Abstract
Problem
A significant rate of spontaneous abortion is observed in cattle pregnancies produced by somatic cell nuclear transfer (SCNT). Major histocompatibility complex class I (MHC‐I) proteins are abnormally expressed on the surface of trophoblast cells from SCNT conceptuses.
Method of study
MHC‐I homozygous compatible (n = 9), homozygous incompatible (n = 8), and heterozygous incompatible (n = 5) pregnancies were established by SCNT. Eight control pregnancies were established by artificial insemination. Uterine and trophoblast samples were collected on day 35 ±1 of pregnancy, the expression of immune‐related genes was examined by qPCR, and the expression of trophoblast microRNAs was assessed by sequencing.
Results
Compared to the control group, trophoblast from MHC‐I heterozygous incompatible pregnancies expressed increased levels of CD28, CTLA4, CXCL8, IFNG, IL1A, IL2, IL10, IL12B, TBX21, and TNF, while GNLY expression was downregulated. The MHC‐I homozygous incompatible treatment group expressed increased levels of IFNG, IL1A, and IL2 while the MHC‐I homozygous compatible group did not differentially express any genes compared to the control group. In the endometrium, relative to the control group, MHC‐I heterozygous incompatible pregnancies expressed increased levels of CD28, CTLA4, CXCL8, IFNG, IL10, IL12B, and TNF, while GATA3 expression was downregulated. The MHC‐I homozygous incompatible group expressed decreased amounts of CSF2 transcripts compared with the control group but did not have abnormal expression of any other immune‐related genes. MHC‐I incompatible pregnancies had 40 deregulated miRNAs compared to control pregnancies and 62 deregulated microRNAs compared to MHC‐I compatible pregnancies.
Conclusions
MHC‐I compatibility between the dam and fetus prevented an exacerbated maternal immune response from being mounted against fetal antigens.
Keywords: cattle, cytokines, gene expression, microRNA, miscarriage, pregnancy, somatic cell nuclear transfer
1. INTRODUCTION
Somatic cell nuclear transfer (SCNT) is still a very inefficient process and this has limited the application of this technology in agricultural production and biomedical research. 1 Only 10%–15% of transferred bovine SCNT embryos will develop to term. 2 , 3 , 4 , 5 Although pregnancy rates at 28–32 days of gestation are similar between in vitro and SCNT produced embryos, pregnancy losses are dramatically increased in embryos produced by SCNT. 6 Most of these losses occur in the first trimester of gestation. 7 , 8 Abnormal placentation seems to be the main cause of early SCNT embryonic death. In such cases, a poorly developed placenta with poor vascularization and few placentomes is observed. 6 , 7 The cellular and molecular mechanisms involved in these pregnancy losses and complications remain poorly understood.
The fetus is a semi‐allograft because it contains paternal genetic material. Therefore, maternal tolerance to the allogeneic fetus is essential for fetal development within the maternal uterus in viviparous species. Although there are other causes of pregnancy loss such as chromosomal abnormalities and hormonal imbalances, immunological rejection of the fetus may account for a considerable proportion of first trimester abortions. Immune‐mediated abortions are probably a consequence of breakdown in the mechanisms that normally prevent activation of the maternal immune system against paternal antigens or an unsuccessful induction of maternal tolerance to fetal proteins.
Since classical major histocompatibility complex class I (MHC‐I) proteins are highly polymorphic, one mechanism that promotes maternal tolerance of the conceptus in eutherian mammals is decreased expression of classical MHC‐I proteins by the trophoblast cells of the placenta, particularly during the first trimester of pregnancy. 9 , 10 , 11 , 12 , 13 , 14 , 15 Davies et al. (2000) showed that MHC‐I proteins are not expressed by trophoblast cells in the interplacentomal region of the bovine placenta until the sixth month of pregnancy, while trophoblast cells in the placentomal villous region remain MHC‐I negative throughout gestation. 15 Bovine trophoblast cells express both classical and non‐classical MHC‐I genes, with non‐classical MHC‐I genes accounting for a much greater proportion of MHC‐I transcripts than in peripheral blood mononuclear cells. 16 , 17 Our group 18 and others 7 , 19 have demonstrated that trophoblast cells from bovine SCNT pregnancies express high levels of MHC‐I during the first trimester, which may be the cause of poor fetal survival. In SCNT pregnancies trophoblast cells exhibit increased expression of both classical and non‐classical MHC‐I genes at day 35 of gestation 17 (unpublished data). However, the ratio of classical to non‐classical transcripts in control and SCNT pregnancies appears to be similar, data from day‐35 trophoblast cells showed that greater than 70% of the transcripts present in trophoblast cells from both control and SCNT pregnancies were encoded by classical MHC‐I genes (unpublished data).
Trophoblast MHC expression is also highly regulated in other species of mammals. In humans, trophoblast cells do not express the polymorphic HLA‐A and HLA‐B antigens but invasive, extravillous cytotrophoblast cells express HLA‐C and the non‐classical MHC‐I genes HLA‐E, HLA‐F, and HLA‐G (reviewed by Le Bouteiller 20 ). In mice vacuolated spongiotrophoblast cells express classical MHC‐I antigens H2‐K and H2‐D from day 7.5 of gestation onward but expression patterns for the more than 30, non‐classical MHC‐I genes are unresolved (reviewed by Rutigliano et al. 21 ). There is limited information on MHC‐I expression in the placentas of sheep and goats; one study failed to find evidence of trophoblast MHC‐I expression in samples collected between 9 and 125 days of gestation, while another study documented MHC‐I expression by interplacentomal trophoblast cells at term with increased expression in SCNT pregnancies. 10 , 22 In horses, MHC‐I proteins have not been detected on the non‐invasive trophoblast cells of the allantochorion; however, the invasive trophoblast cells that form the chorionic girdle express classical MHC‐I proteins between days 30 and 45 of pregnancy (reviewed by Rutigliano et al. 21 ).
Most pregnancies in outbred species are MHC mismatched and the vast majority of mismatched conceptuses are accepted. Key elements that influence acceptance or rejection of the conceptus appear to be: (1) the timing of MHC‐I expression, (2) the specific population(s) of trophoblast cells that are expressing MHC‐I, (3) the anatomic location of the MHC‐I expression, (4) the level of MHC‐I expression, and (5) the balance between expression of classical and non‐classical MHC‐I genes. In cattle, trophoblast MHC‐I protein expression late in pregnancy is necessary for maternal immunological recognition and placental expulsion at parturition. 23 , 24 Nevertheless, expression of classical MHC‐I proteins early in pregnancy can cause activation of the maternal immune system and, consequently, a miscarriage. 25
Our group has determined that in SCNT generated pregnancies, MHC‐I compatibility between the dam and the fetus helps prevent a strong immune response against the developing fetus and, consequently, improves embryonic survival. In an earlier paper from the study that we are reporting on here, we investigated MHC‐I expression and endometrial leukocyte populations using immunohistochemistry. 18 The data in our previous paper showed that at day 35 of gestation, MHC‐I heterozygous incompatible SCNT pregnancies had a significantly lower survival rate, a much higher level of trophoblast MHC‐I expression, significantly greater numbers of CD3+, CD4+, CD8+, γ/δ‐TCR+, MHC‐II+, FoxP3+, and NCR1+ leukocytes, and increased amounts of immunoreactive IL12 and TNF in the endometrium, in comparison to control pregnancies established by artificial insemination. In contrast, MHC‐I compatible SCNT pregnancies had only a slightly higher level of MHC‐I expression, and mildly but significantly elevated numbers of CD3+, γ/δ‐TCR+, and MHC‐II+ leukocytes in comparison to control pregnancies. An intriguing finding was that the number of γ/δ‐TCR+ lymphocytes, which co‐express CD3 and probably act as regulatory T cells, was highest in the MHC‐I compatible SCNT pregnancies. The immunohistochemistry data supports increased infiltration of helper and cytotoxic T lymphocytes, natural killer cells, and macrophages in the endometrium of MHC‐I heterozygous incompatible SCNT pregnancies compared to control and MHC‐I compatible SCNT pregnancies. The data strongly suggest that the MHC‐I heterozygous incompatible SCNT conceptuses triggered an exacerbated endometrial immune response that caused their demise. Moreover, the data support our hypothesis that in SCNT pregnancies a positive feedback loop develops where abnormal expression of classical MHC‐I antigens by trophoblast cells triggers a helper T cell mediated endometrial inflammatory response that results in secretion of interferon‐γ (IFNG), which induces upregulation of trophoblast MHC‐I expression.
In the second part of the study, which we are reporting on here, we investigated the transcriptome profile of immune related genes in trophoblast and endometrial tissues collected from cows at 35 days of gestation. We also evaluated the trophoblast miRNA profiles of these pregnancies. This study was based on the hypothesis that in SCNT pregnancies MHC‐I compatibility between the fetus and dam will prevent a maternal immune response to fetal MHC‐I proteins, which is detrimental for trophoblast development and attachment. The goal was to determine if generation of MHC‐I compatible pregnancies would circumvent the uterine inflammatory responses observed in a high proportion of SCNT pregnancies.
2. MATERIALS AND METHODS
2.1. Animals and tissue collection
The Institutional Animal Care and Use Committee at Utah State University approved all procedures used in this study (USU‐IACUC#1171). Recipient females were black Angus cows from the J.R. Simplot Company located in Boise, ID. In order to test the hypothesis that MHC‐I incompatible pregnancies would have increased expression of pro‐inflammatory mediators compared with MHC‐I compatible and control pregnancies, MHC‐I homozygous compatible (n = 9), homozygous incompatible (n = 8), and heterozygous incompatible (n = 5) pregnancies were produced by SCNT as described previously. 26 For establishment of these pregnancies, cattle and SCNT donor cells were typed for MHC‐I and MHC‐II genes by massively parallel pyrosequencing using a Roche 454 Genome Sequencer FLX (Roche Diagnostics, Bradford, CT), as previously described. 24 Typing data from surrogate dams (embryo transfer recipients) and SCNT donor cell lines were used to identify MHC‐I homozygous and heterozygous SCNT donor cell lines as well as to establish pregnancies where the conceptus and surrogate dam were MHC‐I homozygous compatible, MHC‐I homozygous incompatible, and MHC‐I heterozygous incompatible. 18 Major histocompatibility class I compatible pregnancies were pregnancies where the surrogate dam expressed all the MHC‐I isoforms expressed by the conceptus, and MHC‐I incompatible pregnancies were pregnancies where some or all of the MHC‐I isoforms expressed by the conceptus would be recognized as foreign antigens by the immune system of the mother.
Ovaries were collected at a local abattoir for oocyte isolation. Fibroblast cells from Holstein cows at the USU dairy were used as nuclear donor cells. A single, day‐7 embryo was transferred to each of 79 recipient cows synchronized ± 1 day to the stage of the embryos. Control pregnancies (n = 8) were established by artificial insemination. Twenty‐one days after embryo transfer (day 28 of pregnancy), pregnancy was diagnosed by transrectal ultrasonography. Animals were considered pregnant if the presence of a conceptus with a heartbeat was detected. Pregnant animals were humanely sacrificed at day 35 ± 1 of gestation at the Utah State University, USDA‐inspected Meats Laboratory. Immediately after the cows were killed, reproductive tracts were collected, and endometrial and chorionic tissues from the horn ipsilateral to the pregnancy were harvested. Samples for quantitative RT‐PCR analysis and miRNA sequencing were snap frozen in liquid nitrogen and stored at −80°C.
2.2. Quantification of mRNA expression
The RNA isolation, pre‐amplification step and quantitative RT‐PCR procedures used in this study are described in detail elsewhere. 22 Briefly, total RNA was isolated using the TRIzol Plus Purification System (Thermo Fisher Scientific, Waltham, NY, USA). Concentration of total RNA was determined by Nanodrop 1000 spectrophotometry (Thermo Scientific, Waltham, MA, USA). Five micrograms of RNA were used to produce complementary DNA using the SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific, Waltham, NY, USA) according to the manufacturer's instructions.
Eva Green high‐throughput nanoliter volume microfluidic chip quantitative RT‐PCR (96.96 Dynamic Array; Fluidigm, South San Francisco, CA) was used to determine the level of gene expression of the following genes: interleukin (IL)1A, IL2, IL4, IL5, IL6, IL10, IL12B, IL13, IL15, IL17, IL18, IL23A, chemokine (C‐X‐C motif) ligand 8 (CXCL8), IFNG, tumor necrosis factor‐α (TNF), transforming growth factor‐β1 (TGFB1), colony stimulating factor 2 (CSF2), forkhead box P3 (FOXP3), T‐box 21 (TBX21), GATA binding protein 3 (GATA3), interleukin 2 receptor alpha (IL2RA), cluster of differentiation 28 (CD28), and cytotoxic T‐lymphocyte‐associated protein 4 (CTLA4). Glyceraldehyde‐3‐phosphate‐dehydrogenase (GAPDH) and β‐actin (ACTB) were used as reference genes. Four replicates of all samples were run on the same chip. Primer details are shown in Table 1. Using this panel of cytokine, transcription factors, and CD marker primers, it is possible to identify Th1, Th2, and T regulatory responses.
TABLE 1.
Primers used for quantitative reverse transcription polymerase chain reaction (RT‐PCR)
| Gene | GenBank accession no. | Primer sequence (FP, forward; RP, reverse) |
|---|---|---|
| ACTB | AY141970 | FP: GGCCGAGCGGAAATCG |
| RP: GCCATCTCCTGCTCGAAGTC | ||
| GAPDH | U85042, AJ000039, AF022183, J04038 | FP: GAGAAGGCTGGGGCTCACTT |
| RP: GCTGACAATCTTGAGGGTGTTG | ||
| CD28 | X93304 | FP: GGAGGTCTGTGCTGTGAATGG |
| RP: CGGTGCAGTTGAATTCCTTATTT | ||
| CSF2 | U22385 | FP: CAGAAGTGGAAGCTTACCTCACAGA |
| RP: CCTCCAGTGTGAAGATCCTGAGTT | ||
| CTLA4 | X93305 | FP: GCAGCCAGGTGACCGAAGT |
| RP: TCATCCAGGAAGGTTAGCTCATC | ||
| CXCL8 | AF232704, S74436 | FP: GGAAAAGTGGGTGCAGAAGGT |
| RP: GGTGGTTTTTTCTTTTTCATGGA | ||
| FOXP3 | DQ322170 | FP: AAGAGCCCAGGGACAACTTTC |
| RP: GGGTTCAAGGAGGAAGAGGAA | ||
| GATA3 | XM581415, XM864421, XM872964, XM873167, XM873270, XM873370 | FP: CCGTGGTGTCTGTGTTCTCACT |
| RP: TCAATAGGGAATGTGAGTCTGAATG | ||
| GNLY | AY245798 | FP: GACAAGTTGGGAGATCAGCCC |
| RP: ACCTACTGGCTTGCTTTTGCA | ||
| IFNG | M29867, Z54144 | FP: GATAACCAGGTCATTCAAAGGAGC |
| RP: GATCATCCACCGGAATTTGAATC | ||
| IL1A | M37211 | FP: GCCTTCAATAACTGTGGAACCAAT |
| RP: GTATATTTCAGGCTTGGTGAAAGGA | ||
| IL2 | M12791, M13204, X17201 | FP: GCTGGATTTACAGTTGCTTTTGGAG |
| RP: GATGTTTCAATTCTGTAGCGTTAACC | ||
| IL2RA | NM174358 | FP: GCAGGGACCACAAATTTCCA |
| RP: GTACTCAGTGGTAAATATGAACGTATCC | ||
| IL4 | M77120, U14131, U14159, U14160 | FP: GGCGTATCTACAGGAGCCACAC |
| RP: CAAGAGGTCTTTCAGCGTACTTGT | ||
| IL5 | Z67872 | FP: TGGTGGCAGAGACCTTGACA |
| RP: GAATCATCAAGTTCCCATCACCTA | ||
| IL6 | X57317, X62501 | FP: GGCTCCCATGATTGTGGTAGTT |
| RP: GCCCAGTGGACAGGTTTCTG | ||
| IL10 | U00799 | FP: GAGCAAGGCGGTGGAGAAGG |
| RP: GATGAAGATGTCAAACTCACTCATGG | ||
| IL12B | U11815 | FP: GCTGGGAGTACCCTGACACG |
| RP: GGCTGAGGTTTGGTCCATGAAG | ||
| IL13 | AJ132441 | FP: CAGTGTCATCCAAAGGACCAAG |
| RP: CGGACGTACTCACTGGAAACC | ||
| IL15 | U42433 | FP: GGGCTGTATCAGTGCAAGTCTTC |
| RP: ATTGGGATGAGCATCACTTTCAG | ||
| IL18 | AF124789 | FP: ACTGTTCAGATAATGCACCCCAG |
| RP: GAAACAATTTTGTTCTCACAGGAGAG | ||
| IL23A | XM588269 | FP: CCTCCTTCTCCGTCTCAAGATC |
| RP: CGGAGGTCTGGGTGTCATCCT | ||
| TBX21 | XM583748 | FP: GGACACTGAAGCCCAGTTTTATAAC |
| RP: CCAACCTAACGACATTCTTCCTGT | ||
| TGFB1 | M36271 | FP: CTGAGCCAGAGGCGGACTAC |
| RP: TGCCGTATTCCACCATTAGCA | ||
| TNF | Z48808, Z14137 | FP: TCTACCAGGGAGGAGTCTTCCA |
| RP: GTCCGGCAGGTTGATCTCA |
Gene expression data were analyzed using Fluidigm Real‐Time PCR Analysis software version 3.0.2 (Fluidigm, South San Francisco, CA, USA) to determine relative expression values standardized to expression levels of the control group. Relative gene expression was determined by the 2−ΔΔCt method using the average of the Ct values of the reference genes GAPDH and ACTB for normalization. The values presented in this study are the log2 of the fold change in gene expression for each SCNT group compared with the control group.
2.3. Quantification of miRNA expression
Micro‐RNA sequencing was conducted in the control, homozygous compatible, and heterozygous incompatible treatment groups because these three groups were expected to show the greatest differences. Micro‐RNA was extracted from the frozen trophoblast samples using the miRNeasy Mini Kit (Qiagen, Valencia, CA, USA). A miRNA‐enriched fraction and a total RNA fraction were purified separately. A RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA, USA) was used to cleanup and concentrate miRNA samples. Samples were quantified using both the Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and NanoDrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Purified miRNA samples were kept at –80°C for long‐term storage. Micro‐RNA sequencing was carried out by the USU Genomics Core Laboratory on an Ion Torrent Proton sequencer (Life Technologies, Grand Island, NY, USA) using the Ion Total RNA‐Seq v2 Kit (Life Technologies). Libraries were pooled in groups of four for sequencing using the PI chip, which yielded approximately 7 million reads per sample.
2.4. Quality control, mapping, and differential miRNA expression analysis
Trimming of sequencing reads and adapter sequences was performed with cutadapt v.1.2.1 and reads with less than 15 nt and longer than 27 nt were discarded. 27 Reads with less than a 20 Phred quality score were removed using the fastq_quality_filter function in the FASTX‐Toolkit. 28 Bowtie2 v 2.3.4.2 29 was used to map the raw reads on the Bos Taurus genome downloaded from the Ensembl database (https://uswest.ensembl.org/index.html) allowing no mismatches (v = 0) and up to 200 instances of multi‐mapping (m = 200). Identification of miRNA was done using default parameters of miRDeep2 v3. 30
Differential expression analysis including data quality assessment by sample clustering and visualization was performed with the DESeq2 computer program. 31 Differentially expressed miRNAs were filtered for a fold change > 2 and FDR < .05. Venn diagram and heatmaps were generated using in‐house custom R scripts.
2.5. Target prediction and functional annotation
Targets for all the differentially expressed miRNA were identified using miRanda against Bos taurus gene sequences. We also selected 21 genes from Bos taurus and predicted targets against them separately. Gene Ontology (GO) enrichment and KEGG pathway enrichment analysis were performed for predicted targets using in‐house scripts. GO enrichment and KEGG pathway enrichment figures were created based on enrichment score (−log10[p‐value]).
2.6. Statistical analyses
One‐way analysis of variance (ANOVA) models were used to analyze gene expression. The MIXED procedure of SAS (SAS for Windows, version 9.3, SAS Institute Inc., Cary, NC, USA) was used with treatment, embryonic viability, and their interaction as fixed effects and the random effect of animal nested within treatment. Since embryonic viability and its interactions with treatment were not significant, these factors were removed from the model.
The pdiff option and Tukey's adjustment were used to determine significant differences between treatment groups. A p value equal to or below .05 was considered significant. Differentially expressed miRNAs were considered significant when false discovery rate (FDR) was equal to or lower than .05, and there was a fold change of at least 2.
3. RESULTS
3.1. Gross morphology and embryonic survival
The overall day‐28 embryo transfer pregnancy rate was 29% with similar rates for the MHC‐I homozygous compatible, MHC‐I homozygous incompatible, and MHC‐I heterozygous incompatible groups. The embryonic mortality and placental vascularization data were described previously. 18 Fetal mortality rates between day 28 and 35 of gestation were not statistically different between the control, MHC‐I homozygous compatible, and homozygous incompatible treatment groups (12.5%, 11.1%, 12.5%, respectively). However, in the heterozygous incompatible pregnancies embryonic mortality was 100% and significantly different from the other treatment groups. Embryonic weight and crown‐to‐rump length were not statistically different between SCNT and control embryos. 18
3.2. Trophoblast gene expression
Table 2 summarizes the log2 fold change of expression of the different genes analyzed. The quantitative mRNA expression analyses indicated that no genes were significantly different between trophoblast cells derived from the MHC‐I homozygous compatible and control groups. In contrast, the MHC‐I homozygous incompatible pregnancies had three and the MHC‐I heterozygous incompatible pregnancies had 11 differentially expressed genes compared to the control group.
TABLE 2.
Log2 fold change in expression of trophoblast genes
| Genes | Homozygous compatible | Homozygous incompatible | Heterozygous incompatible |
|---|---|---|---|
| CD28 | .35 ± .38 | .54 ± .32 | 1.90 ± .43* |
| CSF2 | −.47 ± .57 | −.34 ± .35 | .15 ± .46 |
| CTLA4 | .64 ± .69 (a) | .99 ± .90 (a) | 4.74 ± .60* (b) |
| CXCL8 | .07 ± .33 (a) | .70 ± .71 (a) | 4.31 ± 1.66* (b) |
| FOXP3 | .28 ± .29 | .23 ± .17 | .90 ± .54 |
| GATA3 | .71 ± .34 | −.76 ± .45 | .46 ± .83 |
| GNLY | .25 ± .62 (a) | −.54 ± .72 (a) | −2.07 ± .70* (b) |
| IFNG | 1.17 ± .72 (a) | 2.29 ± 1.04* (b) | 6.51 ± .42* (c) |
| IL1A | 1.07 ± .69 (a) | 2.12 ± .85* (b) | 6.00 ± 1.36* (c) |
| IL2 | −.44 ± .55 (a) | 1.40 ± .23* (b) | 1.25 ± .50* (b) |
| IL2RA | −1.25 ± .64 (a) | −.35 ± .39 (a, b) | 1.22 ± .82 (b) |
| IL4 | −.13 ± .31 | −.72 ± .32 | .27 ± .54 |
| IL5 | −1.09 ± 1.1 | −.7 ± .7 | −.3 ± .4 |
| IL6 | .03 ± .48 | .23 ± .35 | .20 ± .59 |
| IL10 | 1.06 ± .78 (a) | 1.16 ± 1.00 (a, b) | 1.69 ± .53* (b) |
| IL12B | .15 ± .40 (a) | .54 ± .38 (a) | 3.18 ± 1.16* (b) |
| IL13 | −.02 ± .30 | −.51 ± .33 | 1.5 ± .50 |
| IL15 | −.02 ± .47 | .37 ± .34 | .82 ± .59 |
| IL18 | .74 ± .28 | .73 ± .20 | 1.07 ± .32 |
| IL23A | −.20 ± .19 | .41 ± .21 | .76 ± .28 |
| TBX21 | −.10 ± .58 (a) | .40 ± .52 (a) | 2.90 ± .83* (b) |
| TGFB1 | −.60 ± .28 | .43 ± .25 | .01 ± .50 |
| TNF | .58 ± .28 (a) | .81 ± .61 (a) | 2.76 ± .60* (b) |
Data are means ± SEM. The data of mRNA expression are normalized to GAPDH and ACTB content and calibrated so that the mean value of the control group equals 1.00. Positive values indicate an increase in transcript abundance; negative values indicate decreased transcript abundance. Different letters within rows indicate a statistical difference between treatment groups. ACTB, β‐actin; GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase; SEM, standard error of mean
Statistically different from control group (p < .05).
The trophoblast cell expression of CTLA4A, CXCL8, GNLY, IL12B, TBX21, and TNF in heterozygous incompatible pregnancies was significantly greater compared to the control, homozygous compatible, and homozygous incompatible treatment groups. IFNG and IL1A gene expression was significantly increased in both the MHC‐I homozygous incompatible and the MHC‐I heterozygous incompatible treatment groups relative to the control and the homozygous compatible groups. CD28 expression was increased in the heterozygous incompatible group compared to the control group.
Interleukin 2 gene expression was upregulated in the heterozygous and homozygous incompatible groups compared to the control and the homozygous compatible treatment groups. Interleukin 10 expression was greater in the MHC‐I heterozygous incompatible compared to the control and the homozygous compatible groups and IL2RA expression was greater in the MHC‐I heterozygous incompatible group compared to the homozygous compatible.
3.3. Endometrial tissue gene expression
The gene expression of immune‐related genes in the endometrium ipsilateral to the pregnancy followed a similar trend as the trophoblast gene expression with pro‐inflammatory mediators being upregulated in the heterozygous incompatible group in comparison with the control group. Table 3 summarizes the log2 fold change of expression of the different genes analyzed. The qPCR data analysis shows that out of 23 immune‐related genes, the endometrial tissue derived from MHC‐I heterozygous incompatible pregnancies had eight abnormally expressed genes compared to the control group, while the homozygous incompatible group had one downregulated gene (CSF2) compared to the control group.
TABLE 3.
Log2 fold change in expression of endometrial genes
| Genes | Homozygous compatible | Homozygous incompatible | Heterozygous incompatible |
|---|---|---|---|
| CD28 | .05 ± .34 (a) | .52 ± .65 (a) | 3.01 ± .53* (b) |
| CSF2 | −1.11 ± .45 (a, b) | −1.63 ± .51* (a) | 1.09 ± .88 (b) |
| CTLA4 | .15 ± .54 (a) | .44 ± .76 (a) | 3.37 ± .65* (b) |
| CXCL8 | .13 ± .32 (a) | .79 ± .30 (a) | 1.91 ± .64* (b) |
| FOXP3 | −.16 ± .20 | −.02 ± .18 | .73 ± .68 |
| GATA3 | −.05 ± .17 (a) | −.17 ± .26 (a) | −1.25 ± .48* (b) |
| GNLY | −.25 ± .32 | −.21 ± .31 | .8 ± .26 |
| IFNG | −.32 ± .28 (a) | .51 ± .52 (a) | 2.82 ± .44* (b) |
| IL1A | −.31 ± .34 | .04 ± .15 | 1.04 ± .54 |
| IL2 | −.25 ± .26 | −.06 ± .28 | 1.07 ± .41 |
| IL2RA | .57 ± .22 | .47 ± .26 | .86 ± .22 |
| IL4 | .64 ± .27 | .41 ± .30 | 1.17 ± .50 |
| IL5 | −.14 ± .31 | −.38 ± .32 | −.82 ± .54 |
| IL6 | −.67 ± .41 | −1.16 ± .27 | −.81 ± .58 |
| IL10 | .22 ± .20 (a) | .43 ± .37 (a) | 2.21 ± .58* (b) |
| IL12B | .18 ± .54 (a) | 1.33 ± .68 (a) | 4.17 ± .33* (b) |
| IL13 | −.45 ± .22 | −.61 ± .14 | −.11 ± .49 |
| IL15 | −.17 ± .14 | −.21 ± .06 | .1 ± .24 |
| IL18 | −.01 ± .11 | .37 ± .28 | 1.09 ± .25 |
| IL23A | .30 ± .14 | .11 ± .16 | .33 ± .27 |
| TBX21 | −1.07 ± .59 | −.79 ± .59 | −.04 ± .65 |
| TGFB1 | −.31 ± .14 | −.35 ± .16 | .50 ± .54 |
| TNF | .11 ± .18 (a) | .21 ± .25 (a) | 1.76 ± .51* (b) |
Data are means ± SEM. The data of mRNA expression are normalized to GAPDH and ACTB content and calibrated so that the mean value of the control group equals 1.00. Positive values indicate an increase in transcript abundance; negative values indicate decreased transcript abundance. Different letters within rows indicate a statistical difference between treatment groups. ACTB, β‐actin; GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase; SEM, standard error of mean
Statistically different from control group (p < .05).
Compared to the control and homozygous groups, endometrial tissue collected from the MHC‐I heterozygous incompatible pregnancies had significantly greater levels of expression of CD28, CTLA4, CXCL8, GATA3, IFNG, IL10, IL12B, and TNF. CSF2 expression was significantly downregulated in the homozygous incompatible pregnancies compared to the control and heterozygous incompatible treatment groups. The expression of GATA3 was downregulated in heterozygous incompatible compared to control and homozygous treatment groups. The intra‐assay coefficient of variation (CV) ranged between .3% and 1.3% while the inter‐assay CV ranged between .3% and 1.5% (Supporting Information Table S1).
3.4. Trophoblast miRNA expression
As shown in Figure 1, a total of 40 trophoblast miRNAs were deregulated in the MHC‐I heterozygous incompatible group compared to the control group. Twenty miRNAs were upregulated and 20 miRNAs were downregulated. Similarly, a total of 62 miRNAs were differentially expressed in the MHC‐I heterozygous incompatible compared to MHC‐I homozygous compatible pregnancies. Of these, 30 miRNAs were upregulated and 32 miRNAs were downregulated. Eighteen up‐ and 18 downregulated miRNAs were shared between the MHC‐I heterozygous incompatible to compatible and the MHC‐I heterozygous incompatible to control group comparisons. When MHC‐I homozygous compatible and control pregnancies were compared, no miRNAs were differentially expressed.
FIGURE 1.
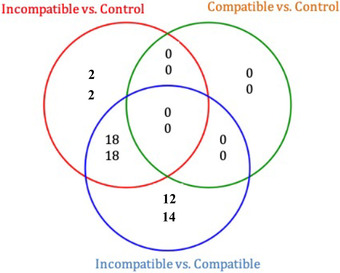
Proportional Venn diagram depicting the number of up‐ (top) and down‐ (bottom) regulated trophoblast miRNAs in day 35 ± 1 pregnancies where fetal and maternal major histocompatibility complex class I (MHC‐I) haplotypes were matched (homozygous compatible, n = 9) and unmatched (heterozygous incompatible; n = 5). Control pregnancies were established by artificial insemination (n = 8)
Among the miRNAs with the most significant upregulation in MHC‐I heterozygous incompatible compared to control pregnancies were miR‐2887, miR‐1246, miR‐2411, miR‐184, and miR‐2904 (Figure 2A). The expression of each of these miRNAs was at least eight times (log2‐fold change ≥ 3) greater in the MHC‐I incompatible pregnancies compared to the control pregnancies. Among the most significantly downregulated miRNAs in MHC‐I heterozygous incompatible compared to control trophoblasts were miR‐133, miR‐99, miR‐451, and miR‐592 (Figure 2A). The expression level of each of these miRNAs was downregulated four‐fold (log2‐fold change ≥ 2) in the MHC‐I incompatible compared to the control pregnancies. Interestingly, all these miRNAs were also among the most deregulated in the MHC‐I incompatible group when compared to the MHC‐I compatible group (Figure 2B).
FIGURE 2.
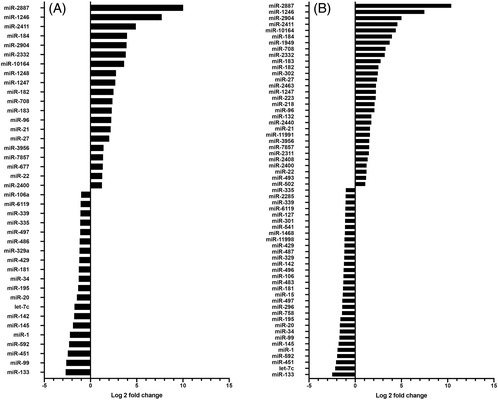
Log2 fold change of differentially expressed miRNAs in trophoblasts of MHC‐I heterozygous incompatible (n = 5) SCNT pregnancies compared to control pregnancies established by artificial insemination (n = 8) (A); and between MHC‐I heterozygous incompatible (n = 5) and MHC‐I homozygous compatible (n = 9) SCNT pregnancies (B) at gestation day 35 ± 1. MHC‐I, major histocompatibility complex class I; SCNT, somatic cell nuclear transfer
Figure 3 shows the 10 most significant GO terms of differentially expressed miRNAs in each category. The GO analysis results divide terms into three categories: molecular function, cellular component, and biological process. When determining the target genes of downregulated miRNAs in trophoblast samples of MHC‐I heterozygous incompatible compared with control pregnancies, the most significant molecular functions were calcium ion binding, GTPase binding, and anion binding. The most significant biological processes were movement of cell or subcellular components, microtubule‐based processes, and supramolecular fiber organization (Figure 3A). For the upregulated miRNAs the main molecular functions were microtubule motor activity, motor activity, and cytoskeletal protein binding; the most significant biological processes were nervous system development, neurogenesis, cell differentiation, and animal organ development (Figure 3B).
FIGURE 3.
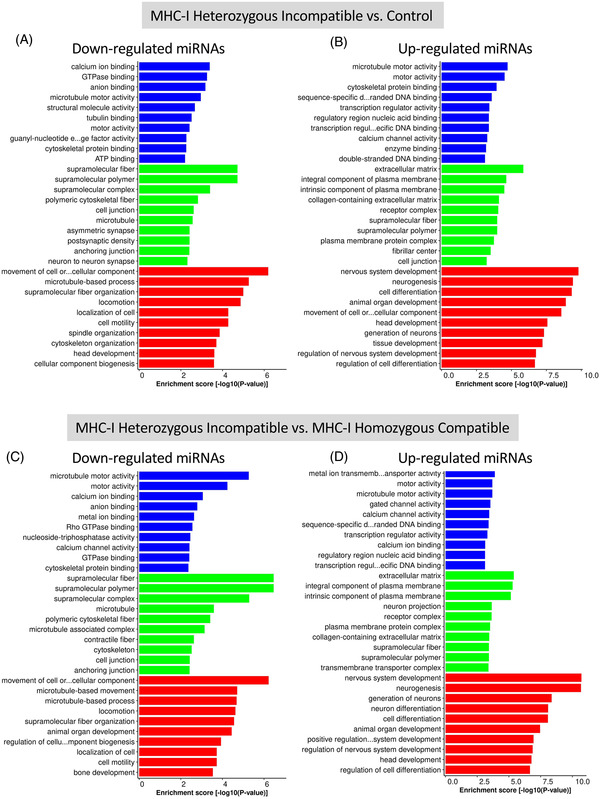
Results of GO analysis of predicted target genes for downregulated miRNAs (A) and upregulated miRNAs (B) between MHC‐I heterozygous incompatible (n = 5) and control trophoblast (n = 8), and predicted target genes for downregulated miRNAs (C) and upregulated miRNAs (D) between MHC‐I heterozygous incompatible (n = 5) and MHC‐I homozygous compatible (n = 9) trophoblast. Blue bars indicate molecular function, green bars indicate cellular components, and red bars indicate biological process. Day 35 ± 1 control pregnancies were established by artificial insemination and MHC‐I heterozygous incompatible and homozygous compatible pregnancies were established by somatic cell nuclear transfer. Enrichment score [−log10(p‐value)] is a measure of how strongly the gene is associated with a specific biological process. GO, Gene Ontology; MHC‐I, major histocompatibility complex class I
In the downregulated miRNAs in MHC‐I heterozygous incompatible compared with the MHC‐I homozygous compatible group, the most important molecular functions were microtubule motor activity, motor activity, and calcium ion binding and the most significant biological processes were movement of cell or subcellular components, microtubule‐based movement, and microtubule‐based process (Figure 3C). In the upregulated miRNAs, the most important molecular functions were metal ion transmembrane transporter activity, motor activity, and microtubule motor activity, while the most significant biological processes were nervous system development, neurogenesis, and generation of neurons (Figure 3D).
KEGG pathway analysis of predicted target genes of downregulated miRNAs in MHC‐I heterozygous incompatible compared with the control group show involvement with N‐glycan biosynthesis, and homologous recombination (Figure 4A). Similarly, for the downregulated miRNAs in the MHC‐I heterozygous incompatible and MHC‐I homozygous compatible group comparison the most significant pathways were N‐glycan biosynthesis, extracellular matrix‐receptor interaction, and homologous recombination (Figure 4C). The most significant pathways associated with upregulated miRNAs in the MHC‐I heterozygous incompatible compared with either the control or MHC‐I compatible pregnancies were axon guidance, mineral absorption, and carbohydrate digestion and absorption (Figure 4B and D). In addition, the upregulated miRNAs were associated with several endocrine pathways.
FIGURE 4.
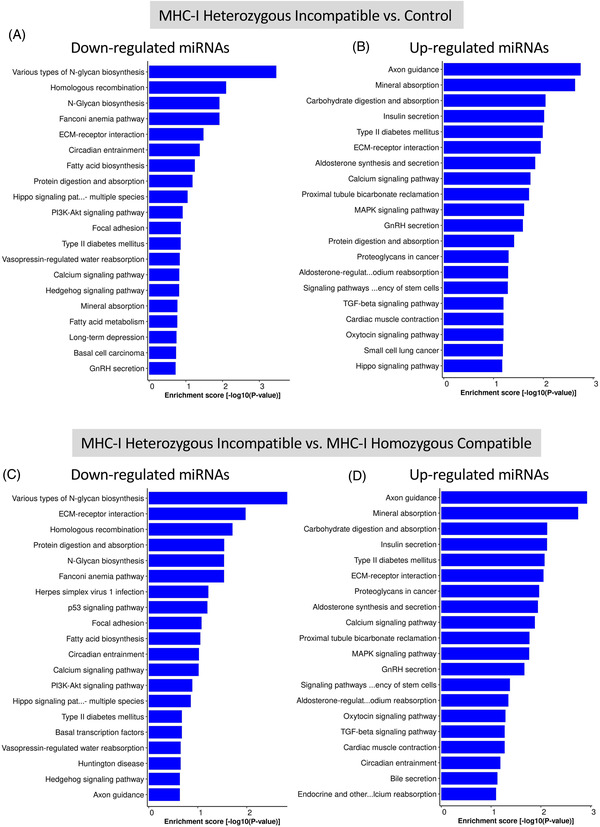
Pathway analysis by KEGG of predicted target genes of downregulated (A) and upregulated (B) miRNAs in bovine trophoblast of MHC‐I heterozygous incompatible (n = 5) compared with control (n = 8) pregnancies, and predicted target genes of downregulated (C) and upregulated (D) miRNAs in bovine trophoblast of MHC‐I heterozygous incompatible (n = 5) compared with MHC‐I homozygous compatible (n = 9) pregnancies at day 35 of gestation. Control pregnancies were established by artificial insemination and MHC‐I heterozygous incompatible and homozygous compatible pregnancies were established by somatic cell nuclear transfer. Enrichment score [−log10(p‐value)] is a measure of how strongly the gene is associated with a specific biological process. KEGG, Kyoto Encyclopedia of Genes and Genomes; MHC‐I, major histocompatibility complex class I
When determining differentially expressed trophoblast miRNAs that could be involved in the regulation of expression of the 23 inflammatory genes assessed in this study by qPCR, it was noticeable that seven miRNAs predicted to regulate three critical genes involved in immune responses were downregulated and two were upregulated. For instance, miR‐1248 was upregulated in MHC‐I heterozygous incompatible compared to control pregnancies and miR‐339 was downregulated in MHC‐I heterozygous incompatible compared to both control and MHC‐I homozygous compatible trophoblasts. In our target gene analysis, miR‐1248 and miR‐339 were predicted to target FOXP3, a transcription factor expressed in regulatory T cells. For IFNG associated miRNAs, miR‐181, miR‐592, and miR‐195 were downregulated in MHC‐I heterozygous incompatible compared to control and MHC‐I homozygous compatible trophoblasts. MiR‐11998, which is predicted to target both IFNG and TNF, was downregulated in MHC‐I heterozygous incompatible compared to MHC‐I homozygous compatible trophoblasts. Additionally, miR‐296, another miRNA predicted to target TNF, was downregulated in MHC‐I heterozygous incompatible compared to MHC‐I homozygous compatible trophoblasts (Table 4).
TABLE 4.
Up‐ and downregulated miRNAs with predicted target genes among the 23 inflammatory genes investigated
| miRNA | Target gene | Target name | Heterozygous incompatible vs. control | Heterozygous incompatible vs. homozygous compatible |
|---|---|---|---|---|
| miR‐1248 | DQ322170.1 | FOXP3 | ↑ | – |
| miR‐339 | DQ322170.1 | FOXP3 | ↓ | ↓ |
| miR‐181 | Z54144.2 | IFNG | ↓ | ↓ |
| miR‐302 | Z54144.2 | IFNG | – | ↑ |
| miR‐592 | Z54144.2 | IFNG | ↓ | ↓ |
| miR‐11998 | Z54144.2 | IFNG | – | ↓ |
| miR‐195 | Z54144.2 | IFNG | ↓ | ↓ |
| miR‐11998 | Z14137.1 | TNF | – | ↓ |
| miR‐296 | Z14137.1 | TNF | – | ↓ |
↓ Indicates downregulation of the miRNA. ↑ Indicates upregulation of the miRNA. – Indicates no change in miRNA expression.
4. DISCUSSION
Although the birth of the first live animal produced by SCNT was reported many years ago, 3 this technique is still very inefficient for producing healthy animals. 2 , 3 , 4 , 5 For instance, in cattle 50%–100% of SCNT generated pregnancies are lost between days 30 and 90. 6 Abnormal placentation appears to be the main cause of these embryonic losses. 6 , 7 , 32
Successful establishment of pregnancy requires normal placental development and an altered maternal immune system that is tolerant to fetal/paternal antigens. One of the mechanisms used by the fetus to prevent an attack from the maternal immune system is the downregulation of trophoblast cell MHC‐I protein expression. In bovine SCNT pregnancies, however, these proteins are expressed on the surface of trophoblast cells as early as day‐35 of gestation. 18 , 19 , 25 In a previous report, we demonstrated that MHC‐I compatibility between an SCNT fetus and dam reduces endometrial lymphocytic infiltration and increases embryonic survival rates compared to MHC‐I incompatible pregnancies. In the MHC‐I incompatible pregnancies, a pronounced T lymphocyte infiltration was present in the endometrium. These lymphocytes were mainly composed by CD4+ T helper (Th) cells, which suggests that the maternal immune system was indirectly recognizing MHC‐I proteins expressed by the trophoblast cells. 18 Here, we show that MHC‐I incompatibility between the dam and the fetus triggers a pronounced pro‐inflammatory cytokine response.
Subpopulations of CD4+ Th cells are classified as Th1 or Th2 cells depending on what cytokines they express. Th1 cells develop in response to IL‐12 and express IFNG, IL2, TNF, and CSF2, which activate macrophages and T cells to promote inflammation and cellular immunity; while Th2 cells produce IL4, IL5, IL6, and IL10, which stimulate antibody production by B cells. 33 The shift from a Th1 to a Th2 response is considered an important factor in the maintenance of pregnancy in humans and mice. 34 , 35 , 36 The cytokine microenvironment dictates the differentiation of naïve Th lymphocytes (Th0) to either a Th1 or a Th2 phenotype. For instance, in the uterus of a non‐pregnant woman there is a balance between Th1 and Th2 responses. 37 In normal pregnancies, this balance is shifted toward a Th2 type response because of the presence of progesterone and placental cytokines. 38 It has been suggested that in cases of pre‐eclampsia this shift does not occur and Th1 responses are not suppressed. 35 , 39 In abortion‐susceptible mouse models, fetal loss has been associated with the expression of Th1 cytokines and deficient expression of Th2 cytokines, 40 , 41 and decidual T cells appear to be defective in producing Th2 cytokines. 42 Additionally, there is strong evidence that a Th1 response is associated with recurrent miscarriages in humans. 43 , 44 Although Th1 cytokines such as IFNG, TNF, and IL2 contribute to placental toxicity and damage, directly or indirectly through the activation of other immune cells, 43 , 45 the production of Th2 cytokines, mainly IL4, IL5, IL6, IL10, and IL13, promotes the growth of trophoblast cells and may be advantageous for the maintenance of pregnancy. 34 , 46 In the present study, a Th1 biased response was observed in the heterozygous incompatible pregnancies where the gene expression of IL2, IL12B, IFNG, and TNF was increased. This Th1 response likely promoted further Th1 lymphocyte differentiation and macrophage activation which contributed to decreased embryonic viability in the MHC‐I heterozygous incompatible group.
Interleukin 10 is produced by macrophages, which are activated by Th1 cytokines, therefore, most likely the increase in IL10 expression observed in the MHC‐I heterozygous incompatible pregnancies was due to increased numbers of macrophages recruited to the maternal–fetal interface in these pregnancies. We have previously demonstrated that the MHC‐I heterozygous incompatible pregnancies present increased numbers of CD68+ macrophages in the bovine placenta in comparison to the MHC‐I homozygous and control groups. 18 Macrophages are recruited to tissues to clean up cell debris and they are also involved in tissue remodeling. In the MHC‐I heterozygous incompatible pregnancies, it is likely that these macrophages are there to remove cell debris from the dying fetuses and placental tissue. It has been proposed that the uptake of trophoblast cell debris from healthy pregnancies decreases the secretion of pro‐inflammatory cytokines like IFNG and TNF and promotes the secretion of anti‐inflammatory cytokines. 47
TBX21 (also known as Tbet) is a transcription factor that when expressed by Th0 cells enables lineage commitment to Th1 cells. Conversely, GATA3 enables commitment of Th0 cells to Th2 cells. An increase in expression of Th1 differentiation marker TBX21 and a decrease in expression of the Th2 differentiation marker, GATA3, indicates the Th1/Th2 ratio is biased to Th1 dominance (reviewed by Zhu et al 48 ). In the present study, we did not observe any changes in GATA3 expression in trophoblastic tissue but GATA3 expression levels were significantly lower in the endometrial tissue of MHC‐I heterozygous incompatible pregnancies compared to the control group and other SCNT groups. TBX21 expression was increased in MHC‐I incompatible trophoblasts compared to the control group. Together the increased trophoblast expression of TBX21 and the decreased endometrial expression of GATA3 in the MHC‐I heterozygous incompatible group are consistent with the Th1 biased cytokine expression profile at the fetal–maternal interface observed in this study.
We observed that CSF2 was downregulated in the endometrium of homozygous incompatible pregnancies. CSF2 inhibits apoptosis, facilitates glucose uptake, and improves the survival of embryonic cells. 49 , 50 Additionally, in cattle, 51 , 52 humans, 53 mice, 49 , 54 and pigs, 55 the addition of CSF2 to embryo culture media has been shown to enhance blastocyst development and increases implantation success. In a study we conducted where term placentas were collected from SCNT and natural breeding‐derived sheep and goat pregnancies, CSF2 expression was increased in goat SCNT cotyledonary tissue. In addition, goat SCNT pregnancies had rates of pregnancy loss and immune related‐gene expression profiles similar to what was observed in goat pregnancies established by natural breeding. 22 In this study, however, endometrial CSF2 expression was downregulated in the “healthier” SCNT pregnancies, MHC‐I homozygous incompatible pregnancies, and numerically downregulated in the MHC‐I homozygous compatible pregnancies. Decreased expression of CSF2, which is a potent proinflammatory factor, is likely due to decreased expression by Th1 lymphocytes and macrophages in the endometrium of healthy MHC‐I homozygous pregnancies. 56
The expression of pro‐inflammatory mediators was increased in the MHC‐I heterozygous incompatible group compared with the other groups. Because all embryos in the MHC‐I heterozygous incompatible treatment were dead, one might think that the increased expression of these cytokines is a consequence, and not the cause, of fetal demise. However, data from our previous study 18 and others 19 demonstrate that the presence of dead embryos in control pregnancies was not associated with increased lymphocytic infiltration in the bovine endometrium. In addition, the death of the concepti in this study likely happened shortly before sample collection since fetal weights and crown‐to‐rump lengths of dead and live embryos were similar. Therefore, our data suggest that the increased expression of these inflammatory mediators is a cause, and not a consequence, of embryonic demise.
Our trophoblast miRNA expression data suggest that MHC‐I compatible SCNT pregnancies are more similar to control pregnancies established by artificial insemination than to MHC‐I incompatible SCNT pregnancies. The lack of differentially expressed miRNAs between the MHC‐I compatible SCNT and control pregnancies further supports our previous observations that matching the MHC‐I haplotype between the fetus and the recipient cow during SCNT pregnancies improves embryonic survival rates and decreases inflammation at the fetal–maternal interface. 18
Previous studies have shown that miR‐2887 is a bovine specific miRNA that is involved in lipid and glucose metabolism. 57 , 58 It has also been shown to have increased expression in media containing degenerated embryos compared to media containing healthy blastocysts in cell culture. 59 In our study, miR‐2887 was significantly increased in trophoblasts of MHC‐I heterozygous incompatible pregnancies which further suggests that this miRNA could be involved in cellular degeneration and apoptosis. MicroRNA‐184 has been found to be overexpressed in the villus trophoblast cells and decidua of recurrent spontaneous abortion human patients, 60 which is in accordance with our results that show that this miRNA is overexpressed in abortion‐prone trophoblast (MHC‐I heterozygous incompatible SCNT). MicroRNA‐1246 targets mRNA of transcription factors for developmentally regulated genes. In our study, this miRNA was the second highest expressed miRNA in trophoblasts of MHC‐I heterozygous incompatible pregnancies. Upregulation of miR‐1246 has been shown to promote trophoblast differentiation in human placentas. 61 It is possible that in cattle, miR‐1246 promotes differentiation of trophoblast cells to become invasive, binucleate trophoblast cells. Increased numbers of binucleate trophoblast cells would increase delivery of trophoblast MHC‐I to the maternal immune system and promote inflammation. This miRNA is downregulated in trophoblast from severe pre‐eclamptic human patients and in human trophoblasts cultured under hypoxic conditions. 61 These results contradict our findings, but because of the promiscuous nature of miRNAs, without a comprehensive pathway analysis, it is difficult to determine the causes or effects of the deregulation of this miRNA. One possible consequence of the upregulation of this miRNA is the premature differentiation of trophoblast cells in the MHC‐I heterozygous incompatible placentas.
MicroRNA‐451 is highly expressed in trophoblast cells and has been shown to be upregulated when these cells are hypoxic. 62 This miRNA inhibits NF‐κB activity and consequently downregulates pro‐inflammatory molecules in several tissues. 63 , 64 , 65 Here, we observed a downregulation of miR‐451 in the MHC‐I heterozygous incompatible pregnancies, which suggests this microRNA is involved in the development of a pro‐inflammatory environment at the fetal–maternal interface. No other hypoxia‐specific miRNAs are among the differentially regulated miRNAs found in this study, suggesting the downregulation of miR‐451 is not due to hypoxia, but rather to other mechanisms.
In our study, we predicted target genes for differentially expressed miRNAs and conducted GO and KEGG analyses. Based on existing knowledge, it is difficult to determine which genes are being regulated by particular miRNAs in any given tissue. However, identification of the dysregulated miRNAs is the first step in this process. Within the up‐ and downregulated miRNA groups, there are significant similarities in GO terms between the MHC‐I heterozygous incompatible versus MHC‐I compatible and the MHC‐I heterozygous incompatible versus control group comparisons.
When determining the target genes of downregulated miRNAs in trophoblasts of MHC‐I heterozygous incompatible versus control pregnancies, and of MHC‐I heterozygous incompatible versus homozygous compatible pregnancies the most significant GO terms were associated with ion transport and binding, motor activity, cellular junction and structure, cellular motility, and bone development. The most significant biological processes associated with upregulated miRNAs in trophoblasts of MHC‐I incompatible compared to control pregnancies, and in trophoblasts of MHC‐I incompatible compared to MHC‐I compatible pregnancies were ion transport and binding, extracellular matrix composition, plasma membrane composition, cellular structure, nervous system function and development, cell differentiation, and organ development.
Pathway analysis showed a dysfunction in genes related to endocrine signaling, mineral handling, nutrient digestion, and absorption in the comparisons between trophoblasts of MHC‐I heterozygous incompatible and control pregnancies, and between trophoblast of MHC‐I incompatible and compatible pregnancies. There was little similarity between the GO and KEGG terms identified in the day 35 ±1 SCNT trophoblast in the present study and the terms identified by Su et al. (2015). 66 In the latter, trophoblast derived from bovine SCNT pregnancies or pregnancies produced by natural breeding were collected via C‐section at term. This discrepancy may be attributed to the difference in gestational age of the trophoblast collected in both studies.
Upon determining differentially expressed trophoblast miRNAs that could be involved in the regulation of expression of the 23 inflammatory genes assessed in this study, it was noticeable that most of these miRNAs were downregulated in the MHC‐I heterozygous incompatible treatment group compared to the MHC‐I homozygous compatible and control groups (Table 4). This may have contributed to the development of a pro‐inflammatory immune response at the fetal–maternal interface in the heterozygous incompatible treatment group.
This is the first study to determine inflammatory mediator mRNA and trophoblast miRNA profiles at the fetal–maternal interface in MHC‐I compatible and incompatible SCNT pregnancies. We observed that the compatibility of MHC‐I haplotypes between the dam and the fetus impacts the expression profiles of inflammatory mediators and miRNAs involved in trophoblast development. We have demonstrated that there is greater expression of pro‐inflammatory mediators in SCNT pregnancies where the dam and the fetus are MHC‐I incompatible compared to MHC‐I compatible or pregnancies established by artificial insemination. Our observations regarding upregulation of immune system genes in the trophoblast cells of SCNT conceptuses, and the dysregulation of trophoblast cell miRNA expression, strongly suggest that trophoblast cells have important immunoregulatory function and likely amplify pro‐inflammatory uterine immune responses. These results help inform our understanding of pregnancy complications and immune‐mediated losses in animals and humans.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
This project was supported by grant 1R01HD055502 from the NIH, National Institute of Child Health and Human Development. Additional support was provided by the Utah Agricultural Experiment Station, Utah State University, which approved this manuscript as journal paper number 9390. The authors thank Dr. Qinggang Meng for micromanipulation of embryos, and Dick Whittier for assisting with tissue collection. The authors also thank Dr. Koji Yoshinaga and the members of the NICHD Interdisciplinary Collaborative Team for Blastocyst Implantation Research for constructive comments regarding this study.
Rutigliano HM, Thomas AJ , Umbaugh JJ, et al. Increased expression of pro‐inflammatory cytokines at the fetal–maternal interface in bovine pregnancies produced by cloning. Am J Reprod Immunol. 2022;87:e13520. 10.1111/aji.13520
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Panarace M, Aguero JI, Garrote M, et al. How healthy are clones and their progeny: 5 years of field experience. Theriogenology. 2007;67(1):142–151. [DOI] [PubMed] [Google Scholar]
- 2. Cibelli JB, Stice SL, Golueke PJ, et al. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science. 1998;280(5367):1256–1258. [DOI] [PubMed] [Google Scholar]
- 3. Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–813. [DOI] [PubMed] [Google Scholar]
- 4. Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full‐term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394(6691):369–374. [DOI] [PubMed] [Google Scholar]
- 5. Wells DN, Misica PM, Tervit HR. Production of cloned calves following nuclear transfer with cultured adult mural granulosa cells. Biol Reprod. 1999;60(4):996–1005. [DOI] [PubMed] [Google Scholar]
- 6. Edwards JL, Schrick FN, McCracken MD, et al. Cloning adult farm animals: a review of the possibilities and problems associated with somatic cell nuclear transfer. Am J Reprod Immunol. 2003;50(2):113–123. [DOI] [PubMed] [Google Scholar]
- 7. Hill JR, Burghardt RC, Jones K, et al. Evidence for placental abnormality as the major cause of mortality in first‐trimester somatic cell cloned bovine fetuses. Biol Reprod. 2000;63:1787–1794. [DOI] [PubMed] [Google Scholar]
- 8. Heyman Y, Chavatte‐Palmer P, LeBourhis D, Camous S, Vignon X, Renard JP. Frequency and occurrence of late‐gestation losses from cattle cloned embryos. Biol Reprod. 2002;66(1):6–13. [DOI] [PubMed] [Google Scholar]
- 9. Hunt JS, Andrews GU, Wood GW. Normal trophoblasts resist induction of class I HLA. J Immunol. 1987;138:2481–2487. [PubMed] [Google Scholar]
- 10. Gogolin‐Ewens KJ, Lee CS, Mercer WR, Brandon MR. Site‐directed differences in the immune response to the fetus. Immunology. 1989;66(2):312–317. [PMC free article] [PubMed] [Google Scholar]
- 11. Loke YW. Trophoblast antigen expression. Curr Opin Immunol. 1989;1(6):1131–1134. [DOI] [PubMed] [Google Scholar]
- 12. Donaldson WL, Zhang CH, Oriol JG, Antczak DF. Invasive equine trophoblast expresses conventional class I major histocompatibility complex antigens. Development. 1990;110:63–71. [DOI] [PubMed] [Google Scholar]
- 13. Low BG, Hansen PJ, Drost M, Gogolin‐Ewens KJ. Expression of major histocompatibility complex antigens on the bovine placenta. J Reprod Fertil. 1990;90:235–243. [DOI] [PubMed] [Google Scholar]
- 14. Kydd JH, Butcher GW, Antczak DF, Allen WR. Expression of major histocompatibility complex (MHC) class 1 molecules on early trophoblast. J Reprod Fert Suppl. 1991;44:463–477. [PubMed] [Google Scholar]
- 15. Davies CJ, Fisher PJ, Schlafer DH. Temporal and regional regulation of major histocompatibility complex class I expression at the bovine uterine/placental interface. Placenta. 2000;21(2‐3):194–202. [DOI] [PubMed] [Google Scholar]
- 16. Davies CJ, Eldridge JA, Fisher PJ, Schlafer DH. Evidence for expression of both classical and non‐classical major histocompatibility complex class I genes in bovine trophoblast cells. Am J Reprod Immunol. 2006;55(3):188–200. [DOI] [PubMed] [Google Scholar]
- 17. Shi B, Thomas AJ, Benninghoff AD, et al. Genetic and epigenetic regulation of major histocompatibility complex class I gene expression in bovine trophoblast cells. Am J Reprod Immunol. 2018;79(1):e12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rutigliano HM, Thomas AJ, Wilhelm A, et al. Trophoblast major histocompatibility complex class I expression is associated with immune‐mediated rejection of bovine fetuses produced by cloning. Biol Reprod. 2016;95(2):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hill JR, Schlafer DH, Fisher PJ, Davies CJ. Abnormal expression of trophoblast major histocompatibility complex class I antigens in cloned bovine pregnancies is associated with a pronounced endometrial lymphocytic response. Biol Reprod. 2002;67(1):55–63. [DOI] [PubMed] [Google Scholar]
- 20. Le Bouteiller P. Major histocompatibioty complex class I unique expression in human trophoblast: facts, questions and controversies. In: Chaouat G, Sandra O, Ledee N, eds. Immunology of Pregnancy 2013. Bentham Science Publishers; 2013. [Google Scholar]
- 21. Rutigliano HM, Thomas AJ, Davies CJ. Expression of MHC‐I proteins by the placenta of domestic and laboratory animals. In: Chaouat G, Sandra O, Ledee N, eds. Immunology of Pregnancy 2013. Bentham Science Publishers; 2013. [Google Scholar]
- 22. Rutigliano HM, Wilhelm A, Hall J, et al. Cytokine gene expression at the maternal‐fetal interface after somatic cell nuclear transfer pregnancies in small ruminants. Reprod Fertil Dev. 2017;29(4):646–657. [DOI] [PubMed] [Google Scholar]
- 23. Joosten I, Sanders MF, Hensen EJ. Involvement of major histocompatibility complex class I compatibility between dam and calf in the aetiology of bovine retained placenta. Anim Genet. 1991;22:455–463. [DOI] [PubMed] [Google Scholar]
- 24. Benedictus L, Thomas AJ, Jorritsma R, Davies CJ, Koets AP. Two‐way calf to dam major histocompatibility class I compatibility increases risk for retained placenta in cattle. Am J Reprod Immunol. 2012;67(3):224–230. [DOI] [PubMed] [Google Scholar]
- 25. Davies CJ, Hill JR, Edwards JL, et al. Major histocompatibility antigen expression on the bovine placenta: its relationship to abnormal pregnancies and retained placenta. Anim Reprod Sci. 2004;82‐83:267–280. [DOI] [PubMed] [Google Scholar]
- 26. Aston KI, Li GP, Hicks BA, et al. The developmental competence of bovine nuclear transfer embryos derived from cow versus heifer cytoplasts. Anim Reprod Sci. 2006;95(3‐4):234–243. [DOI] [PubMed] [Google Scholar]
- 27. Martin M. Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet Journal. 2011;17(1):10–12. [Google Scholar]
- 28. FASTX‐Toolkit. 2020. http://hannonlab.cshl.edu/fastx_toolkit/
- 29. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schlafer DH, Fisher PJ, Davies CJ. The bovine placenta before and after birth: placental development and function in health and disease. Anim Reprod Sci. 2000;60‐61:145–160. [DOI] [PubMed] [Google Scholar]
- 33. Mosmann TR, Sad S. The expanding universe of T‐cell subsets: Th1, Th2 and more. Immunol Today. 1996;17(3):113–123. [DOI] [PubMed] [Google Scholar]
- 34. Lin H, Mosmann TR, Guilbert L, Tuntipopipat S, Wegmann TG. Synthesis of T helper 2‐type cytokines at the maternal‐fetal interface. J Immunol. 1993;151(9):4562–4573. [PubMed] [Google Scholar]
- 35. Saito S, Sakai M, Sasaki Y, Tanebe K, Tsuda H, Michimata T. Quantitative analysis of peripheral blood Th0, Th1, Th2 and the Th1: Th2 cell ratio during normal human pregnancy and preeclampsia. Clin Exp Immunol. 1999;117(3):550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsuda H, Michimata T, Sakai M, Nagata K, Nakamura M, Saito S. A novel surface molecule of Th2‐ and Tc2‐type cells, CRTH2 expression on human peripheral and decidual CD4+ and CD8+ T cells during the early stage of pregnancy. Clin Exp Immunol. 2001;123(1):105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sargent IL, Borzychowski AM, Redman CW. NK cells and human pregnancy – an inflammatory view. Trends Immunol. 2006;27(9):399–404. [DOI] [PubMed] [Google Scholar]
- 38. Piccinni MP, Scaletti C, Maggi E, Romagnani S. Role of hormone‐controlled Th1‐ and Th2‐type cytokines in successful pregnancy. J Neuroimmunol. 2000;109(1):30–33. [DOI] [PubMed] [Google Scholar]
- 39. Saito S, Umekage H, Sakamoto Y, et al. Increased T‐helper‐1‐type immunity and decreased T‐helper‐2‐type immunity in patients with preeclampsia. Am J Reprod Immunol. 1999;41(5):297–306. [DOI] [PubMed] [Google Scholar]
- 40. Chaouat G, Assal Meliani A, Martal J, et al. IL‐10 prevents naturally occurring fetal loss in the CBA x DBA/2 mating combination, and local defect in IL‐10 production in this abortion‐prone combination is corrected by in vivo injection of IFN‐tau. J Immunol. 1995;154(9):4261–4268. [PubMed] [Google Scholar]
- 41. Chaouat G, Menu E, Clark DA, Dy M, Minkowski M, Wegmann TG. Control of fetal survival in CBA x DBA/2 mice by lymphokine therapy. J Reprod Fertil. 1990;89(2):447–458. [DOI] [PubMed] [Google Scholar]
- 42. Piccinni MP, Beloni L, Livi C, Maggi E, Scarselli G, Romagnani S. Defective production of both leukemia inhibitory factor and type 2 T‐helper cytokines by decidual T cells in unexplained recurrent abortions. Nat Med. 1998;4(9):1020–1024. [DOI] [PubMed] [Google Scholar]
- 43. Lim KJ, Odukoya OA, Ajjan RA, Li TC, Weetman AP, Cooke ID. The role of T‐helper cytokines in human reproduction. Fertil Steril. 2000;73(1):136–142. [DOI] [PubMed] [Google Scholar]
- 44. Jenkins C, Roberts J, Wilson R, MacLean MA, Shilito J, Walker JJ. Evidence of a T(H) 1 type response associated with recurrent miscarriage. Fertil Steril. 2000;73(6):1206–1208. [DOI] [PubMed] [Google Scholar]
- 45. Arck P, Dietl J, Clark D. From the decidual cell internet: trophoblast‐recognizing T cells. Biol Reprod. 1999;60(2):227–233. [DOI] [PubMed] [Google Scholar]
- 46. Wegmann TG, Lin H, Guilbert L, Mosmann TR. Bidirectional cytokine interactions in the maternal‐fetal relationship: is successful pregnancy a TH2 phenomenon? Immunol Today. 1993;14:353–356. [DOI] [PubMed] [Google Scholar]
- 47. Abrahams VM, Kim YM, Straszewski SL, Romero R, Mor G. Macrophages and apoptotic cell clearance during pregnancy. Am J Reprod Immunol. 2004;51(4):275–282. [DOI] [PubMed] [Google Scholar]
- 48. Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robertson SA, Sjoblom C, Jasper MJ, Norman RJ, Seamark RF. Granulocyte‐macrophage colony‐stimulating factor promotes glucose transport and blastomere viability in murine preimplantation embryos. Biol Reprod. 2001;64(4):1206–1215. [DOI] [PubMed] [Google Scholar]
- 50. Chin PY, Macpherson AM, Thompson JG, Lane M, Robertson SA. Stress response genes are suppressed in mouse preimplantation embryos by granulocyte‐macrophage colony‐stimulating factor (GM‐CSF). Hum Reprod. 2009;24(12):2997–3009. [DOI] [PubMed] [Google Scholar]
- 51. Loureiro B, Bonilla L, Block J, Fear JM, Bonilla AQ, Hansen PJ. Colony‐stimulating factor 2 (CSF‐2) improves development and posttransfer survival of bovine embryos produced in vitro. Endocrinology. 2009;150(11):5046–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Denicol AC, Block J, Kelley DE, et al. The WNT signaling antagonist Dickkopf‐1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB J. 2014;28(9):3975–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ziebe S, Loft A, Povlsen BB, et al. A randomized clinical trial to evaluate the effect of granulocyte‐macrophage colony‐stimulating factor (GM‐CSF) in embryo culture medium for in vitro fertilization. Fertil Steril. 2013;99(6):1600–1609. [DOI] [PubMed] [Google Scholar]
- 54. Sjoblom C, Roberts CT, Wikland M, Robertson SA. Granulocyte‐macrophage colony‐stimulating factor alleviates adverse consequences of embryo culture on fetal growth trajectory and placental morphogenesis. Endocrinology. 2005;146(5):2142–2153. [DOI] [PubMed] [Google Scholar]
- 55. Lee K, Redel BK, Spate L, et al. Piglets produced from cloned blastocysts cultured in vitro with GM‐CSF. Mol Reprod Dev. 2013;80(2):145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sheng W, PNG CW, Reynolds JM, Zhang Y. T Cell‐derived GM‐CSF, regulation of expression and function. Immunome Res. 2015;11(2):1000098. [Google Scholar]
- 57. Maalouf SW, Liu WS, Albert I, Pate JL. Regulating life or death: potential role of microRNA in rescue of the corpus luteum. Mol Cell Endocrinol. 2014;398(1‐2):78–88. [DOI] [PubMed] [Google Scholar]
- 58. Guo Y, Zhang X, Huang W, Miao X. Identification and characterization of differentially expressed miRNAs in subcutaneous adipose between Wagyu and Holstein cattle. Sci Rep. 2017;7:44026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kropp J, Khatib H. Characterization of microRNA in bovine in vitro culture media associated with embryo quality and development. J Dairy Sci. 2015;98(9):6552–6563. [DOI] [PubMed] [Google Scholar]
- 60. Dong F, Zhang Y, Xia F, et al. Genome‐wide miRNA profiling of villus and decidua of recurrent spontaneous abortion patients. Reproduction. 2014;148(1):33–41. [DOI] [PubMed] [Google Scholar]
- 61. Muralimanoharan S, Kwak YT, Mendelson CR. Redox‐sensitive transcription factor NRF2 enhances trophoblast differentiation via induction of miR‐1246 and aromatase. Endocrinology. 2018;159(5):2022–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mouillet JF, Chu T, Nelson DM, Mishima T, Sadovsky Y. MiR‐205 silences MED1 in hypoxic primary human trophoblasts. FASEB J. 2010;24(6):2030–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li W, Dong M, Chu L, Feng L, Sun X. MicroRNA‑451 relieves inflammation in cerebral ischemia‑reperfusion via the Toll‑like receptor 4/MyD88/NF‑κB signaling pathway. Mol Med Rep. 2019;20:3043–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Okamoto M, Fukushima Y, Kouwaki T, et al. MicroRNA‐451a in extracellular, blood‐resident vesicles attenuates macrophage and dendritic cell responses to influenza whole‐virus vaccine. J Biol Chem. 2018;293(48):18585–18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sun Y, Peng R, Peng H, et al. miR‐451 suppresses the NF‐kappaB‐mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. Mol Cell Endocrinol. 2016;433:75–86. [DOI] [PubMed] [Google Scholar]
- 66. Su J, Liu X, Sun H, et al. Identification of differentially expressed microRNAs in placentas of cloned and normally produced calves by Solexa sequencing. Anim Reprod Sci. 2015;155:64–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


