Summary
Predicting the pathogenicity of acceptor splice-site variants outside the essential AG is challenging, due to high sequence diversity of the extended splice-site region. Critical analysis of 24,445 intronic extended acceptor splice-site variants reported in ClinVar and the Leiden Open Variation Database (LOVD) demonstrates 41.9% of pathogenic variants create an AG dinucleotide between the predicted branchpoint and acceptor (AG-creating variants in the AG exclusion zone), 28.4% result in loss of a pyrimidine at the −3 position, and 15.1% result in loss of one or more pyrimidines in the polypyrimidine tract. Pathogenicity of AG-creating variants was highly influenced by their position. We define a high-risk zone for pathogenicity: > 6 nucleotides downstream of the predicted branchpoint and >5 nucleotides upstream from the acceptor, where 93.1% of pathogenic AG-creating variants arise and where naturally occurring AG dinucleotides are concordantly depleted (5.8% of natural AGs). SpliceAI effectively predicts pathogenicity of AG-creating variants, achieving 95% sensitivity and 69% specificity. We highlight clinical examples showing contrasting mechanisms for mis-splicing arising from AG variants: (1) cryptic acceptor created; (2) splicing silencer created: an introduced AG silences the acceptor, resulting in exon skipping, intron retention, and/or use of an alternative existing cryptic acceptor; and (3) splicing silencer disrupted: loss of a deep intronic AG activates inclusion of a pseudo-exon. In conclusion, we establish AG-creating variants as a common class of pathogenic extended acceptor variant and outline factors conferring critical risk for mis-splicing for AG-creating variants in the AG exclusion zone, between the branchpoint and acceptor.
Keywords: AG exclusion zone, extended acceptor splice-site variants, pre-mRNA splicing, pseudo-exon, minigene, RNA diagnostics
Introduction
Variants that disrupt the process of precursor mRNA (pre-mRNA) splicing are a common cause of genetic disorders, with 38%–50% of pathogenic variants reported to disrupt splicing in various disease cohorts.1, 2, 3 However, correctly classifying a variant as splice disrupting is challenging, necessitating functional studies to accurately characterize mis-splicing events—often requiring difficult to obtain tissue.4,5 Many in silico tools have been developed to predict if a variant will disrupt normal splicing, although these programs are not always accurate.6 Implementation of massively parallel sequencing (MPS) into diagnostic pipelines has led to an exponential increase in identified variants of uncertain significance (VUS), which are clinically unactionable.7 With RNA diagnostic pipelines now emerging into clinical practice,8 identifying variants likely to disrupt the process of pre-mRNA splicing is of great clinical utility.
During the process of pre-mRNA splicing, non-coding introns are removed and coding exons are ligated together to create a mature mRNA transcript, which acts as a genetic blueprint for protein synthesis. This process is catalyzed by five small nuclear ribonucleoprotein particles (snRNPs—U1, U2, U5, and U4/U6) and numerous non-snRNP proteins, which dynamically assemble on the pre-mRNA to form the spliceosome complex.9,10 Recognition of key conserved sequences in the pre-mRNA by the spliceosome complex is vital for accurate splicing. In the early stages of spliceosome assembly, the U1 snRNP binds to the donor (5ʹ) splice-site primarily by base-pairing, followed by the U2 snRNP, which conversely requires multiple splicing factors to recognize and assemble at the branch-site.11 Auxiliary factors U2AF65 and U2AF35 bind to the polypyrimidine tract (PPT) and acceptor (3ʹ) splice-site, respectively, and form a heterodimer, collectively referred to as U2AF, which interacts with the branch-site recognizing splice factor SF1.12 The U2 snRNP is then able to displace SF1 and bind by base-pairing to the pre-mRNA adjacent to the branch-site.12 Variants that disrupt these key sequences (Figure 1Ai) can, therefore, prevent recognition of the pre-mRNA by these splicing factors and disrupt spliceosome assembly.
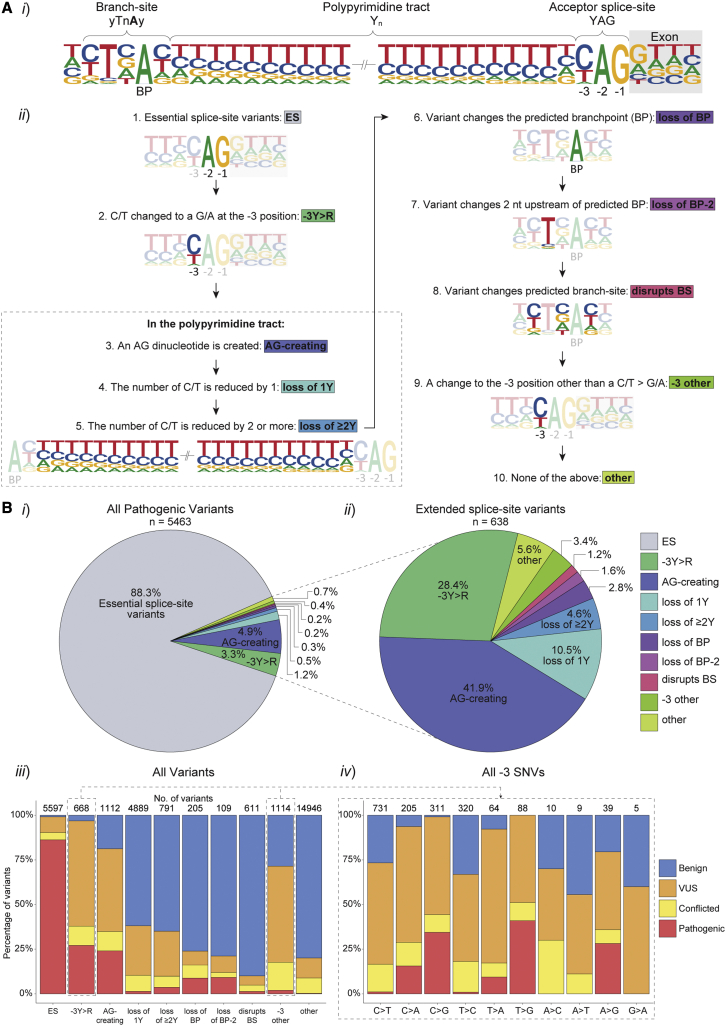
Figure 1.
Intronic acceptor splice-site variants reported in ClinVar and LOVD
(A) Intronic acceptor splice-site variants were assigned to ten variant groups according to the position and consequence of the variant in relation to splicing motifs. (i) Schematic outlining the intronic 3ʹ splicing motifs (acceptor splice-site, PPT and branch-site) in relation to the exon (gray box) in human introns. The comparative heights of the nucleotides (nt) at each position represents the frequency of that nt in 94,874 canonical introns with Branchpointer predicted branchpoints (left) or 185,639 canonical introns (right). (ii) Variants were assigned to one of ten mutually exclusive groups as per the numerical order shown.
(B) Frequencies and clinical classifications of intronic acceptor splice-site variants in the ten different groups. (i) Pie chart of all pathogenic variants depicting the percentage that fall into each group. (ii) Percentage of pathogenic extended splice-site variants in each group (i.e., all pathogenic variants excluding those that disrupt the invariable AG of the acceptor splice-site). (iii) The percentage of 30,042 variants classified as pathogenic (red, n = 5,463), VUS (orange, n = 5,279), benign (blue, n = 16,883), or conflicting classifications (yellow, n = 2,417) in ClinVar and LOVD for each group. (iv) Percentage of clinical classifications for each possible substitution for SNVs at the −3 position of the acceptor splice-site. The number of variants in each group are shown above the bar graphs.
While it is well established that disrupting the invariable AG dinucleotide of the acceptor splice-site interferes with pre-mRNA splicing, the impact of intronic variants in the extended acceptor splice-site region is more challenging to predict, as the PPT and branch-site regions are highly variable.6 There is evidence to suggest that the first AG dinucleotide downstream of the branch-site is selected as the acceptor splice-site for splicing,13,14 with the exception of AG dinucleotides situated close to the branch-site.14,15 This is supported by a natural depletion of AG dinucleotides observed between the acceptor splice-site and branchpoint (BP), termed the AG exclusion zone (AGEZ).16,17 Recently, variants in the AGEZ have been explored in the context of neurofibromatosis type 1 (NF1).18 It was shown that 63% of 91 splice-altering NF1 extended acceptor splice-site variants created an AG in the AGEZ, demonstrating that any variant resulting in a new AG in an AGEZ is highly likely to affect splicing.18
In this study, we further investigate AGEZ variants by analyzing 24,445 extended acceptor splice-site variants reported in ClinVar19 and the Leiden Open Variation Database (LOVD)20 across 2,100 genes associated with clinically relevant monogenic disorders (defined in Dawes et al.21). We describe in detail a clinical example involving a family with a homozygous COL6A2 AG-creating variant and use a minigene construct to model a deep intronic DMD AG-removing variant previously reported to result in pseudo-exon inclusion,22 demonstrating contrasting mechanisms by which variants in the AGEZ may disrupt splicing.
Materials and methods
Extraction of intronic extended acceptor splice-site variants from ClinVar and LOVD
Variants from the databases ClinVar19 and LOVD20 were downloaded (February 2022) and collated into a single dataset. Only SNVs were extracted from LOVD due to historical inconsistencies in the annotation of insertions and deletions (indels), precluding computational processing of large datasets.
Variants were filtered to include only those
-
(1)
In annotated, canonical GRCh37 Ensembl23 transcripts extracted from Ensembl (see web resources) using Bioconductor. Canonical protein-coding transcript annotations were obtained using Ensembl’s Perl API.
-
(2)
In clinically relevant OMIM listed genes with clinically relevant phenotypes (as defined in Dawes et al.21; OMIM gene list downloaded with license from OMIM [see web resources], September 2021).
-
(3)
Located between the annotated acceptor splice-site and predicted branch-site. A recent assessment revealed Branchpointer24 as the best in silico method currently available for predicting BPs25; thus, Branchpointer was selected for use in this study. Branchpointer scores the recommended cutoff of ≥0.5224 was used to predict the likely BPs for each intron in canonical protein-coding transcripts. The closest predicted BP upstream of the acceptor splice-site was selected for each intron. All introns without a predicted BP ≥ 0.52 were excluded from analysis (27.2% of introns). ClinVar and LOVD variants located within/between the branch-site and acceptor splice-site for each remaining intron were extracted for analysis.
-
(4)
Variants were assigned either pathogenic (pathogenic or likely pathogenic), benign (benign or likely benign), VUS, or conflicted or unclassified based on aggregated classifications from all entries in ClinVar and/or LOVD for that variant. Aligning with ClinVar protocols, variants were assigned conflicted if there were at least two opposing classifications (pathogenic versus VUS versus benign). Variants without an American College of Medical Genetics and Association for Molecular Pathology (ACMG/AMP)26 concordant classification (pathogenic, likely pathogenic, VUS, or likely benign or benign) were considered unclassified and removed from analysis.
Additionally, deep intronic variants (defined as > 100 nucleotides (nt) from exon-intron splice junctions27) in canonical Ensembl transcripts of clinically relevant OMIM genes that result in the loss of an AG were curated separately to assess for potential loss of an AG splicing silencer mechanism for pseudo-exon activation. For these variants, Branchpointer was unable to provide a BP prediction and thus branchpoint prediction (BPP)28 was used.
Variants curated via steps 1–4 above were categorized into mutually exclusive groups, according to the position and consequence of the variant in relation to splicing motifs (Figure 1Ai), as per the order outlined in Figure 1Aii. For example, a variant that both created an AG and resulted in the loss of a C/T would be assigned to group 3 (AG-creating) as it precedes group 4 (loss of 1Y). AG-creating SNVs were analyzed separately with their positional context taken into consideration. Each AG-creating SNV was either determined to create an AG (1) closer to the BP, (2) closer to the acceptor splice-site, or (3) directly in the middle of the PPT (Figure 2Ai).
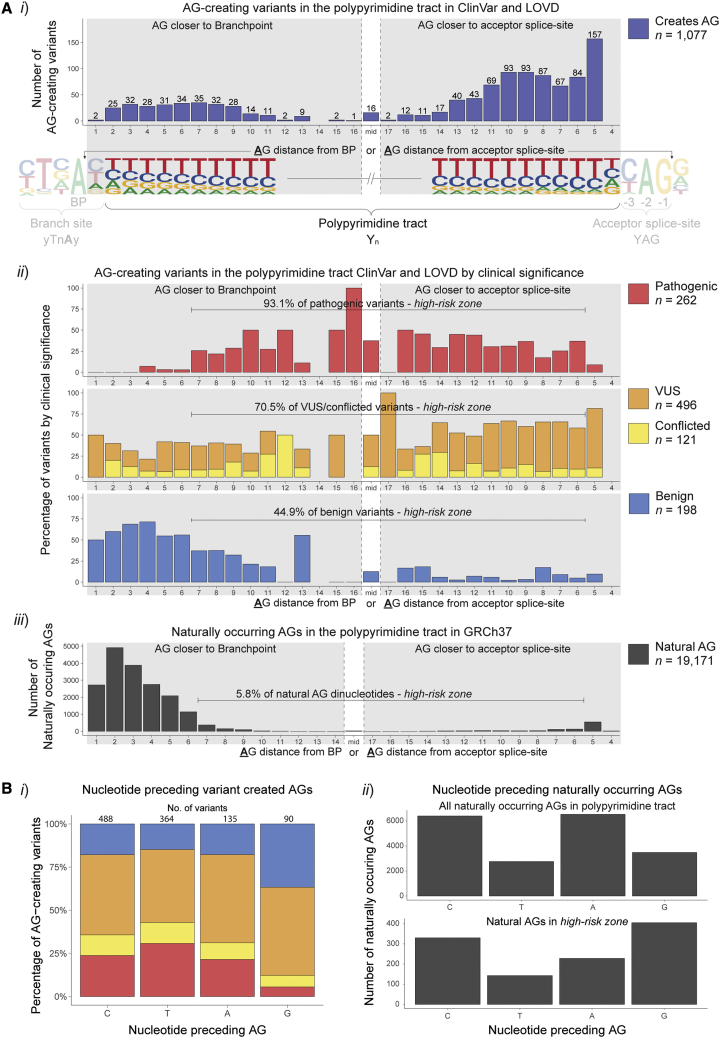
Figure 2.
Variant created and naturally occurring AG dinucleotides in the PPT
(A) Frequency of AG-creating variants in ClinVar and LOVD and naturally occurring AG dinucleotides at each position of the PPT. Numbered positions denote the distance of the A of the AG dinucleotide from either the BP (left) or acceptor splice-site (right), depending on whether the created AG was closer to the BP or acceptor splice-site. AG dinucleotides at equal distance from both the BP and acceptor splice-site were grouped together as mid, due to the varying lengths of PPTs in this dataset. No variants created an AG 4 nt away from the acceptor splice-site due to the rarity of a G at the −3 position of the acceptor motif. (i) The number of all AG-creating variants at each position, with lower numbers in the center due to varying PPT lengths. (ii) The proportion of AG-creating variants with different clinical classifications at each position. (iii) The number of naturally occurring AG dinucleotides in the PPT in the human genome at each position. Only 5.8% of natural AGs fall between >6 nt downstream of the predicted branchpoint or >5 nt upstream from the acceptor splice-site, demonstrating the evolutionary constraint against AGs in this region. Conversely, 93.1% of pathogenic variants fall within this same region, establishing a zone of high-risk for mis-splicing due to AG dinucleotides.
(B) (i) The proportion of clinical classifications for the different nucleotides preceding the variant created AG. (ii) The number of different nucleotides preceding naturally occurring AG dinucleotides.
A recent study revealed SpliceAI29 as the best algorithm for evaluating extended splice-site variants30; thus, SpliceAI was used to assess AG-creating variants in this study. SpliceAI acceptor splice-site scores were obtained for the annotated acceptor splice-site and the relevant dinucleotide containing the AG-creating variant, both without and with the variant change.
Genetic investigations and immunohistochemistry for the COL6A2 family
Ethical approval was obtained from the Human Research Ethics Committees of the Children’s Hospital at Westmead, Australia (10/CHW/45 and 2019/ETH11736), with written, informed consent from all participants. Whole-exome sequencing (WES), whole-genome sequencing (WGS), and muscle RNA sequencing (RNA-seq) was performed and analyzed at the Broad Institute of Harvard and MIT, as previously described.4 Immunohistochemistry (IHC) was performed as previously described.31 Antibodies to the following proteins were used: spectrin (NCL-SPEC1; dilution 1:200; Leica Microsystems) with Alexa Fluor555 conjugated goat anti-mouse secondary antibody (dilution 1:300; Thermo Fisher Scientific); collagen VI (clone 70-XR95, now sold as 70R-CR009X, dilution 1:10,000, Fitzgerald Industries International, MA) with Alexa Fluor555 conjugated goat anti-rabbit secondary antibody (dilution 1:300; Thermo Fisher Scientific) co-stained with perlecan (clone A7L6, MAB1948, dilution 1:40,000; Chemicon International, CA) with Alexa Fluor488 conjugated goat anti-rat secondary antibody (dilution 1:300; Thermo Fisher Scientific). The NM_001849.3(COL6A2):c.2423-22_2423-21insAGCCCGGCCCGGCCC variant was submitted to the Leiden Open Variation Database (individual: 00411326, DB-ID: COL6A2_000503).
Generation of DMDex25-27 minigene constructs
A pCMV6-Entry EGFP-FLAG-DMDex25-27 wild-type construct was created by cloning EGFP-FLAG tagged partial genomic sequences of DMD, encompassing exons 25 to 27 with modified introns 25 and 26, into a pCMV6-Entry expression vector using AsiSI and Mlu1 restriction sites. Subsequently, the wild-type construct was modified by subcloning 466 base pairs (bp) variant sequence gene blocks using NheI and AflII restriction sites. All custom sequences were generated by Integrated DNA Technologies (IDT; Coralville, Iowa, USA) and all constructs were sequence confirmed by Sanger sequencing (Australian Genome Research Facility, Sydney, Australia). Full sequences of wild-type and variant constructs are available in the supplemental materials and methods.
Cell culture and transfection
HEK-293 cells
HEK-293 cells were cultured in Gibco DMEM containing 10% heat-inactivated HyClone fetal bovine serum (FBS; GE Healthcare Life Sciences) and 50 ng/mL Gibco gentamicin. Cells were seeded onto 6-well plates at 30%–40% confluency 16 h prior to transfection with polyethyleneimine (PEI; Polysciences). For each well, 8.31 μL of PEI (1 μg/mL), 200 μL NaCl (0.9%), and 3 μg plasmid DNA were mixed, incubated for 20 min at room temperature, and added to the dish in a dropwise fashion. For each DMDex25-27 minigene plasmid, 4 wells were harvested 48 h after transfection, with 2 wells used each for RNA and protein isolation.
Primary human myoblasts (PHMs)
PHMs from a 37-year-old female control were cultured in Gibco DMEM:Ham’s F-12 supplemented with 10% Gibco AmnioMAX-II, 20% FBS, and 50 ng/mL Gibco gentamicin. Cells were seeded onto 6-well plates coated with 1:50 collagen type 1 (Rat Tail, 3.54 mg/mL, Becton Dickinson) at 50% confluency. When PHMs reached 70%–80% confluency, they were transfected with 2.5 μg plasmid DNA using Lipofectamine 3000 (Invitrogen, Sigma Aldrich), according to the manufacturer’s instructions. Fibroblast cultures were treated with either 100 μg/mL cycloheximide (CHX, 1:300 dilution from 30 mg/mL stock) or 1:300 DMSO for 5 hours prior to RNA extraction.
RNA isolation and RT-PCR
COL6A2 family samples
RNA isolation was performed from 30 × 8 μm thick muscle cryosections (10 mm2 surface area) or from 20 cm2 surface area of fibroblast cultures using Invitrogen TRIzol Reagent according to the product user guide. The RNeasy Mini Kit from QIAGEN was used to clean up the RNA according to the kit protocol. cDNA was synthesized from 1 μg of total RNA with 1:1 oligo-dT and random hexamers using the Invitrogen SuperScript IV First-Strand Synthesis System, according to the manufacturer’s instructions.
DMDex25-27 minigene constructs
Wells were washed 3 times in Dulbecco’s phosphate buffered saline (DPBS) followed by homogenization in RLT plus buffer (QIAGEN) supplemented with 10 μL/mL 2M dithiothreitol (DTT) using a 20-gauge needle and processed using the RNeasy Mini Kit (QIAGEN) by RNA isolation using the RNeasy Plus Kit (QIAGEN), according to manufacturer’s protocol. Purified RNA concentration was measured with a Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized from 1 μg of total RNA using the SuperScript IV First-Strand Synthesis System with oligo (dT) primers (Invitrogen), according to the manufacturer’s instructions.
RT-PCR
PCR was performed on cDNA using Buffer D (Astral Scientific), 1 unit Taq DNA Polymerase (Life Technologies), and 0.3 mM each forward/reverse primer. Primer details for all RT-PCRs are listed in Table S1. Cycling conditions for Taq DNA Polymerase were 95°C for 2 min followed by 25–35 cycles of 95°C for 10 s, 58–64°C for 30 s, and 72°C for 50–120 s (see Table S1). Final extension was done for 8 min at 72°C. The identity of PCR amplicons was confirmed via Sanger sequencing. When multiple bands were detected, individual bands were existed from the gel and purified using the GeneJET Gel Extraction kit (Thermo Scientific) as per manufacturer’s instructions.
Protein isolation and western blot
Cells were washed 3 times in DPBS, scraped off, pelleted by centrifugation, and snap frozen on dry ice. Cell pellets were lysed in 4% SDS, 62.5 mM Tris-HCl pH 6.8, and 1× protease inhibitor cocktai, and sonicated for 10 short bursts, followed by heating to 94°C for 4 min. Protein concentrations were determined via a bicinchoninic acid (BCA) assay (Pierce, Thermo Fisher Scientific), according to the manufacturer’s protocol. Lysates were mixed 3:1 with loading buffer (62.5 mM Tris-HCl pH 6.8, 4% SDS, 0.2 M DTT, 40% glycerol, traces of bromophenol blue, and 1× protease inhibitor cocktail) and heated to 94°C for 1 min; 20 μg protein were electrophoresed on a 10% SDS-polyacrylamide gel (Criterion TGX, Bio-Rad) followed by transfer onto a polyvinylidene difluoride (PVDF) membrane (Merck Millipore Immobilon-P, 0.45 μm) for 1.5 h in Tris-glycine buffer containing 0.075% SDS and 15% methanol. Membranes were blocked for 1 h with 1:1 Intercept Blocking Buffer (LI-COR Biosciences): Tris-buffered saline 0.1% Tween 20 (TBS-T). Primary antibody incubation was performed overnight with mouse anti-FLAG-tag (Clone 12C6c, Developmental Studies Hybridoma Bank). Membranes were washed with TBS-T, blocked for 15 min as above ,and incubated with 1:15,000 IRDye 800CW Donkey anti-Mouse Secondary Antibody (LI-COR Biosciences) for 1 h. Membranes were imaged at 600, 700, and 800 nm for 2 min/channel using the Odyssey Fc Imaging System (LI-COR Biosciences). Western blots were analyzed using Image Studio 5 (LI-COR Biosciences).
Results
Data mining ClinVar and LOVD for intronic acceptor splice-site variants
The 30,042 intronic variants located at the acceptor splice-site, PPT, or branch-site (Figure 1Ai) of annotated Ensembl 23 transcripts were extracted from the variant databases ClinVar and LOVD (see materials and methods). Of these, 18.2% were classified as pathogenic, 56.2% as benign, 17.6% as VUS, and 8.0% as conflicted. Clinical classifications were statistically significantly associated with the assigned variant groups (Pearson’s chi-squared test = 26,785; degrees of freedom [df] = 27; p < 2.2 × 10−16; Figure 1Aii). The majority (88.3%) of reported pathogenic variants impacted the essential AG dinucleotide of the acceptor splice-site and only 0.7% of pathogenic variants did not fall into defined variant groups (other, Figure 1Bi).
Among the 11.7% of pathogenic variants outside the essential AG of the acceptor splice-site (n = 638), 41.9% result in the creation of an AG dinucleotide in the PPT (AG-creating), 28.2% disrupt the pyrimidine at the −3 position of the acceptor splice-site (−3Y > R), and 16.6% result in the loss of one or more pyrimidines (loss of 1Y or loss of ≥2Y, Figure 1Bii). Despite comprising a sizable proportion of pathogenic variants, loss of 1Y or loss of ≥2Y variants were significantly less pathogenic than the AG-creating or −3Y > R variants (Pearson’s chi-squared test = 1,977.5; df = 9; p < 2.2 × 10−16), indicating that the PPT is often tolerant to loss or substitution of a pyrimidine (Figure 1Biii). Comparative analysis of all dinucleotide combinations created by variants revealed increased pathogenicity associated with AG-creation, relative to all other dinucleotide combinations (Pearson’s chi-squared test = 4,630; df = 45; p < 2.2 × 10−16; Figure S1).
VUS and conflicted classifications comprise the highest proportion of classifications for variants at the −3 position (−3Y > R and −3 other), emphasizing increased uncertainty and the necessity for functional testing of pre-mRNA splicing for variants at this position (Figure 1Biii). SNVs that substitute a G nt at the −3 position (i.e., C > G, T > G, and A > G) were more likely to be pathogenic than −3 position variants with other substitutions (odds ratio: 93.7 with 95% CI [47.41, 185.24], Figure 1Biv), consistent with the natural preference of C > T > A > G at the −3 position in the human genome (Figure 1Ai).
Positional risk for pathogenicity for AG-creating variants in the AGEZ
Among 1,077 AG-creating variants within the AGEZ (region between acceptor splice-site and BP), 262 were classified pathogenic, 198 were benign, 496 VUS, and 121 conflicted. Notably, pathogenicity for AG-creating variants was highly dependent upon position of the created AG; 93.1% of pathogenic AG-creating variants fall within >6 nt downstream of the BP and >5 nt upstream of the acceptor splice-site (defined hereafter as the high-risk zone) compared with only 44.9% of benign AG-creating variants (Figure 2Aii). This high-risk zone is complementary to the natural depletion of AG dinucleotides within the AGEZ of human introns; only 5.8% of naturally occurring AG dinucleotides fall within the high-risk zone, indicating evolutionary intolerance of AGs in this region (Figure 2Aiii).
The nt preceding the variant created AG was relevant, with creation of a GAG less likely to be pathogenic than other AGs created (Pearson’s chi-squared test = 43.506; df = 9, p = 1.74 × 10−6; Figure 2Bi), consistent with the increased frequency of naturally occurring GAG trinucleotides in the high-risk zone relative to other trinucleotide combinations for natural AGs in the AGEZ (Figure 2Bii). No SNVs in this dataset created an AG at the −4 position, and very few AGs occur naturally at this position, due to the low frequency of G at the −3 position (to make AG).
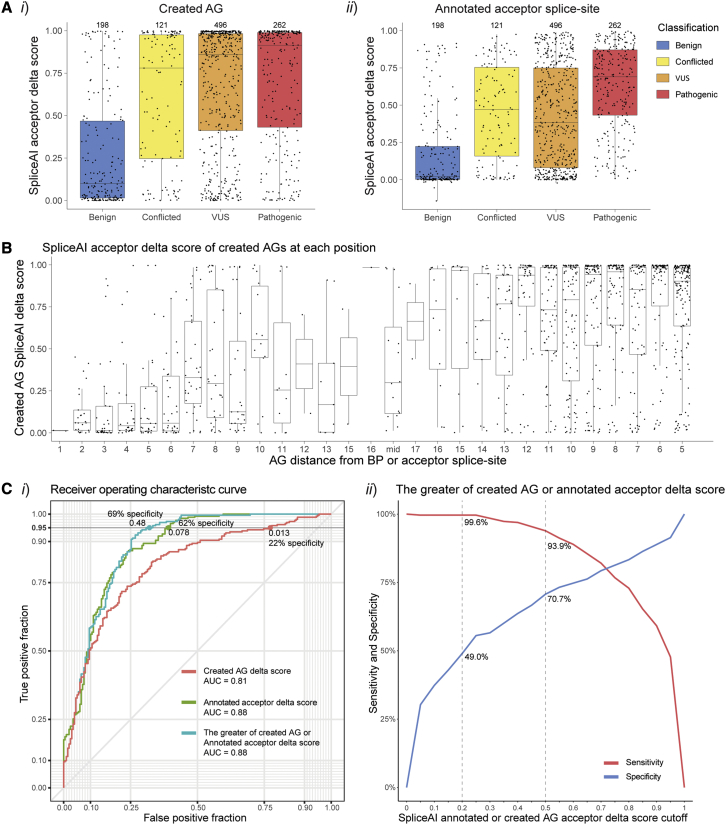
SpliceAI assessment of AG-creating variants
Pathogenic AG-creating variants were strongly predicted by SpliceAI acceptor delta scores to create cryptic acceptor splice-sites, whereas benign AG-creating variants have significantly weaker SpliceAI acceptor delta scores for the created AG (Mann-Whitney U test: U = 9,862; p < 2.2 × 10−16; Figure 3Ai). Further, SpliceAI acceptor delta scores for the annotated acceptor splice-site were significantly higher for pathogenic AG-creating variants than benign AG-creating variants (Mann-Whitney U test: U = 6,467; p < 2.2 × 10−16; Figure 3Aii). SpliceAI acceptor scores are also higher for AGs created closer to the annotated acceptor splice-site than those closer to the BP, consistent with the likely position of a PPT (Mann-Whitney U test: U = 180,518; p < 2.2 × 10−16; Figure 3B). To discriminate pathogenic from benign AG-creating variants with 95% sensitivity, we found that using a SpliceAI acceptor delta cutoff of 0.48 for either the created AG or annotated acceptor splice-site (whichever score is higher) provided the highest specificity at 69%, compared with using either score in isolation (Figure 3C). Together, these data demonstrate SpliceAI as an effective tool for distinguishing pathogenic AG-creating variants from benign AG-creating variants.
Figure 3.
SpliceAI scores for the created AG and annotated acceptor splice-site for AG-creating variants
(A) SpliceAI acceptor delta scores for (i) the AG created by the variant and (ii) the annotated acceptor splice-site.
(B) SpliceAI acceptor delta scores for the created AG for each position of the PPT. Numbered positions denote the distance of the A of the AG dinucleotide from either the BP (left) or acceptor splice-site (right, see Figure 2Ai). AG-creating variants had stronger scores closer to the annotated acceptor splice-site than the BP.
(C) Assessment of SpliceAI’s capability to differentiate pathogenic (n = 262) AG-creating variants from benign (n = 198); (i) receiver operating characteristic (ROC) curve of 3 different SpliceAI acceptor delta scores types; (1) created AG score (red), (2) annotated acceptor splice-site score (green), and (3) the greater of either the created AG score or annotated acceptor splice-site score for each variant (teal). At 95% sensitivity, using the greater of the two SpliceAI scores for each variant resulted in the greatest specificity (69%) using a cutoff of 0.48. (ii) Sensitivity (red) and specificity (blue) at different SpliceAI cutoffs for the greater of the two SpliceAI scores.
Clinical exemplar of pathogenic AG-creating variant in COL6A2 intron-26 high-risk zone
Two affected siblings (VIII:1 and VIII:2) born to distantly consanguineous parents (fourth cousins once removed, Figure 4A) were clinically diagnosed with congenital myopathy. VIII:1 presented at birth with hypotonia and soft skin following an unremarkable pregnancy. She had delayed motor milestones, sitting at 12 months and walking at 3.5 years of age. She had minimally progressive, generalized mild muscle weakness, with proximal muscles slightly weaker than distal. However, from around age 25 years old, her muscle strength slowly declined and, at age 43 years old, she predominantly used a wheelchair, was only able to mobilize short distances with a walker, and required assistance to rise from a chair. She had a long thin face, mild facial weakness, and high arched narrow hourglass-shaped palate. Examination at age 43 years old revealed a Trendelenburg gait with high steppage, elbow flexion and hip contractures, hyperextensible fingers, and mild dysphagia. In addition, she had comorbidities associated with Perrault syndrome, with primary ovarian failure, sensorineural hearing loss, mild intellectual disability, and micrognathia. VIII:2 required neonatal intensive care unit (NICU) admission at birth for respiratory distress, was initially reliant on tube feeding, and then was discharged at four weeks of age on bottle feeds. His milestones were mildly delayed, sitting at 9 months and walking at 22 months. He had a similar pattern of weakness to VIII:1, also presenting with soft skin and hyperextensibility, but did not have symptoms associated with Perrault syndrome. Upon most recent review at age 41 years old, he has had gradual progression in his proximal muscle weakness and has developed contractures with restriction of wrist extension, finger flexion, ankle dorsiflexion, and mild lumbar hyperlordosis. Unlike his sister, he remains ambulant without requiring any assistive devices, though does have difficulty walking on heels and climbing stairs. Their mother, VII:2, had a considerably milder muscle weakness than her children, affecting only her proximal muscles. She had an hourglass palate, retrognathia, hyperextensible elbows, and soft skin.
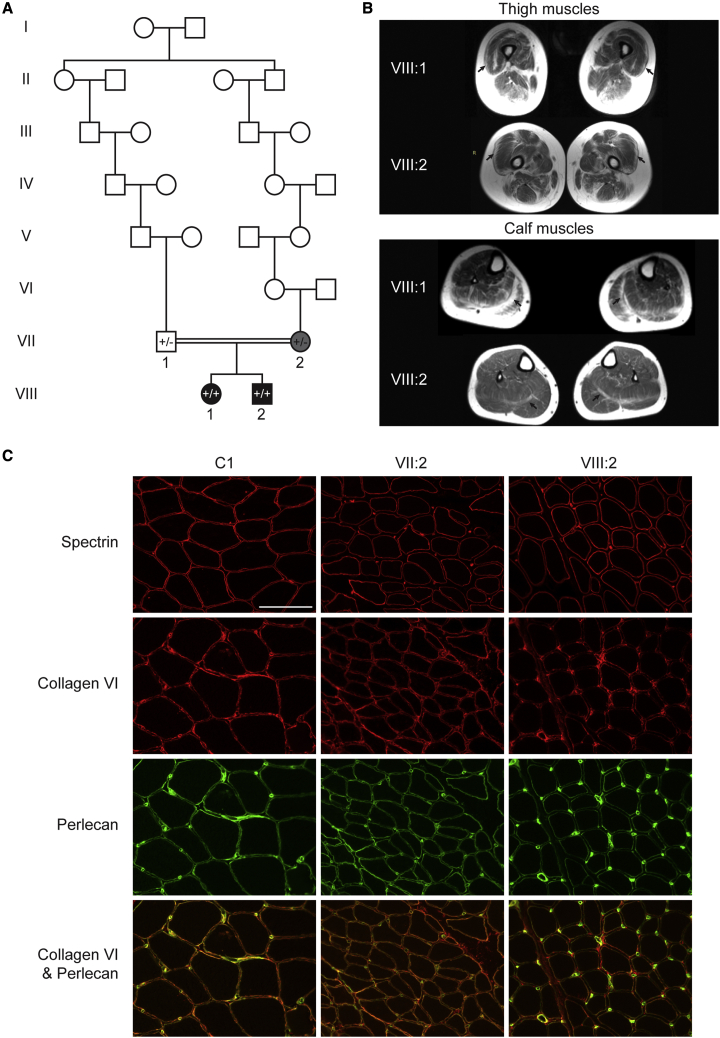
Figure 4.
Pedigree, MRI and immunohistochemical staining for the COL6A2 family
(A) Pedigree of the COL6A2 family. VIII:1 and VIII:2 are homozygous for the COL6A2 NM_001849.3:c.2423-22_2423-21insAGCCCGGCCCGGCCC variant (+/+) and their parents (VII:1 and VII:2) were both heterozygous (+/−). VII:1 and VII:2 are distantly consanguineous.
(B and C) Muscle MRIs for both VIII:1 and VIII:2 show peripheral involvement of thigh and calf muscles, in particular the vastus lateralis (top, arrows) and gastrocnemii (bottom, arrows), typical of those described in collagen-VI-related myopathies.32 (C) Immunohistochemical staining shows reduced, but present, collagen staining in a skeletal muscle biopsy from VIII:2, and present staining in VII:2. Sequential 8 μm thick cryosections of snap frozen skeletal muscle were stained with anti-spectrin, and anti-collagen VI (red) co-stained with anti-perlecan (green) antibodies. In healthy control muscle (C1, 28 years, vastus lateralis), collagen VI stains the muscle sarcolemma as well as the endomysium, whereas perlecan only stains the sarcolemma. In VIII:2 (12 years, quadriceps), collagen VI staining of the sarcolemma is reduced and endomysium staining is normal/increased. In VII:2 (28 years, unknown muscle type), collagen VI staining is within normal limits. Scale bar 100 μm (white line).
Muscle biopsies at age 10 months old for VIII:1 and 12 years old for VIII:2 both showed a myopathic process with fiber-type disproportion (type I fibers smaller than type II fibers), increased internal nuclei, and increased interstitial connective tissue in some areas. A muscle biopsy at 28 years old for VII:2 also revealed small type I fibers. Muscle magnetic resonance images (MRIs) for both VIII:1 and VIII:2 show peripheral atrophy and fatty infiltration of thigh and calf muscles, especially of the vastus lateralis and gastrocnemii, typical of those described in collagen-VI-related myopathies32 (Figure 4B). IHC showed reduced levels of collagen VI staining at the membrane and normal/increased levels in the endomysium for VIII:2 (Figure 4C). Membrane staining in VII:2 was within normal limits (Figure 4C). A muscle biospecimen from VIII:1 was unavailable for IHC.
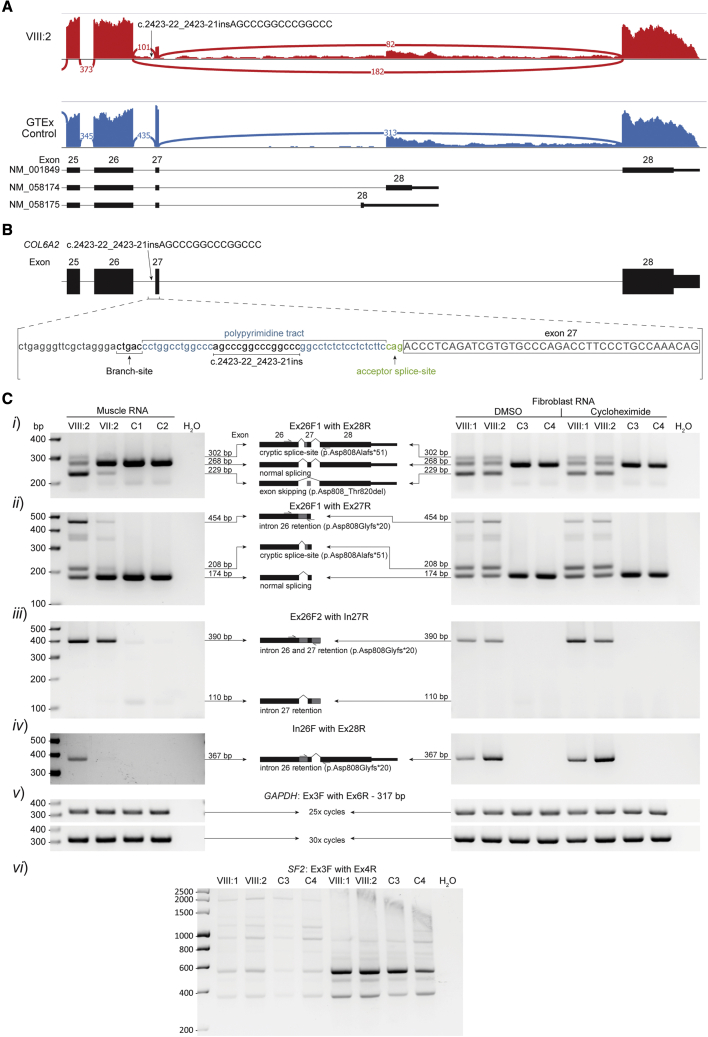
RNA-seq of muscle-derived mRNA from VIII:2 revealed splicing abnormalities in COL6A2 transcripts: exon 27 skipping and increased intron 26 and 27 retention, in addition to canonically spliced COL6A2 transcripts (Figure 5A). On finding this abnormality, WGS data of COL6A2 in this family were re-examined for variants that may cause these mis-splicing events. An extended splice-site variant NM_001849.3(COL6A2):c.2423-22_2423-21insAGCCCGGCCCGGCCC was identified at homozygosity in VIII:1 and VIII:2 and heterozygosity in the parents VII:1 and VII:2 (Figure 5B). The insertion was absent from ClinVar and the Genome Aggregation Database (gnomAD).35 RT-PCR of muscle-derived mRNA from VIII:2 and fibroblast-derived mRNA from VIII:1 and VIII:2 confirmed exon 27 skipping (Figure 5Ci, in-frame), intron retention (Figure 5Cii–iv, encodes a stop codon), and normal splicing and in addition revealed use of the cryptic acceptor inserted by the variant (Figure 5Ci–ii, encodes a stop codon). Increased band intensities for primer pairs Ex26F/In27R and In26F/Ex28R for cycloheximide-treated fibroblasts support nonsense mediated decay (NMD) targeting transcripts with intron 26 retention (Figure 5Ciii–iv). RT-PCR of muscle-derived mRNA from VII:2 showed a similar pattern of mis-splicing to VIII:1 and VIII:2 but with a larger proportion of normally spliced products, consistent with heterozygosity of the COL6A2 splice variant (Figure 5C) in the VII:2 parent. The AG dinucleotide included in the insertion c.2423-22_2423-21insAGCCCGGCCCGGCCC was considered the pathogenic element of the variant as other similar insertions without an AG dinucleotide are reported in ClinVar as likely benign microsatellite variations (ClinVar accession no.: VCV000420369.1, VCV000258324.1, and VCV000420761.1) and one was common in gnomAD (c.2423-18_2423-17insCGGCCCGGCCCGGCC: allele frequency 0.046, 55 homozygotes).
Figure 5.
Mis-splicing arising from the COL6A2 variant
(A) Sashimi plots of COL6A2 exons 25–28 (NM_001849) in RNA-seq data of muscle-derived RNA from VIII:2 (red) compared with a control skeletal muscle sample from GTEx (blue).33 Exon 27 skipping (182 reads) and low levels intron 26 and 27 retention are present in VIII:2 but absent in the GTEx control. Exon 27 skipping was also present in NM_058174 transcripts for VIII:2 (not shown). Normal splicing of exons 26-27-28 can be seen in 25% of reads in VIII:2.
(B) Schematic of the NM_001849.3(COL6A2):c.2423-22_2423-21insAGCCCGGCCCGGCCC variant identified at homozygosity in VIII:1 and VIII:2 in WGS data.
(C) RT-PCR analyses of muscle-derived RNA from VIII:2 (12 years, male, quadriceps), VII:2 (28 years, female, muscle origin not documented), and two controls; C1 (7.5 years, female, muscle origin not documented) and C2 (26 years, female, quadriceps), as well as fibroblast-derived RNA from VIII:1 (37 years, female), VIII:2 (35 years, male), and two controls; C3 (37 years, female) and C4 (age and gender not documented). Primer pairs; (i) Ex26F1 with Ex28R in muscle RNA showed use of the cryptic splice-site created by the variant and exon 27 skipping in both VIII:2 and VII:2, absent in control samples. Normal splicing is reduced in VIII:2 relative to VII:2 and controls; (ii) Ex26F1 with Ex27R in muscle RNA showed intron 26 retention and use of the cryptic splice-site from both VIII:2 and VII:2. Normal splicing is reduced in VIII:2 relative to VII:2 and controls; (iii) Ex26F2 with In27R in muscle RNA showed elevated levels of intron 26 and 27 retention in both VIII:2 and VII:2 relative to controls. Intron 27 retention without intron 26 retention can be seen in very low levels in the controls; (iv) In26F with Ex28R in muscle RNA showed intron 26 retention without intron 27 retention in VIII:2 and VII:2 (low levels), absent in controls; (v) GAPDH demonstrated equal loading; and (vi) SF2 demonstrated NMD inhibition of cycloheximide-treated fibroblasts.34 Fibroblast RNA mis-splicing patterns observed in VIII:1 and VIII:2 were consistent with those observed in muscle RNA in VIII:2, with primer pairs Ex26F2/In27R and In26F/Ex28R showing slightly elevated signal in cycloheximide treated samples.
Pseudo-exons activated by removal of AG splicing silencer in AGEZ
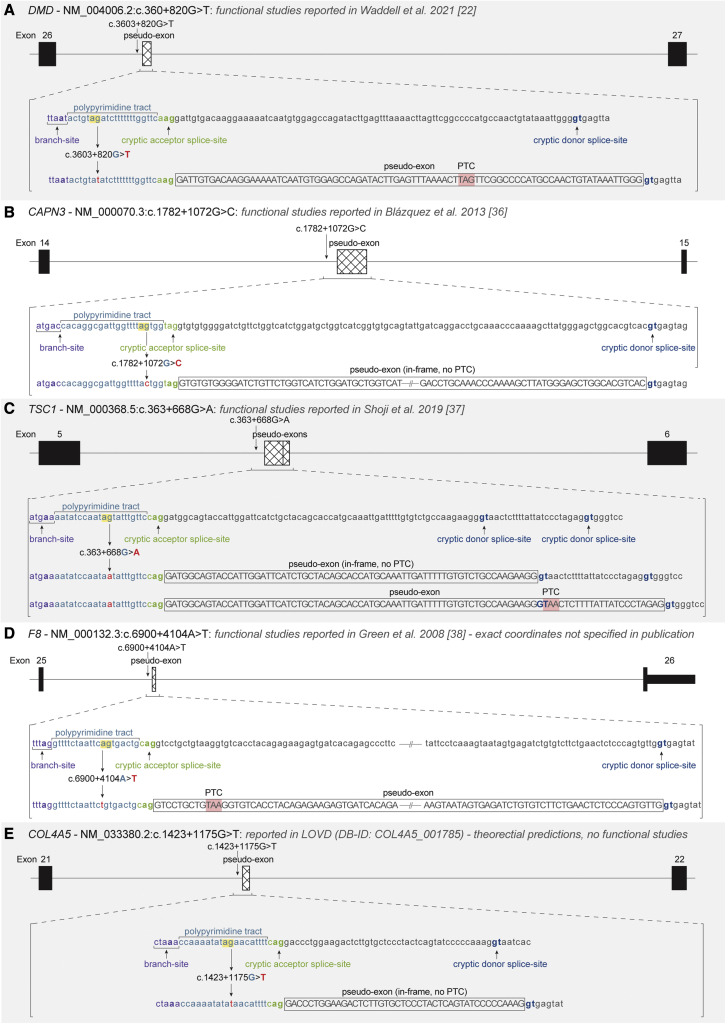
In a previous study, a deep intronic variant (NM_004006.2(DMD):c.3603 + 820G > T) was revealed to disrupt an AG dinucleotide and result in the inclusion of a pathogenic pseudo-exon in intron 26 of DMD, starting 19 nt downstream of the variant22 (Figure 6A). The AG dinucleotide disrupted by the variant was 7 nt downstream of the predicted BP in the AGEZ of the pseudo-exon (high-risk zone), and pseudo-exon inclusion was undetectable in control samples.22 This finding led us to search for other pathogenic AG-disrupting deep intronic variants in the ClinVar and LOVD dataset. Four additional deep intronic SNVs were identified in CAPN3,36 TSC1,37 F8,38 and COL4A5 (LOVD DB-ID: COL4A5_001785), with functional studies confirming pseudo-exon usage in three of the four variants (Figures 6B–6E). All variants result in the loss of an AG dinucleotide in the AGEZ of a pseudo-exon (or predicted pseudo-exon), demonstrating that AG dinucleotides may act as natural splicing silencers to prevent inclusion of damaging pseudo-exons.
Figure 6.
Variants activating pseudo-exons by disrupting AG dinucleotides in AGEZ
(A–E) Schematic of five introns (black lines) with deep intronic variants that activate pseudo-exon inclusion through removal of an AG dinucleotide from the AGEZ; (A) DMD intron 26 NM_004006.2:c.360 + 820G > T22, (B) CAPN3 intron 14 NM_000070.3:c.1782 + 1072G > C34, (C) TSC1 intron 5 NM_000368.5:c.363 + 668G > A35, (D) F8 intron 25 NM_000132.3:c.6900 + 4104A > T36, and (E) COL4A5 intron 21 NM_033380.2:c.1423 + 1175G > T (LOVD DB-ID: COL4A5_001785). Canonical exons (black) and pseudo-exons (hashed) are represented by boxes. Pseudo-exon sequences (as determined by the published functional studies for A–D or SpliceAI predictions for (E) with flanking splicing motifs are shown labeled below, before and after the variant change (red) disrupts the AG dinucleotide (yellow highlight). Branch-sites (purple) were selected as per the strongest BPP score.28
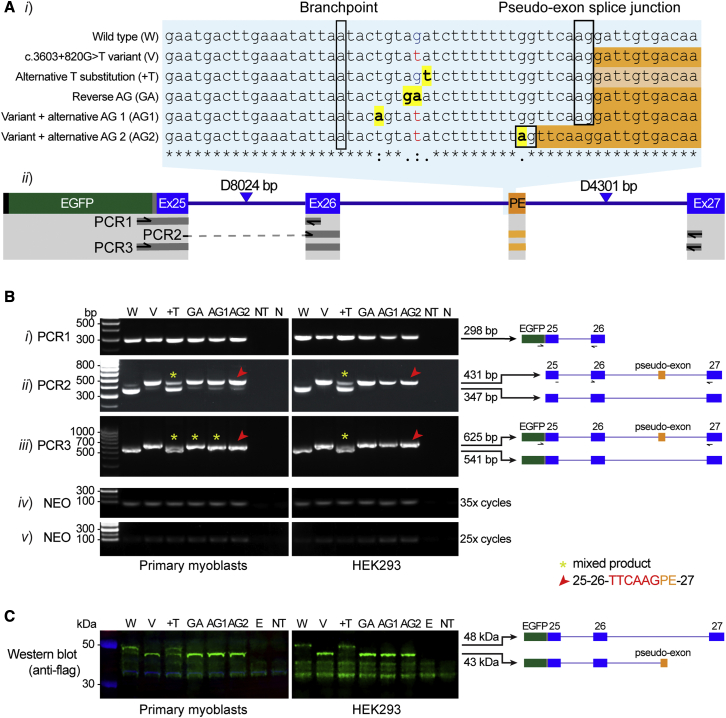
To explore the hypothesis that disruption of AG dinucleotide associated with DMD c.3603 + 820G > T is the specific mechanism activating pseudo-exon inclusion,22 we developed six EGFP-tagged minigene constructs encompassing DMD exons 25-26-27 and intervening (modified) introns 25 and 26 (Figure 7A, see supplemental materials and methods for construct details and sequences). RT-PCR of the wild-type construct (Figure 7Bii–iii, W) shows predominant normal splicing of DMD exons 25-26-27, whereas the c.3603 + 820G > T variant construct (Figure 7Bii–iii, v) results in predominant pseudo-exon inclusion, replicating the splicing pattern previously reported in skeletal muscle.22 As the c.3603 + 820G > T variant also introduces a pyrimidine into the PPT, the +T construct shows that strengthening the PPT (Figure 7Bii–iii) enhances pseudo-exon inclusion (asterisk), though unlike the variant construct (V), maintains robust levels of canonical splicing. Reversing the AG dinucleotide to GA results in predominant pseudo-exon inclusion similar to the variant construct (see GA versus V, Figure 7Bii–iii). From collective results, therefore, inference is that the AG dinucleotide acts as a potent splicing silencer preventing pseudo-exon inclusion, with the +T construct showing that pyrimidine composition of the PPT can also influence pseudo-exon inclusion.
Figure 7.
Minigene constructs modeling the DMD intron 26 pseudo-exon arising from the c.360 + 820G > T variant
(A) (i) Alignment of wild-type and variant DMDex25-27 minigene sequences with modified bases highlighted in yellow. Predicted BP and pseudo-exon (PE) acceptor splice-site are indicated by a black box. The PE sequences for each construct are shown in orange (or light orange to denote reduced levels of PE inclusion). (ii) Schematic of DMDex25-27 minigene construct showing FLAG (black), EGFP (green), linker sequence (gray), canonical exons (Ex; blue), and PE (orange). Note: introns 25 and 26 have been shortened as indicated by blue triangles (8024 and 4301 bp were removed, respectively). Primer locations and area covered by DMDex25-27 PCR 1–3 are shown below.
(B) RT-PCR results of (i) PCR1 (EGFP-Exon 26), (ii) PCR2 (Exon 25/26 junction – Exon 27), and (iii) PCR3 (EGFP-Exon 27). Amplification of the neomycin resistance gene (NEO) for (iv) 35 and (v) 25 cycles served as a transfection control. Red arrows indicate PCR products where alternative splice junction was used and yellow asterisks indicate bands for which Sanger sequencing detected a mixture of products containing and lacking the pseudo-exon.
(C) Western blot of 20 μg protein probed with an anti-flag antibody (green signal) demonstrates a 48 kDa band (protein product of canonically spliced DMDex25-27) or 43 kDa band (protein product of DMDex25-27 when pseudo-exon is included). NT, not transfected; N, no template; E, EGFP.
AG1 and AG2 constructs act as additional controls to probe the impact of the positioning of created AGs within the AGEZ upon pseudo-exon activation (AG1 and AG2, Figure 7Ai). The AG dinucleotide created 4 nt downstream of the BP in AG1 (i.e., outside high-risk zone) does not suppress pseudo-exon inclusion, whereas AG2 8 nt upstream of the pseudo-exon acceptor splice-site (i.e., within high-risk zone) results in pseudo-exon activation via use of the alternative acceptor splice-site created by AG2 (Figure 7Ai and Cii, red arrow). Pseudo-exon activation encodes a premature termination codon that is concordantly detected as a truncated EGFP-FLAG-DMD fusion protein by western blot (Figure 7C).
Discussion
Pathogenic AG-creating variants are common in ClinVar and LOVD
In this study, we demonstrate that AG-creating variants account for 41.8% of pathogenic extended acceptor splice-site variants and 4.9% of all pathogenic acceptor variants reported in ClinVar and LOVD. However, this is likely to be an underestimate of the true prevalence of pathogenic AG-creating variants, considering the bias toward ascertainment of essential splice-site variants in variant databases as they are more readily classified as pathogenic/likely pathogenic.26 We define the region >6 nt downstream of the BP and >5 nt upstream of the annotated acceptor splice-site as a high-risk zone for splice-altering effects of AG-creating variants, compared with other regions of the AGEZ (odds ratio: 14.9, 95% CI [7.99, 27.67]), consistent with our DMDex25-27 minigene studies (Figure 7). In addition, we demonstrate that a SpliceAI acceptor delta score of ≥0.48 for either the annotated acceptor splice-site or variant created AG is a good predictor of pathogenicity (odds ratio: 39.8, 95% CI [21.46, 73.75]). Combining these analyses, we identify 447 VUS and 108 conflicted AG-creating variants that are within the high-risk zone and/or have SpliceAI acceptor delta scores ≥ 0.48, which are likely to result in mis-splicing, and we recommend RNA studies (Table S2) to examine pre-mRNA splicing.
The overall proportions of pathogenic AG-creating, −3Y > R, loss of 1Y, and loss of ≥2Y variants among all disease genes broadly mirrors described proportions among a cohort of 91 extended acceptor splice-site NF1 variants.18 However, due to our larger dataset of 24,445 variants affecting the extended acceptor splice-site region with both pathogenic and benign classifications, we are able to show that the PPT is often tolerant to loss of one or more pyrimidines. Comparative analysis of all dinucleotide combinations created by ClinVar and LOVD variants provides compelling evidence for increased pathogenicity associated with AG-creation (Figure S1), consistent with substitution of AG with GA, resulting in vastly different outcomes in the DMDex25-27 minigene (GA, Figure 7Bii–iii) and in keeping with previously published minigene studies.15,18 Collectively, our data strongly imply acquisition of an AG dinucleotide as the primary pathogenic feature of AG-creating variants rather than coincidental loss of one or more pyrimidines.
However, there are limitations to our dataset that need to be acknowledged. We have assumed that any variant classified as pathogenic will be splice-altering and benign variants will not disrupt splicing. Variant entries in ClinVar and LOVD are not always rigorously reviewed, and evidence for pathogenicity is rarely provided; thus, a subset of variants may have been mis-classified. Further, some benign variants may result in splice-altering outcomes with benign functional impact; particularly AGs created at positions −5 and −8 which would lead to in-frame insertions of 1 or 2 codons if used. This may partially explain the increased percentage of benign variants we observe at positions −5 and −8 (Figure 2Aii) and naturally occurring AGs at the −5 position (Figure 2B), although previous studies have shown that created AGs have reduced splicing efficiency closer to the annotated acceptor splice-site.13 Further, many introns contain multiple branch-sites17,39 and Branchpointer predictions may not correctly identify the predominant branch-site. Experimentally proven BPs39, 40, 41 would be ideal for our analysis; however, Branchpointer allowed for a much larger dataset of variants for analysis. Despite these caveats, the large variant dataset establishes AG-creating variants as a common class of pathogenic variant affecting the splice acceptor in ClinVar and LOVD, indicating the importance of assessing these variants in diagnostic pipelines.
Pathogenic mechanisms for AG dinucleotides in the AGEZ
The obvious mechanism by which an AG dinucleotide introduced into the AGEZ can cause mis-splicing is through creation of a cryptic acceptor. As these pathogenic AGs frequently arise within a PPT in the context of an existing BP at a usable distance upstream, a competitive cryptic acceptor is often created, correlating with SpliceAI acceptor delta scores (Figure 3). However, AG-creating variants not only (or always) create a cryptic acceptor that is used; they can also cause intron retention and exon skipping (as shown for the COL6A2 variant, Figure 5) and/or use of an alternative pre-existing cryptic acceptor.15,18,42 These mis-splicing events suggest that in some instances the created AG acts as a splicing silencer, preventing the annotated acceptor splice-site from being utilized by the spliceosome. Evidence supporting splicing silencer behavior of AGs in the AGEZ is demonstrated by deep intronic AG-disrupting variants that activate pathogenic pseudo-exons flanked by usable splice-sites22,36, 37, 38 and by our DMDex25-27 minigene studies (Figure 7). Additionally, Keegan and colleagues report a small subset of previously published pseudo-exons (8/410) activated by AG-removing variants,43 further demonstrating this mechanism as a rare cause of pseudo-exon activation. These deep intronic AG dinucleotides within the AGEZ of dormant pseudo-exons act as natural splicing silencers, which may have arisen through evolution as one of many mechanisms to suppress undesirable exons from being included in transcripts.
The exact processes by which the spliceosome selects an acceptor splice-site are still unknown. There are at least two inter-related mechanisms explaining disruption of pre-mRNA splicing by AG-creating variants in the AGEZ: (1) competitive binding of spliceosome components to both the AG in the AGEZ and to the annotated acceptor splice-site slowing down the splicing process12—to the extent that neither the annotated nor created cryptic acceptor is used successfully, resulting in exon skipping, intron retention or use of an alternative, more distal, pre-existing cryptic acceptor; and (2) a specific factor binds to the AG in the AGEZ acting as a splicing silencer. Wimmer and colleagues18 suggest that since U2AF35 subunits are able to bind simultaneously to multiple AGs near the acceptor splice-site,12 U2AF35 bound to an AG in the PPT may block U2AF65 from recognizing the PPT, preventing the annotated acceptor splice-site from being used.18 U2AF35 binding to created AGs may explain both plausible mechanisms by simultaneously competing with and silencing the annotated acceptor splice-site. Other splicing regulatory elements may also be acting as splicing silencers, such as the splice--modulating RNA binding protein hnRNPA1 that binds to an RNA motif containing YAG,44 often formed by AG-creating variants.18 hnRNPA1 has been shown to suppress the inclusion of alternatively spliced exons and pseudo-exons by binding downstream of the donor splice-site,44 so it is plausible that hnRNPA1 may also prevent pseudo-exon usage by binding upstream of the acceptor splice-site.
For the COL6A2 c.2423-22_2423-21insAGCCCGGCCCGGCCC variant described in this study, mechanism 1 works well to explain the four different splicing outcomes. Competitive binding of spliceosome factors to the two available acceptor splice-sites (the variant created and annotated acceptors) appears to significantly reduce the efficiency, but not completely prevent, both acceptors from being used. Contrastingly, mechanism 2 would fit better for the DMD c.360 + 820G > T deep intronic AG-removing variant, as the AG dinucleotide silences the pseudo-exon from inclusion in control samples.22 By removing the AG, this pseudo-exon is very efficiently included into almost all DMD transcripts in our minigene assay (Figure 7), acting as a binary switch to include or exclude the exon. Both proposed mechanisms can explain benign AG-creating variants and naturally occurring AG dinucleotides observed within 6 nt of the BP (Figure 2), as AGs in this region (1) are often non-competitive acceptor splice-sites, as they lack a strong PPT and/or usable BP (supported by weak SpliceAI acceptor scores, Figure 3B), and (2) will not sterically prevent U2AF65 binding to the PPT, as most/all of this sequence would still be available for U2AF65 binding. With enough functional data of AG-creating/disrupting variants within introns with experimentally defined branch-sites, it may become possible in the future to predict the nature of mis-splicing (i.e., exon skipping versus cryptic splice-site use versus intron retention), based on AG position and sequence context, improving clinical interpretation for this class of variant.
AG-creating/disrupting variants may induce complete or partial mis-splicing. In the COL6A2 and DMD cases highlighted, remnant normal splicing had significant clinical implications. Trace levels of normal splicing were observed for the individual harboring the intron 26 DMD pseudo-exon,22 resulting in a severe Becker muscular dystrophy phenotype rather than a Duchenne muscular dystrophy presentation that would be expected from a null mutation. Normal splicing of COL6A2 exons 26–27 was identified in VIII:1 and VIII:2 at reduced levels with the homozygous c.2423-22_2423-21insAGCCCGGCCCGGCCC variant. Remnant levels of normally spliced COL6A2 may explain the mild presentation seen in VIII:1 and VIII:2 and their asymptomatic or very mildly affected heterozygous parents (VII:1 and VII:2, respectively).
In conclusion, we establish AG-creating variants as a common class of pathogenic extended acceptor splice-site variant and define a high-risk zone for pathogenicity >6 nt downstream of the BP and >5 nt upstream of the annotated acceptor splice-site. We encourage careful consideration and functional studies (if possible) of any AG-creating extended acceptor splice-site variant and deep intronic AG-disrupting variant, especially if SpliceAI acceptor delta scores are ≥ 0.48 for either the created AG or annotated acceptor, when searching for causative variants in genetic diseases.
Data and code availability
The datasets analyzed during this study are available from ClinVar https://www.ncbi.nlm.nih.gov/clinvar/ and LOVD http://www.dmd.nl/.
Acknowledgments
We thank the families for their invaluable contributions to this research and the clinicians and health care workers involved in their assessment and management. For facilitating clinical discussion that contributed to the diagnoses made, we thank Sarah A. Sandaradura. This study was supported by the National Health and Medical Research Council of Australia (APP1186084, APP1116974, and APP2002640 to S.T.C and APP1121651 to M.Y.). S.J.B. was supported by a Muscular Dystrophy New South Wales PhD scholarship. F.J.E was supported by a US Muscular Dystrophy Foundation Development Grant. R.D was supported by a University of Sydney Research Training Scholarship. WES, WGS, and RNA-seq were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and funded by the National Human Genome Research Institute, National Eye Institute, and National Heart, Lung, and Blood Institute grant UM1 HG008900 to Daniel MacArthur and Heidi Rehm. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health (https://commonfund.nih.gov/GTEx) and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000424.v7.p2.
Declaration of interests
Professor S.T.C. is director of Frontier Genomics Pty Ltd (Australia) and receives no remuneration (salary or consultancy fees) for this role. H.J. offers technology advice to Frontier Genomics Pty Ltd (Australia) and receives no remuneration for this role. Professor S.T.C. and H.J. are named inventors on intellectual property (IP) (Australian patent 2019379868 and PCT/AU2019/000141) owned jointly by the University of Sydney and Sydney Children’s Hospitals Network. This IP relates to splicing variant detection and interpretation and is licensed by Frontier Genomics Pty Ltd. The remaining co-authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2022.100125.
Web resources
ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/
Leiden Open Variation Database: http://www.dmd.nl/
OMIM gene list: https://www.omim.org/downloads/
GRCh37 Ensembl transcripts: http://grch37.ensembl.org/
Supplemental information
References
- 1.Ezquerra-Inchausti M., Barandika O., Anasagasti A., Irigoyen C., López De Munain A., Ruiz-Ederra J. High prevalence of mutations affecting the splicing process in a Spanish cohort with autosomal dominant retinitis pigmentosa. Sci. Rep. 2017;7:1–8. doi: 10.1038/srep39652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ars E., Serra E., García J., Kruyer H., Gaona A., Lázaro C., Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- 3.Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Önengüt S., Tolun A., Chessa L., Sanal Ö., Bernatowska E., Gatti R.A., et al. Splicing defects the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cummings B.B., Marshall J.L., Tukiainen T., Lek M., Donkervoort S., Foley A.R., Bolduc V., Waddell L.B., Sandaradura S.A., O’Grady G.L., et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017;9:eaal5209. doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wai H.A., Lord J., Lyon M., Gunning A., Kelly H., Cibin P., Seaby E.G., Spiers-Fitzgerald K., Lye J., Ellard S., et al. Blood RNA analysis can increase clinical diagnostic rate and resolve variants of uncertain significance. Genet. Med. 2020;22:1005–1014. doi: 10.1038/s41436-020-0766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang R., Prosser D.O., Love D.R. Evaluation of bioinformatic programmes for the analysis of variants within splice site consensus regions. Adv. Bioinformatics. 2016;2016:5614058. doi: 10.1155/2016/5614058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffman-Andrews L. The known unknown: the challenges of genetic variants of uncertain significance in clinical practice. J. Law Biosci. 2017;4:648–657. doi: 10.1093/jlb/lsx038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wai H., Douglas A.G.L., Baralle D. RNA splicing analysis in genomic medicine. Int. J. Biochem. Cell Biol. 2019;108:61–71. doi: 10.1016/j.biocel.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Matera A.G., Wang Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Will C.L., Lührmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkinson M.E., Charenton C., Nagai K. RNA splicing by the spliceosome. Annu. Rev. Biochem. 2020;89:359–388. doi: 10.1146/annurev-biochem-091719-064225. [DOI] [PubMed] [Google Scholar]
- 12.Chen L., Weinmeister R., Kralovicova J., Eperon L.P., Vorechovsky I., Hudson A.J., Eperon I.C. Stoichiometries of U2AF35, U2AF65 and U2 snRNP reveal new early spliceosome assembly pathways. Nucleic Acids Res. 2017;45:2051–2067. doi: 10.1093/nar/gkw860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chua K., Reed R. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith C.W., Chu T.T., Nadal-Ginard B. Scanning and competition between AGs are involved in 3’ splice site selection in mammalian introns. Mol. Cell Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Královičová J., Christensen M.B., Vořechovský I. Biased exon/intron distribution of cryptic and de novo 3′ splice sites. Nucleic Acids Res. 2005;33:4882–4898. doi: 10.1093/nar/gki811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gooding C., Clark F., Wollerton M.C., Grellscheid S.-N.N., Groom H., Smith C.W.J.J. A class of human exons with predicted distant branch points revealed by analysis of AG dinucleotide exclusion zones. Genome Biol. 2006;7:R1. doi: 10.1186/gb-2006-7-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corvelo A., Hallegger M., Smith C.W.J., Eyras E. Genome-Wide association between branch point properties and alternative splicing. PLoS Comput. Biol. 2010;6:e1001016. doi: 10.1371/journal.pcbi.1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wimmer K., Schamschula E., Wernstedt A., Traunfellner P., Amberger A., Johannes Z., Kroisel P., Chen Y., Callens T., Messiaen L. AG-exclusion zones revisited: lessons to learn from 91 intronic NF1 3′ splice site mutations outside the canonical AG-dinucleotides. Hum. Mutat. 2020;41:1145–1156. doi: 10.1002/humu.24005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fokkema I.F.A.C., Taschner P.E.M., Schaafsma G.C.P., Celli J., Laros J.F.J., den Dunnen J.T. LOVD v.2.0: the next generation in gene variant databases. Hum. Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 21.Dawes R., Lek M., Cooper S.T. Gene discovery informatics toolkit defines candidate genes for unexplained infertility and prenatal or infantile mortality. NPJ Genom. Med. 2019;4:8. doi: 10.1038/s41525-019-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waddell L.B., Bryen S.J., Cummings B.B., Bournazos A., Evesson F.J., Joshi H., Marshall J.L., Tukiainen T., Valkanas E., Weisburd B., et al. WGS and RNA studies diagnose noncoding DMD variants in males with high creatine kinase. Neurol. Genet. 2021;7:e554. doi: 10.1212/NXG.0000000000000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yates A.D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Signal B., Gloss B.S., Dinger M.E., Mercer T.R. Machine learning annotation of human branchpoints. Bioinformatics. 2018;34:920–927. doi: 10.1093/bioinformatics/btx688. [DOI] [PubMed] [Google Scholar]
- 25.Leman R., Tubeuf H., Raad S., Tournier I., Derambure C., Lanos R., Gaildrat P., Castelain G., Hauchard J., Killian A., et al. Assessment of branch point prediction tools to predict physiological branch points and their alteration by variants. BMC Genom. 2020;21:86. doi: 10.1186/s12864-020-6484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaz-Drago R., Custódio N., Carmo-Fonseca M. Deep intronic mutations and human disease. Hum. Genet. 2017;136:1093–1111. doi: 10.1007/s00439-017-1809-4. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q., Fan X., Wang Y., Sun M., Shao J., Guo D. BPP: a sequence-based algorithm for branch point prediction. Bioinformatics. 2017;33:3166–3172. doi: 10.1093/bioinformatics/btx401. [DOI] [PubMed] [Google Scholar]
- 29.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 30.Rowlands C., Thomas H.B., Lord J., Wai H.A., Arno G., Beaman G., Sergouniotis P., Gomes-Silva B., Campbell C., Gossan N., et al. Comparison of in silico strategies to prioritize rare genomic variants impacting RNA splicing for the diagnosis of genomic disorders. Sci. Rep. 2021;11:20607. doi: 10.1038/s41598-021-99747-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waddell L.B., Tran J., Zheng X.F., Bönnemann C.G., Hu Y., Evesson F.J., Lek M., Arbuckle S., Wang M.-X., Smith R.L., et al. A study of FHL1, BAG3, MATR3, PTRF and TCAP in Australian muscular dystrophy patients. Neuromuscul. Disord. 2011;21:776–781. doi: 10.1016/j.nmd.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercuri E., Clements E., Offiah A., Pichiecchio A., Vasco G., Bianco F., Berardinelli A., Manzur A., Pane M., Messina S., et al. Muscle magnetic resonance imaging involvement in muscular dystrophies with rigidity of the spine. Ann. Neurol. 2010;67:201–208. doi: 10.1002/ana.21846. [DOI] [PubMed] [Google Scholar]
- 33.Ardlie K.G., DeLuca D.S., Segrè A.V., Sullivan T.J., Young T.R., Gelfand E.T., Trowbridge C.A., Maller J.B., Tukiainen T., Lek M., et al. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun S., Zhang Z., Sinha R., Karni R., Krainer A.R. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat. Struct. Mol. Biol. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 doi: 10.1101/531210. Preprint at. [DOI] [Google Scholar]
- 36.Blázquez L., Aiastui A., Goicoechea M., Martins de Araujo M., Avril A., Beley C., García L., Valcárcel J., Fortes P., López de Munain A. In vitro correction of a pseudoexon-generating deep intronic mutation in LGMD2A by antisense oligonucleotides and modified small nuclear RNAs. Hum. Mutat. 2013;34:1387–1395. doi: 10.1002/humu.22379. [DOI] [PubMed] [Google Scholar]
- 37.Shoji T., Konno S., Niida Y., Ogi T., Suzuki M., Shimizu K., Hida Y., Kaga K., Seyama K., Naka T., et al. Familial multifocal micronodular pneumocyte hyperplasia with a novel splicing mutation in TSC1: three cases in one family. PLoS One. 2019;14:e0212370. doi: 10.1371/journal.pone.0212370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green P.M., Bagnall R.D., Waseem N.H., Giannelli F. Haemophilia A mutations in the UK: results of screening one-third of the population. Br. J. Haematol. 2008;143:115–128. doi: 10.1111/j.1365-2141.2008.07310.x. [DOI] [PubMed] [Google Scholar]
- 39.Mercer T.R., Clark M.B., Andersen S.B., Brunck M.E., Haerty W., Crawford J., Taft R.J., Nielsen L.K., Dinger M.E., Mattick J.S. Genome-wide discovery of human splicing branchpoints. Genome Res. 2015;25:290–303. doi: 10.1101/gr.182899.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taggart A.J., Lin C.-L., Shrestha B., Heintzelman C., Kim S., Fairbrother W.G. Large-scale analysis of branchpoint usage across species and cell lines. Genome Res. 2017;27:639–649. doi: 10.1101/gr.202820.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pineda J.M.B., Bradley R.K. Most human introns are recognized via multiple and tissue-specific branchpoints. Genes Dev. 2018;32:577–591. doi: 10.1101/gad.312058.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vořechovský I. Aberrant 3′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2006;34:4630–4641. doi: 10.1093/nar/gkl535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keegan N.P., Wilton S.D., Fletcher S. Analysis of pathogenic pseudoexons reveals novel mechanisms driving cryptic splicing. Front. Genet. 2022;12:806946. doi: 10.3389/fgene.2021.806946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruun G.H., Doktor T.K., Borch-Jensen J., Masuda A., Krainer A.R., Ohno K., Andresen B.S. Global identification of hnRNP A1 binding sites for SSO-based splicing modulation. BMC Biol. 2016;14:54. doi: 10.1186/s12915-016-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during this study are available from ClinVar https://www.ncbi.nlm.nih.gov/clinvar/ and LOVD http://www.dmd.nl/.