Abstract
The cell membrane system comprises the plasma membrane, endoplasmic reticulum, Golgi apparatus, lysosome, mitochondria, and nuclear membrane, which are essential for maintaining normal physiological functions of cells. The proteins associated with these membrane-organelles are frequently modified to regulate their functions, the most common of which is ubiquitin modification. So far, many ubiquitin E3 ligases anchored in the membrane system have been identified as critical players facilitating intracellular biofunctions whose dysfunction is highly related to cancer. In this review, we summarized membrane-associated E3 ligases and revealed their relationship with cancer, which is of great significance for discovering novel drug targets of cancer and may open up new avenues for inducing ubiquitination-mediated degradation of cancer-associated membrane proteins via small chemicals such as PROTAC and molecular glue.
Keywords: cell membrane system, cancer, E3 ligases, drug targets, PROTAC (proteolysis-targeting chimeric molecule)
Graphical Abstract
Introduction
The cell membrane system comprises the plasma membrane, endoplasmic reticulum (ER), Golgi apparatus, mitochondria, and nuclear membrane, which is essential for maintaining cell morphology and functions. By way of illustration, plasma membrane control the substances’ entrance and exit of cells and protect the integrity of cells and maintain the shape of cells. Other membrane structures provide for the manufacture and packaging of substances within cells. Similarly, other subcellular membranes surround organelles resembling the cell membrane but with different protein and phospholipids compositions. Membrane-bound organelles provide many benefits to eukaryotic cells. Firstly, the membrane system divides the cell into multiple compartments, enabling enzymes to be concentrated in specific compartments, thereby improving the efficiency of biochemical reactions therein. Secondly, Membrane structures can protect the rest of the cell from damage by confining harmful substances to specific compartments. Thirdly, membrane-composed organelles usually rely on vesicles to transport substances and proteins (Dacks and Field, 2018). Remarkably, different membranes are well suited for their functions, primarily thanks to their various molecular composition, especially some anchor proteins. However, the dysfunction of these anchor proteins is closely associated to some human diseases like cancer (Martin and List, 2019), etc.
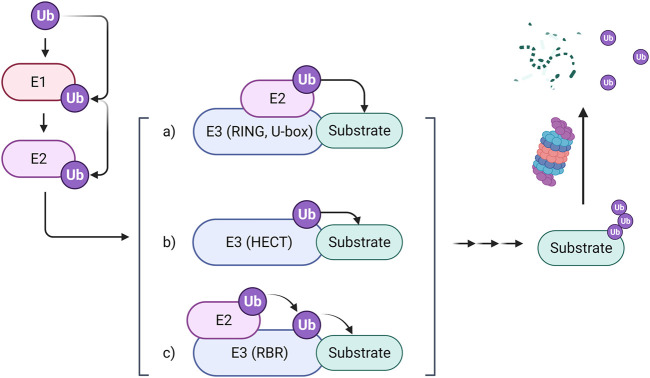
Ubiquitylation regulates protein breakdown and signaling cascades, and hence plays a crucial role in cellular physiology. Three kinds of enzymes are required for the ubiquitylation process: E1, E2, and E3. E3 ligases, in particular, endow ubiquitination with selectivity by promoting ubiquitin transfer from an E2 enzyme to the substrate (Zheng and Shabek, 2017). So far, more than six hundred E3 ubiquitin ligases have been identified in the human proteome, which can be classified into three main types: Really Interesting New Gene (RING) E3s, Homologous to the E6-AP Carboxyl Terminus (HECT) E3s, and RING-between-RING (RBR) E3s. RING E3s are the most abundant ubiquitin ligases. They especially contain RING domains that can bind with an E2-ubiquitin thioester and promote ubiquitin cargo release. RING E3s can also stimulate the transfer of ubiquitin from E2s to the substrate directly (Deshaies and Joazeiro, 2009). The HECT E3s, containing a conserved enzymatic cysteine (Cys), accept ubiquitin from E2s and then transfer it to the substrate by a specific Lys residue (Rotin and Kumar, 2009). Another type of E3 ligases, RBR, play a role in ubiquitination through a HECT-RING hybrid mechanism. Ubiquitin is recruited to RBR via forming a disulfide bond with the crucial cysteine residue on RING2 and then transported to the substrate, forming an isopeptide bond with it (Figure 1). Based on the ubiquitination degradation process, E3 ubiquitin ligases have potential application value in relative researches of some proteins’ structures, functions and distribution. For instance, E3 ligases have been involved in PROteolysis-TArgeting Chimeras (PROTACs) related research to target substrate proteins for degradation. In addition, some E3 ligases were reported as novel biomarkers for some diseases like COVID-19 (Novelli et al., 2021). Furthermore, some preceding works of our group revealed that E3 ligases may participate in regulating carcinogenesis including WD repeat and SOCS box containing 1(WSB1) and Murine double minute-2 (MDM2) (Cao et al., 2015; Ying et al., 2016). These researches inspire us to shine a spotlight on the relationship between E3 ligases and cancer.
FIGURE 1.
The mechanism of the ubiquitin-proteasome system. The ubiquitin-proteasome system comprises three types of enzymes. The ubiquitin-activating enzyme E1 binds with ubiquitin and then transfers it to the ubiquitin-conjugating enzyme E2, which further cooperates with three types of ubiquitin-protein ligase E3 to transport ubiquitin to substrates through different mechanisms. Finally, the labeled substrates are recognized by proteosomes for degradation.
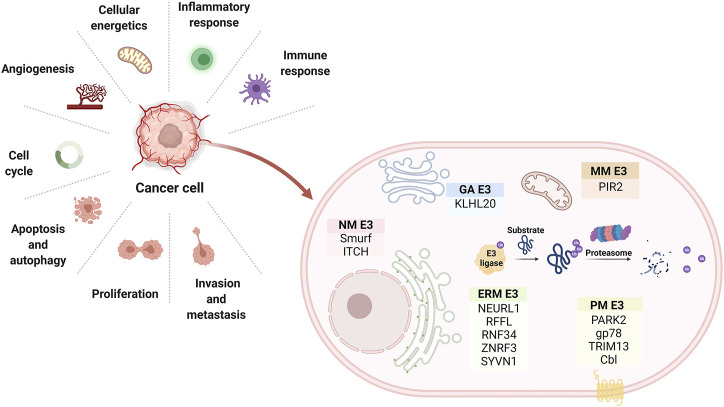
According to the statistics, E3 ligases distribute in a great many organs, cells and subcellular compartments. Compared with other subcellular localized E3 ligases, membrane-associated E3 ligases are much more crucial for the membrane-organelles, which play vital roles in maintaining their morphology and functions. For instance, mainly distributed in the mitochondrion and endoplasmic reticulum, the membrane-associated RING-CH-type finger (MARCH) proteins of E3 ubiquitin ligases positively regulate mitochondrial fission (Xu et al., 2016) and play a crucial role in controlling mitochondrial mass. It can degrade itself in dysfunctional mutants to maintain mitochondrial homeostasis and prevent cellular senescence (Bauer et al., 2017). Another E3 ligase Synoviolin (SYVN1), located in ER, stimulates the degradation of IRE1α through interaction with p53 and maintains ER function by regulating activation of the IRE1α/XBP1 pathway (Namba et al., 2015). However, the membrane-associated E3 ligases remain poorly summarized.
In this review, we summarized the 84 membrane-associated E3 ligases from the following aspects: UniProt ID, gene name, subcellular localization, E3 type and the number of transmembrane segments (Table 1). Moreover, we also described the relationship of some important E3 ligases with carcinogenesis in detail, which is significant for uncovering novel targets of cancer and may provide a new perspective of understanding the ubiquitination process, especially providing inspiration for inducing membrane protein degradation via ubiquitination and proteasome pathway.
TABLE 1.
84 membrane-associated E3 ligases.
| Uniprot ID | Name | Subcellular location | Type | Transmembrane regions |
|---|---|---|---|---|
| Q9NZS9 | BFAR | ERM | RING | 4 |
| Q9UKV5 | AMFR | ERM | RING | 7 |
| Q5T197 | DCST1 | PM | RING | 6 |
| Q9Y4D8 | HECTD4 | Membrane | HECTc | 1 |
| Q96J02 | ITCH | PM, cytoplasm, nucleus, EM | HECTc | NA |
| Q8TDB6 | DTX3L | Cytoplasm, nucleus, EM, LM | RING | NA |
| A6NNE9 | MARCHF11 | VM | RING | 2 |
| Q86UD3 | MARCHF3 | VM, EM | RING | 2 |
| Q9P2E8 | MARCHF4 | GAM | RING | 2 |
| Q9NX47 | MARCHF5 | MM, ERM | RING | 4 |
| Q8TCQ1 | MARCHF1 | GAM, LM, VM, EM, PM | RING | 2 |
| Q86YJ5 | MARCHF9 | GAM, LM | RING | 2 |
| Q9P0N8 | MARCHF2 | ERM, LM, EM | RING | 2 |
| Q5T0T0 | MARCHF8 | VM, LM, EM | RING | 2 |
| Q86YT6 | MIB1 | Cytoplasm, SK, CK, PM | RING | NA |
| Q7L5Y9 | MAEA | Cytoplasm, nucleus, PM, SK | RING | NA |
| O60337 | MARCHF6 | ERM | RING | 14 |
| Q00987 | MDM2 | Cytoplasm, nucleus | RING | NA |
| Q969V5 | MUL1 | MM | RING | 2 |
| Q8WY64 | MYLIP | Cytoplasm, PM | RING | NA |
| P46934 | NEDD4 | Cytoplasm, PM | HECTc | NA |
| O76050 | NEURL1 | Cytoplasm, PM | RING | NA |
| Q6ZNB6 | NFXL1 | Membrane | RING | 1 |
| O60683 | PEX10 | Peroxisome membrane | RING | NA |
| O00623 | PEX12 | Peroxisome membrane | RING | 2 |
| O43164 | PJA2 | Cytoplasm, PM, ERM, GAM | RING | NA |
| Q8WZ73 | RFFL | EM | RING | NA |
| O00237 | RNF103 | ERM | RING | 4 |
| Q9H920 | RNF121 | Membrane | RING | 6 |
| O60260 | PRKN | Cytoplasm, nucleus, ERM, MM, PM | RING | NA |
| Q9H9V4 | RNF122 | GAM, ERM | RING | 1 |
| Q9ULX5 | RNF112 | Cytoplasm, nucleus, VM, PM | RING | 2 |
| Q96EQ8 | RNF125 | GAM | RING | NA |
| P29590 | PML | Cytoplasm, nucleus, ERM | RING | NA |
| Q8WVZ7 | RNF133 | ERM | RING | 1 |
| Q8WU17 | RNF139 | ERM | RING | 12 |
| Q86XS8 | RNF130 | Membrane, cytoplasm | RING | 1 |
| P50876 | RNF144A | PM, VM | RING | 1 |
| Q7Z419 | RNF144B | MM, cytoplasm | RING | 1 |
| Q8WVD5 | RNF141 | Membrane | RING | NA |
| Q8N8N0 | RNF152 | LM | RING | 1 |
| Q8TEB7 | RNF128 | M, cytoplasm, SK, perinuclear region | RING | 1 |
| Q9H6Y7 | RNF167 | M | RING | 1 |
| Q8NC42 | RNF149 | Membrane | RING | 1 |
| Q96MT1 | RNF145 | ERM | RING | 14 |
| Q8N7C7 | RNF148 | Membrane | RING | 1 |
| Q9ULK6 | RNF150 | Membrane | RING | 1 |
| Q96K19 | RNF170 | ERM | RING | 3 |
| Q8N4F7 | RNF175 | Membrane | RING | 5 |
| Q96D59 | RNF183 | ERM, GAM, LM | RING | 1 |
| Q9NXI6 | RNF186 | ERM | RING | 2 |
| Q8N6D2 | RNF182 | Membrane, cytoplasm | RING | 2 |
| Q9Y6U7 | RNF215 | Membrane | RING | 2 |
| Q9NV58 | RNF19A | Membrane, cytoplasm, SK | RING | 2 |
| Q6ZMZ0 | RNF19B | Cytoplasmic granule membrane, ERM | RING | 2 |
| A6NCQ9 | RNF222 | Membrane | RING | 1 |
| E7ERA6 | RNF223 | Membrane | RING | 1 |
| Q96GF1 | RNF185 | MM, ERM | RING | 2 |
| M0QZC1 | RNF225 | Membrane | RING | 1 |
| Q9BY78 | RNF26 | ERM | RING | 5 |
| Q969K3 | RNF34 | PM, nucleus, cytoplasm, cytosol | RING | NA |
| Q9Y225 | RNF24 | GAM | RING | 1 |
| Q8TC41 | RNF217 | Membrane, cytoplasm | RING | 1 |
| Q5M7Z0 | RNFT1 | ERM | RING | 6 |
| Q96EX2 | RNFT2 | Membrane | RING | 4 |
| Q9HCE7 | SMURF1 | Cytoplasm, PM | HECTc | NA |
| A0AVI4 | TMEM129 | ERM | RING | 3 |
| Q9HAU4 | SMURF2 | Nucleus, cytoplasm, PM, membrane raft | HECTc | NA |
| O60858 | TRIM13 | ERM | RING | 1 |
| P36406 | TRIM23 | Cytoplasm, GAM, LM | RING | NA |
| Q86TM6 | SYVN1 | ERM | RING | 6 |
| Q8IWR1 | TRIM59 | ERM | RING | 1 |
| Q6ZMU5 | TRIM72 | PM, sarcolemma, VM | RING | NA |
| Q6ZT12 | UBR3 | Membrane | UBR | 3 |
| Q5T4S7 | UBR4 | Membrane | UBR | 2 |
| Q9H270 | VPS11 | EM, LM, VM, autophagosome | RING | NA |
| P49754 | VPS41 | EM, LM, GAM, VM, clathrin-coated vesicle | RING | NA |
| Q6PJI9 | WDR59 | LM | RING | NA |
| O95159 | ZFPL1 | GAM | RING | 1 |
| Q9H0M0 | WWP1 | Cytoplasm, PM, nucleus | HECTc | NA |
| Q8ND25 | ZNRF1 | VM | RING | NA |
| Q9ULT6 | ZNRF3 | PM | RING | 1 |
| Q8NHG8 | ZNRF2 | EM, LM, PM | RING | NA |
| Q8WWF5 | ZNRF4 | ERM | RING | 1 |
| Q9P253 | VPS18 | EM, LM, VM, autophagosome | RING | NA |
ERM, endoplasmic reticulum membrane; GAM, golgi apparatus membrane; PM, plasma membrane; VM, vesicle membrane; EM, endosome membrane; LM, lysosome membrane; MM, mitochondrion membrane; SK, cytoskeleton and microtubule.
ER-Localized E3
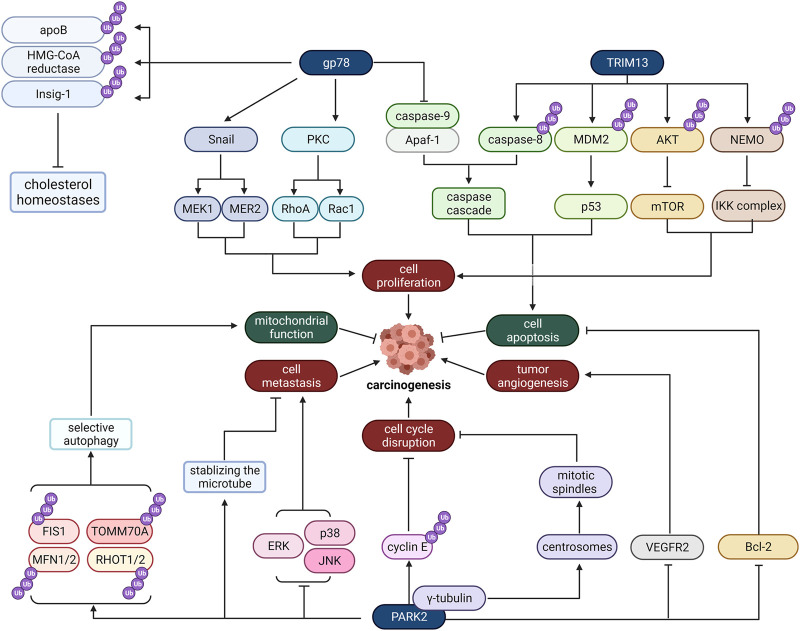
The endoplasmic reticulum functions as the transportation system of the eukaryotic cell and manufactures lipids and proteins. What’s more, the studies presented thus far provide evidence that endoplasmic reticulum stress is closely related to cancer (Yadav et al., 2014). Due to the critical role that E3 ligases play in metabolism occurred in the endoplasmic reticulum, ER-localized E3 ligases are essential for cancer development (Figure 2).
FIGURE 2.
The role of some typical ER-localized E3 ligases in cancer. Gp78, TRIM13, and PARK2 are ER-localized E3 ligases. They regulate carcinogenesis through many distinct pathways, involving cell proliferation, cell apoptosis, tumor angiogenesis, cell cycle disruption, cell metastasis and mitochondrial function.
TRIbM13
TRIM13 is a tumor suppressor gene whose deletion is common in various malignant tumors. It is a tumor suppressor in non-small-cell lung carcinoma (NSCLC) and its mRNA and protein expression was reduced in NSCLC tissues and cell lines (Xu et al., 2019). It has been reported that it is mainly located in the endoplasmic reticulum membrane, mediates the degradation of endoplasmic reticulum-related proteins, and regulates autophagy caused by endoplasmic stress (Tomar et al., 2012). So far, caspase-8, MDM2, Akt, and Nur77 have all been identified as TRIM13 substrates. Furthermore, several studies have suggested that these substrates are all related to tumor cell apoptosis and proliferation.
As for caspase-8, its ubiquitination is critical to proceed downstream caspase cascade, which results in cell apoptosis. TRIM13-induced caspase-8 ubiquitination may lead to its transportation to autophagosomes. Autophagosomes anchor caspase-8 and fusion with lysosomes, providing an environment for cleavage and activation of caspase-8. When the endoplasmic reticulum is stressed, the cell will amplify the downstream caspase cascade and cause cell death (Tomar et al., 2013).
Besides caspase-8, TRIM13 can also form complexes with MDM2, which negatively regulates the tumor suppressor p53 and AKT to provoke cell apoptosis. As a result of their interaction, MDM2 and AKT are ubiquitinated and degraded by proteasomes, thereby increasing the stability of p53, reducing the AKT kinase activity, and inducing cell apoptosis consequently (Joo et al., 2011).
This study suggests that TRIM13 can promote tumor cell apoptosis, which indicates that TRIM13 may be deleted or inactivated in some cancer tissues and cell lines. In 2019, the hypothesis was confirmed by Xu et al. (2019) experimentally. They found that compared with non-cancerous tissues and normal bronchial epithelial cell lines, non-small cell lung cancer tissues and cell lines have reduced TRIM13 mRNA and protein expression. And it is also been found that TRIM13 partly induces the apoptosis of NSCLC cells under the mediation of caspase-3 and exerts an antitumor effect.
TRIM13 can not only promote tumor cell apoptosis but also regulate cell proliferation. In 2014, Tomar and Singh (2014) investigated the influence of TRIM13 on cell growth and proliferation. According to their research, NEMO, a substrate of TRIM13, plays a notable role in the NF-κB signaling pathway, which regulates the expression of several inflammatory cytokines and is associated with cancer. TRIM13 initially interacts with NEMO to induce its ubiquitination and degradation, which may suppress the activity of the IKK complex and thus hinder NF-κB signaling. Ultimately, TRIM13 inhibits the proliferation of cancer cells.
Gp78
Gp78, commonly known as autocrine motility factor receptor (AMFR), was initially found in the B16-F1 melanoma cell line (Nabi and Raz, 1987) and has been discovered to be an AMFR essential for tumor metastasis and migration (Watanabe et al., 1991). The ligand for gp78 is AMF (Nabi et al., 1990), an extracellular tumor cytokine. In response to AMF stimulation, gp78 is activated and then changes cell adhesion, proliferation, movement, and apoptosis activities by activating the downstream signaling pathway. Specifically, the activation of gp78 triggers a protein kinase C-dependent signal cascade and then activates and upregulates RhoA and Rac1 (Kanbe et al., 1994; Tsutsumi et al., 2002), which induce the reorganization of the lesion contact and the formation of stress fibers, resulting in enhanced cell motility and proliferation. The activated gp78 also upregulates the transcription factor SNAIL, leading to loss of cell adhesion (Silletti et al., 1996; Tsutsumi et al., 2004), and phosphorylates MEK1 and MEK2 to activate the MAPK pathway and increases cell proliferation (Torimura et al., 2001). Besides, the expression level of Apaf-1 and caspase-9 is reduced when gp78 is activated, which may inhibit tumor cell apoptosis (Haga et al., 2003).
Other than the function mentioned above of gp78 as a receptor, it can ubiquitinate and degrade some proteins as an E3 ligase, which is associated with its locations. Gp78 is internalized through the endocytosis pathway and directly enters the smooth ER when combined with AMF. Therefore, gp78 is located on the plasma membrane, smooth ER closely connected with mitochondria, and a small amount of rough ER. In ER, gp78 is crucial in proteasomal degradation of ERAD-targeted proteins, and some substrates identified so far contain cholesterol homeostases, such as apolipoprotein B (Liang et al., 2003), HMG coenzyme A reductase (Song et al., 2005) and Insig-1 (Lee et al., 2006). Other gp78 substrates like KAI1, a metastasis inhibitor, are closely correlated with sarcoma metastasis (Tsai et al., 2007).
PARK2
PARK2 is a tumor suppressor gene that plays an essential part in cancer progression. It is related to microtubule stability, cell cycle disruption, mitochondrial homeostasis, cell apoptosis, and metabolism that regulates cell state (Manzanillo et al., 2013). Current studies have confirmed loss of function of PARK2 in various human cancers, including breast (Rodriguez et al., 2000), ovarian (Saito et al., 1996), lung (Kong et al., 2000), and renal cancers (Morita et al., 1991), implying that its inactivation may promote tumor transformation and progression. What’s more, PARK2 protein is negatively regulated in many primary tumors, which may contribute to cancer development. For instance, Parkin deficiency may promote pancreatic tumors through dysregulation of microtubule-dependent mitotic kinesin Eg5 expression and subsequent spindle behavior defects (Sun et al., 2013).
Microtubule stability is critical for cell proliferation, and PARK2 may inhibit tumor migration by stabilizing microtubules through an E3-independent way when paclitaxel is administered as an anticancer drug. This effect is triggered by the binding of three microtubule-binding domains of Parkin (MAPKs), which are mitogen-activated protein kinases, to microtubules. By binding to the external layer of microtubules, Parkin enhances microtubule-paclitaxel interaction as well as the activity of paclitaxel, thereby promoting microtubule stability and assembly, which are two hallmarks of paclitaxel cytotoxicity. Therefore, PARK2 levels may predict paclitaxel treatment outcomes in breast cancer (Wang et al., 2009). In addition, the following activation of protein kinases associated with microtubules such as p38, ERK, and JNK is also blocked, thereby antagonizing the effects of microtubule depolymerization drugs like colchicine (Ren et al., 2009).
Carcinogenesis may be incited by cell cycle disruption induced by Parkin dysfunction. A seminal study of PARK2 by Kyoko Ikeuchi (2009) reports that Parkin help with the ubiquitination and proteasome degradation of cyclin E in human colon cells (Ikeuchi et al., 2009). Furthermore, Shiam-Peng (2010) demonstrated that PARK2 contributes to cell cycle arrest and growth inhibition by specifically upregulating CDK6 mRNA levels in MCF7 breast cancer cells (Tay et al., 2010). Multiple shreds of evidence suggest that PARK2 also modulates centrosomes and mitotic spindles via interacting with γ-Tubulin (Chen et al., 2012). Since centrosomes contribute to mitotic spindles formation, their inactivation may contribute to cell division dysregulation.
Mitochondrial function is usually impaired in cancer, and PARK2 is implicated in mitochondrial functional regulation and turnover. PARK2 combines with mtDNA to enhance mitochondrial transcription mediated by TFAM and restore the expression of PGC-1α, thus promoting mitochondrial production (Kuroda et al., 2006). In addition, it protects mitochondrial genomes from reactive oxygen species-induced damage, and it is also involved in mtDNA repair (Rothfuss et al., 2009). What’s more, activated PARK2 acts as a catalyst for rapid ubiquitination of various mitochondrial proteins, such as FIS1, MFN1/2, TOMM70A, RHOT1/2, etc. (Chan et al., 2011; Narendra et al., 2012; Sarraf et al., 2013), which then recruits adaptor proteins to initiate selective autophagy. Ultimately, PARK2-reliant mitochondrial autophagy maintains healthy mitochondrial populations by selectively degrading damaged mitochondria. Therefore, PARK2 is vital for promoting mitochondrial function and maintaining mitochondrial genome integrity and popularity. As a result, changes in PARK2 can be associated with tumorigenesis.
PARK2 also regulates the activity of several apoptosis-related proteins of the Bcl-2 family, including Bax, Bcl-2, and Mcl1 (Chen et al., 2010; Johnson et al., 2012; Ekholm-Reed et al., 2013), promoting apoptosis of tumor cells. In addition, PARK2 boosts cell apoptosis produced by Microtubule stabilizers and HDAC inhibitors in liver and breast tumor cells, according to studies (Wang et al., 2004; Wang et al., 2009). Furthermore, PARK2 makes HeLa cells susceptible to apoptosis induced by TNF-α (Lee et al., 2012).
PARK2 may influence cancer cell metabolism to some extent. For example, PARK2 is the target gene of p53, which regulates energy metabolism. PARK2 deficiency enhances glycolysis while decreasing mitochondrial respiration, leading to the Warburg effect (Zhang et al., 2011). Moreover, in gliomas, the EGFR-Akt pathway is negatively regulated by PARK2, and PARK2 overexpression can inhibit signal transduction through Akt/mTOR. And the loss of parkin function will enhance the expression of cyclin D1 and Akt-related growth promotion signals and, at the same time, promote the proliferation of glioma cells. Besides, in gliomas PARK2 downregulates VEGFR2, a high-affinity tyrosine kinase receptor involved in tumor angiogenesis. Therefore, it may have the effect of inhibiting tumor angiogenesis (Yeo et al., 2012).
SYVN1
SYVN1 inhibits breast cancer growth and metastasis through the miR-96-5p/SYVN1 axis (Gao et al., 2018). It interacts with IGF-1R and promotes its ubiquitination and degradation through the proteasome. This, in turn, leads to inhibition of the growth, migration and invasion of breast cancer cells (Xu et al., 2015). Tumor metastasis is closely associated with the poor prognosis of hepatocellular carcinoma (HCC). Some studies have found that E3 ubiquitin has been analyzed by proteomics and ubiquitinomics of HCC. Furthermore, there is a correlation between SYVN1 and tumor metastasis. Moreover, SYVN1 interacts with heat shock protein 90 and contributes to the ubiquitination of eukaryotic elongation factor 2 kinase. (Ji et al., 2021).
Tumor cell metabolic abnormalities may cause ER lesion and unfolded protein response (UPR), which maintains ER homeostasis by inducing degradation of unfolded proteins. Typically, proteins not properly folded are perceived by adaptor proteins such as chaperones, ubiquitinated by E3 ubiquitin ligases and degraded by proteasomes. However, ER stress would induce cell apoptosis to exclude them from normal cells. Therefore, ER function is critical for the survival of cancer cells.
Plasma Membrane-Localized E3
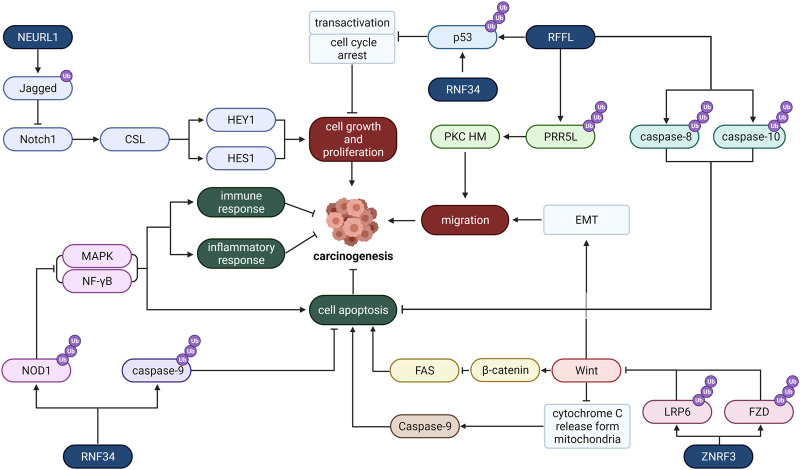
Plasma membrane not only regulates the flow of substances into and out of cells but also protects the integrity of cells and keeps their form. Multitudes of researches have proved that frequent mutations of plasma membrane-associated E3 ligases are engaged in carcinogenesis (Figure 3).
FIGURE 3.
The role of typical Plasma membrane-localized E3 ligases in cancer. RNF34, REFL, RNF34, and NEURL1 are four plasma membrane-localized E3 ligases. They regulate carcinogenesis through various pathways, including cell growth and proliferation, migration, cell metastasis, apoptosis, inflammatory response, and immune response.
ZNRF3
Zinc and ring finger 3 (ZNRF3) is located in the plasma membrane, which is usually correlated to cancer development. In 2014, Assié et al. (2014) reported frequent mutations of ZNRF3 in adrenocortical carcinoma (ACC). Compared to normal surrounding tissues, the ZnRF3 level in gastric tumor tissues has been lower (Zhou et al., 2013), and ZnRF3 mutations are prevalent in pancreatic cancer (Wolpin et al., 2014). Numerous literature has investigated how ZNRF3 regulates the Wnt/β-Catenin signal, which is correlated with cancer (Clevers and Nusse, 2012) because Wnt helps the stabilization and nuclear localization of β-Catenin, which contributes to the formation of TCF/β-Catenin complex and recreation of other co-activators to promote gene activation like c-MYC and cyclin D1 (Ma et al., 2012).
Ubiquitination-mediated Wnt receptor turnover has become a key regulating factor of the Wnt pathway because it affects the sensibility of cells to Wnt ligands. And ZNRF3 precisely regulates Wnt pathway activity by promoting the degradation of Wnt receptors. ZNRF3 inhibits Wnt/PCP and Wnt/β-Catenin signaling pathway by promoting the ubiquitination and degradation of Wnt protein core receptor FZD (Jiang et al., 2015) and Wnt protein co-receptor LRP6 (Hao et al., 2012; Koo et al., 2012). Therefore, ZNRF3 negatively regulates the Wnt pathway and may inhibit cancer cell growth.
It has also been found that ZNRF3 hinders the growth of cancer cells and promotes cell apoptosis by regulating the Wnt/β-Catenin/TCF signaling pathway (Zhou et al., 2013) for the reason that Wnt-1 inhibits apoptosis by blocking mitochondrial cytochrome C release, thereby inhibiting the activity of caspase-9 (Li et al., 2006; Brocardo and Henderson, 2008). And other studies have shown that Wnt signaling inhibits cancer cell apoptosis by impelling NF-κB. Activated β-Catenin may decrease the expression level of FAS, targeted by NF-κB, whence possibly suppressing Fas-mediated apoptosis, leading to tumorigenesis (Ma and Hottiger, 2016).
Furthermore, ZNRF3 may also restrain the migration and invasiveness of some cancer cells like papillary thyroid carcinoma cells by this pathway (Deng et al., 2015). Previous research has established that epithelial-mesenchymal transition (EMT) is a critical step in the migration of cancer cells from the original cancer lesions to nearby organs (Christofori, 2006). The Wnt/β-Catenin pathway is one mechanism for cells to undergo EMT (Thiery et al., 2009). Consequently, ZNRF3 peradventure inhibits cancer cell invasion.
RFFL (CARP-2)
The ring finger and FYVE-like (RFFL) domain-containing E3 ligase is a member of caspase 8/10-associated RING proteins (CARPs), so it is also named CARP-2. Studies have found that RFFL inhibits p53, which is an effective tumor suppressor that tends to be mutated in tumor cells (Bode and Dong, 2004; Olivier et al., 2004) and promotes the growth of cancer cells. RFFL has been reported to have strong functions in colorectal cancer cells (Dong et al., 2013). In contrast, CARP’s silencing upregulates p53 expression and facilitates transcriptional activation and tumor suppression. CARPs negatively regulate p53 through ubiquitination and proteasome degradation of p53 and inhibit transactivation and cell cycle arrest. CARPs can target not only non-phosphorylated p53 but also phospho-p53ser20 and possibly phospho-p53ser15 74, both of which are accumulated after DNA damage and cannot be ubiquitinated by other p53 associated E3s like MDM2. Consequently, CARPs are critical to regulating cancer cell growth through the p53 pathway.
Apart from promoting cancer cell proliferation, CARPs also inhibit cancer cell apoptosis. It helps the ubiquitin-mediated proteolysis of death effector domain (DED) caspase, which induces the damage of cell target during apoptosis and is challenging to be inhibited by regular inhibitors of apoptosis proteins (IAP). It has been found that CARPs interact specifically with caspase-8 and caspase-10 (McDonald and El-Deiry, 2004) and induce their ubiquitination and degradation via the proteasome pathway, thereby impeding cancer cell apoptosis.
And RFFL plays a pivotal part in cell migration. RFFL induces ubiquitination and degradation of PRR5L, which leads to mTORC2-mediated phosphorylation and activation of PKC HM. Continuous activation of PKC is indispensable for maintaining tumor cell migration. Therefore, RFFL may be a potential target to promote tumor migration and metastasis (Gan et al., 2013). Hence, the regulation of RFFL is essential for tumors. Studies have reported that miR-133a can straight bind to RFFL mRNA and inhibit its translation, thereby reducing the level of RFFL protein, enhancing cell apoptosis and inhibiting cell proliferation (Dong et al., 2013).
RNF34 (CARP-1)
Belonging to the CARP family, RNF34 induces the ubiquitination and degradation of death effector domain (DED) caspase to suppress cancer cell apoptosis (McDonald and El-Deiry, 2004). And RNF34 can also inhibit cancer cell apoptosis by regulating the NOD1 pathway. NOD1 can not only induce cell apoptosis but also control cell proliferation, and NOD1 activation can lead to caspase-8 and caspase-9-induced apoptosis (da Silva Correia et al., 2007). Recently there have been reports of missing NOD1 and breast tumor growth (da Silva Correia et al., 2006). RNF34 mainly affects the NOD1 pathway negatively. It interacts directly with NOD1 to induce its ubiquitination and subsequent degradation, therefore reducing the expression level of NOD1 in the cell. As a result, RNF34 hinders NOD1-mediated apoptosis. And it inhibits the activation of NOD1-dependent nuclear factor-κB (NF-γB) along with mitogen-activated protein kinase (MAPK), which results in cytokine, chemokine, antimicrobial peptide production and apoptosis when activated (da Silva Correia et al., 2007; Hasegawa et al., 2008; Park et al., 2007). Therefore, it may inhibit the cellular immune response, inflammatory response and apoptosis in this way.
In addition, RNF34 also stimulates the degradation of p53 and then hinders transactivation, and cell cycle arrests like other CARP members, such as RFFL mentioned above (Yang et al., 2007), which means it also regulates cancer cell cycles and may accelerate cell growth and proliferation.
NEURL1
Neuralized-like protein 1 (NEURL1) is a conserved E3 ligase investigated as a candidate tumor suppressor. The subcellular localization of NEURL1 has not been definitively determined, while some studies demonstrated that overexpressed NEURL1 is localized in the plasma membrane in an N-myristylation-dependent manner (Koutelou et al., 2008). According to studies, NEURL1 is significantly down-regulated in medulloblastoma cells through histone modification and exhibits various tumor suppressor properties, including promotion of cell apoptosis and suppression of tumor growth, angiogenesis, and invasion (Koutelou et al., 2008). These effects are mainly produced through the regulation of the Notch signaling cascade.
NEURL1 can function as an E3 ubiquitin ligase to monoubiquitinate the extracellular domain of a membrane-tethered protein Jagged1 and trigger the endocytosis and degradation of Jagged1 (Koutelou et al., 2008). Decreased Jagged1 level weakens signaling between Jagged1 and its receptor, Notch1, on the surfaces of neighboring cells. This results in less cleavage and detaching of Notch intracellular region (icN1) from the membrane and binding with CSL transcription factor in the nucleus (Sjölund et al., 2005). Consequently, inhibition of the Notch1 signaling pathway downregulates the transcriptional initiation of a variety of cell cycle-related proteins, including HES1 and HEY1 (Teider et al., 2010), suppressing the growth and division of tumor cells.
Other upstream proteins may regulate this effect of NEURL1. Fe65 facilitates the recruitment of NEURL1 to form a stable ternary complex with Jagged1 and is considered a factor negatively regulating the Jagged1/Notch1 pathway (Lee et al., 2015). Neuritin interacted with NEURL1 and down-regulates NEURL1 expression. Therefore, it is considered a positive regulator of the Jagged1/Notch1 pathway (Zhang et al., 2017).
Cbl
Cbl proteins mainly exist on the cell membrane surface and play a significant role in tumorigenesis and antitumor immunity (Liyasova et al., 2015). And it is been reported that Cbl expression correlates with human colorectal cancer (Kumaradevan et al., 2018) and it may inhibit lung cancer and glioblastoma cells’ migration (Lee et al., 2018). First, its activity as an E3 ligase negatively regulates signalings by activated RTKs: this family of proteins attenuates receptor tyrosine kinases by ubiquitinating RTKs, targeting them to lysosomes for degradation (RTK)-related signaling (Ma et al., 2013). Nevertheless, Cbl’s E3 function is lost due to cancer mutations. Furthermore, Cbl-mediated ubiquitination can alter the cellular localization of the protein to modulate its function. In addition, Cbl also act as adaptors to active RTKs by recruiting signaling molecules. For example, it has been shown that Cbl act as adaptors to recruit PI3K to activate RTKs, which subsequently activate the PI3K/AKT pathway (Dombrosky-Ferlan and Corey, 1997; Ueno et al., 1998).
Cbl-b negatively regulates the antitumor function of T cells and natural killer (NK) cells through its E3 ligase activity. Therefore, deletion of Cbl-b will potentiate the NK cell activity. For example, Cbl-b deficient NK cells can suppress oncogene-driven breast cancer. The tyrosine kinase receptor TAM family is the molecular substrate of Cbl-b, including Tyro3, Axl, and Mer. And TAM receptors also negatively regulate NK cells. Therefore, the Cbl-b/TAM receptors inhibit NK cell activation (Paolino et al., 2014), and the consequences of Cbl protein deficiency can lead to malignant tumors or immune dysfunction.
Golgi Localized E3 Ligases
The Golgi apparatus is of particular importance for secretory proteins. Its significant function of regulating metabolism mainly dependents on membrane fluidity and abundant important anchored proteins. And Golgi localized E3 ligases are closely related to tumor hypoxia responses and cell apoptosis, such as KLHL20 (Figure 4).
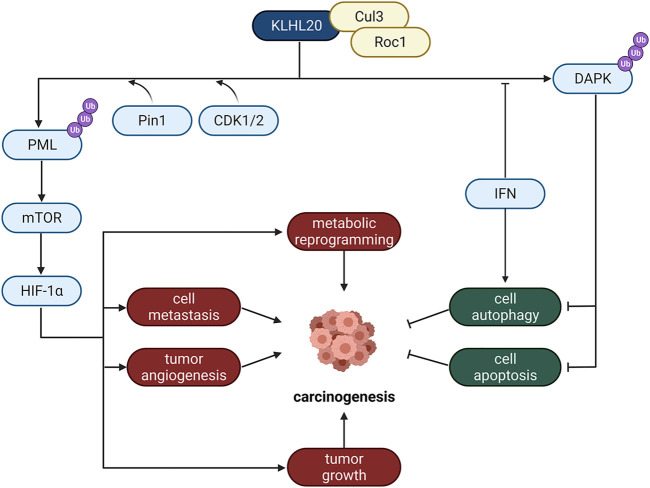
FIGURE 4.
The role of Golgi localized E3 ligases in cancer. KLHL20 is an important Golgi E3 protein, which regulates carcinogenesis through many different pathways, including metabolic reprogramming, cell metastasis, tumor angiogenesis and tumor growth.
KLHL20, a Cullin3 (Cul3) substrate adaptor, belongs to Broad-Complex, Tramtrack, and Bric-a-brac (BTB) family (Pintard et al., 2004). Only if it forms an E3 ligase complex with Cul3 and Roc1 can it induce the ubiquitination of its substrates like PML and DAPK. It promotes tumor growth, migration, angiogenesis and metabolic reprogramming as well as inhibits the processes of autophagy and apoptosis of cancer cells by promoting substrate ubiquitination and degradation, thus promoting prostate cancer progression (Yuan et al., 2011).
KLHL20 mediates hypoxia-induced PML ubiquitination and proteasome-induced degradation. CDK1/2 and Pin1 are also implicated in the regulation of this pathway. CDK1 and CDK2 promote KLHL20-mediated degranulation of PML, and Pin1-mediated proline isomerization promotes the recruitment of PML to KLHL20. Because PML negatively regulates hypoxia-inducible 1α (HIF-1α) protein synthesis by inhibiting mTOR activity (Bernardi et al., 2006). Therefore, degradation of PML would increase HIF-1α content, thereby promoting a variety of HIF-1α-regulated metabolic reprogramming, migration, tumor growth and angiogenesis, etc. (Brahimi-Horn et al., 2007; Finger and Giaccia, 2010; Semenza, 2010).
Studies have shown that KLHL20 also enhances DAPK ubiquitination and proteasome degradation to inhibit cell apoptosis. Death-associated protein kinase (DAPK) is a tumor suppressor. Moreover, interferon (IFN) can suppress tumor cell proliferation and trigger apoptosis, and it has been used in cancer therapy. Because DAPK is a pivotal factor of IFN-induced autophagy (Inbal et al., 2002), KLHL20 impairs its proapoptotic function. It was also found that IFN could reduce the efficiency of KLHL20-DAPK complex formation by sequestering KLHL20, thus reducing DAPK ubiquitination by KLHL20 and stabilizing DAPK protein content; as a result, aiding IFN-induced autophagy (Inbal et al., 2002).
Mitochondria Localized E3
PIR2/RNF144B is a potential targeted biomarker in endometrial cancer to promote proliferation and is mainly localized on the mitochondrion membrane. Experiments in human osteosarcoma cells Saos-2 revealed that the expression of apoptin issues in the activation of proapoptotic TAp73, ending in the upregulation of PUMA and cell death. Thus, PIR2 degrades anti-apoptotic ΔNp73 to increase TAp73 stability to induce apoptosis. In brief, it induces ubiquitination of ΔNp73 for degradation and activates the TAp73-mediated apoptosis pathway. And it was previously shown to regulate the stability of p21WAF1 or p63, which mediate cell growth arrest and epithelial homeostasis (Conforti et al., 2013).
Nuclear Membrane-Localized E3
ITCH
ITCH is localized on the cell and nuclear membranes. It plays a fundamental role in different cellular contexts depending on its different substrates. Its substrates are mainly divided into two categories: transcription factors and cell growth factors, like ErbB-4, a member of the EGFR/ERbB family. These substrates’ ubiquitination and proteasomal degradation affects cell growth, differentiation and apoptosis, and is associated with tumors malignant transformation and chemoresistance (Melino et al., 2008). And it is been found that ITCH is a downstream target of miR-10b which promoted melanoma progression by repressing ITCH (Wang et al., 2019).
Smurf1 and Smurf2
Smad ubiquitin regulatory factor 1 (Smurf1) and Smurf2 are E3 ligases that inhibit the TGF-β pathway through ubiquitination-induced receptors for degrading smads and transforming growth factor-β (TGF-β). Smurf1 may promote ovarian cancer invasion and epithelial-to-mesenchymal transition (EMT) (Fan et al., 2019) and Smurf2 may motivate colon cancer cell proliferation (Yu et al., 2019). However, their substrates are different. Smurf1 binds to Smad1 and Smad5, while Smurf2 binds to Smad1, Smad2, Smad3, Smad6, and Smad7. Smurf2 enhanced the repressive activity of Smad7 while decreasing the transcriptional activity of Smad2. Smurf2 can bind to the transcriptional co-repressor SnoN and degrade it through Smad2 (Bonni et al., 2001). Therefore, Smurf2 may enhance TGF-β signaling beneath certain circumstances. In addition, due to differences in post-translational regulation, the functions of smurf2 and smurf1 also differ to some extent. At the same time, Smurf1 can regulate cell motility by promoting the ubiquitination and degradation of RhoA (Wang et al., 2003), which plays a certain role in the development of cancer. On MDA-MB 231 cells, Smurf2 can ubiquitinate and subsequently degrade Smurf1, but not vice versa. The knockdown of Smurf2 leads to an increase in the protein level of Smurf1, which promotes cell migration in vitro and bone metastasis in vivo (Fukunaga et al., 2008).
Conclusion and Prospects
In summary, membrane-associated E3 ligases are of great significance to the therapeutic improvement of cancer. Lipid alterations in membrane and other cancer syndromes in cancer cells are closely related to E3 ligases (Zalba and ten Hagen, 2017). Thus, the membrane system regulates the proliferation, migration and differentiation of cancer cells partly via E3 ligases. Increasing evidence indicates that specific E3 ligases play a key role in carcinogenesis. What’s more, some membrane-associated E3 ubiquitin ligases are observed to be frequently mutated in many human cancer cell lines, which may induce chemoresistance and cause poor clinic prognosis. These research findings would eventually shed light on a new class of antitumor drugs targeting E3 ubiquitin ligases and the research and development of sensitive biomarkers for cancer diagnosis, treatment, and prognosis, which requires further constant exploration. What’s more, several studies have demonstrated that some E3 ubiquitin ligases are related to the regulation of innate immune response, which provides a solid theoretical basis for expanding the application of E3 ligases to promote tumor immunotherapy.
In the future, the rapid development and breakthrough of PROTAC may largely depend on some research on membrane-associated E3 ligases. Some membrane-anchored proteins, for example, are difficult to target using targeted degradation techniques, but a new strategy utilizing membrane-associated E3 ligases may help the ubiquitination and degradation of these proteins. In addition, the cellular trafficking system may deliver some proteins from the plasma membrane to the trans-Golgi network, thereby chewing up these proteins.
Consequently, the membrane-associated E3 ligases have crucial functions and potential application values in developing new therapeutic strategies for cancer. It is anticipated that membrane E3 ligases will soon be of great significance in tackling medical and biological research problems with more in-depth research and understanding of membrane-associated E3 ubiquitin ligases.
Acknowledgments
The figures were created with BioRender.com
Author Contributions
Concept design: JC, CZ, and XC; XC, LJ, ZZ, and JC wrote the manuscript; JC, QH, and BY directed the study.
Funding
This work was supported by grants from the Zhejiang Provincial Natural Science Foundation of China (No. LR22H310002 to JC and No. LGF21B020001 to CZ), the National Natural Science Foundation of China (No. 81930102 to BY and No. 81872885 to JC), and Zhejiang University K. P. Chao’s High Technology Development Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Assié G., Letouzé E., Fassnacht M., Jouinot A., Luscap W., Barreau O., et al. (2014). Integrated Genomic Characterization of Adrenocortical Carcinoma. Nat. Genet. 46 (6), 607–612. 10.1038/ng.2953 [DOI] [PubMed] [Google Scholar]
- Bauer J., Bakke O., Morth J. P. (2017). Overview of the Membrane-Associated RING-CH (MARCH) E3 Ligase Family. N. Biotechnol. 38, 7–15. 10.1016/j.nbt.2016.12.002 [DOI] [PubMed] [Google Scholar]
- Bernardi R., Guernah I., Jin D., Grisendi S., Alimonti A., Teruya-Feldstein J., et al. (2006). PML Inhibits HIF-1alpha Translation and Neoangiogenesis through Repression of mTOR. Nature 442 (7104), 779–785. 10.1038/nature05029 [DOI] [PubMed] [Google Scholar]
- Bode A. M., Dong Z. (2004). Post-translational Modification of P53 in Tumorigenesis. Nat. Rev. Cancer 4 (10), 793–805. 10.1038/nrc1455 [DOI] [PubMed] [Google Scholar]
- Bonni S., Wang H. R., Causing C. G., Kavsak P., Stroschein S. L., Luo K., et al. (2001). TGF-beta Induces Assembly of a Smad2-Smurf2 Ubiquitin Ligase Complex that Targets SnoN for Degradation. Nat. Cell Biol. 3 (6), 587–595. 10.1038/35078562 [DOI] [PubMed] [Google Scholar]
- Brahimi-Horn M. C., Chiche J., Pouysségur J. (2007). Hypoxia and Cancer. J. Mol. Med. Berl. 85 (12), 1301–1307. 10.1007/s00109-007-0281-3 [DOI] [PubMed] [Google Scholar]
- Brocardo M., Henderson B. R. (2008). APC Shuttling to the Membrane, Nucleus and beyond. Trends Cell Biol. 18 (12), 587–596. 10.1016/j.tcb.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Cao J., Wang Y., Dong R., Lin G., Zhang N., Wang J., et al. (2015). Hypoxia-Induced WSB1 Promotes the Metastatic Potential of Osteosarcoma Cells. Cancer Res. 75 (22), 4839–4851. 10.1158/0008-5472.CAN-15-0711 [DOI] [PubMed] [Google Scholar]
- Chan N. C., Salazar A. M., Pham A. H., Sweredoski M. J., Kolawa N. J., Graham R. L., et al. (2011). Broad Activation of the Ubiquitin-Proteasome System by Parkin Is Critical for Mitophagy. Hum. Mol. Genet. 20 (9), 1726–1737. 10.1093/hmg/ddr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Gao F., Li B., Wang H., Xu Y., Zhu C., et al. (2010). Parkin Mono-Ubiquitinates Bcl-2 and Regulates Autophagy. J. Biol. Chem. 285 (49), 38214–38223. 10.1074/jbc.M110.101469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Fang S. T., Yeh P. C., Yang H. H., Chen S. Y., Chang C. J., et al. (2012). The C-Terminus of PARK2 Is Required for its Self-Interaction, Solubility and Role in the Spindle Assembly Checkpoint. Biochim. Biophys. Acta 1822 (4), 573–580. 10.1016/j.bbadis.2011.12.007 [DOI] [PubMed] [Google Scholar]
- Christofori G. (2006). New Signals from the Invasive Front. Nature 441 (7092), 444–450. 10.1038/nature04872 [DOI] [PubMed] [Google Scholar]
- Clevers H., Nusse R. (2012). Wnt/β-catenin Signaling and Disease. Cell 149 (6), 1192–1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Conforti F., Yang A. L., Piro M. C., Mellone M., Terrinoni A., Candi E., et al. (2013). PIR2/Rnf144B Regulates Epithelial Homeostasis by Mediating Degradation of p21WAF1 and P63. Oncogene 32 (40), 4758–4765. 10.1038/onc.2012.497 [DOI] [PubMed] [Google Scholar]
- da Silva Correia J., Miranda Y., Austin-Brown N., Hsu J., Mathison J., Xiang R., et al. (2006). Nod1-dependent Control of Tumor Growth. Proc. Natl. Acad. Sci. U. S. A. 103 (6), 1840–1845. 10.1073/pnas.0509228103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Correia J., Miranda Y., Leonard N., Ulevitch R. J. (2007). The Subunit CSN6 of the COP9 Signalosome Is Cleaved during Apoptosis. J. Biol. Chem. 282 (17), 12557–12565. 10.1074/jbc.M609587200 [DOI] [PubMed] [Google Scholar]
- Dacks J. B., Field M. C. (2018). Evolutionary Origins and Specialisation of Membrane Transport. Curr. Opin. Cell Biol. 53, 70–76. 10.1016/j.ceb.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Wu B., Xiao K., Kang J., Xie J., Zhang X., et al. (2015). MiR-146b-5p Promotes Metastasis and Induces Epithelial-Mesenchymal Transition in Thyroid Cancer by Targeting ZNRF3. Cell Physiol. Biochem. 35 (1), 71–82. 10.1159/000369676 [DOI] [PubMed] [Google Scholar]
- Deshaies R. J., Joazeiro C. A. (2009). RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 78, 399–434. 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Dombrosky-Ferlan P. M., Corey S. J. (1997). Yeast Two-Hybrid In Vivo Association of the Src Kinase Lyn with the Proto-Oncogene Product Cbl but Not with the P85 Subunit of PI 3-kinase. Oncogene 14 (17), 2019–2024. 10.1038/sj.onc.1201031 [DOI] [PubMed] [Google Scholar]
- Dong Y., Zhao J., Wu C. W., Zhang L., Liu X., Kang W., et al. (2013). Tumor Suppressor Functions of miR-133a in Colorectal Cancer. Mol. Cancer Res. 11 (9), 1051–1060. 10.1158/1541-7786.MCR-13-0061 [DOI] [PubMed] [Google Scholar]
- Ekholm-Reed S., Goldberg M. S., Schlossmacher M. G., Reed S. I. (2013). Parkin-dependent Degradation of the F-Box Protein Fbw7β Promotes Neuronal Survival in Response to Oxidative Stress by Stabilizing Mcl-1. Mol. Cell Biol. 33 (18), 3627–3643. 10.1128/MCB.00535-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Wang Y., Fan J., Chen R. (2019). Deletion of SMURF 1 Represses Ovarian Cancer Invasion and EMT by Modulating the DAB2IP/AKT/Skp2 Feedback Loop. J. Cell Biochem. 120 (6), 10643–10651. 10.1002/jcb.28354 [DOI] [PubMed] [Google Scholar]
- Finger E. C., Giaccia A. J. (2010). Hypoxia, Inflammation, and the Tumor Microenvironment in Metastatic Disease. Cancer Metastasis Rev. 29 (2), 285–293. 10.1007/s10555-010-9224-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga E., Inoue Y., Komiya S., Horiguchi K., Goto K., Saitoh M., et al. (2008). Smurf2 Induces Ubiquitin-dependent Degradation of Smurf1 to Prevent Migration of Breast Cancer Cells. J. Biol. Chem. 283 (51), 35660–35667. 10.1074/jbc.M710496200 [DOI] [PubMed] [Google Scholar]
- Gan X., Wang C., Patel M., Kreutz B., Zhou M., Kozasa T., et al. (2013). Different Raf Protein Kinases Mediate Different Signaling Pathways to Stimulate E3 Ligase RFFL Gene Expression in Cell Migration Regulation. J. Biol. Chem. 288 (47), 33978–33984. 10.1074/jbc.M113.477406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Wang H., Li H., Li M., Wang J., Zhang W., et al. (2018). Long Non-coding RNA CASC2 Inhibits Breast Cancer Cell Growth and Metastasis through the Regulation of the miR-96-5p/SYVN1 Pathway. Int. J. Oncol. 53 (5), 2081–2090. 10.3892/ijo.2018.4522 [DOI] [PubMed] [Google Scholar]
- Haga A., Funasaka T., Niinaka Y., Raz A., Nagase H. (2003). Autocrine Motility Factor Signaling Induces Tumor Apoptotic Resistance by Regulations Apaf-1 and Caspase-9 Apoptosome Expression. Int. J. Cancer 107 (5), 707–714. 10.1002/ijc.11449 [DOI] [PubMed] [Google Scholar]
- Hao H. X., Xie Y., Zhang Y., Charlat O., Oster E., Avello M., et al. (2012). ZNRF3 Promotes Wnt Receptor Turnover in an R-Spondin-Sensitive Manner. Nature 485 (7397), 195–200. 10.1038/nature11019 [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Fujimoto Y., Lucas P. C., Nakano H., Fukase K., Núñez G., et al. (2008). A Critical Role of RICK/RIP2 Polyubiquitination in Nod-Induced NF-kappaB Activation. EMBO J. 27 (2), 373–383. 10.1038/sj.emboj.7601962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K., Marusawa H., Fujiwara M., Matsumoto Y., Endo Y., Watanabe T., et al. (2009). Attenuation of Proteolysis-Mediated Cyclin E Regulation by Alternatively Spliced Parkin in Human Colorectal Cancers. Int. J. Cancer 125 (9), 2029–2035. 10.1002/ijc.24565 [DOI] [PubMed] [Google Scholar]
- Inbal B., Bialik S., Sabanay I., Shani G., Kimchi A. (2002). DAP Kinase and DRP-1 Mediate Membrane Blebbing and the Formation of Autophagic Vesicles during Programmed Cell Death. J. Cell Biol. 157 (3), 455–468. 10.1083/jcb.200109094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji F., Zhou M., Sun Z., Jiang Z., Zhu H., Xie Z., et al. (2021). Integrative Proteomics Reveals the Role of E3 Ubiquitin Ligase SYVN1 in Hepatocellular Carcinoma Metastasis. Cancer Commun. 41 (10), 1007–1023. 10.1002/cac2.12192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Charlat O., Zamponi R., Yang Y., Cong F. (2015). Dishevelled Promotes Wnt Receptor Degradation through Recruitment of ZNRF3/RNF43 E3 Ubiquitin Ligases. Mol. Cell 58 (3), 522–533. 10.1016/j.molcel.2015.03.015 [DOI] [PubMed] [Google Scholar]
- Johnson B. N., Berger A. K., Cortese G. P., LaVoie M. J. (2012). The Ubiquitin E3 Ligase Parkin Regulates the Proapoptotic Function of Bax. Proc. Natl. Acad. Sci. U. S. A. 109 (16), 6283–6288. 10.1073/pnas.1113248109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H. M., Kim J. Y., Jeong J. B., Seong K. M., Nam S. Y., Yang K. H., et al. (2011). Ret Finger Protein 2 Enhances Ionizing Radiation-Induced Apoptosis via Degradation of AKT and MDM2. Eur. J. Cell Biol. 90 (5), 420–431. 10.1016/j.ejcb.2010.12.001 [DOI] [PubMed] [Google Scholar]
- Kanbe K., Chigira M., Watanabe H. (1994). Effects of Protein Kinase Inhibitors on the Cell Motility Stimulated by Autocrine Motility Factor. Biochim. Biophys. Acta 1222 (3), 395–399. 10.1016/0167-4889(94)90046-9 [DOI] [PubMed] [Google Scholar]
- Kong F. M., Anscher M. S., Washington M. K., Killian J. K., Jirtle R. L. (2000). M6P/IGF2R Is Mutated in Squamous Cell Carcinoma of the Lung. Oncogene 19 (12), 1572–1578. 10.1038/sj.onc.1203437 [DOI] [PubMed] [Google Scholar]
- Koo B. K., Spit M., Jordens I., Low T. Y., Stange D. E., Van De Wetering M., et al. (2012). Tumour Suppressor RNF43 Is a Stem-Cell E3 Ligase that Induces Endocytosis of Wnt Receptors. Nature 488 (7413), 665–669. 10.1038/nature11308 [DOI] [PubMed] [Google Scholar]
- Koutelou E., Sato S., Tomomori-Sato C., Florens L., Swanson S. K., Washburn M. P., et al. (2008). Neuralized-like 1 (Neurl1) Targeted to the Plasma Membrane by N-Myristoylation Regulates the Notch Ligand Jagged1. J. Biol. Chem. 283 (7), 3846–3853. 10.1074/jbc.M706974200 [DOI] [PubMed] [Google Scholar]
- Kumaradevan S., Lee S. Y., Richards S., Lyle C., Zhao Q., Tapan U., et al. (2018). c-Cbl Expression Correlates with Human Colorectal Cancer Survival and its Wnt/β-Catenin Suppressor Function Is Regulated by Tyr371 Phosphorylation. Am. J. Pathol. 188 (8), 1921–1933. 10.1016/j.ajpath.2018.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y., Mitsui T., Kunishige M., Shono M., Akaike M., Azuma H., et al. (2006). Parkin Enhances Mitochondrial Biogenesis in Proliferating Cells. Hum. Mol. Genet. 15 (6), 883–895. 10.1093/hmg/ddl006 [DOI] [PubMed] [Google Scholar]
- Lee G. W., Park J. B., Park S. Y., Seo J., Shin S. H., Park J. W., et al. (2018). The E3 Ligase C-CBL Inhibits Cancer Cell Migration by Neddylating the Proto-Oncogene C-Src. Oncogene 37 (41), 5552–5568. 10.1038/s41388-018-0354-5 [DOI] [PubMed] [Google Scholar]
- Lee H. J., Yoon J. H., Ahn J. S., Jo E. H., Kim M. Y., Lee Y. C., et al. (2015). Fe65 Negatively Regulates Jagged1 Signaling by Decreasing Jagged1 Protein Stability through the E3 Ligase Neuralized-like 1. Biochim. Biophys. Acta 1853 (11), 2918–2928. Part A):2918–28. 10.1016/j.bbamcr.2015.08.009 [DOI] [PubMed] [Google Scholar]
- Lee J. N., Song B., DeBose-Boyd R. A., Ye J. (2006). Sterol-regulated Degradation of Insig-1 Mediated by the Membrane-Bound Ubiquitin Ligase Gp78. J. Biol. Chem. 281 (51), 39308–39315. 10.1074/jbc.M608999200 [DOI] [PubMed] [Google Scholar]
- Lee K., Lee M. H., Kang Y. W., Rhee K. J., Kim T. U., Kim Y. S. (2012). Parkin Induces Apoptotic Cell Death in TNF-α-Treated Cervical Cancer Cells. BMB Rep. 45 (9), 526–531. 10.5483/bmbrep.2012.45.9.104 [DOI] [PubMed] [Google Scholar]
- Li F., Chong Z. Z., Maiese K. (2006). Winding through the WNT Pathway during Cellular Development and Demise. Histol. Histopathol. 21 (1), 103–124. 10.14670/HH-21.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. S., Kim T., Fang S., Yamaguchi J., Weissman A. M., Fisher E. A., et al. (2003). Overexpression of the Tumor Autocrine Motility Factor Receptor Gp78, a Ubiquitin Protein Ligase, Results in Increased Ubiquitinylation and Decreased Secretion of Apolipoprotein B100 in HepG2 Cells. J. Biol. Chem. 278 (26), 23984–23988. 10.1074/jbc.M302683200 [DOI] [PubMed] [Google Scholar]
- Liyasova M. S., Ma K., Lipkowitz S. (2015). Molecular Pathways: Cbl Proteins in Tumorigenesis and Antitumor Immunity-Opportunities for Cancer Treatment. Clin. Cancer Res. 21 (8), 1789–1794. 10.1158/1078-0432.CCR-13-2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Hottiger M. O. (2016). Crosstalk between Wnt/β-Catenin and NF-Κb Signaling Pathway during Inflammation. Front. Immunol. 7, 378. 10.3389/fimmu.2016.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Wang R., Fang X., Sun Z. (2012). β-catenin/TCF-1 Pathway in T Cell Development and Differentiation. J. Neuroimmune Pharmacol. 7 (4), 750–762. 10.1007/s11481-012-9367-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K., Kales S. C., Nau M. M., Lipkowitz S. (2013). “Cbl as a Master Regulator of Receptor Tyrosine Kinase Trafficking,” in Vesicle Trafficking in Cancer. Editors Yarden Y., Tarcic G. (New York, NY: Springer; ), 219–244. 10.1007/978-1-4614-6528-7_11 [DOI] [Google Scholar]
- Manzanillo P. S., Ayres J. S., Watson R. O., Collins A. C., Souza G., Rae C. S., et al. (2013). The Ubiquitin Ligase Parkin Mediates Resistance to Intracellular Pathogens. Nature 501 (7468), 512–516. 10.1038/nature12566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C. E., List K. (2019). Cell Surface-Anchored Serine Proteases in Cancer Progression and Metastasis. Cancer Metastasis Rev. 38 (3), 357–387. 10.1007/s10555-019-09811-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald E. R., El-Deiry W. S. (2004). Suppression of Caspase-8- and -10-associated RING Proteins Results in Sensitization to Death Ligands and Inhibition of Tumor Cell Growth. Proc. Natl. Acad. Sci. U. S. A. 101 (16), 6170–6175. 10.1073/pnas.0307459101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G., Gallagher E., Aqeilan R. I., Knight R., Peschiaroli A., Rossi M., et al. (2008). Itch: a HECT-type E3 Ligase Regulating Immunity, Skin and Cancer. Cell Death Differ. 15 (7), 1103–1112. 10.1038/cdd.2008.60 [DOI] [PubMed] [Google Scholar]
- Morita R., Saito S., Ishikawa J., Ogawa O., Yoshida O., Yamakawa K., et al. (1991). Common Regions of Deletion on Chromosomes 5q, 6q, and 10q in Renal Cell Carcinoma. Cancer Res. 51 (21), 5817–5820. [PubMed] [Google Scholar]
- Nabi I. R., Raz A. (1987). Cell Shape Modulation Alters Glycosylation of a Metastatic Melanoma Cell-Surface Antigen. Int. J. Cancer 40 (3), 396–402. 10.1002/ijc.2910400319 [DOI] [PubMed] [Google Scholar]
- Nabi I. R., Watanabe H., Raz A. (1990). Identification of B16-F1 Melanoma Autocrine Motility-like Factor Receptor. Cancer Res. 50 (2), 409–414. [PubMed] [Google Scholar]
- Namba T., Chu K., Kodama R., Byun S., Yoon K. W., Hiraki M., et al. (2015). Loss of P53 Enhances the Function of the Endoplasmic Reticulum through Activation of the IRE1α/XBP1 Pathway. Oncotarget 6 (24), 19990–20001. 10.18632/oncotarget.4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Walker J. E., Youle R. (2012). Mitochondrial Quality Control Mediated by PINK1 and Parkin: Links to Parkinsonism. Cold Spring Harb. Perspect. Biol. 4 (11), a011338. 10.1101/cshperspect.a011338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli G., Liu J., Biancolella M., Alonzi T., Novelli A., Patten J. J., et al. (2021). Inhibition of HECT E3 Ligases as Potential Therapy for COVID-19. Cell Death Dis. 12 (4), 1–18. 10.1038/s41419-021-03513-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M., Hussain S. P., Caron de Fromentel C., Hainaut P., Harris C. C. (2004). TP53 Mutation Spectra and Load: a Tool for Generating Hypotheses on the Etiology of Cancer. IARC Sci. Publ. 157, 247–270. [PubMed] [Google Scholar]
- Paolino M., Choidas A., Wallner S., Pranjic B., Uribesalgo I., Loeser S., et al. (2014). The E3 Ligase Cbl-B and TAM Receptors Regulate Cancer Metastasis via Natural Killer Cells. Nature 507 (7493), 508–512. 10.1038/nature12998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Kim Y. G., McDonald C., Kanneganti T. D., Hasegawa M., Body-Malapel M., et al. (2007). RICK/RIP2 Mediates Innate Immune Responses Induced through Nod1 and Nod2 but Not TLRs. J. Immunol. 178 (4), 2380–2386. 10.4049/jimmunol.178.4.2380 [DOI] [PubMed] [Google Scholar]
- Pintard L., Willems A., Peter M. (2004). Cullin-based Ubiquitin Ligases: Cul3-BTB Complexes Join the Family. EMBO J. 23 (8), 1681–1687. 10.1038/sj.emboj.7600186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Jiang H., Yang F., Nakaso K., Feng J. (2009). Parkin Protects Dopaminergic Neurons against Microtubule-Depolymerizing Toxins by Attenuating Microtubule-Associated Protein Kinase Activation. J. Biol. Chem. 284 (6), 4009–4017. 10.1074/jbc.M806245200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez C., Causse A., Ursule E., Theillet C. (2000). At Least Five Regions of Imbalance on 6q in Breast Tumors, Combining Losses and Gains. Genes Chromosom. Cancer 27 (1), 76–84. [DOI] [PubMed] [Google Scholar]
- Rothfuss O., Fischer H., Hasegawa T., Maisel M., Leitner P., Miesel F., et al. (2009). Parkin Protects Mitochondrial Genome Integrity and Supports Mitochondrial DNA Repair. Hum. Mol. Genet. 18 (20), 3832–3850. 10.1093/hmg/ddp327 [DOI] [PubMed] [Google Scholar]
- Rotin D., Kumar S. (2009). Physiological Functions of the HECT Family of Ubiquitin Ligases. Nat. Rev. Mol. Cell Biol. 10 (6), 398–409. 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- Saito S., Sirahama S., Matsushima M., Suzuki M., Sagae S., Kudo R., et al. (1996). Definition of a Commonly Deleted Region in Ovarian Cancers to a 300-kb Segment of Chromosome 6q27. Cancer Res. 56 (24), 5586–5589. [PubMed] [Google Scholar]
- Sarraf S. A., Raman M., Guarani-Pereira V., Sowa M. E., Huttlin E. L., Gygi S. P., et al. (2013). Landscape of the PARKIN-dependent Ubiquitylome in Response to Mitochondrial Depolarization. Nature 496 (7445), 372–376. 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. L. (2010). Defining the Role of Hypoxia-Inducible Factor 1 in Cancer Biology and Therapeutics. Oncogene 29 (5), 625–634. 10.1038/onc.2009.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silletti S., Paku S., Raz A. (1996). Tumor Autocrine Motility Factor Responses Are Mediated through Cell Contact and Focal Adhesion Rearrangement in the Absence of New Tyrosine Phosphorylation in Metastatic Cells. Am. J. Pathol. 148 (5), 1649–1660. [PMC free article] [PubMed] [Google Scholar]
- Sjölund J., Manetopoulos C., Stockhausen M. T., Axelson H. (2005). The Notch Pathway in Cancer: Differentiation Gone Awry. Eur. J. Cancer 41 (17), 2620–2629. 10.1016/j.ejca.2005.06.025 [DOI] [PubMed] [Google Scholar]
- Song B. L., Sever N., DeBose-Boyd R. A. (2005). Gp78, a Membrane-Anchored Ubiquitin Ligase, Associates with Insig-1 and Couples Sterol-Regulated Ubiquitination to Degradation of HMG CoA Reductase. Mol. Cell 19 (6), 829–840. 10.1016/j.molcel.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Sun X., Liu M., Hao J., Li D., Luo Y., Wang X., et al. (2013). Parkin Deficiency Contributes to Pancreatic Tumorigenesis by Inducing Spindle Multipolarity and Misorientation. Cell Cycle 12 (7), 1133–1141. 10.4161/cc.24215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay S. P., Yeo C. W., Chai C., Chua P. J., Tan H. M., Ang A. X., et al. (2010). Parkin Enhances the Expression of Cyclin-dependent Kinase 6 and Negatively Regulates the Proliferation of Breast Cancer Cells. J. Biol. Chem. 285 (38), 29231–29238. 10.1074/jbc.M110.108241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teider N., Scott D. K., Neiss A., Weeraratne S. D., Amani V. M., Wang Y., et al. (2010). Neuralized1 Causes Apoptosis and Downregulates Notch Target Genes in Medulloblastoma. Neuro Oncol. 12 (12), 1244–1256. 10.1093/neuonc/noq091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009). Epithelial-Mesenchymal Transitions in Development and Disease. Cell 139 (5), 871–890. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Tomar D., Prajapati P., Sripada L., Singh K., Singh R., Singh A. K., et al. (2013). TRIM13 Regulates Caspase-8 Ubiquitination, Translocation to Autophagosomes and Activation during ER Stress Induced Cell Death. Biochim. Biophys. Acta 1833 (12), 3134–3144. 10.1016/j.bbamcr.2013.08.021 [DOI] [PubMed] [Google Scholar]
- Tomar D., Singh R., Singh A. K., Pandya C. D., Singh R. (2012). TRIM13 Regulates ER Stress Induced Autophagy and Clonogenic Ability of the Cells. Biochim. Biophys. Acta 1823 (2), 316–326. 10.1016/j.bbamcr.2011.11.015 [DOI] [PubMed] [Google Scholar]
- Tomar D., Singh R. (2014). TRIM13 Regulates Ubiquitination and Turnover of NEMO to Suppress TNF Induced NF-Κb Activation. Cell Signal 26 (12), 2606–2613. 10.1016/j.cellsig.2014.08.008 [DOI] [PubMed] [Google Scholar]
- Torimura T., Ueno T., Kin M., Harada R., Nakamura T., Kawaguchi T., et al. (2001). Autocrine Motility Factor Enhances Hepatoma Cell Invasion across the Basement Membrane through Activation of Beta1 Integrins. Hepatology 34 (1), 62–71. 10.1053/jhep.2001.25546 [DOI] [PubMed] [Google Scholar]
- Tsai Y. C., Mendoza A., Mariano J. M., Zhou M., Kostova Z., Chen B., et al. (2007). The Ubiquitin Ligase Gp78 Promotes Sarcoma Metastasis by Targeting KAI1 for Degradation. Nat. Med. 13 (12), 1504–1509. 10.1038/nm1686 [DOI] [PubMed] [Google Scholar]
- Tsutsumi S., Gupta S. K., Hogan V., Collard J. G., Raz A. (2002). Activation of Small GTPase Rho Is Required for Autocrine Motility Factor Signaling. Cancer Res. 62 (15), 4484–4490. [PubMed] [Google Scholar]
- Tsutsumi S., Yanagawa T., Shimura T., Kuwano H., Raz A. (2004). Autocrine Motility Factor Signaling Enhances Pancreatic Cancer Metastasis. Clin. Cancer Res. 10 (22), 7775–7784. 10.1158/1078-0432.CCR-04-1015 [DOI] [PubMed] [Google Scholar]
- Ueno H., Sasaki K., Honda H., Nakamoto T., Yamagata T., Miyagawa K., et al. (1998). c-Cbl Is Tyrosine-Phosphorylated by Interleukin-4 and Enhances Mitogenic and Survival Signals of Interleukin-4 Receptor by Linking with the Phosphatidylinositol 3'-kinase Pathway. Blood 91 (1), 46–53. 10.1182/blood.v91.1.46.46_46_53 [DOI] [PubMed] [Google Scholar]
- Wang F., Denison S., Lai J. P., Philips L. A., Montoya D., Kock N., et al. (2004). Parkin Gene Alterations in Hepatocellular Carcinoma. Genes Chromosom. Cancer 40 (2), 85–96. 10.1002/gcc.20020 [DOI] [PubMed] [Google Scholar]
- Wang H., Liu B., Zhang C., Peng G., Liu M., Li D., et al. (2009). Parkin Regulates Paclitaxel Sensitivity in Breast Cancer via a Microtubule-dependent Mechanism. J. Pathol. 218 (1), 76–85. 10.1002/path.2512 [DOI] [PubMed] [Google Scholar]
- Wang H. R., Zhang Y., Ozdamar B., Ogunjimi A. A., Alexandrova E., Thomsen G. H., et al. (2003). Regulation of Cell Polarity and Protrusion Formation by Targeting RhoA for Degradation. Science 302 (5651), 1775–1779. 10.1126/science.1090772 [DOI] [PubMed] [Google Scholar]
- Wang S., Wu Y., Xu Y., Tang X. (2019). miR-10b Promoted Melanoma Progression through Wnt/β-Catenin Pathway by Repressing ITCH Expression. Gene 710, 39–47. 10.1016/j.gene.2019.05.043 [DOI] [PubMed] [Google Scholar]
- Watanabe H., Carmi P., Hogan V., Raz T., Silletti S., Nabi I. R., et al. (1991). Purification of Human Tumor Cell Autocrine Motility Factor and Molecular Cloning of its Receptor. J. Biol. Chem. 266 (20), 13442–13448. 10.1016/s0021-9258(18)98859-9 [DOI] [PubMed] [Google Scholar]
- Wolpin B. M., Rizzato C., Kraft P., Kooperberg C., Petersen G. M., Wang Z., et al. (2014). Genome-wide Association Study Identifies Multiple Susceptibility Loci for Pancreatic Cancer. Nat. Genet. 46 (9), 994–1000. 10.1038/ng.3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wu Q., Zhou X., Wu Q., Fang M. (2019). TRIM13 Inhibited Cell Proliferation and Induced Cell Apoptosis by Regulating NF-Κb Pathway in Non-small-cell Lung Carcinoma Cells. Gene 715, 144015. 10.1016/j.gene.2019.144015 [DOI] [PubMed] [Google Scholar]
- Xu S., Cherok E., Das S., Li S., Roelofs B. A., Ge S. X., et al. (2016). Mitochondrial E3 Ubiquitin Ligase MARCH5 Controls Mitochondrial Fission and Cell Sensitivity to Stress-Induced Apoptosis through Regulation of MiD49 Protein. Mol. Biol. Cell 27 (2), 349–359. 10.1091/mbc.E15-09-0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. M., Wang H. J., Chen F., Guo W. H., Wang Y. Y., Li H. Y., et al. (2015). HRD1 Suppresses the Growth and Metastasis of Breast Cancer Cells by Promoting IGF-1R Degradation. Oncotarget 6 (40), 42854–42867. 10.18632/oncotarget.5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R. K., Chae S. W., Kim H. R., Chae H. J. (2014). Endoplasmic Reticulum Stress and Cancer. J. Cancer Prev. 19 (2), 75–88. 10.15430/JCP.2014.19.2.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Rozan L. M., McDonald E. R., Navaraj A., Liu J. J., Matthew E. M., et al. (2007). CARPs Are Ubiquitin Ligases that Promote MDM2-independent P53 and Phospho-P53ser20 Degradation. J. Biol. Chem. 282 (5), 3273–3281. 10.1074/jbc.M610793200 [DOI] [PubMed] [Google Scholar]
- Yeo C. W., Ng F. S., Chai C., Tan J. M., Koh G. R., Chong Y. K., et al. (2012). Parkin Pathway Activation Mitigates Glioma Cell Proliferation and Predicts Patient Survival. Cancer Res. 72 (10), 2543–2553. 10.1158/0008-5472.CAN-11-3060 [DOI] [PubMed] [Google Scholar]
- Ying M., Zhang L., Zhou Q., Shao X., Cao J., Zhang N., et al. (2016). The E3 Ubiquitin Protein Ligase MDM2 Dictates All-Trans Retinoic Acid-Induced Osteoblastic Differentiation of Osteosarcoma Cells by Modulating the Degradation of RARα. Oncogene 35 (33), 4358–4367. 10.1038/onc.2015.503 [DOI] [PubMed] [Google Scholar]
- Yu L., Dong L., Wang Y., Liu L., Long H., Li H., et al. (2019). Reversible Regulation of SATB1 Ubiquitination by USP47 and SMURF2 Mediates Colon Cancer Cell Proliferation and Tumor Progression. Cancer Lett. 448, 40–51. 10.1016/j.canlet.2019.01.039 [DOI] [PubMed] [Google Scholar]
- Yuan W. C., Lee Y. R., Huang S. F., Lin Y. M., Chen T. Y., Chung H. C., et al. (2011). A Cullin3-KLHL20 Ubiquitin Ligase-dependent Pathway Targets PML to Potentiate HIF-1 Signaling and Prostate Cancer Progression. Cancer Cell 20 (2), 214–228. 10.1016/j.ccr.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Zalba S., ten Hagen T. L. (2017). Cell Membrane Modulation as Adjuvant in Cancer Therapy. Cancer Treat. Rev. 52, 48–57. 10.1016/j.ctrv.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Lin M., Wu R., Wang X., Yang B., Levine A. J., et al. (2011). Parkin, a P53 Target Gene, Mediates the Role of P53 in Glucose Metabolism and the Warburg Effect. Proc. Natl. Acad. Sci. U. S. A. 108 (39), 16259–16264. 10.1073/pnas.1113884108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Luo X., Guo Z., Xiong A., Dong H., Zhang Q., et al. (2017). Neuritin Inhibits Notch Signaling through Interacted with Neuralized to Promote the Neurite Growth. Front. Mol. Neurosci. 10, 179. 10.3389/fnmol.2017.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Shabek N. (2017). Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 86, 129–157. 10.1146/annurev-biochem-060815-014922 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Lan J., Wang W., Shi Q., Lan Y., Cheng Z., et al. (2013). ZNRF3 Acts as a Tumour Suppressor by the Wnt Signalling Pathway in Human Gastric Adenocarcinoma. J. Mol. Histol. 44 (5), 555–563. 10.1007/s10735-013-9504-9 [DOI] [PubMed] [Google Scholar]