Abstract
Pulsed-field gel electrophoresis of restriction endonuclease-digested genomic DNA from a large collection of clinical isolates of Rhodococcus equi, an important pathogen of foals, was used to compare strain distribution between farms and over time. Forty-four strains were found among 209 isolates, with 5 of these accounting for over half the isolates and the 22 strains isolated more than once accounting for 90% of the isolates. The average genotypic diversity on each farm and in each year was found to be less than the genotypic diversity of the isolates taken as a whole, with 5.2% of the total diversity being due to differences between farms and 5.5% to differences between years. A small number of strains on each farm were found to have caused at least half the clinical cases of disease, and these varied between farms and, to a lesser extent, years. Most strains were found on more than one farm, and some very similar restriction patterns were found among isolates from different continents, indicating that strains can be very widespread. Multiple strains were isolated in five of the six cases in which more than one isolate from a single foal was examined, indicating that disease may commonly be caused by simultaneous infection with multiple strains. It was concluded that there are a number of different strains of R. equi which carry the vapA gene, and these strains tend to be widespread, but individual farms tend to have particular strains associated with them.
Rhodococcus equi is an important pathogen of foals worldwide, causing pneumonia mainly in the 2- to 4-month-old age group. Foals typically show signs of purulent bronchopneumonia, with secondary enteritis and enteric lymph node abscessation sometimes seen. Incidences of disease and mortality are quite variable between farms, and this variation is believed to be due to a combination of management factors and the prevalence of virulent strains on each farm.
Virulence of R. equi in foals is associated with a family of plasmids of 85 to 90 kb, which carry a gene encoding VapA, a 15- to 17-kDa lipoprotein shown to be associated with virulence in foals and mice (16, 17). Comparisons of clinical isolates from various species, using ribotyping (6), and of small numbers of isolates from bovine and equine feces (13) and humans and horses (1), using pulsed-field gel electrophoresis (PFGE), have found quite a large degree of diversity among strains. Two surveys of isolates from cases of bovine lymphadenitis (11, 12) found lower levels of diversity, with most isolates appearing to be related. However no studies have examined the genetic diversity of isolates from clinically diseased foals, which almost invariably carry the virulence plasmid.
This study examined the genetic relationships between a large number of clinical isolates of R. equi, most of which possessed the vapA gene, in order to compare isolates from different locations and different years, as well as to investigate any association between strain and site of infection.
MATERIALS AND METHODS
Bacterial isolates.
Isolates of R. equi (n = 212) were obtained from infected foals on Australian thoroughbred horse farms and identified by standard methods (3). Of these, 201 were obtained by the Scone Diagnostic Veterinary Laboratory (Scone, New South Wales, Australia), and the other 11 were from foals presenting to the University of Melbourne Veterinary Clinic and Hospital (Werribee, Victoria, Australia). Almost all isolates obtained by these laboratories from foals born during the 1991 to 1998 foaling seasons were included in the study. The majority of isolates were cultured from tracheal washes, but some were isolated from other sites, as shown in Table 1. Isolates were stored as nutrient agar stab cultures at 4°C for up to 5 years before being grown in brain heart infusion broth (BHIB; Oxoid, Basingstoke, United Kingdom) and stored at −70°C in 50% (vol/vol) glycerol–50% (vol/vol) BHIB. A large number of the isolates had been examined for the presence of the vapA gene by PCR in a previous study (3), and all the remainder were tested by a similar method.
TABLE 1.
Sites of isolation of Australian R. equi isolates
| Site of isolation | No. of isolates | No. of foals surviving/total foals |
|---|---|---|
| Lung-tracheal wash | 163 | 146/164a |
| postmortem | 11 | |
| Submandibular abscess | 7 | 7/7 |
| Subcutaneous abscess/pus | 8 | 7/7b |
| Gut-associated abscesses | 5 | 0/5c |
| Abscesses in other sites | 9 | 9/9 |
| Feces (foals with enterocolitis) | 2 | 2/2 |
| Bone | 1 | 1/1 |
| Peritoneal fluid | 1 | 0/1 |
| Joint fluid | 1 | 1/1 |
| Lymph node | 1 | 0/1 |
| Nasal | 1 | 1/1 |
| Unknown | 2 | 0/2 |
Does not include eight with outcome not known. Some of these isolates came from the same foals.
Does not include 1 with outcome not known.
Four of these foals also had lung isolates taken postmortem.
Isolates were also obtained from Canada (n = 1), the United States (n = 4), Argentina (n = 3), and Brazil (n = 1), courtesy of John Prescott, Department of Pathobiology, University of Guelph, Guelph, Ontario, Canada. These were known to contain virulence plasmids of 85-kb type I (North American isolates) or 87-kb type I (South American isolates) (15). Japanese isolates (n = 10) which contained either the 87-kb type II or 90-kb virulence plasmids (15) and German isolates (n = 6) were also obtained for testing. All these isolates came from infected foals.
Preparation of genomic DNA in agarose blocks.
Each isolate was grown in 5 ml of BHIB at 37°C in a shaking incubator at 225 rpm to a density of approximately 0.95 (measured as A600). Cells from 1 ml of culture were collected by centrifugation at 16,250 × g for 10 min, washed three times in 1 M NaCl–10 mM Tris HCl (pH 7.5), and resuspended in 100 μl of the same solution. The cell suspension was mixed with an equal volume of molten 2% (wt/vol) low-melting-point agarose (SeaPlaque–FMC Bioproducts, Rockland, Maine) and dispensed into 100-μl molds. When set, blocks were incubated in 1 ml of lysis buffer (100 mM Tris HCl, 10 mM EDTA [pH 8], 0.5 M NaCl, 20% [wt/vol] sucrose, and 5 mg of lysozyme/ml) for 24–48 h at 37°C. A volume of 110 μl of 10% sodium N-lauroyl sarcosine was then added and the blocks incubated at 37°C for a further 24 h. The lysis buffer was then replaced with ESP buffer (0.5 M EDTA [pH 9.2], 1% [wt/vol] sodium N-lauroyl sarcosine, and 1 mg of proteinase K/ml), and the blocks were incubated at 50°C for 48–60 h. Blocks were stored at 4°C in this solution until required.
Restriction endonuclease digestion of genomic DNA.
Blocks were equilibrated with TE buffer (10 mM Tris HCl, 1 mM EDTA [pH 8]) before use. Slices about 1 mm thick were placed in a 100-μl digest volume containing 20 U of AsnI (Boehringer Mannheim, Mannheim, Germany) in the buffer provided by the manufacturer, preincubated at 4°C for 16 h, and then incubated at 37°C for 24 h. The remaining portions of the blocks were stored in TE buffer at 4°C.
PFGE.
DNA fragments were separated using clamped homogeneous electric field electrophoresis, using a CHEF DRIII (Bio-Rad Laboratories, Richmond, Calif.). Electrophoresis was performed through a 1% (wt/vol) agarose gel in 0.5× TBE (1× TBE = 89 mM Tris, 98 mM boric acid, 2 mM EDTA) at 6 V/cm for 22 h at 14°C, with an included angle of 120°, and the pulse time was increased linearly from 3 to 90 s. Lambda phage concatemers (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) and HindIII-digested lambda phage DNA were used as size standards for all gels. DNA fragments were detected by staining gels in 0.5 mg of ethidium bromide/liter and examining with UV transillumination.
Testing stability of patterns.
To determine how stable the various restriction patterns were with passage, isolates of the four most common strains (A, C, D, and E) were grown in Luria-Bertani broth and passaged every 24–48 h. Genomic DNA was prepared from the first and tenth passages as detailed above, except that bacteria were grown in Luria-Bertani broth and, because of the smaller numbers of cells, only a single block was made. The DNA was then digested and subjected to PFGE as detailed above, and the patterns from the first and tenth passages were compared.
Southern blotting and ribotyping.
Gels were destained for 3–8 h at room temperature or overnight at 4°C; then, after depurination in 0.25 M HCl, DNA fragments were transferred to nylon membrane (Hybond N+, Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) by capillary transfer (9).
A 32P-labeled probe for ribotyping was made by random primed synthesis (Random Primed DNA labeling kit, Boehringer Mannheim), according to the manufacturer's recommendations. Template DNA was made by amplifying the 16S rRNA gene of R. equi using PCR, and lambda phage DNA was also included in the labeling reaction in order to visualize molecular size markers.
For amplifying the template DNA, the primers used were 5′-GCTTAACACATGCAAGTCGAAC-3′ and 5′-CCGGTACGGCTACCTTGTTA-3′, which were based on the sequence of the 16S rRNA gene of R. equi (8) (GenBank accession no. X80614). R. equi was grown on sheep blood agar for 3 days at 37°C, and one colony was emulsified in the reaction mix, which consisted of 0.4 U of Taq polymerase per 25 μl of reaction mixture in the buffer supplied by the manufacturer (Boehringer Mannheim), 0.4 mM concentrations of each deoxynucleoside triphosphate, and forward and reverse primers at 2 mM each. The reaction was incubated through 40 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1.5 min in a Hybaid thermocycler (Hybaid, Ashford, United Kingdom).
Blots were prehybridized for 2 h at 58°C in prehybridization solution (5× SSC [1× SSC = 150 mM NaCl, 15 mM trisodium citrate, pH 7], 5× Denhardt's solution, 0.5% [wt/vol] sodium dodecyl sulfate, and 20 mg of denatured herring sperm DNA/ml). Denatured probe was added and hybridized to membranes overnight at 58°C. Membranes were then washed three times for 10 min each at 58°C in 2× SSC–0.1% sodium dodecyl sulfate and autoradiographed.
Analysis of restriction patterns.
Isolates were regarded as the same strain if their AsnI restriction patterns were identical or showed differences which would require only one genetic change, such as deletion or insertion of DNA or deletion or creation of a single restriction site (maximum three different bands), as described by Tenover et al. (19).
Analysis of diversity.
The genotypic diversity was calculated for each farm, in total and for each year, as well as the total diversity overall and for each year, using the formula (2, 7, 10)
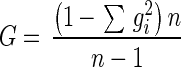 |
where gi is the prevalence of each strain, and n is the total number of isolates. The diversity within and between farms and years was compared using the following formula (2, 7, 10): variation due to s = (Gt − Gs)/Gt, where Gt is the total genotypic diversity, s is the factor of interest, and Gs is the arithmetic mean of the diversity for each category of s. When diversity within and between farms was compared, only isolates from farms where more than one isolate was obtained were included, because diversity for farms where only one isolate was obtained could not be calculated.
Statistical analysis.
Univariate logistic regression analysis (4) was used to test factors including farm, age of foal, date of birth, date of isolation, sex of foal, and site and year of isolation, to see whether any of these were significantly associated with strain type, survival, or site of primary infection (pulmonary or extrapulmonary). Strains A through E were analyzed separately for association with any of the variables being investigated, with isolates of each strain being compared with all other isolates. Strain type (A, B, C, D, E, or “other”) was also tested for association with survival and site of primary infection. Farms were categorized as M, O, W, or “other.” Date of birth was categorized, with the first category being August, then divided into 15- or 16-day (half-month) periods up to the 15th of November, with the last category being later than this. Date of isolation was divided into four categories—September/October, November, December, and January/February. Age was categorized as up to 29 days, then divided into 10-day periods, with the last two categories being 70–89 days and ≥90 days. If there appeared to be a linear correlation between age and a dependent variable when analyzed with age in categories, age was also analyzed linearly with that variable.
All these categories were chosen to give enough groups that differences would not be hidden, but with enough foals in each so that any significant differences could be detected. The end categories had larger ranges in order to keep group sizes more even and large enough, especially since the age and seasonal distribution of cases was so limited. Even though there were few cases from some years, there was no justification for combining cases from different years, and because there were significant associations detected with age, which was the most finely divided category, it was felt that there were sufficient foals in each category to justify the categorizations chosen. The site of isolation for each isolate was classified as pulmonary or extrapulmonary, and each case was classified as primarily either pulmonary (which included those with secondary infections associated with the gastrointestinal tract) or extrapulmonary, if only an extrapulmonary isolate was received, because this was the site of clinical infection, even though pulmonary infection could not be ruled out. Year was taken as the year of birth of the foal, even though some isolates were obtained early in the next calendar year. When more than one isolate had been made from a single foal, the case was counted only once, where pulmonary infection or survival was the dependent variable, by including only the postmortem pulmonary isolate from each foal.
If there were multiple factors which were associated on a univariate analysis with an outcome (P ≤ 0.25), they were used in a multivariate logistic regression to determine the best model for factors affecting that outcome. If age was found to be associated with an outcome using both cateogorical and linear analyses, the more significant of the two was used. The analysis was carried out using a stepwise forward-likelihood ratio method, with a P value for inclusion of ≤0.05 and for exclusion of >0.05. All analyses were carried out using SPSS 6.1.1 for Macintosh.
If more than one variable was found to be significant in the multivariate analysis using SPSS, the analysis was repeated using LogXact 4 for Windows.
RESULTS
PFGE.
Restriction patterns of AsnI digests of R. equi DNA usually contained 13–18 bands, ranging from around 10 kb to around 1 Mb. There were bands of 9.9, 15.7, 31, 56, 67, 96, and 119 kb which appeared to be conserved in most or all of the isolates tested. Three isolates could not be typed, as restriction digests of sufficient quality could not be prepared. The case data for these isolates were, however, left in the analysis when strain was not a factor. It was not determined which bands were genomic DNA and which were plasmid DNA, although one of the virulence plasmids has been found to give two fragments, of 9.9 and 74 kb, when digested with AsnI (5), the smaller of which may be the conserved 9.9-kb fragment noted in this study. No 74-kb band was noted, but the 15.7- and 56-kb bands would add to around 72 kb, so these may be fragments of this larger band. Among the Australian isolates, 22 PFGE strains were found which were represented by at least two isolates, along with 22 isolates which could not be considered the same strain as any others. Some of these unmatched isolates had patterns which were similar to others, indicating that they may be related, but could not be considered the same strain under the criteria used in this study. Numbers of isolates of each PFGE strain from each farm are shown in Table 2, and numbers of isolates and genotypic diversity for each farm in each year are shown in Table 3. It was found that 5.2% of the genotypic diversity could be attributed to variation between farms, and 5.5% to variation between years.
TABLE 2.
Distribution of strains of R. equi on farmsa
| Strain | No. of strains by farm
|
Total | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D | I | J | AA | L | M | N | O | P | R | U | V | W | WBb | X | Otherc | ||
| A | 1 | 1 | 2 | 18 | 4 | 2 | 4 | 1 | 1 | 1 | 35 | ||||||
| B | 6 | 1 | 1 | 3 | 3 | 1 | 15 | ||||||||||
| C | 1 | 10 | 2 | 2 | 1 | 1 | 4 | 2 | 23 | ||||||||
| D | 1 | 9 | 1 | 1 | 2 | 1 | 1 | 16 | |||||||||
| E | 1 | 2 | 11 | 2 | 3 | 1 | 2 | 2 | 24 | ||||||||
| F | 1 | 3 | 1 | 4 | 1 | 2 | 12 | ||||||||||
| G | 1 | 1 | 3 | 1 | 1 | 1 | 8 | ||||||||||
| H | 1 | 3 | 2 | 6 | |||||||||||||
| I | 6 | 6 | |||||||||||||||
| J | 1 | 1 | 1 | 1 | 4 | ||||||||||||
| K | 2 | 2 | 4 | ||||||||||||||
| L | 1 | 1 | 1 | 3 | |||||||||||||
| M | 2 | 1 | 1 | 2 | 6 | ||||||||||||
| N | 1 | 1 | 1 | 1 | 1 | 1 | 6 | ||||||||||
| O | 1 | 1 | 2 | ||||||||||||||
| P | 1 | 1 | 2 | ||||||||||||||
| R | 1 | 1 | 1 | 3 | |||||||||||||
| S | 2 | 2 | |||||||||||||||
| T | 2 | 1 | 3 | ||||||||||||||
| U | 1 | 1 | 2 | ||||||||||||||
| V | 1 | 1 | 2 | ||||||||||||||
| W | 1 | 1 | 1 | 3 | |||||||||||||
| Otherd | 1 | 1 | 4 | 1 | 3 | 1 | 4 | 2 | 2 | 3 | 22 | ||||||
| Total | 4 | 2 | 2 | 5 | 13 | 64 | 8 | 22 | 2 | 14 | 11 | 3 | 30 | 11 | 5 | 13 | 209 |
Farms were assigned random letter codes.
Isolates from The University of Melbourne Veterinary Clinic and Hospital, Werribee, Victoria, Australia.
Farms from which only one isolate was received.
Strains of which one isolate was found.
TABLE 3.
Number and genotypic diversity of R. equi isolates from each farm in each year
| Year | Parameter | No. and diversity (%) of isolates from farm:
|
Totalc | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D | I | J | AA | L | M | N | O | P | R | U | V | W | WBa | X | Otherb | |||
| 1991 | Isolates | 7 | 4 | 1 | 9 | 3 | 3 | 27 | ||||||||||
| Diversity | 0.81 | 0.83 | NMd | 0.86 | 0.66 | NM | 0.883 | |||||||||||
| 1992 | Isolates | 2 | 7 | 1 | 1 | 3 | 2 | 16 | ||||||||||
| Diversity | 1 | 0.90 | NM | NM | 1 | NM | 0.908 | |||||||||||
| 1993 | Isolates | 1 | 1 | 15 | 2 | 1 | 1 | 1 | 3 | 1 | 2 | 28 | ||||||
| Diversity | NM | NM | 0.79 | 1 | NM | NM | NM | 0.66 | NM | NM | 0.87 | |||||||
| 1994 | Isolates | 1 | 25 | 2 | 2 | 11 | 1 | 1 | 3 | 46 | ||||||||
| Diversity | NM | 0.87 | 1 | 1 | 0.87 | NM | NM | NM | 0.92 | |||||||||
| 1995 | Isolates | 2 | 2 | 1 | 8 | 4 | 4 | 3 | 3 | 1 | 7 | 1 | 2 | 1 | 39 | |||
| Diversity | 1 | 0 | NM | 0.89 | 0.5 | 1 | 1 | 1 | NM | 1 | NM | 1 | NM | 0.945 | ||||
| 1996 | Isolates | 2 | 1 | 2 | 1 | 5 | 6 | 1 | 2 | 1 | 10 | 1 | 1 | 1 | 34 | |||
| Diversity | 1 | NM | 1 | NM | 0.6 | 0.86 | NM | 1 | NM | 0.93 | NM | NM | NM | 0.923 | ||||
| 1997 | Isolates | 3 | 1 | 3 | 1 | 1 | 3 | 1 | 1 | 14 | ||||||||
| Diversity | 0.66 | NM | 0.66 | NM | NM | 1 | NM | NM | 0.879 | |||||||||
| 1998 | Isolates | 5 | 5 | |||||||||||||||
| Diversity | 0.7 | 0.7 | ||||||||||||||||
| Totale | Isolates | 4 | 2 | 2 | 5 | 13 | 64 | 8 | 22 | 2 | 14 | 11 | 3 | 30 | 11 | 5 | 13 | 209 |
| Diversity | 1 | 0 | 1 | 0.9 | 0.92 | 0.85 | 0.82 | 0.92 | 1 | 0.96 | 0.96 | 1 | 0.92 | 0.93 | 1 | n/d | 0.93 | |
Isolates from The University of Melbourne Veterinary Clinic and Hospital, Werribee, Victoria, Australia.
Farms from which only one isolate was received.
Total numbers and diversity for each year.
Not meaningful for single isolates.
Total numbers and diversity for each farm.
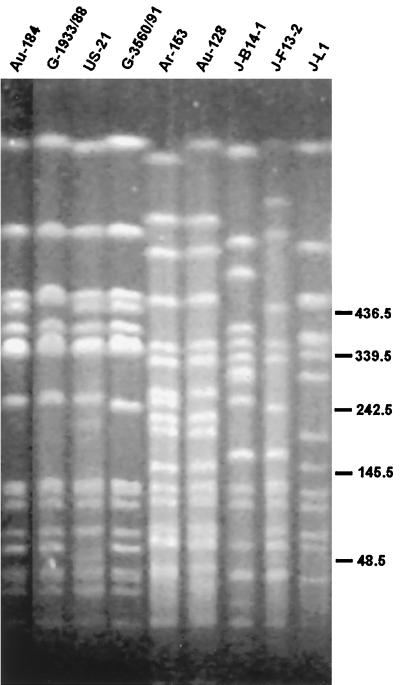
Most of the PFGE strains were fairly widespread, with isolates of most strains coming from a number of different farms and years. However there were some exceptions, such as strain I, which was represented by six isolates from the same farm, four of which were isolated in the same breeding season. Since the two laboratories supplying Australian isolates serve geographically separate horse-breeding areas it was notable that of the isolates from the University of Melbourne, all but two belonged to strains also found among isolates from the Scone laboratory. The three Argentinean isolates had identical restriction patterns, which differed by two bands from a pattern found among the Australian isolates. The six German isolates had almost identical restriction patterns, which differed by between one and four bands from two of the American isolates and some Australian isolates. The Canadian isolate and the Brazilian isolate had restriction patterns which were similar to some Australian isolates, but they could not be considered the same strain. Four of the Japanese isolates had identical restriction patterns, and these and one other isolate had patterns similar to but more than three bands different from Australian isolates. Figure 1 shows some of the isolates with similarity to Australian isolates, as well as three of the Japanese isolates.
FIG. 1.
Restriction patterns of selected R. equi isolates. Isolate numbers are as designated by the source laboratory, with prefixes added to indicate the country of origin. Prefix Ar indicates Argentina, Au indicates Australia, G indicates Germany, J indicates Japan, and US indicates the United States.
In six cases, two or three isolates from one foal were available for comparison. In one case, in which the isolates were the same PFGE strain, one pulmonary antemortem and two postmortem isolates, one from the lung and one from a mesenteric lymph node, were examined. In two cases, an antemortem pulmonary isolate and a postmortem pulmonary isolate from each foal were examined and found to be different strains. In two other cases, one pulmonary and one extrapulmonary isolate, both taken postmortem, were examined and also found to be different strains, while in the other case, two isolates of one strain and one of a different strain were found among one extrapulmonary and two pulmonary isolates taken postmortem.
Six isolates were found to be vapA negative by PCR. Five of these were respiratory isolates, the other being isolated from a submandibular abscess. Two PFGE strains were found to have two vapA-negative isolates, as well as a number of vapA-positive isolates, and one strain had one vapA-negative and two vapA-positive isolates, while no other isolates of the same strain as the other vapA-negative isolate were found.
Ribotyping.
Ribotyping showed that clonal strains as determined by restriction pattern analysis with PFGE generally had consistent ribotypes (data not shown). It was found that the bands distinguishing the PFGE restriction patterns of some subtypes within PFGE strains contained a 16S rRNA gene, resulting in the subtypes differing in ribotype within these strains. Similarly, the PFGE restriction patterns of some PFGE strains differed only in bands which did not contain a 16S rRNA gene, and thus ribotyping did not distinguish these PFGE strains. Ribotyping did not provide any benefit over direct visualization of restriction fragments for strain typing of R. equi with PFGE.
Stability of restriction patterns.
When the restriction patterns of the first and tenth passages of isolates from the four most common PFGE strains were compared, no differences were noted except one band of around 21 kbp which was present in the tenth but not the first passage of isolate 33 (strain A).
Univariate analysis.
It was found that different factors were associated with the prevalences of different strains on univariate analysis, as shown in Table 4. Date of isolation and farm were significantly associated with strain A. The odds of this strain being isolated were found to be 3.7-fold higher on farm M than the “other farms,” nearly 1.5-fold higher in December than in November, and lower at other times. The odds of strain A being isolated from a foal increased to 40 days of age, then remained fairly constant until 70 days, after which they dropped. The variation with date of birth showed no particular pattern. The odds of strain B being isolated were 21.5-fold higher in 1998 than in 1994, as well as showing quite a degree of variation in other years. Odds of isolation of strain C showed significant variation between years, as well as varying with date of birth of infected foals. This strain was also more likely to be isolated from pulmonary infections than extrapulmonary sites. The odds of isolating strain D varied between the four farm categories, this strain being nearly threefold more likely to be isolated on farm M than any of the other farms. Odds of isolating strain D also varied through the breeding season. Odds of isolating strain E varied between farms, with this strain being at least 1.7-fold more likely to be isolated on farm M than any other farm. Odds of this strain also varied between years and with date of birth and age, although with no noticeable pattern, and this strain was over twice as likely to be isolated from pulmonary infections than infections in other sites.
TABLE 4.
Factors associated (P ≤ 0.25) with each strain on univariate analysis
| Strain | Factor | Category | No. of cases | Odds ratio | 95% confidence interval | P value |
|---|---|---|---|---|---|---|
| A vs not A | Date of isolation | Sept./Oct. | 4/43 (9.3%) | 0.47 | 0.14, 1.53 | 0.04 |
| Novembera | 13/72 (18%) | 1 | ||||
| December | 16/65 (25%) | 1.48 | 0.65, 3.38 | |||
| Jan./Feb. | 1/23 (4.3%) | 0.21 | 0.03, 1.49 | |||
| Farm | Farm O | 4/22 (18%) | 0.57 | 0.17, 1.91 | 0.03 | |
| Farm W | 4/30 (13%) | 0.39 | 0.12, 1.29 | |||
| Farm Ma | 18/64 (28%) | 1 | ||||
| Other | 9/93 (9.7%) | 0.27 | 0.11, 0.66 | |||
| Age | <30 days | 1/11 (9.1%) | 0.36 | 0.04, 3.27 | 0.21 | |
| 30–39 days | 7/39 (18%) | 0.79 | 0.26, 2.46 | |||
| 40–49 days | 12/53 (23%) | 1.06 | 0.39, 2.92 | |||
| 50–59 daysa | 8/37 (22%) | 1 | ||||
| 60–69 days | 4/18 (22%) | 1.04 | 0.27, 4.03 | |||
| 70–89 days | 1/15 (6.7%) | 0.26 | 0.03, 2.28 | |||
| ≥90 days | 0/12 (0%) | 0.001 | <10−6, >106 | |||
| Date of birth | August | 4/37 (11%) | 0.31 | 0.09, 1.13 | 0.20 | |
| Sept. 1–15 | 2/26 (7.7%) | 0.21 | 0.04, 1.09 | |||
| Sept. 16–30 | 7/29 (24%) | 0.81 | 0.26, 2.56 | |||
| Oct. 1–15a | 9/32 (28%) | 1 | ||||
| Oct. 16–31 | 6/27 (22%) | 0.73 | 0.22, 2.40 | |||
| Nov. 1–15 | 2/20 (10%) | 0.28 | 0.05, 1.48 | |||
| After Nov. 15 | 4/14 (29%) | 1.02 | 0.25, 4.11 | |||
| B vs not B | Year of birth | 1991 | 0/27 (0%) | 0.0005 | <10−6, >106 | 0.01 |
| 1992 | 1/16 (6.3%) | 0.96 | 0.09, 9.9 | |||
| 1993 | 4/28 (14%) | 2.39 | 0.49, 11.6 | |||
| 1994a | 3/46 (6.5%) | 1 | ||||
| 1995 | 3/39 (7.7%) | 1.19 | 0.23, 6.29 | |||
| 1996 | 1/34 (2.9%) | 0.43 | 0.04, 4.37 | |||
| 1997 | 0/14 (0%) | 0.0005 | <10−6, >106 | |||
| 1998 | 3/5 (60%) | 21.5 | 2.53, 182 | |||
| C vs not C | Year of birth | 1991 | 0/27 (0%) | 0.0002 | <10−6, >106 | 0.007 |
| 1992 | 4/16 (25%) | 2.22 | 0.54, 9.2 | |||
| 1993 | 7/28 (25%) | 2.22 | 0.66, 7.46 | |||
| 1994a | 6/46 (13%) | 1 | ||||
| 1995 | 2/39 (5.1%) | 0.36 | 0.07, 1.90 | |||
| 1996 | 4/34 (12%) | 0.89 | 0.23, 3.43 | |||
| 1997 | 0/14 (0%) | 0.0002 | <10−6, >106 | |||
| 1998 | 0/5 (0%) | 0.0002 | <10−6, >106 | |||
| Date of birth | August | 4/37 (11%) | 0.53 | 0.13, 2.06 | 0.14 | |
| Sept. 1–15 | 0/26 (0%) | 0.0004 | <10−6, >106 | |||
| Sept. 16–30 | 3/29 (10%) | 0.50 | 0.11, 2.22 | |||
| Oct. 1–15a | 6/32 (19%) | 1 | ||||
| Oct. 16–31 | 4/27 (15%) | 0.75 | 0.19, 3.01 | |||
| Nov. 1–15 | 4/20 (20%) | 1.08 | 0.26, 4.44 | |||
| After Nov. 15 | 1/14 (7.1%) | 0.33 | 0.04, 3.07 | |||
| Site of infection | Lung | 21/172 (12%) | 2.36 | 0.53, 10.6 | 0.21 | |
| Other | 2/36 (5.6%) | 1 | ||||
| D vs not D | Farm | Farm O | 1/22 (4.5%) | 0.29 | 0.03, 2.44 | 0.16 |
| Farm W | 1/30 (3.3%) | 0.21 | 0.02, 1.75 | |||
| Farm Ma | 9/64 (14%) | 1 | ||||
| Other | 5/93 (5.4%) | 0.35 | 0.11, 1.09 | |||
| Date of isolation | Sept./Oct. | 5/43 (12%) | 1.45 | 0.41, 5.06 | 0.25 | |
| Novembera | 6/72 (8.3%) | 1 | ||||
| December | 2/65 (3.1%) | 0.35 | 0.07, 1.80 | |||
| Jan./Feb. | 3/23 (13%) | 1.65 | 0.38, 7.20 | |||
| E vs not E | Farm | Farm O | 0/22 (0%) | 0.0005 | <10−6, >106 | 0.05 |
| Farm W | 2/30 (6.7%) | 0.34 | 0.07, 1.66 | |||
| Farm Ma | 11/64 (17%) | 1 | ||||
| Other | 10/93 (11%) | 0.58 | 0.23, 1.46 | |||
| Year of birth | 1991 | 3/27 (11%) | 0.51 | 0.13, 2.09 | 0.03 | |
| 1992 | 0/16 (0%) | 0.0004 | <10−6, >106 | |||
| 1993 | 4/28 (14%) | 0.69 | 0.19, 2.48 | |||
| 1994a | 9/46 (20%) | 1 | ||||
| 1995 | 6/39 (15%) | 0.75 | 0.24, 2.32 | |||
| 1996 | 1/34 (2.9%) | 0.13 | 0.02, 1.04 | |||
| 1997 | 0/14 (0%) | 0.0004 | <10−6, >106 | |||
| 1998 | 0/5 (0%) | 0.0004 | <10−6, >106 | |||
| Date of birth | August | 3/37 (8.1%) | 0.62 | 0.13, 2.99 | 0.17 | |
| Sept. 1–15 | 4/26 (15%) | 1.27 | 0.29, 5.67 | |||
| Sept. 16–30 | 0/29 (0%) | 0.0007 | <10−6, >106 | |||
| Oct. 1–15a | 4/32 (13%) | 1 | ||||
| Oct. 16–31 | 3/27 (11%) | 0.88 | 0.18, 4.30 | |||
| Nov. 1–15 | 4/20 (20%) | 1.75 | 0.38, 7.97 | |||
| After Nov. 15 | 1/14 (7.1%) | 0.54 | 0.05, 5.31 | |||
| Site of infection | Lung | 20/172 (12%) | 2.23 | 0.50, 10.0 | 0.25 | |
| Other | 2/36 (5.6%) | 1 | ||||
| Age | <30 days | 0/11 (0%) | 0.0004 | <10−6, >106 | 0.09 | |
| 30–39 days | 7/39 (18%) | 2.48 | 0.59, 10.4 | |||
| 40–49 days | 7/53 (13%) | 1.72 | 0.42, 7.16 | |||
| 50–59 daysa | 3/37 (8.1%) | 1 | ||||
| 60–69 days | 2/18 (11%) | 1.42 | 0.22, 9.33 | |||
| 70–89 days | 0/15 (0%) | 0.0004 | <10−6, >106 | |||
| ≥90 days | 0/12 (0%) | 0.0004 | <10−6, >106 |
This category was the reference category.
The odds of infected foals having primarily pulmonary infection varied significantly between farms, the odds being nearly 29-fold higher on farm M than on the pooled other farms as shown in Table 5. The odds of primarily pulmonary infection also decreased with age and varied between years and with date of birth of foals. These results may, however, have been affected by sampling bias, because the likelihood of a particular case being cultured would probably differ between pulmonary and extrapulmonary infections and between farms.
TABLE 5.
Factors associated (P ≤ 0.25) with pulmonary infection on univariate analysis
| Factor | Category | No. of cases | Odds ratio | 95% confidence interval | P value |
|---|---|---|---|---|---|
| Farm | Farm O | 18/20 (90%) | 0.14 | 0.01, 1.67 | <5 × 10−5 |
| Farm W | 29/31 (94%) | 0.23 | 0.02, 2.64 | ||
| Farm Ma | 63/64 (98%) | 1 | |||
| Other | 61/89 (69%) | 0.03 | 0.005, 0.26 | ||
| Age | Linear/10 days | 184 | 0.76 | 0.65, 0.90 | 0.001 |
| Age | <30 days | 11/11 (100%) | 1,010 | <10−6, >106 | 0.005 |
| 30–39 days | 38/39 (97%) | 10.5 | 1.24, 88.6 | ||
| 40–49 days | 46/52 (88%) | 2.11 | 0.67, 6.72 | ||
| 50–59 daysa | 29/37 (78%) | 1 | |||
| 60–69 days | 14/18 (78%) | 0.97 | 0.25, 3.76 | ||
| 70–89 days | 10/15 (67%) | 0.55 | 0.15, 2.08 | ||
| ≥90 days | 8/12 (67%) | 0.55 | 0.13, 2.31 | ||
| Date of birth | August | 27/36 (75%) | 0.81 | 0.26, 2.49 | 0.20 |
| Sept. 1–15 | 24/26 (92%) | 3.23 | 0.61, 17.1 | ||
| Sept. 16–30 | 25/29 (86%) | 1.68 | 0.44, 6.46 | ||
| Oct. 1–15a | 26/33 (79%) | 1 | |||
| Oct. 16–31 | 25/27 (93%) | 3.36 | 0.64, 17.8 | ||
| Nov. 1–15 | 19/20 (95%) | 5.11 | 0.58, 45.0 | ||
| After Nov. 15 | 10/13 (77%) | 0.90 | 0.19, 4.17 | ||
| Year of birth | 1991 | 23/26 (88%) | 0.94 | 0.21, 4.27 | 0.23 |
| 1992 | 13/15 (87%) | 0.79 | 0.14, 4.58 | ||
| 1993 | 26/28 (93%) | 1.59 | 0.29, 8.78 | ||
| 1994a | 41/46 (89%) | 1 | |||
| 1995 | 29/36 (81%) | 0.51 | 0.15, 1.75 | ||
| 1996 | 26/33 (79%) | 0.45 | 0.13, 1.58 | ||
| 1997 | 9/15 (60%) | 0.18 | 0.05, 0.73 | ||
| 1998 | 4/5 (80%) | 0.49 | 0.05, 5.27 |
This category was the reference category.
It was found that there was a significant difference between months in the survival of infected foals, with foals diagnosed in January or February being nearly sixfold less likely to survive than those diagnosed in November, as shown in Table 6. There was also a strong association between age and survival, with older foals being less likely to survive. There were significant differences between farms in mortality, with foals on farm W being at least 2.5-fold more likely to survive than foals on farm O or the “other farms” and those on farm M more likely to survive than those on any of the other farms. Odds of survival also varied between strains, with foals from which strain A or C was isolated being more likely to survive than foals infected with other strains.
TABLE 6.
Factors associated (P ≤ 0.25) with survival on univariate analysis
| Factor | Category | No. of cases | Odds ratio | 95% confidence interval | P value |
|---|---|---|---|---|---|
| Age | Linear/10 days | 179 | 0.73 | 0.59, 0.92 | 0.01 |
| Age | <30 days | 10/10 (100%) | 749 | <10−6, 106 | 0.10 |
| 30–39 days | 39/39 (100%) | 749 | <10−6, >106 | ||
| 40–49 days | 47/50 (94%) | 0.44 | 0.04, 4.36 | ||
| 50–59 daysa | 36/37 (97%) | 1 | |||
| 60–69 days | 16/18 (89%) | 0.22 | 0.02, 2.63 | ||
| 70–89 days | 12/14 (86%) | 0.17 | 0.01, 2.01 | ||
| >90 days | 9/11 (82%) | 0.13 | 0.01, 1.54 | ||
| Farm | Farm O | 17/20 (85%) | 0.39 | 0.06, 2.58 | 0.0002 |
| Farm Wa | 29/31 (94%) | 1 | |||
| Farm M | 63/63 (100%) | 1,860 | <10−6, >106 | ||
| Other | 66/81 (81%) | 0.30 | 0.07, 1.41 | ||
| Strain | Aa | 32/33 (97%) | 1 | 0.08 | |
| B | 14/15 (93%) | 0.44 | 0.03, 7.50 | ||
| C | 22/22 (100%) | 310 | <10−6, >106 | ||
| D | 13/16 (81%) | 0.14 | 0.01, 1.42 | ||
| E | 18/22 (82%) | 0.14 | 0.01, 1.36 | ||
| Other | 75/85 (88%) | 0.23 | 0.03, 1.91 | ||
| Date of isolation | Sept./Oct. | 40/41 (98%) | 3.81 | 0.44, 32.8 | 0.009 |
| Novembera | 63/69 (91%) | 1 | |||
| December | 56/62 (90%) | 0.89 | 0.27, 2.91 | ||
| Jan./Feb. | 11/17 (65%) | 0.18 | 0.05, 0.64 |
This category was the reference category.
Multivariate analysis.
All dependent variables except strain B were found to have multiple variables associated with them in the univariate analysis, so these were used in multivariate analyses. With strain D as the dependent variable, none of the variables remained in a multivariate analysis. When strain A or site of primary infection was the dependent variable, farm was the only variable to remain, and when strain C or E was the dependent variable, only year remained; thus the results were identical to those of the univariate analyses for these variables. With survival as the dependent variable, farm, strain, and date of isolation were all significant in the multivariate analysis, as shown in Table 7. When the analysis was repeated using LogXact, the results were very similar, although the P value for strain was 0.08, so strain could no longer be considered significantly associated with mortality.
TABLE 7.
Factors associated (P ≤ 0.05) with survival on multivariate analysis
| Factor | Category | No. of cases | SPSS results
|
LogXact results
|
||||
|---|---|---|---|---|---|---|---|---|
| Adjusted odds ratio | 95% confidence interval | P value | Adjusted odds ratio | 95% confidence interval | P value | |||
| Farm | Farm O | 17/20 (85%) | 0.23 | 0.02, 3.06 | 0.0006 | 0.26 | 0.004, 4.15 | 0.0012 |
| Farm Wa | 29/30 (97%) | 1 | 1 | |||||
| Farm M | 63/63 (100%) | 15,700 | <10−6, >106 | 3.97 | 0.1, infinity | |||
| Other | 60/74 (81%) | 0.31 | 0.03, 3.13 | 0.33 | 0.006, 3.35 | |||
| Strain | Aa | 31/32 (97%) | 1 | 0.048 | 1 | 0.083 | ||
| B | 12/12 (100%) | 1,840 | <10−6, >106 | 0.15 | 0.004, infinity | |||
| C | 22/22 (100%) | 6,150 | <10−6, >106 | 0.78 | 0.02, infinity | |||
| D | 13/16 (81%) | 0.11 | 0.006, 2.03 | 0.15 | 0.002, 3.71 | |||
| E | 18/22 (81%) | 0.08 | 0.005, 1.15 | 0.11 | 0.001, 1.84 | |||
| Other | 73/83 (88%) | 0.49 | 0.05, 4.57 | 0.50 | 0.01, 4.63 | |||
| Date of isolation | Sept./Oct. | 40/40 (100%) | 31,600 | <10−6, >106 | 0.005 | 8.11 | 1.07, infinity | 0.013 |
| Novembera | 62/68 (91%) | 1 | 1 | |||||
| December | 56/62 (90%) | 0.93 | 0.23, 3.8 | 0.94 | 0.19, 4.54 | |||
| Jan./Feb. | 11/17 (65%) | 0.45 | 0.09, 2.19 | 0.48 | 0.08, 2.84 | |||
This category was the reference category.
DISCUSSION
From these data it appears that a number of strains of R. equi carry the vapA gene and thus have the capacity to cause disease in foals. Although 44 different PFGE strains were found among the Australian isolates, the 5 most common strains accounted for over half the isolates studied, and half the strains accounted for almost 90% of the isolates, the remainder being isolates which were single representatives of strains. On each farm from which more than one isolate was obtained, the two strains which were most common on that farm always accounted for at least one-third of the isolates from that farm, as compared to just over a quarter of the isolates taken as a whole belonging to the two most common strains overall. It was also found that the most common strains varied between farms. Over the 10 farms from which multiple isolates of at least one strain were received, eight different strains were represented among the most common (or were the most common in two cases) from each. These findings imply some degree of clustering of isolates on farms, although strains also tend to be rather widespread, and this was supported by the finding that 5.2% of the overall genotypic diversity was due to between-farm rather than within-farm diversity. There also appeared to be lower genotypic diversity within years compared to the total, although, since the numbers of isolates from each farm varied between years, it was hard to separate out the effects of the two variables.
The significantly high prevalence of strain A on farm M compared with the other farms was interesting given that this farm had a high incidence of R. equi pneumonia despite close observation and prompt treatment of foals. One explanation would be that strain A is particularly infectious and so causes a high incidence of disease. This would result in the environment becoming increasingly contaminated with this strain, as infected foals shed large numbers of bacteria. Despite the higher levels of disease, there was no mortality associated with R. equi on this farm, indicating that either detection and treatment methods were effective in eliminating mortality or strain A was not as virulent as some other strains. In the last 3 years under study, it was noted that the prevalence of strain A on farms other than farm M was increasing. The reason for this increase is unknown, but it may be due to farm-to-farm transmission by horse movement. It is also interesting that one of the other strains which was more prevalent on farm M than the other farms was strain D, as this strain had a similar restriction pattern to strain A, indicating that the two may have been closely related.
It was not surprising to find that the proportion of foals with pulmonary manifestations varied between farms and years in the univariate analysis. However, since we could not look at the entire foal populations on the farms, it cannot be determined whether the differences found were due to differences in the number of foals with pulmonary manifestations, sampling bias as mentioned above, or real differences in the proportion of foals which had pulmonary manifestations. Since the incidences of pulmonary and nonpulmonary infections would be affected by different factors, the two would be expected to vary fairly independently. The finding that older foals were less likely to have pulmonary manifestations, especially obvious when age is treated as a continuous variable, is most probably due to a decrease in the incidence and therefore proportion of primary pulmonary infections as foals get older. This is borne out by the absolute numbers of extrapulmonary isolates remaining similar across the age groups, while the number of pulmonary isolates dropped. Since farm was the only variable to remain in a multivariate analysis, this appears to be the most important factor in determining the odds of pulmonary manifestations, but without further information on the whole foal population of these farms, it is impossible to know how to interpret this. It is most likely that this result simply reflects the different incidences of pulmonary disease on different farms.
The increase in mortality through the breeding season and with age was likely to be due to foals diagnosed at an older age, or later in the breeding season, being further advanced in the course of disease and therefore less likely to survive. Since this cannot be controlled for, it is difficult to draw any conclusions regarding the importance of age or time of year in whether infected foals survive.
The differences in survival between foals on different farms was not at all surprising, given the expected differences in management practices and vigilance of farm staff in detecting infected foals. It may also have been confounded by variation between farms in how ill foals were before samples were taken, but this is impossible to ascertain.
The difference between isolates from the same cases warrants further investigation. It would be useful to be able to compare isolates from multiple pulmonary abscesses from a series of cases to determine how many strains are found in a typical infected foal and whether isolates from each abscess are of the same strain. The occurrence of two different strains in lesions from five of six foals suggests that the initial infection may have consisted of a mixture of strains, and the observation is supported by the finding of Takai et al. (18) that isolates from different abscesses in the lung of one infected foal had different ribotypes. This would also explain why isolates from sites other than the lung can be different from pulmonary isolates from the same foal. It is believed that enteritis associated with R. equi pneumonia is due to the large numbers of bacteria expectorated and swallowed (14), which would imply that the same strains should be causing infection in both sites. However, the finding that multiple strains can be isolated from one diseased lung means that lung and enteric isolates from a given foal may not be the same strain if only one or two isolates are examined, as was the case in this study. It also means that the isolates studied may not be a true reflection of the strains present in any one foal. Since a large number of foals were sampled, the isolates examined should give an accurate reflection of the disease-causing strains present on farms, if not in each particular foal, at the time the foals were infected, allowing conclusions about strain epidemiology to be drawn.
Although it is generally accepted that isolates causing disease in foals are vapA positive (3, 14), it is not unusual to find the occasional isolate which is not, as was the case for the isolates used in this study. It is possible that the isolates lost the gene in culture, that these particular isolates were not involved with the disease process, or that coinfection with vapA-positive isolates somehow enables vapA-negative isolates to become involved in causing disease. The fact that two isolates of two strains were found which were vapA negative may imply that these strains are more able to cause disease without the plasmid or alternatively that the plasmid is less stable in these strains so the isolates lost it in culture.
Transfer of strains between farms is likely to be horse associated, either via diseased foals or by carriage as commensal organisms in the digestive tract of mares or foals. Local spread could, however, quite easily occur by wind, especially in hot, dry conditions, which lead to a dramatic increase in the numbers of airborne R. equi. Finding almost identical isolates from different continents implies that strains can be very widespread, which would be consistent with horse-associated spread, given the high level of international horse movement.
Conclusions.
We found that there is quite a degree of genetic variability between strains of R. equi which cause disease in foals, although a relatively restricted number of strains accounted for a majority of the isolates studied. Strains appeared to be widespread, with some isolates from different continents appearing to be closely related, if not the same strains. It also appeared that different farms had particular strains present, with the two most common strains on each farm accounting for at least one-third of the isolates and one strain showing a significant difference in prevalence between farms. It was also found that the odds of pulmonary infection varied between farms and that survival of infected foals varied between farms and through the breeding season.
ACKNOWLEDGMENTS
This work was supported by a grant from the Rural Industries Research and Development Corp.
We thank Angela Irving, Veterinary Clinical Centre, The University of Melbourne, and John Prescott, University of Guelph, for providing a number of isolates for this study.
REFERENCES
- 1.Fuhrmann C, Lämmler C. Charakterisierung von Rhodococcus equi-Isolaten von Pferd und Mensch. Berl Muench Tieraerztl Wschr. 1997;110:54–59. [PubMed] [Google Scholar]
- 2.Gordon D M. The genetic structure of Escherichia coli populations in feral house mice. Microbiology. 1997;143:2039–2046. doi: 10.1099/00221287-143-6-2039. [DOI] [PubMed] [Google Scholar]
- 3.Haites R E, Muscatello G, Begg A P, Browning G F. Prevalence of the virulence-associated gene of Rhodococcus equi in isolates from infected foals. J Clin Microbiol. 1997;35:1642–1644. doi: 10.1128/jcm.35.6.1642-1644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosmer D W, Lemeshow S. Applied logistic regression. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 5.Kanno T, Asawa T, Ito H, Takai S, Tsubaki S, Sekizaki T. Restriction map of a virulence-associated plasmid of Rhodococcus equi. Plasmid. 1993;30:309–311. doi: 10.1006/plas.1993.1065. [DOI] [PubMed] [Google Scholar]
- 6.Lasker B A, Brown J M, McNeil M M. Identification and epidemiological typing of clinical and environmental isolates of the genus Rhodococcus with use of a digoxigenin-labeled rDNA gene probe. Clin Infect Dis. 1992;15:223–233. doi: 10.1093/clinids/15.2.223. [DOI] [PubMed] [Google Scholar]
- 7.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rainey F A, Burghardt J, Kroppenstedt R M, Klatte S, Stackebrandt E. Phylogenetic analysis of the genera Rhodococcus and Nocardia and evidence for the evolutionary origin of the genus Nocardia from within the radiation of Rhodococcus species. Microbiology. 1995;141:523–528. [Google Scholar]
- 9.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning—a laboratory manual. 2nd ed. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 10.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soedarmanto I, Oliveira R, Lämmler C, Dürrling H. Further studies on Rhodococcus equi isolated from bovine lymphadenitis. Med Sci Res. 1998;26:835–837. [Google Scholar]
- 12.Soedarmanto I, Oliveira R, Lämmler C, Dürrling H. Identification and epidemiological relationship of Rhodococcus equi isolated from cases of lymphadenitis in cattle. Zentbl Bakteriol. 1997;286:457–467. doi: 10.1016/s0934-8840(97)80047-3. [DOI] [PubMed] [Google Scholar]
- 13.Soedarmanto I, Zhicai W, Setyamahanani A, Lämmler C. Pheno- and genotyping of Rhodococcus equi isolated from faeces of healthy horses and cattle. Res Vet Sci. 1998;64:181–185. doi: 10.1016/s0034-5288(98)90121-7. [DOI] [PubMed] [Google Scholar]
- 14.Takai S. Epidemiology of Rhodococcus equi infections: a review. Vet Microbiol. 1997;56:167–176. doi: 10.1016/s0378-1135(97)00085-0. [DOI] [PubMed] [Google Scholar]
- 15.Takai S, Fortiner G, Pronost S, Rahal K, Becu T, Begg A, Browning G, Prescott J F. Genetic analysis of virulent Rhodococcus equi based on restriction fragment length polymorphism of virulence plasmids: a molecular approach for epidemiology of virulent R. equi worldwide. In: Werney U, Wade J F, Mumford J A, Kaaden O-R, editors. Equine infectious diseases VIII: Proceedings of the Eighth International Conference. R &W Publications. 1998. pp. 587–588. Newmarket, United Kingdom. [Google Scholar]
- 16.Takai S, Koike K, Ohbushi S, Izumi C, Tsubaki S. Identification of 15- to 17-kilodalton antigens associated with virulent Rhodococcus equi. J Clin Microbiol. 1991;29:439–443. doi: 10.1128/jcm.29.3.439-443.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takai S, Sekizaki T, Ozawa T, Sugawara T, Watanabe Y, Tsubaki S. Association between a large plasmid and 15- to 17-kilodalton antigens in virulent Rhodococcus equi. Infect Immun. 1991;59:4056–4060. doi: 10.1128/iai.59.11.4056-4060.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takai S, Takeda K, Nakano Y, Karasawa T, Furugoori J, Sasaki Y, Tsubaki S, Higuchi T, Anzai T, Wada R, Kamada M. Emergence of rifampin-resistant Rhodococcus equi in an infected foal. J Clin Microbiol. 1997;35:1904–1908. doi: 10.1128/jcm.35.7.1904-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenover F C, Arbeit R D, Goeging R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed–field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]