2): p. 385–392. [DOI] [PubMed] [Google Scholar]
- 43.Golani LK, et al. , Tumor Targeting with Novel 6-Substituted Pyrrolo [2,3-d] Pyrimidine Antifolates with Heteroatom Bridge Substitutions via Cellular Uptake by Folate Receptor α and the Proton-Coupled Folate Transporter and Inhibition of de Novo Purine Nucleotide Biosynthesis. Journal of Medicinal Chemistry, 2016. 59(17): p. 7856–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García-Cañaveras JC, et al. , SHMT inhibition is effective and synergizes with methotrexate in T-cell acute lymphoblastic leukemia. Leukemia, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Motoshima H, et al. , AMPK and cell proliferation--AMPK as a therapeutic target for atherosclerosis and cancer. The Journal of physiology, 2006. 574(Pt 1): p. 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan MR, et al. , Methionine synthase is essential for cancer cell proliferation in physiological folate environments. bioRxiv, 2020: p. 2020.06.12.149005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cantor JR, et al. , Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell, 2017. 169(2): p. 258–272.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vande Voorde J, et al. , Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Science Advances, 2019. 5(1): p. eaau7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon YK, et al. , A domino effect in antifolate drug action in Escherichia coli. Nat Chem Biol, 2008. 4(10): p. 602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allegra CJ, et al. , Inhibition of phosphoribosylaminoimidazolecarboxamide transformylase by methotrexate and dihydrofolic acid polyglutamates. Proceedings of the National Academy of Sciences of the United States of America, 1985. 82(15): p. 4881–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allegra CJ, et al. , Evidence for direct inhibition of de novo purine synthesis in human MCF-7 breast cells as a principal mode of metabolic inhibition by methotrexate. J Biol Chem, 1987. 262(28): p. 13520–6. [PubMed] [Google Scholar]
- 52.Stover P. and Schirch V, 5-Formyltetrahydrofolate polyglutamates are slow tight binding inhibitors of serine hydroxymethyltransferase. Journal of Biological Chemistry, 1991. 266(3): p. 1543–1550. [PubMed] [Google Scholar]
- 53.Matthews RG, Drummond JT, and Webb HK, Cobalamin-dependent methionine synthase and serine hydroxymethyltransferase: targets for chemotherapeutic intervention? Advances in enzyme regulation, 1998. 38: p. 377–392. [DOI] [PubMed] [Google Scholar]
- 54.Walling J, From methotrexate to pemetrexed and beyond. A review of the pharmacodynamic and clinical properties of antifolates. Investigational New Drugs, 2006. 24(1): p. 37–77. [DOI] [PubMed] [Google Scholar]
- 55.Gonen N. and Assaraf YG, Antifolates in cancer therapy: Structure, activity and mechanisms of drug resistance. Drug Resistance Updates, 2012. 15(4): p. 183–210. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Z, et al. , Mechanism-based design, synthesis and biological studies of N⁵-substituted tetrahydrofolate analogs as inhibitors of cobalamin-dependent methionine synthase and potential anticancer agents. Eur J Med Chem, 2012. 58: p. 228–36. [DOI] [PubMed] [Google Scholar]
- 57.Tang C, et al. , Two newly synthesized 5-methyltetrahydrofolate-like compounds inhibit methionine synthase activity accompanied by cell cycle arrest in G1/S phase and apoptosis in vitro. Anticancer Drugs, 2008. 19(7): p. 697–704. [DOI] [PubMed] [Google Scholar]
- 58.Banks EC, et al. , Inhibition of cobalamin-dependent methionine synthase by substituted benzo-fused heterocycles. Febs j, 2007. 274(1): p. 287–99. [DOI] [PubMed] [Google Scholar]
- 59.Chen L, et al. , An LC-MS chemical derivatization method for the measurement of five different one-carbon states of cellular tetrahydrofolate. Anal Bioanal Chem, 2017. 409(25): p. 5955–5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L, et al. , Peak Annotation and Verification Engine for Untargeted LC-MS Metabolomics. Anal Chem, 2019. 91(3): p. 1838–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu W, et al. , Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem, 2010. 82(8): p. 3212–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su X, Lu W, and Rabinowitz JD, Metabolite Spectral Accuracy on Orbitraps. Anal Chem, 2017. 89(11): p. 5940–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author upon request.