Abstract
The All of Us (AoU) Research Program aggregates electronic health records (EHR) data from 300,00+ participants spanning 50+ distinct data sites. The diversity and size of AoU’s data network result in multifaceted obstacles to data integration that may undermine the usability of patient EHR. Consequently, the AoU team implemented data quality tools to regularly evaluate and communicate EHR data quality issues at scale. The use of systematic feedback and educational tools ultimately increased site engagement and led to quantitative improvements in EHR quality as measured by program- and externally-defined metrics. These improvements enabled the AoU team to save time on troubleshooting EHR and focus on the development of alternate mechanisms to improve the quality of future EHR submissions. While this framework has proven effective, further efforts to automate and centralize communication channels are needed to deepen the program’s efforts while retaining its scalability.
Introduction
Overview of the All of Us Research Program
Clinical data research networks (CDRNs) enable researchers to access electronic health records (EHR) from thousands of patients and vast territories in an effort to answer questions in translational research. CDRNs have existed for many years, with each containing sets of patients from distinct populations, ages, or locations1.
Datasets used to inform clinical research, however, often lack the racial, ethnic, geographic, and gender diversity needed to advance personalized medical treatment for generalized patient populations2,3. Consequently, the National Institutes of Health launched the All of Us Research Program, which aims to collect health questionnaires, genetic data, physical measurements, digital health information, and EHR from at least 1 million persons in the United States4. The All of Us Research Program is nearly unparalleled in its diversity; as of June 2020, the program had over 224,000 participants in its workbench, with seventy-seven percent of its cohort identified as underrepresented in biomedical research5. With All of Us’s broadly-accessible dataset4, researchers from across the world will have access to robust information that can power the future of precision medicine.
Overview of the Data Network and Transfer
The All of Us Research Program’s data network is expansive; there are over 50 health provider organizations (HPOs) that coordinate the enrollment and EHR transfer of the program’s several hundred thousand participants. Firstly, HPOs must extract information from the local EHR and transform said data into the OMOP Common Data Model (CDM). This OMOP CDM is used to ensure that EHR, regardless of their original format, are put into a standardized set of data structures to enable end-users to index the data in a systematic and scalable fashion6. Finally, HPOs must load the data into a central repository of Google Cloud Buckets. In order to receive funding, HPOs are required to transfer EHR at least once a quarter after they are officially onboarded by the Data Research Center (DRC). The DRC is the receiving end of the central repository and develops a variety of tools, such as data cleaning rules, to ensure EHR are sufficiently de-identified and processed before they are provided to researchers. There are several protocols to ensure EHR data from HPOs meets basic conformance standards for the OMOP CDM. Outside of this short list of checks, however, sites originally received little to no feedback upon provision of EHR to the repository.
Data Quality Issues
The decentralized approach of All of Us’s collection process comes with large-scale data quality issues. Similar to other CDRNs, the aggregation of healthcare information from sites with different data storage techniques, distinct EHR systems, and varying resources available to dedicate to healthcare informatics inevitably leads to differences in data quality across sites7,8. Upon investigation, this phenomenon – combined with the aforementioned lack of systematic reviews of HPO submissions – resulted in a number of problems in submissions to the All of Us Research Program. These issues included, but not limited to, infrequent data uploads from HPOs, the omission of required fields in patient records, and the failure of sites to translate from native EHR codes to ‘standard’ OMOP codes. These issues are not uncommon to decentralized data environments and have led many within the scientific community to question if EHR data is suitable for research9.
While existing CDRNs implemented a number of mechanisms to assess the quality of their EHR data8-10, few have created systems to track the aggregate quality of data across all of their partner sites in a longitudinal fashion or implemented a systematic approach to provide feedback to their contributors. The scale of the All of Us program - with its 50 and counting HPO data partners and over 300,000 enrolled participants - provides a unique opportunity to implement a robust approach to decentralized EHR data quality reporting and communication.
In this study, we will detail the means by which the All of Us Research Program developed tools to run data quality checks on EHR submissions from enrollment sites. This paper will also detail All of Us’s communication practices to illustrate how findings were disseminated in a scalable manner and ultimately increased data partner engagement. We will then show that these protocols enabled the program to improve EHR data quality from HPOs as measured by metrics developed by the DRC and existing third-party informatics tools. Finally, we will detail means by which All of Us can incorporate additional tools and practices to further strengthen its data quality assessments.
Methods
Creating Data Quality Metrics
In order to assess data quality issues, the All of Us Research Program investigated the three-category framework described by Data Quality Harmonization Framework (DQHF) in 201611. DQHF’s first category is conformance, which refers to adherence to technical and syntactic constraints. Completeness is the second category, which validates that expected data values are present as one may expect within an observational context. The final tenet is plausibility, which ensures that data values provided are consistent with real-world applications of clinical medicine. DQHF was chosen as a means to establish data quality metrics and communicate said issues with sites given the expertise of its developers and its application in a variety of other informatics contexts9, 12,13.
With this framework, the DRC created a variety of metrics and goals to quantify the conformance, completeness, and plausibility of the EHR at both the site- and program levels. The inspiration for these assessments came from a variety of sources, all of which were guided by real-world EHR research applications. Firstly, All of Us program leadership explicitly specified minimum requirements regarding EHR data transfer and quality for HPOs to maintain their grants. Leadership chose these metrics as experts deemed them as foundational pillars to ensure that the EHR dataset would be suitable for medical research. Consequently, all of the program-defined metrics were included in the analyses run by the EHR Operations team. The second source of data quality metrics came from data interaction. The analyst responsible for developing the metrics previously took part in several projects in the All of Us research program and knew of the obstacles faced by the data’s end-users. The analyst also continued data exploration on both intra- and inter-site levels to identify issues that may arise when researchers engage with EHR. These experiences enabled the creation of metrics that could guide sites on how to address issues that obstruct the research usability of the EHR. Finally, investigations of the cleaning rules implemented by the DRC allowed the EHR Operations team to identify points of extensive data elimination as EHR moved through the pipeline. These evaluations allowed us to identify what types of data errors could be resolved prior to submission and thereby improve the ultimate completeness and usability of the All of Us dataset. All of the data quality metrics were approved by program personnel before implementation and dissemination to sites. If needed, the EHR Operations team consulted physicians to determine the relevance and expected frequency of specific clinically-based metrics.
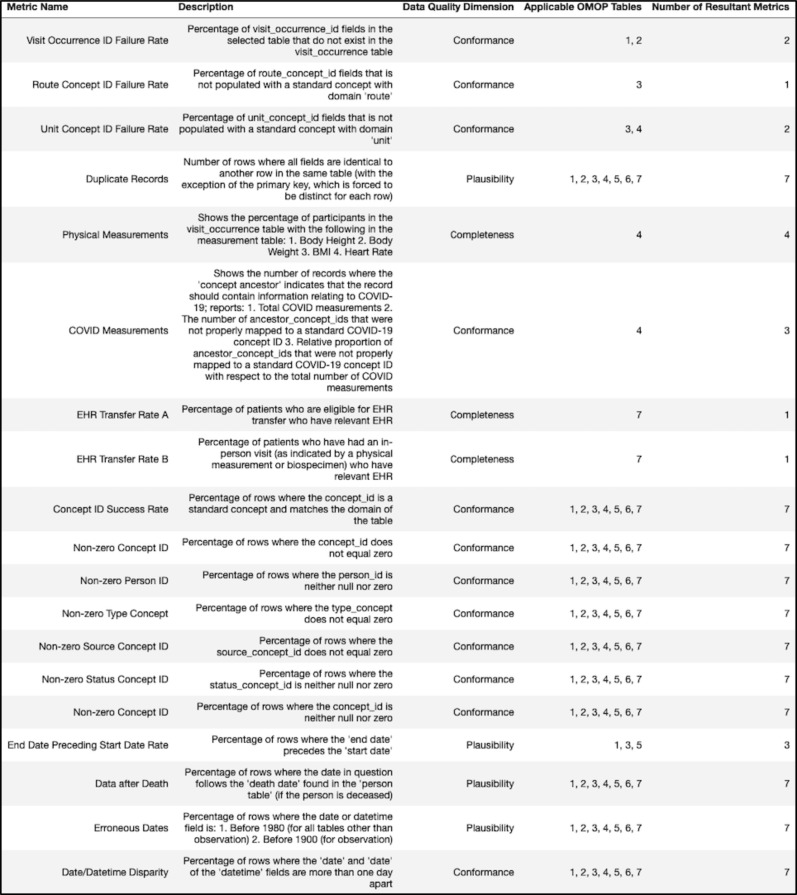
In total, these SQL queries were intended to evaluate 19 distinct metrics relating to the conformance, completeness, and plausibility of the EHR submissions provided by sites. Most of the metrics were applied across multiple tables - with some metrics containing an additional weighted average across six of the OMOP tables (condition_occurrence, observation, procedure_occurrence, measurement, drug_exposure, and visit_occurrence) - raising the total number of metrics to 94. All of the metrics discussed can be found in the figure below (Figure 1).
Figure 1.
Different data quality metrics with their descriptions, relevant data quality dimension, and applicable OMOP table(s). Table key: 1 = condition_occurrence, 2 = procedure_occurrence, 3 = drug_exposure, 4 = measurement, 5 = visit_occurrence, 6 = observation, 7 = tables 1-6 (aggregate metric).
Organization, Calculation, and Presentation of Metrics
After defining data quality metrics, we retrieved data quality attributes on the OMOP-transformed tables provided by sites by using queries constructed on Google BigQuery. This query method construction enabled fast and scalable searches while maintaining the general syntax of Structured Query Language (SQL)14. These metrics were calculated for each site and the results would be output to a Microsoft Excel file.
After creation of the Excel files, another Python script was developed to reorganize the data and create aggregate data quality metrics. This program provided multiple views of seeing the data quality over time. For each metric, the program created the following:
One Excel sheet for every OMOP table. In said sheet, each site occupied a row. The bottom (final) row was the ‘aggregate’ data quality metric, which was an average across all sites with each site being weighted by its number of rows. The columns of said sheet represented different dates when the data quality metric was calculated. This sheet was created for each of the OMOP tables for which the data quality metric was calculated.
One Excel sheet for every data partner. In said sheet, each OMOP table occupied a row. The bottom (final) row was the ‘aggregate’ data quality metric, which was an average across all OMOP tables with each table being weighted by its relative number of rows. The columns of said sheet represented different dates when the data quality metric was calculated. This sheet was created for each of the HPOs for which the data quality metric was calculated. One additional Excel sheet was created to create an ‘aggregate’ data quality metric across all of the data partners. The format of said additional sheet, including the meanings of the rows as OMOP tables and columns as dates, was otherwise identical.
After the creation of these sheets, the information generated was fed into a Jupyter notebook, in which the Python- based Seaborn package created heat maps for said data quality metrics. These heat maps enabled visualization for sites to understand the progression of their data quality over time and the precise areas where they could resolve errors in their submissions.
Finally, another script generated messages to send to data stewards via email. The emails sent to the sites communicated which data quality metrics were not passing the DRC’s standards and provided direct links to resources – such as video tutorials, sample queries, and text explanations - where sites could learn about the exact meaning of these data quality metrics. These emails would contain the aforementioned heatmaps so sites could precisely locate the source of their data quality issues and see how their data quality trended over time. These emails and the corresponding heatmaps would be sent to sites on the first Monday of every month to ensure that sites were routinely notified about their data quality and held accountable for their submissions.
All of the aforementioned scripts are publicly available online within the All of Us GitHub at the following link: https://github.com/noahgengel/curation/tree/develop/data_steward/analytics.
Automating Data Quality Reports
In an effort to improve the scalability and timeliness of data quality reports, the DRC implemented an interactive data visualization tool from Tableau Software®. Starting in June 2021, the Tableau software automatically ran the previously-defined queries on Google BigQuery every day and calculated the corresponding data quality metrics. Once these metrics were calculated, the program created a series of data visualizations – including bar graphs, stacked bar graphs, line graphs, and heatmaps – so data stewards and program leadership could log into their Tableau accounts and receive feedback within 24 hours of each submission. This framework also enabled the DRC to integrate new data quality metrics into the dashboard after testing a metric on several volunteer sites.
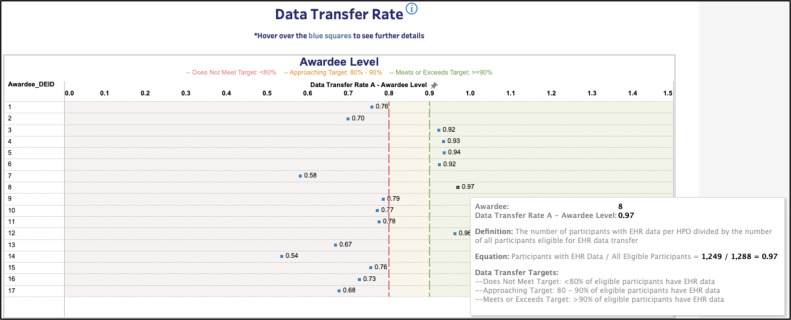
The visualization tools were implemented with versatility, readability, and context in mind. These visualizations were separated into five dashboard tabs and contained interactive filters by which sites could select the metrics they wanted to prioritize and the exact calculations of each metric. Similar to past iterations of the data quality reports, Tableau provided the option to trend a data partner’s metrics in a longitudinal fashion so data stewards could assess their sites’ progression. Finally, unlike past reports, the data quality dashboard displayed the arithmetic mean of each regional consortium’s data partners for each data quality metric. This display (Figure 2) allows data stewards to compare their site’s progress to the status of other sites and see how each consortium compares to the DRC’s targets.
Figure 2.
Example screenshot (sites de-identified) from the interactive data visualization tool. Screenshot also contains a pop-up that data stewards could bring up to better understand the data quality metric at hand.
Additionally, in an effort to validate EHR submissions against proven informatics tools, the DRC leveraged ACHILLES. ACHILLES is a free-to-use and open-source program that characterizes and visualizes data quality rules regarding patient demographics, clinical observations, conditions, procedures, and drugs on OMOP- conformant observational health databases16. By leveraging ACHILLES, the DRC could analyze and tabulate a robust set of potential errors in each EHR submission for each site. ACHILLES served as an appropriate means to externally validate data quality as many of its checks were completely distinct from the DRC’s reported data quality checks. Therefore, reductions in the absolute number of ACHILLES errors over time could signify that improvements in EHR data quality were not confined to specific data quality errors targeted by the DRC.
After each submission, the All of Us pipeline would generate a results.html output file within the next 24 hours. The results.html file would appear in the bucket where sites submitted their data. This file contained information about the most recent upload, including a report on the errors, notifications, and warnings generated by ACHILLES. Sites could then view the output on these files and adjust their subsequent submissions to resolve or reduce the number of errors in the results.html reports.
Mass Communication and Resource Creation
Beyond reporting on data quality metrics, the DRC created means through which it could disseminate data quality information at scale. Firstly, the DRC hosted bi-weekly calls in which data stewards from all sites would join and learn about newly-implemented data quality checks, initiative-wide metrics, and issues that may pertain to their specific EMR systems. Next, in 2019 Q3, the DRC set up regular video conferences in which we would discuss data quality issues with individuals who oversaw EHR transfer from multiple sites within the same regional consortium. The DRC also created a series of data steward email threads based on commonalities between said data stewards – such as EMR systems – to ensure that we could efficiently send out information to pertinent parties. Finally, in 2021 Q2, the DRC leveraged ZenDesk - a customer service management system – to ensure data partners’ questions would be effectively stored, accessible, and organized when compared to traditional email.
The DRC also wanted sites to independently understand and potentially resolve many of their data quality issues. The DRC uploaded all of the code that calculated the data quality metrics – with the exception of any potentially- identifying information – to a public GitHub repository. Pending the technical abilities of each data steward, this transparency allowed partners to recapitulate the DRC’s metrics and identify example rows of EHR data that negatively impacted their data quality. In an effort to improve user experience, said GitHub links – along with definitions of each data quality metric and a means by which sites could provide feedback to the DRC– were incorporated into the Tableau dashboard (Figure 2). To facilitate understanding of the metrics, the DRC created several iterations of knowledge repositories. These repositories allowed users to easily identify a particular metric and find a page to help them recapitulate the DRC’s results. Depending on the iteration of the repository, each page would contain information such as sample SQL queries, text descriptions of the metrics, EMR system-specific information, common points of failure, or an instructional video recorded by the DRC.
Results
Impact on Site Engagement
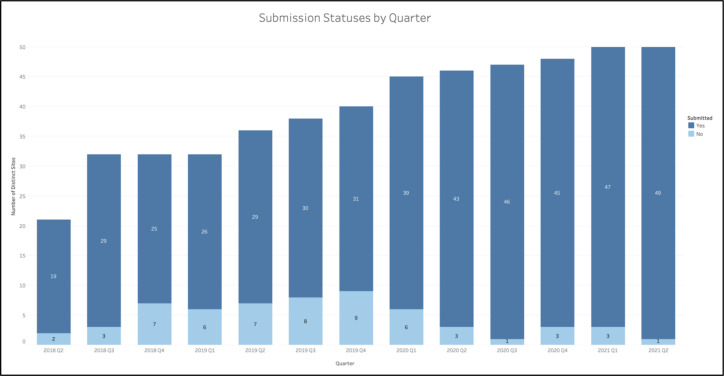
Prior to implementation of regular data quality communications, sites frequently failed to comply with quarterly data transfer requirements. For instance, in the six quarters prior to the systematic communications, there were two quarters where over 20% of the sites did not transfer any EHR to the DRC (Figure 3).
Figure 3.
Number of sites (both total and those with a successful submission) for each quarter.
After implementation of regular communications regarding data quality in Q3 2019, sites were more likely to make at least one submission per quarter. In the first half of 2019, about 81% of sites were compliant with EHR transfer. The following half-years, however, saw average compliance rates of 78%, 90%, 96%, and 96%, respectively. Of note, once the data quality communications were largely established in Q2 2020, the compliance rate for a given quarter never fell below 93% - a rate higher than any of its preceding quarters (Figure 3).
During this same timeframe, the DRC expanded the number of participating HPOs. Between 2020 Q2 and 2021 Q2, the DRC started accepting submissions from 14 new sites, representing a 39% expansion of the program when compared to 2019 Q2 (Figure 3). The sites added during this time frame represented a diverse array of data partners – ranging from federally qualified health centers (FQHCs) to large academic medical centers.
Improvements with Respect to Internal Data Quality Standards
Given that conformance serves as the foundation for the DQHF11, many of the DRC’s high-priority metrics target the adherence of submissions to the OMOP Common Data Model (CDM). One set of metrics evaluated the ‘concept_id’ field for each of six the widely-used OMOP tables previously mentioned to ensure the ‘domain_id’ of the concept_id matched the destination table and that the ‘concept_id’ was a ‘standard’ non-zero value. This metric serves as a proxy to determine whether data partners successfully and completely converted information stored in their native EHR systems to the OMOP CDM. This metric intentionally excludes the first quarter of submissions for each site in order to allow data partners to become familiar with OMOP specifications and data transfer protocols.
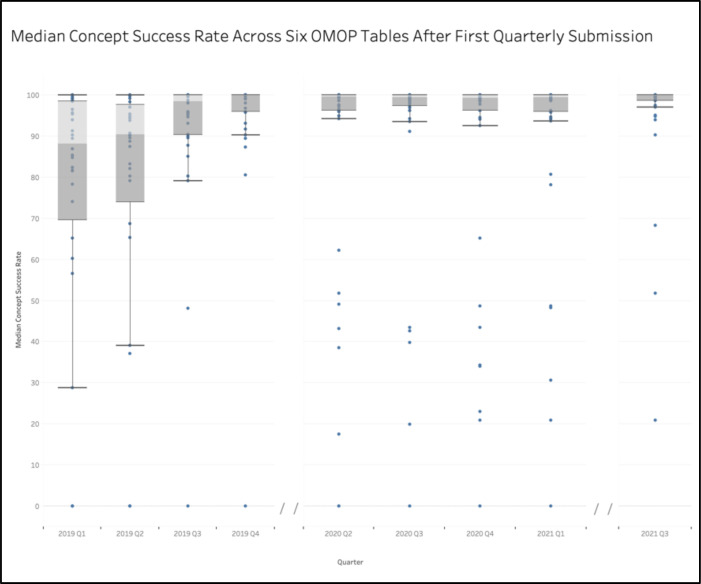
In 2019 Q1, the median concept success rate for each site across the aforementioned tables was below the DRC’s target concept success rate of 90%. The spread of the median concept success rates also proved large, with a standard deviation of 32.9% and 25th-75th percentile range of 20.8%. Upon implementation of the data quality communications in 2019 Q3, the median concept success rate jumped up to 98.5%, with a standard deviation of 19.5% and a 25th-75th percentile range of 9.7%. This trend of increasing OMOP adherence and decreasing site variability in data quality persisted for all subsequent quarters, which all had median values over 99.2%, and high standard deviation and interquartile ranges of 27.4% and 4.07, respectively. The most recent calculations, which reflect 2021 Q3, show an initiative-wide median concept success rate for the six tables of 99.98% with a standard deviation of 13.8% and a 25th-75th percentile range of 1.27% (Figure 4). This trend of improvement in data quality emerged despite the fact that the number of rows nearly tripled from over 1.1 billion rows in 2019 Q1 to nearly 3.3 billion rows in 2021 Q3.
Figure 4.
Median conformance rate for designated OMOP tables across all sites for each quarter. No data available for 2020 Q1 or 2020 Q2.
Improvements with Respect to External Data Quality Standards
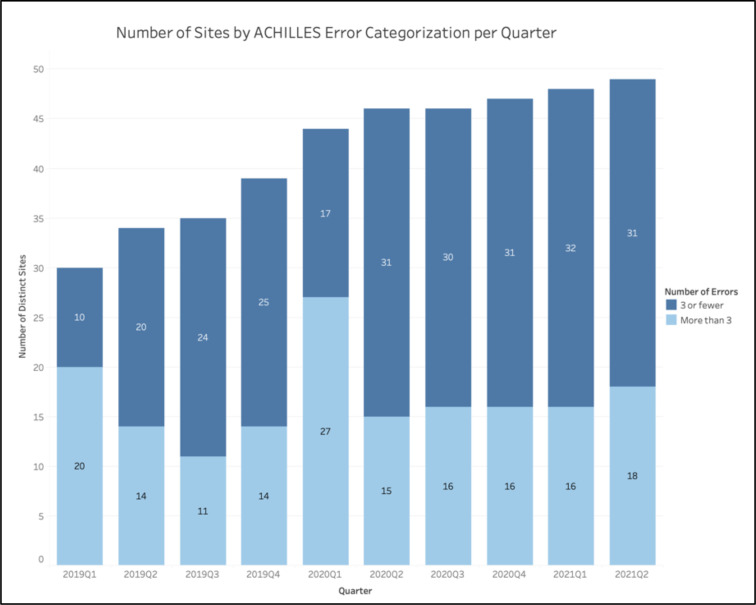
The ACHILLES analyses similarly showed an improvement in data quality across a large number of sites. During the first implementation of the ACHILLES checks in 2019 Q1, of the 30 sites with at least one submission at that time, 67% of sites had over three ACHILLES errors in their final submission of the quarter. While this percentage dropped to 41% of sites in 2019 Q2, it continued to fall and eventually bottomed out to around its most recent value of 35% of sites with over three ACHILLES errors as of the first half of 2021. This relative stabilization in terms of the number of sites with few ACHILLES errors came despite the continual addition of new sites, indicating an increase in the total number of sites with fewer than three ACHILLES errors (Figure 5).
Figure 5.
Number of sites (both total and those with more than three ACHILLES errors) for each quarter.
The only exception to the aforementioned flatline was in 2020 Q1, in which 61% of sites reported three or more ACHILLES errors (Figure 5).
Discussion
Ultimately, the implementation of data quality metrics and regular communications from the DRC to its data partners increased HPO engagement as measured by compliance with data transfer requirements. The implementation of regular communications from the DRC, regardless of each site’s submission status, reversed the trend of many sites not meeting their requirements for quarterly data transfer (Figure 3). Of note, this increase in site submissions arose despite the fact that the DRC took no efforts to further encourage submissions from HPOs. These findings indicate that merely implementing automated reports provided our data stewards with incentive to invest in dedicating their time and resources to the All of Us program. Consequently, the implementation of these protocols should allow the initiative to continue expanding its number of partner sites without experiencing an overwhelming number of non-compliant data partners who would require manual interventions from the DRC and leadership.
The results from this study also indicate that these practices hold potential to improve the quality of EHR data used for research purposes. As indicated by checks on ‘concept success rates’, consistent reporting on a particular issue resulted in a substantial decrease in the number of sites with suboptimal data quality and led to decreased variability in data quality across sites. While these results appear intuitive, they show that initiative-led communications can correct potential data errors without manual interventions on behalf of the DRC.
Equally promising are the results from the ACHILLES analyses. While some overlap existed between the metrics reported by the DRC and the messages displayed by ACHILLES, the ACHILLES tool contained far more checks. It should also be of note that the DRC’s communications with HPOs mostly focused on the initiative-defined metrics and that the ACHILLES reports were oftentimes considered of low-priority to sites, with many data stewards reporting that they seldom or never accessed the reports due to the long processing time of the results.html files. The results from our study, however, indicate that the implementation of data quality checks and communications reduced the relative frequency of sites with more than three ACHILLES errors and an absolute increase in the number of sites with three or fewer ACHILLES errors (Figure 5). The only exception to the trends previously discussed arose in 2020 Q1. This exception likely occurred, however, because data partners were instructed to prioritize increasing their frequency of EHR transfer over resolving potential data quality issues in March 2020 as program leadership wanted recent data in the midst of the COVID-19 pandemic. As a whole, these findings indicate that the process of engaging sites and creating data quality metrics to encourage sites to interrogate their EHR had off-target improvements in data quality as measured by a third-party informatics tool.
The outlined EHR data quality workflow has opportunities for expansion and improvement. As previously mentioned, the DRC specifically targeted issues relating to conformance as this dimension of data quality forms the foundation of the DQHF.11 Future iterations of the workflow, however, should expand the number of metrics calculated to increase the means by which we assess conformance and expand our emphases to the additional dimensions of completeness, plausibility, and recency of the data. Furthermore, the DRC could benefit by working with All of Us researchers to understand the issues faced by users and how it could prioritize existing metrics for our data partners. Finally, the DRC’s checks only work on OMOP-conformant EHR data – limiting the ability of non- OMOP conformant sites to investigate their data quality or the ability of other CDRNs to replicate our protocols.
The DRC could also expand the robustness of its data quality metrics. For instance, future metrics could create notifications if a site’s values for a particular measurement or procedure – such as patient heights or frequency of the flu shot – fall well below the values found in other HPOs. These analyses are already conducted by the DRC but could benefit from automation in order to improve the communication and frequency of these findings. Analyses against external datasets from other institutions could similarly assess the readiness of EHR data for research purposes. The DRC could also leverage data quality tools other than ACHILLES, such as DQe-c8, in order to assess the quality of site submissions against a battery of analyses known to work with OMOP-conformant clinical data.
Finally, the DRC hopes to integrate other information about the processing of the data, such as the amount of data eliminated by its data cleaning rules, into its visualization tools. This information would ensure data stewards can appreciate the importance of data quality and allow leadership to assess if certain sites are disproportionately affected by cleaning.
The DRC could also borrow practices established by other CDRNs in order to improve its data quality workflow. Similar studies implemented a diversity of ways in which they analyzed and quantified data quality interactions – such as characterizing the cause of the issue, quantifying the number of interactions with different sites, and creating an overall quality ‘score’ for each submission to reflect the relative prioritization of different data quality errors15. Past studies could also provide the DRC with insights as to potential future data quality metrics and the relative frequency with which particular issues may arise9.
In total, the current workflow for All of Us has and will continue to undergo many iterations. Future analyses of the effectiveness of the DRC’s workflows should expand upon and strengthen the findings of this study. These additional findings would have the potential to further elucidate the benefit of creating a regular, automated, and approachable means of reporting on data quality within large CDRNs.
Conclusion
Large decentralized data networks inevitably face challenges in their ability to reconcile data from disparate sources and ensure resultant EHR is of sufficient quality for research. All of Us provides a strong case study as to how networks could implement a systematized approach to reporting data quality at a nearly unprecedented scale. Thus far, these protocols have proven effective at increasing the engagement of partner sites and decreasing data quality issues as measured by internal and external assessments. Going forward, the All of Us program could expand on its current efforts in order to further quantify the effectiveness of its data reporting processes and improve the usability of its EHR for use in research.
Figures & Table
References
- 1.Fleurence R. L., Curtis L. H., Califf R. M., Platt R., Selby J. V., Brown J. S. Launching PCORnet, a national patient-centered clinical research network. Journal of the American Medical Informatics Association. 2014. [DOI] [PMC free article] [PubMed]
- 2.Landry L. G., Ali N., Williams D. R., Rehm H. L., Bonham V. L. Lack of diversity in genomic databases is a barrier to translating precision medicine research into practice. Health Affairs. 2018. [DOI] [PubMed]
- 3.Kneale L., Demiris G. Lack of diversity in personal health record evaluations with older adult participants: a systematic review of literature. Journal of Innovation in Health Informatics. 2017. [DOI] [PubMed]
- 4.All of Us Research Program Investigators Denny J. C., Rutter J. L., Goldstein D. B., Philippakis A., Smoller J. W., Jenkins G., Dishman E. The “All of Us” Research Program. The New England journal of medicine. 2019;381(7):668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramirez A. H., Sulieman L., Schlueter D. J., Halvorson A., Qian J., Ratsimbazafy F., Roden D. M. The All of Us Research Program: data quality, utility, and diversity. MedRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 6.Stang P. E., Ryan P. B., Racoosin J. A., Overhage J. M., Hartzema A. G., Reich C., Woodcock J. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Annals of Internal Medicine. 2010;153(9):600–606. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 7.Brown J. S., Kahn M., Toh D. Data quality assessment for comparative effectiveness research in distributed data networks. Medical Care. 2013. [DOI] [PMC free article] [PubMed]
- 8.Estiri H., Stephens K. A., Klann J. G., Murphy S. N. Exploring completeness in clinical data research networks with DQe-c. Journal of the American Medical Informatics Association. 2018. [DOI] [PMC free article] [PubMed]
- 9.Callahan T. J., Bauck A. E., Bertoch D., Brown J., Khare R., Ryan P. B., Kahn M. G. A comparison of data quality assessment checks in six data sharing networks. EGEMs (Generating Evidence & Methods to Improve Patient Outcomes) 2017. [DOI] [PMC free article] [PubMed]
- 10.Klann J. G., Joss M. A. H., Embree K., Murphy S. N. Data model harmonization for the All Of Us Research Program: Transforming i2b2 data into the OMOP common data model. PLoS ONE. 2019. [DOI] [PMC free article] [PubMed]
- 11.Kahn M. G., Callahan T. J., Barnard J., Bauck A. E., Brown J., Davidson B. N., Schilling L. A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data. EGEMs (Generating Evidence & Methods to Improve Patient Outcomes) 2016. [DOI] [PMC free article] [PubMed]
- 12.Sengupta S., Bachman D., Laws R., Saylor G., Staab J., Vaughn D., Bauck A. Data quality assessment and multi-organizational reporting: tools to enhance network knowledge. EGEMS (Washington, DC) 2019;7(1):8. doi: 10.5334/egems.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Q., Han Z., Yu P., An J., Lu X., Duan H. Application of openEHR archetypes to automate data quality rules for electronic health records: a case study. BMC Medical Informatics and Decision Making. 2021;21(1):113. doi: 10.1186/s12911-021-01481-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bisong E. Building Machine Learning and Deep Learning Models on Google Cloud Platform: A Comprehensive Guide for Beginners. Berkeley, CA: Apress; 2019. Google BigQuery; pp. 485–517. [Google Scholar]
- 15.Kahn M. G., Raebel M. A., Glanz J. M., Riedlinger K., Steiner J. F. A pragmatic framework for single-site and multisite data quality assessment in electronic health record-based clinical research. Medical Care. 2012. [DOI] [PMC free article] [PubMed]
- 16.Huser V., DeFalco F. J., Schuemie M., Ryan P. B., Shang N., Velez M., Park R. W., Boyce R. D., Duke J., Khare R., Utidjian L., Bailey C. Multisite Evaluation of a Data Quality Tool for Patient-Level Clinical Data Sets. EGEMS (Washington, DC) 2016;4(1):1239. doi: 10.13063/2327-9214.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]