Abstract
In this invited article, we explain various technical aspects of the LCMV system, providing an updated overview of a prior contribution by Matthias von Herrath and J. Lindsay Whitton. We provide an explanation of the LCMV infection models, highlighting the importance of selecting an appropriate route and viral strain. We also describe how to quantify virus-specific immune responses, followed by an explanation of the challenge models and transgenic systems. All of the reagents and tools listed here are commercially available, or can be obtained through the European Virus Archive (EVA). The corresponding author, Pablo Penaloza, will be able to answer additional technical questions about the LCMV system. Specifically, our article will focus on the following protocols.
Basic Protocol 1: LCMV infection routes
Basic Protocol 2: Measurement of T cell and antibody responses
Basic Protocol 3: Challenge models
Basic Protocol 4: Transgenic models
Keywords: LCMV model, immune memory, immune exhaustion, acute viral infection, chronic viral infection
INTRODUCTION
LCMV was discovered in 1933 by Charles Armstrong in patients with aseptic encephalitis. It is common in house mice and it has been extensively used for solving various questions in immunology. LCMV is an ambisense RNA virus with a non-cytolytic life cycle, and thus, most of the disease caused by LCMV is mediated by the host T cell response. The LCMV system is especially well-poised to answer questions about T cell and B cell regulation. Although there are many strains of LCMV, including the WE, Docile, Pasteur, UBC and Traub strains, we will focus on the most widely used strains, which include the acutely cleared Armstrong strain, and the chronic Clone 13 (Cl-13) strain. A useful mnemonic is to remember the first letter of each viral strain (Armstrong is acutely cleared, whereas Cl-13 is chronic). These two strains differ by only two mutations, including a mutation in the RNA polymerase gene and another one in the glycoprotein (Ahmed, Salmi, Butler, Chiller, & Oldstone, 1984a; Bergthaler et al., 2010; Smelt et al., 2001). These mutations do not affect the T cell epitopes between these two strains, allowing elegant comparison of functional versus dysfunctional T cell responses specific for the same antigen.
For example, a prior study comparing virus-specific CD8 T cells of the same specificity (following Armstrong versus Cl-13 infection) identified PD-1 as a critical regulator of T cell exhaustion (Barber et al., 2006; Wherry et al., 2007). This immune checkpoint was later demonstrated to also be increased on exhausted T cells of humans chronically infected with HIV (Day et al., 2006), demonstrating that findings with chronic LCMV could be widely generalizable to other chronic viral infections in humans.
In this invited article, we will describe several techniques that are commonly used in the LCMV system to measure adaptive immune responses. We provide specific catalog numbers for some reagents that work well in our hands, but reagents from other vendors may work too. In summary, this article includes protocols for infecting mice with LCMV (Basic Protocol 1) and measuring adaptive immune responses (Basic Protocol 2). Protocols to assess immune protection in vivo are also included (Basic Protocol 3). Finally, we list several transgenic mouse models to evaluate LCMV-specific immunity (Basic Protocol 4).
Biosafety Considerations
LCMV belongs to the family Arenaviridae, which are bisegmented ambisense RNA viruses. These viruses are classified into Old World and New World viruses, based on geographic and genetic differences. LCMV is an Old World arenavirus. Other Old World arenaviruses include Lujo and Lassa virus, which causes severe hemorrhagic infections in humans. LCMV may pose health concerns in immunocompromised individuals and pregnant women, and it is transmitted by direct contact of mouse fluids and droppings with mucous membranes or breaks in the skin, or through inhalation. In case of accidental exposure, such as a needlestick accident, the relevant institutional safety office must be notified. In normal immunocompetent humans, LCMV infection caused by accidental laboratory exposure is typically resolved within days or weeks without the need of Ribavirin treatment, which is sometimes avoided due to toxic side effects. LCMV infection could be suspected if the investigator experiences flu-like symptoms, usually within 1–2 weeks of exposure.
Additional precautions must be taken when concentrating the virus and when working with newly isolated strains of LCMV. Many mouse LCMV strains, including the Armstrong and Cl-13 strains, are categorized as BSL-2. Since LCMV may increase its virulence in hamsters, BSL-3 conditions are recommended if working with infected hamsters. However, propagation of LCMV is typically done in hamster cells (BHK21), without increase in virulence. LCMV can be inactivated by heat (temperatures higher than 56°C), by exposure to extremes of pH, or by irradiation. It is recommended to inactivate virus by disposing it in 10% bleach.
Basic Protocol 1: LCMV infection routes in mice
The outcome of an LCMV infection varies with the viral strain, dose, and route of infection. The optimal viral strain and infection route would depend on the experimental question. For example, if the experimental question pertains to immune memory, it is customary to immunize mice intraperitoneally with LCMV Armstrong at a dose of 2×105 plaque forming units (PFU). This would result in a rapidly controlled infection lasting a week.
However, if the experimental question pertains to immune exhaustion, it is customary to utilize LCMV Cl-13 at a dose of 2×106 PFU, administered intravenously. Deviations from the above recommendations are sometimes useful. For instance, if one wants to evaluate acute control of an LCMV Armstrong infection, it is recommended to infect mice with a high intravenous dose (2×106 PFU). Using this high intravenous dose of LCMV Armstrong would still result in viral clearance (albeit delayed 1–2 days, with mice exhibiting higher acute viral loads). This would allow for greater “resolution” and consistency in viral load quantification, as compared to the 2×105 PFU dose.
All protocols using live mice must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) or must conform to governmental regulations.
| Materials: |
|---|
| LCMV viral stock of known titer (for example 107 PFU/mL) (see Support Protocols 1, 2.1 and 2.2) |
| RPMI or Phosphate-buffered saline (PBS) (Gibco, Cat#14190–144) |
| Adult (6–8-week-old) C57BL/6 mice or any appropriate strain |
| Anesthetic: e.g. ketamine/xylazine (if performing intracranial infections) |
| 1-ml syringes and 25- to 27-G needles |
| Mouse Restrainer (if performing intravenous infections) |
Preparing virus working stocks for mouse infections
-
Thaw an aliquot of viral stock (1 ml containing 10^7 to 10^8 PFU/mL viral concentration.) in a 37°C water bath, and place on ice immediately.
Any unused stock virus should be discarded in accordance with biosafety requirements; it should not be refrozen, as viral titer would be substantially reduced upon secondary thawing. LCMV is labile and titers may decline even if the virus is left on ice for several hr.
-
Dilute viral stock accordingly, using either PBS or plain RPMI as diluent.
For example, if an LCMV Armstrong stock is at 107 PFU/mL, and the goal is to infect mice intraperitoneally with 2×105 PFU, one could simply dilute 1 mL of the viral stock in a total volume of 25 mL to generate a 4×105 PFU/mL stock. Then, one could inject each mouse with 500 μL of this diluted stock (2×105 PFU). Using smaller volumes for intraperitoneal or intravenous injections may result in greater variability in terms of viral loads. Normally, mice can excrete this high volume within 1 hr.
Keep virus on ice at all times and inject all mice within 1–2 hr to prevent decline in viral titer.
Perform LCMV infections, depending on the desired routes of infection.
CAUTION: Do not recap used needles for safety reasons.
Intravenous infections
Chronic viral infection and subsequent T cell exhaustion are induced by a high intravenous dose of LCMV Cl-13 (2×106 PFU). Alternatively, a medium intravenous dose of LCMV Cl-13 (2×105 PFU) can be used to model lethal immunopathology (Waggoner, Cornberg, Selin, & Welsh, 2012). In this context, virus-specific T cells will not undergo full exhaustion, so this would result in severe tissue damage. If 2–3-week-old mice are used for chronic LCMV Cl-13 infections, immune exhaustion becomes more exaggerated.
The model of intravenous LCMV Cl-13 infection, using 2×106 PFU is frequently used to interrogate the efficacy of novel immunotherapies. PD-1-based immunotherapy for the treatment of chronic infection was first shown in the chronic LCMV Cl-13 model (Barber et al., 2006), and was then shown to also be effective in the SIV model in macaques (Velu et al., 2009). Many studies on immune regulation have utilized the LCMV system (Bhattacharyya et al., 2017b; Bhattacharyya & Penaloza-MacMaster, 2017, 2018; Im et al., 2016; Kamphorst et al., 2017; Pablo Penaloza-MacMaster et al., 2015; Pablo Penaloza-MacMaster, Kamphorst, et al., 2014; Pablo Penaloza-MacMaster, Masopust, & Ahmed, 2009; P. Penaloza-MacMaster, Provine, Blass, & Barouch, 2015; Pablo Penaloza-MacMaster, Teigler, et al., 2014; Pablo Penaloza-MacMaster et al., 2011; Vezys et al., 2011; Wang et al., 2019; Erin E. West et al., 2013; E. E. West et al., 2011).
Draw up 500 μL of virus working stock in a 1-ml syringe 27-G needle. Keep the diluted virus stock on ice until ready to inject.
-
Place mouse cage under a heat lamp for 5–10 minutes to dilate the lateral tail veins (and facilitate intravenous injections).
Note: Make sure that the mice do not reach heat shock. Heat lamp should be at least a foot away from the cage to avoid overheating the mice. Animals should be monitored during the heat-warm and also for the next 24 hr following injection.
Gently restrain the mouse in a mouse restrainer in a way that it can breathe easily. Spray 70% ethanol on the tail or use alcohol swab, and inject virus into the lateral vein. Before performing intravenous infections, ensure that there are no bubbles in the syringe, as this would kill mice. Tail vein injections require a considerable amount of practice.
In most cases, intravenous infections are indicated when the LCMV Cl-13 model is used. There are two main models of chronic LCMV Cl-13 infection, colloquially referred as the “Conventional” Cl-13 Model and the “CD4-depleted” Cl-13 Model.
The Conventional Cl-13 Model
This is also referred to as the “straight” Cl-13 model, in which mice are infected intravenously via the lateral tail vein without any other acute manipulation (steps 4–6 above), resulting in a systemic infection lasting a month, with long-term infection restricted to brain and kidney (E. J. Wherry, J. N. Blattman, K. Murali-Krishna, R. van der Most, & R. Ahmed, 2003). However, virus in kidney and brain may be substantially reduced (and sometimes cleared) in some mice after ~200 days (our unpublished data).
Thus, the conventional Cl-13 model has a narrow window for therapeutic intervention. For instance, if one wants to test the effect of an immunotherapy (e.g. PD-1 blockade) using the convention Cl-13 model, one must administer the immunotherapy at 2–3 weeks of infection, a time when immune responses are already exhausted and viral loads are still high. After the fourth week, mice begin to control virus in blood and tissues (with high level of variability), rendering it difficult to obtain a consistent or statistically significant viral load difference after a therapeutic intervention.
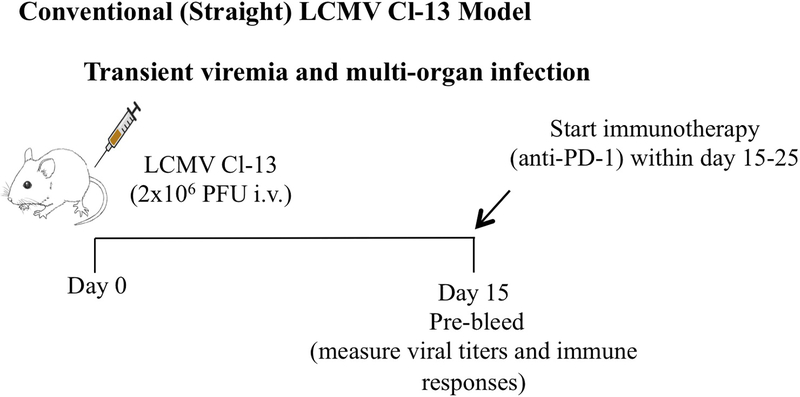
Figure 1 shows an experimental outline for how immunotherapy studies can be conducted using the Conventional Cl-13 model.
Figure 1. Experimental outline for evaluating immunotherapies using the conventional Cl-13 model.
In this model of chronic LCMV infection, most of the long-term persistence is limited to brain and kidney. Viremia is cleared after a month, and thus, the optimal time to perform immunotherapy (e.g. PD-1 blockade) in this model is between days 15–25 post-infection.
The CD4-depleted Cl-13 Model
This model results in a high titer systemic infection lasting for the life of the mouse, with persistent infection in every single tissue. The only technical difference between the conventional model (steps A, B, C above) and the CD4-depleted model is that in the latter, mice are treated intraperitoneally with 500 μg of CD4-depleting antibody (clone GK1.5) on day −1 and day 0 of infection, resulting in lack of CD4 T cells at the time of infection. Thus, the CD4-depleted Cl-13 Model mimics certain features of HIV infection, including suboptimal CD4 T cell help (thus the model is also referred to as the helpless model).
The CD4-depleted Cl-13 Model induces a more protracted immune exhaustion, relative to the Conventional Cl-13 Model, and is associated with suboptimal virus-specific B cell responses. Although CD4 T cells are reconstituted after 2–3 weeks of CD4-depletion, virus-specific CD4 T cells are never generated in this protracted infection model. The CD4-depleted Cl-13 Model allows a wider window of possible therapeutic intervention, since viral loads are maintained at high levels. If one wants to test the effect of an immunotherapy, for example PD-1 blockade, using the CD4-depleted Cl-13 Model, one should administer the immunotherapy within 40–90 days of infection. After this time, the level of T cell exhaustion will be too profound that the mice may not respond well to immunotherapy (P. Penaloza-MacMaster, N. M. Provine, et al., 2015).
Finally, if LCMV Cl-13 is being used to evaluate an immunotherapy, it is critical to pre-bleed mice and perform virologic quantification (e.g. plaque assay) and CD8 T cell stains (tetramer) prior to treatment, since variability between different mice is common. This would allow proper distribution of mice into future treatment groups. Immunotherapies, such as PD-1/PD-L1/LAG-3 blocking antibodies, can be administered intraperitoneally, every 3 days (200 μg in 500 μL), 5 times. This dose is typically used in many experiments, because it blocks all receptors for many inhibitory pathways (Blackburn et al., 2009; Pablo Penaloza-MacMaster, Kamphorst, et al., 2014; P. Penaloza-MacMaster, N. M. Provine, et al., 2015; Vezys et al., 2011; Wang et al., 2019; Erin E. West et al., 2013). However, if testing a novel monoclonal antibody in vivo titration may be needed to determine the saturating dose.
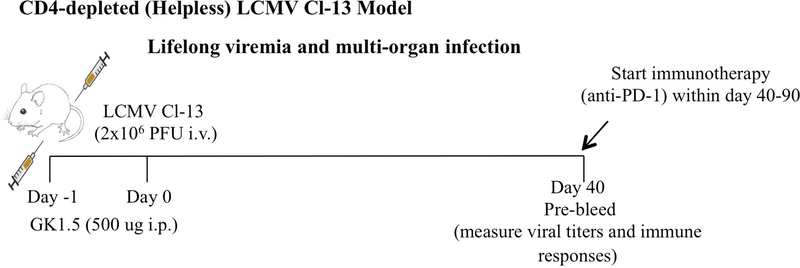
Figure 2 shows an experimental outline for how immunotherapy studies can be conducted using the CD4-depleted Cl-13 model.
Figure 2. Experimental outline for evaluating immunotherapies using the CD4-depleted Cl-13 model.
In this model of chronic LCMV infection caused by depleting CD4 T cells prior to infection, mice exhibit high-titer, lifelong multi-organ infection. The optimal time to perform immunotherapy (e.g. PD-1 blockade) in this model is between days 40–90 post-infection. After day 90, mice will exhibit deep T cell exhaustion, and suboptimal response to immunotherapy. Thus, the CD4-depleted Cl-13 model also allows one to interrogate different phases of T cell exhaustion, including early (<day 40) and late (>day 90) exhaustion.
II. Intraperitoneal infection
Acutely controlled LCMV infections, lasting only a week, are typically induced by intraperitoneal inoculation of LCMV Armstrong, at a dose of 2×105 PFU or lower in a volume of 500 μL. However, low doses of LCMV Cl-13 (102 PFU intraperitoneally) would also result in acutely controlled infection.
Draw up 500 μL of the virus working stock in a 1-ml syringe 27-G needle. Keep the diluted virus stock on ice until ready to inject.
Before injection, spray 70% ethanol on the right or left side of the abdomen. Firmly restrain the mice with one hand and inject the virus using the other hand.
III. Subcutaneous, footpad, and intramuscular infections
These three routes of infection result in more localized acute viral infection, relative to the intraperitoneal route discussed above. These 3 routes could be used when one wants to model antigen draining to local lymph nodes. For example, one can infect mice intramuscularly with LCMV Armstrong, and then quantify dendritic cells (DC) presenting viral antigen in draining lymph nodes at day 3 post-infection.
As few as 102 PFU of LCMV Armstrong are sufficient to reproducibly induce acute infection and adaptive immune responses (after day 7). In the case of footpad infection, there will be a characteristic swelling, reflecting the local T cell response to the virus (Lehmann-Grube, Lohler, Utermohlen, & Gegin, 1993).
Draw up 50 or 100 μL of the virus working stock in a 1-ml syringe 27-G needle. Keep the diluted virus stock on ice until ready to inject. Subcutaneous and intramuscular infections are in 100 μL, footpad infection is in 50 uL.
Before injection, spray 70% ethanol or use alcohol swab on the respective area, firmly restrain the mice, and inject the virus slowly to avoid back-flow of virus. If the infection route is subcutaneous, ensure that a “skin bubble” forms after injection. If the infection route is intramuscular, divide the 100 μL into the 2 quadriceps (50 μL per leg).
IV. Intracranial infection
Following an LCMV Armstrong challenge, there is transient dissemination of virus to most organs, followed by complete viral clearance within a week (E. John Wherry, Joseph N. Blattman, Kaja Murali-Krishna, Robbert van der Most, & Rafi Ahmed, 2003). Within this period of acute LCMV infection, dissemination of virus to the brain is restricted. However, direct intracranial injection of LCMV bypasses the blood brain barrier (BBB) and results in high viral replication in the brain, followed by death of mice around day 7 post-infection. This model of lethal choriomeningitis is exquisitely mediated by virus-specific T cells, and T cells of other specificities are unable to mediate lethal choriomeningitis (Pablo Penaloza-MacMaster et al., 2009). Irradiated mice reconstituted with OVA specific T cells (OT-I) do not die of choriomeningitis, whereas irradiated mice reconstituted with LCMV specific T cells (P14) die of choriomeningitis after intracranial LCMV infection (Pablo Penaloza-MacMaster et al., 2009).
In mice already immune to LCMV, intracranial infection is rapidly cleared by the pre-existing memory response, preventing lethal immunopathology. Thus, intracranial infection can be used as a model for anamnestic immune protection when it is used in immunized mice, and also as a model for T cell-mediated immunopathology when it is used in unimmunized mice.
Below, we provide a protocol for intracranial infection:
Draw up 5 μL of the virus working stock in a small 29-G needle. Keep the diluted virus stock until ready to inject. Intracranial injection of greater volumes may result in death of the animal. Any dose of LCMV can be used to induce lethal choriomeningitis. As little as 1 PFU will cause lethal choriomeningitis in adult immunocompetent mice in about a week. We typically inject 5×104 PFU of LCMV Cl-13 or Armstrong when doing intracranial infections, using 5 μL of undiluted viral stock that is at 107 PFU/mL. Both Armstrong and Cl-13 result in comparable lethality when injected into the brain.
Anesthetize mice using ketamine/xylazine. Anesthesia is absolutely required for this procedure.
Rub the head with an alcohol swap to disinfect the area and to expose the area where the virus will be injected. Render the injection site clearly visible, making a hairline in the place of the skull where the syringe will be inserted.
-
Grip the mouse’s head laterally with thumb and forefinger, gently, but firmly enough to immobilize it. Gently insert the needle ∼5 mm dorsal to the eyes, into the midline of the cranium (through the central suture, where bones of the skull unite; the needle should pass through this suture with minimal pressure. Slowly inject 5 μL of inoculum (injection should take ∼10 sec). Slowly withdraw syringe.
Note: Make sure to restrict the depth of penetration. It is advisable to use 4–6-week-old mice, as their cranium would be softer for intracranial injections. One should not use older mice (>8-week-old) for intracranial injections due to complete ossification of the cranium, which could result in fracture of the skull upon needle insertion. Bleeding should be minimal. The animal should recover from anesthesia within ∼5 min and resume normal locomotor behavior.
Mice must be monitored daily and evaluated for signs of immunopathology, including hunched posture, ruffled hair, blepharitis, emaciation, lethargy, weight loss, and tremors. The mice should be handled as little as possible after intracranial infection, since even gentle handling can precipitate lethal seizures. For this reason, it is advisable to avoid cage changes between days 5 to 9.
The intracranial infection protocol above is used for modeling immunopathology in adult immunocompetent mice. However, intracranial infection of neonate (< 2-day old) mice does not result in immunopathology. Instead, the mice become infected for life, and they transmit the virus to their offspring (vertically). This refers to the model of “congenital LCMV infection,” which is characterized by complete absence of LCMV-specific T cell responses and a high-titer persistent infection. The complete lack of T cell responses in this model is thought to be mediated by thymic negative selection, since LCMV replicates extensively in the thymus, altering T cell selection (King et al., 1992). The chronic LCMV Cl-13 strain was first isolated by Rafi Ahmed from the lymphoid tissue of a mouse that was congenitally infected with the Armstrong strain (Ahmed et al., 1991; Ahmed & Oldstone, 1988; Ahmed, Salmi, Butler, Chiller, & Oldstone, 1984b). In summary, the congenital model of LCMV infection, induced by intracranial infection of neonate mice, can be useful to study viral evolution in the absence of adaptive immunity, as well as the specific effect of novel antiviral drugs in the complete absence of virus-specific adaptive immunity. However, this model is not poised to study therapies that revert T cell exhaustion (e.g. PD-1 blockade), since congenitally infected mice lack exhausted T cells or any type of virus-specific immunity.
Support Protocol 1: Preparation of LCMV stocks
Virus stocks are prepared by propagating the original stock of LCMV on BHK21 cells. LCMV is a BSL-2 agent, so all procedures should be carried out in a certified biosafety cabinet.
| Materials: |
|---|
| Seed stock of LCMV (BEI Resources or European Virus Archive, SKU:006v-EVA1372) |
| BHK21 cells (ATCC® CCL10™) |
| T175 flasks (or T182 cm2 Tissue Culture Flasks, Dot Scientific, Cat#667351) |
| PBS (Gibco, Cat#14190–144) |
| Penicillin/Streptomycin (Pen/Strep) (Gibco, Cat#15140–122) |
| L-Glutamine 200 mM (Gibco, Cat#25030–081) |
| 10% DMEM (supplemented with 10% FBS, 1% Pen/Strep, 1% L-Glutamine) |
| 1% DMEM (supplemented with 1% FBS) |
| 0.25% Trypsin-EDTA (Gibco, Cat#25200–056) |
| Cryogenic storage tubes (2ml) for storing viral stock (Thermo Scientific, Cat# 50000050) |
Thaw and passage BHK21 cells (10×106 cells/mL) in 10% FBS DMEM media (25 mL) in a T175 flask. When cells are 70% confluent (15×106 cells/mL) (usually after 3 days), trypsinize cells and split BHK21 cells again for at least two times after thawing.
-
When BHK21 are ~70% confluent in all T175 flasks, aspirate media and gently add virus at a multiplicity of infection (MOI) of 0.1 in a volume of 6 mL in 1% DMEM and allow total dispersion among the cell monolayer.
For example, for infecting BHK21 cells in a T175 flask with 0.1 MOI, one could assume that there are 107 cells per T175 flask, so one would add 106 PFU of LCMV per flask in a volume of 6 mL of 1% DMEM. Using higher than 1% FBS would decrease initial viral adsorption onto the monolayer. Note: Handle 5 flasks at a time, so that monolayer does not dry out after aspirating media.
Close the lids of the flasks tightly and gently rock flasks to ensure that the virus inoculum is evenly distributed over the BHK21 monolayer. Keep all flasks in a 37°C 5% CO2 incubator for 1 hr, gently rocking flasks every 10 minutes to prevent monolayer from drying.
Note: Keep one 70% confluent BHK21 T175 flask without infection (as a “negative control”).
-
After incubation, aspirate media and add 25 mL of fresh pre-warmed 10% FBS DMEM media. Incubate flasks in 37°C 5% CO2 incubator for 48–72 hr.
Note: Under the microscope, LCMV infection typically produces minimal to no cytopathic effect in infected cells, and sometimes it is difficult to distinguish LCMV-infected cells from uninfected cells. One can only rigorously be able to determine if the virus was produced after performing plaque assay (Support Protocol 2.1).
After 48–72 hr, transfer supernatant from all T flasks into sterile 50 mL tubes and centrifuge tubes at 1000 rpm for 5 minutes at 4°C to remove floating cells and debris.
After centrifugation, transfer the supernatant into a new tube and discard cell pellet. Keep supernatant on ice at all times.
-
Quickly aliquot into 2 mL cryovials and freeze at −80°C. Stocks will be approximately 107 PFU/mL, but further quantification by plaque assay is needed to obtain exact titer.
Note: If higher concentration (>108 PFU/mL) is desired, perform ultra-centrifugation. Transfer supernatant from step 7 into new Sorvall RC2B tubes and perform ultra-centrifugation at 15,000 rpm (in Beckman SW-41 rotor) for 2 hr at 4°C. Discard supernatant. A milky white virus band will be clearly visible. Dissolve pellet in a small volume of cold sterile PBS. For example, 0.5 mL of PBS could be added to only the first tube, mixing well, and using the same suspension to dissolve the rest of the cell pellets in the remaining tubes one by one (assuming a total of 5 tubes). Pool the final suspension in a cryovial for an ultra-concentrated LCMV stock, and freeze at −80°C.
LCMV titer is reduced by freeze/thaw, so viral stocks should not be re-frozen after thawing.
Perform plaque assay to determine viral titer (Support Protocol 2). Typically, one can achieve viral titers between 107 to 108 PFU/mL at this step.
Support Protocols 2: Assays to measure LCMV titers
Support Protocol 2.1: Plaque assay
Infectious viral titers can be determined by counting plaques using the naked eye. Different dilutions of virus (from a new viral stock, or from infected sera or tissue homogenate) are absorbed on Vero e6 cells that generate plaques in 5 days. Vero cell monolayers are fixed in agar overlay and visualized by staining with neutral red. Single plaques originate from a single infectious virus, and plaques are prevented from coalescing with other plaques by virtue of the agar overlay. The sensitivity of the plaque assay is limited by the surface area of the tissue culture plates utilized, the large assay volumes utilized (up to 200 μl), and also by the ability of the human eye to discern individual plaques, which in the case of LCMV are particularly heterogeneous.
| Materials: |
|---|
| Vero e6 cells (ATCC #CRL-1586). |
| Complete EMEM (ATCC 30–2003, supplemented with 10% FBS, 1% Pen/Strep, 1% L-Glutamine) |
| 1% DMEM (supplemented with 1% FBS) |
| Infected samples (sera or tissues of LCMV infected mice from basic protocol 1) or viral stock (from Support Protocol 1) |
| 2×199 media (See reagents and solution) |
| 1% Neutral Red (Fisher Chemical, Cat# N129–25) (see reagents and solution) |
| 1% Agarose (SeaKem ME, Lonza; Cat# 50014) (see reagents and solution) |
| Round Bottom 14 mL tubes with snap cap (if quantifying viral loads from tissues). Make sure the tubes have a wide round bottom to allow the homogenizer probe to reach the bottom of the tube. |
| 6-well tissue culture plates (Dot Scientific; Cat# 667106) |
| 37°C water bath |
| Single-probe homogenizer of TissueRuptur (Qiagen). |
| Transluminator |
| Ice |
-
Determine the number of 6-well plates needed. Typically, one 6-well plate is enough for tittering two samples. Depending on the number of samples, prepare 6-well plates following step 2.
Note: In most of the cases, one 6-well plate is enough for determining viral titers in two mouse samples using three dilutions (10−1, 10−2 and 10−3). However, we recommend using a whole 6-well plate using 6 dilutions if tittering a new viral stock of unknown concentration or a mouse sample at the peak of an LCMV Cl-13 infection (~week 2), since these samples may contain high viral titers.
-
Seed 5×105 Vero e6 cells per well in 6-well plates in 10% EMEM media. Monolayer should be 90–100% confluent after 24–48 hr. Make sure that the Vero e6 cells are not overly confluent (>100%), as this may reduce plaque formation.
Note: If one wants to determine the viral load in tissues such as spleen or liver, perform plaque assay following all these steps. If performing plaque assay from liquid samples, for example a new viral stock or sera of infected mice, skip to step 11. Sera are harvested by centrifuging blood at 15,000 rpm for 10 min at 4°C, harvesting the top clear layer, and storing at −80°C.
If using tissues, determining tissue weight is very important for calculating viral titer per gram; therefore tubes containing 1 ml of 1% DMEM media are weighted before and after adding tissue. Follow steps 4 to 6 for tissue weight before starting the actual plaque assay procedure.
Before harvesting tissue, label 14 mL snap-cap tubes, and add exactly 1 mL of 1% FBS DMEM for each tissue sample (use a P1000 pipette for accuracy).
Weigh each snap-cap tube on an analytical scale up to four decimal places and record this pre-weight value. Make sure to snap close the caps tightly to avoid evaporation, and wipe off any condensation on the outside of the tube before weighing; precise weighing is very important to obtain accurate PFU/g of tissue).
Transfer tissue (e.g. ¼ spleen or liver) from LCMV infected mice (from Basic Protocol 1) to the 14 mL snap-cap tube. Weigh each tube on an analytical scale up to four decimal places and record this post-weight value. Determine net tissue weight by subtracting the pre-weight from the post-weight value.
Freeze tissues at −80°C, or leave at −80°C for at least 2 hr before performing the next step. Samples could also be left frozen for several weeks.
Thaw tissues in a 37°C water bath, and place tissues on ice immediately. Quickly centrifuge samples at 1000 rpm for 1 min at 4°C to ensure all contents are collected at the bottom of the tube.
Homogenize tissues at maximum speed for 15–20 seconds, using a standard homogenizer or a TissueRuptur. An advantage of using the TissueRuptur is that one could simply place a new sterile homogenizing probe after each sample, without having to perform multiple washes on the same probe. This may save time relative to using standard single built-in probe homogenizers. The TissueRuptur probes can be placed on 10% bleach, then washed and autoclaved for re-use. If working with tough tissues, such as kidney or lung, clarify the homogenate using a Falcon 100 μm cell strainer.
Place all samples on ice.
Add 225 μL of 1% DMEM onto all the wells of a 96-well plate for making serial 10-fold dilutions. Then, add 25 μL of tissue homogenate (from step 6) to the first dilution (10-fold dilution). Alternatively, if quantifying viral loads from sera or virus stock, add 25 μL of this to the first dilution.
Make the subsequent dilutions by pipetting up and down 10 times, changing tips, and transferring 25 μL to the next well. Keep on ice.
Quickly invert the 6-well plate containing the Vero e6 cells (from step 2) to discard the media onto a container containing 10% bleach. There will be residual media in the wells. Although inverting the 6-well plate to remove the media may seem rudimentary, it works better than using aspiration. Aspirating all media out is not recommended, as the monolayers would quickly dry out, causing cell death.
Slowly and dropwise add 200 μL of each diluted sample on top of the monolayer (swirling to allow maximal adsorption of virus throughout the monolayer), starting from the lowest to the highest concentration. Only take out 5 plates at a time from the incubator.
Manually rock the plates, and place in a 37°C 5% CO2 incubator for 1 hr, gently rocking every 10 minutes to disperse the virus on the monolayer. This will prevent the monolayer from drying and will maximize virus adsorption.
-
During the 1 hr incubation, melt 1% agarose in a microwave, leaving the lid of the 1% agarose bottle slightly open (it should be bubbling). Mix the melted 1% agarose 1:1 with 2X199 media (see below.
Note: It is recommended to prepare an excess of 50 mL of this 1:1 mixture. For example, if one has one 6-well plate, the volume of mixture needed is 4 mL/well x 6 wells = 24 mL, plus 50 mL extra = 74 mL. So combine 37 mL of 1% agarose plus 37 mL of 2X199 media. Place a sterile thermometer inside the mixture.
-
Wait until the 1:1 agarose/2×199 mixture reaches 40°C, and dispense 4 mL very slowly against the wall of the well, as to not disturb the monolayer. Most technical problems with the plaque assay occur at this stage (adding the mixture too hot, too cold, too fast, or on top of the monolayer).
Note: If the agarose/2×199 mixture gets colder it will clump, and it should be discarded and prepared again from scratch. If you have many plates, consider using a 37°C water bath to keep this mixture from clumping. Discard the leftover mixture in a biohazard bag and not in the sink or vacuum aspirator, as it will solidify and clog the tubing. Leave the plates at room temperature for 10 min to allow the overlay to solidify, and transfer plates back to the 37°C 5% CO2 incubator.
Four days later, add a 1:50 of a 1% neutral red solution to a 1:1 agarose/2×199 mixture (similar to the mixture in step 14). For example, if one has one 6-well plate, the volume of mixture needed is 2 mL/well x 6 wells = 12 mL, plus 50 mL extra = 62 mL. So combine 31 mL of 1% agarose plus 31 mL of 2X199 media, and 1.24 mL of a 1% neutral red solution. Place a sterile thermometer inside the mixture. Wait until the red mixture reaches 40°C, and add 2 mL per well. Leave the plates at room temperature for 10 min to allow the agar overlay to solidify, and transfer plates back to the 37°C 5% CO2 incubator.
Count plaques the next morning using a transluminator. Plaques may be visualized without removing the overlay, but are more easily visualized after the agar overlay is removed (using forceps). To remove the agar overlay, invert the 6-well plate and insert the forceps in the agar, against the wall of the well. Slowly push the forceps toward the center of the well to detach the agar from the monolayer, without disrupting the monolayer. This may require practice.
Virus titer in sera (or virus stock) is reported as PFU/mL. For example, if you count 10 PFU in the well that received 200 μL of a 10-fold dilution, then the PFU / mL is calculated as follows:
PFU x dilution factor x 5,
PFU is 10, and dilution factor is 10.
10 PFU x 10 × 5 = 500 PFU/mL.
Virus titer in tissues is reported as PFU/g. This titer is calculated similar to the titer for sera (above), but then it would be divided by the net weight of the tissue.
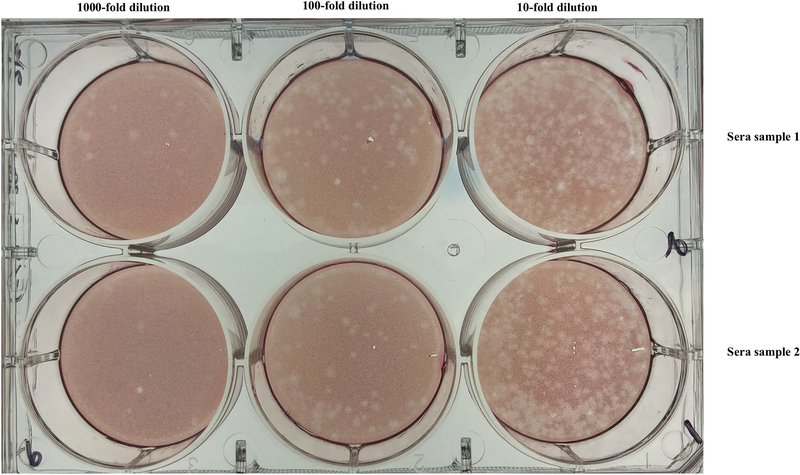
Figure 3 shows a representative plaque assay plate.
Figure 3. LCMV plaque assay plate.
Each row represents a different sample. Each column represents a different dilution (1000-fold, 100-fold and 10-fold, from left to right, respectively). Data are from sera of mice infected intravenously with LCMV Cl-13 (day 12 post-infection). It is advisable to use the wells that have ~50 PFU to calculate viral titer.
Support protocol 2.2: Immunofluorescence focus assay (IFA) to measure LCMV titer
IFA is a modification of conventional plaque assay and a difference is the use of rat anti-LCMV monoclonal antibody VL4, which binds to the nucleoprotein of LCMV in infected cells. The primary VL4 antibody can be visualized using a fluorescently-tagged secondary antibody. Advantages of the IFA over the plaque assay are that it results in easier and high-throughput viral quantification. Moreover, the IFA gives results in only 1 day, whereas the plaque assay gives results in 5 days. However, a disadvantage is that IFA only measures viral antigen incorporated into the cell, and not infectious virus per se.
| Materials: |
|---|
| Vero e6 cells (ATCC #CRL-1586). |
| Infected sample (tissue, sera or stock) |
| Primary antibody: LCMV nucleoprotein-specific monoclonal antibody, Rat IgG, VL4 (6 mg/ml) (BioXcell) |
| Secondary antibody: Anti-Rat IgG-Alexa Fluor568, 2 mg/ml (Abcam, Cat#Ab175475) |
| BSA (Sigma, Cat#A7906–500G) |
| 10% DMEM media (10% FBS, 1% Pen/Strep, L-Glutamine). |
| FBS (Sigma, Cat# F0926–500 mL) |
| Triton X (Sigma Aldrich, Cat# 9002–93-1) |
| PBS (Gibco, Cat#14190–144) |
| 4% Paraformaldehyde solution (Thermo Scientific, Cat# AAJ19943K2) |
| 1% DMEM: (1%FBS+ 1% Pen/Strep+ 1% L-Glutamine) |
| Dilution Buffer: 3% BSA, 0.3% Triton X in PBS |
| Blocking Solution: Dilution buffer with 10% FBS |
| 96-well flat bottom plate (Falcon, Cat#353072) |
| Fluorescence microscope |
-
On day −1, seed 3×104 Vero cells per well in a 96-well flat bottom plate using 10% DMEM media (10% FBS, 1% Pen/Strep, L-Glutamine).
Note: 24-well plates can also be used, but this will reduce the throughput.
On day 0, thaw infected samples in 37°C water bath, and immediately place on ice. If doing IFA using infected tissues, follow steps 1–7 from the plaque assay protocol (tissue homogenization) before following the steps below. If doing IFA using sera or viral stock, follow steps below:
-
Prepare 10-fold sample dilutions. Start by adding 180 μL 1% DMEM in all wells of a 96-well plate (round bottom). Then, transfer 20 μL of the infected sample into the first well, mix 10x and discard the tips. Make the subsequent dilutions by mixing 10 times, changing tips, and transferring 20 μL to the next well. Keep on ice.
Note: The number of dilutions would depend on the expected viral titer of the sample. For example, if quantifying titers in a new viral stock, it is recommended to do at least 6 serial 10-fold dilutions (10−1, 10−2, 10−3, 10−4, 10−5, 10−6). For most tissues, it is customary to do at least 3 serial 10-fold dilutions (10−1, 10−2, 10−3), but more dilutions may be necessary if doing IFA from high titer tissues (such as kidney), especially if the tissues are harvested around weeks 2–3 of chronic LCMV infection.
Remove media from the 96-well plate containing Vero e6 cells, and gently rinse with 1% DMEM media.
Add 100 μL of viral dilutions to the monolayer and rock the plate to allow even distribution of virus. Immediately keep plate in a 37°C 5% CO2 incubator for 24 hr.
The next day, aspirate the supernatant and add 100 μL of 4% PFA in each well to fix the cells, incubate for 30 minutes at room temperature.
Discard supernatant, wash well 2X with 200 μL PBS and decant plate.
Dispense 100 μL blocking buffer in each well and incubate for 1hr at RT.
-
Decant blocking buffer and add 30 μL of the primary antibody (VL4) diluted in dilution buffer (3% BSA, 0.3% Triton X in PBS). Incubate for 1 hr at room temperature in a humidity chamber.
Note: Dilute primary antibody 1: 1000 in dilution buffer.
-
Decant plate, wash 2X with PBS using 100 μL/well. Add 30 μL of the secondary antibody diluted in dilution buffer. Incubate for 40 min at room temperature in a humidity chamber.
Note: Dilute secondary antibody 1: 1000 in dilution buffer.
Discard supernatant and wash 3X with PBS. Finally, add 50 μL PBS to each well.
After completion of the assay, cover the plate with aluminum foil and place at 4°C before reading. Focus forming units (FFU) should be counted soon after staining, using a fluorescence microscope.
A more sensitive technique to quantify LCMV is by quantitative PCR. However, quantitative PCR may detect residual nucleic acid present days after viral antigen has been controlled, which may not represent bona fide infection. A detailed description of this PCR-based quantification protocol is listed in a prior paper (McCausland & Crotty, 2008).
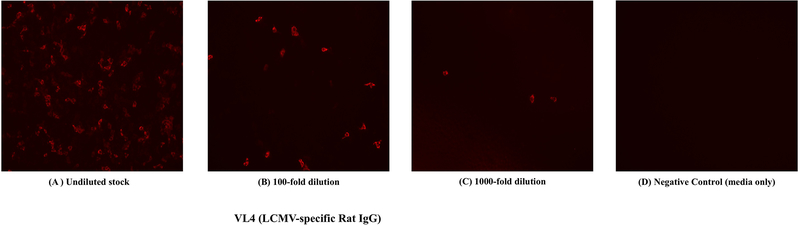
Figure 4 shows representative IFA for detecting LCMV nucleoprotein.
Figure 4. LCMV immunofluorescence images.
LCMV+ cells are represented as fluorescent foci. Monolayer of Vero e6 cells were infected with an LCMV Armstrong stock at different dilutions, and after 24 hr, cells were incubated with a primary VL4 antibody, followed by staining with a secondary antibody (Anti-Rat IgG-Alexa Fluor568).
Basic Protocol 2: Measurement of T- and B-cell responses to LCMV infection
There are many ways to detect virus-specific immune responses in mice. Typically, LCMV-specific antibody responses are detected using ELISA, but there are other assays to specifically detect antibody secreting cells (ASC) or memory B cells. Analyses of LCMV-specific T cell responses commonly require flow cytometry protocols, such as tetramer stains and intracellular cytokine stains. In addition, chromium release assay continues to be a gold standard for quantifying T cell cytotoxicity, due to its extraordinary sensitivity, but radioactivity limits its wide use. In chromium release assays, MC57 mouse fibroblast cell targets are coated with viral peptides, and labeled with 350 μCi 51-Cr. Target cells are then incubated for 6 hr with different amounts of effector CD8 T cells from spleen. Absolute numbers of viral-specific CD8 T cells are retroactively calculated and plotted in terms of effector to target ratio. MC57 cells washed with 1% Triton X are used as positive control (representing total cell killing or total release), and MC57 cells without effectors are used to calculate spontaneous or background release. Chromium-release assay has been extensively explained throughout decades of prior literature and thus, we will not discuss this technique in detail.
Multiple LCMV-specific T-cell epitopes have been identified (Table 1). T cell responses to these epitopes can be detected 7 days after infection, using MHC tetramers or intracellular cytokine staining. Immunodominant responses are indicated in bold.
Table 1:
MHC Class I and II restricted LCMV epitopes.
| Mouse strain | MHC molecule | Protein | Position | Amino acid sequence |
|---|---|---|---|---|
| MHC-I | ||||
| C57BL/6 | D b | GP | 33–41 | KAVYNFATC |
| *K b | GP | 34–43 | AVYNFATCGI | |
| D b | GP | 276–286 | SGVENPGGYCL | |
| Db | GP | 92–101 | CSANNSHHYI | |
| Kb | GP | 70–77 | GVYQFKSV | |
| Kb | GP | 118–125 | ISHNFCNL | |
| D b | NP | 396–404 | FQPQNGQFI | |
| Kb | NP | 205–212 | YTVKYPNL | |
| Db | NP | 166–175 | SLLNNQFGTM | |
| Kb | NP | 235–243 | NISGYNFSL | |
| BALB/c | Ld | NP | 118–126 | RPQASGVYM |
| Ld | NP | 313–322 | WPYIACRTSI | |
| Kd | GP | 283–291 | MGYCLTKWMI | |
| Kd | GP | 99–108 | HYISMGTSGL | |
| MHC-II | ||||
| C57BL/6 | I-Ab | GP | 61–80 | GLNGPDIYKGVYQFKSVEFD |
| I-Ab | GP | 6–20 | TMFEALPHIIDEVIN | |
| I-Ab | GP | 126–140 | TSAFNKKTFDHTLMS | |
| I-Ab | NP | 311–325 | EGWPYIACRTSIVGR | |
T cell response to these epitopes can be identified using intracellular cytokine staining (ICS) or tetramer binding assays. Immunodominant CD8 or CD4 epitopes are shown in boldface. *The Db restricted GP33–41 epitope has an overlapping epitope sequence, also known as the GP34 epitope, which is Kb restricted. When performing ICS using only GP33–41 peptide, both the DbGP33 and the KbGP34 specific CD8 T cells are stimulated, resulting in a higher cytokine to tetramer ratio. There are various altered peptide ligands (APL) derived from GP33, including a 9-mer GP33–41 peptide (KAVYNFATM), which carries a methionine (M) residue at position 41 instead of the cysteine (C) residue.
The GP70–77 CD8 epitope is embedded within the GP61–80 CD4 epitope. Thus, stimulation of LCMV-immune splenocytes with GP61–80 results in stimulation of both CD8 and CD4 T cells (P. Penaloza-MacMaster, D. L. Barber, et al., 2015). All of these peptides could be custom-made (e.g. through GenScript), and some may be purchased already pre-made (e.g. through Anaspec). There are also multiple CD8 epitopes identified in the Z and L proteins of LCMV.
Basic Protocol 2.1 Triple tetramer staining for detection of LCMV-specific CD8 T cells
LCMV-specific CD8+ T cells could be phenotyped by staining lymphocytes with MHC-I tetramers. It is possible to combine several MHC-I tetramers on the same antibody cocktail for assessment of multiple T cell responses at once. We provide an example of a flow cytometry panel, but the markers would vary depending on the investigator’s needs.
| Materials: |
|---|
| MHC-I tetramers conjugated to different fluorophores (MBL International, or NIH tetramer facility). For example, NP396 (APC), GP33 (PE), GP276 (Pacific Blue). |
| Antibodies: |
| CD8α-PerCP-Cy5.5 (Clone 53–6.7, BD Pharmingen, Cat# 551162) |
| CD44-FITC (Clone IM7, BD Pharmingen, Cat# 553133) |
| PD-1-PECy7 (Clone RMP130, Biolegend, Cat# 109110) |
| LIVE/DEAD® Fixable Red Dead Cell Stain (Invitrogen Cat # L34976) |
| Fetal Bovine Serum (FBS) (Sigma, Cat#F0926–500 mL) |
| FACS Buffer (PBS with 1% FBS) |
| PBS (1X) (Gibco, Cat#14190–144) |
| BD Cytofix/Cytoperm (BD Biosciences, Cat#51–2090KZ) |
| 96-well round-bottomed plates, Costar (Corning Incorporate, Cat# 3799) |
| Multichannel pipette |
| Centrifuge |
| Flow cytometer |
| FlowJo (Treestar) |
Plate 100 μL of splenic lymphocytes (10×106 cells/ml splenic cell suspension from the Support Protocol 3) into a 96-well round bottom plate that is equal to 106 splenocytes per well.
Centrifuge plate at 2200 rpm 4°C for 2 min. Flick off plate to remove supernatant.
Wash samples by adding 200 μL of FACS buffer, and centrifuge plate at 2200 rpm 4°C for 2 min. Flick off plate to remove supernatant. Repeat this washing step one more time.
Prepare the antibody cocktail using the aforementioned antibodies/tetramers in FACS buffer (antibodies, tetramers and live dead stain are typically diluted 1:100, but prior titration may be necessary to determine optimal staining conditions). In our laboratory, we typically use 2 μg/mL antibody concentration to stain 106 cells.
Add 50 μL of antibody cocktail per well. Mix well (10 times) and incubate plate in the dark for 30 min at 4° C.
Wash samples by adding 200 μL of FACS buffer, and centrifuge plate at 2200 rpm 4°C for 2 min. Flick off plate to remove supernatant and vortex for 5 seconds. Repeat this step twice.
Vortex plate again to resuspend the cell pellet.
Fix cells by resuspending in 100 μL of Cytofix/Cytoperm and incubate for 15–20 min in dark at 4°C. Mix 10X using a multichannel pipette. Do not leave for longer than 20 minutes, as this may result in background signal. Alternatively, one can fix cells with 4% PFA made in-house, but we have observed that this results in higher background signals.
Add 150 μL cold PBS and centrifuge at 2200 rpm 4°C for 2 min. Flick off plate to remove supernatant. Wash one more time with 200 μL PBS.
After the final PBS wash, add 200 μL PBS, and keep the plates at 4°C in the dark. Acquire samples using a flow cytometer within 3 days.
Note: Washing off the fixative with PBS reduces background noise.
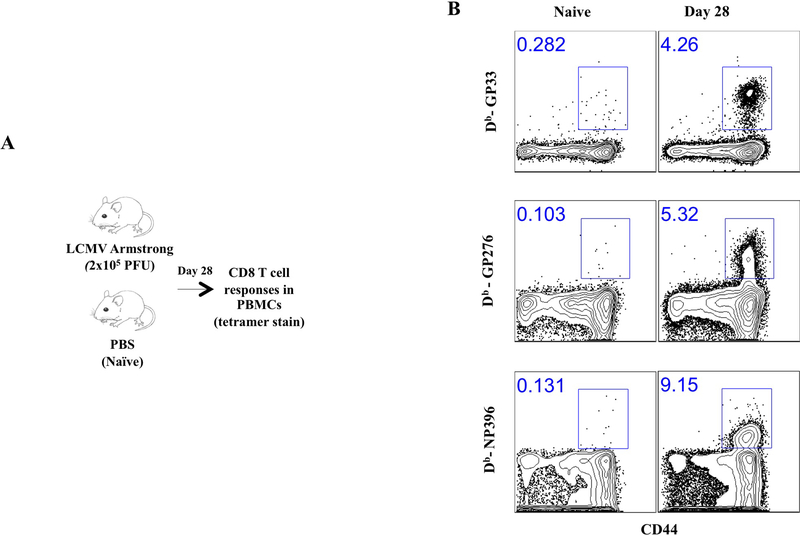
Figure 5 shows representative FACS plots showing tetramer+ CD8 T cell responses following LCMV Armstrong infection.
Figure 5. Representative FACS plots showing tetramer+ CD8 T cell responses following LCMV Armstrong infection.
A) Experimental outline. B) Representative FACS plots showing the frequencies of CD8 T cells that are LCMV-specific in PBMCs at day 28.
Basic Protocol 2.2 Intracellular Cytokine Staining (ICS) for detection of LCMV-specific T cells
This is a method of quantifying the functionality of T-cells in vitro. Lymphocytes are stimulated in the presence of viral peptides. An inhibitor of protein transport is used to retain the cytokines within the T cell. Intracellular cytokines are measured by flow cytometry after staining with fluorophore-tagged antibodies. We provide an example of the antibodies that could be used, but these would vary depending on the experimental question.
| Materials: |
|---|
| Splenocytes (See Support Protocol 3) |
| R10 media (RPMI, 10% FBS, 0.1% Beta mercaptoethanol, 1% Pen-Strep, 1% L-glutamine) |
| GP33 peptide stock (0.4mg/ml) (e.g. GenScript, Cat# RP20257) |
| Brefeldin A (GolgiPlug, BD Biosciences, Cat# 51–2301KZ) |
| Monensin (GolgiStop, BD Biosciences, Cat# 51–2092KZ) |
| Viral peptide stock (e.g. GP33 peptide stock at 0.4mg/ml in DMSO, 2000x) |
| BD Cytofix/Cytoperm (BD Biosciences, Cat#51–2090KZ) |
| BD Perm Wash (BD Biosciences, Cat# 347692). Stock comes as 10X, and must be diluted to 1x with Milli-Q-grade H2O. Sterile filter after diluting to remove precipitate. |
| IL2-PE (BD Pharmingen, Cat# 554428) |
| IFNy-APC (BD Pharmingen, Cat# 554413) |
| TNF-PECy7 (Clone MP6-XT22, BD Pharmingen, Cat# 557644) |
| CD8α-PerCP-Cy5.5 (Clone 53–6.7, BD Pharmingen, Cat# 551162) |
| CD44-FITC (Clone IM7, BD Pharmingen, Cat# 553133) |
| LIVE/DEAD® Fixable Cell Stain (Invitrogen Cat # L34976) |
-
Prepare single cell suspension of splenic lymphocytes (See Support Protocol 3) at 20×106 cells/mL concentration in R10.
Note: It is recommended to include lymphocytes from naïve mouse as negative control, and to have unstimulated (DMSO) controls, as well as PMA/Ionomycin (positive) controls.
Add 100 μL splenic suspension (2×106 cells) per well in a round bottom 96-well plate.
-
Prepare master stimulation cocktail containing viral peptide diluted in R10 (e.g. 0.4 ug/mL of GP33 peptide). Add 1:500 of GolgiStop and 1:500 of GolgiPlug.
For example, for 8 samples (making 2 extra), one could add 1000 μL R10 + 2 μL GolgiStop + 2 μL GolgiPlug + 1 μL GP33 peptide stock. Add 100 μL of this master mix into every well (final volume in the well will be 200 μL), and mix 10X using a multichannel pipette. Final GP33 peptide concentration in the well is 0.2 μg/mL.
Incubate plate in a 37°C 5% CO2 incubator for 5 hr.
Centrifuge plate for 2 min at 2200 rpm 4°C. Discard supernatant by flicking off plate.
Wash samples by adding 200 μL of FACS buffer, and centrifuge plate at 2200 rpm 4°C for 2 min. Flick off plate to remove supernatant. Repeat this washing step one more time.
Prepare the antibody cocktail using the aforementioned antibodies (e.g. CD8, CD44) and live dead stain. These are typically diluted 1:100, but prior titration may be necessary to determine optimal staining conditions).
Add 50 μL of antibody cocktail per well. Mix well (10 times) and incubate plate in the dark for 30 min at 4° C.
Wash samples by adding 200 μL of FACS buffer, and centrifuge plate at 2200 rpm 4°C for 2 min. Flick off plate to remove supernatant and vortex for 5 seconds. Repeat this step twice.
Vortex plate again to resuspend the cell pellet.
Fix cells by resuspending in 100 μL of Cytofix/Cytoperm and incubate for 15–20 min in dark at 4°C. Mix 10X using a multichannel pipette. Do not leave for longer than 20 minutes, as this may result in background.
Wash samples by adding 150 μL Perm Wash, and centrifuge plate for 2 min at 2200 rpm at 4°C. Discard supernatant by flicking plate and wash again 2X with Perm Wash.
-
Add the intracellular antibody cocktail diluted in Perm Wash (e.g. IFNγ, IL-2 and TNFα). Mix well ten times. Incubate for 30 min at 4°C temperature in the dark. These intracellular antibodies are typically diluted 1:100, but prior titration may be necessary to determine optimal staining conditions).
Note: Most companies provide pre-titrated antibodies, so one can follow their recommended dilution. Pre-titration by each laboratory is also suggested.
Add 200 μL of Perm Wash, and centrifuge for 2 min at 2200 rpm 4°C. Repeat again 2X with Perm Wash.
Add 200 μL of PBS, and centrifuge for 2 min at 2200 rpm 4°C. Repeat PBS wash one additional time.
Fix cells by resuspending in 100 μL of Cytofix/Cytoperm and incubate for 15–20 min in dark at 4°C. Mix 10X using a multichannel pipette. Do not leave for longer than 20 minutes, as this may result in background. Alternatively, one can fix cells with 4% PFA made in-house, but this may result in slightly higher background levels.
Add 150 μL cold PBS and centrifuge at 2200 rpm 4°C for 2 min. Flick off plate to remove supernatant. Wash one more time with 200 μL PBS.
After the final PBS wash, add 200 μL PBS, and keep the plates at 4°C in the dark. Analyze samples using a flow cytometer within 2 days.
Note: Washing off the fixative with PBS reduces background noise.
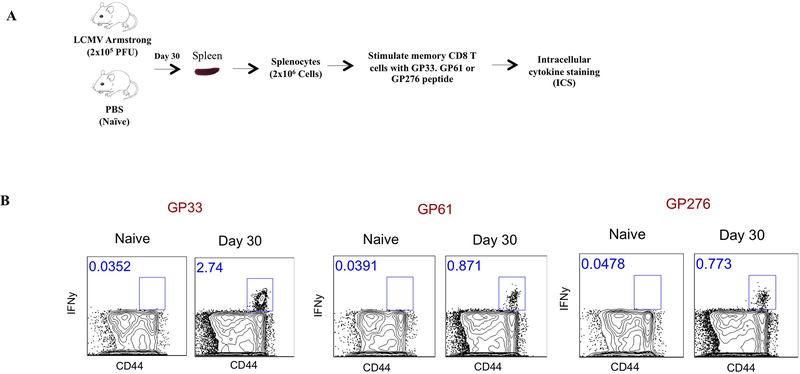
Figure 6 shows representative FACS plots showing CD8 T cell responses by ICS following LCMV Armstrong infection.
Figure 6. Representative FACS plots showing LCMV-specific CD8 T cell responses by ICS following LCMV Armstrong infection.
A) Experimental outline. B) Representative FACS plots showing the frequencies of CD8 T cells that are LCMV-specific in spleen at day 28. All plots are gated from total CD8 T cells. Note that the GP61–80 peptide (known to be a CD4 epitope) also induces CD8 T cell responses.
Basic Protocol 2.3 Enumeration of direct ex-vivo LCMV-specific antibody secreting cells (ASC)
This protocol is adapted from a protocol by Shane Crotty. ASC are typically quantified in spleen or bone marrow. Plasma cells that secrete antibody can be readily detected by this method.
| Materials: |
|---|
| LCMV infected BHK21 cell lysate (For LCMV-specific antibody detection). (See Support Protocol 4). |
| Lymphocytes from Spleen of LCMV infected mice (Basic protocol 1 & see Support Protocol 3) |
| Goat anti-mouse IgG+IgM+IgA (For total antibody detection) |
| 96-well Multiscreen HA filtration plate (Millipore, Cat# MSHAN 4B50) |
| Biotinylated anti-IgGϒ (CALTAG, Cat # M30115, 0.87mg/ml) |
| HRP conjugated avidin-D (Vector Laboratories, Cat#A-2004, Vector, 5ug/ml) |
| PBS-T: PBS + 0.05% Tween-20 |
| PBS-T-FCS: PBS +0.05% Tween-20 + 1% FCS |
| Blocking buffer: R10 |
| Enzyme chromogen substrate (See Reagents and Solutions) |
| Sodium acetate (0.1 M) (Sigma-Aldrich, Cat#S2889) (See Reagents and Solutions) |
| AEC working solution (See Reagents and Solutions) |
Coat plates
-
1
Transfer the 1.5 mL of BHK21-LCMV lysate (See Support Protocol 4) into a 50 mL tube and sonicate at level 13 for 30 seconds. It would be enough for coating one 96-well plate.
-
2
Dilute the lysate 1 : 6.7 (1.5 mL lysate + 8.5 mL PBS) in sterile PBS and coat 96-well multiscreen plate (Millipore, MSHAN 4B50) with 100 μL /well and incubate for 48 hr at RT. Plates coated in sterile conditions can be incubated for up to 4 days, but not longer.
-
3
Alternatively, for total antibody detection (not LCMV specific) coat plates with 100 μL/well of 6.0 μg /ml goat anti-mouse IgG+IgM+IgA (60 μL /10 mL PBS).
Block Plates
-
4
Empty the lysate by flicking plates in biohazard waste and wash once again with 200 μL /well of PBS-T followed by 3x with PBS. Tap out the residual liquid. Washing with PBS removes PBS-T and ensures that cells are not lysed.
-
5
Add 200 μL of freshly prepared blocking medium (R10) to each well and incubate for 2 hr at RT.
Incubate with cells
-
6
Replace wells with 100 μL of blocking medium for diluting the cells.
-
7
Resuspend splenic lymphocytes at 10×106 cells/mL (See Support Protocol 3). Add 50 μL (0.5×106) of this single cell suspension to the first row. Make serial three-fold dilutions by pipetting up and down 8X and then transferring 50 μL to the next row (either across or down the plate). Discard 50 μL from the last dilution so that the final volume in all the wells is 100 μL.
-
8
Incubate the plates for 8 hr in a 37°C 5% CO2 incubator. Do not disturb the plate as it may result in duplicated spots.
Incubate with Biotin antibody
-
9
After the incubation, dump off cells and wash 3X with PBS followed by 3X with PBS-T. Washing with PBS ensures that remnant cells are not lysed when PBS-T is used.
-
10
Add 100 μL of biotinylated anti-IgGϒ (Caltag cat # M30115, 0.87mg/ml) or anti-Ig isotype of interest, diluted 1:1000 in PBS-T-FCS. Incubate 2 days at 4°C.
Incubate with HRP-avidin
-
11
Dump off the antibody and then wash the plate 4X with PBS-T.
-
12
Add 100 μL /well HRP-conjugated avidin-D (A-2004, Vector, 5ug/ml) diluted 1:1000 in PBS-T-FCS. Incubate precisely for 60 min at room temp.
Develop spots with chromogen substrate
-
13
Prepare AEC working solution in acetate buffer (See Reagents and Solutions).
-
14
Dump plate and wash 3X with PBS-T and 3X with PBS.
-
15
Add 100 μL enzyme chromogen substrate and incubate for ~8 min (until can visualize red spots and background coloring is just beginning to get significant). Stop reaction by thoroughly washing plate with running tap water. Also remove plastic base and wash underside of plate. Blot dry and store in dark away from moisture and other chemicals to prevent excess fading (colour will fade over time).
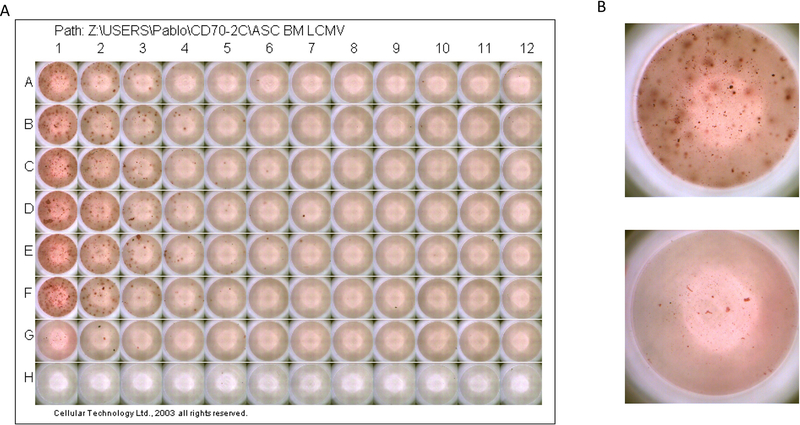
Figure 7 shows representative ASC responses following LCMV Armstrong infection at day 52 post-infection.
Figure 7. Antibody secreting cell (ASC) assay using bone marrow from LCMV Armstrong infected mice.
Data are from day 52 post-infection. Rows A-F represent six different LCMV-immune mice. Row G represents a naïve mouse. Row H received only PBS (no cells). A) 96-well plate at the end of the assay. B) Top: enlarged image of well A1 (LCMV-immune bone marrow); Bottom: enlarged image of well G1 (naïve bone marrow).
Basic Protocol 2.4 Limiting Dilution Assay (LDA) for Detection of LCMV-specific Memory B cells
Another useful method for detecting humoral responses is the LDA for quantification of memory B cells. For a descriptive version of this protocol, refer to a prior publication by Slifka and Ahmed (Slifka & Ahmed, 1996).
Basic Protocol 2.5 ELISA for quantification of LCMV-specific IgG antibody
ELISA is an easy way to quantify antibody responses in sera.
| Materials: |
|---|
| LCMV infected BHK21 cell lysate (For LCMV-specific antibody detection) (See Support Protocol 4). |
| 96 well microplate flat bottom clear (Sigma cat# CLS3358) |
| 96 well microplate sealers or parafilm |
| PBS-T: PBS + 0.5% Tween-20 |
| Blocking solution: PBS-T-FCS: PBS +0.2% Tween-20 + 10% FCS |
| Goat anti-mouse IgG-HRP (Southern Biotech cat# 1030–05) or isotype of interest |
| Microwell peroxidase substrate, SureBlue TMB (SeraCare cat# 5120–0077) |
| Stop solution (1M HCl) (SeraCare cat# 5150–0021) |
| Optical density plate reader |
Coating Plates
-
1
Transfer the 1 mL BHK21-LCMV lysate (See Support Protocol 4) into a 50 mL tube and sonicate at level 13 for 30 seconds. It would be enough for coating of one 96-plate.
-
2
Dilute lysate 1:10 using sterile PBS (1 mL of lysate + 9 mL of sterile PBS, for one 96 well plate).
-
3
Add 100 μL of diluted LCMV lysate to all wells in a 96 well plate. Cover plates with 96 well microplate sealer.
-
4
Incubate at room temperature for 2–4 days, but not longer.
Blocking plates
-
5
Empty wells and wash plates 3X with 200 μL/well of PBS-T.
-
6
After last wash, tap plates down on absorbent paper towels to remove all liquid.
-
7
Immediately proceed to block plates by adding 200 μL of blocking solution to very well. Cover plates with 96 well microplate sealer or parafilm to prevent evaporation or leakage.
-
8
Incubate for 2 hr at room temperature.
Adding samples to plates
-
9
Empty plates and tap plates down on absorbent paper towels to remove all liquid.
-
10
Add 145μL of blocking solution to the first column (A1-H1). Add 100 μL of blocking solution to the rest of the wells. Label wells (one row per sample).
-
11
Add 5μL of your sera sample to the corresponding well in the first column (A1-H1). Mix well by pipetting up and down 10 times. Change tips.
-
12
Perform 3 fold dilutions of your samples by taking 50μL from the first column and adding it to next, mixing 10 times, and changing tips. Repeat, performing 12 serial dilutions. Discard the last 50μL in order to have 100 μL in all wells. Cover plates with 96 well microplate sealer or parafilm.
-
13
Incubate at room temperature for 1.5 hr.
Incubate with detection antibody
-
14
Empty wells and wash plates 3X with 200 μL/well of PBS-T
-
15
After last wash, gently tap plates facing down on absorbent paper towels to remove all liquid.
-
16
Dilute goat anti-mouse IgG HRP 1:5000 in blocking solution (make 10 mL per plate). Mix well.
-
17
Add 100 μL per well. Cover plates with 96 well microplate sealer or parafilm.
-
18
Incubate for 1.5 hr at room temperature.
Adding substrate solution, stop solution, and reading
-
19
Empty wells and wash plates 3X with 200 μL/well of PBS-T
-
20
After last wash, gently tap plates facing down on absorbent paper towel to remove all liquid.
-
21
Add 100 μL of microwell peroxidase substrate solution to each well. Incubate for around 8 minutes or until sufficient color development. Do not overdevelop.
-
22
Add 100 μL of stop solution to each well.
-
23
Tap the plate gently to ensure the stop solution is mixed well. Quickly measure optical density at 490 nm using a microwell plate reader.
Note: To calculate the cut-off value, take the average of the 12 serial dilutions in the naïve sample and multiply by 2. The endpoint titer is then calculated by recording the first dilution above the cut-off value.
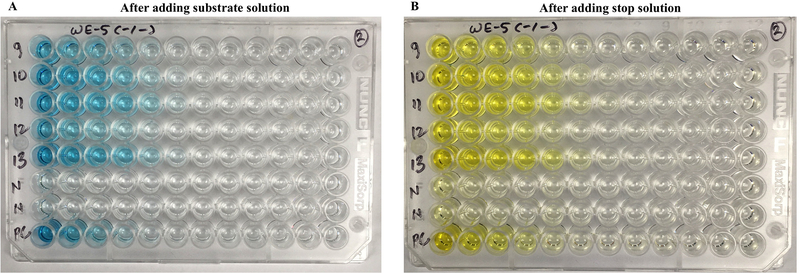
Figure 8 shows representative ELISA plates, using sera from LCMV-immune mice.
Figure 8. ELISA plates for detection of LCMV-specific IgG.
5 μL of sera samples from LCMV-immune mice (9–13), naïve control mice (N), and one Armstrong positive control (PC) was added on the first column of wells (1:30 dilution), followed by 12 serial 3-fold dilutions across the plate. (A) After adding TMB substrate. (B) After adding stop solution. Absorbance was measured in a spectrophotometer. As shown in these images, antibody responses are detected by the development of color in the wells. It is important to sonicate the BHK21-LCMV lysate for accurate results. In this experiment, C57BL/6 Ifnar1−/− mice were immunized with 2×106 PFU of a recombinant LCMV Cl-13 expressing WE glycoprotein (day 12 post-infection).
Support Protocol 3: Preparation of Splenic Lymphocytes
Splenocytes can be isolated from intact spleen harvested from an infected mouse (Basic Protocol 1). Splenocytes are harvested in R10 media, and then used for flow cytometry and other assays.
| Materials: |
|---|
| Spleen from LCMV infected mice (from Basic Protocol 1) |
| PBS (1X) (Gibco, Cat#14190–144) |
| RPMI1640 (Gibco, Cat# 11875–093) |
| Fetal Bovine Serum (Sigma, Cat#F0926–500ml) |
| R5 media (RPMI, 5% FBS, 0.1% Beta merceptoethanol, 1% Pen-Strep, 1% L-glutamine) |
| R10 media (RPMI, 10% FBS, 0.1% Beta merceptoethanol, 1% Pen-Strep, 1% L-glutamine) |
| ACK lysis buffer (See Reagents and Solutions) |
| Trypan Blue solution, 0.4% (Gibco, Cat# 15250061) |
| 1.5 mL Microcentrifuge tubes (Eppendorf, Cat# 05–402-31) |
| 96 well microplate flat bottom clear (Sigma cat# CLS3358) |
| Cell strainer, 100 μM pore size (Dot Scientific, Cat# 667485) |
| Cell counting Glasstic Slide 10 with Grids (Kova International; Cat# 87144) |
| Microscope |
-
Harvest spleen from infected mice.
Note: After harvesting spleen, keep the tubes on ice.
Take a 50 mL conical tube and place a cell strainer over it. Strain the tissue using the back of the sterile syringe. Use R5 media to wash the strainer.
-
Discard the cell strainer and close the lid of the 50 mL conical tube. Pellet cells by spinning tubes at 2,000 rpm for 5 min at 4 °C. Decant supernatant and add 2 mL ACK lysis buffer, vortex briefly, wait 2 min at room temperature (RT).
Note: Do not leave the samples in ACK lysis buffer for longer periods of time, as this may reduce splenocyte viability.
Add ~20 mL R5 media in each tube to neutralize the lysis buffer and spin at 2,000 rpm for 5 min at 4 °C. Decant the tubes. Resuspend pellet in 10 mL complete R10 media and count the cells using a cell counting slide.
For counting, mix 90 μL trypan blue with 10 μL cells (1:10) and then use 10 μL of this mixture for counting.
Spin cells again and resuspend at 10–20 ×106 cells/mL. After this, splenocytes can be used for several immunological assays.
Support Protocol 4: Making BHK21-LCMV lysate
B cell assays require one to coat plates with LCMV antigen. Many laboratories use lysates of LCMV-infected cells as coating antigen for ELISA plates or ASC plates. To prepare LCMV lysate, follow these instructions:
| Materials: |
|---|
| Seed stock of LCMV, 1.5 × 107 PFU/mL (European Virus Archive, SKU:006v-EVA1372) |
| BHK21 cells (ATCC® CCL10™) |
| PBS (Gibco, Cat#14190–144) |
| Fetal Bovine Serum (Sigma, Cat#F0926–500ml) |
| Penicillin/Streptomycin (Pen/Strep) (Gibco, Cat#15140–122) |
| L-Glutamine 200 mM (Gibco, Cat#25030–081) |
| 10% DMEM (supplemented with 10% FBS, 1% Pen/Strep, 1% L-Glutamine) |
| 1% DMEM (supplemented with 1% FBS) |
| 0.25% Trypsin-EDTA (Gibco, Cat#25200–056) |
| Beta 2-mercaptoethanol (Sigma Aldrich, Cat#M6250–100ML) |
| Cryotubes (2ml) for storing viral stock |
| T175 flasks (or T182 cm2 Tissue Culture Flasks, Dot Scientific, Cat#667351) |
| Water bath |
| CO2 Incubator |
Thaw and passage BHK21 cells in 10% FBS DMEM media in a T175 flask. When cells are 70% confluent (usually after 3 days), trypsinize cells and split to propagate BHK21 cells at least twice after thawing. One can use ~ ten T175 flasks for making viral lysates.
When BHK21 are ~70% confluent in all T175 flasks, aspirate media and gently add virus at a multiplicity of infection (MOI) of 0.01 in a volume of 6 mL (1% DMEM) and allow total dispersion among the cell monolayer. Use 1% DMEM containing only 1% FBS. Using higher than 1% FBS would decrease initial viral infection. Note: Handle 5 flasks at a time, so that monolayer does not dry out after aspirating media.
Close the lids of the flasks tightly and rock flasks to ensure that the inoculum is evenly distributed over the BHK21 monolayer. Keep all flasks in a 37°C 5% CO2 incubator for 1 hr, gently rocking flasks every 10 minutes to prevent monolayer from drying.
After incubation, aspirate media and add 25 mL of fresh pre-warmed 5% FBS DMEM media containing 10% FBS, 0.1% Beta 2-mercaptoethanol, 1% Pen-Strep, 1% L-glutamine. Incubate flasks in 37°C 5% CO2 incubator for 48–72 hr.
Rinse the monolayer with cold PBS and discard the supernatant.
Add 3.5 mL of PBS and scrape the cells. Collect the cell suspension into a 50 mL tube with a 5 mL pipette (keep tubes on ice). Add 1.5 mL of PBS again and collect the remaining scraped cells. For all the flasks use the same 5 mL pipette as it minimizes the loss of cell suspension.
Sonicate cells twice at level 13 for 30 seconds without any bubbles.
Dispense the lysate in aliquots and freeze at −80 °C.
It is important to repeat the sonication for 30 seconds before diluting it for coating the plates.
Important notes
BHK21 cells: should be maintained at low passage (<10)
Ensure that the LCMV stock is free of Mycoplasma contamination.
Freshly prepared media should be used.
DMEM (Fisher Scientific, 15–013-CM) must be with sodium pyruvate.
Basic Protocol 3: Challenge models
There are various in vivo challenge models to test immune protection by LCMV-specific immune responses, and selection of a specific challenge model would depend on the experimental question. LCMV Armstrong immune mice are protected from lethal intracerebral LCMV challenge (choriomeningitis). In addition, LCMV Armstrong immune mice exhibit sterilizing immune protection following LCMV Cl-13 challenges. Thus, acute LCMV infection is a model for effective vaccination, protecting animals from lethal LCMV challenge or chronic LCMV challenge. This extraordinary immune protection is conferred by high numbers of memory CD8 T cells of multiple specificities.
Since the level of immune protection in the Armstrong prime / Cl-13 challenge setting is so robust, it does not have the resolution to evaluate small differences between experimental groups. Therefore, some investigators measure anamnestic protection by adoptively transferring low numbers of memory T cells (e.g. 103) into a different host, and then performing LCMV Cl-13 challenge, or a heterologous challenge containing the epitope of interest (E. E. West et al., 2011; E. J. Wherry, V. Teichgraber, et al., 2003). For example, one may first immunize mice with LCMV Armstrong. Then, one would purify GP33-specific CD8 T cells and transfer these cells at low numbers into naïve recipient mice, followed by intravenous challenge with chronic LCMV Cl-13. This type of experimental approach is used to examine how a single memory T cell response contributes to anamnestic protection, in the absence of other memory immune responses.
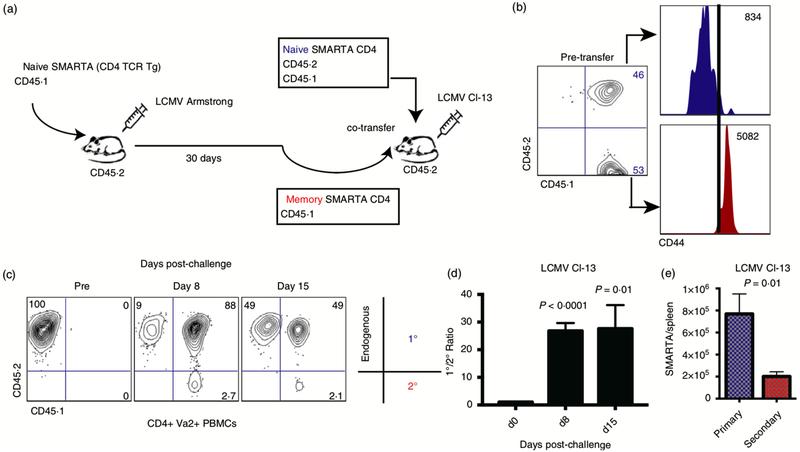
The process of isolating memory CD8 T cells for adoptive transfer studies typically includes MACS negative selection, followed by FACS-based separation using tetramers, which results in a highly pure population of memory virus-specific CD8 T cells. Then, the number of transferred cells is normalized between experimental groups, followed by challenge one day after. In Protocol 4, we will explain how TCR transgenic cells (P14 and SMARTA) could also be utilized in adoptive transfer studies to evaluate anamnestic protection by memory T cells.
Arguably, a caveat of adoptively transferring memory cells is that they are taken out of their original host and transferred at low numbers into a naive host devoid of other memory responses. Therefore, some investigators opt to perform prime immunizations and challenges in the same host. For example, one may first immunize mice with Listeria monocytogenes expressing LCMV GP33 (LM-GP33), and after 30–60 days, challenge mice with LCMV Cl-13. This would allow evaluation of a single epitope-specific CD8 T cell responses (GP33) in an unmanipulated host (P. Penaloza-MacMaster, D. L. Barber, et al., 2015). In this context, LM-GP33 immune mice will clear an LCMV Cl-13 challenge within a few days, allowing the investigator to detect differences in viral control between different experiment groups. However, a potential caveat of using the same host for immunization / challenge studies is variability in the number of memory (GP33-specific) cells, and the fact that the immune host may harbor memory cells with different phenotypes and localization. To control for these variables, investigators can perform adoptive transfer of memory T cells of a specific phenotype and from a single tissue (usually spleen, because it gives high number of cells). This approach would also allow the investigator to be consistent in terms of selecting the specific number of memory cells that each mouse will receive.
Basic Protocol 4: Transgenic models
TCR and BCR transgenic models are commonly used in challenge studies, and also to interrogate the effect of specific immune pathways. An advantage of using TCR and BCR transgenic models is that they allow the investigator to cross the mice to other strains. For example, P14 mice (which contain LCMV GP33-specific CD8 T cells) may be crossed to Ifnar1−/− mice to interrogate the effect of type I interferon (IFN-I) in the CD8 T cell response. An important consideration for adoptive transfers is that the donor and recipient mice are backcrossed to the same C57BL/6 vendor strain. Note that P14 cells backcrossed on the Jackson C57BL/6 background (#000664) are rejected if transferred into Taconic C57BL/6 mice. This may be due to small genetic differences between C57BL/6 from different vendors. The number of adoptively transferred cells would depend on the experimental question. For example, if one wants to interrogate early events in CD8 T cell activation (e.g. day 3 post-infection), it is advisable to transfer a high number of P14 cells to increase detection (106 cells). However, if one wants to interrogate late events in CD8 T cell differentiation (e.g. ≥ day 7 post-infection), it is advisable to transfer a low, physiological number of P14 cells (102 cells). These cells could also be labeled with CFSE or CTV to measure proliferation.
| Mouse | Description |
|---|---|
| P14 (Jackson laboratories # 37394) | CD8 TCR Tg (GP33-specific CD8 T cells). (Pircher, Burki, Lang, Hengartner, & Zinkernagel, 1989) |
| SMARTA (Jackson laboratories # 030450) | CD4 TCR Tg (GP61-specific CD4 T cells). (Oxenius, Bachmann, Zinkernagel, & Hengartner, 1998) |
| KL25 or KL25-H (European virus Archive) | BCR Tg (GP-specific B cells). (Hangartner et al., 2003; Seiler et al., 1998) |
P14 and SMARTA cells could be used with LCMV Armstrong and Cl-13 strains, but the KL25 (or KL25-H) must be used with the LCMV WE strain, or a recombinant LCMV Cl-13 expressing the WE glycoprotein (GP), both available from the European Virus Archive.
Below, see an example of an adoptive transfer experiment to investigate the role of IFN-I on CD8 T cell responses:
Transfer of P14 cells to interrogate the role of IFN-I on CD8 T cell responses
| Materials: |
|---|
| P14 mice (Jackson laboratories #37394) |
| Ifnar1−/− mice (Jackson laboratories #32045) |
| C57BL/6, Thy1.2+ mice (Jackson laboratories #000664). |
| LCMV Armstrong (107 PFU/mL) (See support protocol 1) |
| EasySep™ Mouse CD8+ T Cell Isolation Kit (STEMCELL Technologies, Cat#19853) |
| Fetal Bovine Serum (Sigma, Cat#F0926–500ml) |
| Complete RPMI 1640 (Gibco, Cat#11875–093) |
| L-Glutamine 200 mM (Gibco, Cat#25030–081) |
| VV-GP, Vaccinia Virus Expressing LCMV Glycoprotein (NR-15497, BEI Resources) |
| Ad5-expressing LCMV-GP (Provided by Dr. John Mascola, NIH) |
| LM-GP33, Listeria monocytogenes expressing LCMV GP (Provided by Dr. Rafi Ahmed, Emory University) |
| PR8-GP33 (Provided by E. John Wherry, University of Pennsylvania) |
| MHV-GP33 (Provided by Dr. Susan Weiss, University of Pennsylvania) |
| Round Bottom FACS tubes with cap (Falcon, Cat#352054) |
| FACS tubes without cap (Falcon, Cat#352008) |
| 96-well round bottom plates, Costar (Corning Inc., Cat#3799) |
Backcross P14 mice to Ifnar1−/− mice (Jackson laboratories) until a homozygous population is obtained. The wild type P14 mice could be backcrossed to the Thy1.1 congenic marker, whereas the Ifnar1−/− P14 mice could contain both the Thy1.1 and Thy1.2 congenic markers to allow the investigator to distinguish these populations in vivo.
Harvest a spleen from a P14 mouse and an Ifnar1−/− P14 mouse in separate tubes containing complete RPMI media supplemented with 1% FBS. Prepare single cell suspension of splenocytes (See Support Protocol 3).
To isolate highly purified CD8+ T-cells from above splenocytes suspension, perform MACS negative selection procedure following the manufacturers protocol of EasySep™ CD8+ T Cell Isolation Kit. This kit gives 95% purity of CD8 T-cells.
-
Co-transfer 102 wild type P14 cells (Thy1.1+) and 102 Ifnar1−/− P14 cells (Thy1.1+ Thy1.2+) intravenously into recipient C57BL/6 Thy1.2+ mice (Jackson laboratories #000664). This low number of GP33-specific CD8 T cells is similar to the endogenous frequency in naïve mice (Blattman et al., 2002), and thus, the P14 transfer will not substantially affect acute viral control.
Note: Typically, 6–8 week-old P14 mice are used to isolate LCMV-GP33 specific CD8 cells from spleen. Both donor and recipient mice should be sex matched for adoptive transfer experiments. One mouse can provide ~5 million P14 cells from each P14 spleen.
After 24 hr, infect mice intraperitoneally with 2×105 PFU LCMV Armstrong. Alternatively, one could infect mice with any vector expressing the whole LCMV glycoprotein or GP33, for instance, VV-GP, Ad5-GP, LM-GP33, PR8-GP33 or MHV-GP33.
Bleed after a week and perform tetramer staining in PBMCs to detect expansion of donor GP33-specific (P14) cells using antibodies against Thy1.1 and Thy1.2 to distinguish between the 2 donor populations and the endogenous population (see Basic Protocol 2.1 for a general description of the tetramer staining protocol).
Sample data can be seen in prior papers (Kolumam, Thomas, Thompson, Sprent, & Murali-Krishna, 2005).
Note: The protocol above could be modified, but crossing the Ifnar1−/− mice with SMARTA mice instead, which would allow analysis of how IFN-I affects CD4 T cell expansion (Havenar-Daughton, Kolumam, & Murali-Krishna, 2006). Note that the SMARTA cells downregulate Thy1.1 after viral infection, and thus, it is best to utilize the CD45.1 or CD45.2 congenic markers when using the SMARTA system. Adoptively transferred SMARTA cells do not undergo much expansion or persistence after viral infection relative to P14 cells, and they quickly cull down after 3 weeks. Therefore, it is advisable to transfer between 104 −105 SMARTA cells to be able to visualize them after viral infection. A critical consideration is that injection of >106 naive SMARTA cells can result in immunopathology after chronic LCMV Cl-13 challenge (Bhattacharyya et al., 2017a).
Alternatively, the protocol above can also be modified, crossing the Ifnar1−/− mice with KL25 mice for analyzing the role of IFN-I on B cell expansion. In this setting, it is critical to utilize LCMV WE or rLCMV Cl-13 expressing the WE glycoprotein, since KL25 B cells recognize the WE glycoprotein, but not the Cl-13 or Armstrong glycoprotein. Also, CD45.1 and CD45.2 should be used as congenic markers for these experiments involving KL25 B cells. A more physiological model may be the KL25-H model, in which transgenic B cells contain only the antibody heavy chain gene specific for LCMV, which allows for endogenous light chain pairing and broader diversity in terms of LCMV-specific antibody responses. A critical consideration is that tracking KL25 B cells (or KL25H B cells) is technically difficult, as these cells do not exhibit much expansion after viral infection and may even undergo progressive decimation in vivo, especially in the setting of chronic LCMV infection (Fallet et al., 2016; Moseman, Wu, de la Torre, Schwartzberg, & McGavern, 2016; Sammicheli et al., 2016). Therefore, it is advisable to adoptively transfer high numbers (>5×106) of KL25 B cells to allow their detection in vivo.
As mentioned earlier, adoptive transfer of transgenic cells could also be used to interrogate immune protection by memory lymphocytes. For example, we previously interrogated whether memory CD4 T cells would exhibit enhanced expansion relative to naïve CD4 T cells. To answer this simple question, we performed the following experiment from our prior publication (Bhattacharyya et al., 2017a).
Comparing the expansion of naive versus memory CD4 T cells following chronic viral challenge
| Materials: |
|---|
| CD45.1+ SMARTA mice (Jackson laboratories #030450) |
| CD45.2+ C57BL/6 mice (Jackson laboratories #000664 |
| LCMV Armstrong (107 PFU/mL) (see Basic protocol 1 or support protocol 1) |
| LCMV Cl-13 (107 PFU/mL) |
| EasySep™ Mouse CD4+ T Cell Isolation Kit (STEMCELL Technologies, Cat#19852) |
| Complete RPMI 1640 (Gibco, Cat#11875–093) |
| Fetal Bovine Serum (Sigma, Cat#F0926–500ml) |
| L-Glutamine 200 mM (Gibco, Cat#25030–081) |
| 96-well round bottom plates, Costar (Corning Inc., Cat#3799) |
| Round Bottom FACS tubes with cap (Falcon, Cat#352054) |
| FACS tubes without cap (Falcon, Cat#352008) |
Harvest a spleen from a CD45.1+ SMARTA mouse (Jackson laboratories) and prepare a single cell suspension of splenocytes at 10×106 cells/mL concentration (See Support Protocol 3)
Purify CD4+ T-cells from splenocytes suspension by performing MACS negative selection procedure following the manufacturers recommendations of EasySep™ Mouse CD4+ T Cell Isolation Kit. This step gives 95% purity of CD4 T-cells.
Inject ~105 purified naïve CD45.1+ SMARTA CD4 T cells intravenously into CD45.2+ C57BL/6 recipient mice (Jackson laboratories),.
After 24 hr, infect recipient mice LCMV Armstrong (2×105 PFU, intraperitoneally).
After 30 days, purify CD4 T cells using the same MACS protocol from step 2.
Perform FACS-sorting to purify memory SMARTA CD4 T cells. FACS-sorting to ≥ 98% purity can be achieved by staining CD4-enriched cells with CD45.2 and CD8 antibodies, resulting in untouched memory SMARTA cells for adoptive transfers (CD45.2- and CD8-).
Co-transfer 105 memory CD45.1+ SMARTA CD4 T cells, with 105 naive CD45.1+CD45.2+ SMARTA CD4 T cells into naive CD45.2+ C57BL/6 recipient mice (Jackson laboratories).
After 24 hr, infect recipient mice LCMV Cl-13 (2×106 PFU, intravenously).
Bleed mice weekly and evaluate naïve and memory SMARTA cell expansion in PBMCs.
Note: sample data shown in Figure 9 was obtained from our prior publication (Bhattacharyya et al., 2017a). Paradoxically, memory SMARTA cells expand less than naïve SMARTA cells, when co-transferred at equal numbers into the same host. One could also transfer memory and naïve SMARTA cells separately into different mice, and then compare immune responses and protection following LCMV Cl-13 challenge. In this context, memory CD4 T cells conferred improved help to adaptive immune responses, relative to naïve CD4 T cells (Bhattacharyya et al., 2017a).
Finally, one could also knock down specific genes in transgenic T cells prior to adoptive transfer, using a retroviral RNAi system; see detailed protocol in the following publication (Araki & Konieczny, 2012).
Figure 9. Comparing memory versus naïve SMARTA cell expansion following chronic LCMV challenge.
A) Experimental outline. B) CD44 expression on naive and memory SMARTA CD4 T cells before injection to corroborate antigen experience. C) Representative FACS plots comparing the frequencies of primary and secondary SMARTA effectors (gated on total CD4 T cells). D) Ratio of primary and secondary SMARTA responses. E) Number of SMARTA cells in spleen at day 8 after lymphocytic choriomeningitis virus (LCMV) Cl-13 infection. In these experiments, 105 naive and 105 memory SMARTA cells (day 30 post-immunization) from spleen were co-transferred at (1:1 ratio) into congenically distinct mice, followed by LCMV Cl-13 challenge 1 day after CD4 T-cell transfer. SMARTA cells were evaluated in PBMCs in all panels unless indicated otherwise. Data are from two experiments, n = 4–5 mice/group per experiment. Error bars indicate SEM.
REAGENTS AND SOLUTIONS
ACK lysis buffer
NH4Cl (Amresco, Cat# 0621–1KG)
KHCO3 (Sigma Aldrich, Cat# 298–14-6)
Na2EDTA.2H2O (Invitrogen, Cat# 15576–028)
Stock solution (10X): Dissolve 80.2 gm NH4Cl, 10.0 gm KHCO3 and 0.37 gm Na2EDTA.2H2O in 500 mL distilled water and make up to 1000 mL. Adjust pH at 7.4, filter it and store at 4°C.
Working ACK solution (1X): To prepare 100 mL of working solution, dilute 10 mL from ACK stock (10X) in 90 mL sterile distilled water.
1% Agarose
Agarose powder (SeaKem ME, Cat# Lonza, 50014)
Dissolve 4 gm of Agarose in 400 mL Milli-Q-grade water, autoclave and store at room temperature.
Enzyme chromogen substrate
AEC [3 amino-9-ethyl-carbozole (MP Biochemical, Cat# 195039)
N, N-dimethylformamide (Sigma-Aldrich, Cat# 227056)
0.1 M Na-Acetate buffer (pH 4.8–5.0) (Sigma-Aldrich, Cat# S2889)
3% H202 (Sigma-Aldrich, Cat# H1009–100 mL)
AEC Stock Solution: Make a 20 mg/mL stock solution of AEC (3 amino-9-ethyl-carbozole) in N, N-dimethylformamide. Store at 4°C in the dark for up to 1 month.
AEC working solution: It should be prepare freshly. Dilute AEC stock solution 1 : 67 into 0.1 M Na-Acetate buffer (pH 4.8–5.0), giving a final concentration of 0.3 mg/mL (For example use 150 uL stock AEC per 10 mL acetate buffer). Filter this solution through a 0.2 micron filter before use.
To make Enzyme chromogen substrate, add 100 μL 3% H202 per 10 mL of AEC substrate (working solution). It should be prepared immediately before use.
Note: Alternatively, a pre-made enzyme chromogen substrate could be purchased. MP biochemical AEC seems to give clearer spots and filters better than AEC from Sigma.
2×199 Media
Medium 199 (Gibco, Cat# 31100–027)
NaHCO3 (Fisher Scientific, Cat# S233–500)
Dissolve two bags of Medium 199 and 4.4 gm NaHCO3 in 1L-distilled water.
Mix well and take out 800 mL in another sterile bottle and add 200 mL FBS, 20 mL L-Glutamine, and 20 mL Pen/Strep. Filter the medium and keep at 4°C.
1% Neutral Red
Neutral red powder (Fisher Chemical, Cat# N129–25)
Dissolve 4 gm of Neutral red powder in 400 mL Milli-Q-grade water and autoclave. Cover the bottle with aluminum foil to protect from light and store at 4°C.
Sodium acetate (0.1 M)
Sodium acetate (Sigma-Aldrich, Cat# S2889)
Dissolve 6.804 gm Sodium acetate in 300 mL of water, adjust pH to 4.8–5.0, make it to 500 mL with water. Ensure that all liquid biological waste is decontaminated in a 10% bleach reservoir prior to disposal.
COMMENTARY
BACKGROUND INFORMATION
The protocols and tools used in the LCMV model have been generated by multiple laboratories. Concerted efforts by Zinkernagel, Ahmed, Oldstone and others, have contributed to the success of this model. Other viral models can also be used to study antiviral immunity, but an advantage of the LCMV model is the availability of a diverse toolkit to study immune responses in a natural host. This toolkit includes many mapped epitopes, transgenic systems, and various assays to evaluate antiviral immunity in detail.
CRITICAL PARAMETERS and TROUBLESHOOTING
It is critical that intravenous LCMV Cl-13 infections are performed in a high volume (500 μL), since this will result in more consistent results. We recommend the tail vein injection method, as the investigator would be able to ensure that the whole bolus of virus is administered. Performing LCMV infections retro-orbitally may result in incomplete intravascular dissemination if these are not done well.
All of the assays described here are reproducible and straightforward, except for the LCMV plaque assay, which often stops working, even in the hands of experienced investigators. If the LCMV plaque assays stop working, consider these common sources of error:
Using high-passage Vero e6 cells? Purchase new Vero e6 cells.
Adding the agar overlay too hot and too abruptly? Be gentle when overlaying the Vero e6 monolayer.
Handling too many plates at once and letting the monolayer dry out? It is critical to manually rock the plates every 10 minutes to avoid the monolayer from drying out.
Vero e6 monolayer does not have the right confluency? Ideally, the monolayer should be 95–100% confluent. If less than 70% confluent, this may result in empty areas in the monolayer with detached morphology. If >100% confluent, this may result in lower PFU.
Are the plaques too faint? This could happen if the neutral red solution is expired.
If the plaque assays stop working, also go back to the shelf and check if new reagents are being used. Certain lots of FBS (or FCS) could cause cells to die or not grow well. In addition, due to variability in the plaque assay, it is recommended that pre-treatment PFU and post-treatment PFU are determined on the same day (if longitudinally evaluating viral loads in sera).
Considerations for maintaining and phenotyping P14, SMARTA, and KL25 mice
The colonies of P14 mice and SMARTA mice should always be backcrossed to wild type C57BL/6 mice. For example, a P14 male mouse can be bred with 2 wild type C57BL/6 females harboring a relevant congenic marker (e.g. Jackson laboratories mice #000406, which is Thy1.1). Alternatively, a SMARTA male mouse can be bred with 2 wild type C57BL/6 females harboring the CD45.1 congenic marker (e.g. Jackson laboratories mice #002014, which is also known as Pep mouse).
Backcrossing to wild type mice is important because homologous pairing of these mice (P14 male x P14 female; or SMARTA male x SMARTA female) do not breed well.
To identify the pups that carry the P14 TCR transgene, one could simply perform a tetramer stain using DbGP33 tetramer or antibodies against the Vα2 TCR. ~99% CD8 T cells in P14 mice should be DbGP33+ (or Vα2+). To identify the pups that carry the SMARTA TCR transgene, one could use antibodies against the Vα2 TCR. >90% CD4 T cells in SMARTA mice should be Vα2+, but this frequency may decline as the mouse ages, for reasons that are not clear. This may be due to time-dependent downregulation of the TCR in CD4 T cells. PCR-based techniques using appropriate primers for detecting TCR genes can also be used for genotyping P14 and SMARTA mice.
The colonies of KL25 mice are much easier to maintain. These mice can be crossed with themselves without losing fertility (KL25 male x KL25 female), although laboratories sporadically backcross them to wild type C57BL/6 from their vendor of choice to keep the colony similar to wild type and prevent rejection in adoptive transfer studies. Offspring could be phenotyped using the IIIC4.8 antibody, which recognizes the KL25 BCR heavy chain. All B220+ cells in KL25 (or KL25-H mice) should stain positive for IIIC4.8. The IIIC4.8 antibody clone and the KL25 mice are available thorough the European Virus Archive.
UNDERSTANDING RESULTS
We provide examples of anticipated results and explanation of the data at the end of the protocols.
TIME CONSIDERATIONS
Important time considerations are indicated with each protocol.
Basic Protocol 1: LCMV infection routes
Most routes of infection are fast, involving only a few seconds per mouse, except for the intravenous route. This route of infection involves the pre-warming of the mice, which may take approximately 5–10 minutes, followed by tail vein injection. Time would depend on the experience and dexterity of the investigator.
Basic Protocol 2: Measurement of T cell and antibody responses
Evaluation of CD8+ T cells/CD4+ T-cells by tetramer staining takes 4–5 hours depends upon number of samples and it includes blood collection, PBMC isolation and staining of cells. Intracellular cytokine assay takes little longer time than tetramer staining, where most critical step is to induce lymphocytes in presence of LCMV-specific peptides that need 5 hours incubation. Lower incubation will results insufficient induction of cells and affects end results while incubation more than 5 hours affects the viability of cells. LCMV specific memory cells develop after 30 days post infection, so it take more than 1 month to detect these cells after infection. For quantification of antibody secreting cells by ELISPOT, total 3 more days are needed after establishing of memory cells (after 30 days post infection). Other method of detecting memory cells by Limiting dilution assays takes 6 additional days, after mice develop memory cells by day 30 post-infection.
Basic Protocol 3: Challenge models
The process of giving immunization and developing immune memory takes more than one moth and after challenging mice and to measure immune response need 2 weeks. In total, 1.5–2 months are needed to check the vaccine efficacy in vivo.
Basic Protocol 4: Transgenic models
Overall process require one day for Isolating LCMV specific CD4+/CD8+ T-cells from spleen of chimeric mice and injecting into recipient mice. After adoptive transfer of immune cells and infecting mice, mice should be monitored for 7 days to proliferate donor cells and proliferation of these cells can be measured by specific tetramer staining.
Support Protocol 1 Preparation of LCMV stock
Seeding of flask with BHK cells and to reach 60–70% confluency of cells take 3–4 days, followed by infection with viral dose (2 hours), and incubation (2–3 days) and harvesting/purification (1 day). Overall, the viral stock can be prepared in 7 days.
Support Protocol 2 Assay to measure LCMV titers
In conventional plaque assay, preparation of seed flask with 100% confluent vero cells takes 2–3 days and splitting of these cells into appropriate 6-well plates takes another 3 days. The whole process of preparing plates, infecting monolayers, plaques development and staining takes two weeks. IFA complete within 3 days that includes propagating cells (1 day), infecting monolayer (24 hrs incubation) and staining (1 day), which is less time consuming than conventional plaque assay and require less sample volume.
Support Protocol 3 Preparation of Splenic Lymphocytes
From harvesting spleen to isolation of splenocytes, it takes 3–4 hours depends upon the number of samples. Processing and staining of isolated lymphocytes takes 1–2 hours.
Support Protocol 4 Making BHK21-LCMV lysate
Propagation of BHK21 cells up to 70% confluency takes 3–4 days. After infecting monolayer with LCMV, it takes 2–3 days to propagate virus and 1 hour to prepare LCMV lysate.
SIGNIFICANCE STATEMENT.
The Lymphocytic Choriomeningitis Virus (LCMV) model is a powerful tool to evaluate immune responses in mice. Many important discoveries in immunology were first identified using the LCMV system, including the discovery of MHC restriction (Zinkernagel & Doherty, 1974), immune exhaustion (Moskophidis, Lechner, Pircher, & Zinkernagel, 1993; Zajac et al., 1998), and immune memory (Murali-Krishna et al., 1999; Slifka, Antia, Whitmire, & Ahmed, 1998; Wherry, Barber, Kaech, Blattman, & Ahmed, 2004). A reason why this viral model is so useful in immunology is because it has many experimental tools to study adaptive immune responses.
Although there are many antigen models in immunology, the LCMV model has the advantage of being a natural infection, with more than a dozen different T cell epitopes mapped, allowing detailed characterization of immunodominant and sub-immunodominant T cell responses. The LCMV model also has various TCR and BCR transgenic systems that facilitate adoptive transfer studies. LCMV is highly immunogenic in its natural host, the mouse. A striking feature of this virus is its rapid evolution and diversity, which has resulted in the isolation of various acute and chronic strains. This can allow well-controlled comparison of immune responses in the context of viral clearance or viral persistence. Overall, the LCMV system has been (and continues to be) an important research tool to understand adaptive immunity.
ACKNOWLEDGEMENTS
NIH (1R21AI132848-01A1) to Pablo Penaloza.
These studies were funded by a grant from the National Institutes of Health (1R21AI132848-01A1) to P.P.M.
LITERATURE CITED
- Ahmed R, Hahn CS, Somasundaram T, Villarete L, Matloubian M, & Strauss JH (1991). Molecular basis of organ-specific selection of viral variants during chronic infection. J Virol, 65(8), 4242–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed R, & Oldstone MB (1988). Organ-specific selection of viral variants during chronic infection. J Exp Med, 167(5), 1719–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed R, Salmi A, Butler LD, Chiller JM, & Oldstone MB (1984a). Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. The Journal of experimental medicine, 160(2), 521–540. doi:papers3://publication/uuid/A32D2154–4C81–4A9A-B77C-A2819D08E8A9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed R, Salmi A, Butler LD, Chiller JM, & Oldstone MB (1984b). Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med, 160(2), 521–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, & Konieczny BT (2012). Utilizing a retroviral RNAi system to investigate in vivo mTOR functions in T cells. Methods Mol Biol, 821, 305–316. doi: 10.1007/978-1-61779-430-8_19 [DOI] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, … Ahmed R (2006). Restoring function in exhausted CD8 T cells during chronic viral infection. Nature, 439(7077), 682–687. doi: 10.1038/nature04444 [DOI] [PubMed] [Google Scholar]
- Bergthaler A, Flatz L, Hegazy AN, Johnson S, Horvath E, Lohning M, & Pinschewer DD (2010). Viral replicative capacity is the primary determinant of lymphocytic choriomeningitis virus persistence and immunosuppression. Proc Natl Acad Sci U S A, 107(50), 21641–21646. doi: 10.1073/pnas.1011998107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya M, Madden P, Henning N, Gregory S, Aid M, Martinot A, … Penaloza-MacMaster P (2017a). Regulation of CD4 T cells and their effects on immunopathological inflammation following viral infection. Immunology. doi: 10.1111/imm.12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya M, Madden P, Henning N, Gregory S, Aid M, Martinot AJ, … Penaloza-MacMaster P (2017b). Regulation of CD4 T cells and their effects on immunopathological inflammation following viral infection. Immunology, 152(2), 328–343. doi: 10.1111/imm.12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya M, & Penaloza-MacMaster P (2017). T regulatory cells are critical for the maintenance, anamnestic expansion and protection elicited by vaccine-induced CD8 T cells. Immunology, 151(3), 340–348. doi: 10.1111/imm.12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya M, & Penaloza-MacMaster P (2018). Dynamics of Lymphocyte Reconstitution After Hematopoietic Transplantation During Chronic Lymphocytic Choriomeningitis Virus Infection. AIDS Res Hum Retroviruses, 34(5), 430–438. doi: 10.1089/AID.2017.0251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, … Wherry EJ (2009). Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol, 10(1), 29–37. doi: 10.1038/ni.1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattman JN, Antia R, Sourdive DJ, Wang X, Kaech SM, Murali-Krishna K, … Ahmed R (2002). Estimating the precursor frequency of naive antigen-specific CD8 T cells. J Exp Med, 195(5), 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, … Walker BD (2006). PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature, 443(7109), 350–354. doi: 10.1038/nature05115 [DOI] [PubMed] [Google Scholar]
- Fallet B, Narr K, Ertuna YI, Remy M, Sommerstein R, Cornille K, … Pinschewer DD (2016). Interferon-driven deletion of antiviral B cells at the onset of chronic infection. Sci Immunol, 1(4). doi: 10.1126/sciimmunol.aah6817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangartner L, Senn BM, Ledermann B, Kalinke U, Seiler P, Bucher E, … Hengartner H (2003). Antiviral immune responses in gene-targeted mice expressing the immunoglobulin heavy chain of virus-neutralizing antibodies. Proc Natl Acad Sci U S A, 100(22), 12883–12888. doi: 10.1073/pnas.2135542100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Kolumam GA, & Murali-Krishna K (2006). Cutting Edge: The direct action of type I IFN on CD4 T cells is critical for sustaining clonal expansion in response to a viral but not a bacterial infection. J Immunol, 176(6), 3315–3319. doi: 10.4049/jimmunol.176.6.3315 [DOI] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, … Ahmed R (2016). Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature, 537(7620), 417–421. doi: 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, … Ahmed R (2017). Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science, 355(6332), 1423–1427. doi: 10.1126/science.aaf0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CC, Jamieson BD, Reddy K, Bali N, Concepcion RJ, & Ahmed R (1992). Viral infection of the thymus. J Virol, 66(5), 3155–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolumam GA, Thomas S, Thompson LJ, Sprent J, & Murali-Krishna K (2005). Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med, 202(5), 637–650. doi: 10.1084/jem.20050821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann-Grube F, Lohler J, Utermohlen O, & Gegin C (1993). Antiviral immune responses of lymphocytic choriomeningitis virus-infected mice lacking CD8+ T lymphocytes because of disruption of the beta 2-microglobulin gene. J Virol, 67(1), 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCausland MM, & Crotty S (2008). Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. J Virol Methods, 147(1), 167–176. doi: 10.1016/j.jviromet.2007.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseman EA, Wu T, de la Torre JC, Schwartzberg PL, & McGavern DB (2016). Type I interferon suppresses virus-specific B cell responses by modulating CD8(+) T cell differentiation. Sci Immunol, 1(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskophidis D, Lechner F, Pircher H, & Zinkernagel RM (1993). Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature, 362(6422), 758–761. doi: 10.1038/362758a0 [DOI] [PubMed] [Google Scholar]
- Murali-Krishna K, Lau LL, Sambhara S, Lemonnier F, Altman J, & Ahmed R (1999). Persistence of memory CD8 T cells in MHC class I-deficient mice. Science, 286(5443), 1377–1381. doi:7955 [pii] [DOI] [PubMed] [Google Scholar]
- Oxenius A, Bachmann MF, Zinkernagel RM, & Hengartner H (1998). Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur J Immunol, 28(1), 390–400. doi: [DOI] [PubMed] [Google Scholar]
- Penaloza-MacMaster P, Barber DL, Wherry EJ, Provine NM, Teigler JE, Parenteau L, … Barouch DH (2015). Vaccine-elicited CD4 T cells induce immunopathology after chronic LCMV infection. Science (New York, N.Y.), 347(6219), 278–282. doi:papers3://publication/doi/ 10.1126/science.aaa2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster P, Barber DL, Wherry EJ, Provine NM, Teigler JE, Parenteau L, … Barouch DH (2015). Vaccine-elicited CD4 T cells induce immunopathology after chronic LCMV infection. Science, 347(6219), 278–282. doi: 10.1126/science.aaa2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster P, Kamphorst AO, Wieland A, Araki K, Iyer SS, West EE, … Ahmed R (2014). Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. The Journal of experimental medicine, 211(9), 1905–1918. doi:papers3://publication/doi/ 10.1084/jem.20132577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster P, Masopust D, & Ahmed R (2009). T-cell reconstitution without T-cell immunopathology in two models of T-cell-mediated tissue destruction. Immunology, 128(2), 164–171. doi:papers3://publication/doi/ 10.1111/j.1365-2567.2009.03080.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster P, Provine NM, Blass E, & Barouch DH (2015). CD4 T Cell Depletion Substantially Augments the Rescue Potential of PD-L1 Blockade for Deeply Exhausted CD8 T Cells. J Immunol, 195(3), 1054–1063. doi: 10.4049/jimmunol.1403237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster P, Teigler JE, Obeng RC, Kang ZH, Provine NM, Parenteau L, … Barouch DH (2014). Augmented replicative capacity of the boosting antigen improves the protective efficacy of heterologous prime-boost vaccine regimens. Journal of Virology, 88(11), 6243–6254. doi:papers3://publication/doi/ 10.1128/JVI.00406-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster P, Ur Rasheed A, Iyer SS, Yagita H, Blazar BR, & Ahmed R (2011). Opposing effects of CD70 costimulation during acute and chronic lymphocytic choriomeningitis virus infection of mice. Journal of Virology, 85(13), 6168–6174. doi:papers3://publication/doi/ 10.1128/JVI.02205-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircher H, Burki K, Lang R, Hengartner H, & Zinkernagel RM (1989). Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature, 342(6249), 559–561. doi: 10.1038/342559a0 [DOI] [PubMed] [Google Scholar]
- Sammicheli S, Kuka M, Di Lucia P, de Oya NJ, De Giovanni M, Fioravanti J, … Iannacone M (2016). Inflammatory monocytes hinder antiviral B cell responses. Sci Immunol, 1(4). doi: 10.1126/sciimmunol.aah6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler P, Kalinke U, Rulicke T, Bucher EM, Bose C, Zinkernagel RM, & Hengartner H (1998). Enhanced virus clearance by early inducible lymphocytic choriomeningitis virus-neutralizing antibodies in immunoglobulin-transgenic mice. J Virol, 72(3), 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifka MK, & Ahmed R (1996). Limiting dilution analysis of virus-specific memory B cells by an ELISPOT assay. J Immunol Methods, 199(1), 37–46. doi: 10.1016/s0022-1759(96)00146-9 [DOI] [PubMed] [Google Scholar]
- Slifka MK, Antia R, Whitmire JK, & Ahmed R (1998). Humoral immunity due to long-lived plasma cells. Immunity, 8(3), 363–372. doi: 10.1016/s1074-7613(00)80541-5 [DOI] [PubMed] [Google Scholar]
- Smelt SC, Borrow P, Kunz S, Cao W, Tishon A, Lewicki H, … Oldstone MB (2001). Differences in affinity of binding of lymphocytic choriomeningitis virus strains to the cellular receptor alpha-dystroglycan correlate with viral tropism and disease kinetics. J Virol, 75(1), 448–457. doi: 10.1128/JVI.75.1.448-457.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velu V, Titanji K, Zhu B, Husain S, Pladevega A, Lai L, … Amara RR (2009). Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature, 458(7235), 206–210. doi: 10.1038/nature07662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezys V, Penaloza-MacMaster P, Barber DL, Ha SJ, Konieczny B, Freeman GJ, … Ahmed R (2011). 4–1BB signaling synergizes with programmed death ligand 1 blockade to augment CD8 T cell responses during chronic viral infection. J Immunol, 187(4), 1634–1642. doi: 10.4049/jimmunol.1100077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner SN, Cornberg M, Selin LK, & Welsh RM (2012). Natural killer cells act as rheostats modulating antiviral T cells. Nature, 481(7381), 394–398. doi: 10.1038/nature10624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chung YR, Eitzinger S, Palacio N, Gregory S, Bhattacharyya M, & Penaloza-MacMaster P (2019). TLR4 signaling improves PD-1 blockade therapy during chronic viral infection. PLoS Pathog, 15(2), e1007583. doi: 10.1371/journal.ppat.1007583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West EE, Jin H-T, Rasheed A-U, Penaloza-MacMaster P, Ha S-J, Tan WG, … Ahmed R (2013). PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. The Journal of clinical investigation, 123(6), 2604–2615. doi:papers3://publication/doi/10.1172/JCI67008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West EE, Youngblood B, Tan WG, Jin HT, Araki K, Alexe G, … Ahmed R (2011). Tight regulation of memory CD8(+) T cells limits their effectiveness during sustained high viral load. Immunity, 35(2), 285–298. doi: 10.1016/j.immuni.2011.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Barber DL, Kaech SM, Blattman JN, & Ahmed R (2004). Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc Natl Acad Sci U S A, 101(45), 16004–16009. doi:0407192101 [pii] 10.1073/pnas.0407192101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, & Ahmed R (2003). Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. Journal of Virology, 77(8), 4911–4927. doi:papers3://publication/doi/ 10.1128/JVI.77.8.4911-4927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, & Ahmed R (2003). Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol, 77(8), 4911–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, … Ahmed R (2007). Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity, 27(4), 670–684. doi: 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, … Ahmed R (2003). Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol, 4(3), 225–234. doi: 10.1038/ni889 [DOI] [PubMed] [Google Scholar]
- Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, & Ahmed R (1998). Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med, 188(12), 2205–2213. doi: 10.1084/jem.188.12.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel RM, & Doherty PC (1974). Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature, 248(5450), 701–702. [DOI] [PubMed] [Google Scholar]