Abstract
Including social determinants of health (SDoH) data in health outcomes research is essential for studying the sources of healthcare disparities and developing strategies to mitigate stressors. In this report, we describe a pragmatic design and approach to explore the encoding needs for transmitting SDoH screening tool responses from a large safety-net hospital into the National Covid Cohort Collaborative (N3C) OMOP dataset. We provide a stepwise account of designing data mapping and ingestion for patient-level SDoH and summarize the results of screening. Our approach demonstrates that sharing of these important data - typically stored as non-standard, EHR vendor specific codes - is feasible. As SDoH screening gains broader use nationally, the approach described in this paper could be used for other screening instruments and improve the interoperability of these important data.
Introduction
Including social determinants of health (SDoH) data in health outcomes research is essential for studying the sources of healthcare disparities, identifying exposure and behavioral risk factors, and developing strategies to mitigate stressors.1 SDoH are the risk factors related to how people live, grow, and learn and they may confer advantages or disadvantages towards development of health outcomes.1,2 These psychosocial complexities (e.g., housing instability, food insecurity, social isolation, chronic stress, financial insecurity) are the social needs that contribute powerfully to racial/ethnic disparities in outcomes, which are highly influential factors towards worsening quality of life, increased disability, exacerbations of illness, and premature mortality.2–7 Screening for these factors have been crucial for understanding the complex hardships that patients experience, especially during the COVID-19 pandemic.
Despite their importance, routine collection of patient-level SDoH and facilitated uses of such information from electronic health records (EHRs) have significant data sharing barriers in obtaining and maintaining information through the data life-cycle. First, few health systems have policies to incentivize consistent screening and collection of patient-level SDoH. More and more, these policies are mandated from State or Federal funding agencies, like for Medicaid populations. Even without policy mandates, some health systems have a rapidly advancing mission to improve quality reporting, health equity, and to detect and address patient social needs. Although policies and incentives help to drive change, the institutional mission is also a key driver to collect patient-level SDoH. Health systems would need time and resource investments to modify operational workflows, develop staff capacities, and have documentation plans in place for information storage and retrieval. Studies have reported that observations about patient social determinants of health learned during clinical encounters may be documented within disparate areas within EHRs,8,9 often written from the provider perspective as unstructured clinical notes, which require significant extraction capacities for reuse.9,10 It is unclear if these observations are limited to certain clinical visit scenarios or occur under non-random conditions, or encompass what aspects of social need. More importantly, while cross-referencing between the patient and provider perspective would add context to understand the social needs, datasets describing social needs that a patient experiences should be reported from the patient and maintained as close to the source as possible. Equitable care is only possible with clear and active participation from the patients.
A growing number of institutions, particularly those serving traditionally marginalized populations, are routinely screening and supporting patients with unmet SDoH needs using structured screening instruments like PRAPARE,11 WellRx,12 WeCare,13–16 and BMC-THRIVE (or THRIVE).17 These screening tools inquire about housing instability, food insecurity, education attainment, access to care issues, and various other social risk factors as a short questionnaire. Structure screening instruments provide a flexible, low-barrier method for patients and providers to engage in dialogue on social needs with the care needs.18 Screening tools can be low-fidelity paper questionnaires or electronic survey instruments completed in the waiting-room or prior to the clinical encounter. Once collected, providers can record the instance of diagnostic screening or findings of social need for further referrals.19 In addition, national efforts such as the Gravity Project20 and UCSF Siren21 are developing national platforms to systematically organize and represent the data in these structured screening tools and other EHR data using data standards. The groups are also working to fill gaps in mapping when identified, providing a pathway for representing patient-level SDoH in research sourced from the patient responses to the screening tools.
While consensus mappings of standard concepts are emerging, the way to implement them across the data life-cycle is unclear. Specifically, screening tools are works-in-progress and may evolve over time with different questions and answer options. The studies have generally reported that screening tool data may be predominantly stored within EHR FlowSheet tables.8 Patient-level SDoH in FlowSheets means that the data are compartmentalized within a tabular data structure, relationally mapped to associate the survey, each question, the responses to each question, and the instance of data collections.22 Common data models (CDMs) are often the conduits for making clinical knowledge portable and accessible for research applications and secondary data use in Public Health. As FlowSheet records are localized implementations and may contain text-based responses, additional ETL (extract-transform-load) logic would need to engineer the translation between what each FlowSheet measurement means and the corresponding standardized concept representation.23
Finally, with data harmonization, CDMs may transform between models allowing for data sets to be compared across standards that were previously limited by choice of CDM. One of the main hurdles to constructing FlowSheet mappings is the lack of personnel with the technical access and domain expertise. As such, services like TriNetX24] and consortia like Observational Health Data Sciences Institute (OHDSI) provide technical capacities that facilitate adoption and creation of de-identified or limited datasets.25 However, to analyze across multiple institutions operating with different CDMs, either the source institutions may invest to change CDM instances or midstream data harmonization may be applied to get the data into comparable format. In the National COVID Cohort Collaborative (N3C), data submission may occur using any of a select set of CDMs with the intention to harmonize data into Observational Medical Outcomes Partnership (OMOP) during the data ingestion process.26 There remains a paucity of research that describes this process of engineering patient-level SDoH and harmonizing the information across CDMs. Harmonization may increase the potential for data loss during translation, and loss of question set relationships. The process of translating patient-level SDoH across CDMs would benefit from intentional design during the screening tool FlowSheet mapping development.
In this report, we describe a pragmatic design and approach that leverages previous work related to SDoH Data Engineering.22 In collaboration with Boston Medical Center (BMC), TriNetX, and N3C, we explored the encoding needs for transmitting THRIVE screening tool responses and the technical hurdles experienced in data ingestion and harmonization (DI&H) into the N3C OMOP dataset.
Methods
Objective
We provide the step-wise account of designing data mapping and ingestion for patient-level SDoH. We describe 1) the conceptual model of information flow from clinical encounter to submission to the N3C, 2) the upstream data mapping needed for i2b2 to TriNetX, and 3) the midstream data harmonization considerations for TriNetX to OMOP. We generated concept sets reflective of the screening tool questions and answers to support inspection of the data records. The record counts were returned to the upstream data stewards as confirmatory validation of patient-level SDoH record counts in the data flow through TriNetX dataset and the N3C OMOP limited dataset.
Data source
Boston Medical Center (BMC) is the largest safety-net hospital in New England and began institution-wide screening for SDoH in 1999. As of July 1, 2021 over 200,000 patients had been screened for at least one SDoH domain. BMC has implemented screening for social needs using the THRIVE screening tool and stored the information in FlowSheet tables. The FlowSheet includes some questions and responses that are incorporated into standard instruments that are represented by LOINC, including PRAPARE, but is not itself a standardized instrument. Data is stored within Flowsheets in the Epic EHR,17supporting the referral and routing of social needs detected from patient reporting.
Research instruments
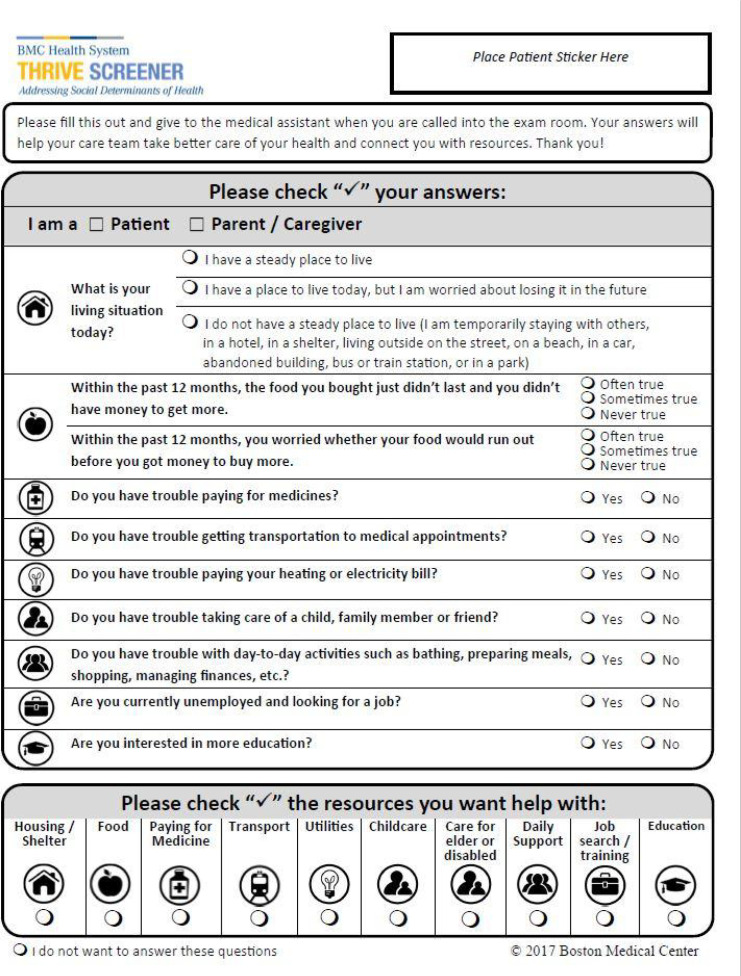
The THRIVE screening tool, displayed in Figure 1, is a one-page, 8 question screening instrument with questions related to homelessness, food insecurity, trouble paying for utilities, trouble paying for medication, transportation difficulties, child/elder care challenges, and desire for additional education. Each category is screened using two different questions (Figure 1).17 The questions used in THRIVE were a subset of questions from national survey tools for use in routine clinical practice. Minor changes were made during the first three years of use. Multiple versions of the THRIVE screening tool were released (2017, 2018, and 2020), so patients may have responded to different versions of the screening. In most settings, the screening was administered via paper and pencil on a clipboard and the data entered by Medical Assistants within EHR FlowSheet measurement records. At BMC, routine use of THRIVE was preceded by a single question related to homelessness.
Figure 1:
The THRIVE screener (circa 2020).
BMC-to-TriNetX-to-N3C mapping overview
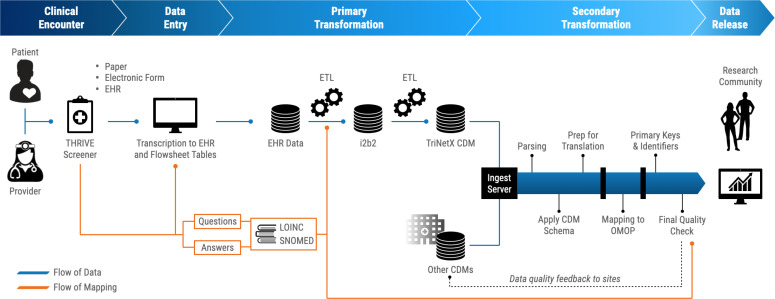
At the start of the engineering process, multiple collaborative discussions within the N3C SDoH domain team were aimed to conceptualize the process of getting patient-level SDoH data into the N3C OMOP dataset (Figure 2). The overall workflow to get data released can be simplified into four phases: 1) Clinical encounter, 2) Data entry (Transcription into EHR FlowSheet tables), 3) Primary transformations (EHR to CDM), and 4) Secondary transformations (CDM to CDM).
Figure 2:
Overall workflow for SDoH screening tool data integration into N3C. Clinical encounter: A patient completes the SDoH screening tool, either on their own, or administered by a healthcare provider, and per institutional protocol. The screening tool itself may be a paper form, an electronic form, or entered directly into the EHR. Data entry: The patient’s responses are entered into the EHR, storing FlowSheet records for the responses to the screening. Primary Transformation: Before transformations may happen, subject matter experts mapped the THRIVE questions and possible answers to LOINC and SNOMED codes. The EHR data goes through an ETL to be converted into an i2b2 data instance and LOINC and SNOMED codes are incorporated during this transformation. The i2b2 data instance is ingested by TriNetX, one of the acceptable N3C CDMs for data submission. Secondary Transformation: TriNetX generates the dataset for upload to N3C data ingestion pipeline, which gets parsed, applied to CDM schema, then mapped to OMOP. A final quality check provides feedback to the contributing sites, prior to being published to the cloud-based FedRamp N3C Enclave for use by the research community.
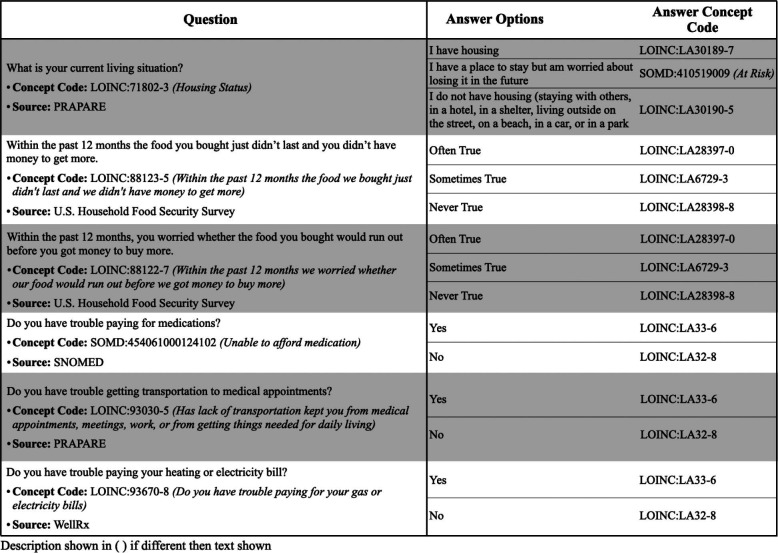
During clinical encounters, BMC clinical information as well as responses to the THRIVE screening are collected then transcribed into the EHR systems. The clinical information and screening responses were first ETL’d into an ACT/i2b2 CDM. To do this, we initially reviewed the THRIVE questions (circa 2020) and generated standardized OMOP concept mappings for the questions and answers using LOINC and SNOMED. Since the data were not directly transformed from the EHR to OMOP, the mapping encodings needed to refer to the vocabulary and concept code to be transmitted to i2b2. Once we compared with the BMC FlowSheet measurements for the THRIVE questions, questions-answers from the prior versions of THRIVE were included and for further mapping. We used PDFs of the THRIVE screening tool to cross-reference the interpretation of the questions and answer options. Most questions-answers were not represented verbatim within LOINC, so we mapped to the nearest LOINC concept with minimal loss of information or change in interpretation. If the concept was not found, we would map to the closest SNOMED (using prefix “SOMD:”) term. After three iterations of review, the mappings represented the closest semantic representation of the question-answer using LOINC and SNOMED concepts. Mappings are shown below in Table 1.
Table 1:
Mappings for SDoH questions related to homelessness, food insecurity, trouble paying for medication, transportation or utilities. For each THRIVE question, the original text is presented, along with the selected concept code, the code description if different than the question, and the source (the instrument or vocabulary that originated the concept code). For each THRIVE answer, the original text is presented along with the corresponding standard concept code in LOINC or SNOMED.
BMC electronic data warehouse creates an i2b2 data instance27 containing the mapped FlowSheet information for THRIVE, which gets ingested by TriNetX. Thereafter, TriNetX filters for patients that meet the N3C COVID Phenotype criteria and generates an N3C-compliant payload for BMC to submit. The LOINC and SNOMED concept codes are used to form concept sets used in the data quality checks.
Data ingestion and midstream harmonization transformations
The N3C DI&H pipeline has been developed in Python and SQL and implemented in the NIH’s instance of the Palantir Foundry Platform. N3C ETL jobs are added as stepwise pipeline tasks and managed using a unified framework within Foundry to ingest and harmonize all incoming COVID-19 EHR data from the participating data partners. Participating data partners can submit their data in one of the five known Common Data Model used by the Clinical and Translational Science Award (CTSA) hubs; OMOP, ACT, TriNetX, PCORnet or PEDSnet. Data partners submit their datasets through a highly secured sFTP location. The N3C DI&H pipeline transforms the submitted data to the OMOP model before merging the dataset to cloud based FedRamp N3C Enclave. The transformation pipeline steps include parsing flat data files, data conformance checks against the native CDM format, primary key checks, domain mapping, and semantic vocabulary translations for all terminologies that exist in the source data, i.e. ICD-10-CM, ICD-10-PCS, LOINC, RxNorm, HCPCS and CPT4.
The THRIVE dataset is submitted in TriNetX CDM format. This dataset can be utilized in N3C to crossmap SDoH observational codes in LOINC or SNOMED to OMOP concept_ids. More specifically, all of the distinct SDoH observational codes submitted in BMC datasets are added to the value set mapping table such that this enriched crosswalk mapping table can be used to translate all other incoming SDoH related codes from other N3C participating data partners. The SDoH concept can be either mapped to the OMOP Observation domain or the Measurement domain. This “map to” information is specified in the domain_id column of the OMOP vocabulary tables. Therefore, based on the domain id, the SDoH observational data are either inserted in the Observation domain or the Measurement domain with translated OMOP codes as observation_concept_id or the measurement_concept_id, respectively. The string value result or the answer to the SDoH observation codes are mapped to corresponding concept ids and inserted in the value_as_concept_id field along with the value_source_value, the verbatim string value from the source data representing the result/answer of the SDoH concept. By convention, concepts that do not correspond to an existing term in the OMOP vocabulary are added to an instance of the OMOP CDM and assigned a concept ID larger than 2000000.
Results
As of July 1, 2021, 76,900 patients including their prior clinical findings since 2018-01-01 were included in the BMC N3C data extract submission. All data were successfully ingested into the N3C enclave. 50,400 (65.5%) had at least one THRIVE SDoH assessment, 49,880 (64.9 %) assessed about homelessness, 21,790 (28.3%) assessed about food insecurity, 20,440 (26.6%) assessed about trouble paying for utilities, and 19,120 (24.9%) assessed about trouble paying for medications. Among respondents, 13.5% were homeless or had unstable housing, 26.4% were experiencing food insecurity, 15.2% reported having trouble paying for utilities, and 13.1% reported having trouble paying for medications.
The N3C enclave is downstream from a number of harmonization and mapping steps, which increases the complexity of provenance tracking, but allows N3C to leverage earlier data cleaning and mapping steps. Data was initially shared as i2b2 data with TriNetX and submitted to N3C in TriNetX CDM format. One of the N3C DI&H pipeline steps is semantic translation from source value sets to OMOP concept ids. The semantic translation utilizes an N3C value set mapping table (analogous to OMOP SOURCE_TO_CONCEPT_MAP) to translate all of the source column field values for a given domain table from the source data to a corresponding OMOP concept_id.
In the TriNetX CDM, the SDoH concept questions are captured in the lab_code column field and the SDoH concept answers are captured in the text_result_value column field. Due to the fact that data partners may collect and submit various enumerations of the answer string for any given SDoH answer concepts in the text_result_val field, N3C opted to extend the OMOP source to concept mapping tables with custom entries in order to harmonize these variations. For example, the text values transferred to N3C included “LOINC:LA30189-7 (I have a steady place to live)” and “LOINC:LA30189-7 (I have housing)”. These LOINC codes were assigned to the parenthetical strings before data were transferred to N3C. While the response “I have housing” is an exact match to the LOINC code LA30189-7, “I have a steady place to live” does not have an assigned LOINC code. When harmonizing data N3C mapped both strings to OMOP concept_id 37079501 (i.e. LOINC:LA30189-7). An alternative strategy would involve late-binding for mappings “I have a steady place to live”, e.g. extending the OMOP Vocabulary to create a custom concept id for “I have a steady place to live” and apply the CONCEPT RELATIONSHIP table at the time of analysis to group these into equivalent analysis concepts. Using either approach, provenance and the original answer text is retained.
In the 2020 version of THRIVE, the screening tool introduced a checkbox for the patient to choose “I do not want to answer the questions” (i.e., “SOMD:31021000119100 (Screening declined)”). In total, 7,530 of the 76,900 patients had elected that they do not voluntarily want to answer the questions. Separately, BMC had incorporated FlowSheet measurement to indicate whether the patient had acknowledged they could not answer the screening tool due to language barrier. Over 1,370 patients had indications of language barriers annotated during transcription. These two types of questions are not about the patient’s SDoH per se, but psychometric assessment markers to provide evidence of validity and reliability of the respondent results to all questions in that screening instance. Through inclusion of these FlowSheet entries, the survey responses of “No” can be clarified as a 1) Patient Endorsement of Answer option “No”, 2) “I don’t understand” as inferred based on the Provider observation about linguistic barrier, 3) a question unintentionally left blank, or 4) “No, I don’t want to answer.” Only in the first option can the responses be taken for immediate value.
Discussion
In this report, we describe a pragmatic approach to mapping structured SDoH screening data to standard vocabularies and demonstrate that sharing of these important data - typically stored as non-standard, EHR vendor specific codes - is feasible. National efforts to organize and share SDoH data like the Gravity Project20 and UCSF Siren28 are progressing quickly; however, in our review of the literature, we only found a few published studies on standards-based representation of SDoH screening data and none that demonstrated that these standards could be used with real-world data for data sharing between sites using different CDMs, highlighting the novel nature of this work.
There is broad acceptance in clinical domains that screening and intervention related to SDoH should be part of routine clinical care. At present, the practice has not become widespread. Some health systems are driven by an institutional mission to improve care, equity, and address the non-clinical social needs that impact the patients’ well-being. Others have adopted screening methods earlier as part of State or Federal mandates. With the COVID pandemic, patient data on SDoH and data to inform of patient cohorts already experiencing the burden of social needs was in high demand.
As SDoH screening gains broader use nationally, the approach described in this paper could be used for other screening instruments and improve the interoperability of these important data. For sites that serve traditionally marginalized or under-resourced populations such as the health system in this project, capturing these data and being able to include them in research has the potential to more fully describe the life experience and health determinants for their patients. When mapped to standard terminologies, these data can be shared on a national scale to help ensure that the broadest possible array of people and their data are represented in national collaborations, like N3C.
Several important challenges were encountered during this project. First, the THRIVE screener evolved over the first three years of use. Versioning metadata was not available in the submitted data set. In several cases, only the answer values changed even though the question text had been edited and the variable identifier remained the same. Correctly mapping values was possible, but required meeting with the developers and clinical teams to better understand the changes, and selecting among inexact matches. In the example shared above, the THRIVE instrument includes a single item that is almost identical to the PRAPARE question which has a pre-assigned LOINC/OMOP code (“What is your living situation today?”). However, rather than presenting responses with three possible values for housing status in THRIVE, PRAPARE has two sequential questions and LOINC codes. The second question asks separately in a Yes/No format “Are you worried about losing your housing?” (https://forms.loinc.org/93025-5), whereas THRIVE simply presents “I have a place today, but I am worried…”. While this PRAPARE question and their “Yes”‘ and “No” responses have standard LOINC codes, answers are not meaningful without the context of a question. In other words, the multiple-choice option in THRIVE is represented as two separate questions in PRAPARE, thus in order to correctly analyze and harmonize data, both question and answer values must be considered in the mapping process. This changes the standard architecture of SOURCE_TO_ CONCEPT_MAP (or CONCEPT_RELATIONSHIP) to require joining two fields in the source data (question and response values) to properly assign the adjudicated value of “at risk” record. Notably, LOINC now includes richer representation of instruments, including information about the instrument, the questions, and the answers for survey data. The OMOP common data model includes “Standardized Derived Data Elements” where there is consensus on complex but consistent logical algorithms for deriving data elements from standardized facts. A similar approach for logical representations may be useful to derive common data elements from heterogeneous survey-based data elements measuring the same or similar concepts using distinct questions and answers.
One limitation of this study is that it reflects one site’s approach to collecting the information. Several steps may be unique to the site, though the insights learned may benefit other sites addressing data silos and patient-level SDoH data representations. The ability to map to LOINC codes improved the continuity of the information and it didn’t appear affected by the multiple transformation steps. This case study applied concept mappings post hoc to extract data that has already been collected into FlowSheets, where the mapping decisions may vary by site and require local technical expertise. FlowSheet records that produce derived summary scores and panel indices, assessments requiring clinical context to understand their non-random occurrence or missingness, and assessment observations based on conditional responses (e.g., probing questions) represent limitations to the FlowSheet extraction presented. As the source vocabularies change, updating the concept mappings will be a sustainability and versioning hurdle. To reduce site variability in FlowSheet mapping decisions, a better approach would be to have metadata maps designed, vetted, and maintained by the instrument developers, such that downstream instrument adopters can incorporate the instruments and compare across sites with fewer technical and interpretation barriers. Our workflow focused on data harmonization into OMOP where other CDM endpoints may be desired. We acknowledge that instrument registration, secondary data-use consideration, and harmonization as part of the data life-cycle may be out-of-scope for instrument developers. However, this remains a major challenge in summarizing data across instruments as many survey instruments do not have controlled vocabulary representations. We encourage instrument developers towards this early strategic planning as it enables downstream comparisons and analyses.
In summary, there is a pressing need to better understand and include SDoH in research and clinical data repositories. There are heterogeneous mechanisms for data capture and standardization, which may result in duplicative efforts where data are captured with commonly used instruments that are not standardized at the time of data capture or ETL. While this case-study was a targeted, pragmatic intervention to extract patient-level SDoH from FlowSheet records, this approach was intended to be scalable and reusable, though not comprehensive. The vast majority of EHR implementations have their own bespoke survey implementations in flowsheets, yet extraction remains a laborious and technical endeavor. Within the well-described domain of structured screeners like PRAPARE, AHC, and THRIVE, mapping locally represented data to standards is feasible and can be used within a variety of CDMs like OMOP, i2b2, and TriNetX. And, here, we’ve shown the utility to translate across these CDMs. As routine screening increases and more data become available, the approach described in this paper as well as the work of many others should be used to ensure that these important data can be shared within and between systems. Understanding the value of undertaking this effort is a critical next step to convincing health systems to invest time and effort in doing so.
Funding Acknowledgements
Jimmy Phuong was partially funded by the National Center for Data to Health (CD2H) grant [NIH/NCATS U24TR002306], supplemental funding from the National COVID Cohort Collaborative [NIH/NCATS U24TR002306-04S3]. Juan Espinoza and Daniella Meeker have received funding from grants UL1TR001855 and UL1TR000130, and a subaward from grant U24TR002306 from the National Center for Advancing Translational Science (NCATS) of the U.S. National Institutes of Health. William Adams has received funding from UL1TR001430. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Figures & Table
References
- 1.Braveman P, Gottlieb L. The Social Determinants of Health: It’s Time to Consider the Causes of the Causes. Public Health Rep. 2014;129:19–31. doi: 10.1177/00333549141291S206. [Internet]. Available from: [cited 2021 Aug 26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuels-Kalow ME, Ciccolo GE, Lin MP, Schoenfeld EM, Camargo CA. The terminology of social emergency medicine: Measuring social determinants of health, social risk, and social need. J Am Coll Emerg Physicians Open. 2020;1:852–6. doi: 10.1002/emp2.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cramm JM, Nieboer AP. Relationships between frailty, neighborhood security, social cohesion and sense of belonging among community-dwelling older people. Geriatr Gerontol Int. 2013;13:759–63. doi: 10.1111/j.1447-0594.2012.00967.x. [DOI] [PubMed] [Google Scholar]
- 4.Marshall GL, Kahana E, Gallo WT, Stansbury KL, Thielke S. The price of mental well-being in later life: the role of financial hardship and debt. Aging Ment Health. 2021;25:1338–44. doi: 10.1080/13607863.2020.1758902. [DOI] [PubMed] [Google Scholar]
- 5.Tucker-Seeley RD, Li Y, Subramanian SV, Sorensen G. Financial hardship and mortality among older adults using the 1996-2004 Health and Retirement Study. Ann Epidemiol. 2009;19:850–7. doi: 10.1016/j.annepidem.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Despard M, Grinstein-Weiss M, Guo S, Taylor S, Russell B. Financial Shocks, Liquid Assets, and Material Hardship in Low- and Moderate-Income Households: Differences by Race. J Econ Race Policy. 2018;1:205–16. [Internet]. Available from: [cited 2021 Aug 26] [Google Scholar]
- 7.Marshall GL, Tucker-Seeley R. The association between hardship and self-rated health: does the choice of indicator matter? Ann Epidemiol. 2018;28:462–7. doi: 10.1016/j.annepidem.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell EK, Dambrun K, Cowburn S, Mossman N, Bunce AE, Marino M, et al. Variation in Electronic Health Record Documentation of Social Determinants of Health Across a National Network of Community Health Centers. Am J Prev Med. 2019;57:S65–73. doi: 10.1016/j.amepre.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Hatef E, Rouhizadeh M, Tia I, Lasser E, Hill-Briggs F, Marsteller J, et al. Assessing the Availability of Data on Social and Behavioral Determinants in Structured and Unstructured Electronic Health Records: A Retrospective Analysis of a Multilevel Health Care System. JMIR Med Inform. 2019;7:e13802. doi: 10.2196/13802. [cited 2021 Jun 21]Available from: http://medinform.jmir.org/2019/3/e13802/ [Internet]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lybarger K, Ostendorf M, Yetisgen M. Annotating social determinants of health using active learning, and characterizing determinants using neural event extraction. J Biomed Inform. 2021;113:103631. doi: 10.1016/j.jbi.2020.103631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.PRAPARE. 2021. [Internet]. NACHC. [cited Aug 26]. Available from: https://www.nachc.org/research-and-data/prapare/ [Google Scholar]
- 12.Page-Reeves J, Kaufman W, Bleecker M, Norris J, McCalmont K, Ianakieva V, et al. Addressing Social Determinants of Health in a Clinic Setting: The WellRx Pilot in Albuquerque, New Mexico. The Journal of the American Board of Family Medicine. 2016;29:414–8. doi: 10.3122/jabfm.2016.03.150272. [cited 2021 Jul 6]Available from: http://www.jabfm.org/cgi/doi/10.3122/jabfm.2016.03.150272. [Internet]. [DOI] [PubMed] [Google Scholar]
- 13.Garg A, Toy S, Tripodis Y, Silverstein M, Freeman E. Addressing Social Determinants of Health at Well Child Care Visits: A Cluster RCT. PEDIATRICS. 2015;135:e296–304. doi: 10.1542/peds.2014-2888. [cited 2021 Jul 6]Available from: http://pediatrics.aappublications.org/cgi/doi/10.1542/peds.2014-2888. [Internet]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg A, Boynton-Jarrett R, Dworkin PH. Avoiding the Unintended Consequences of Screening for Social Determinants of Health. JAMA. 2016;316:813. doi: 10.1001/jama.2016.9282. [cited 2021 Jul 6]Available from: http://jama.jamanetwork.com/article.aspx?doi=10.1001/jama.2016.9282. [Internet]. [DOI] [PubMed] [Google Scholar]
- 15.Garg A, Butz AM, Dworkin PH, Lewis RA, Serwint JR. Screening for basic social needs at a medical home for low-income children. Clin Pediatr. 2009;48:32–6. doi: 10.1177/0009922808320602. [DOI] [PubMed] [Google Scholar]
- 16.Garg A, Butz AM, Dworkin PH, Lewis RA, Thompson RE, Serwint JR. Improving the management of family psychosocial problems at low-income children’s well-child care visits: the WE CARE Project. Pediatrics. 2007;120:547–58. doi: 10.1542/peds.2007-0398. [DOI] [PubMed] [Google Scholar]
- 17.Buitron de la Vega P, Losi S, Sprague Martinez L, Bovell-Ammon A, Garg A, James T, et al. Implementing an EHR-based Screening and Referral System to Address Social Determinants of Health in Primary Care. Med Care. 2019;2:S133–9. doi: 10.1097/MLR.0000000000001029. ;57 Suppl 6 Suppl. [DOI] [PubMed] [Google Scholar]
- 18.Byhoff E, Cohen AJ, Hamati MC, Tatko J, Davis MM, Tipirneni R. Screening for Social Determinants of Health in Michigan Health Centers. J Am Board Fam Med. 2017;30:418–27. doi: 10.3122/jabfm.2017.04.170079. [DOI] [PubMed] [Google Scholar]
- 19.Truong HP, Luke AA, Hammond G, Wadhera RK, Reidhead M, Joynt Maddox KE. Utilization of Social Determinants of Health ICD-10 Z-Codes Among Hospitalized Patients in the United States, 2016-2017. Med Care. 2020;58:1037–43. doi: 10.1097/MLR.0000000000001418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Gravity Project. [Internet]. [cited 2021 Aug 26]. Available from: https://confluence.hl7.org/display/GRAV/The+Gravity+Project. (Phila). [Google Scholar]
- 21.Quiñones-Rivera A, Wing HE, Barr-Walker J, Yee M, Harrison JM, Gottlieb LM. Provider Impacts of Socioeconomic Risk Screening and Referral Programs: A Scoping Review. J Am Board Fam Med. 2021;34:820–31. doi: 10.3122/jabfm.2021.04.210039. [DOI] [PubMed] [Google Scholar]
- 22.Phuong J, Zampino E, Dobbins N, Espinoza J, Meeker D, Spratt H, et al. AMIA Annual Symposium. 2021. Extracting Patient-level Social Determinants of Health into the OMOP Common Data Model. [PMC free article] [PubMed] [Google Scholar]
- 23.Winden TJ, Chen ES, Monsen KA, Melton GB. AMIA Summits on Translational Science Proceedings. 2018. Evaluation of Flowsheet Documentation in the Electronic Health Record for Residence, Living Situation, and Living Conditions; pp. 236–45. [PMC free article] [PubMed] [Google Scholar]
- 24.Topaloglu U, Palchuk MB. Using a Federated Network of Real-World Data to Optimize Clinical Trials Operations. JCO Clin Cancer Inform. 2018;2:1–10. doi: 10.1200/CCI.17.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banda JM. Fully connecting the Observational Health Data Science and Informatics (OHDSI) initiative with the world of linked open data. Genomics Inform. 2019;17:e13. doi: 10.5808/GI.2019.17.2.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haendel MA, Chute CG, Bennett TD, Eichmann DA, Guinney J, Kibbe WA, et al. The National COVID Cohort Collaborative (N3C): Rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2021;28:427–43. doi: 10.1093/jamia/ocaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy SN, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, et al. AMIA Annu Symp Proc. 2007. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside; pp. 548–52. [PMC free article] [PubMed] [Google Scholar]
- 28.UCSF Social Interventions Research & Evaluation Network (SIREN) [Internet] 2020. Social Needs Screening Tool Comparison Table. [Internet]. [cited 2021 Mar 10]. Available from: https://sirenetwork.ucsf.edu/SocialNeedsScreeningToolComparisonTable . [Google Scholar]