Abstract
Percentage is widely used to describe different results in food microbiology, e.g., probability of microbial growth, percent inactivated, and percent of positive samples. Four sets of percentage data, percent-growth-positive, germination extent, probability for one cell to grow, and maximum fraction of positive tubes, were obtained from our own experiments and the literature. These data were modeled using linear and logistic regression. Five methods were used to compare the goodness of fit of the two models: percentage of predictions closer to observations, range of the differences (predicted value minus observed value), deviation of the model, linear regression between the observed and predicted values, and bias and accuracy factors. Logistic regression was a better predictor of at least 78% of the observations in all four data sets. In all cases, the deviation of logistic models was much smaller. The linear correlation between observations and logistic predictions was always stronger. Validation (accomplished using part of one data set) also demonstrated that the logistic model was more accurate in predicting new data points. Bias and accuracy factors were found to be less informative when evaluating models developed for percentage data, since neither of these indices can compare predictions at zero. Model simplification for the logistic model was demonstrated with one data set. The simplified model was as powerful in making predictions as the full linear model, and it also gave clearer insight in determining the key experimental factors.
Microbial data expressed as percentages have been modeled for many years. Percentage data may have very different biological meanings and expressions. In 1971, Genigeorgis et al. initiated the concept of probability for one cell to grow and produce toxin, presented as the ratio of RG over RI, where RG is the number of cells initiating growth, and RI is the number of cells in the inoculum (14). In a time-to-turbidity model, Whiting and Oriente (32) described the maximum probability of growth with the parameter Pmax, this value being obtained from fitting the growth curve with the logistic equation. Chea et al. modeled the extent of spore germination using the plateau value of the germination curve (6). The percent-growth-positive parameter describes the maximum proportion of wells that exhibited growth under various environmental conditions in a study using microplates inoculated with Clostridium botulinum spores (33).
A conventional approach applied to modeling percentage data is to use linear regression with polynomial terms. This method usually results in moderate (R2 < 0.9) (6, 9, 10, 17, 26, 31, 33) to poor (R2 < 0.5) (32) goodness of fit. Generally, the accuracy of linear models for modeling bounded variables (e.g., percentage data) is not as good as for other unbounded variables obtained in the same experiment, and the resulting linear model also predicts poorly at values close to 0 and 1 (6, 32, 33). An insurmountable limitation of the linear approach is that the model can predict percentages outside the probability range, i.e., values of <0 or >1 (6, 26, 31, 33). Generally, all predicted negative values are forced to 0, and those >1 are forced to 1. Even without this modification, it is not meaningful to compare these conditions. For example, 120% cannot be interpreted as a higher percent germination than 101%.
Logistic regression has been widely used in medical research (1, 5, 18, 19, 22, 30). In the field of predictive food microbiology, logistic models have been developed to describe the bacterial growth/no growth interface (4, 21, 24, 25). In these models, the data were presented in the 0-1 format, as in a typical binomial data set. Genigeorgis et al. first presented the concept of the probability that one cell could grow in a specific environment (14). Later, this probability was modeled in various systems using logistic regression combined with a linear regression of the lag period (3, 11, 12, 15, 16, 20). Roberts et al. used a similar concept and the regression approach to model toxin production by C. botulinum in pasteurized pork slurry (27). Cole et al. modeled the probability of growth of spoilage yeast in a model fruit drink by directly relating the logit of probability with the environmental factors (7). In these studies, probability (a continuous number between 0 and 1) instead of a dichotomous variable (i.e., 0, 1) was modeled. As pointed out by Ratkowsky and Ross (25), the response modeled by logistic regression at a given combination of limiting factors can either have a value of 0 or 1 or be a probability. Probability, generally expressed by dividing the number of successes by the total number of trials, is simply a summarization of binomial data and thus can be approximated by a logistic general linear model (8).
In this study, we compared the goodness of fit of linear regression to logistic regression for modeling percentages. We modeled data from our own research and from the literature (including publications from our group) and developed models using both the logistic and linear approaches in exactly the same manner. Five different approaches were used to compare the goodness of fit of the two models. In almost all cases, the logistic models displayed greater accuracy and resulted in less biased predictions.
MATERIALS AND METHODS
Data collection.
Four different sets of percentage data were collected from previous experiments (6, 26, 32, 33). Each set had its own unique biological meaning and was collected with a different method.
Weight is the degree of emphasis a model puts on an observation. The weight for a percentage datum point is the total number of observations associated with this percentage (2). For example, when 10 of 40 tubes turn turbid, the percentage is 25% (10/40) and the weight for this percentage is 40. The assignment of weights was determined differently for each data set, as described below.
Data set I: data for percent-growth-positive were collected by Zhao et al. (33). This data set contained the exact numbers of wells that showed growth and no growth. The total number of wells in each condition is the same, so the weight assigned for each condition is the same. Environmental factors studied were pH, sodium chloride concentration, and inoculum size in a complete 3 by 3 factorial design with a total of 27 different conditions.
Data set II: extent of germination data were collected by Chea et al. (6). The total number of spores studied for each condition was between 200 and 300. The small difference in the total number in each condition is negligible, and equal weight for all the data points was assumed in logistic regression. Environmental factors studied were pH, sodium chloride concentration, and temperature in a complete 3 by 3 factorial design with a total of 27 different conditions.
Data set III: Razavilar and Genigeorgis studied the probability of one cell of Listeria monocytogenes to grow, as affected by sodium chloride concentration, time, and temperature (26). Weights were not obtainable, so this parameter was assumed to be the same in each case.
Data set IV: Pmax was the parameter used to indicate the maximum fraction of positive tubes inoculated with C. botulinum (32). It was obtained by fitting the experimental data with a logistic equation. The total number of tubes varied by condition and was used as the weight in logistic regression. Four environmental factors, pH, sodium chloride concentration, temperature, and inoculum size, were studied in a total of 103 different conditions. A subset, containing 22 data points at 19°C, was not used to develop models; instead, these data points were used later to validate the models developed from the remaining 81 points.
Modeling with linear and logistic regression.
Both linear and logistic models were developed in S-plus (MathSoft, Inc., Seattle, Wash.) for an objective comparison. The generalized linear modeling (“glm”) function was used for both methods. The link function for logistic regression is “binomial” and for linear regression is “gaussian.” The full models generated by each approach, with the same number of terms in the same format, were used to ensure the validity of the comparison.
The linear model with three predictor variables has the following general format:
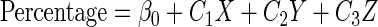 |
1 |
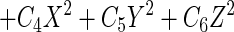 |
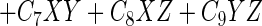 |
where Percentage is the observed percentage, β0 is the intercept, X, Y, and Z are the predictor variables, and Cis are the coefficients.
The logistic model with three predictor variables has the following general format:
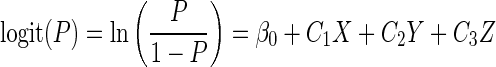 |
2 |
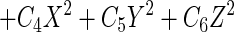 |
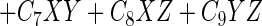 |
where P is the probability that the event would occur according to the model and the remaining symbols have the same meaning as in equation 1.
Model comparison. (i) Adjustment with predictions from linear models.
Predictions from linear models can be greater than 1 or less than 0. In practice, these predictions are generally forced to be 1 and 0, respectively (6, 26, 32, 33). To make the comparison of the two models fairer, predictions from linear models were forced into the range of 0 to 1 in this manner. For all comparisons, the modified predictions from linear models were used, except as noted below.
(ii) Methods to compare model predictions.
Out-of-range predictions from linear models were counted in Method 1. The number of predictions from logistic regression that were closer to the observed values was also calculated. For this calculation, the absolute value of the difference (predicted minus observed) was used. We excluded some observations whose linear regression predictions were out of range in the calculation of the percentage of closer predictions. This is required because logistic regression predicts strictly between 0 and 1. By forcing out-of-range linear predictions to be 0 or 1, we may inappropriately make some linear predictions seem better. For example, if the observation is 1, the logistic prediction is 0.999, and the linear prediction is 1.235, if we force the linear prediction to be 1, it will falsely be judged better.
In Method 2, we compared the ranges of the differences between the predicted and the observed values. Point summaries of the differences (predicted minus observed), i.e., minimum, first quarter, median, mean, third quarter, and maximum, were obtained, and the range and interquarter range (IQR) were calculated.
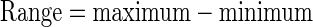 |
3 |
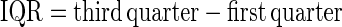 |
4 |
The smaller the values of the range and IQR, the closer the predictions are to the observations. The range is sensitive to outlying points whose predicted and observed values are very different, while the IQR is not affected as much.
For Method 3, the deviation of the model from observations was calculated as follows:
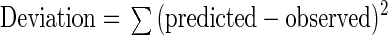 |
5 |
The smaller the deviation, the closer the model predictions were to the observations. Method 1 cannot detect predictions that are far from the observations. Method 2 allows for detection of these wide deviations by measuring the range of the differences between the predicted and observed values, but it is unable to indicate which model results in a greater number of predictions closer to the observed. Method 3 takes both considerations into account.
Method 4 used graphs of the observed values (x axis) versus predicted values (y axis) from both models. A simple linear regression was fitted to the points, and the intercept, the slope, and R2 were obtained. If the predictions are in perfect agreement with the observed values, the intercept should be 0, the slope should be 1, and R2 should be 1. The closer the intercept is to 0, the slope is to 1, and R2 is to 1, the better is the general predictive power of the model. A slope of less than 1 indicates that the model underpredicts the observation.
Method 5 used bias and accuracy values as a quantitative way to measure the goodness of fit of the models (6, 28). The bias factor indicates by how much, on average, a model overpredicts (bias factor > 1) or underpredicts (bias factor < 1) the observed data.
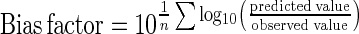 |
6 |
The accuracy factor indicates by how much the predictions differ from the observed data.
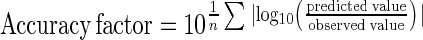 |
7 |
In both equations, n is the number of observations used in the calculation. In a perfect model, both the bias and accuracy factors are equal to 1.
Simplification of the logistic model.
Data from Chea et al. (6) were used to demonstrate the procedure for reducing the number of parameters in the logistic model and to show how better physiological insight into the experiment might be derived from the reduced model.
RESULTS
Data set I: percent-growth-positive.
Thirty percent of the predictions from linear regression are out of the 0 to 1 range (Table 1). Fifteen predictions from the logistic model are closer to the observed. Seven linear predictions were inaccurately made better by forcing predictions over 1 to 1, and one condition was made falsely better by forcing the prediction lower than 0 to 0. The percentage better predicted by logistic regression is calculated by excluding these data points:
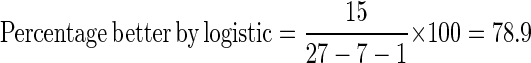 |
8 |
The range of the differences (predicted minus observed) from logistic regression is more than 2.5 times smaller than that from linear regression. The IQR from logistic regression is about one-third of that from linear regression. The deviation value of the logistic model is more than 8 times smaller.
TABLE 1.
Comparison of results for linear and logistic regressions with five different methods in four different data sets
| Method no. | Parameter | Data set I (n = 27)
|
Data set II (n = 27)
|
Data set III (n = 28)
|
Data set IV-model (n = 81)
|
Data set IV-validation (n = 22)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Linear | Logistic | Linear | Logistic | Linear | Logistic | Linear | Logistic | Linear | Logistic | ||
| 1 | No. of predictions >1 | 8 | 0 | 3 | 0 | 4 | 0 | 8 | 0 | 2 | 0 |
| No. of predictions <0 | 1 | 0 | 4 | 0 | 5 | 0 | 9 | 0 | 0 | 0 | |
| No. of predictions of logistic model closer to observed | NAa | 15 | NA | 20 | NA | 19 | NA | 52 | NA | 18 | |
| % of predictions of logistic model closer to observed | NA | 78.9 | NA | 87.0 | NA | 100 | NA | 78.8 | NA | 81.8 | |
| 2b | Mininum | −0.239 | −0.079 | −0.244 | −0.071 | −0.333 | −0.009 | −0.467 | −0.503 | −0.455 | −0.255 |
| 1st quarter | −0.037 | −0.016 | −0.090 | −0.017 | −0.055 | 0.000 | −0.162 | −0.093 | −0.119 | −0.064 | |
| Median | 0.000 | 0.000 | 0.002 | 0.005 | 0.000 | 0.000 | 0.000 | −0.000 | 0.013 | 0.018 | |
| Mean | −0.010 | 0.000 | 0.002 | 0.000 | 0.008 | 0.000 | −0.021 | −0.019 | 0.057 | 0.041 | |
| 3rd quarter | 0.026 | 0.006 | 0.079 | 0.012 | 0.082 | 0.000 | 0.098 | 0.019 | 0.276 | 0.178 | |
| Maximum | 0.205 | 0.092 | 0.368 | 0.109 | 0.414 | 0.010 | 0.475 | 0.382 | 0.589 | 0.396 | |
| Range | 0.444 | 0.171 | 0.612 | 0.180 | 0.747 | 0.019 | 0.942 | 0.885 | 1.044 | 0.651 | |
| IQR | 0.063 | 0.022 | 0.169 | 0.029 | 0.137 | 0 | 0.26 | 0.112 | 0.395 | 0.242 | |
| 3c | Deviation | 0.310 | 0.038 | 0.491 | 0.029 | 0.899 | 0.000 | 3.385 | 1.551 | 1.454 | 0.665 |
| 4d | Intercept | 0.153 | 0.016 | 0.105 | 0.006 | 0.122 | 0.000 | 0.123 | 0.056 | 0.326 | 0.133 |
| Slope | 0.795 | 0.980 | 0.812 | 0.990 | 0.736 | 0.999 | 0.736 | 0.862 | 0.510 | 0.833 | |
| R2 | 0.906 | 0.987 | 0.902 | 0.994 | 0.874 | 1 | 0.750 | 0.899 | 0.620 | 0.819 | |
| 5 | Bias | 1.022 | 0.996 | 0.993 | 0.990 | 1.385 | 0.893 | 1.288 | 0.891 | 1.005 | 1.012 |
| Accuracy | 1.133 | 1.046 | 1.204 | 1.053 | 1.735 | 1.141 | 1.320 | 0.944 | 1.344 | 1.288 | |
| % 0s in observation | 0.074 | 0.259 | 0.357 | 0.309 | 0.273 | ||||||
NA, not applicable.
Differences of values (predicted − observed) were summarized; range = maximum − minimum; IQR = third quarter − first quarter.
Deviation of the model from observation was calculated as Σ(predicted − observed)2.
Linear regression was done between predicted and observed values. A perfect model would have an intercept value of 0, a slope of 1, and an R2 value of 1.
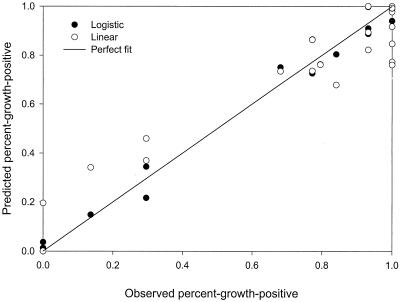
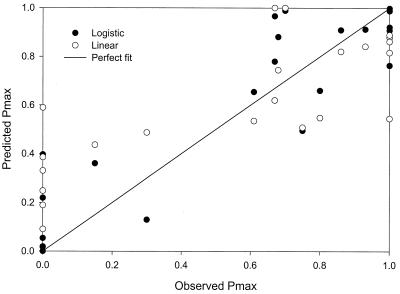
Predictions from logistic regression are much better than those from linear regression over the entire range and especially at points closer to 1 and 0 (Fig. 1). Three predictions by the linear model, each with an observation of 1, are 0.761, 0.773, and 0.848, while the logistic predictions are much better: 0.941, 0.990, and 0.999. The two 0 observations were predicted by the linear model to be 0.195 and <0, while the logistic predictions were 0.036 and 0.013. Another observation at the lower range, 0.136, was predicted to be 0.341 and 0.148 by linear regression and logistic regression, respectively. The fitted line for the predicted values from logistic regression was very close to a perfect fit (Table 1). The fitted line for the linear model predictions versus the observations was considerably worse, with a slope of about 0.8, suggesting systematic underprediction. The bias and accuracy factors for logistic regression are slightly closer to 1 than those for linear regression (Table 1).
FIG. 1.
Goodness of fit of linear regression and logistic regression for C. botulinum percent-growth-positive (Data set I) from Zhao et al. (33).
Data set II: germination extent.
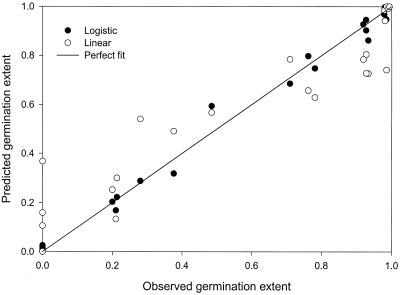
Approximately 26% of the linear predictions are out of range (Table 1). Approximately 87% of the predictions from logistic regression are closer to the observed values. The range of the differences (predicted minus observed) from the logistic model is less than one-third that from the linear model, and the IQR is almost one-sixth that from the linear model. The deviation value of the logistic model is 17 times smaller.
The line fitted to the predicted values from the logistic model compared to observed values is very close to the perfect fitting line (Fig. 2, Table 1). The fitted line for predictions from the linear model had a slope of only 0.812, suggesting underprediction (Table 1). Three of seven observed values of 0 had higher linear predictions, at 0.106, 0.159, and 0.368, while the remaining four predictions were negative. All seven logistic predictions are very close to 0, with the largest being 0.025. Three higher observations, 0.927, 0.933, and 0.985, were predicted to be 0.805, 0.727, and 0.742 by the linear model, while logistic regression produced much more accurate predictions at 0.946, 0.862, and 0.949, respectively. The bias factors for the two models are almost the same, and the logistic model is slightly more accurate as judged by the accuracy factor (Table 1).
FIG. 2.
Goodness of fit of linear regression and logistic regression for C. botulinum germination extent (Data set II) from Chea et al. (6).
Data set III: probability of one cell of Listeria monocytogenes to grow.
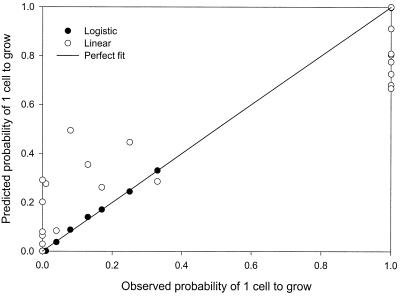
This is a very special data set since 21 of 28 data points are either 0 or 1. Results demonstrated that logistic regression is a much more powerful tool when modeling this type of data set.
Approximately 32% of the linear predicted values are out of range. All observations are predicted better by logistic regression. The range of the differences (predicted minus observed) from the logistic model is more than 39-fold smaller. The IQR and deviation value from the logistic model are also much smaller than those from the linear model (Table 1).
The fitting parameters for the predicted versus the observed values from logistic regression are very good (Table 1). Predictions from the linear model are worse. The majority of the linear predictions fall far from the perfect fit line, indicating substantial deviation of predicted values from observed values (Fig. 3). The bias and accuracy factors from logistic regression are better, but not substantially so, as indicated by the other comparison approaches.
FIG. 3.
Goodness of fit of linear regression and logistic regression for probability of one Listeria monocytogenes cell to grow (Data set III) from Razavilar and Genigeorgis (26).
Data set IV: maximum fraction of positive tubes.
There are 103 observations in total (32). Eighty-one data points were used to develop the models, and the remaining 22 (all at 19°C) were used to test the predictive power of the models for new data.
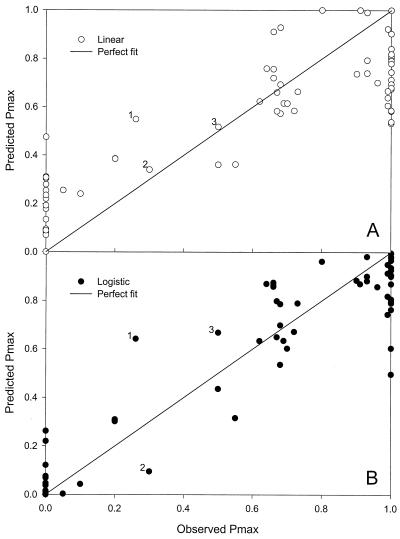
Approximately 21% of the predictions from the linear model are out of range. Seventy-eight percent of the predictions from logistic regression are closer to the observed values. The minimum of the differences (predicted minus observed) from the logistic model is slightly lower. The range from the two models is approximately the same, while the IQR from the logistic model is less than half of that from the linear model. This suggests that both models have predictions far away from the observed values, but if these outlying values are excluded, the logistic model has much better predictive power. The deviation value from the logistic model is less than half of that from the linear model (Table 1).
Although it is not immediately clear from a visual inspection of Fig. 4, the logistic model is better, as evidenced by an intercept closer to 0, a slope closer to 1, and a much higher R2 (Table 1). A closer examination of Fig. 4 shows that at low (0.0 to 0.2) and high (0.8 to 1.0) values, predictions from the logistic model are closer to observations in general. For 0 observations, most predictions (22 of 25) from the logistic model were <0.1, while 11 predictions from the linear model were >0.1, and 9 predictions were <0. Of the 22 observations that resulted in a value of 1, 17 logistic predictions were >0.8, while 11 from the linear model were <0.8 and 8 were >1. In the middle range (0.2 to 0.8), predictions from the two models were comparable, with linear predictions occasionally closer to the observed. In only 3 (labeled 1, 2, 3) of the 19 conditions in the middle range were the linear predictions substantially better than the corresponding logistic predictions (Fig. 4). Bias and accuracy factors from logistic regression are closer to 1 than those from the linear model (Table 1).
FIG. 4.
Goodness of fit of linear regression and logistic regression for maximum fraction of positive tubes inoculated with C. botulinum (Data set IV) from Whiting and Oriente (32). (A) Linear model. (B) Logistic model. Three points in the 0.2 to 0.8 range, numbered in both panels, were substantially better predicted by linear model.
The above analysis shows that logistic models are almost always better at predicting the values used to develop that model. It is more important, however, to be able to predict new observations not used to create the model. Models should not be used for conditions outside the range studied (23). Based on this principle, we picked one temperature (19°C) from the seven temperatures studied (5 to 28°C) and did not use it in the model development. We then used the models to predict the data at the 19°C temperature.
Two of 22 predictions from the linear model are out of range. More than 80% of the values were better predicted by logistic regression. Both the range and the IQR from logistic regression were more than 1.5-fold smaller. The parameter derived from fitting the predicted to the observed values also indicated that logistic regression was much better at predicting data not included in developing the model. The deviation value of the logistic model is less than half that of the linear model (Fig. 5; Table 1). Bias and accuracy factors for the two models exhibit little difference.
FIG. 5.
Validation of linear regression and logistic regression for maximum fraction of positive tubes inoculated with C. botulinum (Data set IV) from Whiting and Oriente (32). Both models were validated by 22 of 103 conditions not used in model development.
Reducing the logistic model.
The logistic model developed for Data set II (6) was simplified using stepwise selection in S-plus 2000. Two parameters, pH and sodium chloride concentration, were retained. Approximately 58% of the time, this model predicts closer to the observed value than does the full linear model. The differences (predicted minus observed) from this reduced model have a slightly larger range but a smaller IQR. This model has a slightly smaller R2 but an intercept closer to 0 and a slope closer to 1. In general, the reduced 2-parameter logistic model has a predictive power very similar to that of the full 9-parameter linear model.
DISCUSSION
Limitation of bias and accuracy factors in evaluating goodness of fit.
We used five different methods to compare the goodness of fit of the models. Our analysis shows good agreement among the first four methods. Bias and accuracy factors on the other hand sometimes indicate a close degree of goodness of fit between the two models, while the other four approaches indicate substantially better fit by logistic regression (Data set I, Data set II, and Data set IV-validation). Bias and accuracy factors also indicate that logistic regression is only moderately good in the case of Data set III (bias factor = 0.89, accuracy factor = 1.14), while the other four methods indicate the clear superiority of the logistic model.
The discrepancy between bias and accuracy factors with the other four methods is due to differences in the treatment of observation values of 0 in the data set. The first four methods all take 0 observations into consideration. However, because the observed value is in the denominator of both equation 6 and equation 7, neither bias factor nor accuracy factor can be used to compare predictions when 0 is observed. Zero was frequently observed in percentage data: 7% in Data set I, 26% in Data set II, 36% in Data set III, 31% in Data set IV-model development, and 27% in Data set IV-validation (Table 1). One major disadvantage of the linear model is that it predicts far less accurately at values closer to 0 or 1. Due to the limitation of their formulae, bias and accuracy factors cannot reflect this. As a result, bias and accuracy factors show that logistic regression is only slightly better or equal to linear regression for the four data sets studied. Similarly, the bias factor loses its meaning of an overall measurement of overprediction or underprediction for percentage data. If the bias factor is 1, a conclusion as to whether or not the model is good cannot be drawn without first checking predictions at 0.
Predictions at the two extremes.
In addition to always making biologically meaningful predictions, logistic regression makes it possible to compare conditions at very low and very high ranges. Linear regression often makes out-of-range predictions at these ranges. For example, a germination probability of −0.999 cannot be interpreted as less likely than a probability of −0.001. On the other hand, out-of-range prediction is not a problem for logistic regression, which, by definition, predicts strictly between 0 and 1. For linear models, predictions greater than 1 occasionally occur when observations are in the middle range as well as when very high. The two >1 predictions for Data set IV-validation had observed values of 0.67 and 0.70.
Reducing the logistic model.
Many times, especially when the data set is not very large, a reduced model is more desirable than the full model (13). Moreover, a reduced model can give better (or at least more straightforward) insight into the physiological factors influencing the experiment. Logistic models can be reduced in a fashion similar to that of linear models (29). As an example, the logistic model developed for Data set II (6) was simplified using the stepwise method in S-plus 2000.
The reduced model gives clear insight as to which environmental factors influence the germination extent most under the conditions studied: pH and salt concentration. As a comparison, the reduced linear model has terms for pH, NaCl, temperature, pH2, and interaction between temperature and pH (6), which results in a far less clear picture.
Why logistic regression is better.
We have demonstrated using four different data sets that logistic regression is better than linear regression for modeling percentage data. This approach can be extended to any data set that is presented as percentages. This is true because percentage is a simple and convenient way to present binomial data, and logistic regression, not linear regression, should be used for binomial data.
When linear regression is used for binary data, three problems arise (23): the variance of the error term is not constant, the error term is not normally distributed, and there is no restriction requiring the prediction to fall between 0 and 1. The first problem can be handled by using weighted least-square regression. When the sample size is very large, the method of least squares provides estimators that are asymptotically normal under fairly general regulations, even when the distribution of the error term is far from normal. The third problem is insurmountable (23).
Logistic regression may seem much more complicated than its linear counterpart. Yet most statistical software packages can do logistic regression with no more effort than linear regression. However, it is not as easy and straightforward to interpret the coefficients and test for goodness of fit of logistic models. There is no R2 associated with a logistic model, since a residual in the commonly accepted sense does not exist. In linear regression, R2 can be easily obtained and is often used to evaluate the goodness of fit of models (6, 23), but R2, despite its usefulness, is not a “gold standard.” Approaches such as the deviance test and graphical tools such as index plot and half-normal plot can be readily employed to evaluate the goodness of fit of logistic models (23).
In conclusion, logistic regression was demonstrated to be a better approach than linear regression to model percentage. It has the inherent advantage of always making biologically meaningful predictions, and in most of the cases it also predicts closer to the observations. We believe that logistic and not linear regression should be used whenever the observations are presented as percentages, even though under certain circumstances linear models may give acceptable goodness of fit.
ACKNOWLEDGMENTS
We thank Thomas J. Montville and Kristin Jackson in Department of Food Science, Rutgers University, for help and support with the manuscript. Thanks also to Minge Xie, Department of Statistics, Rutgers University, for suggestions and help with the statistical discussion section of this paper.
This work was supported by funds from the USDA National Research Initiative (96-35201-3458), the U.S. Hatch Act, and the state of New Jersey.
REFERENCES
- 1.Abbott R D. Logistic regression in survival analysis. Am J Epidemiol. 1985;121:465–471. doi: 10.1093/oxfordjournals.aje.a114019. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. Splus 2000, guide to statistics. Vol. 1. Seattle, Wash: MathSoft; 1999. [Google Scholar]
- 3.Baker D A, Genigeorgis C A, Glover J, Razavilar V. Growth and toxigenesis of C. botulinum type E in fishes packaged under modified atmospheres. Int J Food Microbiol. 1990;10:269–290. doi: 10.1016/0168-1605(90)90075-g. [DOI] [PubMed] [Google Scholar]
- 4.Bolton L F, Frank J F. Defining the growth/no-growth interface for Listeria monocytogenes in Mexican-style cheese based on salt, pH, and moisture content. J Food Prot. 1999;62:601–609. doi: 10.4315/0362-028x-62.6.601. [DOI] [PubMed] [Google Scholar]
- 5.Brazer S R, Pancotto F S, Long III T T, Harrell F E, Jr, Lee K L, Tyor M P, Pryor D B. Using ordinal logistic regression to estimate the likelihood of colorectal neoplasia. J Clin Epidemiol. 1991;44:1263–1270. doi: 10.1016/0895-4356(91)90159-7. [DOI] [PubMed] [Google Scholar]
- 6.Chea F P, Chen Y, Montville T J, Schaffner D W. Modeling the germination kinetics of Clostridium botulinum 56A spores as affected by temperature, pH and sodium chloride. J Food Prot. 2000;63:1071–1079. doi: 10.4315/0362-028x-63.8.1071. [DOI] [PubMed] [Google Scholar]
- 7.Cole M B, Franklin J G, Keenan M H J. Probability of growth of the spoilage yeast Zygosaccharomyces bailii in a model fruit drink system. Food Microbiol. 1987;4:115–119. [Google Scholar]
- 8.Dobson A J. An introduction to statistical modelling. London, United Kingdom: Chapman and Hall Ltd.; 1990. [Google Scholar]
- 9.Dodds K L. Combined effect of water activity and pH on inhibition of toxin production by Clostridium botulinum in cooked, vacuum-packed potatoes. Appl Environ Microbiol. 1989;55:656–660. doi: 10.1128/aem.55.3.656-660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodds K L. An introduction to predictive microbiology and the development and use of probability models with Clostridium botulinum. J Ind Microbiol. 1993;12:139–143. [Google Scholar]
- 11.Garcia G, Genigeorgis C A. Quantitative evaluation of Clostridium botulinum nonproteolytic types B, E, and F growth risk in fresh salmon tissue homogenates stored under modified atmosphere. J Food Prot. 1987;50:390–397. doi: 10.4315/0362-028X-50.5.390. [DOI] [PubMed] [Google Scholar]
- 12.Garcia G W, Genigeorgis C A, Lindroth S. Risk of growth and toxin production by Clostridium botulinum non-proteolytic types B, E, and F in salmon fillets stored under modified atmospheres at low and abused temperatures. J Food Prot. 1987;50:330–336. doi: 10.4315/0362-028X-50.4.330. [DOI] [PubMed] [Google Scholar]
- 13.Gauch H G. Prediction, parsimony and noise. Am Sci. 1993;81:468–478. [Google Scholar]
- 14.Genigeorgis C, Martin S, Frantti C E, Riemann H. Initiation of staphylococcal growth in laboratory media. Appl Microbiol. 1971;21:934–939. doi: 10.1128/am.21.5.934-939.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genigeorgis C A, Meng J, Baker D A. Behavior of nonproteolytic Clostridium botulinum type B and E spores in cooked turkey and modeling lag phase and probability of toxigenesis. J Food Sci. 1991;56:373–379. [Google Scholar]
- 16.Ikawa J Y, Genigeorgis C A. Probability of growth and toxin production by nonproteolytic Clostridium botulinum in rockfish fillets stored under modified atmospheres. Int J Food Microbiol. 1987;4:167–181. doi: 10.4315/0362-028X-50.4.330. [DOI] [PubMed] [Google Scholar]
- 17.Jensen M J, Genigeorgis C A, Lindroth S. Probability of growth of Clostridium botulinum as affected by strain, cell and serologic type, inoculum size and temperature and time of incubation in a model system. J Food Safety. 1987;8:109–126. [Google Scholar]
- 18.Kellett J. Early diagnosis of acute myocardial infarction by either electrocardiogram or a logistic regression model: portability of a predictive instrument of acute cardiac ischemia to a small rural coronary care unit. Can J Cardiol. 1997;13:1033–1038. [PubMed] [Google Scholar]
- 19.Lin K K, Reschke M F. The use of the logistic model in space motion sickness prediction. Aviat Space Environ Med. 1987;58:A9–A15. [PubMed] [Google Scholar]
- 20.Lindroth S E, Genigeorgis C A. Probability of growth and toxin production by nonproteolytic Clostridium botulinum in rockfish stored under modified atmospheres. Int J Food Microbiol. 1986;3:167–181. [Google Scholar]
- 21.Lopez-Malo A, Guerrero S, Alzamora S M. Probabilistic modeling of Saccharomyces cerevisiae inhibition under the effects of water activity, pH, and potassium sorbate concentration. J Food Prot. 2000;63:91–95. doi: 10.4315/0362-028x-63.1.91. [DOI] [PubMed] [Google Scholar]
- 22.Lu W, Bailey J M. Reliability of pharmacodynamic analysis by logistic regression—a computer stimulation study. Anesthesiology. 2000;92:985–992. doi: 10.1097/00000542-200004000-00015. [DOI] [PubMed] [Google Scholar]
- 23.Neter J, Kutner M H, Nachtsheim C J, Wasserman W. Applied linear regression models. Chicago, Ill: The McGraw-Hill Companies, Inc.; 1996. [Google Scholar]
- 24.Presser K A, Ross T, Ratkowsky D A. Modelling the growth limits (growth/no growth interface) of Escherichia coli as a function of temperature, pH, lactic acid concentration, and water activity. Appl Environ Microbiol. 1998;64:1773–1779. doi: 10.1128/aem.64.5.1773-1779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratkowsky D A, Ross T. Modeling the bacterial growth/no growth interface. Lett Appl Microbiol. 1995;20:29–33. [Google Scholar]
- 26.Razavilar V, Genigeorgis C. Prediction of Listeria spp. growth as affected by various levels of chemicals, pH, temperature and storage time in a model broth. Int J Food Microbiol. 1998;40:149–157. doi: 10.1016/s0168-1605(98)00014-2. [DOI] [PubMed] [Google Scholar]
- 27.Roberts T A, Gibson A, Robinson A. Prediction of toxin production by Clostridium botulinum in pasteurized pork slurry. J Food Technol. 1981;16:337–355. [Google Scholar]
- 28.Ross T. Indices for performance evaluation of predictive models in food microbiology. J Appl Bacteriol. 1996;81:501–508. doi: 10.1111/j.1365-2672.1996.tb03539.x. [DOI] [PubMed] [Google Scholar]
- 29.Venables W N, Ripley B D. Modern applied statistics with S-plus. New York, N.Y: Springer-Verlag New York, Inc.; 1999. [Google Scholar]
- 30.Virtanen A, Gomari M, Kranse R, Stenman U. Estimation of prostate cancer probability by logistic regression: free and total prostate-specific antigen, digital rectal examination, and heredity are significant variables. Clin Chem. 1999;45:987–994. [PubMed] [Google Scholar]
- 31.Whiting R C, Call J E. Time of growth model for proteolytic Clostridium botulinum. Food Microbiol. 1993;10:295–301. [Google Scholar]
- 32.Whiting R C, Oriente J C. Time-to-turbidity model for non-proteolytic type B Clostridium botulinum. Int J Food Microbiol. 1997;36:49–60. doi: 10.1016/s0168-1605(96)01244-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhao L, Montville T J, Schaffner D W. Inoculum size of Clostridium botulinum 56A spores influence time-to-detection and percent growth-positive sample. J Food Sci. 2000;65:1369–1375. [Google Scholar]