Abstract
Lack of large quantities of annotated data is a major barrier in developing effective text mining models of biomedical literature. In this study, we explored weak supervision to improve the accuracy of text classification models for assessing methodological transparency of randomized controlled trial (RCT) publications. Specifically, we used Snorkel, a framework to programmatically build training sets, and UMLS-EDA, a data augmentation method that leverages a small number of labeled examples to generate new training instances, and assessed their effect on a BioBERT-based text classification model proposed for the task in previous work. Performance improvements due to weak supervision were limited and were surpassed by gains from hyperparameter tuning. Our analysis suggests that refinements to the weak supervision strategies to better deal with multi-label case could be beneficial. Our code and data are available at https://github.com/kilicogluh/CONSORT-TM/tree/master/weakSupervision.
Introduction
Incomplete reporting and lack of transparency are common problems in biomedical publications and may reduce the credibility of the findings of a study. These problems can have serious consequences, particularly in clinical research publications, since the evidence from these studies inform patient care and healthcare policy. In clinical research, randomized controlled trials (RCTs) are the most robust kind of primary research evidence regarding the effectiveness of therapeutic interventions1 and are a cornerstone of evidence-based medicine2. RCTs are expensive, and if inadequately designed, conducted, or reported, they lead to poor health outcomes and significant research waste3.
Reporting guidelines have been proposed to improve transparency and completeness of reporting for various types of biomedical studies. For example, the CONSORT statement focuses on RCT reporting1,4, and consists of a 25-item checklist and a flow diagram. While endorsed by many high-impact medical journals, adherence to CONSORT remains inadequate1,5 and difficult to enforce in practice, due to substantial workload it involves for journals. Manual CONSORT compliance checks before peer review have been shown to improve reporting quality6; however, they are difficult to scale and require significant domain expertise.
In previous work, we presented a corpus of 50 RCT publications manually annotated at the sentence level with fine-grained CONSORT checklist items and proposed a text mining approach to automate the task of transparency (reporting quality) assessment7. As a first step toward full transparency assessment, we developed sentence classification models to categorize sentences in the Methods sections of RCT publications into 17 methodology-related check-list items (e.g., Eligibility Criteria, Outcomes, Sequence Generation, Allocation Concealment). The best-performing model, based on BioBERT pretrained language model8, yielded reasonable performance on some items, particularly those that are commonly discussed in RCT Methods sections and thus are well-represented in the dataset. However, the results overall suffered from the relatively small size of the dataset and largely failed on the checklist items that are infrequently reported in RCT publications (e.g., Changes to Outcomes).
Annotated data is critical in training modern natural language processing and text mining (NLP) algorithms. In particular, deep neural network architectures heavily depend on large quantities of training data for learning model parameters. While recent pretrained language models, such as BERT9 and its variants, exhibit better sample efficiency and often work well even with relatively small datasets, the importance of annotated data has not diminished. High performance of BERT-based models in NLP tasks and the resulting standardization of architectures arguably underlines data scarcity as the primary bottleneck in NLP10. In response, weak supervision techniques have become increasingly popular, as they offer cheaper or more efficient ways for generating training data10.
In this study, we investigated whether weak supervision techniques can be used to effectively label additional data and improve our sentence classification models for transparency assessment of RCT publications. More specifically, we focused on weak supervision using the Snorkel framework10 and data augmentation based on the UMLS-EDA algorithm11 and used the labels that they generated as additional data for our previously reported BioBERT-based model7. The results show that weak supervision has limited effectiveness on our dataset, while at the same time indicating that hyperparameter tuning can have a more significant impact on model performance.
Related Work
Weak supervision
Weak supervision seeks to use domain knowledge and subject matter expertise in opportunistic ways to assign (some-what noisy) labels to unlabeled data or generate synthetic data. Several general approaches to weak supervision exist. One well-known technique is distant supervision12, based on using domain knowledge in external knowledge bases. While often used for relation extraction12,13, it has also been used for classification tasks applied to RCT publications14,15. For example, risk of bias judgements in the Cochrane database of systematic reviews were used to automatically label sentences in RCT publications and train models for assessing risk of bias in the publications14.
Another related approach is data augmentation, the goal of which is to increase a model’s generalizability by generating realistic data from a limited number of existing examples. First proposed in computer vision research16, it has more recently been adopted in NLP research as well11,17. For example, simple transformations of individual sentences (e.g., synonym replacement, random insertion/deletion) were used to generate additional data and improve modeling accuracy with small datasets11. Similar approaches have been adapted to biomedical domain, for tasks ranging from medical abbreviation recognition18 to named entity recognition19.
Snorkel has been proposed as a general weak supervision framework10. Based on data programming paradigm, Snorkel relies on user-defined labeling functions (LFs), which are heuristic methods that can noisily label large quantities of unlabeled data, learns a generative model over the labeling functions to estimate their accuracy and correlations, and generates probabilistic labels that can be used to train machine learning models. Snorkel has been applied to several biomedical text mining tasks, outperforming distant supervision baselines and approaching manual supervision10. Other weak supervision approaches have also been developed for biomedical NLP tasks, including smoking status classification from clinical notes20, semantic indexing21, and clinical entity classification22.
Text mining on RCT publications
Text mining on RCT literature has mostly focused on annotating and extracting study characteristics relevant for systematic reviews and evidence synthesis23,24. PICO elements received most attention; several corpora have been developed at the sentence and span levels15,25,26, and a variety of traditional and deep machine learning models have been developed to extract these elements from abstracts or full text15,26–28. There is less research on non-PICO elements. Most notably, RobotReviewer14 focuses on risk of bias assessment and classifies RCT publications as high or low risk on several risk categories, including sequence generation and allocation concealment. ExaCT29 identifies 21 elements in clinical trial publications including sample size and drug dosage. Recently, we constructed a corpus of 50 RCT publications (named CONSORT-TM) annotated at the sentence level with 37 fine-grained CONSORT checklist items to assist with transparency assessment7. We also developed baseline NLP models to recognize 17 methodology-specific CONSORT items: two rule-based methods (one keyword-based and another section header-based) as well a linear SVM classifier and a BioBERT-based model. The BioBERT model performed best overall (micro precision: 0.82, recall: 0.63, and F1: 0.72), although it failed to recognize infrequent items, which partly motivated this study.
Materials and Methods
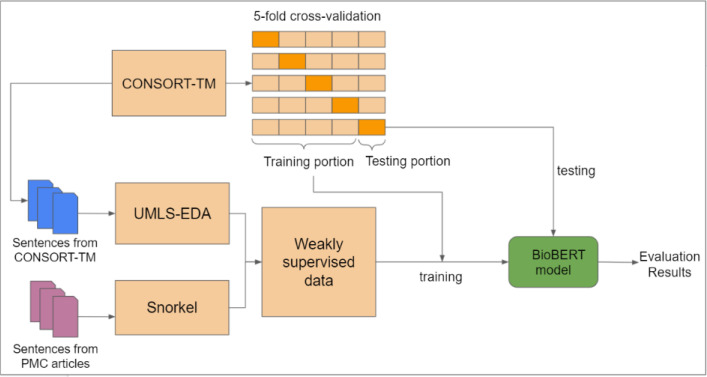
We explored weak supervision to improve the classification performance of our best-performing BioBERT model7. In this section, we first describe the collection and pre-processing of unlabeled RCT data from PubMed Central (PMC) for weak supervision. Second, we provide a brief description of the baseline BioBERT models. Third, we discuss our methodology for generating labels using Snorkel framework as well as the UMLS-EDA algorithm. Lastly, we provide evaluation details. The overall procedure is illustrated in Figure 1.
Figure 1:
Training and evaluation with weakly supervised data.
Data collection and pre-processing
We followed the data collection strategy used in previous work7 to obtain a large set of RCT articles. Cochrane precision-maximizing search queryb was used on 1/15/2021 to search PMC Open Access subset (PMC-OA) for RCT articles published between 1/1/2011 and 12/31/2020c. The results were further limited to articles that have full-text XML in PMC-OA. To get a more reliable RCT subset (since publication types in PubMed can be inaccurate), we filtered the results through RCT Tagger30, a machine learning model that determines whether a publication is a RCT or not. Its accuracy was found to be 99.7% in predicting RCT studies included in Cochrane systematic reviews31. Lastly, we eliminated publications with the word protocol in their title (generally study protocol publications).
We used NCBI e-utilities APId to retrieve publications in XML format, and split them into sentences using our in-house sentence splitter32. Only sentences that belong to Methods section of the publications were taken into account. Stanford CoreNLP package was used for tokenization and part-of-speech tagging33. We eliminated the sentences meeting the following criteria from further consideration, since they are unlikely to indicate CONSORT methodology items: a) contains fewer than five tokens; b) contains numbers only; and c) is a section header or a table/figure caption.
Baseline models
Our best-performing classifier in previous work7 was a BioBERT-based sentence classification model, which uses the BioBERT pretrained language model8 as a sentence encoder, considers the model’s output for the [CLS] token as the sentence representation, and trains a sigmoid layer for multi-label classification of 17 CONSORT methodology items. The input to the model is the raw sentence text prepended with its subsection header. The classifier was implemented using simpletransformers packagee. We refer to this model as BASELINE below.
In this study, we used the huggingfacef BERT implementation. While mostly using the same hyperparameters as BASELINE (batch size: 4, number of epochs: 30, optimizer: Adam, dropout: 0.1), we modified two hyperparameters. First, we used adaptive learning rate instead of a fixed learning rate to optimize the algorithm with different rates based the model performance during training. Second, we set the gradient accumulation steps to 1 (16 for BASELINE), which increases the frequency of model parameter updates. We refer to this optimized model as BASELINE OPT below.
Generating weak labels using Snorkel
Snorkel10 generates weak labels in three steps: a) LF construction; b) creation of a generative model to capture label agreements/disagreements; and c) generation of probabilistic labels for sentences. Input for Snorkel pipeline are unlabeled sentences from RCT publications from PMC-OA.
LFs are expert-defined heuristic rules that can be used to label sentences. For NLP tasks, these can be based on text patterns, syntactic structure, or external knowledge bases. In general, LFs that have high coverage and low overlap are desirable. Such LFs apply to many instances in the dataset yet are unique enough to distinguish instances with different labels. In this study, we used three LF approaches to label CONSORT items: keyword-based, section header-based, and sentence similarity-based. 17 individual LFs were created for each approach (one corresponding to each label).
Keyword-based LFs. These LFs mimic the keyword-based method used in previous work7. Each CONSORT item is associated with a set of keywords or phrases (e.g., power to detect with Sample Size Determination (7a)g). A total of 232 phrases are used. Each LF checks whether an input sentence contains one of its keyphrases, and if so, returns the corresponding label as a weak label (or NO-LABEL, if the sentence does not contain any relevant keyword/phrase).
Section header-based LFs. These LFs also mimic a baseline method from earlier work7. In this case, common subsection headers in Methods sections are associated with CONSORT labels. 48 section header keywords/phrases are mapped to CONSORT items (e.g., the word concealment to the item Allocation Concealment (9)). These LFs check whether the header of the section to which the sentence belongs matches one of the relevant key phrases.
Sentence similarity-based LFs. These LFs assign weak labels to unlabeled sentences based on their similarity to a set of “ground truth” sentences (95 sentences provided as examples for checklist items in the CONSORT Explanation and Elaboration document1 and the CONSORT websiteh). We used BioBERT to generate vector representations of these sentences. Given an unlabeled sentence, we calculate its cosine similarity with every ground truth sentence and consider two labels based on similarity scores: the label of the sentence with the highest similarity and the label that appears most frequently for the top 10 most similar ground truth sentences. If two labels are the same, we use it as the sentence label. Manual checks showed this combination to be more accurate than the most similar sentence label only.
Snorkel applies all LFs to generate a LF matrix that shows the coverage, overlaps, and conflicts between the LFs. Coverage information indicates the fraction of the dataset to which a particular LF is applied. Overlap shows the fraction of dataset where a particular LF and at least one other LF agree. Conflict indicates the fraction of dataset where a particular LF and at least one other LF disagree. Snorkel pools noisy signals from the these three features into a generative model to learn the agreements and disagreements of the LFs, thus assessing the weights of accuracy for each LF. The model then takes into account these accuracies to make a final label prediction for each sentence.
Generating synthetic data using UMLS-EDA
Several CONSORT items are infrequently reported, as they are contingent upon changes in the trial, which may or may not occur (e.g., Changes to Trial Design (3b)). In previous work, text mining methods yielded poor results for these classes7, as may be expected. BASELINE model, although it performed best overall in terms of micro-averaging, yielded no predictions for five labels (out of 17) and less than 0.5 F1 score for 11 items. We do not expect Snorkel to provide significant number of examples for infrequently reported items, since they are also likely to be rare in the unlabeled dataset and Snorkel’s generative model relies on LF agreement, also likely to be uncommon for such labels.
Therefore, we sought to improve the classification performance for such infrequently reported labels using data augmentation. Specifically, we used UMLS-EDA19 and leveraged UMLS34 synonyms to generate sentences that are similar to CONSORT-TM training instances. We define a class as rare if the class frequency in the original dataset (f ) is under a pre-determined threshold (t). In generating instances, we make up the difference between the frequency in the original dataset and the threshold (i.e., t - f instances generated) to make the distribution of the training dataset more uniform. If a class is not rare in the original dataset (i.e., f >= t), no sentences are generated for that label.
UMLS-EDA uses five operations to augment data. Synonym replacement using WordNet randomly chooses n words from the given sentence that are not stopwords and replaces each with a synonym randomly chosen from WordNet. Random insertion inserts random WordNet synonyms of n words in the sentence in random positions. Random swap randomly swaps the position of two words and repeats this n times. Random deletion samples and deletes n words according to a uniform distribution. Synonym replacement using UMLS identifies all the UMLS concepts in the sentence and randomly replaces n words in the sentence with a UMLS synonym, also randomly selected. Operations of the UMLS-EDA data augmentation are illustrated on an example sentence in Table 1. The parameter n is determined dynamically based on the sentence length (l) and the operation type (n = 0.5*l for synonym replacement with UMLS at most and n = 0.2*l for others). While UMLS-EDA aims to generate t-f instances, in most cases, a larger number of instances are generated using these operations and we subsample from the generated instances to reach the threshold.
Table 1:
Example of data augmentation using UMLS-EDA. Bold words indicate modifications made by UMLS-EDA. The label of the original sentence is Eligibility Criteria (4a).
| Operation | Sentence |
|---|---|
| Original | children were excluded if they had impaired fasting glucose, were diabetic, or reported a diagnosed renal, or hepatic disease that might alter body weight. |
| Synonym replacement (WordNet) | children were leave off if they had impaired fast glucose, were diabetic, or account a diagnosed renal, or liverwort disease that mightiness alter body weight. |
| Random insertion | children mightiness were excluded if they had impaired fasting mightiness leave off glucose, were diabetic, or reported a diagnosed child renal, or hepatic disease that might alter body weight. |
| Random swap | children were diagnosed if they had impaired fasting glucose, might excluded or a reported renal, diabetic, hepatic disease that were alter body weight. |
| Random deletion | children were excluded if they had impaired fasting glucose, were diabetic, or reported a diagnosed renal, or hepatic disease that might alter body weight. |
| Synonym replacement (UMLS) | children were excluded if they had impaired fasting glycaemia, were diabetic, or informing a diagnose nephros gastric, or liver disease that might alter body weight. |
Evaluation
To evaluate whether weak supervision generated labels useful for improving sentence classification performance, we compared the results obtained with BASELINE model on the CONSORT-TM dataset using 5-fold cross validation to results obtained when weakly labeled examples from different strategies are added to the training portion of the folds in cross validation (Figure 1). In this setup, data used for validation and testing in each fold remain the same for all the models. As in previous work, we used precision, recall, and their harmonic mean, F1 score, and calculated 95% confidence intervals. In addition to calculating these measures per CONSORT item, we also report micro- and macro-averaged results and the area under ROC curve (AUC).
Results
Weak supervision using Snorkel
Our search strategy retrieved 608K RCTs from PubMed, 155,183 of which have XML full text in PMC. RCT Tagger predicted 71,948 of these as RCTs. Considering only those predicted with a confidence score over 0.95 reduced the dataset to 14,534 publications. Further eliminating publications with protocol in the title, we obtained a set of 11,988 papers. A total of 721,948 sentences from these publications was reduced to 551,936 sentences after filtering.
We processed 551,936 unlabeled sentences using the Snorkel model, which generated 17 probabilities for each sentence. We empirically set a probability threshold of 0.8 to predict the final weak labels for the unlabeled sentences. If no label was predicted with a probability higher than 0.8, no label was assigned. The distribution of weak labels generated by Snorkel are shown in Table 2. Most weak labels corresponded to items that are already relatively well-represented in the dataset; thus, we limited the number of weakly labeled examples for each CONSORT item to a pre-determined threshold in our classification experiments and randomly sampled these examples. We report the results with the threshold that performed best in our experiments (500).
Table 2:
The frequency of each methodology item in CONSORT-TM and the augmented data generated by Snorkel and UMLS-EDA. Numbers in bold correspond to the cases when the CONSORT item was considered rare and augmented for the given threshold t.
| CONSORT Item | Snorkel | UMLS-EDA | |||
|---|---|---|---|---|---|
| Original | t=50 | t=100 | t=200 | ||
| Trial Design (3a) | 3,932 | 55 | 66 | 151 | 312 |
| Changes to Trial Design (3b) | 0 | 10 | 60 | 124 | 249 |
| Eligibility Criteria (4a) | 17,182 | 129 | 132 | 140 | 222 |
| Data Collection Setting (4b) | 740 | 32 | 50 | 110 | 227 |
| Interventions (5) | 11,415 | 199 | 212 | 250 | 336 |
| Outcomes (6a) | 24,104 | 535 | 537 | 551 | 575 |
| Changes to Outcomes (6b) | 0 | 4 | 54 | 109 | 219 |
| Sample Size Determination (7a) | 6,674 | 93 | 93 | 100 | 203 |
| Interim Analyses / Stopping Guidelines (7b) | 124 | 14 | 50 | 100 | 200 |
| Sequence Generation (8a) | 7 | 35 | 70 | 174 | 395 |
| Randomization Type (8b) | 2,915 | 40 | 66 | 165 | 358 |
| Allocation Concealment (9) | 274 | 13 | 55 | 116 | 236 |
| Randomization Implementation (10) | 1,785 | 42 | 71 | 174 | 377 |
| Blinding (11a) | 525 | 47 | 59 | 127 | 266 |
| Similarity of Interventions (11b) | 3 | 15 | 54 | 124 | 255 |
| Statistical Methods for Outcomes (12a) | 45,353 | 215 | 217 | 227 | 258 |
| Statistical Methods for Other Analyses (12b) | 49 | 65 | 65 | 100 | 200 |
| NO LABEL | 436,854 | ||||
Weak supervision using UMLS-EDA
We used thresholds 50, 100, and 200 to generate 246, 844, and 2217 additional examples, respectively, using UMLS-EDA. Data augmentation was implemented as part of 5-fold cross-validation; and therefore, number of examples between folds differ. The numbers of instances for each label in the original dataset and the augmented datasets (for one of the folds) are shown in Table 2. A label can be considered rare or not at different threshold values and may or may not be augmented. For example, while the item Trial Design (3a) is not rare when the threshold is 50, it is considered rare for the threshold 100 and, therefore, augmented (Table 2). The number of rare class instances generally exceed to the threshold slightly, because it is possible to label an augmented example with more than one class.
Classification results
We evaluated BASELINE and BASELINE OPT models on CONSORT-TM using 5-fold cross-validation. In other experiments, we used various sizes of weakly supervised data from Snorkel and UMLS-EDA for additional training. For brevity, we only report the weak supervision results for the best-performing model-data size combinations. For Snorkel, this is BASELINE OPT model augmented with maximum 500 examples per label. For UMLS-EDA, it is the same model augmented with UMLS-EDA data with a threshold of 50. The results are provided in Table 3 (due to space constraints, we provide precision and recall as supplementary material on the project GitHub repository). The results show that hyperparameter tuning (BASELINE OPT) makes a significant difference in performance (7% increase in micro-F1 and 63% in macro-F1), while the impact of weak supervision strategies seems minor; Snorkel data leads to a slight performance degradation, while UMLS-EDA data increases micro-F1 by one percentage point and AUC with 1.6 points, with practically no change in macro-F1.
Table 3:
Classification results using CONSORT-TM and weakly supervised data. SNORKEL(500) uses BASE-LINE OPT with additional 500 instances per label from Snorkel data. UMLS-EDA(50) uses BASELINE OPT with additional instances from UMLS-EDA to add up to at least 50 instances for each label. 3a: Trial Design; 3b: Changes to Trial Design; 4a: Eligibility Criteria; 4b: Data Collection Setting; 5: Interventions; 6a: Outcomes; 6b: Changes to Outcomes; 7a: Sample Size Determination; 7b: Interim Analyses/Stopping Guidelines; 8a: Sequence Generation; 8b: Randomization Type; 9: Allocation Concealment; 10: Randomization Implementation; 11a: Blinding Procedure; 11b: Similarity of Interventions; 12a: Statistical Methods for Outcomes; 12b: Statistical Methods for Other Analyses. P: precision; R: recall; F: F1 score; CI: confidence interval; AUC: Area Under ROC Curve.
| CONSORT Item | BASELINE | BASELINE OPT | SNORKEL(500) | UMLS-EDA(50) | ||||
|---|---|---|---|---|---|---|---|---|
| F1 | [CI] | F1 | [CI] | F1 | [CI] | F1 | [CI] | |
| 3a | 0.63 | [0.46, 0.80] | 0.82 | [0.69, 0.95] | 0.75 | [0.63, 0.88] | 0.78 | [0.72, 0.84] |
| 3b | 0.00 | [0.00, 0 00] | 0.00 | [0.00, 0.00] | 0.00 | [0.00, 0.00] | 0.00 | [0.00, 0.00] |
| 4a | 0.85 | [0.76, 0.95] | 0.89 | [0.82, 0.96] | 0.88 | [0.82, 0.94] | 0.90 | [0.85, 0.95] |
| 4b | 0.36 | [0.06, 0.65] | 0.87 | [0.74, 1.00] | 0.79 | [0.61, 0.97] | 0.81 | [0.68, 0.94] |
| 5 | 0.72 | [0.66, 0.78] | 0.75 | [0.68, 0.81] | 0.73 | [0.66, 0.81] | 0.75 | [0.68, 0.83] |
| 6a | 0.81 | [0.74, 0.88] | 0.82 | [0.75, 0.89] | 0.83 | [0.72, 0.87] | 0.83 | [0.74, 0.91] |
| 6b | 0.00 | [0.00, 0.00] | 0.00 | [0.00, 0.00] | 0.00 | [0.00, 0.00] | 0.00 | [0.00, 0.00] |
| 7a | 0.84 | [0.76, 0.92] | 0.88 | [0.87, 0.90] | 0.90 | [0.86, 0.94] | 0.90 | [0.87, 0.93] |
| 7b | 0.00 | [0.00, 0.00] | 0.70 | [0.47, 0.94] | 0.70 | [0.17, 1.22] | 0.78 | [0.51, 1.05] |
| 8a | 0.38 | [0.15, 0.60] | 0.88 | [0.77, 1.00] | 0.86 | [0.60, 0.91] | 0.88 | [0.79, 0.98] |
| 8b | 0.38 | [0.10, 0.67] | 0.73 | [0.53, 0.93] | 0.67 | [0.51, 0.83] | 0.75 | [0.60, 0.90] |
| 9 | 0.00 | [0.00, 0.00] | 0.45 | [0.35, 0.54] | 0.40 | [0.03, 0.76] | 0.43 | [0.26, 0.60] |
| 10 | 0.24 | [0.05, 0.43] | 0.53 | [0.36, 0.71] | 0.50 | [0.22, 0.77] | 0.52 | [0.32, 0.72] |
| 11a | 0.42 | [0.12, 0.71] | 0.66 | [0.59, 0.74] | 0.59 | [0.46, 0.72] | 0.66 | [0.54, 0.77] |
| 11b | 0.00 | [0.00, 0.00] | 0.45 | [0.06, 0.85] | 0.41 | [0.04, 0.77] | 0.44 | [0.04, 0.84] |
| 12a | 0.75 | [0.69, 0.81] | 0.77 | [0.69, 0.85] | 0.78 | [0.69, 0.87] | 0.78 | [0.72, 0.84] |
| 12b | 0.04 | [-0.06, 0.14] | 0.32 | [0.27, 0.38] | 0.24 | [0.07, 0.40] | 0.31 | [0.26, 0.37] |
| Micro-average | 0.72 | [0.66, 0.76] | 0.77 | [0.71, 0.84] | 0.76 | [0.69, 0.82] | 0.78 | [0.72, 0.83] |
| Macro-average | 0.38 | [0.34, 0.41] | 0.62 | [0.55, 0.69] | 0.58 | [0.48, 0.68] | 0.62 | [0.55, 0.69] |
| AUC | 0.812 | 0.876 | 0.875 | 0.892 | ||||
Discussion
Weak supervision with Snorkel
Approximately 21% of unlabeled sentences were weakly labeled by Snorkel. The number of weak labels reflected to some extent the distribution of labels in the original dataset. Many sentences were weakly labeled with common labels (e.g., Outcomes (6a)). On the other hand, Snorkel failed to weakly label any sentences with the two least frequent labels (Table 2). The quality of Snorkel labels depends largely on the quality of LFs. We used two LFs based on heuristics explored in previous work. Micro-F1 for both methods were found to be around 0.50 in previous work (0.50 for keyword-based and 0.45 for section header-based). More accurate LFs could improve Snorkel results.
To better understand the quality of Snorkel-generated weak labels, we sampled 318 sentences and two authors of this paper (LH and HK) independently labeled the sentences, without access to Snorkel labels. We calculated the agreement of these annotations with Snorkel-generated labels, using Krippendorff’s α with the distance metric MASI35 which accounts for partial agreement in the case of multiple labels. α agreements between Snorkel and each annotator were found to be 0.46 and 0.61, respectively. Inter-annotator agreement was 0.59. Interestingly, agreement between Snorkel and simple majority vote was 0.93. These results suggest that Snorkel may converge to this simple heuristic in some cases, and that it behaves more or less like another annotator in the process.
We found that a large percentage of annotator disagreement with Snorkel came from randomization-related labels (items 8a, 8b, 9, and 10). These items often appear in the same sentence and the clues for them can be overlapping, making it a challenge to label them accurately for both humans and automated methods. In previous work, we found inter-annotator agreement for these items to be somewhat low as well (α=0.62, 0.48, 0.34, 0.35, respectively)7. Snorkel tends to pick a single label for sentences, and this was especially problematic for randomization-related sentences.
Weak supervision using UMLS-EDA
Data augmentation is expected to reduce overfitting and help with model robustness16. While generating data using UMLS-EDA is relatively cheap, the resulting sentences are generally not meaningful, making it difficult to assess the quality of the augmented data (in contrast to Snorkel), aside from the downstream model performance that it produces. We make several observations based on our examination of the augmented data. One of the data augmentation operations (synonym replacement with UMLS) may need to be refined. UMLS synonyms that are used to replace the original words/phrases are sometimes different from the original only in trivial ways (acronyms or swapped tokens), and strategies that only allow more significant replacements could be beneficial. For example, it might be worthwhile to limit the replacement only to terms of particular semantic types or part-of-speech tags. Similar observations were made for synonym replacement with WordNet, as well. Some replacements involved functional words, which may not be as beneficial as replacing content words (nouns, adjectives).
Effect of weak supervision and model hyperparameters on classification performance
We did not observe significant improvements in classification performance due to weakly supervised data. Neither strategy led to any correct predictions for the two least frequent labels (3a, 6a). While this was not unexpected in the case of Snorkel (as no additional examples were labeled with these items), it was more surprising in the case of UMLS-EDA, which seemed to generate sufficient number of examples for these items. We observed AUC improvement with UMLS-EDA (0.892 vs. 0.876 with BASELINE OPT), which may indicate that UMLS-EDA does help with robustness and generalizability. As UMLS-EDA approach is cheap, additional refinements may be promising as a future direction.
Somewhat to our surprise, we found that model hyperparameters made a much more significant difference in model performance. BASELINE OPT model yielded about 7% improvement in micro-F1 and 63% improvement in macro-F1 over the BASELINE model, with improvements in almost all labels. To assess how hyperparameters interacted with weak supervision, we also measured performance when BASELINE model (instead of BASELINE OPT) was trained with weakly supervised data. Using Snorkel for weak supervision in this scenario improved micro-F1 from 0.72 to 0.75, suggesting that hyperparameter optimization may, in some cases, obviate the need for additional (noisy) data.
Limitations
Our investigation was limited to one relatively small corpus. The findings regarding weak supervision (as well as Snorkel and UMLS-EDA specifically) may not be generalizable to other corpora. We used few heuristics with modest performance as LFs and Snorkel label quality is likely to be improved with with additional more accurate LFs; however, this requires significant domain expertise. While we performed some hyperparameter tuning, we did not do an exhaustive search, and it is possible that more optimal hyperparameters can improve results further.
Conclusions and future work
We investigated the impact of two weak supervision strategies on multi-label sentence classification models of RCT publications. We did not observe a clear positive impact of weak supervision on the specific task we studied. More experiments would be needed to determine whether this is a corpus-specific finding or more general. Various forms of weak supervision has been shown to improve classification performance11,15, mostly in multi-class cases; therefore, it is possible that our weak supervision strategies need more refinement for the multi-label case.
In future work, we plan to refine our approach. For example, in UMLS-EDA, we can devise methods to generate more contextually appropriate synonyms from WordNet and UMLS. Snorkel would benefit from more accurate LFs. Other semi-supervised learning approaches (e.g., self-training36, few-shot learning37) can also be investigated as alternatives.
Footnotes
Equal contribution.
The start date is chosen based on the most recent publication of CONSORT guidelines (2010)1.
We use the item numbers used in CONSORT guidelines, as well, hereafter.
Figure & Tables
References
- 1.Moher D, Hopewell S, Schulz KF, Montori V, Gøtzsche PC, Devereaux PJ, et al. CONSORT 2010 Explanation and Elaboration: updated guidelines for reporting parallel group randomised trials. BMJ. 2010;340:c869. doi: 10.1136/bmj.c869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sackett DL, Rosenberg WMC, Gray JAM, Haynes RB, Richardson WS. Evidence based medicine: what it is and what it isn’t. BMJ. 1996;312(7023):71–2. doi: 10.1136/bmj.312.7023.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalmers I, Glasziou P. Avoidable waste in the production and reporting of research evidence. The Lancet. 2009;374(9683):86–9. doi: 10.1016/S0140-6736(09)60329-9. [DOI] [PubMed] [Google Scholar]
- 4.Schulz KF, Altman DG, Moher D. CONSORT 2010 Statement: updated guidelines for reporting parallel group randomised trials. BMJ. 2010;340:c332. doi: 10.1136/bmj.c332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner L, Shamseer L, Altman DG, Schulz KF, Moher D. Does use of the CONSORT Statement impact the completeness of reporting of randomised controlled trials published in medical journals? A Cochrane review. Systematic Reviews. 2012;1(1):60. doi: 10.1186/2046-4053-1-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandis N, Shamseer L, Kokich VG, Fleming PS, Moher D. Active implementation strategy of CONSORT adherence by a dental specialty journal improved randomized clinical trial reporting. Journal of Clinical Epidemiology. 2014;67(9):1044–8. doi: 10.1016/j.jclinepi.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Kilicoglu H, Rosemblat G, Hoang L, Wadhwa S, Peng Z, Malicˇki M, et al. Toward assessing clinical trial publications for reporting transparency. Journal of Biomedical Informatics. 2021;116:103717. doi: 10.1016/j.jbi.2021.103717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–40. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devlin J, Chang MW, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers). Minneapolis, Minnesota: Association for Computational Linguistics; 2019. p. 4171-86.
- 10.Ratner A, Bach SH, Ehrenberg H, Fries J, Wu S, Ré C. Snorkel: rapid training data creation with weak supervision. The VLDB Journal. 2020;29(2):709–30. doi: 10.1007/s00778-019-00552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei J, Zou K. 2019. EDA: Easy data augmentation techniques for boosting performance on text classification tasks. arXiv preprint arXiv:190111196.
- 12.Mintz M, Bills S, Snow R, Jurafsky D. Distant supervision for relation extraction without labeled data. In: Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP; 2009. p. 1003-11.
- 13.Quirk C, Poon H. Distant Supervision for Relation Extraction beyond the Sentence Boundary. In: Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics: Volume 1, Long Papers; 2017. p. 1171-82.
- 14.Marshall IJ, Kuiper J, Wallace BC. RobotReviewer: evaluation of a system for automatically assessing bias in clinical trials. Journal of the American Medical Informatics Association. 2015:193-201. [DOI] [PMC free article] [PubMed]
- 15.Wallace BC, Kuiper J, Sharma A, Zhu M, Marshall IJ. Extracting PICO Sentences from Clinical Trial Reports Using Supervised Distant Supervision. Journal of Machine Learning Research. 2016;17(132):1–25. [PMC free article] [PubMed] [Google Scholar]
- 16.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems; 2012. p. 1097-105.
- 17.Kobayashi S. Contextual Augmentation: Data Augmentation by Words with Paradigmatic Relations. In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 2 (Short Papers). New Orleans, Louisiana: Association for Computational Linguistics; 2018. p. 452-7.
- 18.Skreta M, Arbabi A, Wang J, Brudno M. Training without training data: Improving the generalizability of automated medical abbreviation disambiguation. In: Machine Learning for Health Workshop. PMLR; 2020. p. 233-45.
- 19.Kang T, Perotte A, Tang Y, Ta C, Weng C. UMLS-based data augmentation for natural language processing of clinical research literature. Journal of the American Medical Informatics Association. 2021;28:812–23. doi: 10.1093/jamia/ocaa309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Sohn S, Liu S, Shen F, Wang L, Atkinson EJ, et al. A clinical text classification paradigm using weak supervision and deep representation. BMC medical informatics and decision making. 2019;19(1):1–13. doi: 10.1186/s12911-018-0723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nentidis A, Krithara A, Tsoumakas G, Paliouras G. Beyond MeSH: Fine-grained semantic indexing of biomedical literature based on weak supervision. Information Processing & Management. 2020;57(5):102282. [Google Scholar]
- 22.Fries JA, Steinberg E, Khattar S, Fleming SL, Posada J, Callahan A, et al. Ontology-driven weak supervision for clinical entity classification in electronic health records. Nature communications. 2021;12(1):1–11. doi: 10.1038/s41467-021-22328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Mara-Eves A, Thomas J, McNaught J, Miwa M, Ananiadou S. Using text mining for study identification in systematic reviews: A systematic review of current approaches. Systematic Reviews. 2015;4(1):5. doi: 10.1186/2046-4053-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonnalagadda SR, Goyal P, Huffman MD. Automating data extraction in systematic reviews: a systematic review. Systematic Reviews. 2015;4(1):78. doi: 10.1186/s13643-015-0066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SN, Martinez D, Cavedon L, Yencken L. Automatic classification of sentences to support evidence based medicine. BMC Bioinformatics. 2011;12(2):1–10. doi: 10.1186/1471-2105-12-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nye B, Li JJ, Patel R, Yang Y, Marshall I, Nenkova A, et al. A Corpus with Multi-Level Annotations of Patients, Interventions and Outcomes to Support Language Processing for Medical Literature. In: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Melbourne, Australia: Association for Computational Linguistics; 2018. p. 197-207. [PMC free article] [PubMed]
- 27.Brockmeier AJ, Ju M, Przybyła P, Ananiadou S. Improving reference prioritisation with PICO recognition. BMC Medical Informatics and Decision Making. 2019;19(1):256. doi: 10.1186/s12911-019-0992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin D, Szolovits P. Advancing PICO element detection in biomedical text via deep neural networks. Bioinformatics. 2020;36(12):3856–62. doi: 10.1093/bioinformatics/btaa256. [DOI] [PubMed] [Google Scholar]
- 29.Kiritchenko S, De Bruijn B, Carini S, Martin J, Sim I. ExaCT: automatic extraction of clinical trial characteristics from journal publications. BMC Medical Informatics and Decision Making. 2010;10(1):56. doi: 10.1186/1472-6947-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen AM, Smalheiser NR, McDonagh MS, Yu C, Adams CE, Davis JM, et al. Automated confidence ranked classification of randomized controlled trial articles: an aid to evidence-based medicine. Journal of the American Medical Informatics Association. 2015;22(3):707–17. doi: 10.1093/jamia/ocu025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider J, Hoang L, Kansara Y, Cohen A, Smalheiser NR. 2021. Evaluation of publication type tagging as a strategy to screen randomized controlled trial articles in preparing systematic reviews. JAMIA Open. [DOI] [PMC free article] [PubMed]
- 32.Kilicoglu H, Demner-Fushman D. Bio-SCoRes: A Smorgasbord Architecture for Coreference Resolution in Biomedical Text. PLOS ONE. 2016;11(3):1–38. doi: 10.1371/journal.pone.0148538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manning CD, Surdeanu M, Bauer J, Finkel JR, Bethard S, McClosky D. The Stanford CoreNLP natural language processing toolkit. In: Proceedings of 52nd annual meeting of the association for computational linguistics: system demonstrations; 2014. p. 55-60.
- 34.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004;32(Database issue):267-70. [DOI] [PMC free article] [PubMed]
- 35.Passonneau R. Measuring Agreement on Set-valued Items (MASI) for Semantic and Pragmatic Annotation. In: Proceedings of the Fifth International Conference on Language Resources and Evaluation (LREC’06). Genoa, Italy: European Language Resources Association (ELRA); 2006.
- 36.Xie Q, Luong MT, Hovy E, Le QV. Self-training with Noisy Student improves ImageNet classification. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2020. p. 10687-98.
- 37.Bao Y, Wu M, Chang S, Barzilay R. Few-shot Text Classification with Distributional Signatures. In: International Conference on Learning Representations; 2019.