Abstract
Imaging spectroscopy provides the opportunity to incorporate leaf and canopy optical data into ecological studies, but the extent to which remote sensing of vegetation can enhance the study of belowground processes is not well understood. In terrestrial systems, aboveground and belowground vegetation quantity and quality are coupled, and both influence belowground microbial processes and nutrient cycling. We hypothesized that ecosystem productivity, and the chemical, structural and phylogenetic‐functional composition of plant communities would be detectable with remote sensing and could be used to predict belowground plant and soil processes in two grassland biodiversity experiments: the BioDIV experiment at Cedar Creek Ecosystem Science Reserve in Minnesota and the Wood River Nature Conservancy experiment in Nebraska. We tested whether aboveground vegetation chemistry and productivity, as detected from airborne sensors, predict soil properties, microbial processes and community composition. Imaging spectroscopy data were used to map aboveground biomass, green vegetation cover, functional traits and phylogenetic‐functional community composition of vegetation. We examined the relationships between the image‐derived variables and soil carbon and nitrogen concentration, microbial community composition, biomass and extracellular enzyme activity, and soil processes, including net nitrogen mineralization. In the BioDIV experiment—which has low overall diversity and productivity despite high variation in each—belowground processes were driven mainly by variation in the amount of organic matter inputs to soils. As a consequence, soil respiration, microbial biomass and enzyme activity, and fungal and bacterial composition and diversity were significantly predicted by remotely sensed vegetation cover and biomass. In contrast, at Wood River—where plant diversity and productivity were consistently higher—belowground processes were driven mainly by variation in the quality of aboveground inputs to soils. Consequently, remotely sensed functional, chemical and phylogenetic composition of vegetation predicted belowground extracellular enzyme activity, microbial biomass, and net nitrogen mineralization rates but aboveground biomass (or cover) did not. The contrasting associations between the quantity (productivity) and quality (composition) of aboveground inputs with belowground soil attributes provide a basis for using imaging spectroscopy to understand belowground processes across productivity gradients in grassland systems. However, a mechanistic understanding of how above and belowground components interact among different ecosystems remains critical to extending these results broadly.
Keywords: biodiversity, ecosystem processes, hyperspectral data, imaging spectroscopy, microbial biomass, phylogenetic‐functional groups, plant traits, productivity, remote sensing, soil enzyme activity, soil processes, vegetation chemistry
Introduction
Monitoring biodiversity and understanding its consequences for ecosystem functions and global processes are critical challenges in the face of rapid global change. Remote sensing has proven useful for observing ecosystem functions such as total biomass production (Gamon et al. 1999, Williams et al. 2020) across spatial scales and temporal resolutions because reflectance and absorption of light by vegetation canopies is strongly influenced by their structural, biochemical, physiological, and phenological characteristics (Ustin and Gamon 2010, Schmidtlein et al. 2012). The functional variation in vegetation can be detected using imaging spectroscopy (aka hyperspectral imagery) (Asner and Martin 2009, Asner et al. 2011, Wang et al. 2016, 2019, Schneider et al. 2017, Schweiger et al. 2017, Williams et al. 2020), therefore enabling hyperspectral remote sensing as an approach to test the drivers of ecosystem processes that can be detected aboveground.
In contrast with aboveground processes and attributes, detecting belowground processes remotely remains technically challenging and enigmatic. Nevertheless, the composition, function and diversity of plant assemblages are well known to affect belowground processes (Hooper and Vitousek 1998, Eviner and Chapin 2003, Meier and Bowman 2008, Bardgett and van der Putten 2014, Hobbie 2015). Plants synthesize a wide variety of chemical and structural compounds to support physiological functions, and the abundance and chemical composition of plant tissues and plant exudates influence soil microbial diversity and abundance (Meier and Bowman 2008, Hobbie 2015). Given that plant reflectance spectra are aggregate indicators of chemistry, composition, and abundance of plants within communities (Schmidtlein 2005, Townsend et al. 2013, Serbin et al. 2014, Singh et al. 2015, Schweiger et al. 2018), it is appropriate to ask whether we can remotely sense attributes of vegetation that predict belowground processes (Madritch et al. 2014, Cavender‐Bares et al. 2017).
Both quantity and quality of inputs from above to belowground may influence belowground processes. The total quantity of inputs to the soil from above and belowground primary productivity is expected to influence microbial processes by providing energy to fuel soil microbes (Zak et al. 1994, Hättenschwiler and Jørgensen 2010). Therefore, higher rates of plant inputs (above and belowground) to soils are often associated with higher rates of soil respiration, enzyme activity, and nutrient mineralization (Zak et al. 1990, Cline et al. 2018).
The chemistry (“quality”) of organic matter inputs to the soil can also influence the abundance of different microbial organisms with contrasting metabolic capacities (Degens and Harris 1997, Wardle et al. 2004, Cline et al. 2018). Variation in quality of inputs results from variation in plant foliar and root chemistry because live and dead plant parts include a diversity of organic molecules—such as soluble sugars, cellulose, hemicellulose, lignin, and tannins—which vary greatly in how readily they can be broken down by microbes (Degens and Harris 1997, Meier and Bowman 2008, Freschet et al. 2012). Variation in plant nutrient concentrations, therefore, influences organic matter decomposition and nutrient dynamics (Parton et al. 2007, Cornwell et al. 2008, Fornara et al. 2009).
Plant chemical diversity is in turn linked to physiological function associated with functional groups that often have a phylogenetic basis due to shared ancestry (Cadotte et al. 2009, Kothari et al. 2018), such as nitrogen fixation in legumes and C4 photosynthesis (Hooper and Vitousek 1998, Craine et al. 2002). Plants can, therefore, be meaningfully categorized into phylogenetic‐functional groups (Kothari et al. 2018) that have a high potential to be remotely detected (Cavender‐Bares et al. 2016, Schweiger et al. 2018, Wang et al. 2019, Meireles et al. 2020) as a basis to predict belowground processes (Madritch et al. 2014). Even differentiation between monocots and dicots can be informative. For example, despite high variability, inputs from forbs and legumes tend to be characterized by high nitrogen content and soluble sugars (Adams et al. 2016), which may favor microorganisms with high hydrolytic capacity. In contrast, inputs from grasses (monocots), particularly C4 grasses, tend to be characterized by high cellulose and hemicellulose but low nitrogen content (Craine et al. 2001), which may favor microorganisms with high cellulose degrading activity. Therefore, variation in plant composition—measured in terms of plant chemistry and function or in terms of variation in phylogenetic‐functional group representation—is expected to influence microbial and soil processes, independent of the total quantity of inputs.
The capability of airborne imaging spectroscopy to accurately detect plant chemistry and productivity provides an important means to estimate both the quality and the quantity of aboveground inputs to the soil over large spatial scales (Kokaly et al. 2009, Asner et al. 2011, Serbin et al. 2014, Singh et al. 2015, Wang et al. 2019). Remotely sensed information may be useful in understanding soil processes both because aboveground inputs are significant sources of organic matter to soils and/or because above and belowground attributes (total net primary productivity [NPP], and chemical constituents) may be coupled. For example, in aspen (Populus tremuloides) stands, variation in canopy chemistry among genotypes—including condensed tannins, lignin, and nitrogen concentrations—was associated with foliar spectra and correlated with belowground processes (Madritch et al. 2014). In Hawaii, airborne spectroscopy detected the major source of variation in canopy nitrogen associated with both planted native nitrogen fixing trees (Acacia koa) and invading N fixers (Myrica faya) (Asner et al. 2008, Vitousek et al. 2009), which alter chemical inputs to soil microorganisms and influence soil processes (Vitousek 2004). Unlike in these forest systems, organic matter inputs to soils in many grasslands are dominated by belowground productivity (Hui and Jackson 2006). Nevertheless, aboveground processes such as photosynthesis are essential for providing belowground resources in grasslands, and the coupling of the drivers of aboveground primary production (quantity and chemistry) to those belowground could enable prediction of belowground processes and properties using remote sensing.
Here we investigated the extent to which we could remotely sense aboveground productivity, plant function, phylogenetic‐functional group composition and spectral diversity at the scale of individual plant communities to predict belowground microbial (fungal and bacterial) biomass, composition, diversity and extracellular enzyme activity; net nitrogen mineralization rates; soil respiration; and soil carbon and nutrient concentrations. This work was conducted at the long‐term prairie diversity experiment (BioDIV) at the Cedar Creek Ecosystem Science Reserve (CCESR) in central Minnesota (Tilman et al. 2001) and a more recent prairie diversity experiment at Wood River in central Nebraska established by the Nature Conservancy (TNC) (Nemec et al. 2013). The two experiments occurred on different parts of the diversity–productivity gradient found in natural, degraded and restored prairie systems (Jelinski et al. 2011), with the BioDIV experiment falling at the lower end of the gradient and Wood River at the higher end.
Previous work in the BioDIV experiment at CCESR showed strong linkages between aboveground plant composition and diversity to soil carbon and nutrients and microbial diversity and composition (Zak et al. 2003, Waldrop et al. 2006, Fornara et al. 2009, Steinauer et al. 2015, Cline et al. 2018, Yang et al. 2019). In particular, higher fungal richness was associated with increasing aboveground plant biomass—and therefore total plant‐derived substrate inputs to soils—while fungal community composition and soil carbon and nitrogen cycling and pools were associated with plant functional group diversity, and the relative abundance of C4 grasses and legumes (Fornara et al. 2009, Cline et al. 2018). Linkages between above and belowground processes have not yet been examined in the Wood River experiment, nor have linkages between remotely sensed variables and belowground processes in either experiment.
Our primary goals were (1) to decipher the mechanisms linking above and belowground processes in two contrasting grassland systems that differ in productivity, diversity, and soil type, and (2) to test the extent to which remotely sensed aboveground productivity, chemical characteristics, and phylogenetic‐functional community composition can reveal belowground processes, microbial diversity and soil attributes. In pursuing these goals, we examined how well remotely sensed imaging spectroscopy could characterize aboveground functional and chemical composition, diversity and productivity and the strength and nature of the linkages among aboveground and belowground components and processes.
By comparing two experimental prairie systems that differed in the degree and range of plant diversity and productivity, but had similar functional group composition, we sought to understand whether relationships determined in one system applied to another system and what aspects of the linkages between aboveground and belowground processes must be understood to use remote sensing as a means to predict changes in belowground properties and processes. We hypothesized that the ecosystem productivity and the types of morpho‐physiological and chemical properties of vegetation that vary among phylogenetic‐functional groups would influence belowground productivity, composition and diversity of microbial communities and drive belowground ecosystem processes. We also hypothesized that the two systems would differ in the relative importance of the quantity (aboveground productivity) and the quality (chemical composition and phylogenetic‐functional group composition) of inputs to the soil in driving belowground processes. In BioDIV—where productivity was relatively low—we expected effects of substrate quantity to overwhelm those of substrate quality, whereas in Wood River—where productivity was comparatively high—we expected the effects of substrate quality to be more apparent (Fig. 1). In both systems, we predicted that plant diversity would have a direct influence on productivity but would influence belowground processes primarily through its influence on productivity and the chemistry of plant inputs to soils. We tested these hypotheses using structural equation models (SEMs).
Fig. 1.
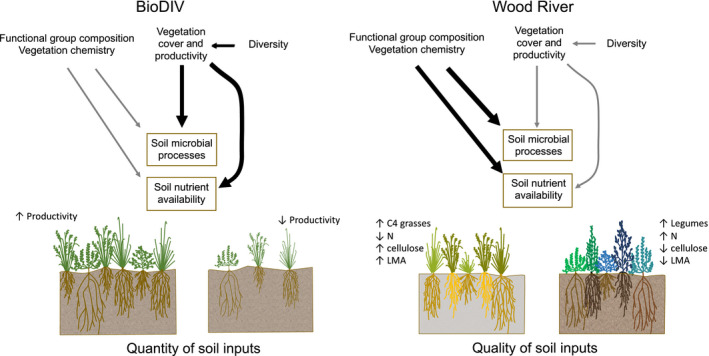
Both the quantity (productivity) and quality (phylogenetic, functional and chemical composition) of inputs from aboveground vegetation into the soil are important in influencing belowground microbial processes and nutrient cycling. However, the relative importance of these characteristics of inputs varies among systems. Shown are distinct hypotheses for how attributes of vegetation that can be remotely sensed influence and are therefore useful in predicting soil microbial processes and nutrient availability in two contrasting prairie grassland diversity experiments. The BioDIV experiment at Cedar Creek varies in species density from one to 16 species per 81 m2, which has led to high variation among plots in annual productivity over time (from 25 to 283 g/m2). In this system, productivity (the quantity of inputs) is hypothesized to be the dominant driver of variation in belowground processes. The Wood River experiment has higher overall species density (one to 13 species per 1 m2 or 10–46 species per 1,200 m2) and productivity (285–1,079 g/m2). As a consequence, the total quantity of inputs is less likely to be limiting, and functional group composition and the resulting variation in vegetation chemistry (quality of inputs) are hypothesized to play a more dominant role in driving belowground processes. These conceptual diagrams are the basis for our structural equation models presented in Fig. 7.
Our study, therefore, investigates how the quality and quantity of aboveground inputs influence belowground processes and attributes using imaging spectroscopic data to detect plant productivity, chemical and functional composition, and taxonomic, phylogenetic and functional diversity based on foliar chemical traits. Using this integrated approach, we determine the extent to which belowground processes can be inferred from imaging spectroscopy once mechanisms linking above and belowground processes have been characterized.
Methods
Study areas
The study was conducted at CCESR (East Bethel, Minnesota) in the long‐term BioDIV experiment and at the Wood River prairie restoration experiment maintained by TNC near Wood River, Nebraska (Appendix S1: Fig. S1). The sites differ in soil type, plot size, plot number, species composition, and the range of species of richness and aboveground productivity within plots.
The BioDIV experiment (45°24′11″N, 93°11′21″W) was established in 1994 (Tilman 1997, Tilman et al. 2001). Cedar Creek is located on a sandy glacial outwash plain and the soils where BioDIV is located are Typic Udipsamments (Grigal et al. 1974). Prior to the establishment of the experiment, topsoil was removed. In the full experiment, 1, 2, 4, 8, 16 or 32 perennial grassland species were planted from a pool of 34 species (eight species, each, of C4 grasses, C3 grasses, legumes, non‐legume forbs; two species of woody plants) in a total of 342 9 × 9 m plots, separated by 1.5 m of grass or bare soil. Of these, 154 have species composition and species richness levels maintained by annual weeding. The plots are also burned annually—such that annual aboveground biomass is a measure of aboveground productivity—with the consequence that litterfall has limited impact on soil organic matter buildup. We collected data in a subset of plots covering planted diversity levels ranging from 1 to 16 species per plot. The number of plots sampled and the specific measurements taken in each year are given in Appendix S2: Table S1.
The Wood River TNC experiment is a prairie restoration study along the Platte River, 10 km south of Wood River, Nebraska (40°44′37′′N, 98°35′26′′W). The site is located within the Platte River floodplain and soils are of loamy alluvium or sandy alluvium parent material (NCSS 2010). The site is characterized by a high water table, poor drainage, and high soil organic content (Jelinski and Currier 1997) and was farmed as a corn and soybean rotation before being converted into an experimental prairie (Nemec et al. 2013). The TNC study area includes 36 60 × 60 m plots across two fields. The site therefore has much larger plot sizes but fewer plots than BioDIV. As described in Nemec et al. (2013) and Gholizadeh et al. (2019), one set of 12 plots was seeded in 2010 (“young plots”) at low, medium and high species richness levels—with measured species richness per plot in 2017 ranging from 11 to 48 species. An earlier set of 24 plots was seeded in 2006 (“old plots”) with two diversity levels and two seeding rates. Plots are not weeded, and measured species richness in the old plots in 2017 varied from 28 to 46 species per plot. We collected leaf‐level spectral data, soil samples, and annual aboveground biomass in all of the young plots and in the 12 old plots with low seeding rate at both low and high diversity levels. Our study therefore includes a total of 24 3600‐m2 plots. Given the large size of the plots, and heterogeneity within them, including gradients in hydrology toward the Platte River, we sampled eight 6‐m2 subplots within each plot for a total of 192 subplots. Like the BioDIV, experiment, the site is managed with prescribed fires, although less frequently, and was last burned in late March 2015, two years prior to our study.
Several previous studies at each site used the airborne imagery, some of the soil data, and some of the leaf functional trait data (Appendix S2: Table S1). This study goes beyond previous work by measuring soil processes at Wood River, adding new microbial data to BioDIV, quantifying leaf‐level functional traits using spectroscopy and mapping these traits as well as phylogenetic‐functional groups at the landscape scale in Wood River, and by examining the associations between remotely detected spectroscopic information from aboveground vegetation and belowground processes. In combination, these studies represent a synthesis of our efforts to decipher the spectral signatures of plants and how they relate to ecosystem processes, with a focus on links between aboveground and belowground states and processes.
Soil sampling
In the BioDIV experiment, soil was sampled to quantify microbial respiration from year‐long incubations, net nitrogen mineralization, extracellular enzyme activity, and soil carbon and nitrogen concentrations. Soil was sampled to a depth of 10 cm at 1 m from the plot edge in four corners as well as the center (2 cm diameter, 5 cores/plot) and combined into a single sample per plot. In July 2014, 35 plots were sampled. Additional cores were taken in early August 2014 and washed over a 1 mm sieve to collect roots. In July 2015, 125 plots (including the 35 plots from the 2014 sampling) were sampled. Soil carbon and nitrogen concentrations were measured in 154 plots using the same sampling approach in 2014, 2015, and 2016; means across years are presented here.
In the Wood River experiment, in 2017 soil was sampled to a depth of 10 cm in the center of each 6‐m2 subplot located along transects approximately 20–22 m from the western and eastern borders of each plot and starting 12, 24, 36 and 48 m from the southern borders (192 subplots in total). From two aggregated soil cores per subplot, soil carbon and nitrogen, extracellular enzyme activity, potential net mineralization rate, and microbial biomass were measured.
Soils were composited by plot (BioDIV) or subplot (Wood River) and sieved (2 mm). Subsamples were dried (65°C, 48 h), milled, and total soil carbon and nitrogen measured by combustion using a Costech ECS4010 element analyzer (Valencia, California, USA). Subsamples of 2015 fresh soils from BioDIV were transported on ice and stored at −80°C for molecular characterization of bacteria and fungi. Note that soil analyses from BioDIV in 2015 were presented previously in Cline et al. (2018). We include those data as part of new analyses here.
In both experiments, soils were analyzed for microbial biomass carbon using a chloroform fumigation direct extraction procedure (Brookes et al. 1985). Within one to two weeks after collection and storage at 4°C, subsamples of sieved soil were extracted with 0.5 mol/L K2SO4 (unfumigated) or extracted following fumigation in a chloroform atmosphere for 3 d (fumigated). Total dissolved carbon in extracts was determined on a TOC/TN analyzer (Shimadzu TOC‐V, Shimadzu Corporation, Kyoto, Japan). Soil microbial biomass carbon and nitrogen were calculated as the difference between extractable carbon and nitrogen in the fumigated and unfumigated samples.
Soil process rates
Total soil respiration rate was measured in the BioDIV experiment in 2015 as the accumulation of CO2 from 50 g of soil in airtight 1 L Mason jars during 24–48 h intervals on 16 dates throughout a year‐long aerobic incubation (Cline et al. 2018). Cumulative carbon respired (mgfnd="ER"> CO2‐C·[g soil]−1) was calculated as the sum of the average respiration rate between adjacent measurement dates multiplied by the time interval between measurements and divided by the initial soil mass. We calculated net nitrogen mineralization rates (μg N·[g soil]−1·d−1) as the difference between initial and final 2 mol/L KCl‐extractable concentrations of nitrogen as and (Horwáth 2003) from 30‐d incubations of field‐moist soils in the laboratory, measured using microplate salicylate and sulfanilamide methods, respectively, on a BioTek Synergy H1 microplate reader (Winooski, Vermont, USA).
Root chemistry
Roots were oven dried at 65°C for biomass determination. Root carbon fractions (cell solubles, hemicellulose and bound proteins [hemicellulose, from this point forwards], cellulose, and acid non‐hydrolyzable residue, a measure of lignin plus recalcitrants [lignin, from this point forwards]) were determined with sequential digestion using an ANKOM fiber analyzer (ANKOM Technology, Macedon, New York, USA). Carbon and nitrogen concentration (% dry mass) were determined using combustion–reduction elemental analysis (TruSpec CN Analyzer; LECO, St. Joseph, Michigan, USA).
Fungal and bacterial sampling
Fungal and bacterial composition and diversity were only determined in the BioDIV experiment. Soil samples were collected for the same 35 plots used for other analyses in 2014 and DNA extracted using the Fas‐tDNA SPIN Kit (MP Biomedical, Solon, Ohio, USA). Fungal DNA analyses were reported in Cline et al. (2018). Briefly, polymerase chain reaction (PCR) amplification of the ITS1 gene region was conducted using primers ITS1F and ITS2 (Smith and Peay 2014). Sequencing was performed on the MiSeq platform (Illumina, San Diego, California, USA) with 250 paired‐end reads at West Virginia University’s Genomic Core Facility. For bacterial DNA analyses, which were not previously reported, 16S rRNA genes were amplified by PCR and sequenced at the West Virginia Genomics Core Facility in Morgantown, West Virginia, for sequence analysis with the Illumina MiSeq platform (Illumina, San Diego, California, USA) with 250 paired‐end reads at West Virginia University’s Genomic Core Facility. Detailed methods are provided in the supplemental material (Appendix S3). Aligned and screened sequences were clustered at 97% similarity and operational taxonomic unit (OTU) classification of unique sequences was completed using the Ribosomal Database Project (RDP) taxonomic database (release 9; Cole et al. 2009) and rarified to account for variation in the number of sequence reads per sample. Fastq files are stored with NCBI (SRA accession: 108802). Total OTU richness and inverse Simpson diversity are reported for fungi and bacteria; fungal OTU richness was previous published (Cline et al. 2018). We used non‐metric multidimensional scaling (NMDS) analyses to describe the fungal and bacterial communities within each BioDIV plot using metaMDS in the “vegan” package in R (Oksanen et al. 2018). We used an unconstrained NMDS approach because our goal was to describe the microbial communities with as few assumptions as possible rather than determine how much of the variation in microbial community structure was driven by environmental variables (Kuczynki et al. 2010).
Extracellular microbial enzyme activity
In both the BioDIV and Wood River sites, we estimated the hydrolytic and oxidative (lignolytic) enzyme activity of soil communities using extracellular enzyme assays as described in Cline et al. (2018). We measured activity of α‐glucosidase (AG, EC 3.2.1.20), β‐1,4‐glucosidase (BG, EC 3.2.1.21), cellobiohydrolase (CBH, EC 3.2.1.91), β‐1,4‐xylosidase (BX, EC 3.2.1.37), and N‐acetyl‐β‐glucosaminidase (NAG, EC 3.1.6.1), using methylumbellyferyl (MUB)‐linked substrates (German et al. 2011). A 25‐mmol/L l‐dihydroxy‐phenylalanine substrate was used to assay phenol oxidase (PO, EC 1.10.3.2) and peroxidase (PX, EC 1.11.1.7) activity. Hydrolytic enzyme potential was calculated as the sum of the activities of CBH, AG, BG, BX, and NAG, and oxidative activity was calculated as the sum of PX and PO.
Aboveground biomass and species sampling
At the BioDIV site, we quantified plot productivity and the relative dominance of plant functional groups by collecting aboveground plant biomass (g/m2) within the plots that are maintained for species richness levels 1–16 (2014, n = 121; 2015, n = 154; 2016, n = 154) in a 9 m × 6 cm strip clip in late‐July of each year. These plots include those in which soils were sampled. Aboveground biomass was sorted by plant species, dried at 60°C for 48 h and weighed. Aboveground biomass was assigned to 79 plant species and four plant phylogenetic‐functional groups, including C3 grasses, C4 grasses, forbs and legumes, as well as the two major phylogenetic groups, monocots and dicots. We quantified total aboveground biomass of each plot, as well as the relative proportion of biomass (i.e., relative dominance) made up by individual plant functional groups. Vegetation cover was estimated using the point‐quadrat method in 35 plots in 2014 (the same plots in which soil samples were collected that year).
At the Wood River site, we sampled aboveground biomass and percent cover of species within two 0.5 m2 areas within each subplot. These metrics were used to determine species richness and abundance at the subplot scale. In addition, we recorded presence of species along two 1‐m wide transects, each centered approximately 20 m from the eastern and western borders of the plots. These data were used to determine species richness at the whole‐plot scale.
Foliar sampling
At the BioDIV site, spectral sampling at the leaf level, the collection of foliar samples, chemical assays and the prediction of foliar traits from spectra were described in detail in Schweiger et al. (2018). An SVC full‐range (400–2,500 nm) portable spectrometer (Spectra Vista Corporation, Poughkeepsie, New York, USA) with a leaf clip and tungsten‐halogen light source (LC‐RP PRO; Spectra Vista Corporation) was used to obtain foliar spectra and develop leaf trait models. Leaf‐tissue samples of 130 individuals from 62 species were collected together with leaf spectra in the summers of 2015 and 2016 at the CCESR, flash frozen in liquid nitrogen and stored at −80°C for subsequent pigment analysis. An additional set of 130 samples of 61 species were spectrally sampled and oven dried at 65°C for chemical analyses of non‐labile constituents. Pigment concentration and area‐based pigment content was determined on flash‐frozen samples using high‐performance liquid chromatography (HPLC, Agilent 1200 Series; Agilent Technologies, Santa Clara, California, USA). The pigments included chlorophyll a, chlorophyll b, one carotene pigment (β‐carotene), and five xanthophylls (lutein, zeaxanthin, violaxanthin, antheraxanthin, and neoxanthin). The total carotenoid pigment pool was calculated as the sum of five xanthophyll pigments plus β‐carotene (Croft and Chen 2018). Carbon fraction concentrations (% dry mass) were determined with sequential digestion, as described for roots above, for soluble cell contents, hemicellulose, cellulose, and lignin. Carbon and nitrogen concentration (% dry mass) were determined using combustion–reduction elemental analysis (TruSpec CN Analyzer; LECO, St. Joseph, Michigan, USA). Bivariate trait relationships are shown in the correlation matrix in Appendix S1: Fig. S2.
At the BioDIV site, leaf‐level spectra were measured in four to eight 1‐m2 subplots per plot, as described in Schweiger et al. (2018). At the Wood River site, leaf‐level spectra were collected at regular intervals of 2 m along two transects starting at 2 m from the southern border and ending at 58 m, yielding a total of 29 individual plants measured per plot. Foliar trait models for all pigments, all carbon fractions, C and N were developed using partial least square regression (PLSR; Martens and Naes 1989, Wold et al. 1983) implemented in R (R Development Core Team 2018) using the R package “plsRglm” (Bertrand et al. 2014) and are reported in Schweiger et al. (2018). Model performance ranged between 0.53 and 0.84 R 2. The models were applied to all 1,129 leaf‐level spectra collected in the BioDIV experiment and all 1,399 leaf‐level spectra collected at Wood River. A PLSR model built from 892 grass and forb samples with concurrent SVC spectra collected during peak growing season in grassland ecosystems near Madison, Wisconsin was used to predict leaf mass per area (LMA, g/m2) for each individual plant sampled in the BioDIV (Wang et al. 2019) and Wood River experiments.
Species mean leaf‐level traits were scaled to the plot level based on relative biomass at the BioDIV experiment (Wang et al. 2019). At Wood River, species mean traits were scaled to the subplot level using percent cover. The same was done for phylogenetic‐functional groups. The whole‐plot‐level and whole‐subplot‐level trait estimates, respectively, were matched to spectra extracted from airborne imagery to develop trait maps, as described below.
Airborne spectroscopic data collection, trait and phylogenetic‐functional group mapping
Imaging spectroscopy data from the Airborne Visible/Infrared Imaging Spectrometer—Next Generation (AVIRIS‐NG, Hamlin et al. 2011) were collected at BioDIV with a spatial resolution of 0.9 m on the ground by the National Aeronautics and Space Administration (NASA) on August 25, 2014, August 30, 2015 and August 31, 2016 (Wang et al. 2019). The AVIRIS‐NG reflectance data comprise 432 spectral bands and span 380 nm to 2,510 nm with a spectral resolution of 5 nm. At Wood River, airborne imaging spectroscopy data were collected on August 23, 2017, with a spatial resolution of 1 m on the ground by an AISA Kestrel (Specim, Oulu, Finland) sensor operated by the University of Nebraska’s Center for Advanced Land Management Information Technologies (CALMIT) (Gholizadeh et al. 2019). The AISA Kestrel reflectance data comprised 178 spectral bands and spanned 400 nm to 1,000 nm with a spectral resolution of approximately 3.5 nm (Gholizadeh et al. 2020). We used PLSR (Wold et al. 1983) as implemented in the R package “pls” (Mevik et al. 2018) to model and predict biomass, functional group composition and foliar functional traits from imaging spectroscopy data, an approach widely used for the retrieval of plant traits and vegetation parameters (Asner and Martin 2008, Serbin et al. 2014, Asner et al. 2015, Singh et al. 2015, Wang et al. 2019).
In BioDIV, plot‐level trait data were linked to the spectroscopic images as reported in Wang et al. (2019). In short, models for predicting functional traits from remotely sensed spectra were developed based on AVIRIS‐NG spectra and leaf trait models (Schweiger et al. 2018) scaled to the plot level with PLSR using plsregress in MATLAB (The MathWorks, Inc., Natick, Massachusetts, USA) (Wang et al. 2019). We modeled vegetation cover as a function of the mean spectral angles between soil and vegetation spectra per plot using logistic regression (see Serbin et al. 2015) and predicted vegetation cover for all the pixels in the three images based on the resulting regression equation. We used a 7 × 7 pixel window centered in the 9 m × 9 m plots, and used mean spectra and trait estimates by plot as model inputs. Pixels with vegetation cover of less than 20% were excluded from analysis. For trait mapping, we applied PLSR coefficients to all pixels in the AVIRIS‐NG images. For analysis with soils data, we used the predicted mean vegetation traits per plot for the central 7 × 7 pixel window and again excluded pixels with <20% cover.
In Wood River, similar to the approach used for modeling functional traits in BioDIV, we used plot‐level and subplot‐level trait estimates and airborne spectra extracted from the same plots and subplots to model biomass and functional group composition and functional traits, including foliar soluble cell contents and the contents of carbon, nitrogen, hemicellulose and lignin with PLSR; here we used the R package“pls” (Mevik et al. 2018). For modeling biomass, phylogenetic‐functional group composition and chemical traits, 25% of the full dataset was set aside for independent validation of the models; 75% was used to develop the models. Of this portion of the data, we used, depending on the number of observations, 75% or 80% of the data for model calibration and the rest for validation over 500 model runs. To avoid overfitting, the number of components used in PLSR was selected by minimizing the mean root mean square error (RMSE) and the cross‐validated Prediction Residual Error Sum of Squares (PRESS) (Chen et al. 2004). The mean model coefficients with significant fits (P < 0.05) were then applied to the spectral images to predict pixelwise biomass, functional group composition, and chemical traits. Model performance was assessed based on the R 2 for linear regressions between measured and predicted values, the RMSE, %RMSE and model bias (see Appendix S2: Table S2). As the plots at the Wood River site had no significant soil exposure, pixels were not filtered based on vegetation cover.
Diversity metrics
Phylogenetic species richness (PSR) (Helmus 2007) was calculated using the phylogeny from Smith and Brown (2018), pruned to include the measured species in the BioDIV experiment and the WoodRiver experiment separately. Absent species were added within the correct genus in a randomly assigned location using the congeneric merge function in “pez” in R (Pearse et al. 2015). Functional diversity was calculated using Scheiner’s functional dispersion [FD(qDTM)] (Scheiner et al. 2017) using biomass or percent cover (at Wood River) of each species within each plot to weight the species following Schweiger et al. (2018). Remotely sensed spectral diversity using AISA airborne data in Wood River was calculated as Euclidean distances among vector‐normalized reflectance values for all wavelengths among all pixels per plot, using the dist function in R.
Structural equation modeling
The causal diagrams (Fig. 1) outline our hypotheses regarding how aboveground vegetation and soil processes are connected. We tested the degree of support for these hypotheses for the BioDIV and Wood River experiments, respectively, with SEMs as implemented in the R package “lavaan” (Rosseel 2012). Measured variables were used as indicators of each of the constructs in the model. We used aboveground biomass as an indicator of aboveground vegetation quantity and total organic inputs to soil, given it strongly predicts belowground root biomass in BioDIV and other prairie systems (Fornara and Tilman 2008, Appendix S1: Fig. S3). Root biomass per m−2 is nearly five times greater than aboveground biomass, on average, and is likely to be the primary source of inputs belowground. We used aboveground vegetation traits—including foliar chemical composition and specific leaf area (SLA)—and functional group abundance (proportion or biomass) as indicators of vegetation quality. Hydrolytic enzyme activity, microbial biomass and cumulative soil respiration were used as indicators of soil microbial activity. Net nitrogen mineralization rates and soil carbon and nitrogen concentration were used as indicators of soil nutrient availability. To avoid model overfitting, we selected those chemical vegetation traits and functional groups we considered to be the most important factors influencing vegetation quality in our study systems. Although more complex relationships are theoretically possible, we used linear specifications for our variables, which were also supported by the univariate regression analyses.
Results
Consistency between phylogenetic relationships, functional groups, and functional traits
In both BioDIV and Wood River, the functional groups that the diverse set of species represents largely correspond to monophyletic phylogenetic lineages, nested within the monocots or the dicots (eudicots)—the two major angiosperm lineages found in these ecosystems (Fig. 2). Within the monocots, C3 and C4 grasses in these experimental systems each fall within individual monophyletic lineages. The legumes and the forbs also each fall within distinct phylogenetic clades and—together with the small number of woody plants occasionally in the plots—are collectively nested within the dicot lineage. As a consequence, we refer to the functional groups as phylogenetic‐functional groups.
Fig. 2.
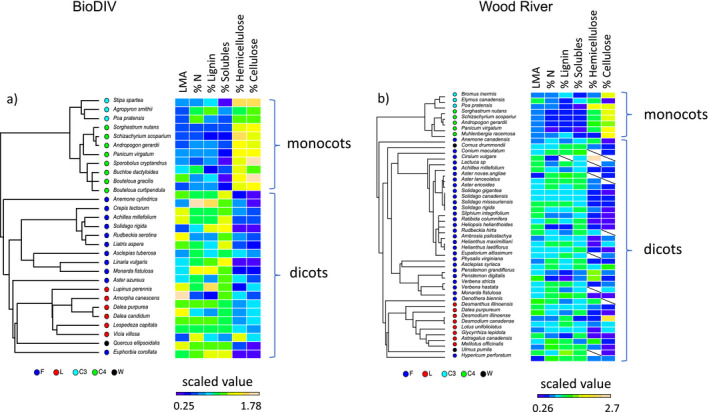
Phylogenetic relationships among species, their functional group categories (forbs, F; legumes, L; C3 grasses, C3; C4 grasses, C4; and woody species, W) and their foliar functional and chemical composition (leaf mass per area, LMA; % N, % lignin; % cell solubles; % hemicellulose, and % cellulose) in (a) the BioDIV experiment at Cedar Creek and (b) the Wood River experiment in Nebraska. Note that F, L and W groups are nested within the dicot lineage and C3 and C4 grasses are nested within the monocots. Functional trait values are scaled—centered to their means and divided by their standard deviations—and color‐coded with blue colors indicating low values and yellow or salmon colors indicating high values. Missing values are indicated by a slash.
The morphological and chemical traits of species also tended to correspond to their phylogenetic‐functional group identities. Differences in vegetation chemistry were consistently pronounced between monocots and dicots. In both systems, monocots (C3 and C4 grasses) had lower LMA, foliar nitrogen and lignin concentrations, lower cell soluble concentrations, and higher hemicellulose and cellulose concentrations compared with the forb and legume groups within the dicots (Table 1). The C4 grasses, in particular, tended to have high cellulose and hemicellulose concentrations. In contrast, the dicots tended to have lower foliar hemicellulose and cellulose concentrations and somewhat higher foliar nitrogen and lignin concentrations than the monocots (Fig. 2, Table 1). Differences between groups within these two major lineages depended on the compound and to some extent, the site. Legumes had higher foliar nitrogen and lignin concentrations than the other groups at the Wood River site but only significantly higher foliar nitrogen and lignin than C4 grasses in BioDIV (Table 1). C4 grasses had lower concentrations of foliar nitrogen, lignin and solubles but higher concentrations of cellulose and hemicellulose than other functional groups, particularly at the Wood River site.
Table 1.
Phylogenetic‐functional group means ± SE for chemical trait values derived from leaf level spectra for monocots, dicots, C3 grasses, C4 grasses, forbs and legumes.
| Monocots | Dicots | C3 | C4 | Forbs | Legumes | ||
|---|---|---|---|---|---|---|---|
| BioDIV | LMA | 57.05 ± 3.69a | 86.56 ± 5.37b | 54.23 ± 4.93a | 58.11 ± 4.85a | 80.01 ± 4.85b | 93.99 ± 4.85b |
| % solubles | 26.03 ± 2.28a | 61.27 ± 1.98b | 30.46 ± 6.49a | 24.37 ± 2.06a | 65.27 ± 2.06b | 54.62 ± 2.06c | |
| % hemicellulose | 34.34 ± 1.21a | 16.70 ± 1.51b | 31.64 ± 3.97a | 35.35 ± 0.81a | 13.72 ± 0.81b | 21.33 ± 0.81c | |
| % cellulose | 34.57 ± 1.78a | 14.07 ± 1.16b | 31.78 ± 3.98a | 35.62 ± 1.99a | 11.38 ± 1.99b | 19.04 ± 1.99c | |
| % ADL | 6.28 ± 0.45a | 9.02 ± 0.60b | 7.95 ± 0.76a | 5.66 ± 0.37b | 9.53 ± 0.37ac | 8.07 ± 0.37abc | |
| % C | 45.60 ± 0.22a | 45.07 ± 0.56a | 45.35 ± 0.24a | 45.69 ± 0.29a | 45.44 ± 0.29a | 43.95 ± 0.29a | |
| % N | 1.41 ± 0.10a | 1.87 ± 0.09b | 1.76 ± 0.17a | 1.28 ± 0.09b | 1.94 ± 0.09ac | 1.73 ± 0.09ac | |
| C:N | 34.45 ± 2.39a | 25.43 ± 1.28b | 26.49 ± 2.97a | 37.43 ± 2.37b | 24.49 ± 2.37ac | 27.02 ± 2.37ac | |
| Wood River | LMA | 75.57 ± 5.30a | 93.18 ± 2.80b | 85.75 ± 12.55ab | 69.47 ± 5.30a | 93.38 ± 3.14b | 90.76 ± 7.77ab |
| % solubles | 35.64 ± 4.46a | 70.22 ± 1.66b | 41.23 ± 12.40a | 32.28 ± 4.46a | 71.67 ± 1.51b | 62.20 ± 4.81c | |
| % hemicellulose | 20.09 ± 2.44a | 12.82 ± 1.34b | 22.21 ± 6.01a | 18.82 ± 2.44a | 13.20 ± 1.77ab | 11.80 ± 1.79b | |
| % cellulose | 39.07 ± 4.27a | 15.69 ± 1.51b | 31.24 ± 10.50a | 43.77 ± 4.27a | 14.70 ± 1.25b | 19.78 ± 5.16b | |
| % ADL | 9.22 ± 0.94a | 13.19 ± 0.47b | 9.38 ± 1.91ab | 9.12 ± 0.94a | 12.81 ± 0.51c | 13.38 ± 1.11bc | |
| % C | 45.83 ± 0.59a | 45.52 ± 0.47a | 46.49 ± 1.24a | 45.44 ± 0.59a | 45.57 ± 0.57a | 44.82 ± 0.82a | |
| % N | 1.27 ± 0.11a | 2.00 ± 0.06b | 1.54 ± 0.11a | 1.11 ± 0.11b | 2.00 ± 0.07c | 2.06 ± 0.18ac | |
| C:N | 42.03 ± 4.93a | 24.98 ± 1.21b | 32.65 ± 2.16ab | 47.65 ± 4.93a | 25.17 ± 1.53b | 24.07 ± 2.30b |
For BioDIV, trait values were averaged across all individuals of each of 29 species (Schweiger et al. 2018); for Wood River, trait values were averaged across all individuals of each of 49 species; phylogenetic‐functional group means were averages of species means. Significant differences between monocots and dicots (left) or among the C3 grass, C4 grass, forb and legume groups (right) based on Tukey’s HSD tests are shown with different letters, P cutoff = 0.05.
Measured biodiversity‐productivity relationships
In the BioDIV experiment, measured species richness (using 2014 data and counting only species that were originally planted in the experiment as the other ones are annually removed), strongly predicted aboveground biomass (R 2 = 0.45, N = 153, P < 0.0001, Fig. 3a), as has been reported numerous times previously (Reich et al. 2012). Productivity was also significantly predicted by PSR, a measure of phylogenetic diversity that accounts for shared ancestry and evolutionary distance among species (R 2 = 0.15, N = 152, P < 0.0001, Fig. 3b), and leaf‐level functional diversity [FD(qDTM)] calculated using mean traits per species and their abundances, based on biomass (R 2 = 0.39, N = 153, P < 0.0001, Fig. 3c). The strengths of the relationships in different years were very similar. In the Wood River experiment, measured species richness was a weaker predictor of biomass than in the BioDIV experiment. At the subplot scale (6 m2), species richness explained 9% of the variation in aboveground biomass (R 2 = 0.09, N = 191, P < 0.0001, Fig. 3d). At the plot scale, species richness explained 16% in aboveground biomass (R 2 = 0.16, N = 24, P = 0.05, Fig. 3g). PSR significantly but weakly predicted aboveground biomass at both the subplot scale (R 2 = 0.07, N = 191, P < 0.0001, Fig. 3e) and the plot scale (R 2 = 0.20, N = 24, P = 0.026, Fig. 3h). Abundance‐weighted functional diversity [FD(qDTM)] also weakly predicted biomass at the subplot scale (R 2 = 0.09, df = 190, P < 0.0001, Fig. 3f) and plot scale (R 2 = 0.15, N = 24, P = 0.066, Fig. 3i).
Fig. 3.
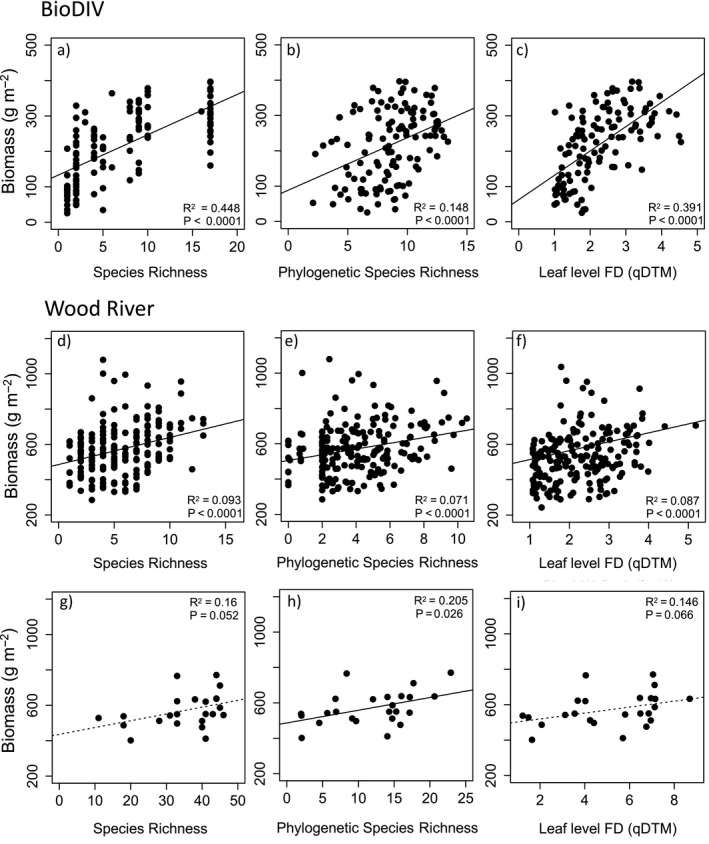
Biodiversity–Ecosystem Function relationships in the BioDIV and Wood River experiments. Shown are regressions between plant diversity metrics and annual aboveground biomass for BioDIV from 2014, N = 124 (a–c) and Wood River at the subplot scale from 2017, N = 192 (d–f) and plot scale, N = 24 (g–i). Diversity metrics include measured species richness (SR), phylogenetic species richness (PSR) and abundance‐weighted functional diversity [FD(qDTM)] measured at the leaf level.
Remote sensing of biomass (annual productivity)
Annual aboveground plant biomass, which is a measure of annual aboveground productivity in grasslands, varied considerably across the plots in the BioDIV experiment, ranging from approximately 19 to 740 g/m2 and is strongly associated with vegetation cover (Appendix S1: Fig. S3b–d). PLSR models developed for each of the three years using AVIRIS‐NG spectra predicted plot‐level biomass well (Fig. 4a for 2014, Appendix S2: Table S2). Independent validation results in 2014 (R 2 = 0.75, 22 components, %RMSE = 12.05, P < 0.0001), 2015 (R 2 = 0.47, 18 components, %RMSE = 16.4, P < 0.0001) and 2016 (R 2 = 0.64, 18 components, %RMSE = 15.13, P < 0.0001) were consistent (Appendix S2: Table S2). Remotely sensed vegetation cover reported in Wang et al. (2019) (R 2 = 0.68, %RMSE = 11)—based on the soil spectral angle calculated between the averaged soil spectrum and each pixel in the airborne image for the plot (Serbin et al. (2015)—also predicted biomass for the three years using linear regression (Fig. 4b, R 2 = 0.63 for 2014, 0.70 for 2015, 0.55 for 2016, N = 153, P < 0.0001 all years, Appendix S2: Table S2), serving as an accurate proxy of productivity in this system.
Fig. 4.
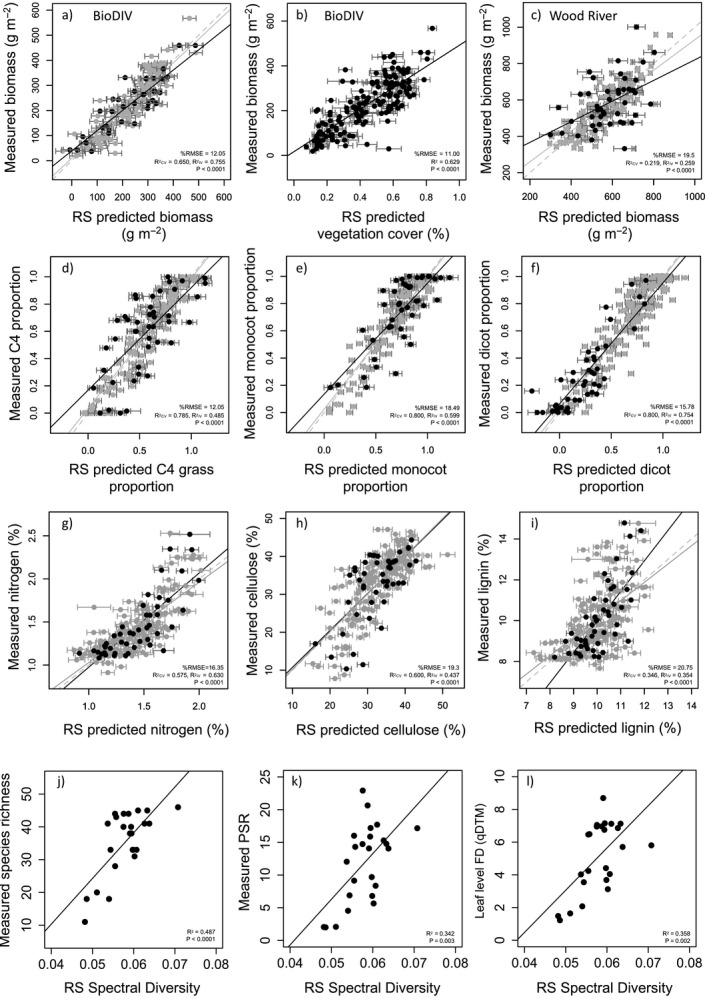
Detection of aboveground plant biomass (BioDIV and Wood River) and phylogenetic‐functional groups, foliar traits and plant diversity (Wood River) from airborne remotely sensed (RS) imagery. In the BioDIV experiment, in situ measured biomass was accurately predicted from AVIRIS NG spectroscopic imagery using either a partial least squares regression (PLSR) model (a) or the vegetation cover model reported in Wang et al. (2019) (b). In the Wood River experiment, biomass was predicted from AISA Kestrel spectroscopic imagery using PLSR (at the 6‐m2 subplot scale) (c). Airborne data also predicted phylogenetic‐functional group composition in the Wood River experiment—shown here for C4 (d), monocot (e) and dicot (f) proportions—and functional traits, including foliar nitrogen (g), cellulose (h) and lignin (i) concentrations. Both cross‐validation (CV, gray circles) and independent‐validation (IV, black circles) % root mean square error (RMSE) and R 2 are shown for PLSR models. In (j–l), remotely sensed spectral diversity—the mean of the vector‐normalized spectral distances among pixels within a plot—predicts measured species richness, phylogenetic species richness (PSR) and abundance‐weighted functional diversity [FD(qDTM)] (at the 3600‐m2 plot scale).
In the Wood River experiment, PLSR models also significantly predicted biomass at the subplot scale (R 2 = 0.426, 11 components, %RMSE = 19.50, P < 0.0001) (Fig. 4c, Appendix S2: Table S2). When predicted values of biomass were applied to every pixel in the plots and averaged, predicted biomass at the plot scale was strongly associated with the average measured biomass in each plot (R 2 = 0.76, N = 24, P < 0.0001, Appendix S1: Fig. S5). In contrast with the BioDIV experiment, vegetation cover was 100% throughout the experimental landscape (Gholizadeh et al. 2019) and was not associated with aboveground biomass (Appendix S1: Fig. S3d). Consequently, we did not attempt to develop a PLSR model to predict vegetation cover because it could not be used as a proxy for biomass at Wood River. Rather, we used the PLSR model for biomass to predict aboveground biomass.
Detecting plant phylogenetic‐functional groups from airborne spectroscopic imagery
In the Wood River experiment, the proportion of each phylogenetic‐functional group within a subplot was significantly predicted from airborne imagery (Fig. 4d–f, Appendix S2: Table S2). Independent validation results from PLSR models at Wood River using AISA Kestrel for monocot, dicot, legume, forb, C4 grass, and C3 grass proportions in each subplot show that all are well‐predicted although with somewhat higher accuracy at broader phylogenetic levels—monocots, dicots—than for the groups within those (Appendix S2: Table S2). In BioDIV, PLSR models predicting the biomass of individual phylogenetic‐functional groups were inconsistent but most accurate and consistent for monocot biomass across years (Appendix S2: Table S2). They were generally not able to predict the proportion of these groups likely due to the high exposed soil/vegetation cover in this experiment (Wang et al. 2018) and particularly to the limited number of plots with a high proportion of any given functional group that also had low exposed soil area.
Functional trait models for Wood River
We developed PLSR models to map functional traits using AISA data at Wood River (Appendix S2: Table S2), following a similar approach as reported in Wang et al. (2019) for mapping traits in the BioDIV experiment from AVIRIS‐NG data using % cover to calculate community weighted mean traits. Independent validation results for predicted foliar concentrations of nitrogen (R 2 = 0.58, components = 5, %RMSE = 16.35, P < 0.0001, Fig. 4g), cellulose (R 2 = 0.42, components = 4, %RMSE = 19.30, P < 0.0001, Fig. 4h), lignin (R 2 = 0.35, components = 3, %RMSE = 20.75, P < 0.0001, Fig. 4i) and cell solubles (R 2 = 0.35, components = 4, %RMSE = 19.56, P <0.0001), as well as LMA (R 2 = 0.44, N = 191, components = 3, %RMSE = 19.05, P < 0.0001) show that that traits could be reliably mapped in Wood River using AISA data (Appendix S2: Table S2).
Detecting plant diversity from remotely sensed spectra
We calculated spectral diversity from airborne data as the mean of the vector‐normalized spectral distances among all pixels per plot (or subplot), a metric that does not require site‐specific model development. At Wood River, based on AISA Kestrel data at the plot scale, spectral diversity readily predicted all measures of plant diversity, including species richness (R 2 = 0.497, P < 0.0001, Fig. 4j), PSR (R 2 = 0.342, P < 0.003, Fig. 4k), and leaf‐level functional diversity [FD(qDTM)] (R 2 = 0.358, P < 0.002, Fig. 4l). These relationships did not hold at the subplot scale, however, perhaps because the number of pixels per subplot (6) was insufficient to capture the variability or because there was limited variability at this scale, or both. In BioDIV, spectral diversity based on 0.9 m spatial resolution AVIRIS‐NG data did not predict any metric of plant diversity, including measured species richness. Prior studies at this site have shown that accuracy in predicting plant diversity is highly dependent on spatial resolution and the high soil fraction/vegetation cover fraction poses difficulties for predicting plant diversity from remotely sensed measures of spectral diversity at a spatial resolution of ˜1 m (Wang et al. 2018, Gamon et al. 2020, Gholizadeh et al. 2018).
Remotely sensing the biodiversity‐productivity relationship in either experiment was challenging due to the inability to predict plant diversity metrics from airborne data in BioDIV, and due to the relatively weak measured associations between plant diversity and biomass in Wood River at either the subplot (Fig. 3d–f) or plot scale (Fig. 3g–i). Nevertheless, there was a weak relationship between remotely sensed spectral diversity and remotely sensed biodiversity at the subplot scale (Appendix S1: Fig. S5c), although this was not significant at the plot scale (Appendix S1: Fig. S5b). Remotely sensed spectral diversity at the plot scale was strongly associated with foliar nitrogen concentration, which may indicate that in ecosystems that are more diverse, plants are able to acquire more nutrients from the soil and allocate it to leaves (Williams et al. 2020), but this same pattern did not emerge at the subplot scale (Appendix S1: Fig. S5d,e).
Associations between total aboveground inputs and belowground processes and attributes
In the BioDIV experiment, the quantity of total aboveground inputs—or productivity, measured as annual aboveground biomass or vegetation cover—but not their quality, measured as the chemical or phylogenetic‐functional composition, were strongly and positively associated with hydrolytic enzyme activity but not oxidative enzyme activity. Vegetation cover and biomass were also positively associated with microbial biomass carbon and nitrogen, net nitrogen mineralization rate, cumulative respiration rate and soil carbon concentration, but not soil nitrogen concentration (Fig. 5; Appendix S2: Table S3). Consequently, remotely sensed vegetation cover significantly predicted cumulative soil respiration rates, hydrolytic enzyme activity, microbial biomass carbon, net nitrogen mineralization rates and soil carbon concentration (Fig. 6a–e). In the Wood River experiment, the quantity of total aboveground inputs—measured directly as aboveground biomass or remotely sensed based on PLSR models—were not associated with any of the measured belowground processes and attributes (Fig. 5). In contrast, foliar nitrogen concentration at Wood River measured on the ground (Appendix S2: Table S3) or mapped from remotely sensed data significantly predicted microbial biomass, hydrolytic enzyme activity, net nitrogen mineralization rate, and total soil nitrogen and carbon (Fig. 6). Foliar nitrogen concentration in the BioDIV experiment was not associated with any of these soil characteristics (Fig. 5, Appendix S2: Table S3).
Fig. 5.
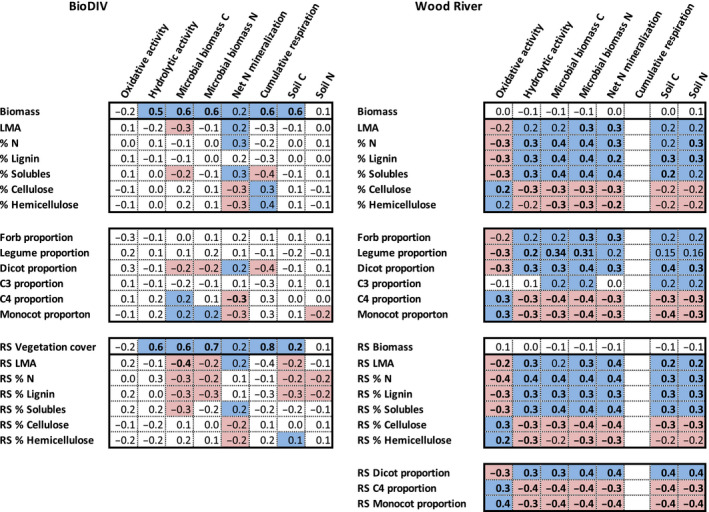
Belowground processes tended to be predicted by in situ or remotely sensed measures of biomass (including vegetation cover) in BioDIV but by in situ or remotely sensed measures of foliar function and chemical composition in Wood River. Shown are correlation coefficients (r values) at the plot level (BioDIV) or subplot level (Wood River) between remotely sensed (RS) traits, directly measured leaf traits calculated as community weighted means, or functional group composition (proportion or biomass) and soil processes, including oxidative enzyme activity (nmol·g−1·h−1), hydrolytic enzyme activity (nmol·g−1·h−1), microbial biomass carbon (mg C·[g soil]−1), nitrogen net mineralization rate (mg N·[g soil]−1·d−1), cumulative respiration rate (mg CO2‐C·[g soil−1·d−1]), and soil carbon and nitrogen concentrations (%). Shading indicates that the correlation is significant at P < 0.05 or if bolded at P < 0.001. Blue shading indicates the correlation is positive, pale red indicates the correlation is negative. No shading indicates that we found no relationship between the variables, and blank squares indicate no data.
Fig. 6.
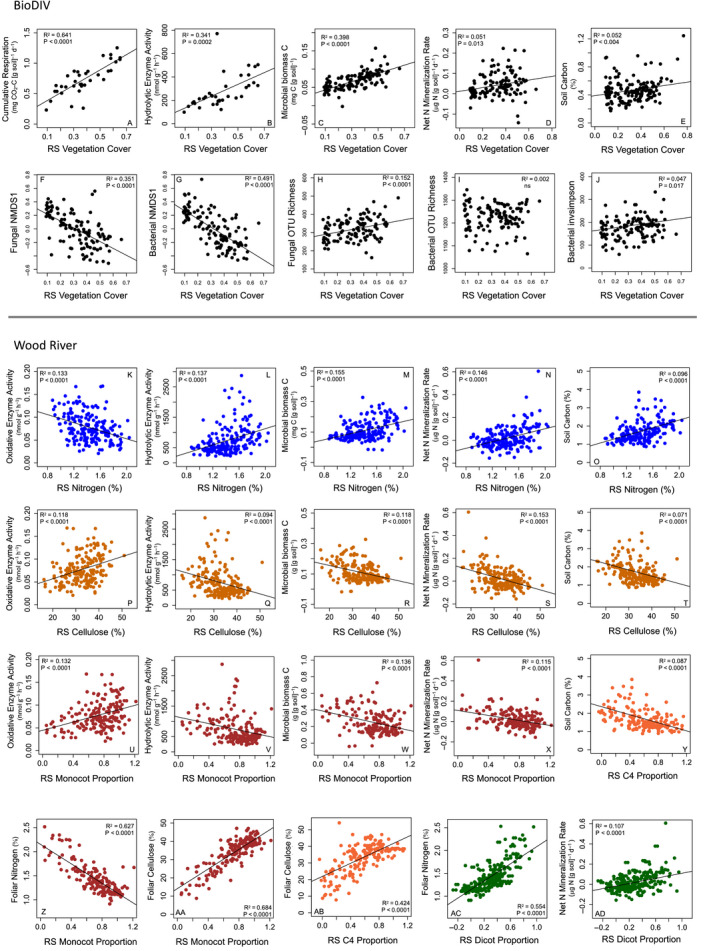
In the BioDIV experiment, remotely sensed vegetation cover predicted (a) cumulative respiration rate, (b) hydrolytic enzyme activity, (c) microbial biomass carbon, (d) nitrogen net mineralization rate, (e) soil carbon, (f) the first NMDS1 axis of the fungal community data and (g) the first NMDS1 axis of the bacterial community data as well as (h) fungal operational taxonomic unit (OTU) richness but not (i) bacterial OTU richness. (j) Bacterial diversity measured by the inverse Simpson metric is weakly predicted. Data are from 2015, N = 119. In the Wood River experiment, remotely sensed vegetation nitrogen concentration in the vegetation (RS Nitrogen) predicted (k) oxidative enzyme activity, (l) hydrolytic enzyme activity, (m) microbial biomass carbon, (n) net nitrogen mineralization rate and (o) soil carbon. Remotely sensed (RS) cellulose concentration and RS monocot proportion positively predicted (p, u) oxidative enzyme activity, and negatively predicted (q, v) hydrolytic enzyme activity, (r, w) microbial biomass carbon, (s, x) net nitrogen mineralization rate and (t, y) soil carbon. RS monocot proportion was negatively associated with (z) foliar nitrogen (%) and positively associated with (AA) foliar cellulose concentration (%), as was (AB) C4 grass proportion, while dicot proportion was positively associated with (AC) foliar nitrogen and (AD) soil net nitrogen mineralization rate.
Plant productivity was strongly associated with soil fungal and bacterial composition and to a lesser degree fungal and bacterial diversity in BioDIV. Remotely sensed vegetation cover significantly predicted the first axis of NMDS ordination analyses, for both the fungal (Fig. 6f) and bacterial communities (Fig. 6g). These results indicate that the composition of microbial communities is associated with plant productivity (Fig. 6f–j). Remotely sensed vegetation cover weakly but significantly predicted fungal diversity, measured as OTU richness (Fig. 6h), but not inverse Simpson diversity (not shown). Remotely sensed vegetation cover also weakly but significantly predicted bacterial diversity, measured as inverse Simpson diversity (Fig. 6j), but not as OTU richness (Fig. 6i).
Predicting belowground processes and attributes from leaf chemistry and functional group composition
BioDIV
In the BioDIV experiment, directly measured or remotely sensed functional traits showed relatively few associations with belowground properties, in contrast with the strong and consistent associations between total aboveground inputs and belowground properties (Fig. 5; Appendix S2: Table S3). However, some relationships were still found. In BioDIV, net nitrogen mineralization rates were positively associated with directly measured LMA, foliar nitrogen concentration, and foliar concentration of cell solubles—measured at the leaf level—but negatively associated with foliar cellulose and hemicellulose concentration. Net nitrogen mineralization rates were also predicted by remotely sensed LMA and foliar concentration of cell solubles, cellulose and hemicellulose, although remotely sensed nitrogen did not show a significant relationship. Cumulative soil respiration was positively associated with directly measured foliar cellulose and hemicellulose concentration and negatively associated with cell soluble concentration. Microbial biomass carbon was negatively associated with LMA and cell soluble concentration—measured at the leaf level or remotely sensed—and with foliar nitrogen concentration and lignin concentration measured at the leaf level. However, most of the measured belowground processes in BioDIV, including oxidative enzyme activity, hydrolytic enzyme activity, microbial biomass nitrogen and total soil carbon and nitrogen concentration were not associated with any of the functional traits measured at the leaf level. Some remotely sensed traits were negatively associated with soil carbon and nitrogen concentration, including foliar nitrogen and lignin concentration, and remotely sensed foliar hemicellulose concentration was significantly but weakly associated with soil carbon concentration. In general, however, functional traits were not strong or consistent predictors of belowground processes in the BioDIV experiment.
Consistent with these findings, phylogenetic‐functional group proportion or biomass explained relatively few belowground processes in BioDIV (Fig. 5; Appendix S2: Table S3). Dicot proportion was negatively associated with microbial biomass carbon and nitrogen and cumulative respiration, while monocot and C4 grass proportion were positively associated with microbial biomass carbon and negatively associated with net nitrogen mineralization rate. Biomass of these phylogenetic‐functional groups showed trends that were similar to both proportion and total biomass relationships with soil attributes. Monocot and C4 grass biomass positively predicted hydrolytic enzyme activity, microbial biomass carbon and nitrogen, cumulative respiration rate and soil carbon, and negatively predicted net nitrogen mineralization rate, similar to total aboveground biomass (Appendix S2: Table S3). Forb biomass strongly predicted microbial biomass nitrogen (but not microbial biomass carbon), and net nitrogen mineralization rate. C3 grass and forb biomass also weakly predicted soil carbon concentration (Appendix S2: Table S3).
Wood River
In contrast with BioDIV, at the Wood River site, foliar and remotely sensed plant functional traits were strongly and consistently associated with all belowground processes and attributes, summarized in Fig. 5 and the strength of the regression coefficients were very similar (Appendix S2: Table S3). Specifically, both leaf‐level and remotely sensed foliar nitrogen (Fig. 6k–o) were negatively associated with oxidative enzyme activity and positively associated with hydrolytic enzyme activity, microbial biomass carbon and nitrogen, net nitrogen mineralization rate and soil nitrogen and carbon, as were leaf‐level and remotely sensed lignin, cell soluble concentration and LMA (Appendix S2: Table S3). Both leaf‐level and remotely sensed foliar cellulose (Fig. 6p–t) were positively associated with oxidative enzyme activity and negatively associated with microbial biomass carbon and nitrogen, net nitrogen mineralization rate and soil nitrogen and carbon concentrations, as were leaf‐level and remotely sensed hemicellulose concentration (Appendix S2: Table S3).
The sign of these relationships was largely mirrored by phylogenetic‐functional group proportion. Monocot proportion—which is strongly negatively associated with remotely sensed foliar nitrogen (Fig. 6z) and positively associated with cellulose concentration (Fig. 6aa)—was positively associated with oxidative enzyme activity and negatively associated with hydrolytic enzyme activity, microbial biomass carbon, net nitrogen mineralization rate (Fig. 6p–x), and soil carbon (Appendix S2: Table S3). C4 grass proportion showed these same patterns although C3 grass proportion showed weakly contrasting patterns or no relationship at all (Fig. 5). Specifically, C3 grass proportion was positively associated with microbial biomass carbon and nitrogen as well as soil carbon and nitrogen concentration, but not any of the other soil processes. Dicot proportion—which is strongly associated with remotely sensed nitrogen concentration (Fig. 6ac) and negatively associated with cellulose concentration—was negatively associated with oxidative enzyme activity, and positively associated with hydrolytic enzyme activity, microbial biomass carbon, net nitrogen mineralization rate (Fig. 6ad). The proportion of variation explained in any of these plant functional trait–belowground process relationships was relatively low, ranging from approximately 5% to 15%. However, the consistency in the associations between soil properties and directly measured plant chemical traits, remotely sensed chemical traits or directly measured and remotely sensed phylogenetic‐functional groups is striking in the Wood River system (Fig. 5). We were able to predict soil properties from any of the ways that we measured vegetation quality, from plant functional traits to phylogenetic‐functional group proportions, measured directly or remotely sensed (Fig. 6K–X, Appendix S2: Table S3).
Structural equation models
Our SEM results confirm the hypotheses illustrated in our conceptual models (Fig. 1) and are consistent with the bivariate relationships in Figs. 5, 6. The analysis synthesizes these findings and shows that vegetation quantity and quality influenced belowground attributes and processes and their interrelationship in the BioDIV and Wood River experiments differently. In BioDIV, the quantity of aboveground inputs to the soil, measured as aboveground biomass, was an important positive predictor of the activity and abundance of soil microbes, while vegetation quality, measured as the carbon to nitrogen ratio in aboveground vegetation, was not (Fig. 7a). The quantity of aboveground inputs also was positively associated with soil nutrient availability, measured as net nitrogen mineralization rate (Nmin), while vegetation quality was not.
Fig. 7.
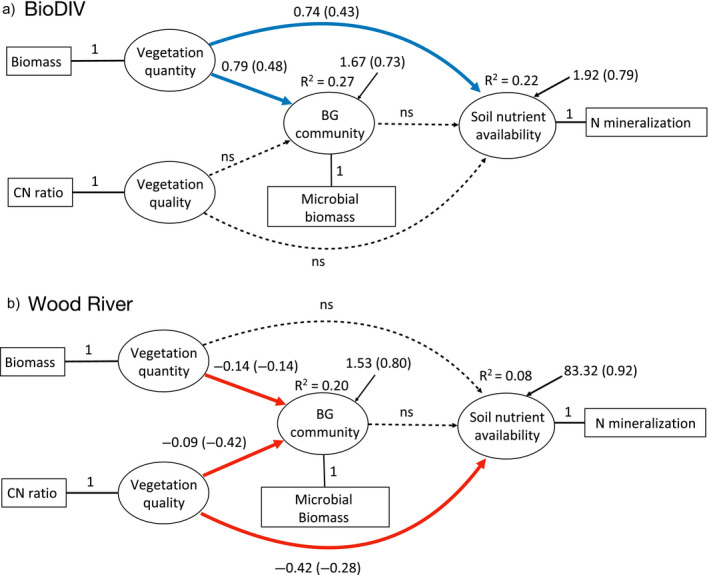
Structural equation models reveal the relative importance of aboveground quantity and quality of vegetation inputs for belowground microbial processes and soil nutrient availability and their interrelations in the BioDIV (chi‐squared = 0.30, df = 1, P = 0.59) (a) and Wood River (chi‐squared = 0.02, df = 1, P = 0.89) (b) experiments. In each model, the latent variable constructs (ovals) are represented by measured variables (rectangles). Vegetation quantity and quality are represented by aboveground biomass (“Biomass”) and vegetation carbon to nitrogen ratio (“CN ratio”), respectively. The latent construct of belowground microbial community functions (“BG community”) is represented by microbial biomass carbon (“Microbial biomass”), while the soil nutrient availability is represented by net nitrogen mineralization rate (“N mineralization”). Blue arrows indicate significant positive relationships. Red arrows indicate significant negative relationships. Dashed lines non‐significant relationships. Coefficients and standardized coefficients (in brackets, one unit change in the predictor variable causes the amount of one standardized coefficient change in the dependent variable) are given next to the arrows. Predictor variables were converted the following way: one unit of biomass in the model equals measured dry plant biomass (g/m2)/100, one unit net nitrogen mineralization rate in the model equals (1 + measured N (mg)·[g soil]−1·d−1) × 100, and one unit microbial biomass carbon in the model equals for BioDIV (measured microbial biomass C·[g soil]−1) × 100 and for Wood River (1 + measured microbial biomass C·[g soil]−1) × 10.
In contrast, in the Wood River system, vegetation quality, measured as the carbon to nitrogen ratio of aboveground vegetation, was strongly and negatively associated with soil nutrient availability, measured as net nitrogen mineralization rate (Fig. 7b), while the quantity of aboveground inputs to the soil, measured as aboveground biomass, was not. In addition, in this system both vegetation quality and vegetation quantity, measured as aboveground biomass, was negatively associated with the abundance of microbes in the soil, such that greater amounts of vegetation nitrogen relative to carbon increased microbial biomass.
Discussion
Belowground biodiversity and processes are critical to essential ecosystem functions such as soil fertility and carbon sequestration that maintain the Earth’s life support systems. Yet they are difficult to study across large spatial scales. Here we demonstrate the potential for using spectroscopic imagery across broad spatial extents to predict belowground properties and functions in grassland systems that varied in diversity and productivity. In a relatively low productivity system, with low soil carbon and nitrogen concentrations (BioDIV), the supply of substrates and therefore total vegetation biomass limited microorganisms and their functions in the soil, influencing microbial community composition and nutrient dynamics. In a more productive system with higher soil carbon and nitrogen concentrations (Wood River), where substrate supply was less limiting to soil microbes, the chemical composition of vegetation had a predominant influence on microbial processes and nutrient dynamics.
The IPBES 2018 Americas Assessment documented that 35% of grassland systems in the Americas have been converted to pasture or grazing lands and only a small fraction of intact grassland systems remains (Cavender‐Bares et al. 2018). In North America alone, a large fraction of grasslands has been altered by overgrazing and agricultural conversion, reducing their biodiversity and ecosystem functions relative to pre‐European settlement (Bultena et al. 1996, Jones 2000, Sala et al. 2013, Wick et al. 2016). The range of grassland systems—from those more heavily altered by humans to those that are relatively intact—has been simulated to some extent by these diversity experiments in grassland systems, both at the low end of a diversity–productivity gradient (BioDIV in central Minnesota) and toward the higher end of a diversity–productivity gradient (Wood River in central Nebraska).
In such experiments, we show that remotely sensed variables of prairie grassland ecosystems related to both the quantity and quality of aboveground inputs predicted belowground processes. Airborne spectroscopic data at 0.9 m or 1 m resolution predicted aboveground biomass, foliar chemistry and function, and where vegetation cover was high, phylogenetic‐functional group composition. These aboveground metrics, in turn, predicted belowground microbial and soil processes, as well as nutrient availability in both systems. However, the relative importance of the total quantity of vegetation biomass—or total energy inputs—compared with the functional and chemical quality of these inputs in driving microbial communities and belowground processes varied between the two experimental systems. In BioDIV, the quantity of vegetation inputs to the soil was the most critical predictor for microbial abundance and soil processes, while the quality of inputs was of secondary importance. The greater importance of quantity over quality of aboveground inputs likely occurred because of high variation in productivity and the very low productivity associated with low diversity levels in BioDIV. Weeding to maintain the low diversity treatments greatly reduces plant cover causing the low diversity plots to have very low productivity compared with most grasslands, even compared with heavily influenced grasslands (Knapp et al. 2001, Hui and Jackson 2006, Fay et al. 2015). In Wood River, which had a higher plant density resembling more typical prairie, the quality of vegetation inputs was the most important predictor for microbial abundance and soil processes.
In grassland systems, detecting taxonomic composition and diversity levels at pixel sizes of ˜1 m or greater remains complicated due to a mismatch between pixel size and the size of individual plants (Wang et al. 2018, Gholizadeh et al. 2018). While plant biodiversity was readily predicted by spectral diversity from airborne imagery at Wood River (Fig. 4j–l, see also Gholizadeh et al. 2019), accurate detection in BioDIV was possible only with very high spatial resolution (<10 cm) (Wang et al. 2018) using a ground‐based robotic tram (Gamon et al. 2006). Using a different metric of spectral diversity—the mean distance between vector‐normalized spectra among pixels—we again found at Wood River that spectral diversity predicts plant diversity, including species richness, PSR and leaf‐level functional diversity (qDTM) at the plot scale but not at the subplot scale. Remotely sensed spectral diversity was not significantly associated with remotely sensed biomass at the plot scale (Appendix S1: Fig. S5B), and directly measured metrics of plant diversity were only weakly associated at this scale (Fig. 3g–i). At the subplot scale these relationships were significant—both directly measured (Fig. 3d–f) and remotely sensed (Appendix S1: Fig. S5c)—but spectral diversity explained a low proportion of the variance in biomass. In BioDIV, where the relationships between measures of plant diversity and productivity are stronger, remotely sensed spectral diversity did not predict metrics of plant diversity at 1 m resolution. Nevertheless, Schweiger et al. (2018) found that spectral diversity, measured both at the leaf level and remotely using 1 cm tram data, accurately predicted plant functional diversity and phylogenetic diversity in BioDIV, and that spectral diversity strongly predicted biomass. These results highlighted the need for close attention to spatial scale, both in terms of grain size of measurement and consideration of the spatial extent at which relationships between biodiversity and ecosystem functions can be observed (Gamon et al. 2020).
The ability to remotely map functional traits and recover the same relationships with soil processes as measured trait values demonstrates promise for current and forthcoming satellite missions and airborne detection efforts of a range of ecosystem processes through mapping of functional traits with spectroscopy. Where canopy conditions permit—which we hypothesized to depend on high vegetation cover—mapping of phylogenetic‐functional groups is also likely to be predictive of belowground processes. Applying this approach to other systems will require an understanding of the extent to which substrate quantity limits microbial processes and the relative importance of vegetation quality in driving belowground dynamics. Vegetation cover and biomass can be remotely sensed with high accuracy using imaging spectroscopy and have the potential to inform where on the diversity–productivity continuum an ecosystem resides, and therefore whether total substrates are a limiting factor or whether chemical and phylogenetic‐functional groups are likely to be more predictive of belowground processes and attributes. Targeted field campaigns to test the expected relationships between vegetation inputs and belowground processes, informed by cover, biomass and trait maps, will be important.
Relationships between vegetation cover, productivity and belowground attributes and processes
Total organic matter inputs to soils are likely to be much lower in BioDIV than in Wood River, for several reasons, and therefore likely to limit soil microbes at BioDIV (Zak et al. 1994). In BioDIV, the diversity treatments (1–16 species/plot, drawn from a pool of 32) created 16‐fold variation in biomass (25–283 g/m2 in 2015) and therefore in the quantity of substrate inputs to the soil. Even the most diverse and productive plots have modest productivity relative to the original tall grass prairie of the region. At Wood River, with higher species richness/plot and a total species pool of c. 80 species, biomass ranged from 285 to 1,079 g/m2 (in 2017), such that the least productive communities at this site are comparable with the most productive communities in BioDIV. Aboveground biomass approximates the aboveground productivity in these herbaceous systems, where all aboveground biomass senesces each year; aboveground and belowground biomass are tightly correlated in BioDIV (Fornara and Tilman 2008, Appendix S1: Fig. S3) and presumably in Wood River; and root production is likely to be proportional to root biomass. Consequently, total biomass production—the primary source of organic matter inputs to soil—must be lower in BioDIV than in Wood River. Annual to semi‐annual burning further reduces organic inputs to soils in BioDIV; Wood River is less frequently burned (approximately every three years). While root turnover and rhizodeposition provide an important source of substrate inputs to the soil in both systems, aboveground litter inputs are likely to have a greater contribution to total organic matter inputs to soils at Wood River, given the more frequent burning at BioDIV than at Wood River. In addition, higher productivity at Wood River was likely to have arisen in part from a warmer climate, finer‐textured soils with deposits from the Platte River, and therefore higher soil fertility.
Lower productivity at BioDIV for reasons mentioned above are likely to lead to stronger microbial substrate limitations there than at Wood River. Previous studies have documented that, when substrate quantity is limiting, it is positively related to microbial biomass and fungal abundance (Wardle 1992, Zak et al. 1994, Cleveland and Liptzin 2007, Whitaker et al. 2014) and to soil carbon, which has been previously documented in BioDIV (Fornara and Tilman 2008, Yang et al. 2019). Higher microbial biomass and soil carbon are therefore expected to be associated with high soil respiration (Whitaker et al. 2014). In systems in which energy inputs primarily limit microbial processes, the nutrient composition of soil inputs is secondary (Hättenschwiler and Jørgensen 2010, Whitaker et al. 2014, Cline et al. 2018). A similar pattern appears to be the case in BioDIV, where chemical composition of the above ground vegetation is relatively less important and less predictive than total energy inputs to microbial abundance, composition, activity and the soil processes they drive (Zak et al. 2003).
Relationships between vegetation chemistry, foliar functional traits, phylogenetic‐functional groups and belowground attributes and processes
Phylogenetic‐functional group composition—associated with changes in vegetation chemistry and root chemistry—was a stronger influence on microbial composition, biomass and activity in Wood River than in BioDIV. In the Wood River system, remotely sensed foliar chemistry and plant functional‐phylogenetic group composition, as well as in situ measures of these same traits and phylogenetic‐functional group proportion, were stronger predictors of belowground enzyme activity, net nitrogen mineralization rates, microbial biomass, and soil carbon and nitrogen compared with BioDIV. Nevertheless, the proportion of the major phylogenetic lineages—monocots and dicots—as well as individual functional groups, particularly C4 grasses, to varying extents, remained predictors of microbial biomass carbon and nitrogen, cumulative soil respiration and net nitrogen mineralization rates in BioDIV, as did some aspects of chemical composition, consistent with past studies at BioDIV and the nearby BioCON experiment (Dijkstra et al. 2006, Fornara et al. 2009, Mueller et al. 2013, Wei et al. 2019).
In Wood River, oxidative enzyme activity was negatively predicted by leaf level and remotely sensed LMA, foliar nitrogen and lignin concentration, and foliar cell soluble concentration and positively predicted by leaf level and remotely sensed foliar cellulose and hemicellulose concentration. In contrast, hydrolytic enzyme activity, microbial biomass carbon and nitrogen, and soil carbon were positively predicted by foliar nitrogen, cell solubles, lignin, and LMA. Foliar nitrogen and lignin concentration are highly correlated in both of these grassland systems, such that teasing apart their independent relationships with other factors is problematic. Nitrogen, both when applied externally and in leaf litter, has been shown to inhibit oxidative enzyme activity (Fenn et al. 1981, Carreiro et al. 2000, DeForest et al. 2004, Zak et al. 2008, Edwards et al. 2011, Hobbie et al. 2012), which may explain why these enzymes showed reduced activity in plots with higher foliar nitrogen, despite higher lignin inputs. Greater soil carbon is expected to support higher microbial biomass (Wardle 1992, Zak et al. 1994, Cleveland and Liptzin 2007), which helps to explain the higher hydrolytic enzyme activity in plots with higher foliar nitrogen (Whitaker et al. 2014).
Nitrogen in substrates can control the composition and activity of microbial communities and the processes they drive (Hättenschwiler and Jørgensen 2010). If litter‐feeding organisms and decomposer communities are limited by the relative availability of nitrogen, their activity is expected to increase with substrates higher in N. We found that both remotely sensed foliar nitrogen concentration, as well as the proportion of legumes, positively predicted microbial biomass carbon and nitrogen and their hydrolytic enzyme activity and nitrogen net mineralization rates, consistent with results from BioCON, adjacent to BioDIV (Dijkstra et al. 2006, Mueller et al. 2013, Wei et al. 2019). Similarly, the proportion of C4 grasses and of grasses, in general—which have low tissue N concentrations and high cellulose and hemicellulose concentrations—was a negative predictor of these same processes. These results demonstrated that remotely sensed traits mapped at landscape scales, as well as phylogenetic‐functional group composition, can be predictive of belowground processes and attributes. However, in situ characterization of the systems is necessary to understand how vegetation inputs drive soil processes, as well as to determine the extent to which traits or phylogenetic‐functional groups can be robustly mapped.
Relevance of phylogenetic‐functional groups for predicting belowground processes and attributes using imaging spectroscopy
Our results showed that airborne data can provide critical information relevant to belowground ecosystem processes, both in terms of quantity and quality of inputs to soil that are not easily obtained from the ground. Chemical sampling of vegetation is time‐consuming and costly, particularly over large spatial extents. Airborne information, coupled with increasingly available vegetation trait and biomass models, will eventually provide this kind of information with ultimately lower cost and less time. However, our work shows that modeling how the quality and quantity of inputs influence belowground processes requires characterizing systems in advance. The relative importance of total aboveground inputs to soil processes compared with the chemical composition of those inputs is expected to vary along the diversity–productivity gradient, given that carbon is not likely to be limiting at the high productivity end of this gradient. Understanding where a given ecosystem falls along the plant diversity–productivity gradient is likely to be critical for determining whether remotely sensed measures of productivity or vegetation chemistry are most predictive of belowground microbial processes and soil nutrients.
Based on our results from two contrasting experimental grassland systems, we can anticipate that belowground processes of overgrazed and degraded grasslands that are low in biodiversity and productivity may be driven by input quantity. In contrast, belowground processes in less disturbed prairie systems may be more driven by vegetation composition and quality. If these relationships can be established generally at large spatial scales across grassland systems, we will be in a position to advance our understanding of belowground ecosystem processes in these globally threatened ecosystems, facing land‐use change, increasing grazing pressure and a range of other anthropogenic factors.
Supporting information
Appendix S1
Appendix S2
Appendix S3
Acknowledgments
The project was funded by NSF/NASA DEB‐1342872; the NSF Biology Integration Institute ASCEND, DBI: 2021898; and the Cedar Creek Long Term Ecological Research Program through NSF DEB 1831944. AKS acknowledges support by the University Research Priority Program Global Change and Biodiversity of the University of Zurich. We thank Ian Carriere, Brett Fredericksen, Jesús Pinto‐Ledezma, Chris Buyarski, Troy Mielke, Shan Kothari, and numerous Cedar Creek research interns for field and laboratory assistance, Laura Williams for assistance with scripting and Lauren Cline for organization of and access to fungal and soil data in BioDIV.
Cavender‐Bares, J. , Schweiger A. K., Gamon J. A., Gholizadeh H., Helzer K., Lapadat C., Madritch M. D., Townsend P. A., Wang Z., and Hobbie S. E.. 2022. Remotely detected aboveground plant function predicts belowground processes in two prairie diversity experiments. Ecological Monographs 92(1):e01488. 10.1002/ecm.1488
Corresponding Editor: Daniel B. Metcalfe.
Open Research
Data and novel code (Cavender‐Bares et al. 2021) are accessible through a Creative Commons license for non‐commercial use on DRUM, the Data Repository of the University of Minnesota, at https://conservancy.umn.edu/handle/11299/220311 and at the Cedar Creek Ecosystem Science Reserve Long‐Term Ecological Research data repository: https://www.cedarcreek.umn.edu/research/data.
Literature Cited
- Adams, M. A. , Turnbull T. L., Sprent J. I., and Buchmann N.. 2016. Legumes are different: Leaf nitrogen, photosynthesis, and water use efficiency. Proceedings of the National Academy of Sciences of the United States of America 113:4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asner, G. P. , Anderson C. B., Martin R. E., Tupayachi R., Knapp D. E., and Sinca F.. 2015. Landscape biogeochemistry reflected in shifting distributions of chemical traits in the Amazon forest canopy. Nature Geoscience 8:567. 10.1038/ngeo2443 [DOI] [Google Scholar]
- Asner, G. P. , Knapp D. E., Kennedy‐Bowdoin T., Jones M. O., Martin R. E., Boardman J., and Hughes R. F.. 2008. Invasive species detection in Hawaiian rainforests using airborne imaging spectroscopy and LIDAR. Remote Sensing of Environment 112:1942–1955. [Google Scholar]
- Asner, G. P. , and Martin R. E.. 2008. Spectral and chemical analysis of tropical forests: Scaling from leaf to canopy levels. Remote Sensing of Environment 112:3958–3970. 10.1038/ngeo2443 [DOI] [Google Scholar]
- Asner, G. P. , and Martin R. E.. 2009. Airborne spectranomics: mapping canopy chemical and taxonomic diversity in tropical forests. Frontiers in Ecology and the Environment 7:269–276. [Google Scholar]
- Asner, G. P. , Martin R. E., Knapp D. E., Tupayachi R., Anderson C., Carranza L., Martinez P., Houcheime M., Sinca F., and Weiss P.. 2011. Spectroscopy of canopy chemicals in humid tropical forests. Remote Sensing of Environment 115:3587–3598. [Google Scholar]
- Bardgett, R. D. , and van der Putten W. H.. 2014. Belowground biodiversity and ecosystem functioning. Nature 515:505–511. [DOI] [PubMed] [Google Scholar]
- Bertrand, F. , Meyer N., and Maumy‐Bertrand M.. 2014. Partial least squares regression for generalized linear models R package version 1.1.1. https://CRAN.R‐project.org/package=plsRglm
- Brookes, P. C. , Landman A., Pruden G., and Jenkinson D. S.. 1985. Chloroform fumigation and the release of soil nitrogen: A rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biology and Biochemistry 17:837–842. 10.1016/0038-0717(85)90144-0 [DOI] [Google Scholar]
- Bultena, G. L. , Duffy M. D., Jungst S. E., Kanwar R. S., Menzel B. W., Misra M. K., Singh P., Thompson J. R., van de Valk A., and Willham R. L.. 1996. Effects of agricultural development on biodiversity: lessons from Iowa. Pages 80–94 in Srivastava J. P., Smith N. G. H., and Forno D. A., editors. Biodiversity and agricultural intensification. World Bank, Washington D.C., USA. https://core.ac.uk/download/pdf/212844124.pdf [Google Scholar]
- Cadotte, M. W. , Cavender‐Bares J., Tilman D., and Oakley T. H.. 2009. Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLOS ONE 4:e5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreiro, M. , Sinsabaugh R., Repert D., and Parkhurst D.. 2000. Microbial enzyme shifts explain litter decay responses to simulated nitrogen deposition. Ecology 81:2359–2365. [Google Scholar]
- Cavender‐Bares, J. , et al. 2016. Associations of leaf spectra with genetic and phylogenetic variation in oaks: prospects for remote detection of biodiversity. Remote Sensing 8:221. [Google Scholar]
- Cavender‐Bares, J. , et al. 2018. Status and trends of biodiversity and ecosystem functions underpinning nature’s benefit to people. Regional and subregional assessments of biodiversity and ecosystem services: regional and subregional assessment for the Americas. IPBES Secretariat, Bonn, Germany. https://ipbes.net/sites/default/files/2018_americas_full_report_book_v5_pages_0.pdf [Google Scholar]
- Cavender‐Bares, J. , Gamon J., Gholizadeh H., Kimberly H., Lapadat C., Madritch M., Townsend P. A., Wang Z., and Hobbie S. E.. 2021. Remotely detected aboveground plant function predicts belowground processes in two prairie diversity experiments. Data Repository for the University of Minnesota. https://conservancy.umn.edu/handle/11299/220311 [DOI] [PMC free article] [PubMed]
- Cavender‐Bares, J. , Gamon J. A., Hobbie S. E., Madritch M. D., Meireles J. E., Schweiger A. K., and Townsend P. A.. 2017. Harnessing plant spectra to integrate the biodiversity sciences across biological and spatial scales. American Journal of Botany 104:1–4. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Hong X., Harris C. J., and Sharkey P. M.. 2004. Spare modeling using orthogonal forest regression with PRESS statistic and regularization. IEEE Transaction on Systems, Man and Cybernetics 34:898–911. [DOI] [PubMed] [Google Scholar]
- Cleveland, C. , and Liptzin D.. 2007. C:N:P stoichiometry in soil: is there a "Redfield ratio" for the microbial biomass? Biogeochemistry 85:235–252. 10.1007/s10533-007-9132-0 [DOI] [Google Scholar]
- Cline, L. C. , Hobbie S. E., Madritch M. D., Buyarski C. R., Tilman D., and Cavender‐Bares J. M.. 2018. Resource availability underlies the plant‐fungal diversity relationship in a grassland ecosystem. Ecology 99:204–216. [DOI] [PubMed] [Google Scholar]
- Cole, J. R. , et al. 2009. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Research 37:D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornwell, W. K. , et al. 2008. Plant species traits are the predominant control on litter decomposition rates within biomes worldwide. Ecology Letters 11:1065–1071. [DOI] [PubMed] [Google Scholar]
- Craine, J. M. , Froehle J., Tilman D. G., Wedin D. A., and Chapin I. F. S.. 2001. The relationships among root and leaf traits of 76 grassland species and relative abundance along fertility and disturbance gradients. Oikos 93:274–285. 10.1034/j.1600-0706.2001.930210.x [DOI] [Google Scholar]
- Craine, J. M. , Tilman D., Wedin D., Reich P., Tjoelker M., and Knops J.. 2002. Functional traits, productivity and effects on nitrogen cycling of 33 grassland species. Functional Ecology 16:563–574. [Google Scholar]
- Croft, H. , and Chen J. M.. 2018. Leaf pigment content. Pages 117–142 in Liang S., editor. Comprehensive remote sensing. Elsevier, Oxford, UK. 10.1016/B978-0-12-409548-9.10547-0 [DOI] [Google Scholar]
- DeForest, J. L. , Zak D. R., Pregitzer K. S., and Burton A. J.. 2004. Atmospheric nitrate deposition, microbial community composition, and enzyme activity in northern hardwood forests. Soil Science Society of America Journal 68:132–138. [Google Scholar]
- Degens, B. P. , and Harris J. A.. 1997. Development of a physiological approach to measuring the catabolic potential of soil microbial communities. Soil Biology and Biochemistry 29:1309–1320. 10.1016/S0038-0717(97)00076-X [DOI] [Google Scholar]
- Dijkstra, F. A. , Hobbie S. E., and Reich P. B.. 2006. Soil processes affected by sixteen grassland species grown under different environmental conditions. Soil Science Society of America Journal 70:770–777. [Google Scholar]
- Doane, T. A. , and Horwáth W. R.. 2003. Spectrophotometric determination of nitrate with a single reagent. Analytical Letters 36:2713–2722. [Google Scholar]
- Edwards, I. , Zak D., Kellner H., Eisenlord S., and Pregitzer K.. 2011. Simulated atmospheric N deposition alters fungal community composition and suppresses ligninolytic gene expression in a northern hardwood forest. PLOS ONE 6:e20421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eviner, V. T. , and Chapin F. S.. 2003. Functional matrix: A conceptual framework for predicting multiple plant effects on ecosystem processes. Annual Review of Ecology Evolution and Systematics 34:455–485. 10.1146/annurev.ecolsys.34.011802.132342 [DOI] [Google Scholar]
- Fay, P. A. , et al. 2015. Grassland productivity limited by multiple nutrients. Nature Plants 1:15080. [DOI] [PubMed] [Google Scholar]
- Fenn, P. , Choi S., and Kirk T.. 1981. Ligninolytic activity of Phanerochaete chrysosporium: physiology of suppression by NH4+ and l‐glutamate. Archives of Microbiology 130:66–71. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1388442/pdf/hw1833.pdf [Google Scholar]
- Fornara, D. A. , and Tilman D.. 2008. Plant functional composition influences rates of soil carbon and nitrogen accumulation. Journal of Ecology 96:314–322. [Google Scholar]
- Fornara, D. A. , Tilman D., and Hobbie S.. 2009. Linkages between plant functional composition, fine root processes and potential soil N mineralization rates. Journal of Ecology 97:48–56. 10.1111/j.1365-2745.2008.01453.x [DOI] [Google Scholar]
- Freschet, G. T. , Weedon J. T., Aerts R., van Hal J. R., and Cornelissen J. H. C.. 2012. Interspecific differences in wood decay rates: insights from a new short‐term method to study long‐term wood decomposition. Journal of Ecology 100:161–170. 10.1111/j.1365-2745.2011.01896.x [DOI] [Google Scholar]
- Gamon, J. A. , Cheng Y., Claudio H., MacKinney L., and Sims D. A.. 2006. A mobile tram system for systematic sampling of ecosystem optical properties. Remote Sensing of Environment 103:246–254. 10.1016/j.rse.2006.04.006 [DOI] [Google Scholar]
- Gamon, J. , Qiu H.‐L., and Sanchez‐Azofeifa A.. 1999. Ecological applications of remote sensing at multiple scales. Pages 805–846 in Pugnaire F., and Valladares F., editors. Handbook of functional plant ecology. Marcel Dekker Inc, New York, New York, USA. http://www.esalq.usp.br/lepse/imgs/conteudo_thumb/Functional-Plant-Ecology-by-Francisco-I--Pugnaire-and-Fernando-Valladares--2007-.pdf [Google Scholar]
- Gamon, J. , Wang R., Gholizadeh H., Zutta B., Townsend P. A., and Cavender‐Bares J.. 2020. Consideration of scale in remote sensing of biodiversity. Pages 425–447 in Cavender‐Bares J., Gamon J., and Townsend P. A., editors. Remote sensing of plant biodiversity. Springer Verlag. New York, New York, USA. https://link.springer.com/chapter/10.1007%2F978-3-030-33157-3_16 [Google Scholar]
- German, D. P. , Weintraub M. N. A., Grandy A. S. A., Lauber C. L. A., Rinkes Z. L. A., and Allison S. D. A.. 2011. Optimization of hydrolytic and oxidative enzyme methods for ecosystem studies. Soil Biology & Biochemistry 43:1387–1397. [Google Scholar]
- Gholizadeh, H. , Gamon J., Helzer C., and Cavender‐Bares J.. 2020. Airborne hyperspectral reflectance Wood River Nebraska multi‐day 1 m V001. NASA EOSDIS Land Processes DAAC. 10.5067/Community/Airborne/AEHYPWRNE1M.001 [DOI]
- Gholizadeh, H. , Gamon J. A., Townsend P. A., Zygielbaum A. I., Helzer C. J., Hmimina G. Y., Yu R., Moore R. M., Schweiger A. K., and Cavender‐Bares J.. 2019. Detecting prairie biodiversity with airborne remote sensing. Remote Sensing of Environment 221:38–49. 10.5067/Community/Airborne/AEHYPWRNE1M.001 [DOI] [Google Scholar]
- Gholizadeh, H. , Gamon J. A., Zygielbaum A. I., Wang R., Schweiger A. K., and Cavender‐Bares J.. 2018. Remote sensing of biodiversity: Soil correction and data dimension reduction methods improve assessment of alpha‐diversity (species richness) in prairie ecosystems. Remote Sensing of Environment 206:240–253. [Google Scholar]
- Grigal, D. F. , Chamberlain L. M., Finney H. R., Wroblewski D. V., and Gross E. V.. 1974. Soils of the cedar creek natural history area. Agricultural Experiment Station, University of Minnesota, Minneapolis, Minnesota, USA. https://agris.fao.org/agris-search/search.do?recordID=US201300444514 [Google Scholar]
- Hamlin, L. , Green R. O., Mouroulis P., Eastwood M., Wilson D., Dudik M., and Paine C.. 2011. Pages 1–7. In AERO’11, imaging spectrometer science measurements for terrestrial ecology: AVIRIS and new developments. Proceedings of the 2011 IEEE Aerospace Conference, Big Sky, Montana, USA. [Google Scholar]
- Hättenschwiler, S. , and Jørgensen H. B.. 2010. Carbon quality rather than stoichiometry controls litter decomposition in a tropical rain forest. Journal of Ecology 98:754–763. [Google Scholar]
- Helmus, M. R. , Bland T. J., Williams C. K., and Ives A. R.. 2007. Phylogenetic measures of biodiversity. American Naturalist 169:E68–E83. [DOI] [PubMed] [Google Scholar]
- Hobbie, S. E. 2015. Plant species effects on nutrient cycling: revisiting litter feedbacks. Trends in Ecology & Evolution 30:357–363. [DOI] [PubMed] [Google Scholar]
- Hobbie, S. , Eddy W., Buyarski C., Adair E., Ogdahl M., and Weisenhorn P.. 2012. Response of decomposing litter and its microbial community to multiple forms of nitrogen enrichment. Ecological Monographs 82:389–405. [Google Scholar]
- Hooper, D. U. , and Vitousek P. M.. 1998. Effects of plant composition and diversity on nutrient cycling. Ecological Monographs 68:121–149. [Google Scholar]
- Hui, D. , and Jackson R. B.. 2006. Geographical and interannual variability in biomass partitioning in grassland ecosystems: a synthesis of field data. New Phytologist 169:85–93. [DOI] [PubMed] [Google Scholar]
- Jelinski, D. E. , and Currier P. J.. 1997. Ecological risk assessment case study: Middle Platte River Flood plain, Nebraska. U. S. Environmental Protection Agency, Washington, D.C., USA. [Google Scholar]
- Jelinski, N. A. , Kucharik C. J., and Zedler J. B.. 2011. A test of diversity–productivity models in natural, degraded, and restored wet prairies. Restoration Ecology 19:186–193. 10.1111/j.1526-100X.2009.00551.x [DOI] [Google Scholar]
- Jones, A. 2000. Effects of cattle grazing on North American arid ecosystems: A quantitative review. Western North American Naturalist 60:155–164. https://scholarsarchive.byu.edu/cgi/viewcontent.cgi?article=1152&context=wnan [Google Scholar]
- Knapp, A. , Briggs J., and Koelliker J. K.. 2001. Frequency and extent of water limitation to primary production in a mesic temperate grassland. Ecosystems 4:19–28. [Google Scholar]
- Kokaly, R. , Asner G., Ollinger S., Martin M., and Wessman C.. 2009. Characterizing canopy biochemistry from imaging spectroscopy and its application to ecosystem studies. Remote Sensing of Environment 113:S78–S91. [Google Scholar]
- Kothari, S. , Cavender‐Bares J., Bitan K., Verhoeven A. S., Wang R., Montgomery R. A., and Gamon J. A.. 2018. Community‐wide consequences of variation in photoprotective physiology among prairie plants. Photosynthetica, 56:455–467. [Google Scholar]
- Kuczynki, J. , Liu Z., Lozupone C., McDonald D., Fierer N., and Knight R.. 2010. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nature Methods 7:813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madritch, M. D. , Kingdon C. C., Singh A., Mock K. E., Lindroth R. L., and Townsend P. A.. 2014. Imaging spectroscopy links aspen genotype with below‐ground processes at landscape scales. Philosophical Transactions of the Royal Society B: Biological Sciences 369:20130194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, H. , and Naes T.. 1989. Multivariate calibration. John Wiley & Sons, Ltd, Chichester, UK. 441 pp. [Google Scholar]
- Meier, C. L. , and Bowman W. D.. 2008. Links between plant litter chemistry, species diversity, and belowground ecosystem function. Proceedings of the National Academy of Sciences of the United States of America 105:19780–19785. 10.1073/pnas.0805600105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles, J. E. , O'Meara B., and Cavender‐Bares J.. 2020. Linking leaf spectra to the plant tree of life. Pages 155–172 in Cavender‐Bares J., Gamon J. A., and Townsend P. A., editors. Remote sensing of plant biodiversity. Springer. https://link.springer.com/chapter/10.1007/978‐3‐030‐33157‐3_7 [Google Scholar]
- Mevik, B.‐H. , Wehrens R., and Liland K. H.. 2018. Partial least squares and principal component regression. R package version 2.7‐0. https://www.jstatsoft.org/article/view/v018i02
- Mueller, K. E. , Hobbie S. E., Tilman D., and Reich P. B.. 2013. Effects of plant diversity, N fertilization, and elevated carbon dioxide on grassland soil N cycling in a long‐term experiment. Global Change Biology 19:1249–1261. [DOI] [PubMed] [Google Scholar]
- Nemec, K. T. , Allen C. R., Helzer C. J., and Wedin D. A.. 2013. Influence of richness and seeding density on invasion resistance in experimental tallgrass prairie restorations. Ecological Restoration 31:168–185. [Google Scholar]
- NCSS . 2010. Web soil survey, Hall County, Nebraska. In, editor. National Cooperative Soil Survey, U.S. Department of Agriculture. websoilsurvey.nrcs.usda.gov/app/ [Google Scholar]
- Oksanen, J. , et al. 2018. vegan: community ecology package. R package version 2.5‐3. https://CRAN.R‐project.org/package=vegan
- Parton, W. , et al. 2007. Global‐scale similarities in nitrogen release patterns during long‐term decomposition. Science 315:361. [DOI] [PubMed] [Google Scholar]
- Pearse, W. D. , Cadotte M. W., Cavender‐Bares J., Ives A. R., Tucker C. M., Walker S. C., and Helmus M. R.. 2015. pez: phylogenetics for the environmental sciences. Bioinformatics 31:btv277. 10.1093/bioinformatics/btv277 [DOI] [PubMed] [Google Scholar]
- R Development Core Team . 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R‐project.org [Google Scholar]
- Reich, P. B. , Tilman D., Isbell F., Mueller K. M., Hobbie S. E., Flynn D. F. B., and Eisenhauer N.. 2012. Impacts of biodiversity loss escalate through time as redundancy fades. Science 336:589–592. [DOI] [PubMed] [Google Scholar]
- Rosseel, Y. 2012. lavaan: An R package for structural equation modeling. Journal of Statistical Software, 48:1–36. https://www.jstatsoft.org/article/view/v048i02 [Google Scholar]
- Sala, O. E. , Vivanco L., and Flombaum P.. 2013. Grassland ecosystems. Pages 1–7 in Levin S. A., editor. Encyclopedia of biodiversity. Second edition. Academic Press, Waltham, Massachusetts, USA. [Google Scholar]
- Scheiner, S. M. , Kosman E., Presley S. J., and Willig M. R.. 2017. Decomposing functional diversity. Methods in Ecology and Evolution 8:809–820. [Google Scholar]
- Schmidtlein, S. 2005. Imaging spectroscopy as a tool for mapping Ellenberg indicator values. Journal of Applied Ecology 42:966–974. 10.1111/j.1365-2664.2005.01064.x [DOI] [Google Scholar]
- Schmidtlein, S. , Feilhauer H., and Bruelheide H.. 2012. Mapping plant strategy types using remote sensing. Journal of Vegetation Science 23:395–405. [Google Scholar]
- Schneider, F. D. , Morsdorf F., Schmid B., Petchey O. L., Hueni A., Schimel D. S., and Schaepman M. E.. 2017. Mapping functional diversity from remotely sensed morphological and physiological forest traits. Nature Communications 8:1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger, A. K. , Cavender‐Bares J., Townsend P. A., Hobbie S. E., Madritch M. D., Wang R., Tilman D., and Gamon J. A.. 2018. Plant spectral diversity integrates functional and phylogenetic components of biodiversity and predicts ecosystem function. Nature Ecology & Evolution 2:976–982. [DOI] [PubMed] [Google Scholar]
- Schweiger, A. K. , Schütz M., Risch A. C., Kneubühler M., Haller R., and Schaepman M. E.. 2017. How to predict plant functional types using imaging spectroscopy: linking vegetation community traits, plant functional types and spectral response. Methods in Ecology and Evolution 8:86–95. [Google Scholar]
- Serbin, S. P. , Singh A., Desai A. R., Dubois S. G., Jablonski A. D., Kingdon C. C., Kruger E. L., and Townsend P. A.. 2015. Remotely estimating photosynthetic capacity, and its response to temperature, in vegetation canopies using imaging spectroscopy. Remote Sensing of Environment 167:78–87. [Google Scholar]
- Serbin, S. P. , Singh A., McNeil B. E., Kingdon C. C., and Townsend P. A.. 2014. Spectroscopic determination of leaf morphological and biochemical traits for northern temperate and boreal tree species. Ecological Applications 24:1651–1669. [DOI] [PubMed] [Google Scholar]
- Singh, A. , Serbin S. P., McNeil B. E., Kingdon C. C., and Townsend P. A.. 2015. Imaging spectroscopy algorithms for mapping canopy foliar chemical and morphological traits and their uncertainties. Ecological Applications 25:2180–2197. [DOI] [PubMed] [Google Scholar]
- Smith, D. P. , and Peay K. G.. 2014. Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLOS ONE 9:e90234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. A. , and Brown J. W.. 2018. Constructing a broadly inclusive seed plant phylogeny. American Journal of Botany 105:302–314. [DOI] [PubMed] [Google Scholar]
- Steinauer, K. , Tilman D., Wragg P. D., Cesarz S., Cowles J. M., Pritsch K., Reich P. B., Weisser W. W., and Eisenhauer N.. 2015. Plant diversity effects on soil microbial functions and enzymes are stronger than warming in a grassland experiment. Ecology 96:99–112. [DOI] [PubMed] [Google Scholar]
- Tilman, D. 1997. Community invasibility, recruitment limitation, and grassland biodiversity. Ecology 78:81–92. [Google Scholar]
- Tilman, D. , Reich P. B., Knops J., Wedin D., Mielke T., and Lehman C.. 2001. Diversity and productivity in a long‐term grassland experiment. Science 294:843–845. [DOI] [PubMed] [Google Scholar]
- Townsend, P. A. , Serbin S. P., Kruger E. L., and Gamon J. A.. 2013. Disentangling the contribution of biological and physical properties of leaves and canopies in imaging spectroscopy data. Proceedings of the National Academy of Sciences of the United States of America 110:E1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustin, S. L. , and Gamon J. A.. 2010. Remote sensing of plant functional types. New Phytologist 186:795–816. [DOI] [PubMed] [Google Scholar]
- Vitousek, P. M. 2004. Nutrient cycling and limitation: Hawai'i as a model system. Princeton University Press, Princeton, New Jersey, USA. https://press.princeton.edu/books/paperback/9780691115801/nutrient-cycling-and-limitation [Google Scholar]
- Vitousek, P. , Asner G. P., Chadwick O. A., and Hotchkiss S.. 2009. Landscape‐level variation in forest structure and biogeochemistry across a substrate age gradient in Hawaii. Ecology 90:3074–3086. [DOI] [PubMed] [Google Scholar]
- Waldrop, M. P. , Zak D. R., Blackwood C. B., Curtis C. D., and Tilman D.. 2006. Resource availability controls fungal diversity across a plant diversity gradient. Ecology Letters 9:1127–1135. [DOI] [PubMed] [Google Scholar]
- Wang, R. , Gamon J. A., Cavender‐Bares J., Townsend P. A., and Zygielbaum A. I.. 2018. The spatial sensitivity of the spectral diversity–biodiversity relationship: an experimental test in a prairie grassland. Ecological Applications 28:541–556. [DOI] [PubMed] [Google Scholar]
- Wang, R. , Gamon J. A., Montgomery R. A., Townsend P. A., Zygielbaum A. I., Bitan K., Tilman D., and Cavender‐Bares J.. 2016. Seasonal variation in the NDVI–species richness relationship in a prairie grassland experiment (Cedar Creek). Remote Sensing 8:128. https://www.mdpi.com/2072-4292/8/2/128 [Google Scholar]
- Wang, Z. , Townsend P. A., Schweiger A. K., Couture J. J., Singh A., Hobbie S. E., and Cavender‐Bares J.. 2019. Mapping foliar functional traits and their uncertainties across three years in a grassland experiment. Remote Sensing of Environment 221:405–416. [Google Scholar]
- Wardle, D. A. 1992. A comparative assessment of factors which influence microbial biomass carbon and nitrogen levels in soil. Biological Reviews 67:321–358. 10.1111/j.1469-185X.1992.tb00728.x [DOI] [Google Scholar]
- Wardle, D. A. , Bardgett R. D., Klironomos J. N., Setala H., van der Putten W. H., and Wall D. H.. 2004. Ecological linkages between aboveground and belowground biota. Science 304:1629–1633. [DOI] [PubMed] [Google Scholar]
- Wei, X. , Reich P. B., and Hobbie S. E.. 2019. Legumes regulate grassland soil N cycling and its response to variation in species diversity and N supply but not CO2. Global Change Biology 25:2396–2409. [DOI] [PubMed] [Google Scholar]
- Whitaker, J. , Ostle N., Nottingham A. T., Ccahuana A., Salinas N., Bardgett R. D., Meir P., McNamara N. P., and Austin A.. 2014. Microbial community composition explains soil respiration responses to changing carbon inputs along an Andes‐to‐Amazon elevation gradient. Journal of Ecology 102:1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick, A. F. , Geaumont B. A., Sedivec K. K., and Hendrickson J. R.. 2016. Grassland degradation. Pages 257–276 in Shroder J. F., and Sivanpillai R., editors. Biological and environmental hazards, risks, and disasters. Academic Press, Boston, Massachusetts, USA. 10.1016/B978-0-12-394847-2.00016-4 [DOI] [Google Scholar]
- Williams, L. J. , Cavender‐Bares J., Townsend P. A., Couture J. J., Wang Z., Stefanski A., Messier C., and Reich P. B.. 2020. Remote spectral detection of biodiversity effects on forest biomass. Nature Ecology & Evolution 5:46–54. [DOI] [PubMed] [Google Scholar]
- Wold, S. , Martens H., and Wold H.. 1983. Matrix pencils pp. Pages 286–293 in Ruhe A., and Kagstrom B., editors. Lecture notes in mathematics. Springer, Heidelberg. 10.1007/BFb0062108 [DOI] [Google Scholar]
- Yang, Y. , Tilman D., Furey G., and Lehman C.. 2019. Soil carbon sequestration accelerated by restoration of grassland biodiversity. Nature Communications 10:718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak, D. R. , Grigal D. F., Gleeson S., and Tilman D.. 1990. Carbon and nitrogen cycling during old‐field succession: Constraints on plant and microbial biomass. Biogeochemistry 11:111–129. https://link.springer.com/article/10.1007/BF00002062 [Google Scholar]
- Zak, D. , Holmes W., Burton A., Pregitzer K., and Talhelm A.. 2008. Simulated atmospheric NO3‐ deposition increases soil organic matter by slowing decomposition. Ecological Applications 18:2016–2027. [DOI] [PubMed] [Google Scholar]
- Zak, D. R. , Holmes W. E., White D. C., Peacock A. D., and Tilman D.. 2003. Plant diversity, soil microbial communities, and ecosystem function: Are there any links? Ecology 84:2042–2050. [Google Scholar]
- Zak, D. R. , Tilman D., Parmenter R. R., Rice C. W., Fisher F. M., Vose J., Milchunas D., and Martin C. W.. 1994. Plant production and soil microorganisms in late‐successional ecosystems: A continental‐scale study. Ecology 75:2333–2347. 10.2307/1940888 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Appendix S2
Appendix S3
Data Availability Statement
Data and novel code (Cavender‐Bares et al. 2021) are accessible through a Creative Commons license for non‐commercial use on DRUM, the Data Repository of the University of Minnesota, at https://conservancy.umn.edu/handle/11299/220311 and at the Cedar Creek Ecosystem Science Reserve Long‐Term Ecological Research data repository: https://www.cedarcreek.umn.edu/research/data.


