Abstract
We investigated whether individual populations of freshwater bacteria in mixed experimental communities may exhibit specific responses to the presence of different bacterivorous protists. In two successive experiments, a two-stage continuous cultivation system was inoculated with nonaxenic batch cultures of the cryptophyte Cryptomonas sp. Algal exudates provided the sole source of organic carbon for growth of the accompanying microflora. The dynamics of several 16S rRNA-defined bacterial populations were followed in the experimental communities. Although the composition and stability of the two microbial communities differed, numerous members of the first assemblage could again be detected during the second experiment. The introduction of a size-selectively feeding mixotrophic nanoflagellate (Ochromonas sp.) always resulted in an immediate bloom of a single phylotype population of members of the class Actinobacteria (Ac1). These bacteria were phylogenetically affiliated with an uncultured lineage of gram-positive bacteria that have been found in freshwater habitats only. The Ac1 cells were close to the average size of freshwater bacterioplankton and significantly smaller than any of the other experimental community members. In contrast, no increase of the Ac1 population was observed in vessels exposed to the bacterivorous ciliate Cyclidium glaucoma. However, when the Ochromonas sp. was added after the establishment of C. glaucoma, the proportion of population Ac1 within the microbial community rapidly increased. Populations of a beta proteobacterial phylotype related to an Aquabacterium sp. decreased relative to the total bacterial communities following the addition of either predator, albeit to different extents. The community structure of pelagic microbial assemblages can therefore be influenced by the taxonomic composition of the predator community.
Bacterivorous protists such as nanoflagellates and ciliates are omnipresent components of aquatic microbial food webs and are probably the major consumers of bacterioplankton biomass (34, 36). Grazing mortality is believed to substantially influence community features such as total bacterial abundance, secondary production, and cell morphologies, and predation may also control the frequencies of the metabolically most active or dividing cells (4, 35, 39, 46).
At least two mechanisms can potentially explain the typically small average cell size of freshwater and marine bacterioplankton. First, there is ample evidence that the cell volume of aquatic bacteria is reduced during long-term substrate or nutrient depletion (24). Second, a small average community cell size also can be the consequence of the selective grazing behavior of protistan predators (11, 39). There are apparent upper and lower limits to the size of particles that can be handled and consumed by many free-living protists (13, 19). Within the edible size range, protists predominantly forage size-selectively, preferentially ingesting larger bacteria or artificial prey surrogates (11, 39).
Protistan grazing not only influences bacterioplankton size structure, but it also causes shifts in the composition of aquatic microbial communities. This has been shown in model systems, experimental assemblages, and field studies (13, 20, 41, 43). Size-selective grazing mortality should be disadvantageous for the larger bacterial species and should favor those small but actively growing bacteria (28, 32) that are common and dominant members of freshwater (or marine) systems. Recent evidence partially supports this hypothesis. The addition of a bacterivorous flagellate to freshwater experimental microbial assemblages resulted in a phenotypic shift towards small cells with high DNA synthesis rates (32). Introduction of the same flagellate to another type of model assemblage again caused a size shift towards smaller cells, but it remained unresolved whether those small bacteria were indeed related to phylogenetic lineages commonly found in lakes (12).
While there is evidence that certain bacterial strains are resistant to grazing by one particular predator (14, 38), it is unclear how aquatic microbes in mixed assemblages will respond to the presence of various bacterivore species. Protistan predators may differ not only in their foraging selectivity but also in their utilization and recycling of limiting nutrients (33), thus simultaneously interfering with several aspects of bacterial interspecific competition. Individual bacterivores might, therefore, suppress or support the growth of particular microbial populations to a variable extent. So the composition of the predator community would influence the bacterioplankton community structure.
We performed two separate experiments in a two-stage continuous cultivation system that allowed us to split one experimental bacterial community into several second-stage vessels. We subjected these second-stage assemblages to predation by two protists with contrasting prey uptake selectivity and investigated if any bacterial population responded specifically to the presence of one or both predators. The effects of the individual predation scenarios on microbial productivity and community size structure of the first experiment are described in detail elsewhere (32). Here we present an analysis of community composition by molecular methods (1, 2, 25), cell size measurements on single populations (27), and the relative contributions of different bacteria during experimental perturbations.
MATERIALS AND METHODS
Continuous flow systems.
Two experiments with comparable cultivation setups were performed in February 1998 and April 1999. The design of the continuous culture systems was adapted from that of S̆imek et al. (43) and is described in detail elsewhere (32). For the first stage of the systems, a glass vessel was aerated by an opening at the bottom (volume, 2.3 liters; dilution rate for 1998, 0.38 day−1; dilution rate for 1999, 0.42 day−1). At the top of this first-stage vessel were openings for de-aeration, for inflow of the medium, for a 20-cm-long steel needle to inoculate and sample the culture, and for the connecting lines to the second-stage vessels. A lateral opening in the upper third of the bottle allowed for outflow of excess medium. The 10-liter medium vessel and the cultivation vessels were de-aerated via a sterile glass tube filled with sterile cotton as a bacterial and dust trap. Special pear-shaped bacterial traps were used to disrupt flows between stages to avoid a possible immigration of organisms into the medium containers from the second to the first stage or from the waste vessels back into the main bottles.
The outflow of the first stages was fed into parallel second-stage vessels (volume, 750 ml; dilution rate for 1998, 0.25 day−1; dilution rate for 1999, 0.27 day−1). The vessels for the second stage were again aerated from the bottom and had a lateral outflow. All vessels were closed with silicon stoppers, and two glass tubes penetrating the stoppers were used for sterile de-aeration and for inflow of the medium from the first stage. Sampling was performed through steel needles (20 cm long) that had been pierced through the stoppers before initial sterilization of the system. The Cryptomonas sp. and accompanying microflora from a batch culture line that has been maintained in our laboratories for the past 5 years were introduced to the first-stage vessels of the systems. Algae were grown at inorganic phosphorus concentrations of 200 μg liter−1. The reactors were operated with 24 h of light. Several days after inoculation, when changes in algal densities indicated that a steady state had been reached, two second-stage vessels were inoculated with prerinsed subsamples (5 ml) from batch cultures of either a mixotrophic flagellate, an Ochromonas sp., or a bacterivorous ciliate, Cyclidium glaucoma. A third second-stage reactor was left unmanipulated and served as a predator-free control. The Ochromonas sp. originated from an axenic culture and had been maintained in batch cultures of the Cryptomonas sp. that also served as the inoculum of the first stage. The C. glaucoma strain had also been cocultivated with the Cryptomonas sp. for several weeks prior to the experiments. Details of the inoculation procedure are given in the work of S̆imek et al. (43). Community changes were studied during a period of 12 and 16 days in the first and second experiment, respectively. In the second experiment, C. glaucoma was introduced into two parallel second-stage vessels. On day 7, one of these vessels (referred to as C. glaucoma II) was inoculated with 5 ml from the vessel that contained the Ochromonas sp.
Selected samples (from days 8, 10, and 12) from two earlier experiments of Cryptomonas sp.-associated experimental microbial communities in comparable two-stage continuous cultivation systems (43; K. S̆imek, unpublished data) were available for partial analysis. In those investigations we used a different predator species, Bodo saltans. An experimental second-stage reactor containing this heterotrophic flagellate was also run during the 1998 investigation. It was excluded from a more detailed analysis because no replication of this treatment was performed in 1999.
Diversity analysis.
To obtain information about the phylogenetic composition of the microbial communities, denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA gene segments was performed for samples from the different stages at four timepoints (days 0, 1, 9, and 12) in 1998 and two timepoints (days 0 and 12) in 1999. Cell lysis and DNA extraction from cells concentrated on polycarbonate filters were performed as previously described (8). Oligonucleotide primers 341F and 907R (25) were used to amplify an approximately 550-nucleotide segment of the 16S rRNA genes by PCR. DGGE was carried out as previously described (25), and bands from different vertical positions in the gel were excised under UV illumination after ethidium bromide staining. DNA fragments were subsequently eluted from the individual gel bands with distilled water, reamplified by PCR, and sequenced on an ABI377 sequencer (PE Applied Biosystems, Norwalk, Conn.). Several PCR products consisting of a mix of DNA fragments were cloned using the pGEM T Easy Vector system (Promega, Madison, Wis.) according to the manufacturer's instructions. Ten clones from each set were then screened by digestion with the endonucleases Sau3AI and HaeIII (Promega), and inserts showing different restriction patterns were sequenced.
To increase the fraction of diversity detected by cultivation-independent methods, we additionally isolated bacterial strains from the different stages during the 1999 experiment. Cultivation was performed on the sterilized outflow from the second-stage vessels and amended with 1% prewashed agar, either directly or after a dilution series and several days of enrichment in microtiter plates. Isolates were prescreened by fluorescent in situ hybridization (FISH) with group-specific probes, and the 16S ribosomal DNA (rDNA) gene of selected representatives was amplified and sequenced as previously described (5).
Phylogenetic analysis.
Comparative sequence analysis was performed using the program package ARB (http://www.mikro.biologie.tu-muenchen.de [Lehrstuhl für Mikrobiologie, Technische Universität München]). A phylogenetic tree was reconstructed for the almost full-length 16S rDNA sequences of isolates by maximum parsimony of all sequences of >1,400 nucleotides in the ARB database, using neighbor joining and maximum likelihood analyses on various subsets for the evaluation of topologies. Alignment positions at which less than 50% of the sequences of the entire data set shared the same residues were excluded from the calculations. Partial sequences from DGGE and other partial sequences from the ARB database that were most closely affiliated to those sequences were added to the tree by maximum parsimony analysis.
Cell numbers, probe design, and FISH.
Subsamples for total bacterial and protistan counts were fixed with 2% (wt/vol) formaldehyde, and bacteria and protists were enumerated by staining with 4′,6′-diamidino-2′-phenylindole (DAPI) and epifluorescence microscopy (30, 43).
Oligonucleotide probe design for sequences derived from DGGE or cultivation was carried out with the ARB software, and potential positions for probes were evaluated by the 16S rRNA accessibility information provided by Fuchs et al. (7). Stringent conditions for FISH were established by analysis of fluorescence intensities of the target cells from hybridizations at increasing concentrations of formamide in hybridization buffer (26) (Table 1).
TABLE 1.
Probe sequences used in this study
| Probe | rRNA positionsa | Sequence (5′-3′) | % Formamideb | Target(s) | Accession no.c |
|---|---|---|---|---|---|
| ALF1-645 | 645–663 | CTC ACA CTC AAG ACA TCC | 35 | DGGE sequence ALF1 | |
| Isolate A1a | |||||
| Caulobacter sp. strain FWC14d | M83800 | ||||
| BET3-447 | 447–464 | AGC GCA GAC CGT TTC TTC | 40 | DGGE sequence BETA3 | |
| CF1-853 | 853–873 | TAA TGC TTT CGC TCA GAC AC | 35 | DGGE sequence CF1 | |
| Isolate CF1 | |||||
| Flavobacterium ferrugineum | M28237 | ||||
| Flexibacter elegans | M58782 | ||||
| Flexibacter sancti | M28057 | ||||
| Cytophaga arvensicola | D12657 | ||||
| Ac1-847 | 847–865 | TTA GCT GCG TCG CAC AAA C | 20 | DGGE sequence Ac1 |
Escherichia coli numbering (3).
Indicates the appropriate formamide concentration in hybridization buffer for each probe.
GenBank accession numbers from the National Center for Biotechnology Information.
Sequence similarity of >99% to DGGE sequence ALF1.
Samples for FISH were prefixed with alkaline Lugol's solution followed by formaldehyde fixation for at least 30 min and were decolorized by the addition of sodium thiosulfate in order to avoid the rupture of Cryptomonas sp. cells (37). Five to 10 ml of sample was filtered onto white membrane filters (type GTTP; diameter, 50 mm; pore size, 0.2 μm; Millipore, Bedford, Mass.), rinsed with distilled water, and stored frozen until further processing. FISH of filter sections with specific Cy3-labeled oligonucleotide probes (Interactiva, Ulm, Germany) and subsequent microscopic evaluation were carried out as previously described (9). At least 1,000 DAPI-stained cells per sample were inspected. The mounting medium Citifluor (Citifluor Ltd., Kent, United Kingdom) was amended with ca. 20% VectaShield (Vector Laboratories, Burlingame, Calif.). This significantly reduced bleaching of the probe signal, which was particularly useful for the measurement of cell sizes.
Cell size determination.
In samples from days 9, 10, and 12 of experiment 1, size measurements of all DAPI-stained bacteria and of cells hybridizing with the specific probes Ac1-847 and BET3-447 were carried out essentially as previously described (27, 31). In the community that formed with the Ochromonas sp., this was only feasible for the Ac1 population and for total DAPI-stained cells due to the low relative contributions of the other populations. Cell dimensions of hybridized cells were inferred from DAPI staining to allow comparison with results for the total community size structure. Double images of Cy3 and DAPI fluorescence of individual cells in microscopic preparations were captured using the PC-based image analysis software MetaMorph 3.5 (Universal Imaging, West Chester, Pa.) in combination with a SPOT slow-scan cooled charge-coupled device camera (resolution, 1,033 by 1,315 pixels; Diagnostic Instruments, Sterling Heights, Mich.) mounted on a Zeiss Axioplan 2 epifluorescence microscope (Carl Zeiss, Jena, Germany). Image acquisition at two excitation wavelengths was facilitated by a software-controlled motorized filter wheel. Between 500 and 1,000 hybridized cells were analyzed per sample (corresponding to 5 to 30 image pairs). Biomass size distributions, i.e., the fractions of total population biomass within different classes of cell length, were calculated as described previously (29) and averaged for the sampling dates.
Statistical evaluation.
Comparisons between treatments were performed separately for each experiment by a nonparametric simultaneous test procedure for multiple pairwise comparisons based on the Mann-Whitney-Wilcoxon U test (44). This procedure was chosen rather than an analysis of variance model because the data sets did not meet the assumption of normal distribution (Kolmogorov-Smirnov test; P < 0.05), as is frequently observed when using ratios (44). The following hypotheses were evaluated: (i) there are significant differences in the relative community contributions of populations Ac1 and BETA3 between the individual predator and control treatments, and (ii) there are significant differences in the relative community contributions of populations Ac1 and BETA3 between the predator treatments.
Nucleotide sequence accession numbers.
Sequences obtained in this study have been deposited in the GenBank database under accession no. AF361179 through AF361191.
RESULTS
Microbial diversity.
The exudates from a phosphorus-limited population of the Cryptomonas sp. maintained a phylogenetically diverse microbial assemblage (Fig. 1). Samples from all stages at day 0 and/or day 1 were identical in their DGGE banding patterns for both experiments (data not shown). However, the banding patterns poorly reflected compositional or diversity changes of the bacterial assemblages, since the patterns were always dominated by two strong bands. After PCR reamplification of DNA from these bands, partial 16S rDNA sequences were retrieved that phylogenetically affiliated with plastids of the Cryptomonas sp. and the Ochromonas sp. rather than with bacteria. This may reflect the predominance of plastid DNA or could be a bias caused by selective DNA extraction and/or PCR amplification (45).
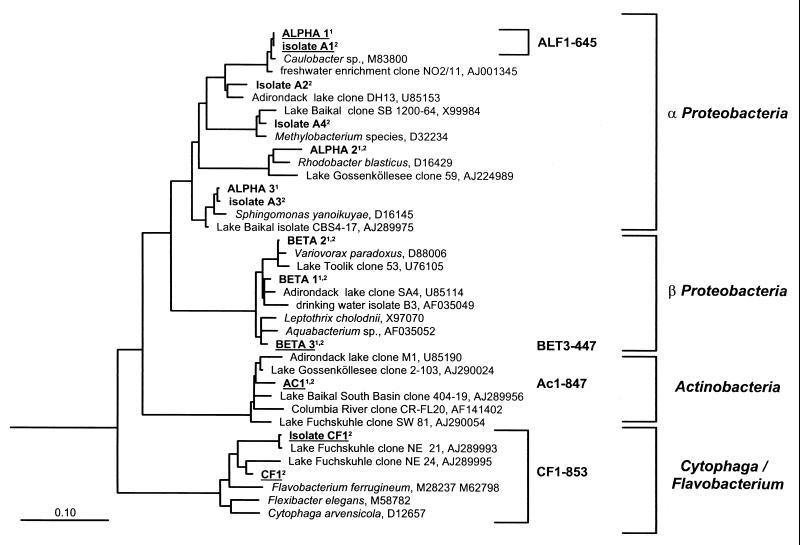
FIG. 1.
Phylogenetic affiliation of the partial 16S rDNA sequences retrieved from various second-stage vessels of the cultivation system by DGGE and affiliation of selected bacterial isolates and of bacteria targeted by probe CF1-853. Populations that were monitored by FISH are underlined. The bar indicates 10% estimated sequence divergence. Populations detected in the 1998 and 1999 experiments are indicated with a superscript 1 and 2, respectively.
Altogether, eight different DGGE bands could be discerned and excised for further analysis in the 1998 experiment, and six positions could be discerned and excised in 1999. Only one DGGE band was clearly indicative of a change in community composition. It appeared during the first experiment, in samples from the Ochromonas sp. stage at days 9 and 12. The sequence of reamplified rDNA from this band associated with a cluster within the class Actinobacteria (gram-positive bacteria with a high G+C content in DNA), which is known exclusively from freshwater samples (10). Three different DGGE-derived sequences were found to affiliate with the beta proteobacteria, three were affiliated with the alpha proteobacteria, and one was affiliated with members of the Cytophaga-Flavobacterium-Bacteroides (CFB) group (Fig. 1).
Of 48 isolated strains, 39 were affiliated with alpha proteobacteria, 6 with the CF1 lineage of the CFB phylum (10), and 3 with other CFB members. Several strains with different affiliations and colony morphologies were selected for 16S rDNA sequencing (Fig. 1).
Community composition and population dynamics.
Sequence-specific probes were designed for most partial sequences derived from DGGE and applied for FISH, but only three were used for further monitoring of population dynamics. Several other phylotypes depicted in Fig. 1 formed populations that constituted between <1 and 5% of the total abundances, as determined by FISH with the respective specific probes (data not shown). Probe sequences used in this study, their targets, and the stringent hybridization conditions for FISH are given in Table 1. The fourth FISH probe, CF1-853, was originally designed to cover a small, well-defined phylogenetic branch within the Cytophaga-Flavobacterium cluster containing 16S rDNA sequences from freshwater clone libraries (10). It matches the sequences of one isolate and of one DGGE band that were both retrieved from chemostat samples in 1999 (Fig. 1).
The four specific probes used in this study (Table 1) identified 64% of the DAPI-stained cells within the various communities (range, 41 to 94%) in the 1998 experiment and 63% (range, 28 to 112%) in 1999 (Fig. 2; Table 2). A summed detection rate of >100% was observed twice in 1999, which is most likely the result of cumulative error propagation. Most of the identified representatives of the microbial community from the 1998 experiment were again detected in 1999 by FISH (DGGE sequences ALPHA1, ALPHA2, BETA1, BETA2, BETA3, and Ac1), isolation (isolates A1 and A3 had >99% identity with DGGE sequences ALPHA1 and ALPHA3), or sequencing of DGGE bands (DGGE sequences ALPHA1, ALPHA3, BETA1, and BETA3) (Fig. 1).
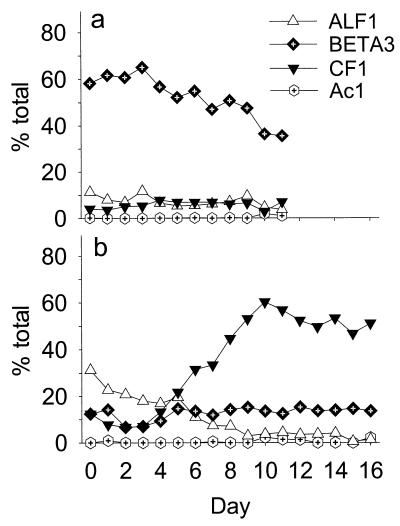
FIG. 2.
Relative contribution of the four bacterial populations, BETA3, CF1, Ac1, and ALF1, to the microbial assemblages in the first stages of the cultivation system. (a) Experiment in 1998. (b) Experiment in 1999.
TABLE 2.
Relative contributions of bacteria detected by probes CF1-853 and ALF1-645 in the different second-stage vessels of the cultivation systems
| Treatment | Mean % (range [minimum–maximum]) of DAPI-stained bacteria with probe:
|
|||
|---|---|---|---|---|
| CF1-853
|
ALF1-645
|
|||
| 1998 | 1999 | 1998 | 1999 | |
| Ochromonas sp. | 3 (<1–12) | 22 (3–46) | 8 (4–14) | 15 (4–33) |
| C. glaucoma | 10 (3–25) | 28 (7–43) | 12 (5–16) | 19 (4–33) |
| Control | 6 (4–8) | 33 (6–55) | 8 (3–12) | 15 (4–26) |
| C. glaucoma IIa | 15 (7–27) | 23 (11–33) | ||
| C. glaucoma II plus Ochromonas sp.b | 38 (28–53) | 12 (5–21) | ||
Before the addition of the Ochromonas sp. to the C. glaucoma II treatment (days 0 to 7).
After the addition of the Ochromonas sp. to the C. glaucoma II treatment (days 8 to 16).
There were clear differences in the compositions of the communities that formed with the Cryptomonas sp. in the first-stage vessels during successive experiments (Fig. 2). In 1998 the beta-proteobacterial phylotype BETA3 constituted 52% of the total cell counts in the first stage (mean ± standard deviation, 4.35 × 106 ± 0.84 × 106 cells ml−1) and 47% of the total in the unmanipulated control stage (5.68 × 106 ± 0.74 × 106 cells ml−1), averaged over the whole course of the experiment. The relative contribution of the BETA3 population to the total community gradually declined from 60 to 40% in the first- stage vessel (Fig. 2). Bacteria targeted by probes CF1-853 and ALF1-645 played a minor role (Fig. 2; Table 2) and remained rather constant. For the second experiment, the mean relative contributions of the BETA3 population in the first-stage and the control-stage vessels ranged around 13 and 12%, and the populations reached cell densities that were comparable to those in the previous investigation (3.1 × 106 ± 0.7 × 106 and 4.0 × 106 ± 1.5 × 106 cells ml−1, respectively). There was a clear shift in community composition during the course of the second experiment. A Caulobacter sp. targeted by probe ALF1-645 continuously decreased in relative abundance during the first week and remained in low abundance thereafter. In contrast, members of the lineage covered by probe CF1-853 increased in abundance from <10 to >50% of the total community between days 3 and 9 (Fig. 2).
Predator and bacterial population dynamics in coculture.
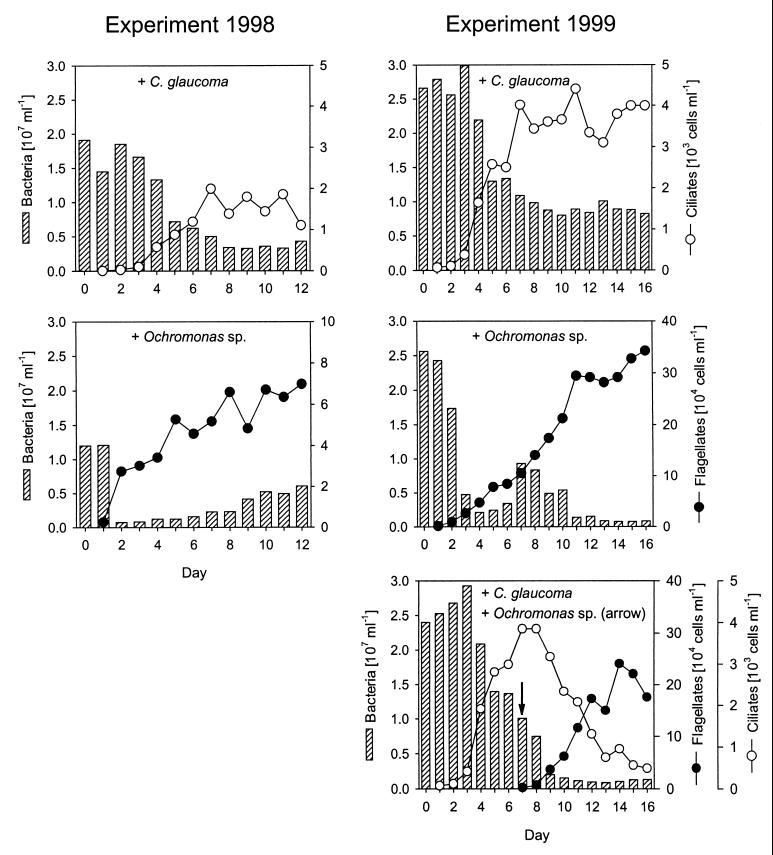
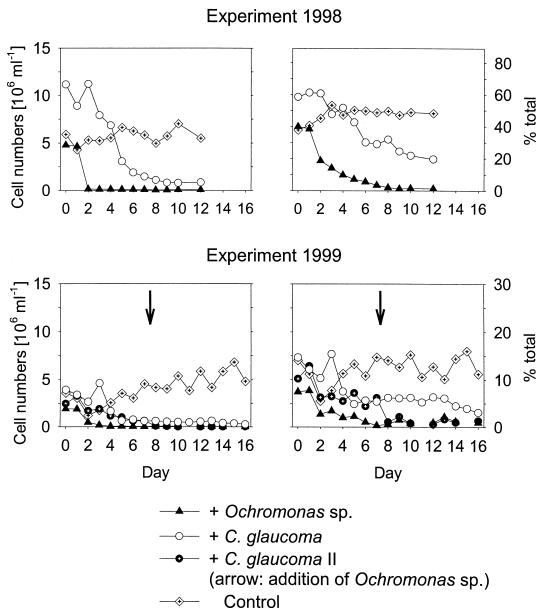
In both experiments the Ochromonas sp. and C. glaucoma rapidly formed dense populations after inoculation (Fig. 3) that were accompanied by a pronounced decline in bacterial abundance.
FIG. 3.
Total bacterial and protistan abundance in different second-stage cultures of the cultivation systems (1998 experimental data are from reference 32). The arrow indicates the inoculation of the Ochromonas sp. into a parallel reactor with C. glaucoma.
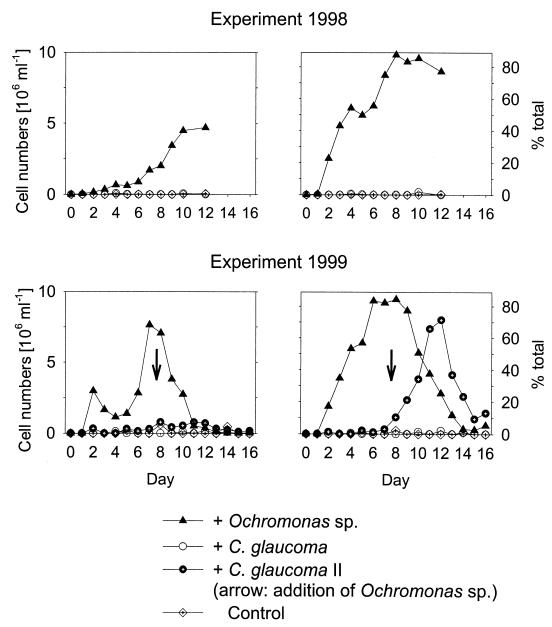
Bacteria hybridizing with probe Ac1-847 initially were detectable in low abundance (104 to 105 cells ml−1) during all stages for both experiments and formed <1% of the total communities. At the end of the experiments these bacteria were still present in low numbers in the control cultures. Concomitant with the rise of the Ochromonas sp. populations, the Ac1 population quickly became the dominant community member in both experiments and the relative abundance of the Ac1 population was significantly higher in the treatment cultures than in controls and in reactors containing C. glaucoma (Fig. 4). For the 1999 experiment, the relative abundance of the Ac1 population declined after several days, and the majority of bacteria were found to form aggregates of variable sizes. During this phase, bacteria targeted by probe CF1 dominated in all of the grazed and ungrazed communities (Table 2). No significant increase of the Ac1 population was observed in the community coexisting with C. glaucoma (Table 3). In the 1999 experiment a temporary rise in the relative abundance of the Ac1 population was also observed after the inoculation of the Ochromonas sp. to a parallel vessel containing a C. glaucoma population (Fig. 4). In order to support the assignment with probe Ac1-847, a second probe targeted to all members of the recently described freshwater lineage of the class Actinobacteria (HG1-840) (10) was applied to selected samples. Results were comparable to those of the strain-specific probe Ac1-847 (data not shown).
FIG. 4.
Abundance and relative community contribution (% DAPI-stained cells) of population Ac1 in the second stages of the experimental assemblages. Arrows indicate the inoculation of the Ochromonas sp. into a parallel reactor with C. glaucoma.
TABLE 3.
Nonparametric multiple comparison of the relative contributions of populations Ac1 and BETA3 between predator-exposed reactors and control stages and between different predator reactorsa
| Experiment | Population | No. of samples (days sampled) | Control stage vs Ochromonas sp.
|
Control stage vs C. glaucoma
|
Ochromonas sp. vs C. glaucoma
|
|||
|---|---|---|---|---|---|---|---|---|
| Usb | P | Us | P | Us | P | |||
| 1998 | Ac1 | 9 (3–12) | 81 | <0.01 | 48.5 | NSc | 81 | <0.01 |
| BETA3 | 9 (3–12) | 81 | <0.01 | 71 | <0.05 | 81 | <0.01 | |
| 1999 | Ac1 | 14 (3–16) | 196 | <0.01 | 106 | NS | 196 | <0.01 |
| BETA3 | 14 (3–16) | 196 | <0.01 | 183 | <0.01 | 195 | <0.01 | |
For details of the statistical procedure, see the text.
Us, test parameter.
NS, not significant at P values of <0.05.
A significant decrease in the absolute and relative abundance of the BETA3 population compared to the control cultures was observed in communities after the addition of either predator (Fig. 5; Table 3) to both experiments. This decrease was also observed in samples from two earlier investigations and in the 1998 experiment after the addition of a different heterotrophic flagellate, B. saltans (Table 4). The relative contributions of the BETA3 population were significantly lower in treatments containing the Ochromonas sp. than in treatments containing C. glaucoma (Table 3). No significant differences in the relative contributions of the Ac1 and BET3 populations were found between the parallel treatments of C. glaucoma (1999 experiment; Fig. 4 and 5) before inoculation of the Ochromonas sp. (Mann-Whitney U test; n = 6).
FIG. 5.
Abundance and relative community contribution (% DAPI-stained cells) of population BETA3 in the second stages of the experimental assemblages. Arrows indicate the inoculation of the Ochromonas sp. into a parallel reactor with C. glaucoma.
TABLE 4.
Relative contributions of population BETA3 to the first and second stages of four continuous cultivation experiments with Cryptomonas sp.-associated bacterial assemblages (1995–1999) using different predators
| Experiment | Mean % (range [minimum–maximum]) of DAPI-stained bacteriaa
|
||||
|---|---|---|---|---|---|
| First stage | Second stage
|
Control | |||
| Ochromonas sp. | B. saltans | C. glaucoma | |||
| 1995 | 24 (18–30) | 7 (4–10) | 36 (25–47) | ||
| 1996 | 28 (24–32) | 12 (9–15) | 30 (25–35) | ||
| 1998 | 41 (32–50) | 2 (2–2) | 19 (8–30) | 24 (17–31) | 49 (48–50) |
| 1999 | 14 (13–15) | >1 (0–1) | 6 (6–6) | 14 (13–15) | |
Mean values were of three timepoints (days 8, 10, and 12).
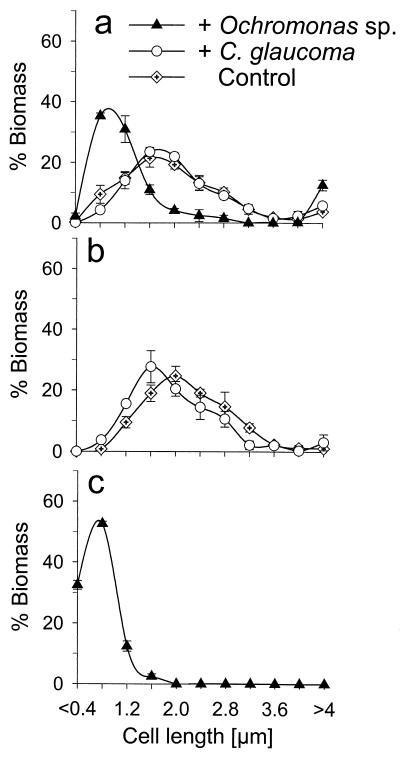
Cell sizes and biomass size distributions.
Figure 6 depicts biomass size distributions of the total microbial community and populations of Ac1 and BETA3 in different experimental reactors. Since the relative abundance of the Ac1 population was <1% in the treatment containing C. glaucoma and in the control vessel, it was not possible to determine the cell sizes of this population in those reactors. This was also the case for population BETA3 in the treatment with the Ochromonas sp. Towards the end of the first experiment (days 9, 10, and 12), the biomass of the bacterial communities in the treatments containing the Ochromonas sp. was allocated in smaller-cell-size classes than in the presence of C. glaucoma or in the control culture (Fig. 6) (32). The BETA3 population phenotype was shifted towards slightly smaller cells in communities grazed by C. glaucoma (mean cell length for days 9, 10, and 12, 1.6 ± 0.1 μm) than in controls (1.9 ± 0.1 μm). In contrast, 97% of the biomass of the gram-positive Ac1 populations was found in cells of <1.2 μm. In the 1999 experiment, the Ac1 population again consisted of small, free-living cells.
FIG. 6.
Biomass size distributions of the total bacterial communities (a), population BETA3 (b), and population Ac1 (c) in different second-stage vessels (experiment 1998, days 8, 10, and 12). Results shown are means, with minimum and maximum values indicated with bars.
DISCUSSION
Composition and stability of model communities.
Our cultivation approach selected for bacteria that were able to compete for a major limiting nutrient (Pi) and a typical planktonic carbon source (phytoplankton exudates) at realistic growth rates. The composition of the communities was thus determined both by the environmental conditions and the species present in the inoculum.
The majority of 16S rDNA sequences from the two communities were affiliated with recently described phylogenetic lineages of freshwater microorganisms (10, 49) or with genera of aquatic bacteria that could be traditionally identified in situ because of their conspicuous morphology, such as Caulobacter (ALF1). The experimental communities were therefore not colonized by typical cultivation-favored bacteria, e.g., by opportunistic gamma proteobacteria selectively isolated on rich media (48). In contrast, a dominance of Vibrio spp. and Pseudomonas spp. was observed in mixed assemblages cultivated on complex NSY medium (12). Our findings suggest that microbes that are phylogenetically more closely related to common lake bacteria can be enriched more readily in coculture with freshwater algae. In addition, several microbial populations were preserved in the Cryptomonas sp. stock culture for at least 1 year or even longer. For example, the BETA3 population could be traced back to experiments performed in 1995 (Table 4).
In both experiments, on average >60% of all bacteria could be identified by four specific probes. Provided that our FISH probes were indeed phylotype specific, many of the 16S rRNA-defined populations were present in both successive experiments (Fig. 1), including the prominent populations ALF1 and BETA3 but also including minor community members such as populations ALF3, BETA2, and Ac1. At least 11 different bacterial phylotypes were found in the assemblages (Fig. 1). An exhaustive coverage of microbial diversity was not, however, attempted in this study. The combination of isolation, DGGE, 16S rDNA sequencing, and FISH was primarily applied for the detection and subsequent monitoring of a few key populations in order to address our main hypothesis of predator-specific responses and to learn about the reproducibility of community structure in the experimental systems.
Our data revealed clear differences in the composition and stability of the microbial assemblages in the two experiments (Fig. 2). Both investigations were carried out a few days after inoculation of the cultivation system. The abundance of the Cryptomonas sp. was stable after day 4 of the first experiment (32) and during the whole course of the second one (data not shown), so it is probable that the algal population reached a steady state. This was not the case for the bacterial assemblages (Fig. 2b), and during the course of the second experiment, a community shift to a CF1 population was observed. It is unclear whether mixed assemblages will ever reach a steady state at all or if fluctuating populations of numerous species competing for a few limiting resources play a role in the maintenance of diversity (16). In a comparable study, two reactors inoculated with subsamples from a mixed microbial assemblage and operated in parallel were found to develop different communities over time (12). Second-stage cultures of microbial assemblages originating from lake water inocula and fed with UV-killed algal cultures clearly differed in community structure between parallel vessels and consecutive samplings (47). This indicates that we do not presently know the mechanisms that determine microbial successions, even in such controlled systems. We are therefore limited to studying community effects that can be reproducibly induced by experimental disturbance.
Growth of members of the class Actinobacteria induced by an Ochromonas sp.
Despite the differences in microbial community composition and stability, a pronounced bloom of bacteria affiliating with the Actinobacteria (i.e., population Ac1) was observed in both experiments after the addition of an Ochromonas sp. Bacteria targeted by probe Ac1-847 rapidly became the dominant population in the grazed communities (Fig. 4), and their absolute cell numbers increased in parallel by 1 to 2 orders of magnitude. Since Ac1 cells were present in low densities in both the first-stage and control reactors throughout the experiment, it is unlikely that these bacteria could have been contaminants introduced with the inoculum of the Ochromonas sp. Moreover, the originally axenic Ochromonas sp. culture had been maintained exclusively on batch cultures of the Cryptomonas sp. and was thus exposed to the same bacterial diversity that was inoculated into the first-stage vessel of the system.
Population Ac1 consisted of significantly smaller cells than population BETA3 (Fig. 6b and c). It increased in relative abundance in the presence of the Ochromonas sp. but not of C. glaucoma, which is in good agreement with the size-selective feeding modes of the two predators. The Ochromonas sp. preferably ingests bacteria of between 0.8 and 4 μm (3), whereas C. glaucoma efficiently feeds on smaller prey also (6, 42; for a compilation of literature data, see Table 1 in Posch et al. [32]). Therefore, the Ac1 population might have strongly profited by the grazing selectivity of the flagellate. The different feeding strategies of the two protists are also reflected in the general biomass shifts towards smaller cells in the Ochromonas sp.-exposed community at the end of the first experiment compared to the ungrazed control culture. In addition, >10% of the total biomass was present in grazing-protected filaments (i.e., cells of >4 μm) with this treatment (Fig. 6a). The size distribution of the community in the presence of the ciliate was only marginally different from that of the control. The observed differences in the communities grazed by the Ochromonas sp. and by C. glaucoma might have also been related to the contrasting nutrient recycling patterns of the heterotrophic and the mixotrophic predator species (33).
During the second experiment, the Ochromonas sp. population was much more dense than during the first investigation (Fig. 3), and consequently grazing rates were much higher. This was most likely because of the higher total bacterial numbers in the first stage of the system and thus a higher input of particulate organic carbon to the second stages. We speculate that higher productivity of the second experimental community might have been the result of small differences in illumination. In spite of this, population Ac1 again clearly responded to the addition of the Ochromonas sp. (Fig. 4b). However, a continuous reduction of the Ac1 population was observed during the second half of the second experiment (Fig. 4). The selectivity against particles of poorer nutritional quality is reduced in food-limited flagellates (18), and therefore we assume that members of the Ac1 population were eventually consumed in the absence of other prey. Since the observation period for the 1998 experiment was shorter than for the 1999 experiment, it cannot be excluded that a similar decline might have eventually occurred at lower Ochromonas sp. densities. The decrease of the Ac1 population during experiment 2 is nevertheless clear evidence that this phylotype was not completely grazing protected or poisonous for the predator.
At the end of the second experiment, the majority of bacteria were associated with the CF1 and ALF1 populations (Table 2) and were located in aggregates of various sizes. Aggregate formation apparently represents even better protection against predation by this protist than does small cell size. During the rise of protistan grazers after food web manipulation of freshwater plankton a prominent fraction of the total bacterial biomass was located in aggregates (20). A similar phenotypic succession during high Ochromonas sp. grazing from large, grazing-vulnerable to small, free-living cells and eventually to aggregated bacteria has recently been observed by other investigators (12). No molecular identification of those small cells was attempted in that study. Therefore, we cannot tell if the maintenance of small cell size during growth represents another phylogenetically widespread antipredator adaptation, as has been shown for facultative filamentous morphotypes (14).
The success of the Ac1 population during heavy size-selective grazing by the Ochromonas sp. clearly indicates the ecological benefit of preserving small cell size during growth in the presence of size-selective predators. A gram-positive cell wall might also have prolonged digestion times by the predator (17). High numbers of similarly sized members of the class Actinobacteria from this lineage have been visualized by FISH and image analysis of samples from a mountain lake (10). Considering the ubiquity of 16S rDNA sequences from this phylogenetic branch of class Actinobacteria in fresh waters (10, 15) and the low numbers of the Ac1 population in the absence of predation (Fig. 2), we suggest that heavy size-selective grazing could be a major factor that promotes the success of this lineage in the environment.
Aquabacterium sp. suppressed by grazing of both predators.
The population dynamics of the BETA3 populations represent a model for high grazing sensitivity. Population BETA3 is closely affiliated with a bacterial isolate that was dominant in drinking water biofilms (21), a habitat that is probably not exposed to high levels of protistan grazing. This isolate has subsequently been described as a member of a novel genus, Aquabacterium (22).
The high frequency of members of population BETA3 in assemblages of first-stage and control vessels in three out of four continuous cultivation experiments with Cryptomonas sp.-associated assemblages (Table 4) shows that it was a successful competitor for limiting nutrients and alga-derived substrates. A reduction in the community contribution of phylotype BETA3 was observed at very different relative abundances in subsequent experimental communities that all followed the same basic concept and were exposed to grazing by different bacterivores (Table 4).
This phylotype formed significantly larger cells than members of population Ac1 did, as determined during the experiment in 1998 (Fig. 6b and c), and most of the biomass was found in the optimal edible size range of both the Ochromonas sp. and C. glaucoma. The size of the total microbial community shifted towards small cells in the presence of the Ochromonas sp. but not of C. glaucoma (Fig. 6a). Members of population BETA3, therefore, were among the largest cells in the assemblage with the Ochromonas sp. but were slightly smaller than average in the community with C. glaucoma (Fig. 6b). Consequently, higher mortality rates of population BETA3 members by size-selective flagellate grazing are expected. This agrees with the observed population dynamics: in both experiments the reduction of the BETA3 population was significantly more pronounced in the presence of the Ochromonas sp. than of C. glaucoma (Fig. 5; Table 3). Moreover, the addition of the Ochromonas sp. to the community that had formed in the presence of C. glaucoma in the 1999 experiment resulted in an immediate further decline of the BETA3 population, to the levels observed in the vessel inoculated with the Ochromonas sp. only (Fig. 5).
Conclusions.
We provide evidence that two protistan predators affect microbial community structure differently and that a member of a typical freshwater phylogenetic lineage only forms larger populations when the assemblages are exposed to size-selective flagellate grazing. Therefore, the taxonomic composition of the predator community may be an important parameter for understanding the abundance of particular microbial populations in the water column. However, cultivation-independent approaches to identify bacterivorous protists, in particular flagellates, in situ are still so limited (23, 40) that presently this issue cannot be satisfactorily addressed in the field.
ACKNOWLEDGMENTS
We thank B. Sonntag and C. Wawer for help during the experiments and in the lab and T. F. Thingstad for fruitful discussions. We also thank D. Kirchman for his helpful comments on an earlier version of the manuscript and M. Hahn for providing cultures of the Ochromonas sp.
This study was supported by a grant from the Austrian National Bank (OENB 6513), by AKTION Austrian-Czech Republic 23p5, (to K.S. and R.P.), by a GA CR research grant (206/99/0028 to K.S.), and by the Max Planck Society.
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chrzanowski T H, Simek K. Prey-size selection by freshwater flagellated protozoa. Limnol Oceanogr. 1990;35:1429–1436. [Google Scholar]
- 4.Del Giorgio P A, Gasol J M, Vaque D, Mura P, Agusti S, Duarte C M. Bacterioplankton community structure: protists control net production and the proportion of active bacteria in a coastal marine community. Limnol Oceanogr. 1996;41:1169–1179. [Google Scholar]
- 5.Eilers H, Pernthaler J, Glöckner F O, Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. 2000;66:3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenchel T. Suspension feeding in ciliated protozoa: functional response and particle size selection. Microb Ecol. 1980;6:1–11. doi: 10.1007/BF02020370. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs B M, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuhrman J A, Comeau D E, Hagström A, Chan A M. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl Environ Microbiol. 1988;54:1426–1429. doi: 10.1128/aem.54.6.1426-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 10.Glöckner F O, Zaichikov E, Belkova N, Denissova L, Pernthaler J, Pernthaler A, Amann R. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl Environ Microbiol. 2000;66:5053–5065. doi: 10.1128/aem.66.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez J M, Sherr E B, Sherr B F. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl Environ Microbiol. 1990;56:583–589. doi: 10.1128/aem.56.3.583-589.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn M W, Höfle M G. Flagellate predation on a bacterial model community: interplay of size-selective grazing, specific bacterial cell size, and bacterial community composition. Appl Environ Microbiol. 1999;65:4863–4872. doi: 10.1128/aem.65.11.4863-4872.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn M W, Höfle M G. Grazing pressure by a bacterivorous flagellate reverses the relative abundance of Comamonas acidovorans PX54 and Vibrio strain CB5 in chemostat cocultures. Appl Environ Microbiol. 1998;64:1910–1918. doi: 10.1128/aem.64.5.1910-1918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn M W, Moore E R B, Höfle M G. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl Environ Microbiol. 1999;65:25–35. doi: 10.1128/aem.65.1.25-35.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiorns W D, Methe B A, Nierzwicki-Bauer S A, Zehr J P. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl Environ Microbiol. 1997;63:2957–2960. doi: 10.1128/aem.63.7.2957-2960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huisman J, Weissing F J. Biodiversity of plankton by species oscillations and chaos. Nature. 1999;402:407–410. [Google Scholar]
- 17.Iriberri J, Azua I, Labirua-Iturburu A, Artolozaga I, Barcina I. Differential elimination of enteric bacteria by protists in a freshwater system. J Appl Bacteriol. 1994;77:476–483. doi: 10.1111/j.1365-2672.1994.tb04390.x. [DOI] [PubMed] [Google Scholar]
- 18.Jürgens K, DeMott W D. Behavioural flexibility in prey detection by bacterivorous flagellates. Limnol Oceanogr. 1995;40:1503–1507. [Google Scholar]
- 19.Jürgens K, Guede H. The potential importance of grazing-resistant bacteria in planktonic systems. Mar Ecol Prog Ser. 1994;112:169–188. [Google Scholar]
- 20.Jürgens K, Pernthaler J, Schalla S, Amann R. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl Environ Microbiol. 1999;65:1241–1250. doi: 10.1128/aem.65.3.1241-1250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalmbach S, Manz W, Szewzyk U. Isolation of new bacterial species from drinking water biofilms and proof of their in situ dominance with highly specific 16S rRNA probes. Appl Environ Microbiol. 1997;63:4164–4170. doi: 10.1128/aem.63.11.4164-4170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalmbach S, Manz W, Wecke J, Szewzyk U. Aquabacterium gen. nov., with description of Aquabacterium citratiphilum sp. nov., Aquabacterium parvum sp. nov. and Aquabacterium commune sp. nov., three in situ dominant bacterial species from the Berlin drinking water system. Int J Syst Bacteriol. 1999;49:769–777. doi: 10.1099/00207713-49-2-769. [DOI] [PubMed] [Google Scholar]
- 23.Lim E L, Amaral L A, Caron D A, DeLong E F. Application of rRNA-based probes for observing marine nanoplanktonic protists. Appl Environ Microbiol. 1993;59:1647–1655. doi: 10.1128/aem.59.5.1647-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morita R Y. Bacteria in oligotrophic environments: starvation-survival lifestyle. Vol. 1. New York, N.Y: Chapman Hall; 1997. [Google Scholar]
- 25.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, Van Elsas J D, De Bruijn F J, editors. Molecular microbial ecology manual. 2nd ed. Vol. 3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- 26.Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Schleifer K H. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62:4329–4339. doi: 10.1128/aem.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pernthaler J, Alfreider A, Posch T, Andreatta S, Psenner R. In situ classification and image cytometry of pelagic bacteria from a high mountain lake (Gossenköllesee, Austria) Appl Environ Microbiol. 1997;63:4778–4783. doi: 10.1128/aem.63.12.4778-4783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pernthaler J, Posch T, Šimek K, Vrba J, Amann R, Psenner R. Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl Environ Microbiol. 1997;63:596–601. doi: 10.1128/aem.63.2.596-601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pernthaler J, Sattler B, Šimek K, Schwarzenbacher A, Psenner R. Top-down effects on the size-biomass distribution of a freshwater bacterioplankton community. Aquat Microb Ecol. 1996;10:255–263. [Google Scholar]
- 30.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 31.Posch T, Pernthaler J, Alfreider A, Psenner R. Cell-specific respiratory activity of aquatic bacteria studied with the tetrazolium reduction method, cyto-clear slides and image analysis. Appl Environ Microbiol. 1997;63:867–873. doi: 10.1128/aem.63.3.867-873.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posch T, Šimek K, Vrba J, Pernthaler S, Nedoma J, Sattler B, Sonntag B, Psenner R. Predator-induced changes of bacterial size-structure and productivity studied on an experimental microbial community. Aquat Microb Ecol. 1999;18:235–246. [Google Scholar]
- 33.Rothaupt K O. Nutrient turnover by freshwater bacterivorous flagellates: differences between a heterotrophic and a mixotrophic chrysophyte. Aquat Microb Ecol. 1997;12:65–70. [Google Scholar]
- 34.Sanders R W, Caron D A, Berninger U G. Relationship between bacteria and heterotrophic nanoplankton in marine and freshwaters: an inter-ecosystem comparison. Mar Ecol Prog Ser. 1992;86:1–14. [Google Scholar]
- 35.Sherr B F, Sherr E B, McDaniel J. Effect of protistan grazing on the frequency of dividing cells in bacterioplankton assemblages. Appl Environ Microbiol. 1992;58:4371–4378. doi: 10.1128/aem.58.8.2381-2385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherr E B, Sherr B F. Bacterivory and herbivory: key roles of phagotrophic protists in pelagic food webs. Microb Ecol. 1994;28:223–235. doi: 10.1007/BF00166812. [DOI] [PubMed] [Google Scholar]
- 37.Sherr E B, Sherr B F. Protistan grazing rates via uptake of fluorescently labelled prey. In: Kemp P, Sherr B F, Sherr E B, Cole J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publications; 1993. pp. 695–701. [Google Scholar]
- 38.Shikano S, Luckinbill L S, Kurihara Y. Changes of traits in a bacterial population associated with protozoal predation. Microb Ecol. 1990;20:75–84. doi: 10.1007/BF02543868. [DOI] [PubMed] [Google Scholar]
- 39.Šimek K, Chrzanowski T H. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl Environ Microbiol. 1992;58:3715–3720. doi: 10.1128/aem.58.11.3715-3720.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Šimek K, Hartman P, Nedoma J, Pernthaler J, Springmann D, Vrba J, Psenner R. Community structure, picoplankton grazing and zooplankton control of heterotrophic nanoflagellates in a eutrophic reservoir during the summer phytoplankton maximum. Aquat Microb Ecol. 1997;12:49–63. [Google Scholar]
- 41.Šimek K, Kojecka P, Nedoma J, Hartman P, Vrba J, Dolan D R. Shifts in bacterial community composition associated with different microzooplankton size fractions in a eutrophic reservoir. Limnol Oceanogr. 1999;44:1634–1644. [Google Scholar]
- 42.Šimek K, Vrba J, Hartman P. Size-selective feeding by Cyclidium sp. on bacterioplankton and various sizes of cultured bacteria. FEMS Microbiol Ecol. 1994;14:157–167. [Google Scholar]
- 43.Šimek K, Vrba J, Pernthaler J, Posch T, Hartman P, Nedoma J, Psenner R. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl Environ Microbiol. 1997;63:587–595. doi: 10.1128/aem.63.2.587-595.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokal R R, Rohlf F J. Biometry. W. H. New York, N.Y: Freeman and Company; 1995. [Google Scholar]
- 45.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thingstad T F, Lignell R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol. 1997;13:19–27. [Google Scholar]
- 47.van Hannen E J, Mooij W, van Agterveld M P, Gons H J, Laanbroek H J. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:2478–2484. doi: 10.1128/aem.65.6.2478-2484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zwart G, Hiorns W, Methe B, Van Agterveld M, Huismans R, Nold S, Zehr J, Laanbroek H. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst Appl Microbiol. 1998;21:546–556. doi: 10.1016/S0723-2020(98)80067-2. [DOI] [PubMed] [Google Scholar]