Abstract
Homologous recombination (HR) is a highly conserved pathway that can facilitate the repair of DNA double-strand breaks (DSBs). Several Deubiquitinases (DUBs) have been implicated as key players in DNA damage repair (DDR) through HR. Here we report USP22, a DUB that is highly overexpressed in multiple cancer types, is necessary for HR through a direct interaction with PALB2 through its C-terminal WD40 domain. This interaction stimulates USP22 catalytic activity in-vitro. Furthermore, we show USP22 is necessary for BRCA2, PALB2, and Rad51 recruitment to DSBs and this is in part through USP22 stabilizing BRCA2 and PALB2 levels. Taken together, our results describe a role for USP22 in DNA repair.
Implications:
This research provides new and exciting mechanistic insights into how USP22 overexpression promotes chemo-resistance in lung cancer. We believe this study, and others, will help aid in developing targeted drugs towards USP22 and known binding partners for lung cancer treatment.
Keywords: USP22, PALB2, homologous recombination, DNA damage response, WD40, lung cancer
Introduction
DNA damage by environmental stressors and agents must be repaired efficiently for the cell to maintain its genomic integrity. Cells employ several repair pathways and mechanisms to rectify damaged DNA. Homologous Recombination (HR) repair comprises a series of pathways in a cell that function to repair DNA double-stranded breaks (DSBs) through template-dependent mechanisms to preserve genomic integrity. Failure to repair DSBs can result in chromosomal loss, carcinogenesis, and apoptosis1. Partner and Localizer of BRCA2 (PALB2) is a key protein involved in HR. It was originally identified as an interacting partner with BRCA2, which, along with BRCA1, are predisposing breast cancer genes2,3. For its role in HR, PALB2 is controlled in part by phosphorylation by ATM, ATR, and several CDK’s during the S/G2 phases4. This phosphorylation, in part, can control its interaction with BRCA1, its ability to assemble Rad51/BRCA2, and its association with damaged DNA5. PALB2 can also recognize ubiquitinated chromatin through its interaction with the E3 ubiquitin ligase RNF168. RNF168 physically couples the HR machinery to Histone H2A ubiquitylation through its association with the C-terminal WD40 domain of PALB26. Conversely, PALB2 also associates with the Deubiquitinase (DUB) USP11 which can antagonize RNF168 ubiquitylation at chromatin and promote PALB2 interaction with BRCA1, which is essential for proper HR to occur at DNA damage sites7. Thus, PALB2 function during HR appears to be regulated by ubiquitination signaling, including the DUB USP11.
We considered that other DUBs might be important for HR, with a focus on USP22. In humans there are nearly 100 DUB proteins that participate in the removal of monoubiquitin and polyubiquitin chains from proteins8. In general, polyubiquitination signals a protein for degradation by the cellular proteasome while a monoubiquitin mark can act as an epigenetic modification8. One example is the monoubiquitylation of Histone H2B at Lysine 120 (H2BK120ub) that has been implicated in nucleosome compaction, gene transcription, DNA damage response, and even centromere integrity9,10,11. This monoubiquitin mark is antagonized by a DUB called Ubiquitin-specific peptidase 22 (USP22)12. USP22 carries out this function through its association with the Spt-Ada-Gcn5-acetyltransferase (SAGA) deubiquitinating module that, together with ENY2, ATXN7, and ATXN7L3, forms a sub complex that can antagonize H2BK120ub13,14. USP22, as part of SAGA, acts in opposition to the E3 ubiquitin ligase complex RNF20/RNF40 that promotes H2Bk120ub, in this way the cell can dynamically regulate H2B ubiquitination status. The H2B ubiquitination and deubiquitination cycle is critical for DNA repair as studies in yeast and humans have shown H2BK120ub promotes chromatin disassembly and recruitment of HR factors around DSBs11,15,16,17. USP22 can also remove polyubiquitin chains from proteins thereby stabilizing and protecting them from proteasome degradation. These proteins include Sirt1, CCND1, and c-Myc18,19,20. Sirt1 is an NAD-dependent histone deacetylase, that through USP22 stabilization, prohibits the transcriptional and proapoptotic functions of the tumor suppressor protein p5318. Previous studies have also suggested SAGA may participate in global nucleotide excision repair at chromatin21 and that H2B deubiquitination is required for repair during transcription after UV damage-induced RNA polymerase II stalling22. Furthermore, USP22 has been shown to promote antibody class switch recombination through non-homologous end joining (NHEJ) and also affected homologous recombination (HR) repair23,24. Although the specific effects of USP22 on the regulation of HR and the DNA damage response (DDR) have remained unclear.
USP22 was identified by microarray analysis as a “Death by cancer” signature gene whose overexpression is associated with poor patient prognosis25. Furthermore, overexpression of USP22 leads to Cisplatin resistance in lung cancer cells 26,27. Lung cancer is the most common cause of cancer death worldwide28 and despite treatment advances, this cancer type is known for developing recurrence and treatment resistance. A major mechanism of treatment resistance is the ability of cancer cells to upregulate DDR after therapies such as chemotherapy. Several drugs have been developed to target the DDR, and therapies targeting DUBs involved in the DDR are being developed. In this study we show that USP22 is necessary for HR, localizes at sites of DNA damage, and is necessary for recruitment of the PALB2-BRCA2-Rad51 complex through stabilizing PALB2 and BRCA2 protein levels at the translational level. Lastly, we show USP22 directly binds to the WD40 domain of PALB2 and this binding stimulates USP22 catalytic activity. These findings provide evidence for a new and novel role of USP22 in the HR pathway.
Materials & Methods
DNA Constructs:
All plasmids were created by PCR and cut-and-paste cloning using restriction enzymes. The mCherry-LacI vector was obtained from Addgene (#18985). All mCherry-LacI constructs were made by cutting insert and vector with KPNI and BamHI and ligating fragments with T4 DNA ligase (NEB). The cDNA for PALB2 and all its subsequent fragments was also obtained from Addgene (#71113) and used as a PCR substrate for ligation into mCherry-LacI. For the mCherry-PALB2 fragments, mCherry-LacI was cut with BglII and BamHI to excise the LacI sequence and the PALB2 fragments were PCRed, cut with the same enzymes and ligated in. FLAG-USP22, GFP-USP22, FLAG-H2B, and FLAG-PALB2 were purchased from Genscript. PMAL-p2X was used as the vector (Addgene #75287) for MBP-PALB2WD40 (amino acids 841-1186) which was cloned in via BamHI and PstI. To construct mCherry-LacI-BRCA2 mCherry-LacI was excised from original plasmid with BamHI and AscI, A plasmid containing SFB-BRCA2 (Addgene #99395) was cut with the same restriction enzymes thereby excising the N-terminal SFB tag and mCherry-LacI was subsequently ligated in to make mCherry-LacI-BRCA2. Mutagenic PCR was performed using a site directed mutagenesis kit and protocol from New England Biolabs, according to manufacturer’s instructions.
Antibodies:
PALB2 (Abcam, ab220861), BRCA2 (Abcam, ab27976), Rad51 (Cell Signaling Technology, 8875S), USP22 (Santa Cruz, sc-390585), FLAG (Sigma, F3165-1MG), β-Actin (Cell Signaling Technology, 4970S), mCherry (Origene, TA180028), yH2A.X (Cell Signaling Technology, 2577S), MBP (New England Biolabs, E8032S), 6X-His (Santa Cruz, sc-8036), ENY2 (Abcam, ab183622), ATXN7L3 (Bethyl, A302-800A), ATXN7 (Bethyl, A302-638A).
Cell Transfection and Immunocytochemistry:
Cell lines were purchased from ATCC and were tested for Mycoplasma contamination using MycoSEQ from Thermo Fisher. Additionally, the media was supplemented with a Mycoplasma preventative antibiotic (Normocin from Invivogen), cells were kept until passage 25 and then thrown away. All cell lines were transfected with Lipofectamine-2000 (Invitrogen) for DNA constructs and lipofectamine RNAi-MAX for siRNA according to the manufacturer’s protocol. USP22 and control siRNA were purchased from Dharmacon (USP22 siRNA: L-006072-03-0010. Control siRNA: D001810-01-20) and used at a final concentration of 20nM. Cells were processed for immunocytochemistry 48 hr after transfection. Cells were kept in DMEM media supplemented with 10% FBS and 1% pen/strep antibiotic along with normocin. Immunocytochemistry and immunoprecipitation experiments were conducted as described previously29. U2OSFOKI cells were a generous gift from Roger Greenberg at the University of Pennsylvania and were kept in complete DMEM media supplemented with 100ug/ml hygromycin and 1ug/ml puromycin. U2OSFOKI cells were induced for DNA damage by supplementing media with Shield-1 ligand (Clontech, 0.5mM stock in ethanol then used at 1:500 final concentration) and 4-OHT (Sigma, 1mM stock in ethanol then used at 1:1000 final concentration) for 6 hours and then cells were subsequently fixed for immunofluorescence. Images of fixed cells were collected using a 40× water immersion objective lens on a Zeiss LSM 880 confocal microscope using Zen Black acquisition software. Images were presented as stacked images. Images within experiments were collected with identical exposure times and scaled equally. U2OS cell lines carrying reporter constructs EJ5-GFP, DR-GFP, and SA-GFP were tested as previously described30. Briefly, cells were transfected with the siRNA described above using RNAiMax, followed, 24 hr later, by co-transfection using lipofectamine-2000 of the I-SceI plasmid (pCBASce30, Addgene #26477) and mCherry (Original plasmid is mCherry-LacI from Addgene #18985. Whole plasmid PCR was performed excluding the LacI sequence and then blunt end ligation was performed) along with more siRNA. Three days after transfection, mCherry+ cells were examined for %GFP+ using BD LSRFortessa Analyzer. 200ng pCBASce, 50ng mCherry, and 20nM siRNA were used, in a total transfection volume 0.6ml. For H1299 cells treated with MG132 (Figure S1c), cells were treated with MG132 in complete DMEM for 6 hours at 15uM then harvested and lysed with sample buffer for western blot analysis.
Computer Modeling:
Using ZDOCK we were able to generate a computer model using the best ZRANK-scored structure31,32 of the WD40 domain of PALB2 (amino acids 846-1186) binding to the DUB domain of USP22 (amino acids 164-502). We were further able to identify tentative amino acid residues on PALB2-WDR that could be necessary for its interaction with USP22 using the ZRANK-scored structure. The protein-protein interface residues were defined as those which had an inter-protein heavy atom distance of 4 Angstroms or less. The most important residues which formed inter-protein hydrogen bonds or large buried surface area were selected as necessary for the interaction. The crystal structure of PALB2 WD40 domain was used (PDB: 2W18). The tentative structure for the DUB domain of USP22 was predicted using the crystal structure of its yeast homolog UBP8 (PDB: 3M99).
Immunoprecipitations:
Mcherry-PALB2 fragments were be co-transfected into H1299 cells along with FLAG-USP22 using lipofectamine 2000 and allowed to express for 48 hours. After 48 hours cells were lysed in RIPA buffer (50mM Tris PH=8.0, 150mM NaCl, 2mM MgCl2, 1mM CaCl2, 0.15% SDS, 0.3% Sodium Deoxycholate, 1% NP-40, 3% glycerol, 5mM BME, 1X Roche Protease inhibitors) on ice for 20 minutes with occasional vortexing. After 20 minutes the lysate was diluted 10X in RIPA buffer without SDS and Sodium Deoxycholate (RIPA-2) and 200 units of benzoase nuclease were added to digest chromatin. The lysates were incubated for 30 minutes at 4°C to allow the benzoase to digest the chromatin. After 30 minutes 1mM EDTA was added to stop the benzoase reaction and the lysates were spun at 20,000xg for 20 minutes to make the final chromatin free extract (CFE). Magnetic RFP-Trap Beads that bind the mCherry tag (chromotek) were incubated with the CFE overnight at 4°C. The next day the beads were washed 5X in RIPA-2 buffer and the beads were boiled in 2X SDS buffer and run on a western blot using antibodies against mCherry (Origene) and FLAG (Sigma) to assess binding.
Recombinant protein production and in-vitro pull downs:
Recombinant proteins (MBP-PALB2WD40 and MBP) were made and expressed in E. Coli Rosetta bugs using the inducible T7 RNA polymerase system33. The Rosetta bugs were grown in LB media to an OD of 0.6 at 37°C and induced at 18°C with 0.1mM IPTG for 16 hr. Bacteria were lysed using a steel Wheaton-Dounce in buffer containing 50mM Tris-HCl pH 7.5, 250mM NaCl, 20mM MgCl2, 0.5mM CaCl2, 10% glycerol, 0.1% NP-40, 5mM BME, and 1X Roche Protease inhibitors. Lysates were centrifuged at 20,000xg for 20 minutes. Supernatants were collected and incubated with amylose resin for 2 hours at 4°C. The resin was washed in the same lysis buffer 5X times and eluted in the same buffer with 10mM maltose added. His-USP22 was purchased from Abnova. The in-vitro pull downs were performed as follows: Proteins were combined for 1 hr at room temperature at 1:1 molar ratio in binding buffer: 50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 20 mM MgCl2, 0.5% NP-40, 10% glycerol, and 5 mM BME. Amylose resin was added after 1 hr for 30 minutes. The amylose beads were washed 6X in 1 ml of binding buffer. The amylose beads were then boiled in SDS sample buffer. Western blots were performed using antibodies against the 6× His tag (Santa Cruz) and MBP tag (NEB).
Fluorescence ubiquitin cleavage:
His-USP22 alone and in combination with MBP-PALB2WD40 or MBP were incubated together in DUB buffer at a 1:1 molar ratio (50mM Tris pH=8.0, 150mM NaCl, 2mM MgCl2, 1mM DTT) at room temperature for 30 minutes to form their respective complexes. Rhodamine-Ubiquitin (BPS Bioscience, #81151) was then added in at a 1:1 molar ratio and incubated with the complexes at 37°C for 1 hr. Fluorescence (denoting cleavage of ubiqutin and thereby enzymatic activity) was assessed using a fluorescence plate reader to assess light emission at 580nm for 0.1s. As a positive control, recombinant UCHL1 protein was used (R&D Systems, #E-340-025) as a positive control.
Cisplatin treatment and cell counting:
H1299WT or H1299FLAG-PALB2 cells were siRNA treated with control or USP22 siRNA for 24 hrs in OptiMEM media (Thermo) with the RNAiMax Transfection Reagent (Thermo). Cells were then washed and put in complete DMEM media supplemented with 3uM Cisplatin for 72 hrs. After this time cells were washed 2X in PBS, trypsinized, and then counted using a hemocytometer. Western blot was performed to assess knock down using antibodies for USP22, β-Actin, FLAG, and PALB2. For IC50 treatment and cell counting H1299 cells were treated with varying doses of Cisplatin 24 hours after siRNA knockdown, 48 hours later cells were washed with 1X PBS and then stained with tryptan blue and counted.
qPCR:
TaqMan probes for USP22, BRCA2, and PALB2 were obtained from Thermo Fisher with the respective product numbers: USP22 (4331182), BRCA2 (4351372), and PALB2 (4351372). QPCR methods used were described previously27.
Results
USP22 Is Necessary for Efficient HR
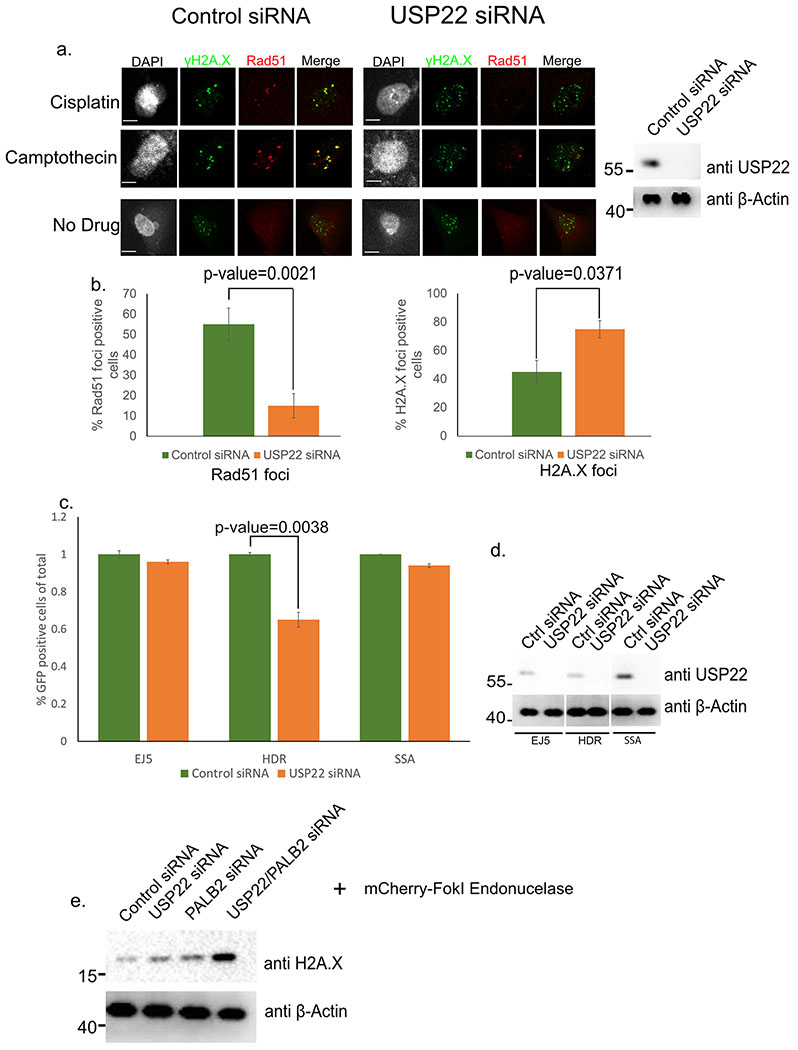
Upon DNA damage, the central HR protein Rad51 is known to form distinct foci at sites of DNA damage34. Given that previous work had shown USP22 to be necessary for HR23, we analyzed if siRNA knockdown (KD) of USP22 in H1299 cells had any effect on Rad51 foci formation after 6-hour treatment with the DNA damaging agents Cisplatin and Camptothecin, two drugs that are used in treatment of lung cancer (Figure 1a). From this analysis, USP22 KD significantly reduced the number of Rad51 foci positive cells compared to control from 55% to 15% (p-value=0.0021) in both drug treatments (Figure 1b, left graph). We hypothesize part of this effect is from the partial G1 arrest USP22 KD can induce in cells19 not allowing them to perform HR since it is a cell cycle regulated process. Conversely, cells containing foci pertaining to phosphorylation at serine 139 of histone H2A.X (γH2A.X), a histone mark found at sites of DNA damage35, were significantly increased upon USP22 KD with both DNA damaging agents from 45% to 75% (p-value=0.0371) (Figure 1b, right graph). Thus, USP22 appears to be important for Rad51 foci formation, and limiting γH2A.X foci, which indicate that this factor is important for DSB repair via HR. To further examine the role of USP22 in DSB repair, we used a set of reporters in U2OS cells to assay for 3 different types of DNA repair; non-homologous end joining (NHEJ, EJ5), a subtype of Rad51-depedent homologous recombination called homology-directed repair (HDR), and single-strand annealing (SSA)30. Each reporter is designed such that repair of an I-SceI-induced DSB by the respective pathway restores GFP+ cells. Cells were depleted of USP22 for 24 hours by siRNA, then a plasmid encoding for I-SceI was transfected in (along with an mCherry plasmid as a transfection efficiency control) to cause the DNA damage event along with more USP22 siRNA. 48 hours later cells were fixed and sorted by FACS (Figure 1c,d). Contrary to the previous work with USP22 in antibody class switch repair24,36 we did not see any noticeable change in the percentage of GFP positive cells with the EJ5 reporter cell line (denoting NHEJ) upon USP22 KD, but we did notice a similar phenotype as the other studies24,37 with our HDR reporter cell line (Figure 1c). Namely, upon USP22 KD the percentage of GFP positive cells in the HDR reporter cell line was markedly reduced by 40% (p=0.0038). On the other hand, we observed no effect in the SSA reporter cell line, which is a Rad51-independent event38. Furthermore, combined separate or combinatorial USP22/PALB2 knockdown in the U2OSFOKI cells showed increased γH2A.X protein by western blot (Figure 1e). Given that γH2A.X is a key marker denoting DNA damage it strongly supports our hypothesis that USP22 is integral for proper DNA repair and without it the DNA damage events persist. Thus, USP22 is important for Rad51 foci formation, and HDR using a reporter assay, indicating that this factor is important for HR.
Figure 1:
USP22 is necessary for efficient HR of DSBs. a. H1299 cells treated with DNA damaging drugs Cisplatin or Camptothecin and fixed with indicated antibodies. (right) Western blot of control versus USP22 siRNA KD of H1299 cells after 48-hour treatment. β-Actin is used as a loading control in the input. Numbers to the left of western blots indicate molecular weight. b. Rad51 and H2A.X foci count per cell for experiment 1a. 120 cells were analyzed for every condition (i.e. 120 cells were analyzed for Rad51 control siRNA, 120 cells were analyzed for Rad51 USP22 siRNA etc.). Experiment was done in triplicate c. FACS analysis results of U2OS cells after USP22 or control siRNA KD and SceI endonuclease cleavage of truncated GFP gene30. EJ5=NHEJ, HDR=Homology-directed repair, and SSA=Single strand annealing. Experiment was performed in triplicate. d. Western blot for experiment 1c with indicated antibodies for indicated cell line noted below. β-Actin is used as a loading control and molecular weight markers are on the left. For all statistical analysis a student’s two-tailed t-test was used to establish p-value. e. Western blot showing gamma-H2A.X is upregulated upon DNA damage in U2OSFokI in the presence of a combined USP22/PALB2 knockdown. Cells were treated with siRNA for 48 hours then FokI treatment was performed for 6 hours before cells were harvested. Numbers to left of western blots indicate molecular weight.
USP22 Localizes at DSBs and is Required for Repair HR Repair Machinery Recruitment
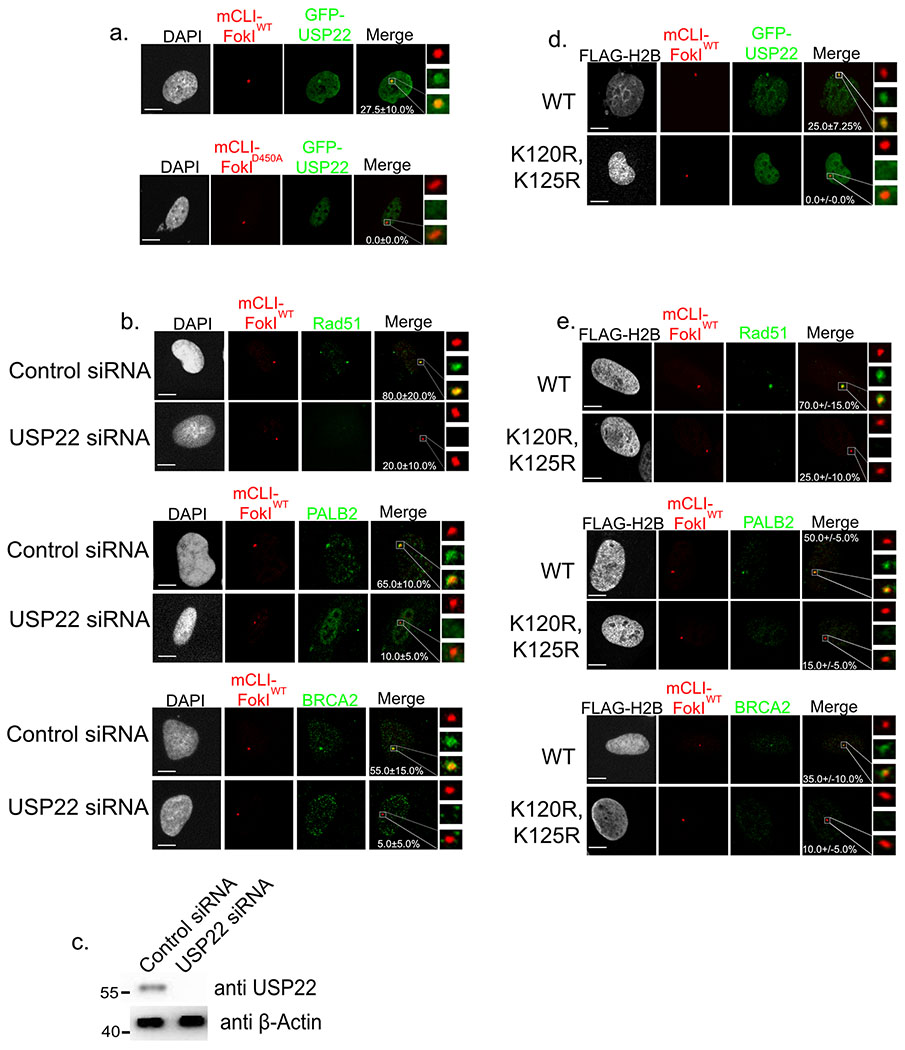
We next considered whether USP22 may localize to DSB sites, and thereby play a direct role in repair. To assay for USP22 recruitment to DSBs we used human U2OS cells with a 256X LacO repeat inserted near the telomere of chromosome 1 (Figure 2a)39. This cell line is also equipped with stable expression of the mCherry-LacI-FokI nuclease fused to a destabilization domain and a modified estradiol receptor. This enables inducible nuclease expression following administration of small molecules Shield1 ligand and 4-OHT39. Cells were transfected with GFP-USP22, and two days later induced to produce DSBs for 6 hours with FokIWT, which was compared to the catalytically inactive mutant (D450A) control. GFP-USP22 was found to localize to LacO array after DSB induction in 27.5+/−10.0% of cells (p-value=0.0089) analyzed while induction with the catalytically dead FokI produced no obvious GFP-USP22 co-localization. Thus, USP22 efficiently localizes DSB sites.
Figure 2:
Both USP22 and H2BK120ub are necessary for efficient recruitment of key HR factors to DSBs. a. U2OSFOKI cells were transfected with GFP-USP22 and treated with 4OHT and SHIELD1 ligand to induce DDR at 256X LacO array by the endonuclease mCherry-LacI-FOKIWT. Catalytically dead D450A version was used as a negative control. Percentage of cells with GFP-USP22 recruitment to array are indicated with +/− SD. White scale bars indicate 5um. 120 cells were counted for each condition for every experimental round. Experiment was done in triplicate. b. U2OSFOKI cells treated with 4OHT and SHIELD1 ligand to induce DDR at the 256X LacO array by the endonuclease mCherry-LacI-FOKIWT with/without USP22 siRNA knockdown for 48 hours. Cells were stained with antibodies against indicated proteins. Percentage of cells with recruitment of indicated protein to the array are indicated with +/−SD. White scale bars indicate 5um. 120 cells were counted for each condition for every experimental round. Experiment was done in triplicate c. Western blot against endogenous USP22 and β-Actin (loading control) after 48-hour siRNA knockdown, refers to cells from 2b. Numbers to left of western blots indicate molecular weight. d. U2OSFOKI cells were transfected with GFP-USP22 and indicated FLAG-H2B type and treated with 4OHT and SHIELD1 ligand to induce DDR at 256X LacO array by the endonuclease mCherry-LacI-FOKIWT. Percentage of cells with GFP-USP22 recruitment to array are indicated with +/− SD. White scale bars indicate 5um. 120 cells were counted for each condition for every experimental round. Experiment was done in triplicate. e. U2OSFOKI cells were transfected with indicated FLAG-H2B type for 48 hours then treated with 4OHT and SHIELD1 ligand to induce DDR at the 256X LacO array by the endonuclease mCherry-LacI-FOKIWT. Cells were stained with antibodies against indicated proteins. Percentage of cells with recruitment of indicated protein to the array are indicated with +/−SD. White scale bars indicate 5um. 120 cells were counted for each condition for every experimental round. Experiment was done in triplicate. For all statistical analysis a student’s two-tailed t-test was used to establish p-value.
Since USP22 localizes to DSBs and is important for Rad51 foci formation, we hypothesized it was upstream of Rad51 and its associated factors and could be necessary for their recruitment to DSBs. Specifically, we sought to examine the influence of USP22 on the localization of Rad51, and associated factors (i.e., PALB2 and BRCA2) to DSBs using the U2OSFOKI cell line system (Figure 2b,c). USP22 KD significantly reduced the recruitment efficiency of Rad51, PALB2, and BRCA2 from 80.0+/−20.0% to 20.0+/−10.0% (p-value=0.0072), 65.0+/−10.0% to 10.0+/−5.0% (p-value=0.0011), and 55.0+/−15.0% to 5.0+/−5.0% (p-value=0.0016), respectively. This suggests USP22 is upstream of these factors in the HR pathway and necessary for their recruitment to DSBs.
USP22 has been shown to deubiquitylate H2BK120ub, and previous work showed this ubiqutin mark on H2B to be necessary for Rad51 and BRCA1 foci formation in MCF7 cells overexpressing myc-H2BWT or the ubiquitin null mutant myc-H2BK120R,K125R, 40. Thus, we sought to test if H2BK120ub is necessary for recruitment of Rad51, PALB2, and BRCA2 to DSBs. To assay for this we used the U2OSFOKI cell line and overexpressed either FLAG-H2BWT or FLAG-H2BK120R, K125R and analyzed recruitment of GFP-USP22, Rad51, PALB2, and BRCA2 after induction of mCherry-LacI-FOKI (Figure 2d,e). Upon overexpression of FLAG-H2BK120R, K125R the recruitment efficiencies of Rad51, PALB2, and BRCA2 were reduced from 70.0+/−15.0% to 25.0+/−10.0% (p-value=0.0124), 50.0+/−5.0% to 15+/−5% (p-value=0.0237), and 35.0+/−10.0% to 10.0+/−5.0% (p-value=0.0179), respectively (Figure 2e). GFP-USP22 showed the same phenotype with no recruitment to the array when FLAG-H2BK120R,K125R was overexpressed (p-value=0.0092). Given that USP22 KD causes moderate increase in global H2Bk120ub and the data show both USP22 and H2BK120ub are clearly important for recruitment of USP22, Rad51, PALB2, and BRCA2 to sites of DSBs, this suggests the modulation and fine tuning of H2Bk120ub is important. Namely, there needs to be both a dynamic process of both ubiquitination and deubiquitination at sites of DSBs for proper recruitment of these key HR factors.
USP22 DUB Domain Interacts with PALB2 WD40 Domain
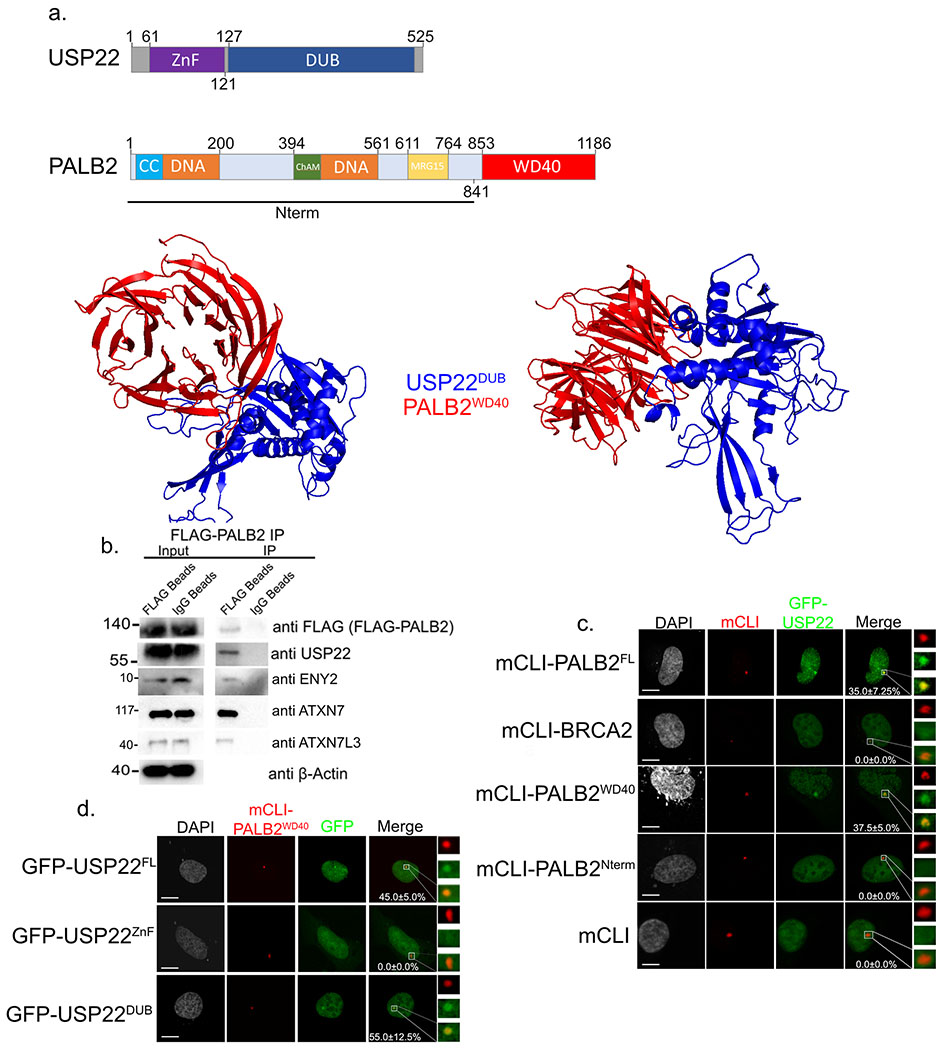
Given that USP22 was affecting BRCA2, PALB2, and Rad51 recruitment to DSBs we wanted to assess if it forms complex with any of these HR proteins. USP22 contains two domains, on its N-terminus is the Zinc Finger (ZnF) domain that is necessary for USP22 to complex with SAGA members ENY2, ATXN7, and ATXN7L3. The C-terminus of USP22 contains the catalytic DUB domain13,14. PALB2 contains several previously characterized domains, which includes its C-terminal WD40 domain (Figure 3a). To begin with, computer modeling was used to assess potential USP22 binding (ZRANK)31,32, since human USP22 has not been crystallized a best fit model of its putative structure was assembled using a crystal structure of the USP22 yeast homolog UBP8 (PDB: 3M99)41. Our in-silico analysis showed an interaction between the C-terminal DUB domain of USP22 and the C-terminal WD40 domain of PALB2 that has been previously crystallized (Figure 3a, lower panels, PDB: 2W18)42. Previous studies have shown there are several DUBs that readily interact with WD40 domains43. To evaluate USP22-PALB2 binding in cells an IP was performed with FLAG-PALB2 in H1299 lung adenocarcinoma cells in which endogenous USP22 was successfully pulled down (Figure 3b). Furthermore, the SAGA components ENY2, ATXN7L3, and ATXN7 that have been previously shown to complex with USP22 also pulled down. Although this shows USP22 can bind both PALB2 and SAGA it does not delineate whether these are two separate populations or whether USP22 can interact with all of these components including PALB2 at the same time. But, based on in-silico model showing the USP22 DUB domain binding PALB2 this should not deter the other SAGA components from binding USP22 as they are known to bind in the N-terminal ZnF region.
Figure 3:
USP22 DUB domain interacts with PALB2 WD40 domain. a. Cartoon of different domains of USP22 and PALB2 (upper). Computer model using ZDOCK predicting USP22 DUB (blue) domain binding to PALB2 WD40 domain (red). b. Immunoprecipitation of FLAG-PALB2 plasmid transiently transfected into H1299 cells. Western blot against FLAG (FLAG-PALB2), USP22, ENY2, ATXN7L3, ATXN7, and β-Actin as a loading control in the input. Numbers to the left of western blots indicate molecular weight. c. U2OSFOKI cells co-transfected with the indicated mCherry-LacI protein to tether it to 256X LacO array along with GFP-USP22. mCherry-LacI alone is used as a negative control. Percentage of cells with recruitment of GFP-USP22 to LacO array are indicated with +/− SD. White scale bars indicated 5um. 120 cells were counted for each condition for every experimental round. Experiment was done in triplicate. d. U2OSFOKI cells co-transfected with mCherry-LacI-PALB2WD40 protein to tether it to 256X LacO array along with indicated GFP-USP22 fragment. Percentage of cells with recruitment of GFP-USP22 to LacO array are indicated with +/− SD. White scale bars indicated 5um. 120 cells were counted for each condition for every experimental round. Experiment was done in triplicate. For all statistical analysis a student’s two-tailed t-test was used to establish p-value.
To deduce what domain of PALB2 USP22 was binding and to ascertain if our in-silico model was correct we used the 256X LacO array in the U2OSFOKI cells. LacI-PALB2FL, LacI-PALB2Nterm (N-terminus lacking the WD40 domain), and LacI-PALB2WD40 (C-terminus containing the WD40 domain) were tethered to the LacO array and recruitment efficiency of GFP-USP22 was analyzed. LacI-BRCA2 and LacI were used as negative controls (Figure 3a,c). The only fragments able to recruit GFP-USP22 to the LacO array were LacI-PALB2FL (35.0+/−7.25%, p-value=0.0013) and LacI-PALB2WD40 (37.5+/−5.0%, p-value=0.0017) containing the WD40 domain which aligns with our computer model prediction. Furthermore, the DUB domain of USP22 (GFP-USP22DUB) was also specifically recruited by LacI-PALB2WD40 (55.0+/−12.5%, p-value=0.0011) while the ZnF domain of USP22 (GFP-USP22ZnF) was not recruited at all (Figure 3a,d) again confirming our in-silico model. Taken together, these results show USP22 is interacting with PALB2 through its WD40 domain.
PALB2 WD40 Domain Directly Binds USP22 and stimulates its Catalytic Activity
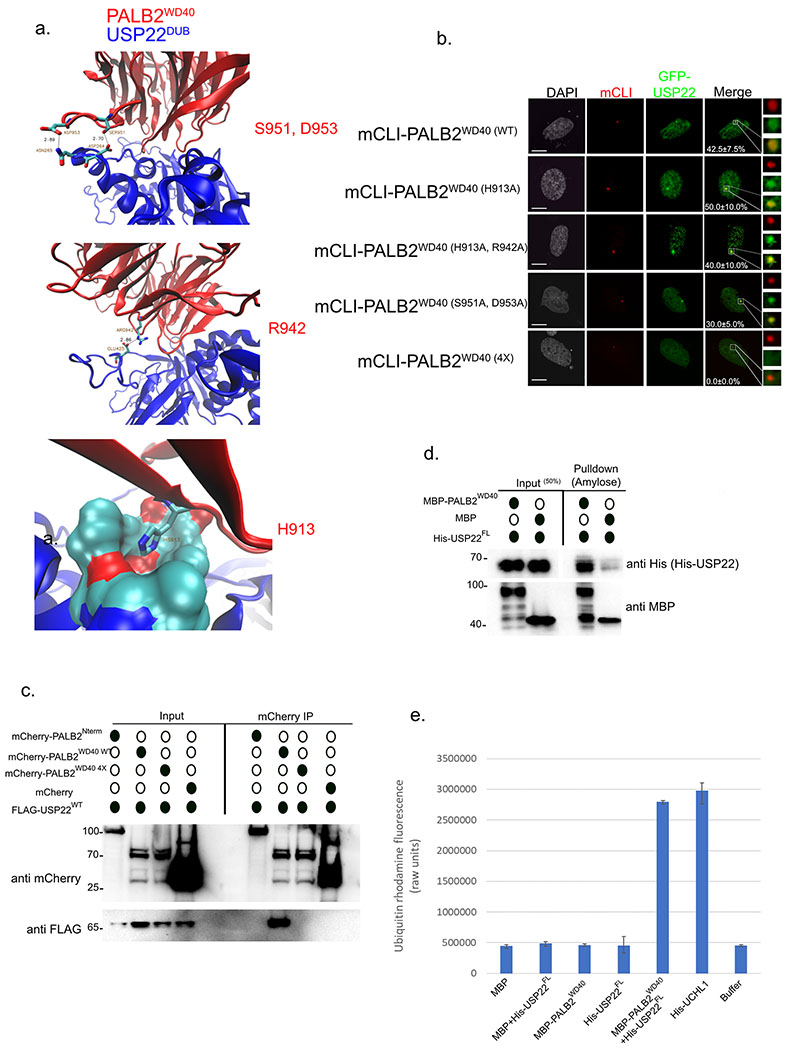
Using our in-silico model of PALB2-USP22 binding (Figure 4a) we predicted several residues on the PALB2 WD40 domain that looked critical for this interaction; S951, N953 (upper panel), R942 (middle panel), and H913 (lower panel). LacI-PALB2WD40 point mutants were made in different combinations, tethered to the LacO array in U2OSFOKI cells, and assayed for their ability to recruit GFP-USP22 (Figure 4b). The LacI-PALB2WD40 (4X) mutant with all four residues (H913, R952, S951, and N953) mutated to Alanine was not able to recruit GFP-USP22 to the array. The WD404x mutant was also assayed for its ability to recruit endogenous BRCA2 and Rad51, two known binding partners of PALB2 that can bind its WD40 domain44,45, to the LacO array (Figure S1A). Both Rad51 and BRCA2 were robustly recruited to the array vy the WD404x mutant in similar frequencies to the WT version, which makes sense as both of these proteins have been shown to bind to different blades of the WD40 region of PALB242,44,46. Using an mCherry tagged version of this mutant (mCherry-PALB2WD40 4X) we performed an IP experiment in H1299 lung adenocarcinoma cells to assess its ability to pull down FLAG-USP22WT (Figure 4c). Consistent with our LacO array recruitment experiment, (Figure 4b) mCherry-PALB2WD40 4X could not pull-down FLAG-USP22WT while mCherry-PALB2WD40 WT robustly bound FLAG-USP22WT. The fragment mCherry-PALB2Nterm that does not contain the WD40 domain was not able to pull-down FLAG-USP22WT validating our previous LacO array data (Figure 3c). To elucidate if USP22 was directly binding PALB2WD40 we performed in-vitro pulldown experiments using MBP-PALB2WD40 as the bait along with His-USP22 (Figure 4d). MBP-PALB2WD40 successfully pulled down His-USP22 demonstrating their interaction is direct and further validating our in-silico model (Figure 3a).
Figure 4:
PALB2WD40 directly binds USP22 and stimulates its catalytic DUB activity. a. In-silico model using ZDOCK of USP22DUB binding PALB2WD40 predicting key residues involved in the interaction. b. U2OSFOKI cells co-transfected with the indicated mCherry-LacI protein to tether it to 256X LacO array along with GFP-USP22. Percentage of cells with recruitment of GFP-USP22 to LacO array are indicated with +/− SD. White scale bars indicated 5um. 120 cells were counted for each condition for every experimental round. Experiment was done in triplicate. c. Immunoprecipitation of mcherry-PALB2 fragments plasmids transiently transfected into H1299 cells. Western blot against mCherry, FLAG (FLAG-USP22), Rad51. mCherry alone is used as a negative control. Numbers to the left of western blots indicate molecular weight. d. In-vitro pulldown using recombinant MBP-PALB2WD40 (prey) and His-USP22FL (bait). MBP was also incubated with His-USP22 as a negative control. Western blot was performed against His (His-USP22) and MBP (MBP-PALB2WD40, MBP). Numbers to the left of western blots indicate molecular weight. e. Ubiquitin cleavage assay using rhodamine-ubiquitin that is quenched and upon cleavage of ubiquitin fluoresces. Proteins indicated below were pre-incubated to form their respective complexes then rhodamine-ubiquitin was added. Units on vertical axis indicate raw fluorescent units at 565nm. Buffer alone was used as a negative control for auto-fluorescence baseline. Error bars indicated +/− SD. Experiment was done in triplicate.
Previous work on UAF1-USP1 binding showed WD40 domain of UAF1 was sufficient to act as an adapter protein and activate the catalytic DUB activity of USP1, which on its own has very little intrinsic catalytic activity in-vitro47. To evaluate if PALB2WD40 had the same effect on USP22 we performed an in-vitro ubiquitin cleavage assay using ubiquitin bound to the rhodamine-red fluorophore (Figure 4e). Upon cleavage of ubiquitin from rhodamine-red, the fluorophore is no longer quenched and fluoresces at roughly 535nm denoting ubiqutin substrate cleavage. His-USP22 was mixed with MBP-PALB2WD40 in stoichiometric amounts to form a complex and then rhodamine-ubiquitin was added in and incubated with the complexes for 30 minutes. UHCL1 was used as a positive control as it is a robust deubiquitinating enzyme that is active in-vitro. Upon addition of MBP-PALB2WD40, USP22 enzymatic activity was robustly activated (Figure 4e) showing that PALB2WDR directly binds USP22 and stimulates its catalytic activity.
USP22 modulates PALB2 and BRCA2 levels to promote chemoresistance in lung adenocarcinoma
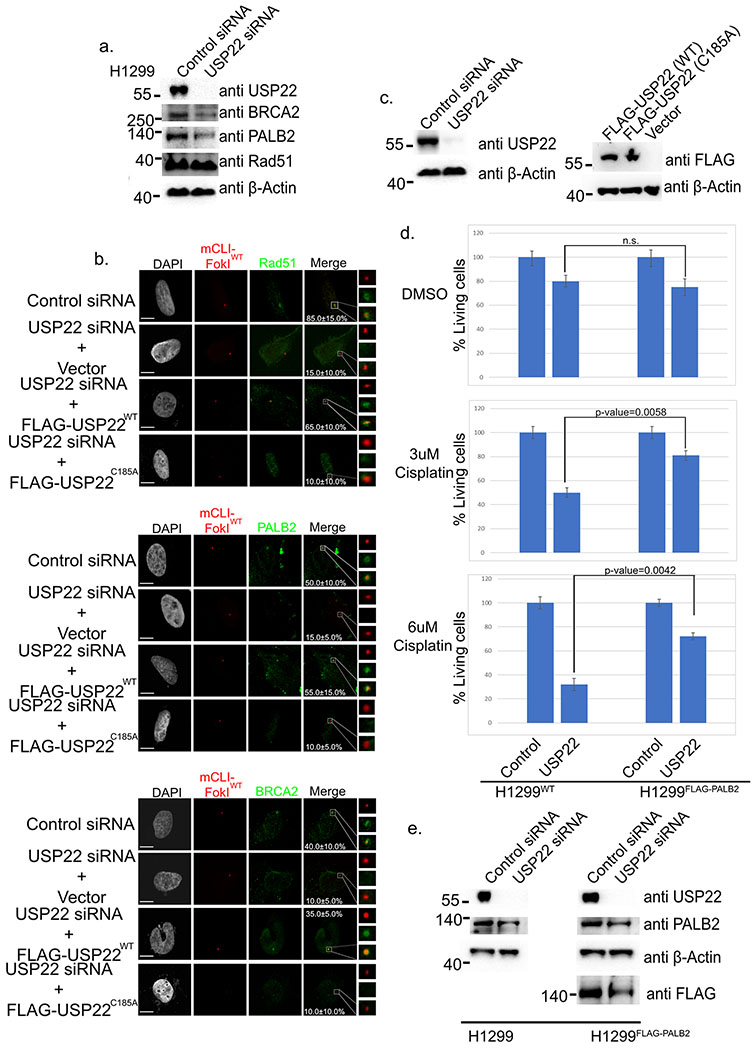
USP22 has been shown to regulate the protein stability of several proteins it binds to including Sirt1, CCND1, and EGFR18,19,48. We postulated that USP22 may also stabilize PALB2 levels. To assess this USP22 KD was performed in H1299 lung adenocarcinoma cells (Figure 5a). Both PALB2 and BRCA2 proteins levels were markedly reduced while Rad51 was not affected. Given that PALB2 and BRCA2 help localize Rad51 to sites of DNA damage their lower levels upon USP22 KD could be the reason for the lack of Rad51 recruitment to sites of DSBs in the U2OSFOKI cells (Figure 2b). We also performed a rescue experiment in the U2OSFOKI cells by overexpressing FLAG- U2OSWT or a catalytically dead FLAG- U2OSC185A mutant after knockdown of endogenous USP22 and induction of DNA damage (Figure 5b,c). As was predicted USP22WT overexpression rescued PALB2, BRCA2, and Rad51 recruitment to the LacO array after DNA damage, but USP22C185A was insufficient to rescue PALB2. BRCA2, and Rad51 recruitment. We previously showed that depleting USP22 led to increased Cisplatin sensitivity27. To determine whether USP22’s role in regulating PALB2 levels plays a part in the association between USP22 expression and chemotherapy resistance, we set out to determine whether PALB2 overexpression rescues USP22 KD cells from Cisplatin sensitivity. We performed USP22 KD in H1299 cells with/without FLAG-PALB2 overexpression with 3uM Cisplatin, 6uM Cisplatin, or DMSO as a control (Figure 5d,e). After 24 hours cells were treated with 3uM or 6uM Cisplatin for 72 more hours and then viable cells were counted. As was previously shown, WT H1299 cells had a 50% and 68% reduction in viable cells with 3uM and 6uM Cisplatin respectively upon USP22 KD. Strikingly, PALB2 overexpression in H1299 cells only had 20% and 28% reduction viable cells with 3uM and 6uM Cisplatin treatment respectively after USP22 KD as compared with control. To determine whether USP22 is modulating PALB2 and BRCA2 protein levels at the transcriptional level we performed qPCR of BRCA2 and PALB2 with and without USP22 knockdown after 48 hours (Figure S1b). The mRNA levels of BRCA2 and PALB2 showed no statistical differences between USP22 knockdown and control conditions showing that USP22 is modulating these two proteins at the translational level. To further validate USP22 was modulating and stabilizing BRCA2 and PALB2 levels at the translational level, cells were depleted of USP22 again with and without treatment of the proteasome inhibitor MG132 to see if this could rescue PALB2 and BRCA2 levels in a USP22 knockdown background (Figure S1c). As was predicted, MG132 was clearly able to rescue PALB2 and BRCA2 protein levels further validating USP22 is positively modulating these proteins at the translational level. Lastly, H1299 cells treated with USP22/PALB2 separately or combinatorialy along with Cisplatin had a significantly decreased IC50 when compared to control (Figure S1d, 5.1uM (control), 2.0uM (PALB2 siRNA), 2.1uM (USP22 siRNA), and 1.4uM (USP22+PALB2 siRNA)
Figure 5:
USP22 modulates PALB2 protein stability aiding in chemoresistance. a. Western blot of H1299 cells against endogenous USP22, BRCA2, PALB2, Rad51, and β-Actin (loading control) after 48-hour siRNA knockdown. Numbers to left of western blots indicate molecular weight. b. U2OSFOKI cells treated with 4OHT and SHIELD1 ligand to induce DDR at the 256X LacO array by the endonuclease mCherry-LacI-FOKIWT with/without USP22 siRNA knockdown for 48 hours and/or transfection of empty vector or FLAG-USP22 plasmids. Cells were stained with antibodies against indicated proteins. Percentage of cells with recruitment of indicated protein to the array are indicated with +/−SD. White scale bars indicate 5um. 120 cells were counted for each condition for every experimental round. Experiment was done in triplicate. c. Western blot against endogenous USP22 and β-Actin (loading control) after 48-hour siRNA knockdown, refers to cells from 5b. Numbers to left of western blots indicate molecular weight. (Right) western blot to deduce expression of indicated FLAG-USP22 construct. Western blot done against FLAG and β-Actin (loading control). Numbers to left of western blots indicate molecular weight. d. H1299 or H1299FLAG-PALB2 cells stably overexpressing indicated protein had siRNA KD performed then 3uM/6uM cisplatin or DMSO was added in for an additional 72 hours. Cells were washed then counted. Graph indicates total % of living cells as they were pertaining to the control for both cell lines. Experiment was done in triplicate. e. Western blot for cells from Figure 5d against endogenous USP22, PALB2, and β-Actin (loading control). FLAG antibody was used to assay for FLAG-PALB2. Numbers to left of western blots indicate molecular weight.
Discussion
In this study we establish USP22 can actively localize to DSBs. We also demonstrate USP22 is necessary for BRCA2, PALB2, and Rad51 localization to sites of DSBs. Moreover, we show H2BK120ub is also important for localization of BRCA2, PALB2, Rad51, and even USP22 to DSBs. We identified a direct interaction between the C-terminal WD40 domain of PALB2 and the DUB domain of USP22 and that this interaction was necessary and sufficient to activate USP22 catalytic DUB activity in-vitro. Lastly, we show USP22 positively regulates BRCA2 and PALB2 levels at the protein level. These findings further our mechanistic understanding of USP22 and show a new role for it in DNA repair in conjunction with the core HR protein PALB2.
USP22 promotes DSB repair via HR
DSBs are repaired by HR in the S/G2 phase of the cell cycle and involves assembly of BRCA1, BRCA2, PALB2, and Rad51. BRCA1 is important for the localization of and accumulation of PALB2 to sites of DSBs (and is also therefore required for BRCA2 and Rad51 localization)49. In this study we show an unexpected role for USP22 in HR. We show USP22 KD impairs HR efficiency in U2OS cells after I-SceI cleavage of a truncated GFP gene. We show USP22 KD impairs BRCA2, PALB2, and Rad51 recruitment to a single DSB in U2OSFOKI cells and impairs Rad51 foci formation in H1299 cells after treatment with the DNA damaging agents Cisplatin and Camptothecin. Furthermore, overexpression of siRNA resistant FLAG-USP22 in U2OSFOKI cells rescues the localization of BRCA2, PALB2, and Rad51 to the array after DNA damage. Taken together, our results show a direct role for USP22 in repair of DSBs through the HR pathway.
PALB2 binding stimulates USP22 DUB activity
Previous work on UAF1, a WD40 domain containing protein, has shown it acts as an adapter protein and can activate the catalytic activity of several DUBs. The WD40 domain of USP1 binds and stimulates the catalytic activity of USP1, which on its own, has very little intrinsic catalytic activity. Two more DUBs, USP12 and USP46, were also found to bind UAF1 which also stimulated their DUB activity50,51,52,53. In fact, many human DUBs are found to interaction with WD40 domain containing proteins43. In this study we show USP22 specifically interacts with the C-terminal WD-40 domain of PALB2 and this interaction is necessary and sufficient to stimulate USP22 DUB activity. This finding is novel because USP22 has been thought to require adaptor proteins ATXN7L3 and ENY2 for catalytic activity12,54. Without these adaptor proteins USP22 has very little intrinsic catalytic activity in-vitro and the binding of adaptor proteins is believed to change the confirmation of USP22 and other DUBs thereby making them catalytically active. This gives the cell control over the deubiquitylation of substrates. The crystal structure of USP22 alone or in conjunction with PALB2WD40 has not been solved so understanding the exact conformational change USP22 takes to become catalytically active is not yet known. Understanding the interaction of USP22-PALB2 and indeed the interaction of other DUBs with WD40 domain containing proteins could present a new way to target DUBs for cancer therapies.
USP22 regulates PALB2 and BRCA2 levels at the translational level
Because DUBs are involved in cleaving ubiquitin off proteins it is no surprise many of them can positively regulate protein stability and counteract proteasome degradation. Many DUBs, including USP22, can also regulate protein expression at the transcriptional level as well55. USP22 for instance is recruited via Myc to several Myc target genes such as CAD and MTA1 positively regulating their transcription56. Our study reveals USP22 positively regulates PALB2 and BRCA2 levels at the translational level. These results explain the lack of PALB2, BRCA2, and Rad51 recruitment to the LacO array after DNA damage in the U2OSFOKI cells as PALB2 is necessary for recruitment of these factors to damaged DNA. Although this is an important and new function of USP22 in DDR this does not rule out other potential roles for USP22 in DNA repair. USP22 is involved in the deubiquitylation of H2BK120ub, a mark that is found at DSB sites, USP22 could be a “reader” for this mark and potentiate the recruitment of other DNA damage factors through this ubiquitin mark. Furthermore, the lysine residues on PALB2 that USP22 could be potentially deubiquitinating have not been mapped by this study or others and is an excellent future direction for further study on the USP22-PALB2 interaction.
Supplementary Material
Acknowledgements
We would like to thank Dr. Roger Greenberg in the Perelman School of Medicine at the University of Pennsylvania for his methods and generous gift of the U2OSFOKI cell line.
Funding:
This work was supported by the V Foundation for Cancer Research (D.J. Raz), the Stop Cancer Foundation (D.J. Raz), the generous support of the Baum Family Foundation, and R01CA197506 (J.M.S.). Research reported in this publication included work performed in the Light Microscopy and Computational Therapeutics Cores supported by the National Cancer Institute of the National Institutes of Health under award number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest
References:
- 1.Hoeijmakers JHJ Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Wooster R et al. Identification of the breast cancer susceptibility gene BRCA2. Nature 378, 789 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Wooster R et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science (80-. ). 265, 2088 LP-2090 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Ahlskog JK, Larsen BD, Achanta K & Sørensen CS ATM/ATR-mediated phosphorylation of PALB2 promotes RAD51 function. EMBO Rep. 17, 671–681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nepomuceno TC et al. The Role of PALB2 in the DNA Damage Response and Cancer Predisposition. Int. J. Mol. Sci 18, 1886 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luijsterburg MS et al. A PALB2-interacting domain in RNF168 couples homologous recombination to DNA break-induced chromatin ubiquitylation. Elife 6, e20922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orthwein A et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Turcu FER, Ventii KH & Wilkinson KD Regulation and Cellular Roles of Ubiquitin-specific Deubiquitinating Enzymes. Annu. Rev. Biochem 78, 363–397 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandrasekharan MB, Huang F & Sun Z-W Histone H2B ubiquitination and beyond: Regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics 5, 460–468 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadeghi L, Siggens L, Svensson JP & Ekwall K Centromeric histone H2B monoubiquitination promotes noncoding transcription and chromatin integrity. Nat. Struct. Mol. Biol 21, 236–243 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Moyal L et al. Requirement of ATM-Dependent Monoubiquitylation of Histone H2B for Timely Repair of DNA Double-Strand Breaks. Mol. Cell 41, 529–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X-Y et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activator-driven transcription and cell cycle progression. Mol. Cell 29, 102–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lang G et al. The Tightly Controlled Deubiquitination Activity of the Human SAGA Complex Differentially Modifies Distinct Gene Regulatory Elements . Mol. Cell. Biol 31, 3734–3744 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y et al. A TFTC/STAGA Module Mediates Histone H2A and H2B Deubiquitination, Coactivates Nuclear Receptors, and Counteracts Heterochromatin Silencing. Mol. Cell 29, 92–101 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Nakamura K et al. Regulation of Homologous Recombination by RNF20-Dependent H2B Ubiquitination. Mol. Cell 41, 515–528 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Kari V, Shchebet A, Neumann H & Johnsen SA The H2B ubiquitin ligase RNF40 cooperates with SUPT16H to induce dynamic changes in chromatin structure during DNA double-strand break repair. Cell Cycle 10, 3495–3504 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Game JC, Williamson MS, Spicakova T & Brown JM The RAD6/BRE1 Histone Modification Pathway in Saccharomyces Confers Radiation Resistance Through a RAD51-Dependent Process That Is Independent of RAD18. Genetics 173, 1951–1968 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Z et al. USP22 Antagonizes p53 Transcriptional Activation by Deubiquitinating Sirt1 to Suppress Cell Apoptosis and Is Required for Mouse Embryonic Development. Mol. Cell 46, 484–494 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Gennaro VJ et al. Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc. Natl. Acad. Sci (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D et al. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J. Cell. Physiol 232, 3664–3676 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Martinez E et al. Human STAGA Complex Is a Chromatin-Acetylating Transcription Coactivator That Interacts with Pre-mRNA Splicing and DNA Damage-Binding Factors In Vivo. Mol. Cell. Biol 21, 6782–6795 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao P, Meas R, Dorgan KM & Smerdon MJ UV damage-induced RNA polymerase II stalling stimulates H2B deubiquitylation. Proc. Natl. Acad. Sci. U. S. A 111, 12811–12816 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramachandran S et al. The SAGA Deubiquitination Module Promotes DNA Repair and Class Switch Recombination through ATM and DNAPK-Mediated γH2AX Formation. Cell Rep. 15, 1554–1565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C et al. The H2B deubiquitinase Usp22 promotes antibody class switch recombination by facilitating non-homologous end joining. Nat. Commun 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X et al. High expression of USP22 predicts poor prognosis and advanced clinicopathological features in solid tumors: a meta-analysis. Onco. Targets. Ther 11, 3035–3046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang A et al. USP22 Induces Cisplatin Resistance in Lung Adenocarcinoma by Regulating γH2AX-Mediated DNA Damage Repair and Ku70/Bax-Mediated Apoptosis. Front. Pharmacol 8, 274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yun X et al. Targeting USP22 Suppresses Tumorigenicity and Enhances Cisplatin Sensitivity Through ALDH1A3 Downregulation in Cancer-Initiating Cells from Lung Adenocarcinoma. Mol. Cancer Res 16, 1161 LP-1171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Services H CDC WONDER On-Line Database, Compiled from Compressed Mortality File 1999-2016. Centers Dis. Control Prev. Natl. Cent. Heal. Stat 2, (2017). [Google Scholar]
- 29.Nardi IK, Zasadzińska E, Stellfox ME, Knippler CM & Foltz DR Licensing of Centromeric Chromatin Assembly through the Mis18α-Mis18β Heterotetramer. Mol. Cell 61, 774–787 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunn A & Stark JM in (ed. Bjergbæk L) 379–391 (Humana Press, 2012). doi: 10.1007/978-1-61779-998-3_27 [DOI] [Google Scholar]
- 31.Pierce B & Weng Z ZRANK: Reranking protein docking predictions with an optimized energy function. Proteins Struct. Funct. Bioinforma 67, 1078–1086 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Pierce BG, Hourai Y & Weng Z Accelerating Protein Docking in ZDOCK Using an Advanced 3D Convolution Library. PLoS One 6, e24657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosano GL & Ceccarelli EA Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol 5, 172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haaf T, Golub EI, Reddy G, Radding CM & Ward DC Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. U. S. A 92, 2298–2302 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogakou EP, Boon C, Redon C & Bonner WM Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol 146, 905–916 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramachandran S et al. The SAGA Deubiquitination Module Promotes DNA Repair and Class Switch Recombination through ATM and DNAPK-Mediated γH2AX Formation. Cell Rep. 15, 1554–1565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramachandran S et al. The SAGA Deubiquitination Module Promotes DNA Repair and Class Switch Recombination through ATM and DNAPK-Mediated γH2AX Formation. Cell Rep. 15, 1554–1565 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pohl TJ & Nickoloff JA Rad51-Independent Interchromosomal Double-Strand Break Repair by Gene Conversion Requires Rad52 but Not Rad55, Rad57, or Dmc1. Mol. Cell. Biol 28, 897 LP-906 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang J et al. Acetylation Limits 53BP1 Association with Damaged Chromatin to Promote Homologous Recombination. Nat. Struct. Mol. Biol 20, 317–325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura K et al. Regulation of Homologous Recombination by RNF20-Dependent H2B Ubiquitination. Mol. Cell 41, 515–528 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Köhler A, Zimmerman E, Schneider M, Hurt E & Zheng N Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141, 606–617 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliver AW, Swift S, Lord CJ, Ashworth A & Pearl LH Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 10, 990–996 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villamil MA, Liang Q & Zhuang Z The WD40-repeat protein-containing deubiquitinase complex: catalysis, regulation, and potential for therapeutic intervention. Cell Biochem. Biophys 67, 111–126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dray E et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Publ. Gr 17, 1255–1259 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buisson R et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Publ. Gr 17, 1247–1254 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J-Y et al. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene 33, 4803 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohn MA et al. A UAF1-Containing Multisubunit Protein Complex Regulates the Fanconi Anemia Pathway. Mol. Cell 28, 786–797 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Zhang H et al. USP22 promotes resistance to EGFR-TKIs by preventing ubiquitination-mediated EGFR degradation in EGFR-mutant lung adenocarcinoma. Cancer Lett. 433, 186–198 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Zhang F et al. PALB2 Links BRCA1 and BRCA2 in the DNA-Damage Response. Curr. Biol 19, 524–529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dharadhar S, Clerici M, van Dijk WJ, Fish A & Sixma TK A conserved two-step binding for the UAF1 regulator to the USP12 deubiquitinating enzyme. J. Struct. Biol 196, 437–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohn MA, Kee Y, Haas W, Gygi SP & D’Andrea AD UAF1 Is a Subunit of Multiple Deubiquitinating Enzyme Complexes. J. Biol. Chem 284, 5343–5351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sowa ME, Bennett EJ, Gygi SP & Harper JW Defining the Human Deubiquitinating Enzyme Interaction Landscape. Cell 138, 389–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murai J et al. The USP1/UAF1 Complex Promotes Double-Strand Break Repair through Homologous Recombination. Mol. Cell. Biol 31, 2462–2469 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lang G et al. The Tightly Controlled Deubiquitination Activity of the Human SAGA Complex Differentially Modifies Distinct Gene Regulatory Elements . Mol. Cell. Biol 31, 3734–3744 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammond-Martel I, Yu H & Affar EB Roles of ubiquitin signaling in transcription regulation. Cell. Signal 24, 410–421 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Zhang X-Y et al. The Putative Cancer Stem Cell Marker USP22 Is a Subunit of the Human SAGA Complex Required for Activated Transcription and Cell-Cycle Progression. Mol. Cell 29, 102–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.