Abstract
Two proteins exhibiting α-l-rhamnosidase activity, RhaA and RhaB, were identified upon fractionation and purification of a culture filtrate from Aspergillus aculeatus grown on hesperidin. Both proteins were shown to be N glycosylated and had molecular masses of 92 and 85 kDa, of which approximately 24 and 15%, respectively, were contributed by carbohydrate. RhaA and RhaB, optimally active at pH 4.5 to 5, showed Km and Vmax values of 2.8 mM and 24 U/mg (RhaA) and 0.30 mM and 14 U/mg (RhaB) when tested for p-nitrophenyl-α-l-rhamnopyranoside. Both enzymes were able to hydrolyze α-1,2 and α-1,6 linkages to β-d-glucosides. Using polyclonal antibodies, the corresponding cDNA of both α-l-rhamnosidases, rhaA and rhaB, was cloned. On the basis of the amino acid sequences derived from the cDNA clones, both proteins are highly homologous (60% identity).
Microorganisms secreting glycosidases are widespread throughout nature and play an important role in hydrolyzing and catabolizing polysaccharides. In this context, filamentous fungi, such as aspergilli and trichoderma species, have been studied in great detail. The filamentous fungi of the genus Aspergillus are important to the food industry due to their ability to produce metabolites, such as organic acids and extracellular glycosidases (1, 2). Particularly important are representatives of the black aspergilli, such as Aspergillus niger, products of which hold the Generally Recognized as Safe status. A. niger is an eminent source for the production of glycosidase activities. Black aspergilli are able to produce a wide variety of enzyme activities in relatively large amounts. For example, the enzyme systems involved in the degradation of xylan and pectin are very well studied (6, 21, 27, 28).
Many microorganisms have been studied for their potential to produce glycosidases. However, little is known about microorganisms that produce α-l-rhamnosidase activity. Most studies have been done using α-l-rhamnosidases from bacterial origin, e.g., those produced by Sphingomonas sp. (11), Bacteroides (13), Pseudomonas paucimobilis (18), and Clostridium stercorarium (31).
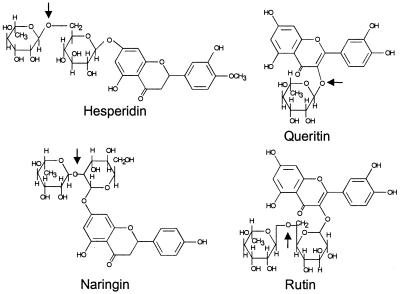
α-l-Rhamnosidases (EC 3.2.1.40) have several potential applications. They have been used for elucidating the structure of biologically important glycosides, polysaccharides, and glycolipids (14, 15). Also, α-l-rhamnosidases were used for the hydrolysis of rhamnosyl residues present in flavonoid glycosides, such as naringin, hesperidin, rutin, and quercitrin. The structures of these compounds are shown in Fig. 1. For instance, the hydrolysis of rutin and quercitrin, the most common flavonoid glycosides in the human diet, by bacterial α-l-rhamnosidases has been reported (3). There are also several technological applications of α-l-rhamnosidases, such as the industrial removal of bitterness from citrus juices caused by naringin (for a review, see reference 22) and the hydrolysis of hesperidin by α-l-rhamnosidases to release l-rhamnose and hesperetin glucoside, which is an important precursor in sweetener production (5). In addition, there is an industrial interest in α-l-rhamnosidases for their action towards terpenyl glycosides in the application of enhancing aroma in grape juices and derived beverages (4, 10, 29). Cloning of α-l-rhamnosidase genes and the production of pure enzyme preparations would allow testing their application in structural studies and biotechnological processes. However, so far only one gene encoding an α-l-rhamnosidase has been cloned; this gene originates from the bacterium C. stercorarium (31).
FIG. 1.
Structure of flavonoid glycosides hesperidin [3′,5,7-trihydroxy-4′-methoxyflavanone-7-α-l-rhamnopyranoside-(1,6)-β-d-glucopyranoside], quercitrin [3,3′,4′,5,7-pentahydroxyflavone-3-α-l-rhamnopyranoside], rutin [3,3′,4′,5,7-pentahydroxyflavone-3-α-l-rhamnopyranoside-(1,6)-β-d-glucopyranoside], and naringin [4′,5,7-trihydroxy-flavanone-7-α-l-rhamnopyranoside-(1,2)-β-d-glucopyranoside]. The arrows indicate the possible linkages hydrolyzed by α-l-rhamnosidases.
α-l-Rhamnosidase of fungal origin has been purified from commercial enzyme preparations of Penicillium (30) and Aspergillus species (17, 19). Recently, α-l-rhamnosidases of Aspergillus terreus (8, 9) and Aspergillus nidulans (20) have been produced and were shown to be of potential oenological interest. Aspergillus aculeatus is a good producer of pectin-degrading enzymes, such as rhamnogalacturonan hydrolase (24) and rhamnogalacturonan acetylesterase (25). The purification of an α-l-rhamnosidase from an A. aculeatus pectinolytic enzyme preparation has been described (19). We used A. aculeatus as a source for the production of α-l-rhamnosidase activity, and here we report the biochemical characterization of two different enzymes showing α-l-rhamnosidase activity. Using polyclonal antibodies, we cloned the corresponding cDNA of both α-l-rhamnosidases from a hesperidin-induced cDNA library.
MATERIALS AND METHODS
Substrates and chemicals.
p-Nitrophenyl-α-l-rhamnopyranoside (pNPR), 4-methylumbelliferyl-α-l-rhamnoside (MUR), hesperidin, naringin, quercitrin, rutin, and bicinchoninic acid protein assay reagent were purchased from Sigma Chemical Co. (St. Louis, Mo.). Molecular mass markers (protein test mixture 4) for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie brilliant blue R-250 and G-250 were obtained from Serva (Heidelberg, Germany). CM Sephadex C-50, S-Sepharose Fast Flow, Mono S HR 5/5, Superose 12, Ampholine polyacrylamide plate gels, and a pI calibration kit for isoelectric focusing were from Pharmacia Biotech (Uppsala, Sweden). N-glycanase F and bovine serum albumin were purchased from Boehringer Mannheim (Mannheim, Germany). Alkaline phosphatase-labeled goat anti-mouse immunoglobulin G was obtained from Bio-Rad (Hercules, Calif.).
Fungal strains, medium, and growth conditions.
A. aculeatus NW240 (Centraalbureau voor Schimmelcultures [CBS] 101.43) was grown in medium containing 0.5 g of KCl/liter, 0.5 g of MgSO4 · 7H2O/liter, 15 g of KH2PO4/liter, and 4 g of NH4Cl/liter. This medium was supplemented with 1 ml of Vishniac solution (26) and with 5 g of yeast extract/liter and 1 g of Casamino Acids/liter and contained hesperidin as the carbon source (5 g/liter). The liquid medium was adjusted to pH 6, inoculated with 106 spores ml−1, and incubated at 30°C in an orbital shaker at 250 rpm for 6 days.
Enzyme activity assays.
α-l-Rhamnosidase activity was determined using pNPR as the substrate as described previously (17). One unit of enzyme activity was defined as the amount of enzyme that releases 1 μmol of p-nitrophenol per min at 30°C in 50 mM McIlvaine buffer (citrate-phosphate buffer), pH 4.5.
Enzyme purification.
All purification steps were performed at 4°C, unless stated otherwise. The fractions collected were screened for protein content (A280) and α-l-rhamnosidase activity.
A culture filtrate (1.7 liters) of A. aculeatus grown on hesperidin for 6 days was adjusted to pH 3.5 with 0.1 M HCl and then diluted by the addition of 5 volumes of distilled water. The secreted proteins were adsorbed batchwise onto CM Sephadex C-50 (2 g [dry wt] equilibrated in 20 mM sodium citrate buffer, pH 3.5). After 2 h of stirring, the resin was transferred to a glass column and the adsorbed proteins were pulse eluted with 10 mM sodium citrate buffer (pH 3.5) containing 1 M NaCl. Fractions containing α-l-rhamnosidase activity were pooled and dialyzed against 10 mM sodium citrate buffer (pH 3.5). The dialyzed enzyme solution was applied to an S-Sepharose Fast Flow column (2.5 by 25 cm), which was preequilibrated with 10 mM sodium citrate buffer, pH 3.5. The column was washed extensively with the same buffer, and proteins were eluted with a linear NaCl gradient of 0 to 1 M in the same buffer (total volume, 750 ml). The α-l-rhamnosidase activity eluted in two peaks, pool A (fractions 20 to 22, 0.20 to 0.27 M NaCl; fraction volume, 10 ml) and pool B (fractions 27 to 31, 0.30 to 0.37 M NaCl; fraction volume, 10 ml). Pools A and B were dialyzed overnight against 10 mM sodium citrate buffer, pH 3.5, and were loaded separately onto a Mono S HR 5/5 column preequilibrated with 10 mM sodium citrate buffer, pH 3.5. The column was extensively washed with this buffer and eluted with a linear gradient of 0 to 0.5 M NaCl in the same buffer (total volume, 60 ml). From pool A, one single peak containing α-l-rhamnosidase activity was obtained (fractions 4 and 5, 0.12 to 0.15 M NaCl; 2-ml fractions). The pooled fractions were dialyzed against 20 mM piperazine/HCl buffer, pH 6, and were used as the purified enzyme preparation RhaA throughout this study. From pool B a single peak containing α-l-rhamnosidase activity was eluted. Active fractions (fractions 7 and 8, 0.18 to 0.22 M NaCl; 2-ml fractions) were pooled and dialyzed overnight against 20 mM piperazine buffer, pH 6. The pooled fractions originating from pool B were further purified by gel filtration on a Superose 12 column (1.5 by 50 cm) preequilibrated with 20 mM piperazine/HCl buffer (pH 6)–100 mM NaCl. Elution was made with the same buffer (total volume, 120 ml), and one single peak containing α-l-rhamnosidase activity was eluted (fractions 25 to 31; 2-ml fractions). The pooled fractions were desalted and used as the purified enzyme preparation RhaB throughout this study.
Preparation of antibodies.
Antibodies against RhaA and RhaB were raised in mice as previously was described (7). The antibodies were tested for cross-reactivity for RhaA and RhaB. By spotting different concentrations of the native protein directly onto a membrane, the specificity of the antisera was tested. Using the anti-RhaA serum, approximately 1 ng of RhaA could be detected, while 900 ng of RhaB was needed to give a reaction. Using the anti-RhaB serum, approximately 3 ng of RhaB could be detected; 700 ng of RhaA gave a reaction with the anti-RhaB serum. From this both sera were considered to be specific for RhaA and RhaB in cDNA screening.
Analytical methods.
Protein concentrations were measured using the commercial bicinchoninic acid protein assay reagent using bovine serum albumin as standard. The purification of the α-l-rhamnosidases was monitored by SDS-PAGE (16), and the proteins separated were stained with Coomassie brilliant blue R-250. For the calculation of the protein molecular masses, an SDS–10% polyacrylamide gel was used and calibrated with protein test mixture 4. Deglycosylation of the enzymes with N-glycanase F was performed according to the supplier's instructions. The enzymes were O deglycosylated by incubation in 0.1 M NaOH for 30 min at room temperature. The pI was determined by isoelectric focusing at 4°C in the pH range of 3.5 to 9.5 using a broad-range pI calibration kit, and the proteins were stained with Coomassie brilliant blue G-250. Detection of α-l-rhamnosidase activity after isoelectric focusing using MUR as the substrate was performed as described previously (17). N-terminal amino acid sequences were analyzed by Eurosequence (Groningen, The Netherlands) using a gas-phase sequencer (model 477; Applied Biosystems, Foster City, Calif.) equipped with a phenylthiohydantoin analyzer.
Enzyme characterization.
The optimum pH for the two α-l-rhamnosidase activities was determined by incubating the enzyme preparation with pNPR in McIlvaine buffers in the pH range of 3 to 8. The pH stability was assessed by preincubating the enzymes in McIlvaine buffers over a pH range of 3 to 5 at 30°C and measuring activities at 22 h using the standard protocol. Thermal stability was measured by preincubation of the enzymes at the optimum pH at different temperatures (30, 40, 55, 65, and 75°C) and following the activity over time. Kinetic experiments were performed at 30°C at the optimal pH. The Michaelis-Menten constants were determined by nonlinear regression using pNPR at concentrations ranging from 0.083 to 6.66 mM. Inhibition studies were performed using l-rhamnose at concentrations ranging from 0 to 16 mM. Substrate specificity studies of the α-l-rhamnosidase activities towards the rhamnoglucosides hesperidin, naringin, quercitrin, and rutin were carried out by high-performance anionic exchange chromatography as described previously (17). The rhamnoglucosides were dissolved at 0.25% (mass/vol) concentration (corresponding to hesperidin and rutin at 4.1 mM, naringin at 4.3 mM, and quercitrin at 5.6 mM) in 50 mM sodium acetate buffer (pH 4.5) and incubated with purified enzymes, at a final concentration in the incubation mixture of 1.1 μg/ml (RhaA) and 1.5 μg/ml (RhaB), for 14 h at 30°C.
Construction of a hesperidin-induced cDNA library and isolation of cDNA clones corresponding to the rhaA and rhaB genes.
A. aculeatus NW240 was cultivated for 24, 48, 72, 96, and 120 h on minimal medium containing 0.5% hesperidin, after which the mycelium was harvested by filtration and washed with sterile saline. The mycelium was subsequently frozen in liquid nitrogen, after which it was powdered using a Microdismembrator (Braun). Total RNA was isolated from the mycelial powder using TriZol (Life Technologies) in accordance with the manufacturer's instructions. Poly(A)+ mRNA was isolated from 2 mg of total RNA by oligo(dT)-cellulose chromatography (23) with the following modification: cDNA was synthesized from 5 μg of poly(A)+ mRNA and was ligated into bacteriophage lambda Uni-ZAP XR by using the ZAP-cDNA synthesis kit (Stratagene) according to the manufacturer's instructions.
To screen the A. aculeatus NW240 cDNA library for the expression of α-l-rhamnosidase, 104 PFU per plate was plated in NZYCM top agarose containing 0.7% agarose on 85-mm-diameter NZYCM (1.5% agar) plates as described (23), using Escherichia coli BB4 (Stratagene) as the plating bacteria. Phages expressing the α-l-rhamnosidase A or B protein were identified by probing the filters with anti α-l-rhamnosidase A or B antiserum and by subsequent detection using an alkaline phosphatase conjugate, according to the procedure described previously (7). After restriction analysis the nucleotide sequence of both cDNA inserts was determined using the Thermosequenase cycle sequencing kit and an ALFexpress sequencer (Amersham-Phamacia Biotech), resulting in the rhaA and rhaB cDNA sequences.
Nucleotide sequence accession number.
The rhaA and rhaB cDNA sequences have been deposited in the GenBank and EMBL sequence databases under accession no. AF284761 and AF284762, respectively.
RESULTS
Purification of α-l-rhamnosidase activity from A. aculeatus.
Two proteins, RhaA and RhaB, showing α-l-rhamnosidase activity were purified to apparent homogeneity from a culture filtrate of A. aculeatus grown on hesperidin, by using cation exchange and gel filtration chromatography. A summary of the purification procedure is presented in Table 1. SDS-PAGE of both RhaA and RhaB revealed two single-protein bands having apparent molecular masses of 92 and 85 kDa, respectively.
TABLE 1.
Purification of α-l-rhamnosidases A and B from A. aculeatus
| Step | Total activity (U) | Total protein (mg) | Vol (ml) | Sp act (U/mg) | Yield (%) |
|---|---|---|---|---|---|
| Culture fluid | 560 | 470 | 1,700 | 1.2 | 100 |
| CM-C50 | 230 | 30 | 200 | 7.7 | 41 |
| S-Sepharose | |||||
| Pool A | 39 | 8.2 | 30 | 4.8 | 7.0 |
| Pool B | 140 | 15 | 50 | 9.3 | 25 |
| Pool A | |||||
| Mono S RhaA | 37 | 4.8 | 4 | 7.7 | 6.6 |
| Pool B | |||||
| Mono S | 110 | 9.7 | 4 | 11 | 20 |
| Superose 12 RhaB | 100 | 7.5 | 14 | 13 | 18 |
General properties of RhaA and RhaB.
After N deglycosylation, the molecular masses of both proteins decreased by about 24 and 15%, respectively, resulting in molecular masses of 70 kDa (RhaA) and 72 kDa (RhaB). Alkali treatment had no influence on the molecular masses (results not shown), which indicates that both enzymes are solely N glycosylated. RhaA has a neutral pI of approximately 6.2, whereas RhaB showed microheterogeneity upon isoelectric focusing. Several protein bands could be seen in the pH range of 5.2 to 5.9, all having α-l-rhamnosidase activity towards 4-methylumbelliferyl-α-l-rhamnoside (results not shown). While the isoelectric focusing results suggested an elution order from the S-Sepharose Fast Flow and Mono S of RhaB (pI 5.2 to 5.9) followed by RhaA (pI 6.2), the enzymes actually eluted in reverse order, which may be due to an uneven distribution of the surface charge of these enzymes.
The optimal pH of the purified RhaA and RhaB was found to be 4.5 to 5 in McIlvaine buffer. RhaA and RhaB showed more than 85% of their maximum activity in the pH ranges of 4 to 5.5 and 3 to 5.5, respectively. Both α-l-rhamnosidase activities were stable in the pH range from 3 to 5. After 20 h of incubation at 30°C, the enzymes retained 80% (RhaA) and 90% (RhaB) of their initial activities over the same pH range. The thermostability of the enzymes was measured at 30, 40, 55, 65, and 75°C. RhaA was stable for 4 h at 40°C, whereas after 4 h at 55 and 65°C, the enzyme retained 87 and 60% of its original activity, respectively. RhaB was stable for 4 h at 40°C, whereas after 4 h at 55, 65, and 75°C the enzyme retained 75, 55, and 50% of its original activity, respectively.
In order to establish whether the two purified proteins are encoded by different genes or by a single gene, part of each of their primary structure was determined. The N-terminal amino acid sequences, comprising 17 amino acid residues, were determined to be VPFEDYILAPQSRTLNF for RhaA and ARVPYREYILAPSSRVI for RhaB. The amino acid sequences showed the two α-l-rhamnosidase forms to be different proteins.
Catalytic properties.
The kinetic behavior of RhaA and RhaB was studied on pNPR. The Michaelis constant (Km) was 2.8 and 0.30 mM for RhaA and RhaB, respectively. The Vmax values were found to be 24 U/mg (RhaA) and 14 U/mg (RhaB). The specificity constants (Vmax/Km) for the hydrolysis of pNPR were calculated as 8.6 (RhaA) and 47 (RhaB).
The inhibition of both enzymes by l-rhamnose was studied using pNPR as a substrate. l-Rhamnose acted as a competitive inhibitor of pNPR hydrolysis with inhibitor constants (Ki) of 4.2 and 1.5 mM for RhaA and RhaB, respectively.
Flavonoids from plant origin, such as hesperidin, naringin, quercitrin, and rutin (Fig. 1), could be natural substrates for α-l-rhamnosidases. From Table 2, it can be seen that RhaA and RhaB were able to release l-rhamnose from hesperidin, naringin, and rutin. Both enzymes were active towards naringin, in which the l-rhamnose residue is α-1,2 linked to the β-d-glucoside, and towards hesperidin and rutin, with α-1,6 linkages to the β-d-glucosides. The enzymes were not able to release l-rhamnose from quercitrin, in which the rhamnosyl residue is linked directly to the aglycon.
TABLE 2.
Substrate specificity of A. aculeatus α-l-rhamnosidases A and B
| Substratea | Rhamnose released from total (%)b
|
|
|---|---|---|
| RhaA | RhaB | |
| Hesperidin | 1.1 | 0.7 |
| Naringin | 25 | 5.4 |
| Rutin | 23 | 11 |
| Quercitrin | ND | ND |
Substrate concentrations: 4.1 mM (hesperidin and rutin), 4.3 mM (naringin), and 5.6 mM (quercitrin).
Percent rhamnose released from total in 14 h at 30°C. Final protein concentrations, 1.1 μg/ml (RhaA) and 1.5 μg/ml (RhaB). ND, not determined.
Isolation and analysis of rhaA and rhaB cDNA clones.
A hesperidin-induced cDNA library was screened using antibodies raised against RhaA and RhaB and resulted in the isolation of three positive RhaA phages and six positive RhaB phages. The cDNA inserts were sequenced, and their identity was confirmed by comparison with the N-terminal amino acid sequences obtained from the purified proteins (Fig. 2). The cDNA insert corresponding to rhaA is 2,361 bp long and encodes a protein of 660 amino acids. The N-terminal amino acid sequence determined from RhaA is found at position 20, the preceding leader being the signal sequence. On the basis of the cDNA sequence, the mature RhaA protein would be expected to have a derived molecular mass of 69 kDa and a theoretical pI of 5.9. The cDNA insert corresponding to rhaB is smaller in size (2,057 bp) and encodes a protein of 597 amino acids. The determined N-terminal amino acid sequence is found at position 17, and cDNA translation results in a derived molecular mass of 62 kDa and a theoretical pI of 6.0.
FIG. 2.
Alignment of amino acid sequences of RhaA (accession no. AF284761) and RhaB (accession no. AF284762) from A. aculeatus and of RamA (accession no. AJ238748) from C. stercorarium using ClustalW and Boxshade (www.CH.EMBNET.ORG). Identical amino acids at conserved positions are boxed in black, while similar residues are boxed in grey.
DISCUSSION
Although α-l-rhamnosidases have several potential biotechnological applications, only a limited number of microbial enzymes have been characterized and only a single gene encoding a bacterial α-l-rhamnosidase has been cloned (31).
A. aculeatus, when grown on hesperidin, produces two proteins (RhaA and RhaB) with α-l-rhamnosidase activity, which are N-glycosylated enzymes, as can be concluded from the reduction in apparent molecular mass to 70 kDa (RhaA) and 72 kDa (RhaB) after N-glycanase treatment and the change from a diffuse (glycosylated) band to a sharp (deglycosylated) band upon SDS-PAGE. Molecular masses similar to those found for RhaA and RhaB have been described for different fungal α-l-rhamnosidases. However, the pIs found for RhaA (6.2) and RhaB (5.2 to 5.85) are slightly higher than those reported for α-l-rhamnosidases of A. niger (85 kDa), 4.5 to 5.2 (17); A. terreus (96 kDa), 4.6 (8); and A. aculeatus (87 kDa), 4.8 (19). The acidic optimal pH found for RhaA and RhaB and the stability at acidic pH values make these enzymes suitable for use in processes operating at low pH values, such as winemaking and citrus juice processing. These are in contrast to bacterial α-l-rhamnosidases, for which neutral and alkaline pH optima have been found (11, 13, 18, 31).
RhaA and RhaB from A. aculeatus are able to hydrolyze pNPR and release l-rhamnose from naringin, hesperidin, and rutin. pNPR is used as a model substrate for rhamnohydrolase activity. Naringin is the main bitter flavanone glycoside of grapefruit juices. Hesperidin is the predominant nonbitter flavanone glycoside in lemons and sweet oranges, and rutin is a flavone glycoside found in many plants. Although able to hydrolyze the four substrates, both α-l-rhamnosidases showed a clear preference for the aryl-rhamnoside pNPR, in which the l-rhamnose residue is directly linked to the aglycon. However, quercitrin, a flavone rhamnoside (Fig. 1) in which the rhamnosyl residue is as well linked directly to the aglycon, was not hydrolyzed, which may be explained by the differences in the aglycon structure. The l-rhamnose residue is α-1,2 linked to a β-glucosidic residue in naringin and α-1,6 linked in hesperidin and rutin (Fig. 1), and from the data collected, both enzymes seemed specific for both kinds of linkages to β-d-glucose. The reason why hesperidin and rutin, which both have an α-1,6 linkage, are hydrolyzed so differently may be explained by steric hindrance due to the attachment of the diglycoside to the aglycon molecule via C7 in hesperidin, whereas the attachment is via C3 in rutin. From these results and those obtained from bacterial α-l-rhamnosidases (31), it seems that the preferred and potentially natural substrate for these glycosidases is still unknown. Similar substrate specificity was described for fungal (17) and bacterial (11, 13) α-l-rhamnosidases.
RhaA and RhaB are encoded by different genes. On the basis of the amino acid sequences derived from the rhaA and rhaB cDNA clones, RhaA and RhaB have N-terminal amino acid extensions compared to the determined amino acid sequences. These N-terminal extensions represent the signal sequences. The signal peptidase cleavage site of RhaB is, however, likelier to be located between residues 17 and 18 than between residues 18 and 19 (12). The determined N-terminal amino acid, therefore, might result from limited proteolysis. Both proteins are highly homologous, 60% of the amino acid sequence being identical. However, there is a marked difference in the central part of both proteins. The amino acid residues 348 to 444 in RhaA are lacking in RhaB and account for most of the difference in the molecular mass of both proteins. However, both proteins have a low degree of identity with the C. stercorarium α-l-rhamnosidase (31); only 11% of the sequence is identical. Despite this low overall identity with this rhamnosidase of prokaryotic origin, there is a significant conservation of certain residues at specific positions (Fig. 2).
Expression of both genes, rhaA and rhaB, will allow the production of pure enzyme preparations. These pure enzyme preparations will then allow the study of their application in biotechnological processes and in structural research.
ACKNOWLEDGMENTS
This work was supported by EC project AIR3-CT94–2193. P. Manzanares was the recipient of a Formación Personal Investigador fellowship from the Spanish government.
REFERENCES
- 1.Barbesgaard P. Industrial enzymes produced by members of the genus Aspergillus. In: Smith J E, Pateman J A, editors. Genetics and physiology of Aspergillus. British Mycological Society Symposium Series. London, United Kingdom: Academic Press; 1977. pp. 391–404. [Google Scholar]
- 2.Bennett J W. Molds, manufacturing and molecular genetics. In: Timberlake W E, editor. Molecular genetics of filamentous fungi. New York, N.Y: Alan R. Liss; 1985. pp. 345–366. [Google Scholar]
- 3.Bokkenheuser V D, Shackelton C H L, Winter J. Hydrolysis of dietary flavonoid glycosides by strains of intestinal Bacteroides from humans. Biochem J. 1987;248:953–956. doi: 10.1042/bj2480953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canal-Llauberes R M. Enhancing the aroma of wines. Aust Grapegrower Winemaker. 1994;368:49–51. [Google Scholar]
- 5.Chase T. Flavour enzymes. Adv Chem Ser. 1974;136:241–266. [Google Scholar]
- 6.Coughlan P M, Hazlewood G P. β-1,4-Xylan-degrading enzyme systems: biochemistry, molecular biology and applications. Biotechnol Appl Biochem. 1993;17:259–289. [PubMed] [Google Scholar]
- 7.Flipphi J A M, Visser J, van der Veen P, de Graaff L H. Cloning of the Aspergillus niger gene encoding α-l-arabinofuranosidase A. Appl Microbiol Biotechnol. 1993;39:335–340. doi: 10.1007/BF00192088. [DOI] [PubMed] [Google Scholar]
- 8.Gallego V M, Piñaga F, Ramón D, Vallés S. Production and characterization of an Aspergillus terreus α-l-rhamnosidase of oenological interest. Z Lebensm-Unters -Forsch. 1996;203:522–527. [Google Scholar]
- 9.Gallego, V. M., F. Piñaga, D. Ramón, and S. Vallés. Purification and characterization of an α-l-rhamnosidase from Aspergillus terreus of interest in winemaking. J. Food Sci., in press.
- 10.Gunata Z, Bitteur S, Brillouet J-M, Bayonove C, Cordonnier R. Sequential enzymic hydrolysis of potentially aromatic glycosides from grape. Carbohydr Res. 1988;184:139–149. [Google Scholar]
- 11.Hashimoto W, Murata K. α-l-Rhamnosidase of Sphingomonas sp. R1 producing an unusual exopolysaccharide of sphingan. Biosci Biotechnol Biochem. 1998;62:1068–1074. doi: 10.1271/bbb.62.1068. [DOI] [PubMed] [Google Scholar]
- 12.Heijne G von. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4691. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang I S, Kim D H. Purification and characterization of α-l-rhamnosidase from Bacteroides JY-6, a human intestinal bacterium. Biol Pharm Bull. 1996;19:1546–1549. doi: 10.1248/bpb.19.1546. [DOI] [PubMed] [Google Scholar]
- 14.Kamiya S, Esaki S, Tanaka R. Synthesis of some disaccharides containing an l-rhamnopyranosyl or l-mannopyranosyl residue, and the substrate specificity of α-l-rhamnosidase from Aspergillus niger. Agric Biol Chem. 1985;49:55–62. [Google Scholar]
- 15.Kamiya S, Esaki S, Tanaka R. Synthesis of certain disaccharides containing a α- or β-l-rhamnopyranosidic group and the substrate specificity of α-l-rhamnosidase from Aspergillus niger. Agric Biol Chem. 1985;49:2351–2358. [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Manzanares P, de Graaff L H, Visser J. Purification and characterization of an α-l-rhamnosidase from Aspergillus niger. FEMS Microbiol Lett. 1997;157:279–283. doi: 10.1111/j.1574-6968.1997.tb12785.x. [DOI] [PubMed] [Google Scholar]
- 18.Miake F, Satho T, Takesue H, Yanagida F, Kashige N, Watanabe K. Purification and characterization of intracellular α-l-rhamnosidase from Pseudomonas paucimobilis FP2001. Arch Microbiol. 2000;173:65–70. doi: 10.1007/s002030050009. [DOI] [PubMed] [Google Scholar]
- 19.Mutter M, Beldman G, Schols H A, Voragen A G J. Rhamnogalacturonan α-l-rhamnopyranohydrolase. A novel enzyme specific for the terminal nonreducing rhamnosyl unit in rhamnogalacturonan regions of pectin. Plant Physiol. 1994;106:241–250. doi: 10.1104/pp.106.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orejas M, Ibañez E, Ramón D. The filamentous fungus Aspergillus nidulans produces an α-l-rhamnosidase of potential oenological interest. Lett Appl Microbiol. 1999;28:383–388. [Google Scholar]
- 21.Pilnik W, Rombouts F M. Pectic enzymes. In: Birch G G, Blakebrough N, Parker K J, editors. Enzymes and food processing. London, United Kingdom: Applied Science Publishers Ltd.; 1981. pp. 105–128. [Google Scholar]
- 22.Puri M, Marwaha S S, Kothari R M, Kennedy J F. Biochemical basis of bitterness in citrus fruit juices and biotech approaches for debittering. Crit Rev Biotechnol. 1996;16:145–155. [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Schols H A, Voragen A G J, Colquhoun I J. Isolation and characterization of rhamnogalacturonan-oligomers, liberated during degradation of pectic hairy regions by rhamnogalacturonase. Carbohydr Res. 1994;256:97–111. doi: 10.1016/0008-6215(94)84230-2. [DOI] [PubMed] [Google Scholar]
- 25.Searle-van Leeuwen J F M, van den Broek L A M, Schols H A, Beldman G, Voragen A G J. Rhamnogalacturonan acetylesterase: a novel enzyme from Aspergillus aculeatus, specific for the deacetylation of hairy (ramified) regions of pectins. Appl Microbiol Biotechnol. 1992;38:347–349. [Google Scholar]
- 26.Vishniac V, Santer M. Thiobacilli. Bacteriol Rev. 1957;21:195–213. doi: 10.1128/br.21.3.195-213.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visser J, Beldman G, Kusters van Someren M A, Voragen A G J. Xylans and xylanases. Amsterdam, The Netherlands: Elsevier; 1992. [Google Scholar]
- 28.Whitaker J R. Microbial pectolytic enzymes. In: Fogerty W M, Kelly C T, editors. Microbial enzymes and biotechnology. London, United Kingdom: Elsevier Applied Science; 1991. pp. 133–175. [Google Scholar]
- 29.Williams P, Strauss C, Wilson B. Studies on the hydrolysis of Vitis vinifera monoterpene precursor compounds and model monoterpene β-d-glucosides rationalizing the monoterpene composition of grapes. J Agric Food Chem. 1982;30:1219–1223. [Google Scholar]
- 30.Young N M, Johnston R A Z, Richards J C. Purification of the α-l-rhamnosidase of Penicillium decumbens and characteristics of two glycopeptide components. Carbohydr Res. 1989;191:53–62. [Google Scholar]
- 31.Zverlov V V, Hertel C, Bronnenmeier K, Hroch A, Kellermann J, Schwarz W H. The thermostable α-l-rhamnosidase RamA of Clostridium stercorarium: biochemical characterization and primary structure of a bacterial α-l-rhamnosidase hydrolase, a new type of inverting glycoside hydrolase. Mol Microbiol. 2000;35:173–179. doi: 10.1046/j.1365-2958.2000.01691.x. [DOI] [PubMed] [Google Scholar]