Summary
The ring-like cohesin complex plays an essential role in chromosome segregation, organization, and double-strand break repair through its ability to bring two DNA double helices together. Scc2 (NIPBL in humans) together with Scc4 functions as the loader of cohesin onto chromosomes. Chromatin adapters such as the RSC complex facilitate the localization of the Scc2-Scc4 cohesin loader. Here, we identify a broad range of Scc2-chromatin protein interactions that are evolutionarily conserved and reveal a role for one complex, Mediator, in the recruitment of the cohesin loader. We identified budding yeast Med14, a subunit of the Mediator complex, as a high copy suppressor of poor growth in Scc2 mutant strains. Physical and genetic interactions between Scc2 and Mediator are functionally substantiated in direct recruitment and cohesion assays. Depletion of Med14 results in defective sister chromatid cohesion and the decreased binding of Scc2 at RNA Pol II-transcribed genes. Previous work has suggested that Mediator, Nipbl, and cohesin connect enhancers and promoters of active mammalian genes. Our studies suggest an evolutionarily conserved fundamental role for Mediator in the direct recruitment of Scc2 to RNA Pol II-transcribed genes.
Keywords: cohesin, cohesin loader, Scc2-Scc4, Mediator, Med14, sister chromatid cohesion, Saccharomyces cerevisiae, chromatin adapters
Graphical abstract
Highlights
-
•
Mediator and Scc2 display physical and genetic interactions
-
•
Mediator recruits Scc2-Scc4 to RNA Pol II-transcribed genes
-
•
Mediator-dependent recruitment of Scc2-Scc4 promotes sister chromatid cohesion
-
•
Several Scc2-chromatin complex interactions are evolutionarily conserved
Mediator, Nipbl, and cohesin are hypothesized to connect enhancers and promoters of mammalian genes. Mattingly et al. reveal a role for Mediator in the recruitment of the Scc2 cohesin loader to RNA Pol II-transcribed genes and in sister chromatid cohesion in yeast, demonstrating evolutionary and functional conservation of these interactions.
Introduction
Cohesin is a vital component in the establishment of sister chromatid cohesion and functions in DNA double-strand break repair and genome organization through chromatin looping. This ring-like complex functions in these roles by the entrapment of one or more DNA helices. The four subunit cohesin complex consists of two structural maintenance of chromosomes (SMC) proteins (Smc1 and Smc3), the kleisin protein (Scc1), and the Huntingtin-elongation factor 3-protein phosphatase 2A-TOR1 (HEAT) repeat subunit Scc3. Smc1 and Smc3 form long coiled coils that dimerize via interactions between their hinge domains.1, 2, 3 The N- and C-terminal ends of Smc1 and Smc3 form globular ATP binding heads bridged by Scc1 bound by Scc3.4, 5, 6, 7, 8 The capture of DNA by the cohesin ring requires ATP hydrolysis by the globular head domains of the SMC subunits.9,10 The loading of cohesin rings is dependent on a loading complex that comprises heterodimer Scc2-Scc4.11 The human ortholog of SCC2, NIBPL, is mutated in the human developmental syndrome known as Cornelia de Lange syndrome (CdLS), with many of the mutations occurring in the evolutionarily conserved HEAT repeats.12,13 CdLS is characterized by limb malformation, facial dysmorphism, cognitive deficiencies, and structural defects in multiple organs. Sister chromatid cohesion during mitosis does not appear to be affected in CdLS patients. Instead, the misregulation of gene expression during development may underlie the clinical features. Much of what we understand about the function of the cohesin loader is based on studies performed in model organisms such as budding yeast.
Biochemical and structural studies have revealed insights into the cohesin loading mechanism. The Scc2-Scc4 loading complex is made up of two functional modules.14, 15, 16, 17 The C-terminal end of Scc2 (Scc2C) forms a hook structure containing multiple contiguous HEAT repeats. Scc2C contacts the cohesin ring at numerous sites and is sufficient to load cohesin onto naked DNA in vitro but is not sufficient for in vivo loading.14,18 Scc4 binds to the N-terminal end of Scc2 forming the module crucial for localizing the loading complex onto chromatin.14,15 Scc4 bound to the N terminus of Scc2 does not bind DNA in vitro, suggesting complex localization is bridged via chromatin adaptors.
The recruitment of the loading complex and loading of cohesin is not sequence specific and instead depends on chromatin structure and chromatin remodelers. A common feature of Scc2 sites is nucleosome depletion.19 In budding yeast, the loading complex interacts with the chromatin remodeler remodels the structure of chromatin (RSC).19,20 RSC generates a nucleosome depleted region required for cohesin loading.20 Additional Scc2 recruitment factors include the histone chaperone and remodeler FACT and the Chd1 chromatin remodeler.21,22 Cohesin loading at budding yeast pericentromeres is enhanced through Scc2-Scc4 recruitment by the inner kinetochore protein Ctf19.23 Replication factors, such as components of the pre-replication complex in Xenopus egg extract and the Mcm2-7 replicative helicase in HeLa cells, may play a role in the recruitment of the cohesin loader.17,24 Localization of the cohesin loader on chromosomes is not fully understood but multiple protein complexes facilitate its recruitment.
The Mediator complex has been implicated in cohesin loading for promoter-enhancer interactions in vertebrates.25 The Mediator complex is a transcriptional co-regulator required for activation and repression of RNA Pol II transcription.26,27 Mediator is a large, evolutionarily conserved, protein complex with four submodules: head, middle, tail, and a reversibly binding Cdk8 kinase module that regulates interaction with RNA Pol II.28,29 In yeast, Mediator consists of 21 core subunits and a four subunit Cdk8 kinase module with the Med14 subunit acting as an assembly scaffold.30 Mediator co-localizes with NIPBL and cohesin at enhancers and promoters of actively expressed genes in mouse embryonic stem cells25 and shows a synergistic effect on gene expression and limb development when partially reduced along with NIPBL in mice and zebrafish.31 In yeast, Mediator shows a similar binding pattern to RSC and the cohesin loading complex, binding at nucleosome depleted regions and promoters of RNA Pol II genes in addition to tRNA and snRNA genes,32 buts it is unclear if Mediator is part of the loading mechanism in genomes lacking promoter-enhancer interactions.
To better understand the loading of cohesin, we investigated the structural and functional domains of Scc2. We demonstrate that mutations in the evolutionarily conserved C-terminal HEAT repeat domains are lethal in budding yeast. A broad range of Scc2 interactors was identified by mass spectrometry. Comparison with previous mass spectrometry for human NIPBL reveals interactions with chromatin complexes, splicing factors and RNA-binding proteins are evolutionarily conserved. Based on mass spectrometry and a high copy genetic suppressor screen, we identified both a genetic and physical interaction with Mediator. An engineered interaction between Med14 and Scc2C is sufficient for cell growth and cohesin loading. The depletion of Med14 results in loss of cohesion and decreased binding of Scc2 at RNA Pol II-transcribed genes. Mediator, NIPBL, and cohesin have been suggested to regulate gene expression by linking enhancers to promoters in higher eukaryotes. Our studies suggest an evolutionarily conserved role for Mediator in direct recruitment of Scc2 to RNA Pol II-transcribed genes and sister chromatid cohesion.
Results
Identification of essential domains in Scc2
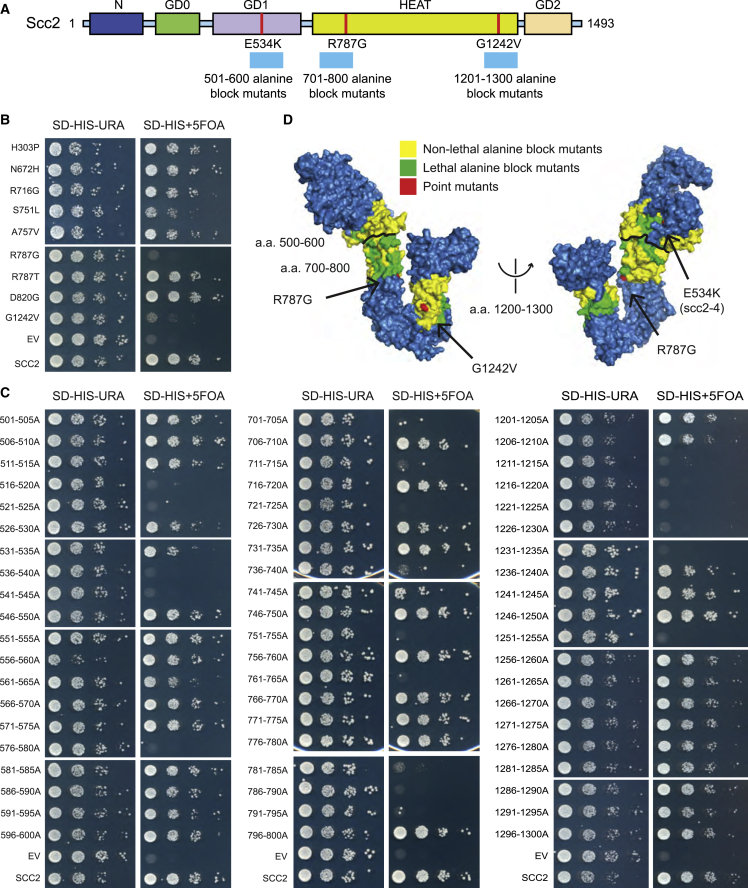
The most common and most severe mutations in CdLS are in the NIPBL gene.13 To further investigate the structure and function of the yeast cohesin loader, Scc2, we sought to identify essential domains. Using an amino acid alignment of Scc2 and NIPBL, as well as cataloged CdLS NIPBL mutations,13 we identified CdLS missense mutations that occur at amino acids conserved between yeast and humans (Figure S1A). We introduced these mutations into an SCC2 plasmid shuffle strain. We identified two mutations in the HEAT domain of SCC2 that result in significantly reduced growth, scc2R787G and scc2G1242V (Figures 1A and 1B). Additional essential residues were identified using an alanine block scanning approach, changing five consecutive amino acids to alanine (Figure 1C). Regions targeted were amino acids 701–800 and 1201–1300, the regions containing scc2R787G and scc2G1242V, respectively, as well as amino acids 501–600 containing the previously studied temperature-sensitive scc2E534K (scc2-4) allele.11,33 A structural model of S. cerevisiae Scc21–1493 was created based on the A. gossypii Scc21–1479 crystal structure34 (Figure 1A). The E534K mutation is in the N-terminal globular domain (GD1) whereas R787G and G1242V mutations are located on opposite ends of the hook-shaped HEAT domain. Mutations were mapped to the crystal structure of Chaetomium thermophilum Scc2385–1840 16 (Figure 1D). The E534K, R787G, and G1242V mutations all occur at surface residues conserved from fungus to humans. Interestingly, the R787G and G1242V mutations occur at or proximal to amino acids that when mutated decreased the ability to interact with and load cohesin.16,34 Additionally, R787G and the 781–795 alanine scan mutations fall within a positive patch of amino acid residues that interact with DNA in the gripping state intermediate of topological cohesin loading onto DNA while G1242V is proximal to the interface between Scc2 and the SMC heads35, 36, 37 (Figure S1B). Our data suggest the gripping domain, along with other regions of the HEAT repeats, are critical for Scc2 function.
Figure 1.
Identification of essential domains in Scc2
(A) Schematic of Ashbya gossipii Scc2 domains aligned to S. cerevisiae Scc2. Point mutants are indicated in red and cyan bars represent regions of the alanine block scan.
(B) Plasmid shuffle assay of CdLS mutations at residues conserved from Nipbl to Scc2. Endogenous SCC2 is deleted and covered by wild-type SCC2 on a URA3 selectable plasmid, whereas a second mutant copy of scc2 is present on a HIS3 selectable plasmid. 10-fold serial dilutions of the indicated strains were spotted on SD-HIS-URA and SD-HIS+5FOA. Growth on 5FOA indicates the viability of mutation. The R787G and G1242V mutations result in reduced growth.
(C) Identification of additional essential residues in Scc2 using the plasmid shuffle assay for alanine block mutants in Scc2.
(D) Mutated amino acids mapped to the C-terminal crystal structure surface map of Chaetomium thermophilum Scc2385–1840. Point mutants are indicated in red, lethal alanine block mutants in green, and non-lethal alanine block mutants in yellow.
See also Figure S1.
Scc2 mutants have misregulated gene expression profiles
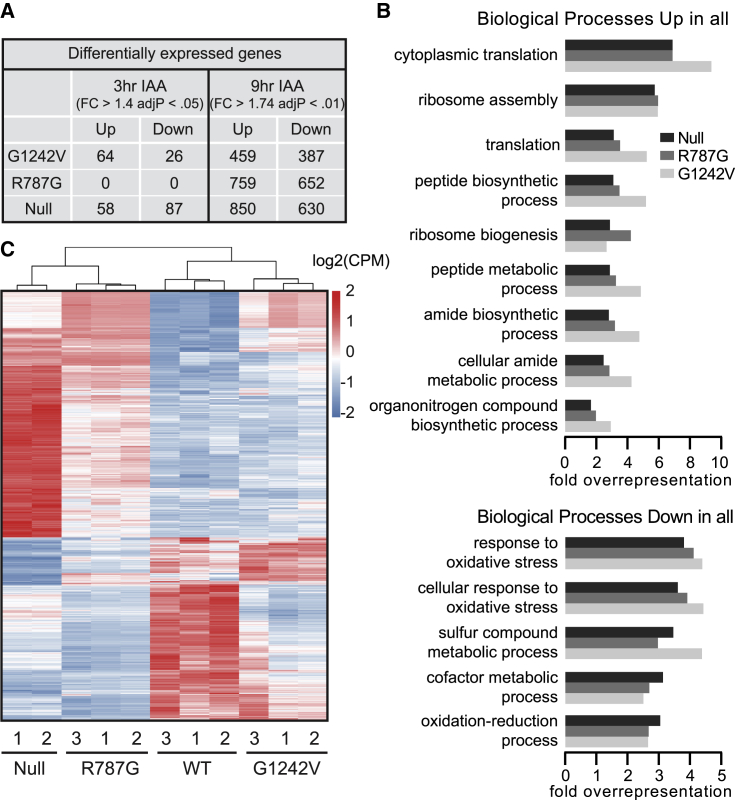
To investigate the biological processes affected in Scc2 mutants, gene expression analysis was performed in the scc2R787G and scc2G1242V mutants. A plasmid containing either SCC2WT, scc2R787G, scc2G1242V, or an empty vector (null) was introduced into an Scc2 auxin-inducible degron (Scc2-AID) background. Scc2-AID degradation was confirmed by growth assay on an auxin, 3-indoleacetic acid (IAA), plate and western blot (Figures S2A and S2B). The Scc2 mutant growth phenotypes in the Scc2-AID background were consistent with the plasmid shuffle (Figure S2A) and the degron tagged Scc2 was no longer detectable by western blot after culturing the cells with auxin (Figure S2B). Strains were grown in triplicate and RNA sequencing was performed after 3 h of auxin treatment. No genes were differentially expressed in the scc2R787G strain, and only modest effects in the scc2G1242V and null strains were observed, suggesting direct effects on gene expression are minimal (Figure 2A). Cell growth was not affected until 6 h of auxin treatment, even though the degron tagged Scc2 was undetectable by western blot after 3 h (Figure S2C). RNA sequencing with a 9-h auxin treatment revealed hundreds of differentially expressed genes in the mutant and null strains (Figure 2A). Indirect effects from Scc2 loss may be more prevalent at this time point relative to the 3-h time point. As expected, the number of differentially expressed genes correlates with the severity of the growth phenotype of each strain, with the most differentially expressed genes in the null strain.
Figure 2.
Scc2 mutants have misregulated gene expression profiles
(A) Few genes were differentially expressed after 3 h of auxin treatment while hundreds of genes were differentially expressed with auxin treatment extended to 9 h. RNA sequencing was performed in Scc2degron strains containing a plasmid copy of WT, null, R787G, or G1242V SCC2.
(B) The gene expression profile of scc2R787G is similar to null, whereas scc2G1242V clusters more closely to WT. Heatmap showing the log2 transformed read counts per million (CPM) of the differentially expressed genes in the null strain after 9 h auxin treatment.
(C) Differentially expressed genes in scc2R787G, scc2G1242V, and null compared with WT are enriched for GO terms translation and ribosome biogenesis for upregulated genes and oxidative stress for downregulated genes.
See also Figure S2.
The gene expression profile of each strain can be visualized in the heatmap showing counts per million reads (CPM) at the genes differentially expressed in the null strain (Figure 2B). The scc2R787G mutant expression profile more closely resembles the null strain whereas the scc2G1242V mutant clusters with the wild type (WT). Gene ontology (GO) term analyses for upregulated genes in both the Scc2 mutants and the null show enrichment for genes involved in translation and ribosome assembly and biogenesis. Downregulated genes are enriched for genes required for response to oxidative stress and the oxidation-reduction process (Figure 2C). Many of the changes observed in gene expression are likely indirect consequences of the absence of functional Scc2. Cytometry shows the R787G and null mutations result in arrest at G2/M of the cell cycle, whereas the G1242V mutation shows less severe defects (Figure S2D). Nonetheless, these results are highly similar to the previously published gene expression profile in a strain bearing the scc2-4 mutation, grown at permissive temperature with minimal cell cycle defects, which also showed upregulation of ribosome biogenesis and rRNA processing genes and downregulation of genes involved in oxidative phosphorylation.33 Upregulation of rRNA processing genes was also present in the GO term analyses of the null and scc2R787G strains. Ribosomal protein genes (RPs) were upregulated in scc2-433 and indeed similar profiles are observed in each of the scc2R787G, scc2G1242V, and null strains compared with WT. Mutations at R787 and G1242 impair interaction with cohesin and cohesin loading is diminished in the scc2-4 mutant.11,16 Therefore, the gene expression profile in the three different Scc2 mutant strains may reflect a consistent transcriptional response to defects in the loading of cohesin. The scc2-4 mutant strain is associated with defects in the production of ribosomal RNA, ribosome assembly, and splicing,33 and similar defects may be present in the R787G and G1242V mutants given the similarity in gene expression profiles. These findings reveal the E534K (scc2-4), R787G, and G1242V mutations all result in similar gene expression profiles and likely lead to defects in similar biological processes.
MED14 is a suppressor of scc2 mutants
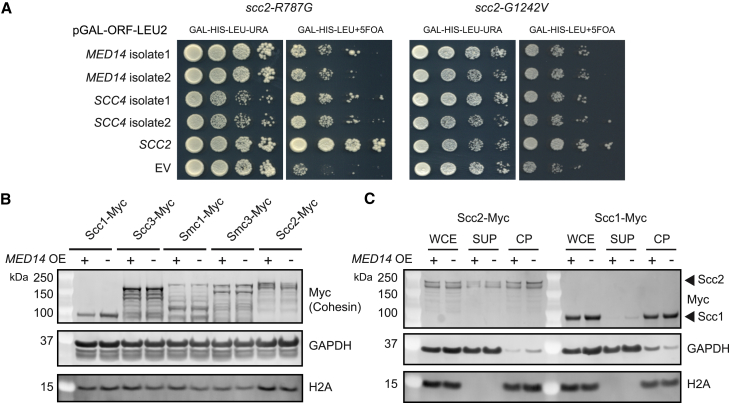
To identify the genetic interactors of SCC2, we performed overexpression screens with strains bearing the scc2R787G and scc2G1242V mutations. The yeast genomic tiling vector collection was transformed into both the scc2R787G and scc2G1242V SCC2 plasmid shuffle strains. The genomic tiling vector collection consists of 1,588 vectors covering 97% of the S. cerevisiae genome with each 2μ LEU2 selectable plasmid containing ∼4–5 genes. Multiple tiling vectors rescued the scc2R787G and scc2G1242V growth phenotypes when overexpressed. Candidate suppressor genes were cloned into a LEU2 selectable galactose inducible vector and tested for suppression in the scc2R787G and scc2G1242V SCC2 plasmid shuffle strains. In addition to SCC2, the two strongest suppressors of the scc2R787G and scc2G1242V mutants were the SCC4 component of the cohesin loading complex and the MED14 subunit of the Mediator complex (Figure 3A). Suppression is specific to the MED14 subunit, as overexpression of other subunits of Mediator (MED1, MED2, MED3, MED8, MED17, MED18, and MED19) did not rescue growth (Figure S3A). Med14 is crucial for the intermodular interactions of the Mediator complex, contacting the three main Mediator modules and acting as a scaffolding protein for assembly of the head, middle, and tail modules.30,38 MED14 overexpression also showed a limited rescue effect in the scc2E534K (scc2-4) mutant (Figure S3B).
Figure 3.
Overexpression of MED14 acts as a growth enhancer in strains bearing point mutations in SCC2
(A) Identification of a genetic interaction between Scc2 and Mediator. A genomic tiling vector screen identified MED14 and SCC4 as possible suppressors of scc2R787G and scc2G1242V mutations. MED14 and SCC4 were cloned into a LEU2 selectable galactose overexpression plasmid and transformed into the scc2R787G and scc2G1242V plasmid shuffle strains. 10-fold serial dilutions of the indicated strains were spotted on Gal-HIS-LEU-URA and Gal-HIS-LEU+5FOA.
(B) Increase in cell growth is observed in the MED14 overexpression strain compared with the empty vector (EV) control on 5FOA (B) MED14 overexpression does not affect protein levels of cohesin or Scc2. Cohesin and Scc2 Myc tagged strains containing the MED14 galactose overexpression plasmid (MED14 OE +) or empty vector (MED14 OE −) were grown in Gal-LEU and levels of cohesin subunits and Scc2 were analyzed by immunoblot. GAPDH and H2A levels are included as loading controls.
(C) Overexpressing MED14 does not increase bulk Scc2 or cohesin on chromatin. Chromatin fractionation was performed on yeast cells and levels of Myc tagged Scc2 or Scc1 in the whole-cell extract (WCE), supernatant (SUP), and chromatin pellet (CP) were visualized by immunoblotting. GAPDH and H2A served as loading controls and positive controls for the SUP and CP fractions, respectively.
See also Figure S3.
MED14 overexpression could potentially upregulate transcription of SCC2 and/or cohesin subunits, thereby rescuing the mutants. To test this possibility, we examined protein levels of Myc epitope-tagged Scc2 and cohesin subunits following MED14 overexpression (Figure 3B). Protein levels of Scc2 and the four core subunits of cohesin are not significantly affected. If the mechanism of rescue is through upregulating transcription, overexpression of MED14 might also be expected to rescue scc4 or cohesin mutations. However, the rescue is specific to mutations in scc2, as no rescue was observed in scc4 or cohesin temperature-sensitive mutants (Figures S3C and S3D). A study in human mammary stem cells showed that increased dosage of the MED14 scaffold stabilized the Mediator complex.39 To test the possibility that increased levels of Mediator stabilize Scc2 on chromatin and therefore increase binding of cohesin, chromatin fractionation was performed (Figure 3C). Overexpressing MED14 did not alter the bulk amount of cohesin or Scc2 associated with chromatin. These findings suggest a genetic interaction between the Scc2 cohesin loader and Mediator subunit Med14. The mechanism of rescue does not appear to be increased transcription leading to higher levels of the loading complex or cohesin. Moreover, MED14 overexpression does not create a global increase in chromatin association of the cohesin loader. These results suggest MED14 overexpression may create more functional binding or support binding at a subset of locations.
Scc2 and Mediator physically interact
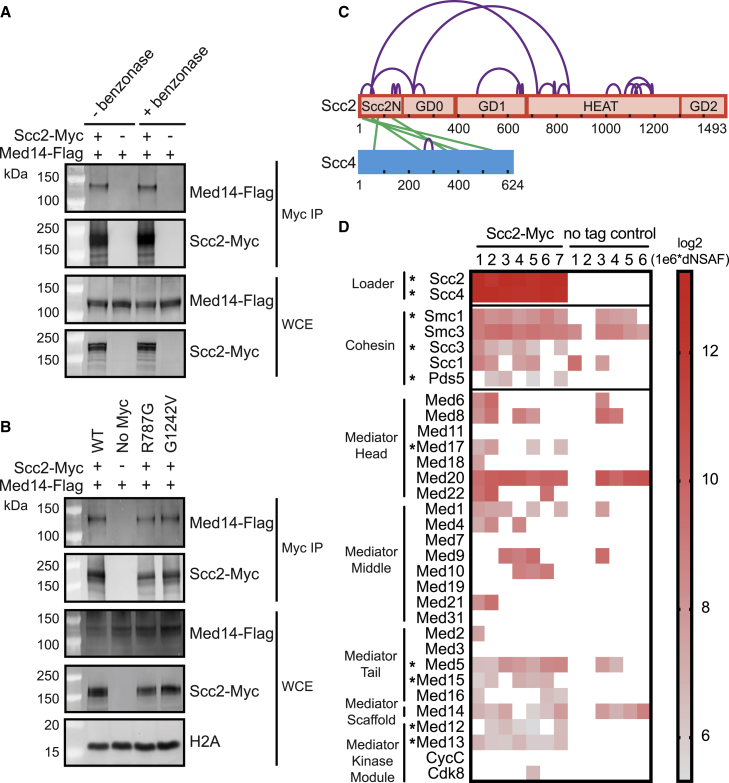
Nipbl and Mediator co-localize at enhancers and promoters and interact in higher eukaryotes.25 After observing a genetic interaction between SCC2 and MED14 in yeast, we tested if Scc2 and Med14 physically interact by co-immunoprecipitation (coIP). Scc2-Myc co-precipitated with Med14-Flag in whole-cell extract (WCE). The coIP of Scc2 and Med14 persists after treating WCE with benzonase, suggesting the interaction is DNA independent (Figure 4A). Further supporting a physical interaction between Mediator and the cohesin loader, Scc2 co-precipitates with Med1 and Med16 subunits of Mediator (Figures S4A and S4B). Mutations in Scc2 did not impact the interaction with Med14 (Figure 4B). Additionally, the point mutations did not impact the coIP of Scc2 with Scc4 or two subunits of cohesin (Figures S4C–S4E). To determine if the interaction between the cohesin loader and Mediator is dependent on the presence of cohesin or the chromatin deposition of Scc2 mediated by Scc4, we performed coIP of Scc2 and Med14 after depletion of auxin-inducible degron tagged Scc1 or Scc4. Depletion of Scc1-AID and Scc4-AID was confirmed by a growth assay (Figure S4F). Neither depletion impacted the interaction between Scc2 and Med14 (Figures S4G and S4H), suggesting the interaction is independent of cohesin and Scc4.
Figure 4.
Scc2 and Mediator physically interact
(A) Scc2 and Med14 interact by co-immunoprecipitation independent of DNA. Myc tagged Scc2 was immunoprecipitated from WCE. Co-precipitation of Flag-tagged Med14 was analyzed by immunoblotting. Benzonase treating WCE to remove DNA did not affect the observed co-precipitation.
(B) R787G and G1242V mutations do not affect the Scc2-Med14 interaction. Immunoprecipitations from the lysate of Myc tagged wild type, no tag, R787G, and G1242V Scc2 were analyzed for coIP of Med14-Flag by immunoblotting. The ratio of Scc2 to Med14 in the Scc2 mutants is comparable with WT in the Myc pulldowns.
(C) Diagram indicating inter- (green) and intra- (purple) protein cross-links of the Scc2-Scc4 complex. Positions of Scc2 domains are marked.
(D) Mediator is detected by mass spectrometry of purified Scc2. Cross-linking mass spectrometry was performed on Myc tagged Scc2 purified from WCE with anti-Myc magnetic beads with on bead disuccinimidyl sulfoxide (DSSO) cross-linking and on bead digestion. The heatmap of 1e6 log2 transformed dNSAF values shows enrichment of Mediator subunits in seven MS runs with purified Scc2-Myc compared with six no tag control runs. The loader and cohesin subunits are included as a positive control. Asterisks indicate statistical significance at p < 0.05.
See also Table S2 and Figures S4 and S5.
To further define the interaction between Scc2 and Mediator we performed cross-linking mass spectrometry analysis of in vivo purified Scc2-Myc endogenously expressed from its native promoter. Although cross-linking data has been reported from recombinant cohesin loading complex,34 this is the first cross-linking proteomics performed with in vivo purified Scc2. Although many proteins were enriched in our proteomic analysis of Scc2, Scc4 was the only protein identified with inter-protein cross-links to Scc2 (Figure 4C). The inter- and intra-protein cross-links observed between Scc2 and Scc4 are consistent with previous reports,15,34 suggesting the method was successful. In addition to previously observed intra-molecular cross-links, cross-links within Scc2 were identified that may contribute to the flexible conformations exhibited by the Scc2 hook and the Scc2N-Scc4 head of the cohesin loading complex observed by electron microscopy.14
Subunits of the Mediator complex, as well as known interactors of Scc2, were detected by MS in the Scc2-Myc purifications. Scc4 was the most abundant protein after Scc2, and all subunits of cohesin were present (Figure 4D). We detected peptides from 19 Mediator subunits in at least one of the Scc2 mass spectrometry runs. The results of Mediator co-purification with Scc2 are summarized in the heatmap (Figure 4D). Five Mediator subunits were significantly enriched, one from the head module (Med17), two from the tail (Med5 and Med15), and two from the kinase module (Med12 and Med13). We conclude that the interaction of the cohesin loader and Mediator complex, previously identified in higher eukaryotes,25 is conserved in yeast.
Evolutionarily conserved interactions between Scc2 and a broad range of chromatin-associated proteins
In addition to Mediator, mass spectrometry identified many chromatin remodelers, proteins implicated in cohesin loading, and chromatin-modifying complexes not previously reported to interact with Scc2. Multiple subunits from the RSC complex, a chromatin remodeler that acts as an adapter for the cohesin loader,20 were enriched in the Scc2 purifications (Figure S5A). We also observed significant enrichment of other complexes implicated in cohesin recruitment such as the FACT and Chd1 chromatin remodelers21,22 and the MCM2-7 and ORC components of the pre-replication complex (pre-RC)17,24 (Figure S5A). The Irc5 and Chl1 helicases, which have been implicated in cohesin deposition,40, 41, 42, 43, 44, 45 were also significantly enriched (Figure S5A). The histone acetyltransferases SAGA and NuA4 and the methyltransferase COMPASS, chromatin modifier complexes involved in the regulation of transcription, were examples of newly discovered and highly enriched interactors, as was the Ccr4-Not complex involved in mRNA regulation and transcription initiation (Figure S5B). In addition to cohesin, subunits from condensin and Smc5/6 complexes were enriched in the Scc2 purifications (Figure S5C), consistent with a role for Scc2 in the loading of these complexes.46
Proteomics of human cohesin and Nipbl revealed interactions with RNA-binding proteins, splicing factors, MCM proteins, and Mediator.47 Many splicing and RNA-binding proteins were likewise found as Scc2 interactors in yeast (Table S2). In addition to identifying protein complexes that could potentially be involved in cohesin localization, our data demonstrate that many interactions are evolutionarily conserved. Mediator is hypothesized to form a complex with Nipbl and cohesin to loop enhancers to promoters in metazoan genomes. The interaction of Mediator and Scc2 in yeast, an organism that lacks long-range enhancer-promoter interactions as well as canonical topologically associating domains (TADs), raises the possibility that this interaction may have been co-opted for genome organization in metazoans but may have a more elemental function.25
Mediator as a chromatin receptor for Scc2C
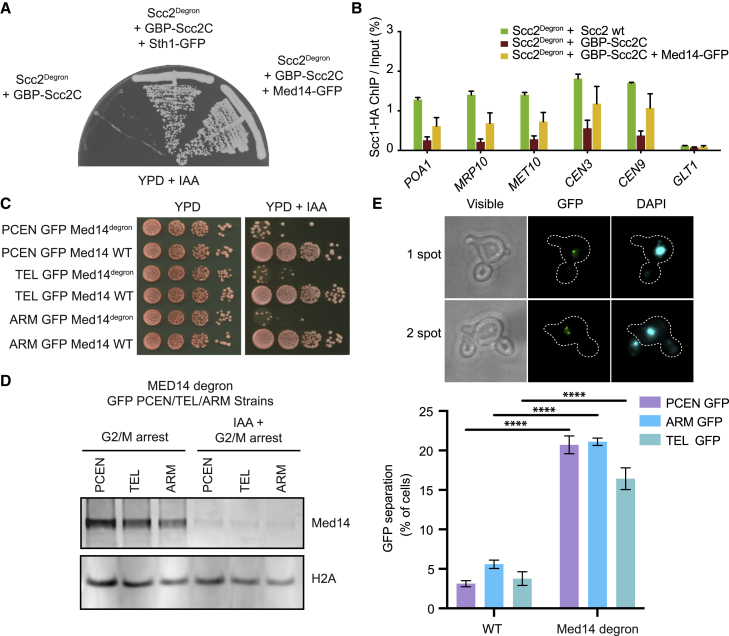
The functional modularity of the cohesin loader has been displayed through structural and biochemical studies.14,15,17 Scc4 binds the N-terminal domain of Scc2 and is required for chromatin targeting in vivo,14 whereas Scc2C is capable of loading cohesin onto naked DNA in vitro.18,48 Tethering of Scc2C to the RSC complex, as well as other chromatin remodelers, bypasses the requirement for Scc4 and is sufficient for cohesin loading.20 We did not detect significant enrichment of remodelers that fail to rescue cell growth in the tethering assay, such as the Ino80 and Swr1 complexes. RSC, Isw1, and Chd1, remodelers that restore viability, were all enriched in our proteomics experiment (Figure S5). To test Mediator as a chromatin receptor for Scc2C, Med14 was tagged with GFP at the C terminus and GFP binding protein (GBP) was fused to the N terminus of Scc2C in an Scc2 auxin-inducible degron background. Strikingly, Med14 is a viable receptor for GBP-Scc2C and sustained robust cell growth upon Scc2 depletion at levels similar to the Sth1 subunit of the RSC complex (Figure 5A). Cohesin levels were measured by ChIP/qPCR at several locations. Binding at chromosome arms and centromere-adjacent regions was diminished in cells expressing only GBP-Scc2C, whereas the tethering of Med14-GFP to GBP-Scc2C restored cohesin at both arms and centromere-adjacent regions to approximately half of the wild type (Figure 5B). This suggests that the Mediator complex can act as an Scc2C receptor and facilitate cohesin loading, similar to RSC.
Figure 5.
Mediator as a chromatin receptor for Scc2C
(A) Tethering of GBP-Scc2C to Med14-GFP bypasses the need for Scc2. Cells expressing the indicated components were streaked on to YPD medium containing 3-indoleacetic acid (IAA) to degrade the Scc2degron. The Sth1 subunit of RSC is included as a positive control.
(B) Tethering GBP-Scc2C to Med14-GFP reconstitutes cohesin loading. Cells were synchronized in G1 with alpha-factor, Scc2degron was depleted with IAA, and cells were released into nocodazole-imposed mitotic arrest. Cohesin levels were assessed by ChIP against HA-tagged Scc1 followed by quantitative real-time PCR at three chromosome arms and two centromere-adjacent cohesin binding sites and a negative control site. Means and SEM of three independent experiments are shown.
(C) Validation of Med14degron in the presence of auxin (IAA). 10-fold serial dilutions of yeast strains used to examine sister chromatid cohesion, containing a lacO-LacI-GFP marked pericentromere, arm, or telomere site and either WT or degron tagged Med14, were spotted onto YPD+IAA to deplete Med14degron. Depletion of Med14 results in diminished cell growth.
(D) Med14 protein is degraded in sister chromatid cohesion strains after the addition of auxin. Cells were synchronized in G1 with alpha-factor, Med14degron was depleted with IAA, and cells were released into nocodazole-imposed G2/M arrest. Cells were pelleted and WCE prepared and analyzed for the depletion of Med14 by immunoblot.
(E) Mediator promotes sister chromatid cohesion. Cells were synchronized in G1 with alpha-factor, Med14degron was depleted with IAA, and cells were released into nocodazole-imposed mitotic arrest. Cells were fixed and DAPI stained, and the percent of large-budded cells with a single DAPI mass containing 2 GFP spots, indicating loss of sister chromatid cohesion, was scored. A representative image of a cell with cohesion (1 spot) and loss of cohesion (2 spots) is shown. Means and SEM of three independent experiments are shown. A minimum of 100 cells were scored for each replicate; WT CEN, ARM, TEL n = 523, 443, 570 and Med14 degron CEN, ARM, TEL n = 603, 576, 637; ∗∗∗∗ p <.0001; two-way ANOVA test.
See also Figure S6.
If Mediator plays a significant role in the recruitment of Scc2 and cohesin, then sister chromatid cohesion may be compromised without Mediator. We evaluated sister chromatid cohesion by monitoring lacO-LacI-GFP-marked pericentromere, arm, or telomere sites in Med14 auxin-inducible degron tagged strains. Degron function and efficient depletion of Med14 was confirmed by growth assay and western blot (Figures 5C and 5D). The strains containing the degron tagged Med14 exhibited markedly reduced growth with the addition of auxin (Figure 5C). Med14 was depleted by adding auxin to cells arrested in G1 using alpha-factor. The cells were then released into a G2/M arrest by nocodazole treatment, depleting Med14 within one cell cycle (Figures S6A and S6B). Degron tagged Med14 was not detectable after auxin treatment (Figure 5D). Med14 depletion resulted in defective sister chromatid cohesion at all loci examined (Figure 5E). Levels of cohesin subunits in WCE were not affected by Med14 depletion (Figure S6C), suggesting the cohesion defects cannot be attributed to decreased levels of cohesin. These results indicate a crucial role of Mediator in facilitating sister chromatid cohesion in yeast.
Mediator facilitates the recruitment of Scc2 to RNA Pol II gene groups
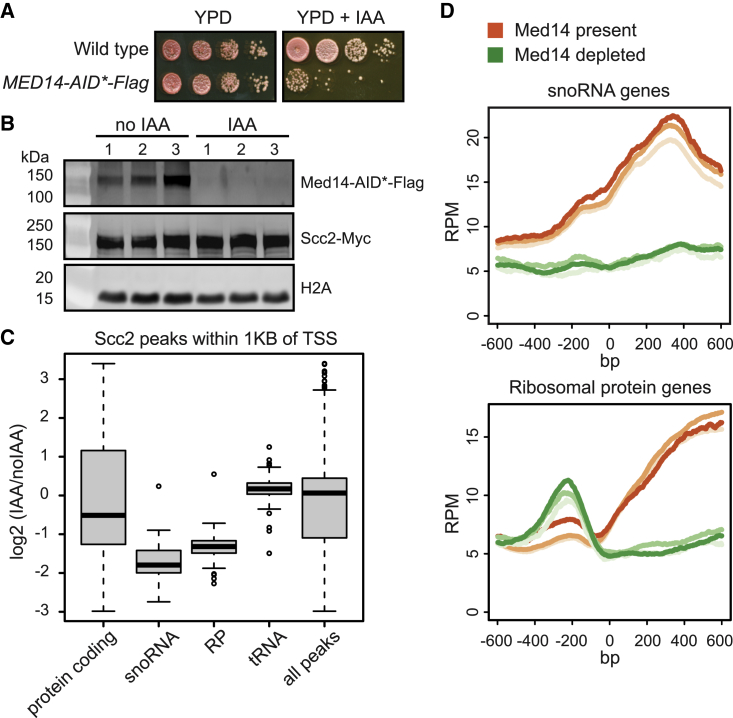
To further define the role of Mediator in the localization of the cohesin loader, we performed calibrated ChIP-seq of Scc2 in a Med14 conditional depletion background. Scc2-Myc was introduced in a Med14-AID-FLAG background. The functionality of the Med14 degron was assessed by the inhibition of cell growth with auxin (Figure 6A). Cultures were arrested in G1, auxin added (if depleting), and released into G2/M arrest (Figures S6D and S6E). Depletion of Med14 was verified by western blot and levels of Scc2 were not affected (Figure 6B). Enrichment of DNA in the ChIP compared with a control with primary antibody omitted was confirmed by polymerase chain reaction (PCR) (Figure S6F).
Figure 6.
Mediator facilitates Scc2 recruitment
(A) Validation of Med14degron in Scc2-Myc tagged strain used for calibrated ChIP-seq. 10-fold serial dilutions were spotted on YPD and YPD+IAA.
(B) Med14degron is depleted in the presence of IAA. Cells grown in triplicate were synchronized in G1 with alpha-factor, Med14degron was depleted with IAA, and cells were released into nocodazole-imposed mitotic arrest. Cells were then cross-linked with formaldehyde. A portion of the culture was removed before cross-linking for gel electrophoresis and levels of Med14 and Scc2 were monitored by immunoblot. H2A served as a loading control.
(C) Scc2 peaks are most affected at snoRNA and RP genes after Med14 depletion. A spike-in of Mis4-Myc tagged S. pombe was added to each culture for calibration, and the Scc2-Myc immunopurified chromatin was extracted for ChIP-seq. The experiment was performed in triplicate with Med14 depleted (IAA) and Med14 present (noIAA). Peaks within 1 kb of transcription start sites were mapped to genes by biotype. The boxplot shows the log2 ratio of Scc2 peaks with Med14 depleted over no depletion (IAA/noIAA).
(D) Metagene analysis of the ChIP-seq data shows that Scc2 peaks decrease at snoRNA and ribosomal protein genes when Med14 is depleted.
See also Figure S6.
Peaks within 1 kb of a transcription start site (TSS) were mapped to genes by biotype and the ratio of peaks without and with Med14 (IAA/noIAA) was examined (Figure 6C). Although the average Scc2 binding at all peaks was not affected, we found that depletion of Med14 results in a modest average decrease in Scc2 peaks at protein coding genes, with the largest decrease at the snoRNA, and ribosomal protein genes. Metagene analysis confirms decreased Scc2 binding at the RNA Pol II-transcribed snoRNA and ribosomal protein genes (Figure 6D). This suggests Mediator facilitates Scc2 recruitment to RNA Pol II gene groups in budding yeast.
Discussion
The co-localization of the cohesin loader and Mediator at gene promoters and enhancers has been observed in multiple studies with their interaction proposed to induce enhancer-promoter interactions.25,51,52 Here, we show the interaction between the cohesin loader and Mediator is conserved in yeast and provide evidence that Mediator acts as a chromatin adapter for the cohesin loader, facilitating the recruitment of Scc2 to RNA Pol II gene groups. Yeast lack enhancers and long-range interactions for gene activation are rare. Our findings suggest a more evolutionarily conserved function for Mediator in cohesin loader recruitment, independent from enhancer-promoter interactions.
Our analysis identified essential domains in the C-terminal HEAT repeat domain of S. cerevisiae Scc2 and a genetic interaction between SCC2 and MED14. Previous studies have highlighted the essential residues of the HEAT domain of the cohesin loader. Mutations in the Scc2 ortholog in Chaetomium thermophilum, corresponding to budding yeast Scc2 R787 and G1242 residues, result in diminished binding with an N-terminal fragment of the Scc1 subunit of cohesin. A budding yeast double mutant with one mutation adjacent to the R787G mutation (K788A R792A) is defective in loading cohesin.16,34 Recent studies in S. pombe show that the C-terminal HEAT domain of Scc2 contains two DNA binding domains, one of which corresponds to a region including S. cerevisiae R787, that are crucial for initial binding of cohesin with DNA in an intermediate of topological loading.35,36 We speculate that the growth phenotypes observed with the R787G and G1242V mutations are due to diminished cohesin loading. The overexpression of Scc4, known to be essential for localization of the loading complex to chromatin,14 rescued the growth of these mutants. Increased levels of Scc4 may increase the localization of the loading complex, allowing for an increase in cohesin loading and account for the increase in cell viability. The observation that the Med14 subunit of Mediator rescues these mutants similar to Scc4, suggests a role for Mediator in cohesin loader recruitment. Increased dosage of MED14 in human cells led to higher protein levels of multiple Mediator subunits due to stabilization of the complex rather than changes in transcription.39 We speculate that the overexpression of the Med14 subunit in yeast similarly drives an increase in the Mediator complex because it is the scaffolding subunit.
Cohesion defects are observed with deficiencies in other purported chromatin adapters of the cohesin loading complex.20,23,53,54 Likewise, we report a cohesion defect upon Mediator depletion. We find that depletion of Mediator decreased cohesion at a pericentromeric, arm, and telomeric site. Therefore, Mediator is important to maintain normal levels of cohesion. A recent study in S. cerevisiae also implicates Mediator in sister chromatid cohesion.55 A strain forced to use the meiosis-specific kleisin subunit of cohesin during mitotic growth selectively accumulated compensatory mutations in the Mediator complex that improved sister chromatid cohesion. Interestingly, mutations that introduced early stop codons in subunits of the kinase module showed the highest increase in cell fitness. This suggests the Mediator complex devoid of the kinase module, which is the version that regulates RNA Pol II transcription, may also promote cohesion. A genetic screen with an S. pombe mutant defective in acetylation of Smc3 identified genetic interactions of cohesin with Mediator and the CCR4-NOT complex, which is also present in our Scc2 purifications.56 Other recent studies in budding yeast found that segmental amplifications containing SCC2 and MED14 were acquired as adaptations to DNA replication stress57 and that an oxidative stress-induced association of Mediator and cohesin plays a role in base excision repair of DNA lesions.58 Evidence is accumulating for a functionally significant and evolutionarily conserved interaction between Mediator and Scc2.
Physical interaction between the cohesin loader, cohesin ring, and Mediator has previously only been observed in higher eukaryotes.25,47,58 Mass spectrometry of Mediator purified from neural stem cells contained NIPBL and both SMC subunits of cohesin as well as other complexes that were present in our Scc2 purification such as subunits of the COMPASS and SWI/SNF complexes.59 Mediator is postulated to interact with transcription factor activation domains through dynamic shape-agnostic “fuzzy” interfaces. Multiple subunits of cohesin are predicted to contain such domains, providing potential Mediator interaction interfaces.60 We show that Scc2-Mediator interaction is present in yeast and that Scc2C fusion to Mediator is sufficient for cohesin loading, similar to the RSC complex.20 We propose Mediator acts as a chromatin receptor for Scc2 at a subset of RNA Pol II gene groups, namely, snoRNA and ribosomal protein genes. Interestingly, both Scc2 and Mediator localize at snoRNA genes and promote proper snoRNA transcription in yeast,33,61 and human Mediator regulates the transcription of snRNA genes.62 We speculate that the evolutionarily conserved functional interaction between Mediator and the cohesin loader occurs at a subset of RNA Pol II- highly transcribed genes.
Nucleosome-depleted DNA is required for the loading of cohesin. Chromatin remodelers that promote the recruitment of Scc2-Scc4 may generate a nucleosome-free region required to load cohesin.20 The human ISWI remodeler has been implicated in cohesin loading,63 fitting with the emerging idea that the Scc2-Scc4 receptor landscape is multifactorial. Mediator may participate in creating or marking the nucleosome-depleted DNA environment where cohesin loading occurs. Mediator interacts with multiple chromatin remodelers such as SWI/SNF, RSC, and Chd132,64, 65, 66 and Mediator itself may contribute to nucleosome displacement in both yeast and humans.67,68 Whether Mediator works together with chromatin remodelers to load cohesin or is sufficient to recruit Scc2-Scc4 and generate nucleosome depleted DNA in the absence of remodelers is an area for future investigation.
Mutations in NIPBL and the human SWI/SNF complex result in congenital developmental disorders with closely related clinical features, known as CdLS and Coffin-Siris syndrome, respectively.12,69 Mutations in Mediator are also linked to developmental syndromes and cognitive defects in humans. Interestingly, a screen for chromosomal rearrangements in clinically diagnosed CdLS patients, lacking mutations in cohesin or the cohesin loader, identified patients with chromosomal imbalances containing Mediator, helicase, and nucleosome remodeler genes.70 Mutations in the Mediator complex result in FG, Lujan, and X-linked Ohdo syndromes as well as schizophrenia.71, 72, 73, 74 Investigation into how Mediator, the cohesin loader, and its chromatin receptors interact to load cohesin and regulate genome architecture and gene expression will deepen our understanding of the etiologies of these human diseases.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mouse IgG HRP conjugated | GE Healthcare | RRID# AB_772210 |
| Anti-Rabbit IgG HRP conjugated | GE Healthcare | Cat# NA934V |
| Anti-c-Myc 9E10 | BioLegend | RRID# AB_2148451 |
| Anti-c-Myc-Peroxidase | Roche | RRID# AB_390910 |
| Monoclonal ANTI-FLAG M2 | Sigma-Aldrich | RRID# AB_259529 |
| Monoclonal ANTI-FLAG M2-Peroxidase | Sigma-Aldrich | RRID# AB_439702 |
| Anti-HA-Peroxidase | Roche | RRID# AB_390917 |
| Histone H2A antibody | Active Motif | RRID# AB_2687477 |
| GAPDH antibody | GeneTex | RRID#AB_11174761 |
| Anti-HA F7 | Santa Cruz | Cat# sc-7392 |
| Anti-HA (12CA5) | Roche | RRID# AB_514505 |
| Chemicals, peptides, and recombinant proteins | ||
| α-factor | Zymo Research | Cat# Y1001 |
| Nocodazole | Sigma-Aldrich | Cat# M1404 |
| Indole-3-acetic acid (IAA) | Sigma-Aldrich | Cat# 45533 |
| Formaldehyde Solution | Sigma-Aldrich | Cat# 252549 |
| Proteinase K | ThermoFisher | Cat# EO0491 |
| Benzonase nuclease | Sigma-Aldrich | Cat# E1014 |
| RNase A | ThermoFisher | Cat# EN0531 |
| Propidium iodide solution | Sigma-Aldrich | Cat# P4864 |
| GelRed Nucleic Acid Gel Stain | Biotium | Cat# 41103 |
| 5-Fluoroorotic Acid | Zymo Research | Cat# F9001 |
| DAPI | ThermoFisher | Cat# 62248 |
| cOmplete Protease Inhibitors Cocktail | Sigma-Aldrich | Cat# 11836145001 |
| Triton X-100 | Sigma-Aldrich | Cat# T9284 |
| NP-40 Alternative | Sigma-Aldrich | Cat# 492016 |
| Sodium Deoxycholate | Sigma-Aldrich | Cat# S1827 |
| 2-Chloroacetamide | Sigma-Aldrich | Cat# C0267 |
| Dithiothreitol | ThermoFisher | Cat# R0861 |
| Sequencing grade modified trypsin | Promega | Cat# V5111 |
| Copper (II) sulfate | Sigma-Aldrich | Cat# C1297 |
| Critical commercial assays | ||
| Pierce ECL 2 western blotting substrate | ThermoFisher | Cat# 80196 |
| Nitrocellulose Blotting Membrane | Sigma-Aldrich | Cat# GE10600002 |
| Dynabeads Protein G | ThermoFisher | Cat# 10004D |
| Pierce Anti-c-Myc Magnetic Beads | ThermoFisher | Cat# 88842 |
| NuPAGE LDS Sample Buffer | ThermoFisher | Cat# NP0007 |
| NuPAGE bis-tris 4-12% gradient protein gel | ThermoFisher | Cat# NP0323BOX |
| Gateway LR Clonase II Enzyme mix | ThermoFisher | Cat# 11791020 |
| Gateway BP Clonase II Enzyme mix | ThermoFisher | Cat# 11789020 |
| TruSeq Stranded Yeast Ribo-Zero Gold Kit | Illumina | Cat# MRZY1324 |
| TruSeq Stranded RNA LT kit with Ribo-Zero Gold | Illumina | Cat# RS-122-2301 |
| KAPA HTP Library Prep Kit for Illumina | Roche | Cat# KK8234 |
| PowerUp SYBR Green Master Mix | ThermoFisher | Cat# A25742 |
| Deposited data | ||
| ChIP sequencing data | This study | GEO: GSE201194 |
| RNA sequencing data | This study | GEO: GSE201194 |
| Mass spectrometry data | This study | MassIVE: MSV000086339, ProteomeXchange: PXD022100 |
| Unprocessed images presented in this manuscript | This study | https://www.stowers.org/research/publications/libpb-1710 |
| Experimental models: Organisms/strains | ||
| All Saccharomyces cerevisiae strains used in this study are listed in Table S1 | Lab stock and this study | N/A |
| Oligonucleotides | ||
| POA1-F AAACGGCCACATCAAATACC | Muñoz et al.20 | N/A |
| POA1-R TCCAAGGGACTCCGAATATG | Muñoz et al.20 | N/A |
| MRP10-F ACCCCCTCTTCCCAGACTAA | Muñoz et al.20 | N/A |
| MRP10-R CCAGCACATTTAGGGCTCAT | Muñoz et al.20 | N/A |
| MET10-F ACTTGTGTGGCCCTACTTGG | Muñoz et al.20 | N/A |
| MET10-R CGACTTTGATGCCTCTTTCC | Muñoz et al.20 | N/A |
| CEN3-F CGCCACTTTAACAAATGTGC | Muñoz et al.20 | N/A |
| CEN3-R GCAGAACCACCGTAGCAGTT | Muñoz et al.20 | N/A |
| CEN9-F TGTCACCTGGCTGTTTTGAG | Muñoz et al.20 | N/A |
| CEN9-R TGGGTAATGTCAGCTGTGGA | Muñoz et al.20 | N/A |
| GLT1-F TTTGACCCCAGCACATGTTA | Muñoz et al.20 | N/A |
| GLT1-F GGGTGTGGAGTTTGTGGTCT | Muñoz et al.20 | N/A |
| CEN3chip-F AAGTCACATGATGATATTTGAT | This study | N/A |
| CEN3chip-R ATTTCTTTTTTAACTTTCGGAA | This study | N/A |
| Recombinant DNA | ||
| Yeast genomic tiling collection | OpenBiosystems | Cat# YSC4613 |
| pRS413-SCC2-7MYC Point and alanine block mutants | This study | N/A |
| pHyg-AID∗-6FLAG | Addgene | Cat# 99519 |
| pHyg-AID∗-6HA | Addgene | Cat# 99520 |
| Software and algorithms | ||
| Fiji (Image J) V2.3.1/1.53f | https://imageJ.net/ | RRID# SCR_002285 |
| FlowJo v9 | FlowJo | RRID# SCR_008520 |
| Prism V9.3.1 | GraphPad | RRID# SCR_002798 |
| PyMOL | http://www.pymol.org/ | RRID# SCR_000305 |
| ProLuCID | Xu et al.75 | N/A |
| DTASelect v1.9 | Tabb et al.76 | N/A |
| NSAF v0.0.1 | Zhang et al.77 | N/A |
| STARTApp | Nelson et al.78 | N/A |
| ReactomePA | Yu and He79 | RRID# SCR_019316 |
| HiSeq Control Software v2.2.58 | Illumina | N/A |
| Illumina Primary Analysis version RTA v2.4.11 | Illumina | N/A |
| Illumina Secondary Analysis bclfastq2 v2.17 | Illumina | N/A |
| R 3.6-4.0 | https://www.r-project.org/ | RRID# SCR_001905 |
| Bioconductor 3.10-3.13 | http://www.bioconductor.org | RRID# SCR_006442 |
| edgeR v3.28.0 | http://www.bioconductor.org/packages/edgeR/ | RRID# SCR_012802 |
| TopHat 2.1.1 | http://ccb.jhu.edu/software/tophat/index.shtml | RRID# SCR_013035 |
| bowtie2 bowtie2-2.4.2 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | RRID# SCR_016368 |
| STAR STAR_2.6.1c | http://code.google.com/p/rna-star/ | RRID# SCR_004463 |
| Macs2 2.1.2 | https://github.com/macs3-project/MACS | RRID# SCR_013291 |
| Ensembl 96 | http://www.ensembl.org/ | RRID# SCR_002344 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by lead contact Jennifer L. Gerton (jeg@stowers.org).
Materials availability
Yeast strains and plasmids generated in this study are available for distribution.
Experimental model and subject details
All Saccharomyces cerevisiae yeast strains used in this study were of the W303 or BY4741 background and are listed in Table S1. Cells were cultured at 30°C. α-factor was used at a concentration of 7.5 μg/ml, nocodazole at 15 μg/ml and indole-3-acetic acid (IAA) at 1 mM.
Method details
Yeast strain construction
To construct the SCC2 mutant plasmid shuffle strains, SCC2-7MYC including the promoter region was amplified from yeast genomic DNA and cloned into pRS413. Plasmid pRS413-SCC2-7MYC point mutations and alanine block mutations were made by site-directed mutagenesis and transformed into the SCC2 plasmid shuffle strain. Endogenous genes were epitope tagged by gene targeting using polymerase chain reaction (PCR) products. The auxin degron strains were constructed using pHYG-AID∗6FLAG (Addgene #99519) or pHYG-AID∗6HA (Addgene #99520) and yeast strain BY26972 from the Japan National BioResource Project (NBRP::Yeast).
SCC2 mutant plasmid shuffle
SCC2 plasmid shuffle yeast strains were grown overnight in synthetic media lacking histidine (SD-HIS). The following day cultures were diluted to OD600 1.0 and 10 fold serial dilutions were spotted on SD-HIS-URA and SD-HIS-URA+5FOA agarose plates and grown at 30°C.
SCC2 point mutants suppressor screen
SCC2 point mutant plasmid shuffle strains R787G and G1242V were transformed with the yeast genomic tiling vector collection (Open Biosystems) as per standard high-throughput 96-well yeast protocols. Transformants were selected on SD-HIS-LEU-URA. Transformants were cultured overnight in SD-HIS-LEU at 30°C in 96 well plates and pinned to SD-HIS-LEU-URA and SD-HIS-LEU+5FOA 96 well format agarose plates. Scc2-R787G strains were pinned at 1:5 and 1:10 dilutions while scc2-G1242V strains were pinned at 1:20 and 1:40 dilutions. Candidate suppressor genes were amplified by PCR and cloned into the LEU2 selectable gal overexpression vector pAG425 using the Gateway (ThermoFisher) cloning system.
RNA sequencing
Scc2 degron strains carrying pRS413(HIS) plasmid with SCC2-WT, R787G, G1242V or empty vector (null) were cultured in triplicate (except for null strain in duplicate) overnight in SD-HIS. The following day, strains were diluted to ∼OD600 = 0.1 in SD-HIS, after 90 minutes, 3-indoleacetic acid (IAA) was added to a concentration of 1mM and cells were cultured for an additional 9 hours to OD600 range of 0.4 – 0.8. Cells were pelleted and RNA was extracted. RNA integrity was checked on a 1.2% agarose gel and run on an Agilent Technologies 2100 Bioanalyzer. Samples were ribo-depleted with TruSeq Stranded Yeast Ribo-Zero Gold Kit (Illumina, MRZY1324) per manufacturer’s instructions. Following depletion, Illumina libraries were generated using the TruSeq Stranded RNA LT kit with Ribo-Zero Gold, 48; Set A (Illumina, RS-122-2301). Libraries were normalized and pooled together with an average base pair size of 290 and sequenced as 50 bp single reads on the HiSeq 2500 (Illumina) using HiSeq Control Software v2.2.58. Following sequencing, Illumina Primary Analysis version RTA 1.18.61 and Secondary Analysis bcl2fastq2 v2.17 were run to demultiplex reads from all libraries and generate FASTQ files. Reads were aligned to version sacCer3 of the Saccharomyces cerevisiae genome using tophat2. The resulting BAM files were analyzed in R. Bioconductor package edgeR was used to perform differential expression analysis. Data has been deposited at GEO: GSE201194.
Whole cell extract co-immunoprecipitation
Cultures were grown to mid-log phase. IAA was added to 1mM for 3 hours for depletion in degron strains. Whole cell extracts were prepared by resuspending cell pellets in lysis buffer (50 mM Tris (pH 7.5), 150 mM NaCl, 0.1% NP-40, 1 mM dithiothreitol (DTT), 10% glycerol and protease inhibitors) and vortexing with glass beads at 4°C. Cell extracts were diluted with dilution/wash buffer (50 mM Tris (pH 7.5), 150 mM NaCl, 0.1% NP-40). Diluted cell extracts were incubated with the Myc 9E10 antibody (BioLegend) overnight followed by 2 h with protein G dynabeads (Invitrogen-10004D) at 4°C. The beads were washed three times with dilution/wash buffer and proteins were eluted in NuPAGE LDS sample buffer at 95°C for 10 min. Immunoprecipitates were subjected to polyacrylamide gel electrophoresis (PAGE) on NuPAGE 4-12% Bis-Tris Protein Gels (Invitrogen) and Western blotting (Myc-HRP, FLAG-HRP, HA-HRP, GAPDH, H2A).
Scc2 purification and mass spectrometry
Yeast culture
For each yeast strain (either Scc2-13Myc in BY4741 or wild-type BY4741), 6L cultures were grown in YPD medium and the cell pellet frozen and stored at -80°C until use.
Myc-Affinity Capture and on-Beads Crosslinking
In brief, after binding to the Myc-beads and two rounds of washes, bound proteins were crosslinked on-beads with 1mM DSSO for 40min, then digested on-beads with trypsin for 30 min. Multiple rounds of Myc-AP-XL were performed from three independent batches of Scc2-Myc cells and one BY4741 (no tag) cell culture. Several technical replicate LC/MS analyses were acquired from the eluted proteins. In all, seven mass spectrometry runs were performed from the Scc2 purification and six runs of the no tag control to identify proteins enriched in the purified Scc2.
Cell pellets were resuspended in a total volume of up to 30 ml in BH0.15 extraction buffer (25 mM HEPES, pH 7.5, 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA-KOH, 15% Glycerol, 0.1% NP-40, 150 mM KCl) with freshly added 100x protease inhibitor and 5mM DTT. Cells were frozen using liquid nitrogen and dry ice and lysed with a Waring Commercial Blender, 250 ml container with 5 small pieces of dry ice blended for 10-15 sec. The frozen cells were transferred to a 600 ml beaker and thawed in a warm water bath. 100 ml of 30 mg/ml heparin and 20 ml of 25U/ul SAN (Salt Active Nuclease; ArcticZyme) were added to the thawed cells, which were rotated at RT for 20 minutes. The extract was clarified by spinning the lysate in a tabletop centrifuge for 15 min at 4000 rpm, 4 °C. Supernatant was saved and the pellets resuspended to 30 ml total volume by adding BH0.15 extraction buffer to repeat the freeze/thaw/incubate steps once more. Extractions from two rounds were combined and clarified by centrifugation for a final time at 19,000 rpm for 30 min using a JA25.50 rotor in a floor centrifuge.
Pierce™ Anti-c-Myc Magnetic Beads (150 μl) were washed with 1 ml of BH0.15 extraction buffer. The supernatant was discarded, and the washed beads were added to the protein lysate and incubated on a rotating wheel overnight at 4°C. Proteins bound to anti-Myc beads were washed twice with 1.5 ml BH0.15. Beads were transferred to a new 1.5 ml tube, washed with 1 ml of pre-elution rinse buffer (50 mM HEPES, pH 7.5, 75 mM KCl, 1 mM EGTA), and the supernatant was discarded. Proteins bound to anti-Myc beads were crosslinked by adding 150 μl pre-elution rinse buffer complemented with protease inhibitor cocktail (Sigma) and DTT (dithiothreitol; Thermo Fisher Scientific) with 0.6 μl of 250 mM disuccinimidyl sulfoxide (DSSO) to a final concentration of 1mM and left to crosslink at room temperature for 40 min. The crosslinking reaction was quenched by adding 7.5 μl of 1 M NH4CO3 (final 50 mM) and rotating at RT for 15 min. The supernatant was transferred to a new tube and saved as flow-through.
On-beads protein digestion
Crosslinked proteins bound to anti-Myc beads were washed twice with 500 μl 10 mM Tris-HCl pH7.5, 150 mM NaCl, then denatured, reduced and digested with 100 ml 50 mM Tris-HCl pH7.5, 2 M urea, 1 mM TCEP (Pierce), 5 μl Trypsin at 0.1 mg/ ml (Sequencing Grade Modified Trypsin; Promega), at 30 °C for 30 min. The supernatant was collected. Another 60 μl of 50 mM Tris-HCl pH7.5, 2 M urea, 5 mM 2-Chloroacetamide (CAM, Sigma) were added to the beads to alkylate free cysteines, and the resulting supernatant combined with the first one. This 2-step process was repeated once more. CAM was added to 2.5 mM and the reaction incubated in the dark at RT for 30 min. CaCl2 was added to 2 mM along with 5 μl of Trypsin, and the digestion was incubated at 37 °C overnight. The digestion was quenched by adding formic acid to 5%. The combined supernatants from these multiple steps constitute the first “elution” (E1). These digestion steps were repeated once, and the collected supernatants combined as the second elution (E2). After digestion, the beads were resuspended in 150 μl of 2% Sodium Deoxycholate (SDS; Sigma) with 50mM HEPES and incubated at 95°C for 30 min. This fraction was labeled as “SDS-E1”. From each of the E1, E2, and SDS-E1 fractions, 14 μl was set aside to analyze protein contents on silver-stained SDS-PAGE.
LC/MS acquisition
Digested peptides were analyzed on an Orbitrap Fusion Lumos mass spectrometer equipped with a FAIMS Pro interface (Thermo Scientific, San Jose, CA) coupled to a Dionex Ultimate 3000 RSCLnano System. Peptides were loaded on an Acclaim™ PepMap™ 100 C18 0.3 mm i.D. x 5 mm length trap cartridge (Thermo Scientific, San Jose, CA) with loading pump at 2 ml/min via autosampler. A 75 μm i.d. analytical microcapillary column was packed in-house with 250 mm of 1.9 μm ReproSil-Pur C18-AQ resin (Dr. Masch GmbH, Germany). AgileSLEEVE (Analytical Sales & Products, Pompton Plains, NJ) was used to maintain column temperature at 40°C. The organic solvent solutions were water/acetonitrile/formic acid at 95:5:0.1 (v/v/v) for buffer A (pH 2.6) and at 20:80:0.1 (v/v/v) for buffer B. The chromatography gradient was a 25 min column equilibration step in 2% B; a 3 min ramp to reach 10% B; 90 min from 10 to 40 % B; 6 min to reach 95% B; a 9 min wash at 95% B; 0.1 min to 2% B; followed by a 12 min column re-equilibration step in 2% B. The nano pump flow rate was set to 180 nL/min. Orbitrap Fusion Lumos was set up with peptide identification method as: full MS1 resolution 120,000; ITMS2 isolation window 1.4 m/z, ITMS2 max ion injection time 50 ms, ITMS2 CID 35% with normal scan. FAIMS compensation voltages (CVs) were set up as -40V, -60V, and -80V.
MS dataset processing
Collected MS/MS spectra were searched with the ProLuCID algorithm75 against a database of 12276 protein sequences combining 6010 non-redundant Saccharomyces cerevisiae proteins (NCBI, 2017-05-16 release), 193 common contaminants, and their corresponding 6138 randomized amino acid sequences. All cysteines were considered as fully carboxamidomethylated (+57 Da statically added), while methionine oxidation was searched as a differential modification. DTASelect v1.976 and swallow, an in-house developed software, were used to filter ProLuCID search results at given FDRs at the spectrum, peptide, and protein levels. Here, all controlled FDRs were less than 1%. All 13 data sets were contrasted against their merged data set, using Contrast v1.9 and in house developed sandmartin v0.0.1. Our in-house developed software, NSAF7 v0.0.1, was used to generate spectral count-based label free quantitation results dNSAF.77 Mass spectrometry files have been deposited to the ProteomeXchange via the MassIVE repository with accession numbers: PXD022100 and MSV000086339.
Calibrated ChIP-seq
250 ml cultures per replicate were grown in YPD to OD600 ∼ 0.2 and arrested 1.5 hrs with alpha-factor. IAA was added to 1mM and cultures were held in alpha-factor arrest for an additional 1.5 hrs before release into YPD containing IAA and nocodazole for 2 hrs until mitotic arrest was reached. 50ml of culture was removed to verify Med14 depletion by Western blot. Mis4-Myc tagged S. pombe cells were added as a spike-in control at a 1:4 OD ratio. Cells were fixed in 1% formaldehyde for 30 min at room temperature, then quenched with glycine (final concentration 0.125 M) for 5 min. Cells were washed twice with cold PBS, frozen in liquid nitrogen, and stored in −80°C until used. Cells were lysed by bead beating at 4°C in FA-Lysis SDS buffer (50 mM Hepes (pH7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.2% SDS, protease inhibitors). The lysate was sonicated using a Covaris SS20 sonicator for 25 min and centrifuged to remove cell debris. 10% of the total chromatin was processed for input and chromatin shearing was verified on a bioanalyzer. Chromatin extract was used to set up three IPs and one no antibody control per replicate and incubated with Myc antibody (BioLegend 626802) at 4°C overnight followed by 4hrs at 4°C with Protein G Dynabeads (Invitrogen 100-04D) pre-washed in FA-Lysis buffer. Beads were washed at room temperature with the following sequence of buffers; FA-Lysis buffer, two washes in FA-Lysis buffer with 500 mM NaCl, TEL buffer (0.25M LiCl, 10 mM Tris–HCl pH 8.0, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate), twice with 1XTE. Dynabeads from the three IPs per replicate and three no antibody controls (one from each replicate) were combined during the final wash. Chromatin fragments were eluted using 200 μl of Elution Buffer (TE + 1% SDS) at 65°C 20 min with agitation. NaCl was added to a final concentration of 250mM, and crosslinks were reversed overnight at 65°C. Elutions were treated with RNAse 1hr at 37°C and proteinase K for 2hr at 55°C. DNA was purified by running the elutions through a Qiagen PCR purification column.
Libraries were prepared with a Sciclone liquid-handling robot using the KAPA HTP Library Prep Kit for Illumina (Roche KK8234) and Bio Scientific NEXTflex DNA barcodes (PerkinElmer). NEXTflex DNA barcode adapters were used at a dilution of 1:125 with 15 cycles of PCR. The resulting libraries were then quantified using a Bioanalyzer (Agilent Technologies) and a Qubit fluorometer (Life Technologies). Libraries were pooled and sequenced on an Illumina NextSeq 500 instrument as High Output 75 bp single read runs. Following sequencing, Illumina Primary Analysis version RTA v2.4.11 was run to demultiplex reads for all libraries and generate FASTQ files. Reads were aligned to version sacCer3 of the yeast genome from UC Santa Cruz using bowtie2 v2.4.2. Concordant alignments were analyzed in R using Bioconductor to generate coverage and normalize the data in rpm. Locations of enrichment for each protein were identified using MACS2 version 2.1.2. ChIP calibration was performed as previously described.49,50 The boxplot was constructed using the top 50th percentile of peaks by P-value quantified by the mean RPM value in a 100 base window at the peak center and peaks within 1 kilobase of transcription start sites were mapped to genes by biotype. Metagene plots were constructed by converting coverage of the Scc2 ChIP into rpm per base and then extracting out coverage for a given set of genes, defined as a range of base pairs upstream and downstream of the transcription start site (TSS), and plotting the average of the gene set at each base pair. Data has been deposited at GEO: GSE201194.
Quantitative ChIP
Chromatin immunoprecipitation was performed as previously described.20 Cells were grown in medium lacking methionine, arrested 1.5 hour with alpha-factor, and shifted to YPD medium containing IAA in addition to alpha-factor for 2 hours, before release to a nocodazole-imposed mitotic arrest. Cohesin levels were assessed by ChIP against HA tagged Scc1 followed by quantitative real-time PCR at three chromosome arm and two centromere cohesin binding sites and a negative control site.
Sister chromatid cohesion assay
Yeast strains containing either wild type MED14 or MED14-AID∗, copper inducible lacI-GFP and lacO repeat arrays inserted at either PERICEN, TEL, or ARM sites. Cells were grown in YPD + 200μM Cu2SO4, arrested in G1 with alpha-factor, IAA added, and released into nocodazole-imposed mitotic arrest. Cells were fixed briefly in 70% ethanol, DAPI stained, and imaged using a Nikon widefield microscope. Z stacks with 15 images at 0.1μm intervals were acquired and merged by maximum intensity projection. Percentage of cells showing 2 separate GFP foci was quantified using ImageJ.
Quantification and statistical analysis
For the Scc2 mass spectrometry the dNSAF log2FoldChange and the p-value between the Scc2-Myc purifications and negative controls acquired in parallel were calculated with the linear model of STARTApp package.78 After applying log2FoldChange >= 1 and p-value <= 0.05 as filtering criteria for proteins significantly enriched in the Scc2-Myc purifications, the gene subset was used for GO enrichment through enrichGO function in the R package ReactomePA.79
DNA from the quantitative ChIP was quantified using the PowerUP SYBR Green Master Mix (ThermoFisher) and a Quant Studio 12K Real-Time PCR System (Thermo Fisher).
For the sister chromatid cohesion assay means and SEM of three independent experiments were used. Percentage of cells showing 2 separate GFP foci was quantified using ImageJ. A minimum of 100 cells were scored for each replicate. Exact number of cells counted (n) is indicated in the figure legend. For statistical analysis, a two-way ANOVA test was performed within GraphPad Prism. ∗∗∗∗ indicates p value <.0001.
Acknowledgments
We thank the Molecular Biology, Bioinformatics, Microscopy, and Media Prep facilities at the Stowers Institute, Kansas City, MO, USA for their assistance. We thank Dan Bradford for his assistance in performing the genomic tiling vector screen. We thank the members of the Gerton Lab at the Stowers Institute for their advice and discussions. For technical support, we thank Cindy Maddera and Rhonda Egidy. The graphical abstract for this manuscript was created with BioRender.com. This study was supported by the Stowers Institute for Medical Research. F.U. received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 670412) and the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001198), the UK Medical Research Council (FC001198), and the Wellcome Trust (FC001198). S.M. was supported by an EMBO long-term fellowship.
Author contributions
M.M. and J.L.G. conceived the study. M.M. carried out the characterization of Scc2 mutants, suppressor screen, Scc2 mutant RNA-seq, analysis of Scc2 proteomics, cohesion assay, and Scc2 ChIP-seq experiments. C.S. performed the computational analysis for the RNA-seq and ChIP-seq experiments. S.M. performed the tethering experiment and cohesin ChIP experiment. Y.H., Y.Z., Z.W., and L.F. performed the Scc2 proteomic experiments. S.M. and F.U. provided discussion and suggestions for writing the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2022.05.019.
Supplemental information
Data and code availability
-
•
RNA-seq data and ChIP-seq data have been deposited at GEO and are publicly available as of the date of publication. All mass spectrometry files have been deposited to the ProteomeXchange via the MassIVE repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original images and data underlying this manuscript can be accessed from the Stowers Original Data Repository (ODR). The ODR link is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Haering C.H., Löwe J., Hochwagen A., Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 2.Hirano M., Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melby T.E., Ciampaglio C.N., Briscoe G., Erickson H.P. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gligoris T.G., Scheinost J.C., Bürmann F., Petela N., Chan K.L., Uluocak P., Beckouët F., Gruber S., Nasmyth K., Löwe J. Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science. 2014;346:963–967. doi: 10.1126/science.1256917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruber S., Haering C.H., Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 6.Huis in ’t Veld P.J., Herzog F., Ladurner R., Davidson I.F., Piric S., Kreidl E., Bhaskara V., Aebersold R., Peters J.M. Characterization of a DNA exit gate in the human cohesin ring. Science. 2014;346:968–972. doi: 10.1126/science.1256904. [DOI] [PubMed] [Google Scholar]
- 7.Roig M.B., Löwe J., Chan K.L., Beckouët F., Metson J., Nasmyth K. Structure and function of cohesin's Scc3/SA regulatory subunit. FEBS Lett. 2014;588:3692–3702. doi: 10.1016/j.febslet.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson D.E., Losada A., Erickson H.P., Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arumugam P., Gruber S., Tanaka K., Haering C.H., Mechtler K., Nasmyth K. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Weitzer S., Lehane C., Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A., Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 12.Krantz I.D., McCallum J., DeScipio C., Kaur M., Gillis L.A., Yaeger D., Jukofsky L., Wasserman N., Bottani A., Morris C.A., et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mannini L., Cucco F., Quarantotti V., Krantz I.D., Musio A. Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome. Hum. Mutat. 2013;34:1589–1596. doi: 10.1002/humu.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao W.C., Murayama Y., Muñoz S., Costa A., Uhlmann F., Singleton M.R. Structural studies reveal the functional modularity of the Scc2-Scc4 cohesin loader. Cell Rep. 2015;12:719–725. doi: 10.1016/j.celrep.2015.06.071. [DOI] [PubMed] [Google Scholar]
- 15.Hinshaw S.M., Makrantoni V., Kerr A., Marston A.L., Harrison S.C. Structural evidence for Scc4-dependent localization of cohesin loading. eLife. 2015;4:e06057. doi: 10.7554/eLife.06057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi S., Borek D.M., Otwinowski Z., Tomchick D.R., Yu H. Crystal structure of the cohesin loader Scc2 and insight into cohesinopathy. Proc. Natl. Acad. Sci. USA. 2016;113:12444–12449. doi: 10.1073/pnas.1611333113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi T.S., Basu A., Bermudez V., Hurwitz J., Walter J.C. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008;22:1894–1905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murayama Y., Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014;505:367–371. doi: 10.1038/nature12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Serra L., Kelly G., Patel H., Stewart A., Uhlmann F. The Scc2-Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat. Genet. 2014;46:1147–1151. doi: 10.1038/ng.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muñoz S., Minamino M., Casas-Delucchi C.S., Patel H., Uhlmann F. A role for chromatin remodeling in cohesin loading onto chromosomes. Mol. Cell. 2019;74:664–673.e5. doi: 10.1016/j.molcel.2019.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boginya A., Detroja R., Matityahu A., Frenkel-Morgenstern M., Onn I. The chromatin remodeler Chd1 regulates cohesin in budding yeast and humans. Sci. Rep. 2019;9:8929. doi: 10.1038/s41598-019-45263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Luis J., Lazar-Stefanita L., Gutierrez-Escribano P., Thierry A., Cournac A., García A., González S., Sánchez M., Jarmuz A., Montoya A., et al. FACT mediates cohesin function on chromatin. Nat. Struct. Mol. Biol. 2019;26:970–979. doi: 10.1038/s41594-019-0307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinshaw S.M., Makrantoni V., Harrison S.C., Marston A.L. The kinetochore receptor for the cohesin loading complex. Cell. 2017;171:72–84.e13. doi: 10.1016/j.cell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng G., Kanchwala M., Xing C., Yu H. MCM2-7-dependent cohesin loading during S phase promotes sister-chromatid cohesion. eLife. 2018;7:e33920. doi: 10.7554/eLife.33920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S., et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y.J., Björklund S., Li Y., Sayre M.H., Kornberg R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 27.Koleske A.J., Young R.A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 28.Dotson M.R., Yuan C.X., Roeder R.G., Myers L.C., Gustafsson C.M., Jiang Y.W., Li Y., Kornberg R.D., Asturias F.J. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. USA. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knuesel M.T., Meyer K.D., Bernecky C., Taatjes D.J. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson P.J., Trnka M.J., Pellarin R., Greenberg C.H., Bushnell D.A., Davis R., Burlingame A.L., Sali A., Kornberg R.D. Molecular architecture of the yeast Mediator complex. eLife. 2015;4:e08719. doi: 10.7554/eLife.08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muto A., Ikeda S., Lopez-Burks M.E., Kikuchi Y., Calof A.L., Lander A.D., Schilling T.F. Nipbl and mediator cooperatively regulate gene expression to control limb development. PLoS Genet. 2014;10:e1004671. doi: 10.1371/journal.pgen.1004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chereji R.V., Bharatula V., Elfving N., Blomberg J., Larsson M., Morozov A.V., Broach J.R., Björklund S. Mediator binds to boundaries of chromosomal interaction domains and to proteins involved in DNA looping, RNA metabolism, chromatin remodeling, and actin assembly. Nucleic Acids Res. 2017;45:8806–8821. doi: 10.1093/nar/gkx491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zakari M., Trimble Ross R., Peak A., Blanchette M., Seidel C., Gerton J.L. The SMC loader Scc2 promotes ncRNA biogenesis and translational fidelity. PLOS Genet. 2015;11:e1005308. doi: 10.1371/journal.pgen.1005308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao W.C., Murayama Y., Muñoz S., Jones A.W., Wade B.O., Purkiss A.G., Hu X.W., Borg A., Snijders A.P., Uhlmann F., Singleton M.R. Structure of the cohesin loader Scc2. Nat. Commun. 2017;8:13952. doi: 10.1038/ncomms13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higashi T.L., Eickhoff P., Sousa J.S., Locke J., Nans A., Flynn H.R., Snijders A.P., Papageorgiou G., O'Reilly N., Chen Z.A., et al. A structure-based mechanism for DNA entry into the cohesin ring. Mol. Cell. 2020;79:917–933.e9. doi: 10.1016/j.molcel.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurokawa Y., Murayama Y. DNA binding by the Mis4(Scc2) loader promotes topological DNA entrapment by the cohesin ring. Cell Rep. 2020;33:108357. doi: 10.1016/j.celrep.2020.108357. [DOI] [PubMed] [Google Scholar]
- 37.Collier J.E., Lee B.G., Roig M.B., Yatskevich S., Petela N.J., Metson J., Voulgaris M., Gonzalez Llamazares A., Löwe J., Nasmyth K.A. Transport of DNA within cohesin involves clamping on top of engaged heads by Scc2 and entrapment within the ring by Scc3. eLife. 2020;9:e59560. doi: 10.7554/eLife.59560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cevher M.A., Shi Y., Li D., Chait B.T., Malik S., Roeder R.G. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol. 2014;21:1028–1034. doi: 10.1038/nsmb.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richart L., Mary-Loup W.,M., Macario M., Aflaki S., Salvador M.A., Wicinski J., Chevrier V., Le Cam S., Kamhawi H.A., et al. 2021. Loss of XIST impairs human mammary stem cell differentiation and increases tumorigenicity Through enhancer and mediator complex hyperactivation. Available at SSRN. [DOI] [PubMed] [Google Scholar]
- 40.Litwin I., Bakowski T., Maciaszczyk-Dziubinska E., Wysocki R. The LSH/HELLS homolog Irc5 contributes to cohesin association with chromatin in yeast. Nucleic Acids Res. 2017;45:6404–6416. doi: 10.1093/nar/gkx240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samora C.P., Saksouk J., Goswami P., Wade B.O., Singleton M.R., Bates P.A., Lengronne A., Costa A., Uhlmann F. Ctf4 links DNA replication with sister chromatid cohesion establishment by recruiting the Chl1 helicase to the replisome. Mol. Cell. 2016;63:371–384. doi: 10.1016/j.molcel.2016.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borges V., Smith D.J., Whitehouse I., Uhlmann F. An Eco1-independent sister chromatid cohesion establishment pathway in S. cerevisiae. Chromosoma. 2013;122:121–134. doi: 10.1007/s00412-013-0396-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudra S., Skibbens R.V. Chl1 DNA helicase regulates Scc2 deposition specifically during DNA-replication in Saccharomyces cerevisiae. PLoS One. 2013;8:e75435. doi: 10.1371/journal.pone.0075435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen D., Skibbens R.V. Chl1 DNA helicase and Scc2 function in chromosome condensation through cohesin deposition. PLoS One. 2017;12:e0188739. doi: 10.1371/journal.pone.0188739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Litwin I., Bakowski T., Szakal B., Pilarczyk E., Maciaszczyk-Dziubinska E., Branzei D., Wysocki R. Error-free DNA damage tolerance pathway is facilitated by the Irc5 translocase through cohesin. EMBO J. 2018;37:e98732. doi: 10.15252/embj.201798732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D'Ambrosio C., Schmidt C.K., Katou Y., Kelly G., Itoh T., Shirahige K., Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J.S., He X., Liu J., Duan Z., Kim T., Gerard J., Kim B., Pillai M.M., Lane W.S., Noble W.S., et al. Systematic proteomics of endogenous human cohesin reveals an interaction with diverse splicing factors and RNA-binding proteins required for mitotic progression. J. Biol. Chem. 2019;294:8760–8772. doi: 10.1074/jbc.RA119.007832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minamino M., Higashi T.L., Bouchoux C., Uhlmann F. Topological in vitro loading of the budding yeast cohesin ring onto DNA. Life Sci. Alliance. 2018;1:e201800143. doi: 10.26508/lsa.201800143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu B., Petela N., Kurze A., Chan K.L., Chapard C., Nasmyth K. Biological chromodynamics: a general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic Acids Res. 2015;43:e132. doi: 10.1093/nar/gkv670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makrantoni V., Robertson D., Marston A.L. Analysis of the chromosomal localization of yeast SMC complexes by chromatin immunoprecipitation. Methods Mol. Biol. 2019;2004:119–138. doi: 10.1007/978-1-4939-9520-2_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Misulovin Z., Schwartz Y.B., Li X.Y., Kahn T.G., Gause M., MacArthur S., Fay J.C., Eisen M.B., Pirrotta V., Biggin M.D., Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuin J., Franke V., van Ijcken W.F., van der Sloot A., Krantz I.D., van der Reijden M.I., Nakato R., Lenhard B., Wendt K.S. A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet. 2014;10:e1004153. doi: 10.1371/journal.pgen.1004153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J., Hsu J.M., Laurent B.C. The RSC nucleosome-remodeling complex is required for cohesin’s association with chromosome arms. Mol. Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 54.Shimada K., Gasser S.M. The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell. 2007;128:85–99. doi: 10.1016/j.cell.2006.11.045. [DOI] [PubMed] [Google Scholar]
- 55.Hsieh Y.P., Makrantoni V., Robertson D., Marston A.L., Murray A.W. Evolutionary repair: changes in multiple functional modules allow meiotic cohesin to support mitosis. PLoS Biol. 2020;18:e3000635. doi: 10.1371/journal.pbio.3000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Z., McCrosky S., Guo W., Li H., Gerton J.L. A genetic screen to discover pathways affecting cohesin function in Schizosaccharomyces pombe identifies chromatin effectors. G3 (Bethesda) 2012;2:1161–1168. doi: 10.1534/g3.112.003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fumasoni M., Murray A.W. Ploidy and recombination proficiency shape the evolutionary adaptation to constitutive DNA replication stress. PLoS Genet. 2021;17:e1009875. doi: 10.1371/journal.pgen.1009875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lebraud E., Pinna G., Siberchicot C., Depagne J., Busso D., Fantini D., Irbah L., Robeska E., Kratassiouk G., Ravanat J.L., et al. Chromatin recruitment of OGG1 requires cohesin and mediator and is essential for efficient 8-oxoG removal. Nucleic Acids Res. 2020;48:9082–9097. doi: 10.1093/nar/gkaa611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quevedo M., Meert L., Dekker M.R., Dekkers D.H.W., Brandsma J.H., van den Berg D.L.C., Ozgür Z., van IJcken W.F.J., Demmers J., Fornerod M., Poot R.A. Mediator complex interaction partners organize the transcriptional network that defines neural stem cells. Nat. Commun. 2019;10:2669. doi: 10.1038/s41467-019-10502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanborn A.L., Yeh B.T., Feigerle J.T., Hao C.V., Townshend R.J., Lieberman Aiden E., Dror R.O., Kornberg R.D. Simple biochemical features underlie transcriptional activation domain diversity and dynamic, fuzzy binding to Mediator. eLife. 2021;10:e68068. doi: 10.7554/eLife.68068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tourigny J.P., Saleh M.M., Schumacher K., Devys D., Zentner G.E. Mediator is essential for small nuclear and nucleolar RNA transcription in yeast. Mol. Cell. Biol. 2018;38 doi: 10.1128/MCB.00296-18. e00296–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi H., Takigawa I., Watanabe M., Anwar D., Shibata M., Tomomori-Sato C., Sato S., Ranjan A., Seidel C.W., Tsukiyama T., et al. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat. Commun. 2015;6:5941. doi: 10.1038/ncomms6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hakimi M.A., Bochar D.A., Schmiesing J.A., Dong Y., Barak O.G., Speicher D.W., Yokomori K., Shiekhattar R. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 64.Lemieux K., Gaudreau L. Targeting of Swi/Snf to the yeast GAL1 UAS G requires the Mediator, TAF IIs, and RNA polymerase II. EMBO J. 2004;23:4040–4050. doi: 10.1038/sj.emboj.7600416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin J.J., Lehmann L.W., Bonora G., Sridharan R., Vashisht A.A., Tran N., Plath K., Wohlschlegel J.A., Carey M. Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev. 2011;25:2198–2209. doi: 10.1101/gad.17554711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma V.M., Li B., Reese J.C. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev. 2003;17:502–515. doi: 10.1101/gad.1039503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kremer S.B., Kim S., Jeon J.O., Moustafa Y.W., Chen A., Zhao J., Gross D.S. Role of Mediator in regulating Pol II elongation and nucleosome displacement in Saccharomyces cerevisiae. Genetics. 2012;191:95–106. doi: 10.1534/genetics.111.135806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nock A., Ascano J.M., Barrero M.J., Malik S. Mediator-regulated transcription through the +1 nucleosome. Mol. Cell. 2012;48:837–848. doi: 10.1016/j.molcel.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parenti I., Teresa-Rodrigo M.E., Pozojevic J., Ruiz Gil S., Bader I., Braunholz D., Bramswig N.C., Gervasini C., Larizza L., Pfeiffer L., et al. Mutations in chromatin regulators functionally link Cornelia de Lange syndrome and clinically overlapping phenotypes. Hum. Genet. 2017;136:307–320. doi: 10.1007/s00439-017-1758-y. [DOI] [PubMed] [Google Scholar]
- 70.Gervasini C., Picinelli C., Azzollini J., Rusconi D., Masciadri M., Cereda A., Marzocchi C., Zampino G., Selicorni A., Tenconi R., et al. Genomic imbalances in patients with a clinical presentation in the spectrum of Cornelia de Lange syndrome. BMC Med. Genet. 2013;14:41. doi: 10.1186/1471-2350-14-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Risheg H., Graham J.M., Jr., Clark R.D., Rogers R.C., Opitz J.M., Moeschler J.B., Peiffer A.P., May M., Joseph S.M., Jones J.R., et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat. Genet. 2007;39:451–453. doi: 10.1038/ng1992. [DOI] [PubMed] [Google Scholar]
- 72.Schwartz C.E., Tarpey P.S., Lubs H.A., Verloes A., May M.M., Risheg H., Friez M.J., Futreal P.A., Edkins S., Teague J., et al. The original Lujan syndrome family has a novel missense mutation (p.N1007S) in the MED12 gene. J. Med. Genet. 2007;44:472–477. doi: 10.1136/jmg.2006.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spaeth J.M., Kim N.H., Boyer T.G. Mediator and human disease. Semin. Cell Dev. Biol. 2011;22:776–787. doi: 10.1016/j.semcdb.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vulto-van Silfhout A.T., de Vries B.B., van Bon B.W., Hoischen A., Ruiterkamp-Versteeg M., Gilissen C., Gao F., van Zwam M., Harteveld C.L., van Essen A.J., et al. Mutations in MED12 cause X-linked Ohdo syndrome. Am. J. Hum. Genet. 2013;92:401–406. doi: 10.1016/j.ajhg.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu T., Park S.K., Venable J.D., Wohlschlegel J.A., Diedrich J.K., Cociorva D., Lu B., Liao L., Hewel J., Han X., et al. ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tabb D.L., McDonald W.H., Yates J.R., 3rd. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y., Wen Z., Washburn M.P., Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 78.Nelson J.W., Sklenar J., Barnes A.P., Minnier J. The START App: a web-based RNAseq analysis and visualization resource. Bioinformatics. 2017;33:447–449. doi: 10.1093/bioinformatics/btw624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu G., He Q.Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA-seq data and ChIP-seq data have been deposited at GEO and are publicly available as of the date of publication. All mass spectrometry files have been deposited to the ProteomeXchange via the MassIVE repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original images and data underlying this manuscript can be accessed from the Stowers Original Data Repository (ODR). The ODR link is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.