Abstract
Sleep apnea (SA) is a common sleep disorder characterized by respiratory disturbance during sleep. Polysomnography (PSG) is the gold standard for apnea diagnosis, but it is time-consuming, expensive, and requires manual scoring. As an alternative to PSG, we investigated a real-time SA detection system using oxygen saturation level (SpO2) and electrocardiogram (ECG) signals individually as well as a combination of both. A series of R-R intervals were derived from the raw ECG data and a feed-forward deep artificial neural network is employed for the detection of SA. Three different models were built using 1-minute-long sequences of SpO2 and R-R interval signals. The 10-fold cross-validation result showed that the SpO2-based model performed better than the ECG-based model with an accuracy of 90.78 ± 10.12% and 80.04 ± 7.7%, respectively. Once combined, these two signals complemented each other and resulted in a better model with an accuracy of 91.83 ± 1.51%.
Introduction
Sleep apnea (SA) is one of the most common forms of sleep disorders which is characterized by respiratory disturbance during sleep and affects about 2%-5% of the total adult population and more than 30% of the elderly population in the US [1]. There are mainly three types of sleep apnea: obstructive sleep apnea (OSA), central sleep apnea (CSA), and complex sleep apnea syndrome. OSA is characterized by the repeated pharyngeal collapse that causes shortness (hypopnea) or cessation (apnea) of breathing during sleep [2]. Apnea is often defined as the cessation of breathing for at least 10 seconds and hypopnea is defined by a significant reduction of airflow for a minimum period of 10 seconds accompanied by either 4% desaturation of blood oxygen level or neurological arousal [3]–[5]. CSA is characterized by recurrent apneic events accompanied by a lack of respiratory effort as the brain does not transmit any stimulus to the breathing muscles [6]. Such disturbances often cause arousal from sleep which results in excessive daytime sleepiness and fatigue. Complex Sleep Apnea Syndrome occurs when a patient has both OSA and CSA. Severe forms of SA can lead to cardiovascular dysfunction, ischemic heart disease, and stroke [3]. It is linked with significant cardiovascular morbidity and is one of the major causes of hypertension [7]. Therefore, accurate and timely diagnosis and treatment of sleep apnea are essential for risk minimization.
Currently, polysomnography (PSG) is considered to be the gold standard for apnea diagnosis, which requires a subject to spend a night or two in a sleep laboratory under the supervision of sleep specialists. Usually, multiple sensors and wires are attached to the subject’s body to record various physiological signals. These signals may include brain waves (electroencephalogram, or EEG), eye movements (electrooculogram, or EOG), blood oxygen level (SpO2), heart rate, and rhythm (electrocardiogram, or ECG). The final diagnosis requires analysis of the recorded data by the specialists. The severity of the SA is addressed by the apnea-hypopnea index (AHI), which is defined as the number of apnea-hypopnea events per hour over the entire sleep period. The process is time-consuming, expensive, and the attachment of multiple sensors and wires causes discomfort to the subjects. As a result, the researchers have made extensive efforts to establish alternatives to PSG with simpler schemes and faster decision-making capabilities.
In the past several years, multiple methods to detect SA have been proposed. These methods differ from one another in terms of the classification methods and the physiological signals used to detect SA. Multiple rule-based techniques have been developed for SA detection [8], [9]. Recently, the machine learning-based approach has become a popular technique for SA detection and monitoring and researchers have a number of machine learning models at their disposal. Models such as support vector machine (SVM), K-nearest neighbor (KNN), and decision tree have been used successfully in several studies [10]–[14]. There are reports of SA detection systems that used a combination of multiple classifiers as well [3], [15]. Recently, deep learning techniques, such as convolutional neural network (CNN) and recursive neural network (RNN), have emerged as very efficient techniques of SA detection [16]–[19]. Among all the physiological signals used in the above methods, the most commonly used ones are SpO2 and ECG. Many models require appropriate feature extraction from these two signals for efficient performance. However, CNN and RNN based models usually do not require any additional feature extraction from the signals, which makes them extremely effective in real-time SA detection systems with minimum signal analysis. For these networks to perform well without manual feature extraction, complex architecture consisting of feature extraction layer, convolutional layer, pooling layer, recurrent layer, etc. are required. The lightweight deep artificial neural network (ANN), without any of those special layers, has been deployed in multiple systems [20], [21]. But the performance of such a network is yet to be investigated if no feature from the signal is provided.
In this study, we investigated the SpO2 and ECG signals for a real-time sleep apnea detection system. A feed-forward artificial neural network is used to evaluate the performance of the signals for detection of SA. Instead of using heavyweight convolutional neural networks, the proposed scheme demonstrates the use of a simple feed-forward neural network for detecting sleep apnea in real-time without any manual feature extraction. Thus, it enables much simpler and faster implementation of apnea detection system. In addition, the proposed scheme works with two different physiological signals and shows that the dual-channel technique of apnea detection performs better than the single-channel technique and thus substantiates the efforts of further exploration of dual-channel approaches.
Methods
Dataset
The proposed scheme employs PhysioNet Apnea-ECG dataset [22], [23]. There are a total of 70 records in this dataset. The entire dataset is divided into a training set of 35 records and a testing set of 35 records. The lengths of the records vary from 7 hours to 10 hours. Each of the recordings includes a digitized ECG signal, a set of apnea annotations derived by human experts, and a set of machine-generated QRS annotations. In addition to the ECG signals, 8 of these recordings have chest and abdominal respiratory effort signal, oronasal airflow signal, and oxygen saturation level signal, SpO2. Since the study aimed to build the ANN models by using ECG and SpO2 signals both individually and as a combination, only those 8 recordings in our analysis were used. The signals were sampled at the rate of 100 samples per second. In each recording, the annotation was placed at the start of every minute, followed by a one-minute interval. The annotation ‘A’ signifies that an apneic event was in progress at the beginning of the associated minute. The annotation ‘N’ means that no apneic event was in progress at the beginning of the associated minute. Figure 1 illustrates the applied annotation criteria in the Apnea-ECG dataset.
Figure 1:
Annotation criterion of Apnea-ECG dataset. Elapsed time is indicated by the distance from the left edge. ‘~’s denote the apneic periods, ‘|’s denote the time of apnea annotation (0, 60, 120, …seconds), and apneic and non-apneic intervals are marked by ‘A’ and ‘N’ annotations, respectively.
Data Segmentation
Since apnea annotations were provided for each one-minute interval, both the SpO2 and the ECG signals were divided into segments with a duration of 1 minute. Figure 1 shows that apnea annotation depends only on the presence or absence of apneic events at the beginning of the associated minute. Such annotation scheme may be misleading, as it can be seen in the 6th interval in Figure 1 where the interval is annotated as ‘N’, although the apneic event was persistent for most of the 1-minute duration. Similarly, the 7th interval is annotated as ‘A’ despite the apneic event lasting for a short time during the associated 1-minute period. To overcome this problem, considered only the 1st 30 seconds of each of the 1-minute segments were considered and the rest were discarded. It made sure that for any ‘N’-annotated 1-minute segment if there were sample points with apneic events, most of these were discarded. Similarly, if there were non-apneic sample points in an ‘A’-annotated segment, most of those were removed. Since an apneic event is marked by abnormal breathing and persistent SpO2 signal for at least 10 seconds, the decision to consider only 30 seconds of each interval is justified as it is more than the required minimum duration.
SpO2 Signal Processing
The first step of SpO2 signal processing was to identify and remove all the artifacts. Any SpO2 value less than 50% is not physiologically possible and was marked as an artifact. Moreover, all changes of SpO2 values greater than 4% were marked as artifacts [7]. Once those artifacts were removed, the signal was resampled at 1 Hz by using a simple moving average filter. After resampling, only 30 sample points from the 1st 30 seconds of each of the 1-minute segments were retained. Before resampling, if the total number of artifacts in the 30-seconds interval was greater than 10% of the total number of sample points in that interval, the entire associated 1-minute segment was discarded. It was done to make sure that none of the intervals with significant information loss, caused by the removal of sample points marked as artifacts, were used to train the ANN model.
ECG Signal Processing
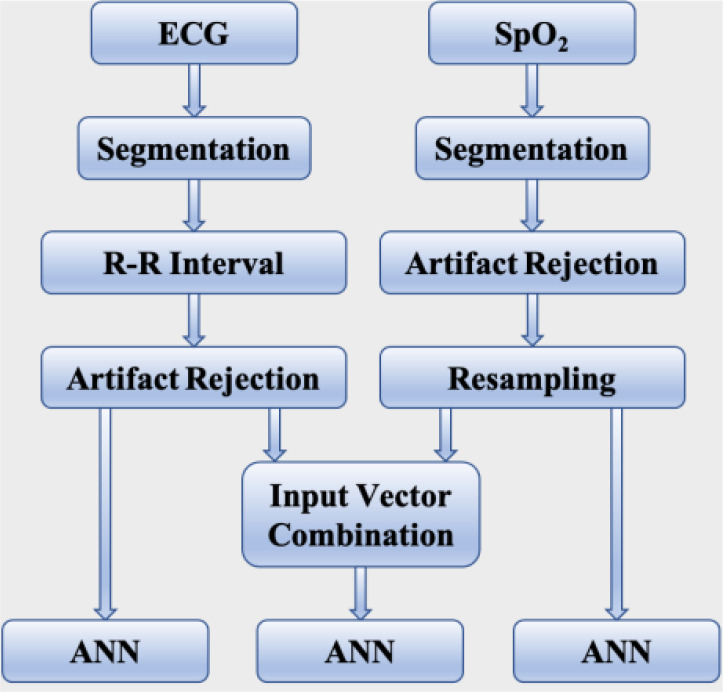
An ECG graph consists of P wave, QRS complex, and T wave. R-peak is the maximum amplitude in the R wave. The machine-generated QRS annotations associated with each recording were used to detect the R-peaks from the ECG signal within the first 30-seconds interval of each 1-minute segment. The QRS detector used in this database is based on the study by Zong et al. [24]. The R-R interval is simply the time interval between two successive R-peaks. A sliding window technique was implemented to remove the ectopic sample points from the R-R interval series. The window length was 5 and any R-R interval value larger than 20% of the average value of all the R-R interval values within the window was marked as ectopic beats and was removed [10]. Following the removal of the artifacts, the entire associated 1-minute interval was discarded if there were less than 30 sample points from the 30-seconds interval. If there were more than 30 R-R interval points, only the first 30 of these points were considered to be consistent with the number of points in each input vector derived from the SpO2 signal for training the ANN model. The flow diagram in Figure 2 shows all the steps involved in the data processing sequentially.
Figure 2:
Illustration of all the data processing steps.
Deep Neural Network
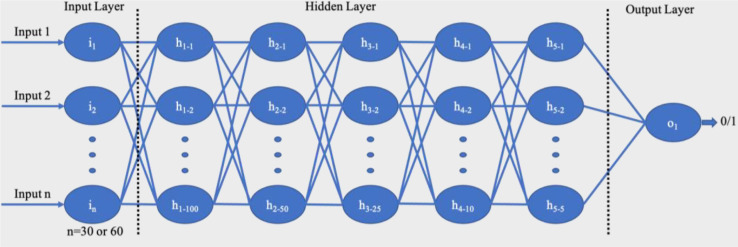
A deep neural network consists of multiple layers of nonlinear processing units where the output of one layer serves as the input of the next one. It is called a feed-forward network since the flow of information is always from the input- end to the output-end [25]. In this study, three deep ANN models were built, one for each SpO2, and ECG signals and one for a combination the two signals. The network architecture is illustrated in Figure 3. Each model had 5 hidden layers in addition to an input and an output layer. The hidden layers had 100, 50, 25, 10, and 5 neurons respectively. The output layer had only one neuron with an output of 0 or 1, denoting the absence or presence of apneic event respectively. The networks, which were built using SpO2 and ECG signals individually, had 30 neurons in the input layer to take in 30×1 input vector. Since the third model was created using the combination of two signals, its input layer had 60 neurons.
Figure 3:
Architecture of the proposed feed-forward deep neural network.
For optimization, the Adam optimization technique [26] was used while in the training phase, mean-squared loss function was used to achieve the model parameters with optimal values. In the proposed feed-forward ANN model each hidden layer had ReLU activation function (eq. 1) which is a piece-wise linear function that sends the input as output if the value is positive or zero and a forced zero for the negative input values. The output layer had Sigmoid activation function (eq. 2) which successfully categorizes between 0 (Normal Condition) and 1 (Apnea Occurred).
The training was performed for 1000 epochs with 10-fold cross-validation on the training set having a mini-batch size of 10. The performance of a model achieved by the cross-validation technique was evaluated based on its accuracy.
| (1) |
| (2) |
After having an estimation of the performance of each model employing the cross-validation technique, it was further analyzed to evaluate the performance of the model in a test set. This test set was separated from the entire set of data and was not used in training the model. On this occasion, the performance of a model was evaluated based on accuracy, precision, recall, and F1-score.
Results
Following the data segmentation and the artifact removal steps, different number of sequences were extracted from each type of signal. From the SpO2 signal, a total of 3,815 sequences were obtained, out of which 2,293 corresponded to normal events and 1,522 corresponded to apnea events. Out of the 2606 sequences extracted from the ECG signal, 1534 were from normal and 1072 were from apnea events. From the combined signal, a total of 2,530 sequences were obtained with 1,512 normal and 1,018 apnea events. Each of the combined sequences had 60 sample points and the rest of the sequences had 30 sample points. For each model, the entire associated set of sequences was split into training and testing sets with a ratio of 3:1. Table 1 shows the number of sequences before and after the split with the numbers of associated normal and apnea events.
Table 1.
Number of Sequences Before and After the Train-Test Split
| Sequence | SpO2 | ECG | Combined | |
|---|---|---|---|---|
| Entire set | Normal | 2293 | 1534 | 1512 |
| Apnea | 1522 | 1072 | 1018 | |
| Total | 3815 | 2606 | 2530 | |
| Train | Normal | 1730 | 1153 | 1136 |
| Apnea | 1131 | 801 | 761 | |
| Total | 2861 | 1954 | 1897 | |
| Test | Normal | 563 | 381 | 376 |
| Apnea | 391 | 271 | 257 | |
| Total | 954 | 652 | 633 | |
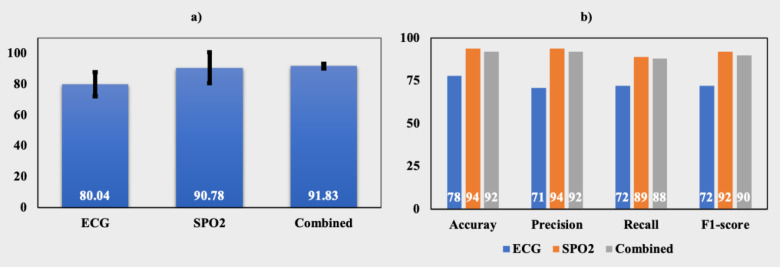
To build the ANN models from each signal, 10-fold cross-validation was performed using the associated training set. The performance of the model was evaluated based on the accuracy achieved after validation. Figure 4-a) shows the comparison of the cross-validation performance among the models. It can be seen that the best-performing model was the one that was built using the combined signal. The cross-validation accuracy of this model was 91.83 ± 1.51%. SpO2 based model was the 2nd best model with an accuracy of 90.78 ± 10.12%. The ECG signal-based model demonstrated an accuracy of 80.04 ± 7.7%, which was the least among the three models.
Figure 4:
a) accuracy (%) of the models after 10-fold cross-validation, b) accuracy, precision, recall, and F1-score of all the models on the test data.
Figure 4-b) shows the performance of the models in the test set, which was separated during the train-test split. For each type of signal, a new model was created using the entire training set, and the performance of the model was evaluated by using accuracy, precision, recall, and F1-score. Although the combined signal provided the best performing model from the cross-validation results, the SpO2-based model outperformed others in predicting the test set. It achieved the highest values of all the performance metrics. It had an accuracy of 94% whereas the ECG and combined signal provided accuracy of 92% and 78%, respectively and its prevision, recall, and F1-score were 94%, 89%, and 92%, respectively. As estimated from the validation result, the ECG-based model had the least values of the performance metrics. The combined signal-based model was the 2nd best model with accuracy, precision, recall, and F1-score of 92%, 92%, 88%, and 90%.
Discussion
In this study, three different types of input signals were used to build the feed-forward ANN model. The objectives of this study were to evaluate how the SpO2 and the ECG signals perform to detect sleep apnea and to observe whether the combination of those two signals can provide better performance. From the cross-validation result, it was evident that the combined signal outperformed the models based on individual signals. But for evaluation of the test data, the SpO2-signal based model performed better than the combined signal-based model. A comparison of the accuracy of these two models from the cross-validation demonstrates that their accuracy values were not significantly different. But the standard deviation of all the folds was significantly high for the SpO2 signal. It was 10.12% for SpO2, whereas it was only 1.51% for the combined signal. Such a low standard deviation value indicates that the combined signal-based model is very consistent in detecting with high accuracy from a set of unknown data. On the other hand, although the SpO2 based model performed better on the test set, it may not be as consistent as the combined signal for detecting apnea from an unknown dataset. Therefore, it can be concluded that the combined signal-based model is the most reliable model with a very high accuracy of 91.83%.
Although the ECG-based model did not yield an accuracy as high as the SpO2-based model, once the ECG sequence was combined with the SpO2 sequence, it complemented the SpO2 sequence and as a result, the performance of the combined sequence was better than the individual SpO2 sequence. This is an indication that instead of a single-channel technique, a dual-channel technique using both the SpO2 and the ECG signals should be a better option for real-time apnea detection. Nowadays, there are multiple studies involving wearable sensor-based real-time apnea detection systems [27][28]. With the advancement in the field of sensor technology, researchers are highly interested in such wearable sensor-based apnea detection systems. The study presented in this work will provide the researchers with a direction towards a dual-channel technique of detection.
In this work, every 1-minute interval of the signal was analyzed, but the classification was performed based on the first 30 seconds of each of the 1-minute intervals. Most of the studies based on the Physionet Apnea-ECG database reported in the literature utilized the entire 1-minute segment. Therefore, these models needed a sequence of 1-minute duration to detect apnea events. On the other hand, the proposed model can generate output by working on a 30-seconds long sequence. Such a shorter processing window makes this model a better contender for a real-time apnea detection system.
Most of the neural network-based apnea detection systems rely on feature extraction from the signal after necessary noise and artifact removal. In this study, a feed-forward neural network model is proposed, which does not need any feature extraction from the SpO2 signal once the noise and artifacts are removed. On the other hand, the proposed ECG-based model takes R-R interval as input. Most of the ANN-based systems using R-R intervals as input require additional features from the R-R interval sequences, such as mean, variance, maximum value, minimum value, etc. Other studies that do not require feature extraction from R-R interval sequence are usually based on more complex networks, such as CNN, RNN, etc. To the best of our knowledge, this study is the first demonstration of a simple feed-forward ANN-based apnea detection system using R-R interval without additional features.
There have been multiple demonstrations of these models yielding higher accuracy than the proposed model [16]. In the era of artificial intelligence embedded hardware systems, simplification of the models is as highly desired as the accuracy for the feasibility of embedding the models on chips. Techniques, such as pruning and quantization can further be applied on the proposed model to make it more suitable for hardware implementation within power constraints. CNN and RNN are too complex to be embedded in hardware even after pruning and quantization.
The major limitation of the study is that the proposed architecture of neural network failed to achieve high values of performance metrics in detecting sleep apnea from the ECG signal. It can be attributed to the erroneous R-R intervals. The R-R interval series was extracted from the raw ECG signal by using the machine-generated QRS annotations. The QRS detector that the database used to generate the annotations was unaudited and contained errors as mentioned in the PyhsioNet website (https://physionet.org/content/apnea-ecg/1.0.0/). A better QRS detector could have detected the R-beats at accurate time points with less error. Moreover, only 8 recordings were used in this study. In our future study, we aim to work with larger dataset and design our own QRS detector, which can lead to better accuracy of the model.
Conclusion
In this study, SpO2 and ECG signals were used both individually and in combination to build a real-time SA detection system with feed-forward ANN. R-R intervals were derived from the raw ECG signal and three models were built using SpO2 and the R-R intervals. The proposed models did not require any additional features from any of those signals for SA detection. According to the results from 10-fold cross-validation, it was evident that the combined signal-based model performed better than the individual signal-based models. Although the accuracy achieved by the ECG-based model was significantly lower than the SpO2-based model, which was attributed to the erroneous QRS detector, in the model employing the combination of SpO2 and ECG the two complemented each other resulting in the highest accuracy among the three models. From this study, it can be concluded that the dual-channel technique is preferable for a real-time apnea detection system and a better performing model can be built by using a more efficient QRS detector.
Figures & Table
References
- 1.Uddin MB, Chow CM, Su SW. Classification methods to detect sleep apnea in adults based on respiratory and oximetry signals: A systematic review. Physiol Meas. 2018. [DOI] [PubMed]
- 2.Chang HY, Yeh CY, Lee C Te, Lin CC. 2020. A sleep apnea detection system based on a one-dimensional deep convolution neural network model using single-lead electrocardiogram. Sensors (Switzerland).
- 3.Xie B, Minn H. 2012. Real-time sleep apnea detection by classifier combination. IEEE Trans Inf Technol Biomed.
- 4.Magalang UJ, Dmochowski J, Veeramachaneni S, Draw A, Mador MJ, El-Solh A, et al. 2003. Prediction of the Apnea-Hypopnea Index from Overnight Pulse Oximetry. Chest. [DOI] [PubMed]
- 5.Dumitrache-Rujinski S, Calcaianu G, Zaharia D, Toma CL, Bogdan M. 2013. The role of overnight pulse-oximetry in recognition of obstructive sleep apnea syndrome in morbidly obese and non obese patients. Maedica (Buchar). [PMC free article] [PubMed]
- 6.Muza RT. 2015. Central sleep apnoea - A clinical review. Journal of Thoracic Disease.
- 7.De Chazal P, Heneghan C, Mcnicholas WT. 2009. Multimodal detection of sleep apnoea using electrocardiogram and oximetry signals. Philos Trans R Soc A Math Phys Eng Sci.
- 8.Lee H, Park J, Kim H, Lee KJ. 2016. New Rule-Based Algorithm for Real-Time Detecting Sleep Apnea and Hypopnea Events Using a Nasal Pressure Signal. J Med Syst.
- 9.Nguyen HD, Wilkins BA, Cheng Q, Benjamin BA. 2014. An online sleep apnea detection method based on recurrence quantification analysis. IEEE J Biomed Heal Informatics.
- 10.Khandoker AH, Palaniswami M, Karmakar CK. 2009. Support vector machines for automated recognition of obstructive sleep apnea syndrome from ECG recordings. IEEE Trans Inf Technol Biomed.
- 11.Khandoker AH, Karmakar CK, Palaniswami M. 2009. Automated recognition of patients with obstructive sleep apnoea using wavelet-based features of electrocardiogram recordings. Comput Biol Med.
- 12.Vimala V, Ramar K, Ettappan M. 2019. An Intelligent Sleep Apnea Classification System Based on EEG Signals. J Med Syst. [DOI] [PubMed]
- 13.Mendez MO, Ruini DD, Villantieri OP, Matteucci M, Penzel T, Cerutti S, et al. 2007. Detection of sleep apnea from surface ECG based on features extracted by an autoregressive model. In: Annual International Conference of the IEEE Engineering in Medicine and Biology - Proceedings. [DOI] [PubMed]
- 14.Ting H, Mai YT, Hsu HC, Wu HC, Tseng MH. 2014. Decision tree based diagnostic system for moderate to severe obstructive sleep apnea. J Med Syst. [DOI] [PubMed]
- 15.Mostafa SS, Mendonça F, Juliá-Serdá G, Morgado-Dias F, Ravelo-García AG. 2020. SC3: self-configuring classifier combination for obstructive sleep apnea. Neural Comput Appl.
- 16.Mostafa SS, Mendonça F, Ravelo-García AG, Morgado-Dias F. 2019. A systematic review of detecting sleep apnea using deep learning. Sensors (Switzerland).
- 17.Erdenebayar U, Kim YJ, Park JU, Joo EY, Lee KJ. 2019. Deep learning approaches for automatic detection of sleep apnea events from an electrocardiogram. Comput Methods Programs Biomed.
- 18.Hafezi M, Montazeri N, Saha S, Zhu K, Gavrilovic B, Yadollahi A, et al. 2020. Sleep Apnea Severity Estimation from Tracheal Movements Using a Deep Learning Model. IEEE Access.
- 19.Mostafa SS, Mendonça F, Morgado-Dias F, Ravelo-García A. 2017. SpO2 based sleep apnea detection using deep learning. In: INES 2017 - IEEE 21st International Conference on Intelligent Engineering Systems, Proceedings.
- 20.Li K, Pan W, Li Y, Jiang Q, Liu G. 2018. A method to detect sleep apnea based on deep neural network and hidden Markov model using single-lead ECG signal. Neurocomputing.
- 21.Elmoaqet H, Eid M, Glos M, Ryalat M, Penzel T. 2020. Deep recurrent neural networks for automatic detection of sleep apnea from single channel respiration signals. Sensors (Switzerland). [DOI] [PMC free article] [PubMed]
- 22.Penzel T, Moody GB, Mark RG, Goldberger AL, Peter JH. 2000. Apnea-ECG database. In: Computers in Cardiology.
- 23.A.L. G, L.A. A, L. G, J.M. H, P.C. I, R.G. M, et al. 2000. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation.
- 24.Engelse WAH, Zeelenberg C. 1979. Single scan algorithm for qrs-detection and feature extraction. In: Computers in Cardiology.
- 25.Liu W, Wang Z, Liu X, Zeng N, Liu Y, Alsaadi FE. 2017. A survey of deep neural network architectures and their applications. Neurocomputing.
- 26.Kingma DP, Ba JL. 2015. Adam: A method for stochastic optimization. In: 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings.
- 27.Lin YY, Wu HT, Hsu CA, Huang PC, Huang YH, Lo YL. 2017. Sleep Apnea Detection Based on Thoracic and Abdominal Movement Signals of Wearable Piezoelectric Bands. IEEE J Biomed Heal Informatics.
- 28.Surrel G, Aminifar A, Rincón F, Murali S, Atienza D. 2018. Online Obstructive Sleep Apnea Detection on Medical Wearable Sensors. IEEE Trans Biomed Circuits Syst. [DOI] [PubMed]