Abstract
The presence of a copper-containing dissimilatory nitrite reductase gene (nirK) was discovered in several isolates of β-subdivision ammonia-oxidizing bacteria using PCR and DNA sequencing. PCR primers Cunir3 and Cunir4 were designed based on published nirK sequences from denitrifying bacteria and used to amplify a 540-bp fragment of the nirK gene from Nitrosomonas marina and five additional isolates of ammonia-oxidizing bacteria. Amplification products of the expected size were cloned and sequenced. Alignment of the nucleic acid and deduced amino acid (AA) sequences shows significant similarity (62 to 75% DNA, 58 to 76% AA) between nitrite reductases present in these nitrifiers and the copper-containing nitrite reductase found in classic heterotrophic denitrifiers. While the presence of a nitrite reductase in Nitrosomonas europaea is known from early biochemical work, preliminary sequence data from its genome indicate a rather low similarity to the denitrifier nirKs. Phylogenetic analysis of the partial nitrifier nirK sequences indicates that the topology of the nirK tree corresponds to the 16S rRNA and amoA trees. While the role of nitrite reduction in the metabolism of nitrifying bacteria is still uncertain, these data show that the nirK gene is present in closely related nitrifying isolates from many oceanographic regions and suggest that nirK sequences retrieved from the environment may include sequences from ammonia-oxidizing bacteria.
Nitrite reduction by ammonia-oxidizing bacteria is intriguing not only because of the uncertain physiological role it plays in these organisms, which require oxygen to oxidize ammonia and generate energy (17), but also because it has been demonstrated that ammonia oxidizers, such as Nitrosomonas europaea, produce the environmentally important trace gases nitric oxide (NO) and nitrous oxide (N2O) by reduction of nitrite (33, 35). In addition to N. europaea, other ammonia-oxidizing bacteria are known to produce NO and N2O (14, 25), although the mechanism is less certain. These bacteria use O2 for two separate functions, first as a substrate in the oxidation of ammonia to hydroxylamine and second as a terminal electron acceptor for their electron transport chain. There is no known substitute for O2 in the first function, but it may be that in low-O2 environments, nitrogen oxides can substitute for O2 as a terminal electron acceptor, that is, perform a denitrification-like respiration (4).
Classical denitrifying bacteria have a series of enzymes, nitrate reductase, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase, which allow them to utilize nitrate (NO3−), nitrite (NO2−), NO, and N2O, respectively, as terminal electron acceptors in anaerobic respiration (51). Nitrite reductase is a central enzyme in the denitrification pathway because it produces the first gaseous product, NO, which is largely unavailable for use by other organisms (48). There are two primary forms of respiratory nitrite reductase, one containing cytochromes c and d1 (cd1-NiR) and the other containing two copper centers (Cu-NiR) at each active site. The Cu-NiR is encoded by the gene nirK, and the cd1-NiR is encoded by the gene nirS. Despite their structural differences (1, 47), the two forms of nitrite reductase perform the same physiological function in denitrifying bacteria.
The process of nitrifier denitrification, the reduction of NO2− to gaseous products (NO and N2O) by ammonia-oxidizing bacteria, was indicated by early biochemical work. A soluble nitrite reductase from N. europaea, the most familiar terrestrial nitrifier, was first described by Hooper (19). This enzyme produced a mixture of NO and N2O from NO2− and hydroxylamine (NH2OH). However, the mechanism of NO and N2O formation under normal physiological conditions, whether through the decomposition of an unstable intermediate in the oxidation of NH2OH to NO2− or purely from NO2− reduction, was unclear until the use of 15N tracers demonstrated that N2O was derived primarily from NO2− (33, 35). The partially purified nitrite reductase from N. europaea was further shown to be a copper-containing enzyme with biochemical similarities (spectroscopic characteristics, inhibition profile, and reaction products) to the copper-containing nitrite reductases from classical heterotrophic denitrifiers (29, 36). In addition, expression of the N. europaea nitrite reductase is enhanced by low O2 levels (29), which is similar to some denitrifying nitrite reductases (21) and is consistent with the demonstration of higher N2O and NO yields at lower oxygen levels by cultures of various ammonia-oxidizing bacteria (14, 25). Taken together, these observations suggest that N. europaea shares a nitrite-reducing mechanism with classical denitrifiers (36). The goals of this research are to understand, at a genetic level, the pathway of N2O production in ammonia-oxidizing bacteria and to determine whether it is related to the typical denitrification pathway.
In the present study, the gene for nitrite reductase (nirK) was discovered in several ammonia-oxidizing bacteria, and the partial sequences obtained were compared to nirKs from denitrifying bacteria. In addition, the phylogeny of nitrite reductase in ammonia-oxidizing bacteria was compared to their phylogenies based on 16S rRNA and ammonia monooxygenase (amoA) genes.
MATERIALS AND METHODS
Bacterial strains and culturing.
Nitrosococcus oceani strain 27 and marine ammonia-oxidizing isolates URW (North Pacific), NO3W (North Pacific), C-45 (Gulf of Maine), C-113a (Red Sea), and TA-921i-NH4 (Chesapeake Bay), characterized by Ward and Carlucci (40), were maintained in the seawater medium of Watson (46). Nitrosomonas marina (B. B. Ward culture collection, obtained from S. Watson, 1977) was maintained in similar medium made up in 50% seawater. N. europaea (Schmidt strain, ATCC 19718), Nitrosomonas eutropha (Schmidt strain), and Nitrosospira briensis C-128 were maintained in freshwater medium (38). Working cultures were maintained in semicontinuous batch culture by periodically replacing half of the culture with fresh medium (44). Nitrifier cultures were grown at room temperature (18°C) in the dark with no agitation. Pseudomonas aureofaciens ATCC 13985 and Alcaligenes faecalis ATCC 8750 were grown in Luria-Bertani (LB) medium with agitation at room temperature, and Alcaligenes xylosoxidans ATCC 15173 was grown in Difco nutrient broth at 26°C.
DNA extraction.
Cells were harvested by filtration and then washed and resuspended in Tris-EDTA buffer. Standard procedures for DNA extraction were followed (2). DNA was tested for PCR inhibition by amplification with universal eubacterial 16S ribosomal DNA (rDNA) primers (24).
nirK PCR amplification.
PCR primers were designed based on conserved regions of the nirK gene, encoding the copper-containing nitrite reductase (Cu-NiR) from P. aureofaciens, A. faecalis, A. xylosoxidans, Rhizobium hedysari, Achromobacter cycloclastes, Bradyrhizobium japonicum, and Rhodobacter sphaeroides obtained from the GenBank database (3). Primers Cunir3 [5′-CGT CTA (C/T)CA (C/T)TC CGC (A/C/G)CC-3′] and Cunir4 [5′-GCC TCG ATC AG(A/G) TT(A/G) TGG-3′] amplify a 537- or 540-bp fragment of the nirK gene, depending on the target organism. The PCRs optimized for Cunir3-Cunir4 amplification used 50 mM KCl, 10 mM Tris base (pH 8.0), 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphates (dNTP), 50 pmol of each primer, 0.5 μl of DNA template (∼50 ng/μl), and 1 U of Taq polymerase in a 50-μl total reaction volume. A touchdown PCR program, similar to that used by Braker et al. (5), was used for the amplification of nirK with the Cunir3 and Cunir4 (Cunir3-4) primers. Denaturation at 95°C for 2 min was followed by 10 cycles of 94°C for 30 s, 45°C (−0.5°C per cycle) for 40 s, and 72°C for 40 s, 20 cycles of 94°C for 30 s, 43°C for 40 s, and 72°C for 40 s, and a final cycle of 94°C for 30 s, 43°C for 40 s, and 72°C for 7 min.
Sequencing nirK products.
Cunir3-4 amplification products of the expected size were extracted from a 1% agarose gel using the Qiaquick gel extraction kit (Qiagen) and cloned using the Topo-TA cloning kit (Invitrogen). Transformants were selected on LB plates with kanamycin (50 μg/ml), spread with 40 μl of isopropylthiogalactopyranoside (IPTG; 100 mM) and 40 μl of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (20 mg/ml in dimethylformamide). White transformant colonies were screened by PCR for inserts of the correct size. Colonies were picked directly into tubes containing 49.5 μl of PCR mix with 50 mM KCl, 10 mM Tris base (pH 8.0), 2.5 mM MgCl2, 200 μM each dNTP, and 3.2 pmol each of T7 and M13 (reverse) primers. The PCR cycling protocol recommended by the manufacturer (Invitrogen) was used for T7-M13 amplification. T7-M13 PCR products were used as templates for cycle sequencing of both strands using T7 and M13 (reverse) primers and the BigDye terminator kit (Perkin-Elmer). Cycle-sequencing products were precipitated according to the manufacturer's instructions and sequenced on the ABI310 genetic analyzer (Perkin-Elmer).
Ammonia monooxygenase PCR amplification and sequencing.
Fragments of the amoA gene were PCR amplified from genomic DNA using the Amo189 and Amo682 primers (18). PCR products from amoA amplifications were cloned and sequenced as described above for nirK PCR products.
Sequence analysis.
T7 and M13 (reverse) sequences from 7 to 10 clones were assembled using AutoAssembler version 1.4.0 (Perkin-Elmer) into a single consensus sequence for each cloning experiment. Sequence Navigator version 1.0.1 (Perkin-Elmer) was used to align homologous regions of 16S rRNA and amoA gene or nirK deduced amino acid sequences from different organisms. These aligned sequences were analyzed by distance matrix methods using DNADIST and PROTDIST programs, respectively, in the phylogenetic inference package PHYLIP 3.572 (13). Distances were calculated using the Kimura two-parameter model for nucleic acid sequences (22) and the Dayhoff PAM 100 matrix for amino acid sequences (12). The input files were each bootstrapped 100 times using the SEQBOOT program of PHYLIP prior to distance matrix analyses. Neighbor-joining trees were produced for each pseudoreplicate analysis. The CONSENSE program was used to compute the majority-rule consensus tree for each set of molecular sequences, and trees were drawn using the DRAWGRAM program from PHYLIP.
Nucleotide sequence accession numbers.
The partial amoA and nirK sequences from N. marina, NO3W, URW, C-45, C-113a, and TA-921i-NH4 have been deposited in the GenBank database under accession numbers AF339038 to AF339043 (amoA) and AF339044 to AF339049 (nirK), respectively. The 16S rRNA gene sequences for NO3W (AF338206), URW (AF338210), C-45 (AF338203), C-113a (AF338200), and TA-921i-NH4 (AF338207) were obtained from M. Voytek.
RESULTS
Primer development.
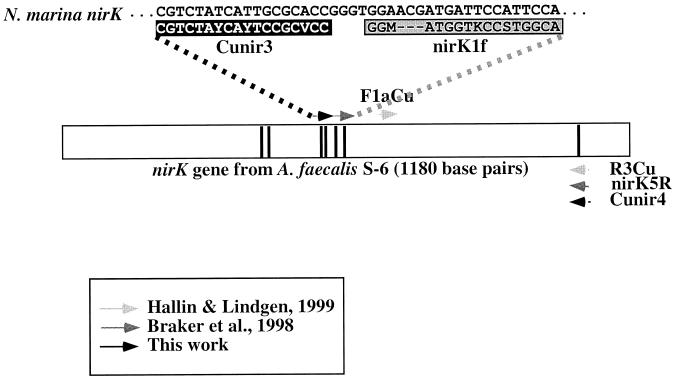
The locations of the target sequences for primers Cunir3 and Cunir4 are compared with those of other primers targeting the nirK gene in Fig. 1. Primers Cunir3 and Cunir4 amplify a larger region than do primers F1aCu and R3Cu (15), similar to primers nirK1F and nirK5R (5). However, Cunir3 and Cunir4 potentially amplify a greater diversity of nirK genes than do primers nirK1F and nirK5R due to the occurrence of three nucleotides within the nirK1F target region of certain sequences which are unaccounted for in that primer sequence (Fig. 1).
FIG. 1.
Locations of PCR primer pairs for the nirK gene relative to the nirK sequence of Alcaligenes faecalis S-6. Vertical bars indicate the locations of the seven amino acids involved in copper binding. Primers F1aCu and R3Cu (15) yield a 473-bp product, while nirK1F and nirK5R (5) and Cunir3 and Cunir4 (this study) yield 514- and 540-bp products, respectively. The sequence of the Cunir3 target site in the N. marina nirK gene was verified from the sequence of an additional 300-bp fragment which overlaps the 5′ end of the Cunir3-4 fragment (results not shown). Note that the target region for nirK1F spans a region with a 3-bp insertion (deletion) in the N. marina (A. faecalis) nirK sequence. Y = C or T; V = A, C, or G; M = A or C; K = G or T; S = G or C.
Conditions for amplifying nirK using primers Cunir3 and Cunir4 were optimized with genomic DNA extracted from the denitrifying bacteria Pseudomonas aureofaciens, Alcaligenes faecalis, and Alcaligenes xylosoxidans, each known to possess the nirK gene (5, 30, 39, 50). PCR optimization of the individual strains yielded different optimal conditions for the three denitrifying strains. Using a touchdown PCR protocol, it was subsequently possible to amplify the correct product from all three under uniform conditions. Although the use of a touchdown protocol resulted in the amplification of nontarget sequences in some cases (Fig. 2), the eventual goal was to detect nirK sequences in ammonia oxidizers which were potentially more divergent. Products of the expected size from Cunir3-4 amplification of P. aureofaciens, A. faecalis, and A. xylosoxidans were cloned and sequenced, verifying that they are sections of the nirK gene by comparison to published sequences.
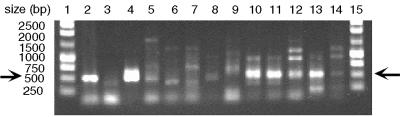
FIG. 2.
Agarose gel visualization of Cunir3-4 PCR products. Lanes: 1 and 15, DNA size standards (Promega); 2, P. aureofaciens; 3, A. xylosoxidans; 4, A. faecalis 8750; 5, N. marina; 6, N. europaea; 7, N. eutropha; 8, N. briensis; 9, N. oceani; 10, NO3W; 11, URW; 12, C-113a; 13, C-45; 14, TA-921i-NH4. Arrows indicate the expected size of the Cunir3-4 product, 540 bp.
Nitrifier Cunir3-4 amplification and sequencing.
Cunir3-4 amplification of genomic DNA from N. marina, N. europaea, N. eutropha, N. oceani, and N. briensis reproducibly yielded multiple products when visualized on 1% agarose gels (Fig. 2). Products of the expected size (approximately 540 bp) were excised from the gel, cloned, and sequenced for each organism. From this first round of screening, nirK was located in N. marina but not in N. europaea, N. eutropha, N. briensis, or N. oceani. The Cunir3-4 products sequenced from these latter organisms were all smaller than 540 bp and proved to be nonspecific amplification products. Because N. europaea was expected to have this gene and the Cunir3-4 fragment could potentially vary in length, additional Cunir3-4 products were sequenced from N. europaea and N. eutropha. Still, nirK products were not obtained from these organisms.
In the next phase, the Cunir3-4 fragment of the nirK gene was successfully amplified from ammonia-oxidizing isolates most similar to N. marina (Fig. 2). Cunir3-4 PCR products of the expected size from isolates URW, NO3W, C-45, C-113a, and TA-921i-NH4 were cloned, and these products yielded nirK sequences for these five additional ammonia-oxidizing strains. The products from N. marina, NO3W, URW, C-45, and C-113a were each 540 bp, the same size as the Cunir3-4 region of B. japonicum and R. sphaeroides. The Cunir3-4 product from TA-921i-NH4 was 537 bp, similar to the same region from P. aureofaciens, A. xylosoxidans, A. faecalis, R. hedysari, and A. cycloclastes. Thus, the Cunir3-4 products that yielded positive matches to nirK were all very similar in length (537 to 540 bp). The nonspecific amplification products did not interfere with detection of the nirK target, but further optimization of the PCR conditions may eliminate nonspecific products.
Alignment of nitrite reductase sequences.
The partial nirK sequences from ammonia-oxidizing bacteria were aligned with the published nirK sequences from denitrifying bacteria and the preliminary nirK sequence from N. europaea (http: //www.jgi.doe.gov/tempweb/JGI microbial/html/index.html) using ClustalW multiple sequence alignment (41). Excluding N. europaea, the similarity between the nirK sequences of ammonia oxidizers and classical denitrifiers was 62 to 75% at the nucleic acid level. The sequence from this section of the putative N. europaea nirK is only 6 to 12% similar to other nirK sequences at the nucleic acid level.
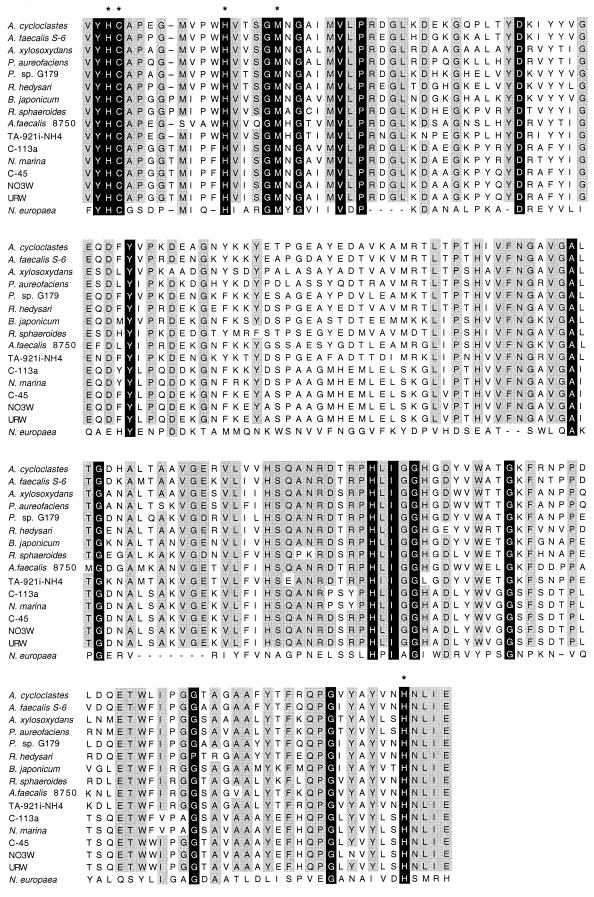
The deduced amino acid sequences from the above nirK sequences were also aligned using ClustalW and subsequently used for phylogenetic analysis. The alignment of NirK deduced amino acid sequences is presented in Fig. 3. The 3-bp difference in certain sequences mentioned earlier corresponds to a single amino acid codon (located between nucleotides 528 and 529 in the nirK gene of A. faecalis) that is lacking in some strains (Fig. 3). Excluding N. europaea, the nitrifier NirK amino acid sequences are 58 to 99% similar to each other and 59 to 72% similar to denitrifier NirK sequences. The available denitrifier NirK sequences are 66 to 85% similar to each other over this region. In addition to the relatively high overall similarity among NirK sequences in the Cunir3-4 region, the regions of highest conservation occur in the vicinity of Cu-binding residues. Most notably, the identity and spacing of each copper-binding residue included in this fragment are strictly conserved across NirK sequences (Fig. 3). In contrast, the N. europaea nirK sequence corresponds to a deduced amino acid sequence only 20 to 26% similar to other NirK sequences over the Cunir3-4 section. Although copper-binding residues are potentially conserved, large gaps are required to align the N. europaea NirK sequence with the others.
FIG. 3.
Alignment of partial NirK sequences using ClustalW multiple sequence alignment. Asterisks indicate locations of the copper-binding residues contained in this section of the NirK protein. Amino acid residues conserved in all sequences are shaded in black, and residues conserved in >80% of sequences are shaded in gray.
NirK phylogeny.
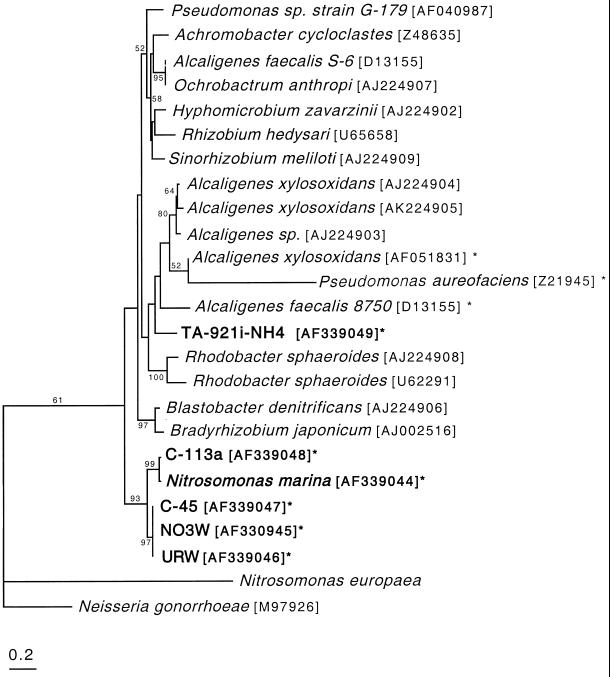
Distance analysis of partial NirK amino acid sequences yields the tree shown in Fig. 4. This analysis was performed using 144 amino acids, the region of overlap between the Cunir3-4 sequences and the nirK1F-nirK5R sequences (5), instead of the longer Cunir3-4 secion (179 amino acids) in order to include these eight additional sequences in the analysis. The NirK sequences cluster into two main groups. The first group, consisting of sequences from a subset of the ammonia-oxidizing bacteria (N. marina, C-113a, C-45, URW, and NO3W), is supported by 93% of bootstrap replicates. The second group, consisting of sequences from the classical heterotrophic denitrifiers plus TA-921i-NH4 (an ammonia-oxidizing isolate), is not well defined. However, the NirK sequence from TA-921i-NH4 rarely groups away from denitrifier sequences. The NirK sequence from N. europaea is very different from the other NirK sequences and always lies outside the two main clusters. Pan1, an anaerobically induced outer membrane protein from Neisseria gonorrhoeae (10), was used as the outgroup (see Discussion). Several additional sequences are available for a shorter section of the nirK gene, which reduces the region of sequence analysis to 111 amino acids (6). These sequences were included in a separate analysis (results not shown) which supports the groupings represented in Fig. 4, with the additional nirK sequences clustering among the established denitrifier groups and the N. marina group remaining isolated.
FIG. 4.
Phylogeny of NirK deduced amino acid sequences based on distance matrix analysis (12) of 144 amino acids. New sequences from this study are in bold type, and sequences which amplified with Cunir3-Cunir4 are indicated with an asterisk. Bootstrap values greater than 50% are shown. The scale bar indicates 0.2 substitutions per amino acid position.
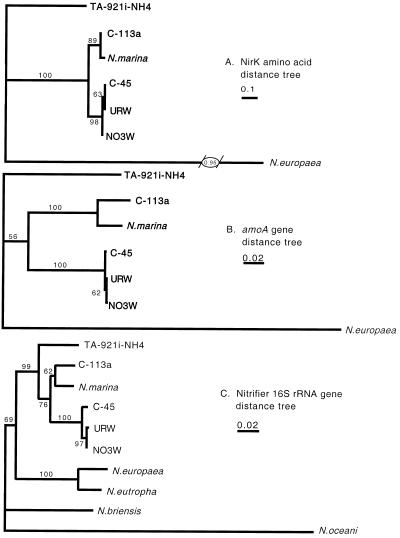
Distance matrix and neighbor-joining analysis of NirK deduced amino acid sequences for the ammonia oxidizers alone yields the tree presented in Fig. 5A. The only branch that is not highly supported is that separating NO3W from C-45 and URW. The NirK sequence for NO3W differs at a single position from C-45 and URW sequences, and thus an evolutionary relationship between these organisms cannot be resolved from this section of their NirK sequences. However, the separate grouping of C-113a and N. marina (bootstrap value 89%) from URW, NO3W, and C-45 (bootstrap value 98%) is robust, as is the separation of these five sequences from TA-921i-NH4 and N. europaea (bootstrap value 100%).
FIG. 5.
Comparison of structural (16S rRNA) and functional (amoA and nirK) gene phylogenies by distance analysis. (A) Deduced amino acid sequences (179 amino acids) from nitrifier nirK genes; (B) amoA gene (525 bp) for ammonia monooxygenase; (C) 16S rRNA gene (∼1,100 bp) from nitrifying bacteria. Distances were calculated from amino acid sequences using Dayhoff PAM 100 matrix (12) and for nucleic acid sequences using the Kimura two-parameter model (22). The 16S rRNA sequences for URW, NO3W, C-45, C-113a, and TA-921i-NH4 are from Voytek (42). Sequences from this study are in bold type. Bootstrap values over 50% are shown. The scale bars indicate substitutions per site.
amoA phylogeny.
A phylogenetic tree based on distance analysis of 525-bp amoA nucleic acid sequences from N. marina, C-113a, C-45, URW, NO3W, and TA-921i-NH4, combined with an amoA sequence for N. europaea obtained from the GenBank database (AF058692), is shown in Fig. 5B. Clustering within the amoA tree is nearly identical to that seen in the NirK tree for the same organisms (Fig. 5A). N. marina, C-113a, NO3W, URW, and C-45 sequences form a coherent cluster, with TA-921i-NH4 falling just outside that group and N. europaea further removed. Again, the grouping of URW and C-45 away from NO3W is not well supported by bootstrapping, although the grouping of URW, C-45, and NO3W is supported by 100% of bootstrap replicates. In this analysis, the placement of TA-921i-NH4 is also poorly constrained. Fifty-six percent of bootstrap replicates place TA-921i-NH4 outside the N. marina group, while 44% of bootstrap replicates place TA-921i-NH4 within that group, on the branch leading to N. marina and C-113a.
DISCUSSION
Understanding the mechanisms of NO and N2O production by nitrifying bacteria and how these processes are regulated may provide new insight into the factors which control N2O production in the environment. Obtaining sequences from the nirK genes in N. marina and related ammonia oxidizers will enable the investigation of nitrite reductase expression in these environmentally relevant organisms. The similarity between the nirK sequences from ammonia oxidizers, from this study, and classical denitrifiers also raises interesting ecological and evolutionary questions regarding the history of nitrite reductase in nitrifying bacteria and bears on the interpretation of nirK sequences amplified directly from the environment. These questions may be best framed through phylogenetic comparisons.
The 16S rRNA gene phylogeny of the ammonia oxidizers included in this study is shown in Fig. 5C. The majority of ammonia-oxidizing nitrifiers form a phylogenetically coherent group in the beta proteobacteria, with the exception of N. oceani strains, which belong to the gamma proteobacteria (16, 40). The original 16S rRNA sequences for isolates C-45, C-113a, URW, NO3W, and TA-921i-NH4 were obtained by Voytek (42) and are available in GenBank under accession numbers AF338200 to AF338214. A diverse set of nitrifiers (N. europaea, N. eutropha, N. briensis, N. marina, and N. oceani), each known to produce N2O (14; unpublished results), was initially screened for nirK by PCR. Because N. marina was the only nitrifier initially screened in which nirK was detected, the study was refined to focus on isolates closely related to N. marina, as indicated by their 16S rRNA sequences: NO3W, C-45, C-113a, URW, and TA-921i-NH4. Sections of the nirK and amoA genes were amplified and sequenced from these five additional nitrifying isolates, and the phylogenies based on these functional genes are discussed below.
amoA phylogeny.
Ammonia monooxygenase is an enzyme that is unique to ammonia-oxidizing nitrifiers, and the amoA gene has been shown to provide fine-scale resolution of the phylogeny of ammonia-oxidizing bacteria (37). The use of amoA nucleic acid sequences rather than amino acid sequences in phylogenetic analysis allows the distinction between closely related nitrifiers, for which few differences are apparent in AmoA amino acid sequences. The amino acid sequences deduced from the amoA genes in this study produce a phylogenetic tree with poor resolution within the N. marina group, which contains very closely related strains. Therefore, amoA gene sequences are used here. One potential complication with using the amoA gene sequences is that each organism may possess multiple copies of the amoA gene. The cloned amoA fragments amplified from a single organism could, therefore, be a mixture of different copies of amoA. For the ammonia-oxidizing bacteria for which this has been documented, the amoA genes within an organism are much more similar to each other than to copies from any other organism (23, 31). If this holds for the nitrifying isolates in this study, the inclusion of multiple copies from each organism would not affect the branching order of the organisms on the amoA tree or the phylogenetic comparisons made here.
Distance analysis of a 525-bp fragment of the amoA gene for the ammonia oxidizers in this study (Fig. 5B) shows that the topology of the amoA tree corresponds closely to the 16S rRNA phylogeny (Fig. 5C). This is expected because ammonia monooxygenase is essential to ammonia-oxidizing bacteria, and earlier studies of amoA gene phylogenies have shown similar results (18, 34, 37). The amoA phylogeny provides an interesting comparison to the phylogeny for ammonia oxidizers based on nitrite reductase, discussed below.
NirK phylogeny.
Several studies have targeted the nitrite reductase gene as a measure of the diversity of denitrifying bacteria because as a group, denitrifiers are widely dispersed in 16S rRNA phylogeny (5, 15, 26, 45). This study expands the known sequence diversity of nirKs with the addition of nitrite reductases from ammonia-oxidizing bacteria. Five of the NirK sequences from ammonia oxidizers cluster separately from the denitrifier sequences, while the TA-921i-NH4 sequence falls within the main denitrifier cluster (Fig. 4). It is likely that the full diversity of Cu-NiRs, as implied from immunological data (11), has not yet been covered by sequencing efforts, and analysis of NirK sequences from additional cultured nitrifiers and denitrifiers may be required to further expand the NirK tree. However, the similarity of nirK sequences from the ammonia oxidizers in this study to denitrifier nirKs suggests that nirK sequences amplified directly from the environment may include a component from nitrifying bacteria. Distinguishing between nitrifier and denitrifier nitrite reductase genes based on sequence alone may prove to be difficult, and further study should establish whether there is a functional difference between nitrifier and denitrifier nitrite reductases.
Comparison of NirK phylogeny to 16S rRNA phylogeny for nitrifiers and denitrifiers yields an interesting contrast. Within the group of NirK sequences which includes the classical denitrifiers (Fig. 4), the phylogenetic relationships do not reflect the 16S rRNA relationships. Organisms from the alpha, beta, and gamma subdivisions of the proteobacteria are interspersed throughout this group within well-supported clusters. For example, the grouping of P. aureofaciens (gamma) with A. xylosoxidans (beta) is supported by a bootstrap value of 100%. Overall, the phylogeny based on NirK sequences indicates an evolutionary history for nitrite reductase in denitrifiers that is distinct from their 16S rRNA phylogeny. This suggests that some classic heterotrophic denitrifiers have acquired nirK genes by lateral gene transfer and supports earlier accounts of such transfer of denitrifying genes (26, 32, 51).
A similar question may be posed regarding the history of nitrite reductase in ammonia-oxidizing bacteria, that is, whether the ammonia-oxidizing bacteria have acquired or distributed nirK through lateral gene transfer. Nitrite reductase may contribute to the ability of ammonia-oxidizing bacteria to tolerate large amounts of nitrite or grow in low-O2 environments and could be important to their success in the environment. Therefore, it would be interesting to understand when in the evolutionary history of ammonia-oxidizing bacteria the ability for nitrite reduction arose. Again, the 16S rRNA gene phylogeny provides the basis by which lateral gene transfer is assessed. The topology of the NirK tree (Fig. 5A) is indistinguishable from that of the 16S rRNA (Fig. 5C) and amoA (Fig. 5B) gene trees. The grouping of C-113a with N. marina and C-45 with NO3W and URW is well supported by bootstrapping in all cases. The separation of TA-921i-NH4 and N. europaea from these nitrifiers in the NirK tree is also consistent with their 16S rRNA and amoA relationships. Therefore, while this data set includes sequences from a limited representation of ammonia-oxidizing bacteria, the close correspondence between the NirK phylogeny and the 16S rRNA and amoA gene phylogenies do not support lateral gene transfer as the mechanism for the nirK distribution within this subset of nitrifying bacteria. Identification of nitrite reductases from more distantly related ammonia-oxidizing bacteria (Nitrosospira types) will be important for extending the implications from this work to a broader evolutionary context.
The choice of an outgroup for the phylogenetic analysis of NirK sequences was not obvious, because the evolutionary origin of this enzyme is unknown and very few proteins show significant sequence similarity to NirK. Aside from other NirK sequences, the closest match for any of the NirK sequences over the Cunir3-4 section is Pan1, an anaerobically induced outer membrane protein of Neisseria gonorrhoeae (10). Pan1 (AniA) was recently shown to be associated with nitrite reductase activity in N. gonorrhoeae (27). This protein aligns well with the NirK sequences, at 29 to 33% amino acid similarity over the CuNir3-4 region, and shares conserved regions around the Cu-binding regions in NirK. After Pan1, the next closest match to NirK is β-amylase, which aligns weakly with the NirK sequences and does not share any of the copper-binding sites. The gene for the heme-containing nitrite reductase (nirS) used as an outgroup by Hallin and Lindgren (15) also does not have significant sequence similarity to nirK despite its close functional similarity.
N. europaea nirK.
The N. europaea nirK was not amplified with the primers described in this work and was obtained only from the preliminary whole-genome sequence (http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html). The reason for the failure of Cunir3-4 amplification of N. europaea nirK became clear upon inspection of its sequence from the Joint Genome Institute (JGI): the primer sites are not conserved in N. europaea. As mentioned earlier, the putative N. europaea nirK sequence is quite different from any other known nirK sequence. It remains unresolved whether the nirK sequence annotated in the N. europaea genome encodes the protein previously studied. However, it is the best candidate for a gene encoding nitrite reductase identified in the N. europaea genome sequence, having the highest overall similarity to published nirK sequences and conserved residues which align to the copper-binding regions of denitrifier NirKs (Fig. 3). More extensive genetic analysis will be required to prove that this putative nirK gene from N. europaea indeed encodes the nitrite reductase that has been studied biochemically (19, 29, 36).
N2O production by nitrifiers.
Nitrite reductase is a central enzyme of the denitrification pathway in that it produces the first gaseous product, NO. This study presents the first genetic evidence for a nitrite reductase that is homologous to the nirK of classical denitrifiers in Nitrosomonas marina and closely related ammonia-oxidizing nitrifiers. These results parallel the finding of nirK in N. europaea by an independent sequencing effort (JGI) and support earlier studies which demonstrate the involvement of a copper-containing nitrite reductase in the production of NO and N2O by N. europaea (19, 28, 35). While nirK homologs were not detected in N. oceani, N. briensis, or N. eutropha using the PCR primers developed in this study, further investigation may reveal nitrite reductases in these organisms which are more divergent from the classical denitrifier nirK, as was the case for N. europaea. Bruns et al. (7) demonstrated low-stringency hybridization of a nirK probe from Pseudomonas sp. strain G-179 to genomic DNA from Nitrosolobus sp. strain 24-C and Nitrosospira sp. strain NpAV, but not to N. europaea or certain other Nitrosospira strains. It is difficult to gauge these results quantitatively, although they suggest further variability in either the distribution of nirK in ammonia oxidizers or their sequence similarity to denitrifier nirKs.
Of course, the presence of this fragment of the nirK gene does not guarantee that it is functional. Direct evidence of nirK expression in nitrifying bacteria will be needed to prove that this gene encodes a functional protein; however, the following evidence suggests that this is the case. In addition to the earlier biochemical characterization of a functional nitrite reductase in N. europaea, the analysis of the amino acid sequence from the active site of NirK in several ammonia oxidizers, presented here, indicates that despite sequence divergence in areas outside the copper-binding regions, the copper-binding residues are highly conserved (Fig. 3). This suggests that there has been selective pressure for this enzyme to remain functional during the course of its evolution.
While N. marina and its close relatives have nitrite reductases similar to the copper-containing nitrite reductase of typical denitrifiers, it cannot immediately be assumed that they can grow under anaerobic conditions at the expense of NO2−, e.g., Rhizobium hedysari HCNT-1 (8, 9). The requirement for ammonia monooxygenase for O2 (17) likely precludes the chemolithotrophic growth of ammonia oxidizers under strictly anaerobic conditions.
Information on the mechanism and regulation of nitrite reduction by ammonia-oxidizing bacteria is needed in order to clarify the role of these bacteria in N2O production in the environment. The expression of nitrite reductase in classical denitrifying bacteria is regulated differently by levels of O2, NO2−, and NO3−. Several investigators have demonstrated that NO and N2O yields from ammonia-oxidizing bacteria are enhanced at low O2 concentrations (14, 25, 49), and the activity of nitrite reductase is also elevated at low O2 concentrations (29). It will be important to demonstrate expression of nirK in N. marina and to explore the conditions under which nirK is expressed in ammonia-oxidizing bacteria in order to understand the molecular details of variations in their production of NO and N2O.
In this study, the nirK gene was identified in several ammonia-oxidizing bacteria. These sequences may be used to detect nirK expression in relation to N2O production by these strains under various environmental conditions. In addition, the Cunir3-4 PCR primers developed here may augment the currently available primers in the study of nirK in additional nitrifier and denitrifier cultures and in natural samples.
ACKNOWLEDGMENTS
We thank M. Voytek for providing access to 16S rRNA sequences prior to publication. Comments on earlier versions of the manuscript from M. Voytek, M. Haygood, and anonymous reviewers were greatly appreciated. We also thank D. Martino for helpful discussion.
Funding was provided by the National Science Foundation (OCE-9617690) and the Center for Environmental Bioinorganic Chemistry.
REFERENCES
- 1.Adman E T, Godden J W, Turley S. The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2− bound and with type II copper depleted. J Biol Chem. 1995;270:27458–27474. doi: 10.1074/jbc.270.46.27458. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Sideman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 3.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock E, Schmidt I, Stuven R, Zart D. Nitrogen loss caused by denitrifying Nitrosomonas cells using ammonium or hydrogen as electron donors and nitrite as electron acceptor. Arch Microbiol. 1995;163:16–20. [Google Scholar]
- 5.Braker G, Fesefeldt A, Witzel K-P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol. 1998;64:3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braker G, Zhou J, Wu L, Devol A H, Tiedje J M. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl Environ Microbiol. 2000;66:2096–2104. doi: 10.1128/aem.66.5.2096-2104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruns M A, Fries M R, Tiedje J M, Paul E A. Functional gene hybridization patterns of terrestrial ammonia-oxidizing bacteria. Microb Ecol. 1998;36:293–302. doi: 10.1007/s002489900116. [DOI] [PubMed] [Google Scholar]
- 8.Casella S, Shapleigh J P, Payne W J. Nitrite reduction in Rhizobium hedysari strain HCNT-1. Arch Microbiol. 1986;146:233–238. [Google Scholar]
- 9.Casella S, Toffanin A, Ciompi S, Rossi N, Payne W J. Metabolism of nitrogen-oxides and hydroxylamine in cells of true denitrifiers and Rhizobium hedysari HCNT-1. Can J Microbiol. 1994;40:1–5. [Google Scholar]
- 10.Clark V L, Campbell L A, Palermo D A, Evans T M, Klimpel K W. Induction and repression of outer membrane proteins by anaerobic growth of Neisseria gonorrhoeae. Infect Immun. 1987;55:1359–1364. doi: 10.1128/iai.55.6.1359-1364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyne M S, Arunakumari A, Averill B A, Tiedje J M. Immunological identification and distribution of dissimilatory heme cd1 and non heme copper nitrite reductases in denitrifying bacteria. Appl Environ Microbiol. 1989;55:2924–2931. doi: 10.1128/aem.55.11.2924-2931.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dayhoff M O. Atlas of protein sequence and structure. 5, suppl. 3. Washington, D.C.: National Biomedical Research Foundation; 1979. [Google Scholar]
- 13.Felsenstein J. Phylip-phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 14.Goreau T J, Kaplan W A, Wofsy S C, McElroy M B, Valois F W, Watson S W. Production of NO2− and N2O by nitrifying bacteria at reduced concentrations of oxygen. Appl Environ Microbiol. 1980;40:526–532. doi: 10.1128/aem.40.3.526-532.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallin S, Lindgren P-E. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl Environ Microbiol. 1999;65:1652–1657. doi: 10.1128/aem.65.4.1652-1657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Head I M, Hiorns W D, Embley T M, McCarthy A J, Saunders J R. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Microbiol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- 17.Hollocher T C, Tate M E, Nicholas D J D. Oxidation of ammonia by Nitrosomonas europaea—definitive O-18 tracer evidence that hydroxylamine formation involves a mono-oxygenase. J Biol Chem. 1981;256:834–836. [PubMed] [Google Scholar]
- 18.Holmes A J, Costello A, Lidstrom M E, Murrell J C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 19.Hooper A B. A nitrite-reducing enzyme from Nitrosomonas europaea: preliminary characterization with hydroxylamine as electron donor. Biochim Biophys Acta. 1968;162:49–65. doi: 10.1016/0005-2728(68)90213-2. [DOI] [PubMed] [Google Scholar]
- 20.Hooper A B, Vannelli T, Bergmann D J, Arciero D M. Enzymology of the oxidation of ammonia to nitrite by bacteria. Antonie van Leeuwenhoek. 1997;71:59–67. doi: 10.1023/a:1000133919203. [DOI] [PubMed] [Google Scholar]
- 21.Ka J-O, Urbance J, Ye R W, Ahn T-Y, Tiedje J M. Diversity of oxygen and N-oxide regulation of nitrite reductases in denitrifying bacteria. FEMS Microbiol Lett. 1997;156:55–60. doi: 10.1111/j.1574-6968.1997.tb12705.x. [DOI] [PubMed] [Google Scholar]
- 22.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 23.Klotz M G, Norton J M. Multiple copies of ammonia monooxygenase (amo) operons have evolved under biased AT/GC mutational pressure in ammonia-oxidizing autotrophic bacteria. FEMS Microbiol Lett. 1998;168:303–311. doi: 10.1111/j.1574-6968.1998.tb13288.x. [DOI] [PubMed] [Google Scholar]
- 24.Liesack W, Weyland H, Stackerbrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 25.Lipschultz F, Zafiriou O C, Wofsy S C, McElroy M B, Valois F W, Watson S W. Production of NO and N2O by soil nitrifying bacteria. Nature. 1981;294:641–643. [Google Scholar]
- 26.Martino D P, Ward B B. Abstracts of the Annual Meeting of the American Society of Limnology and Oceanography. 1999. Diversity of denitrifying bacteria from Tomales Bay, CA, based on ribosomal and nitrite reductase gene sequences; p. 117. Santa Fe, N.Mex. [Google Scholar]
- 27.Mellies J, Jose J, Meyer T F. The Neisseria gonorrhoeae gene aniA encodes an inducible nitrite reductase. Mol Gen Genet. 1997;256:525–532. doi: 10.1007/s004380050597. [DOI] [PubMed] [Google Scholar]
- 28.Miller D J, Wood P M. The soluble cytochrome oxidase of Nitrosomonas europaea. J Gen Microbiol. 1983;129:1645–1650. [Google Scholar]
- 29.Miller D J, Nicholas D J D. Characterization of a soluble cytochrome oxidase/nitrite reductase from Nitrosomonas europaea. J Gen Microbiol. 1985;131:2851–2854. [Google Scholar]
- 30.Nishiyama M, Suzuki J, Kukimoto M, Ohnuki T, Horinouchi S, Beppu T. Cloning and characterization of a nitrite reductase gene from Alcaligenes faecalis and its expression in Escherichia coli. J Gen Microbiol. 1993;139:725–733. doi: 10.1099/00221287-139-4-725. [DOI] [PubMed] [Google Scholar]
- 31.Norton J M, Low J M, Klotz M G. The gene encoding ammonia monooxygenase subunit A exists in three nearly identical copies in Nitrosospira sp. NpAV FEMS Microbiol Lett. 1996;139:181–188. doi: 10.1111/j.1574-6968.1996.tb08200.x. [DOI] [PubMed] [Google Scholar]
- 32.Ohkubo S H, Iwasaki H, Hori H, Osawa S. Evolutionary relationship of denitrifying bacteria as deduced from 5S rRNA sequences. J Biochem. 1986;100:1261–1267. doi: 10.1093/oxfordjournals.jbchem.a121832. [DOI] [PubMed] [Google Scholar]
- 33.Poth M, Focht D D. N-15 kinetic analysis of N2O production by Nitrosomonas europaea—an examination of nitrifier denitrification. Appl Environ Microbiol. 1985;49:1134–1141. doi: 10.1128/aem.49.5.1134-1141.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purkhold U, Pommerening-Roser A, Juretschko S, Schmid M C, Koops H-P, Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie G A F, Nicholas D J D. Identification of the sources of nitrous oxide produced by oxidative and reductive processes in Nitrosomonas europaea. Biochem J. 1972;126:1181–1191. doi: 10.1042/bj1261181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie G A F, Nicholas D J D. The partial characterization of purified nitrite reductase and hydroxylamine oxidase from Nitrosomonas europaea. Biochem J. 1974;138:471–480. doi: 10.1042/bj1380471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rotthauwe J H, Witzel K P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soriano S, Walker N. Isolation of ammonia oxidizing autotrophic bacteria. J Appl Bacteriol. 1968;31:493–497. doi: 10.1111/j.1365-2672.1968.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki E, Horikoshi N, Kohzuma T. Cloning, sequencing, and transcriptional studies of the gene encoding copper-containing nitrite reductase from Alcaligenes xylosoxydans NCIMB 11015. Biochem Biophys Res Commun. 1999;255:427–431. doi: 10.1006/bbrc.1998.9932. [DOI] [PubMed] [Google Scholar]
- 40.Teske A, Alm I, Regan J M, Toze S, Rittmann B E, Stahl D A. Evolutionary relationships among ammonia-oxidizing and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J D, Higgins D G, Gibson T J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voytek M A. Relative abundance and species diversity of autotrophic ammonia-oxidizing bacteria in aquatic systems. Ph.D. thesis. Santa Cruz: University of California; 1996. [Google Scholar]
- 43.Ward B B, Carlucci A F. Marine ammonia-oxidizing and nitrite-oxidizing bacteria—seriological diversity determined by immunofluorescence in culture and in the environment. Appl Environ Microbiol. 1985;50:194–201. doi: 10.1128/aem.50.2.194-201.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward B B. Kinetic studies on ammonia and methane oxidation by Nitrosococcus oceanus. Arch Microbiol. 1987;147:126–133. [Google Scholar]
- 45.Ward B B. Diversity of culturable denitrifying bacteria: limits of rDNA RFLP analysis and probes for the functional gene, nitrite reductase. Arch Microbiol. 1995;163:167–175. [Google Scholar]
- 46.Watson S W. Characteristics of a marine nitrifying bacterium, Nitrosocystis oceanus sp. nov. Limnol Oceanogr. 1965;10:R274–R289. [Google Scholar]
- 47.Williams P A, Fulop V, Garman E F, Saunders N F W, Ferguson S J, Hajdu J. Haem-ligand switching during catalysis in crystals of a nitrogen-cycle enzyme. Nature. 1997;389:406–412. doi: 10.1038/38775. [DOI] [PubMed] [Google Scholar]
- 48.Ye R W, Averill B A, Tiedje J M. Denitrification: production and consumption of nitric oxide. Appl Environ Microbiol. 1994;60:1053–1058. doi: 10.1128/aem.60.4.1053-1058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida N. N-15-depleted N2O as a product of nitrification. Nature. 1988;335:528–529. [Google Scholar]
- 50.Zumft W G, Gotzmann D J, Frunzke K, Viebrock A. Type 1, blue copper proteins constitute a respiratory nitrite-reducing system in Pseudomonas aureofaciens. Eur J Biochem. 1987;168:301–307. doi: 10.1111/j.1432-1033.1987.tb13421.x. [DOI] [PubMed] [Google Scholar]
- 51.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]