Dear Editor
The 2019 dengue outbreak in Bangladesh saw a sharp increase in reported fatalities. In connection to this, the present study was conducted to understand the infection status and genomic characteristics of DENV strains that caused the 2019 outbreak. A total of eight DENV-3 genomes were sequenced (referred to as our samples). Following this, their genomes and proteomes were compared with other available DENV3 sequences from Bangladesh (referred to as past isolates), to identify features that could be linked with pathogenic functions and thus explain the increased death toll caused by the strains of the recent outbreak.
Samples were collected from patients with complaints of acute fever (≥5days) with or without retro orbital pain, muscle/joint/bone pain and rash during the period of January to December 2019 at IEDCR. Samples, positive for dengue virus were further analysed for dengue serotyping by multiplex PCR where 50 pmol (each) of DENV1 and DENV3 specific primers, 25 pmol (each) of DENV2 and DENV4 specific primers, and 9 pmol of each probe were combined in 25 μl total reaction volume. Serotype identification was done using NCBI Blast [1]. This study was initiated on May, 2020; we obtained all previously sequenced DENV3 sequences from Bangladesh, available on the NCBI database at the time. Immunoinformatics analysis was carried out using the NetCtl (predicting number of CD8+ T cell binding fragments) [2] and the IEDB Episcore (predicting number of CD4+ T cell binding fragments) [3] tools. The results from NetCTL and Episcore were subjected to Principle Component Analysis.
The 8 samples we sequenced contained a total of nucleotide 9671 variants, across 6786 genomic positions. A fisher test revealed 5 of the variants to be significantly more common to the older isolates. These variants (3506T > C, 3638C > T, 4049A > G,8738T > C, and 9104C > T) did not occur in our samples at all, despite occurring in almost all of the older isolates.
For amino acid substitutions, our samples contained marginally higher numbers of variants compared to the past samples. The mean number of variants per sample was 77, with an interquartile range of 2.5. The past samples contained, on average, 70 variants, with an interquartile range of 5. The standard deviation was 2.88 for our samples, and 5.95 for the older samples. Once again, the difference in the means of the two groups was higher than each of their standard deviations, suggesting that the difference may be significant.
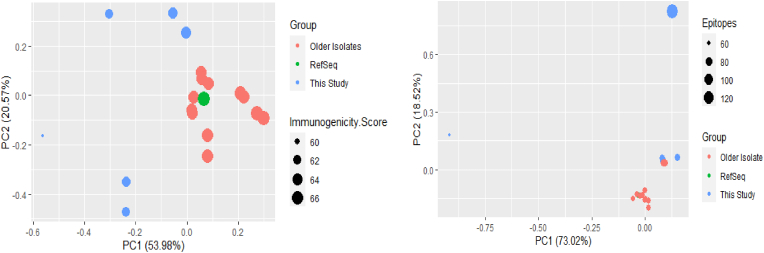
The mean immunogenicity score, calculated by Episcore, for samples from previous years was 65.68, whereas for our samples it was 62.46. The standard deviation within the two groups (our samples and past samples) was 1.52 and 0.67 respectively, meaning the difference in the means (>3) is significant. Our samples displayed lower mean immunogenicity scores compared to past samples, suggesting they have weaker or fewer interactions with CD4+ T cells. A reduction in immune system detection has the potential to lead to more serious symptoms [4], something that would line up with the observed statistics of the 2019 outbreak. Fig. 1 shows the PCA results for the IEDB data.
Fig. 1.
PCA plot of (a) CD4+ T cell binding data, (b) CD8+ T cell binding data.
It can be seen that most of our isolates were clustered together, separate from past isolates. The RefSeq was grouped with the past samples, while two of our samples were scattered quite far from both groups. Nonetheless, the PCA analysis did result in two separate clusters for our samples and the majority of the past isolates. Four of the older isolates however, did group with our samples. Two of these were Bangladeshi travellers were diagnosed with dengue in Guangzou, China, in 2019, and the other two were from the 2017 Bangladesh dengue outbreak.
For the NetCTL analysis (Fig. 1b), our samples did not form any cluster and were scattered across the graph, whereas the past isolates, along with the RefSeq, were placed together in a single cluster. Over 91% of variation was explained by principle components 1 and 2.
The fisher test also revealed two SNPs displayed significantly different occurrence frequencies between the two groups. The fact that these variants occurred in almost all of the past isolates and in none of ours, suggests that these should be investigated for connection to increased pathogenicity as well.
As a result of this study, we have identified CD4+ T cell epitopes, the number of amino acid substitutions, and two specific SNPs that should be the target of more in depth research, with an expanded sample size, as part of efforts to understand the increased fatalities of the 2019 Bangladesh dengue outbreak. as well to better guide efforts to protect against future such outbreaks.
Conflict of interests
None to declare.
Data availability
The sequences are available at GenBank under the following accessions: MW396462, MW396463, MW396464, MW396465, MW396466, MW396467, MW396468 and MW396469.
Acknowledgements
We acknowledge the contribution of Zhen Chen and Wuxiang Guan at Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China for providing sequencing support.
References
- 1.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Pages H., Aboyoun P., Gentlemen R., DebRoy S. Biostrings. 2019 [Google Scholar]
- 3.Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqua M., Alam A.N., Muraduzzaman A.K.M., Tahmina S. NS-1 antigen positive dengue infection and molecular characterization of dengue viruses in a private medical college hospital in Dhaka, Bangladesh. Bangladesh J Med Sci. 2018;17(4):669–673. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences are available at GenBank under the following accessions: MW396462, MW396463, MW396464, MW396465, MW396466, MW396467, MW396468 and MW396469.