Abstract
Aims
The progesterone response of the nuclear progesterone receptor plays an important role in the female reproductive system. Changes in the function of the progesterone receptor gene may increase the risk of reproductive cancer. The present study performed a meta-analysis to examine whether the progesterone receptor gene PROGINS polymorphism was a susceptibility factor for female reproductive cancer.
Materials and methods
We searched the PubMed, Cochrane Library, Web of Science and EMBASE databases for literature on PROGINS polymorphisms and female reproductive cancer published before September 2020. We evaluated the risk using odds ratios [ORs] and 95% confidence intervals via fixed effects models and random-effects models, which were calculated for all five genetic models. We grouped the analyses by race, cancer, and HWE.
Results
Thirty studies comprised of 25405 controls and 19253 female reproductive cancer cases were included in this meta-analysis. We observed that the Alu insertion polymorphism and the V660L polymorphism were significantly associated with female reproductive cancer in the allele and dominant genetic models. The allele genetic model and (Alu-insertion polymorphism: OR = 1.22, 95% CI = 1.02–1.45; V660L polymorphism: OR = 1.02, 95% CI = 1.00–1.13) dominant genetic model (Alu-insertion polymorphism: OR = 1.27, 95% CI = 1.03–1.58; V660L polymorphism: OR = 1.10, 95% CI = 1.011.19) demonstrated a significantly increased risk of female reproductive cancer. A subgroup analysis according to ethnicity found that the Alu insertion was associated with female reproductive cancer incidence in white (Allele model: OR = 1.21, 95% CI = 1.00–1.45; Heterozygous model: OR = 3.44, 95% CI = 1.30–9.09) and Asian (Dominant model: OR = 3.12, 95% CI = 1.25–7.79) populations, but the association disappeared for African and mixed racial groups. However, the V660L polymorphism was significantly associated with female reproductive cancer in the African (Allele model: OR = 2.52, 95% CI = 1.14–5.56; Heterozygous model: OR = 2.83, 95% CI = 1.26–6.35) and mixed racial groups (Dominant model: OR = 1.28, 95% CI = 1.01–1.62). Subgroup analysis by cancer showed that the PROGINS polymorphism increased the risk of cancer in the allele model, dominant mode and heterozygous model, but the confidence interval for this result spanned 1 and was not statistically significant. This sensitivity was verified in studies with HWE greater than 0.5.
Conclusion
Our meta-analysis showed that the progesterone receptor gene Alu insertion and the V660L polymorphism contained in the PROGINS polymorphism were susceptibility factors for female reproductive cancer.
Introduction
Cancer is a major public health problem worldwide. Cancer is a multifactorial disease, and there is a coordinated relationship between genetic and environmental factors [1,2]. Despite extensive research to prevent cancer, cancer cases continue to increase sharply. Data from the American Cancer Society in 2022 predicts 1.9 million new cancer cases in 2022. More than 609,360 Americans die of cancer annually, which is equivalent to greater than 1,700 people dying of cancer daily [3].
Progesterone is a key regulatory factor in the proliferation and differentiation of female reproductive tract cells. Progesterone inhibits the proliferation of reproductive tract cells by excessive estrogen via the progesterone receptor (PgR) [4–6], and excessive estrogen stimulation increases the risk of female reproductive tract cancer [7]. PgR is a member of the nuclear steroid hormone receptor family and is expressed primarily in female reproductive tissues and the central nervous system. It is encoded by a single gene (Gene ID: 5241) located at 11q22–q23 [8], which encodes two isoforms, PgR-B and PgR-A. The two PgR isoforms with different functions come from different transcriptional promoters. PgR-B (114 KDa) is a transcriptional activator and a mediator of cell proliferation, and PgR-A (94 KDa) is a suppressor of transcription. In vitro studies showed that PgR isoforms exhibited different transcriptional regulatory activities. Robert A. et al. [9] found that selective PgR-A knockout induced endometrial epithelial cell proliferation in mice, which suggests that PgR-A is required to control potential adverse reactions of PgR-B. The expression of PgR-A in PgR-B knockout mice was sufficient and necessary to regulate the antiproliferative response of progesterone and estrogen-induced hyperplasia. Prompt changes in the relative expression of these two isoforms or changes in isoform activity or any other genetic mutations may lead to progesterone receptor alienation. The anti-estrogen proliferation effect of progesterone primarily depends on PgR-B, but the excessive expression of PgR-B causes progesterone-dependent proliferation. Progesterone receptor alienation leads to increased susceptibility to female reproductive cancer.
Silencing or mutation of the PgR gene affects the expression of the progesterone receptor. Six variable sites, four polymorphisms, and five common haploids have been detected in the PgR gene. PROGINS contains the Alu insertion in intron 7 of the PgR gene, which is completely linked to the unbalanced linkage (LD) with rs1042838 (V660L in exon 4) and rs1042839 (H770H in exon 5) [10]. The alleles of Alu-insertion alter transcript levels and may contribute to disease risk [11]. The PROGINS polymorphism of the human progesterone receptor diminishes the response to progesterone [12]. The PROGINS allele was significantly associated with decreased serum progesterone levels in patients with polycystic ovary syndrome (PCOS) [13].
The current study considered PROGINS as a risk modifier for gynecological benign and malignant diseases, which indicated that PROGINS may affect PgR function. Alu insertion of the PROGINS allele was inversely associated with breast cancer risk and ovarian cancer risk in certain races [14–16]. However, only two research reports that concluded that PROGINS affected the risk of endometrial cancer [17,18]. The V660L polymorphism is caused by G > T, which causes a valine > leucine substitution in the fourth exon of the PgR gene. No significant association of the PROGINS polymorphism was found in breast or ovarian cancer studies [19,20]. One study on ovarian cancer [21] also failed to find a link, but another study showed that the T allele (leucine) was associated with an increased risk of breast cancer [22]. However, the results of these studies are inconclusive. Therefore, to clarify the role of the PRPGINS PgR polymorphism in female reproductive cancers, we performed a meta-analysis of all eligible case–control studies to derive the overall cancer risk associated with this polymorphism.
Materials and methods
The current study conformed to the checklist for meta-analysis of genetic association studies specified for the PLOS One approach (S1 Table).
Literature search and identification
This meta-analysis adhered to the PRISMA guidelines. PubMed, Cochrane Library, Web of Science and EMBASE were used to perform a comprehensive search of published related documents. The following search keywords were used: “polymorphism, genetic” or “breast cancer” or “ovarian cancer” or “endometrial cancer” or “gynecologic neoplasm” and “PROGINS” or “V660 L” or “rs1042838” or “rs1042839” or “H770H” or “Z49816.1” or “Alu-insertion”. A search strategy was developed (S2). The last search was updated on September 26, 2020.
Inclusion and exclusion criteria
The studies were selected using the following criteria.
The following inclusion criteria were used: (a) case-control or cohort study; (b) assessment of PgR polymorphisms for PROGINS and cancer risk; (c) pathology for diagnosis of cancer patients and confirmation that the control was cancer-free; (d) report odds ratios (ORs) and 95% confidence interval (CIs) values or sufficient data to calculate these values; (e) clearly describe genotyping and statistical methods; (f) participants in the control group were in Hardy-Weinberg Balance (HWE); and (g) no language limitations, regardless of sample size.
The following exclusion criteria were used: (a) case reports, comments, comments, and editorial articles; (b) studies of research progress, severity, treatment response, or survival; (c) when overlapping data from the same case series were included in multiple publications, the most recent or most complete study was selected to perform the meta-analysis, and if no information was available, the study was excluded; and (d) literature with specific requirements for the inclusion of cases or. Any differences in the inclusion of the study were resolved via discussion and subsequent consensus.
Data extraction
Two authors independently extracted the characteristics of the selected study using a standardized protocol, and the third investigator reviewed the results. The following information was extracted from each study: first author, year of publication, study population (country, ethnicity), type of cancer, number of cases and controls, genotype frequencies for cases and controls, and Hardy-Weinberg equilibrium in controls (HWE). We compared key research characteristics, such as location, study time, and authorship, to determine the existence of multiple publications in the same study.
Quality assessment of the studies
We evaluated the quality based on the NOS quality evaluation to determine the quality of the included literature, and low-quality articles with less than 3 points were excluded. Chen Zhou and Xiangman Zou independently performed the literature search and data extraction. Disputes were discussed and resolved by Xiaosha Wen and Zifen Guo.
Statistical analysis
STATA software (version 14.0) was used to synthesize the relevant data, and the odds ratio (OR) and 95% confidence interval (CI) were used to evaluate the relationships between PROGINS gene polymorphisms and female reproductive cancer. Five genetic models were used: T2 vs. T1 (allelic), T1T2+T2T2 vs. T1T1 (dominant), T1T2 vs. T1T1 (heterozygous), T2T2 vs. T1T2+T1T1 (recessive), and T2T2 vs. T1T1 (homozygous). Heterogeneity was evaluated using I2 statistics. When the heterogeneity test found significant heterogeneity (I2 > 50 or P < 0.05), a random-effect model was used. Otherwise, the fixed model was used. When heterogeneity was present in the study, subgroup analysis was performed according to ethnicity, type of disease and HWE of the included cases to examine the sources of heterogeneity. Sensitivity analysis (excluding one study at a time or changing the model) was used to assess the stability of each efficacy index. Begg’s funnel chart and Egger’s test were used to evaluate the publication bias of this study. When P < 0.05, publication bias was present in this study.
Results
Study selection and characteristics
Fig 1 outlines the study selection process in a flowchart following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. A total of 189 articles related to PROGINS polymorphism were retrieved using the retrieval method. Among these articles, 139 articles were excluded after review of the abstracts and unrelated literature, and 11 articles were excluded strictly according to the inclusion and exclusion criteria. Ultimately, 30 articles were included in the meta-analysis (Fig 1) [8,18,19,21–47]. Of the 30 independent studies, 28 studies included genetic frequency analysis of whites, 2 studies included mixed races, 2 studies included Asians and 2 studies included Africans. The types of diseases included breast cancer, ovarian cancer and endometrial cancer. All samples were taken from humans and genotyped using polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP), DNA sequencing, TaqMan and other genotyping methods (Table 1). The quality of the studies is shown in Table 2.
Fig 1. Flowchart showing the meta-analysis literature screening process.
Table 1. Characteristics of the studies included in the meta-analysis.
| Study | Country | Ethnicity | Cancer | Detection method | Sample size (case/control) |
Genotype frequency | Allele frequency | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| case | controls | Case (%) | Control (%) | ||||||||||||
| Alu insertion | T1T1 | T1T2 | T2T2 | T1T1 | T1T2 | T2T2 | T1 | T2 | T1 | T2 | |||||
| Albalawi, I. A | Saudi Arabia | Asian | BC | PCR–RFLP | 100/100 | 81 | 18 | 1 | 93 | 6 | 1 | 10 | 90 | 4 | 96 |
| Donaldson, C J | USA | white | BC | PCR–RFLP | 23/60 | 17 | 5 | 1 | 41 | 16 | 3 | 84.4 | 15.2 | 81.3 | 18.3 |
| Donaldson, C J -2 | USA | African | BC | PCR–RFLP | 61/81 | 56 | 5 | 0 | 73 | 8 | 0 | 95.9 | 4.1 | 95.1 | 4.9 |
| Govindan, S | India | white | BC | PCR–RFLP | 157/108 | 134 | 23 | 0 | 102 | 6 | 0 | 92.7 | 7.3 | 97.2 | 2.8 |
| Junqueira, M. G | Brazil | white | EC | PCR–RFLP | 282/121 | 221 | 61 | 0 | 100 | 18 | 3 | 89.2 | 10.8 | 90.1 | 9.9 |
| Lancaster, J. M -2 | USA | white | BC | PCR–RFLP | 68/101 | 55 | 12 | 1 | 79 | 18 | 4 | 89.7 | 10.3 | 87.1 | 12.9 |
| Lancaster, J. M | USA | white | OC | PCR–RFLP | 309/397 | 219 | 80 | 10 | 285 | 95 | 17 | 83.8 | 16.2 | 83.8 | 16.2 |
| Leite, D.B | Brazil | white | OC | PCR–RFLP | 80/282 | 57 | 12 | 11 | 221 | 61 | 0 | 78.8 | 21.2 | 89.2 | 10.8 |
| Manolitsas, T. P | UK | white | BC | PCR–RFLP | 292/220 | 229 | 61 | 2 | 162 | 54 | 4 | 88.9 | 11.1 | 85.9 | 14.1 |
| Manolitsas, T. P -2 | UK | white | OC | PCR | 231/220 | 173 | 52 | 6 | 162 | 54 | 4 | 86.1 | 13.9 | 85.9 | 14.1 |
| McKenna, N. J | Ireland | white | OC | S-blot | 41/83 | 26 | 15 | 0 | 58 | 21 | 4 | 81.7 | 18.3 | 82.5 | 17.5 |
| McKenna, N. J-2 | Germany | white | OC | S-blot | 26/101 | 17 | 8 | 1 | 88 | 12 | 1 | 80.8 | 19.2 | 93.1 | 6.9 |
| Patricia, G. A | Mexico | white | BC | PCR | 481/209 | 360 | 103 | 18 | 176 | 33 | 0 | 85.6 | 14.4 | 92.1 | 7.9 |
| Runnebaum, I. B | USA | white | OC | PCR | 167/496 | 101 | 60 | 6 | 328 | 153 | 15 | 78.4 | 21.6 | 81.6 | 18.4 |
| Surekha, S | India | white | BC | PCR | 250/249 | 241 | 7 | 2 | 242 | 7 | 0 | 97.8 | 2.2 | 98.6 | 1.4 |
| V660L | GG | GT | TT | GG | GT | TT | G | T | G | T | |||||
| Gabriel, C. A | USA | white | BC | TaqMan | 346/357 | 236 | 101 | 9 | 255 | 92 | 10 | 82.8 | 17.2 | 84.3 | 15.7 |
| Gabriel, C. A -2 | USA | African | BC | TaqMan | 86/327 | 75 | 11 | 0 | 309 | 16 | 2 | 93.6 | 6.4 | 96.9 | 3.1 |
| Clendenen, T | Mix | white | BC | TaqMan | 658/1099 | 846 | 288 | 26 | 1516 | 523 | 54 | 85.5 | 14.5 | 84.9 | 15.1 |
| Fabjani, G | Austria | white | BC | DNA | 155/106 | 119 | 32 | 4 | 78 | 28 | 0 | 87.1 | 12.9 | 86.8 | 13.2 |
| Fernandez, L.P | Spain | white | BC | TaqMan | 550/564 | 354 | 153 | 24 | 375 | 154 | 15 | 81.1 | 18.9 | 83.1 | 16.9 |
| Johnatty, S.E | Australia | white | BC | PCR–RFLP | 1444/583 | 1017 | 380 | 47 | 409 | 160 | 14 | 83.6 | 16.4 | 83.9 | 16.1 |
| Lee, E | USA | MIX | EC | TaqMan | 198/1077 | 170 | 25 | 3 | 954 | 114 | 9 | 92.2 | 7.8 | 93.9 | 6.1 |
| Lee, E -2 | USA | white | EC | TaqMan | 379/836 | 259 | 109 | 11 | 615 | 199 | 22 | 86 | 14 | 90.2 | 9.8 |
| Lundin, E | MIX | white | EC | TaqMan | 391/705 | 281 | 96 | 14 | 540 | 147 | 18 | 84.1 | 15.9 | 87 | 13 |
| O’Mara, T. A | Singapore | Asian | EC | TaqMan | 528/1538 | 414 | 151 | 17 | 1147 | 361 | 30 | 84.1 | 15.9 | 86.3 | 13.7 |
| O’Mara, T. A -2 | UK | white | EC | TaqMan | 1086/1591 | 765 | 294 | 27 | 1123 | 434 | 34 | 84 | 16 | 84.2 | 15.8 |
| O’Mara, T. A -3 | Australia | white | EC | TaqMan | 1220/1354 | 867 | 323 | 30 | 933 | 383 | 38 | 84.3 | 15.7 | 83.1 | 16.9 |
| Pearce, C. L | USA | white | OC | DNA | 267/397 | 173 | 82 | 12 | 279 | 111 | 6 | 80.1 | 19.9 | 84.5 | 15.5 |
| Pearce, C. L-2 | USA | white | BC | DNA | 1715/2505 | 1400 | 252 | 15 | 2025 | 363 | 37 | 91.5 | 8.5 | 91 | 9 |
| Pooley, K. A | Englishman | white | BC | TaqMan | 2345/2281 | 1302 | 517 | 42 | 1461 | 513 | 39 | 83.9 | 16.1 | 85.3 | 14.7 |
| Romano, A | Netherlands | white | BC | PCR–RFLP | 167/31 | 123 | 41 | 3 | 22 | 7 | 2 | 85.9 | 14.1 | 82.3 | 17.7 |
| Romano, A | German | white | BC | PCR–RFLP | 545/443 | 399 | 133 | 14 | 347 | 87 | 9 | 85.4 | 14.6 | 88.1 | 11.9 |
| Romano, A -2 | German | white | OC | PCR–RFLP | 67/443 | 42 | 24 | 1 | 347 | 87 | 9 | 80.6 | 19.4 | 88.1 | 11.9 |
| Spurdle, A. B | Australia | white | OC | PCR–RFLP | 551/298 | 395 | 144 | 12 | 203 | 90 | 5 | 84.8 | 15.2 | 83.2 | 16.8 |
| Terry, K. L | USA | white | OC | TaqMan | 895/939 | 648 | 223 | 25 | 612 | 298 | 29 | 84.9 | 15.1 | 81 | 19 |
| De vivo, I | USA | white | BC | TaqMan | 1252/1660 | 869 | 348 | 35 | 1186 | 434 | 40 | 83.3 | 16.7 | 84.5 | 15.5 |
| Tong, D | Austrian | white | OC | DNA | 226/194 | 167 | 50 | 9 | 141 | 52 | 1 | 85 | 15 | 86.1 | 13.9 |
| Quaye, L | UK/USA | white | OC | TaqMan | 1424/2408 | 1005 | 377 | 42 | 1819 | 526 | 63 | 83.8 | 16.2 | 86.5 | 13.5 |
| Ghali, R. M | Tunisia | white | BC | TaqMan | 183/216 | 127 | 50 | 6 | 172 | 37 | 7 | 83.1 | 16.9 | 88.2 | 11.8 |
PCC, population-based case–control study, HCC, hospital-based case–control study, PCR–RFLP PCR-restriction fragment length polymorphism, BC, Breast cancer, EC, Endometrial cancer, OC, Ovarian cancer, DNA, DNA sequencing, S-blot, Southern blot.
Table 2. Article quality evaluation.
| Study | Adequate case definition | Definition of controls | Comparability | HWE>0.05 | PCC | PMH (Past Medical History) | Unified detection method | Article quality |
|---|---|---|---|---|---|---|---|---|
| Alu insertion | ||||||||
| Albalawi 2020 [47] | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 5 |
| Donaldson 2002 [28] | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 5 |
| Govindan, S 2007 [34] | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 5 |
| Junqueira, M.G 2007 [18] | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 6 |
| Lancaster 1998 [25] | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| Lancaster, J.M 2003 [30] | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 6 |
| Leite, D.B. 2008 [37] | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 5 |
| Manolitsas TP 1997 [24] | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 5 |
| McKenna 1995 [23] | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 3 |
| Patricia Gallegos-Arreola, M 2015 [15] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Runnebaum, I.B 2001 [26] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Surekha 2009 [39] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| V660L | ||||||||
| Gabriel, C. A.2013 [44] | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 6 |
| Clendenen, T 2013 [43] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Fabjani, G 2002 [29] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Fernandez, L.P 2006 [31] | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 6 |
| Johnatty, S.E 2008 [36] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Lee, E 2010 [40] | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 6 |
| Lundin, E 2012 [42] | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 6 |
| O’Mara, T.A 2011 [41] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Pearce, C.L. 2005 [22] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 6 |
| Pooley, K.A 2006 [32] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 6 |
| Romano A 2007 [12] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Romano, A 2006 [33] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 6 |
| Spurdle 2001 [21] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Terry, K.L. 2005 [8] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| De vivo 2004 [19] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 6 |
| Tong, D. 2001 [27] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 6 |
| Quaye 2009 [38] | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 6 |
| Ghali RM 2020 [46] | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 5 |
low quality, <3; Medium quality,3–4; high quality, ≥5.
Alu-insertion polymorphism and the risk of female reproductive cancer
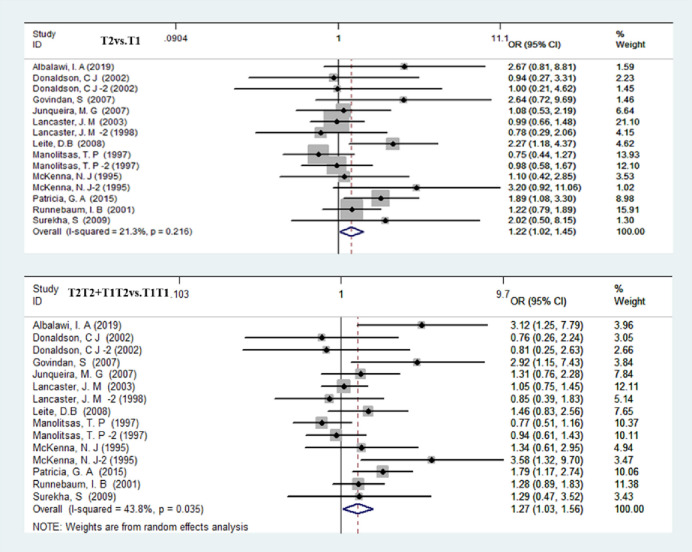
We counted the Alu-insertion locus and the susceptibility to female reproductive cancer in the five models of allele genetic model (T2 vs. T1), homozygous genetic model (T2T2 vs. T1T2), heterozygous genetic model (T1T2 vs. T1T1), dominant genetic model (T1T2+T2T2 vs. T1T1) and recessive genetic models (T2T2 vs. T1T2+T1T1) (Table 3). The meta-analysis showed a significant association between Alu-insertion polymorphisms and the risk of female reproductive cancer in the allele genetic model (OR = 1.22 95% CI = 1.02–1.45), the dominant genetic model (OR = 1.27 95% CI = 1.03–1.58), and the heterozygote genetic model (OR = 1.19 95% CI = 1.03–1.38) (Figs 2 and 3). A significant association was found in the allele genetic model of the white group (OR = 1.21 95% CI = 1.00–1.45) (Table 4).
Table 3. Meta-analysis of the association between the PROGINS polymorphism and female reproductive cancer susceptibility.
| Polymorphism | Genetic model | Case/Control | Test of association | Heterogeneity | Publication bias | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | P | I2(%) | PHet | Model | Egger‘s test p value | Begg’s test p value |
|||
| Alu insertion | T2 vs. T1 | 2568/2828 | 1.22[1.02,1.45] | 0.027 * | 21.3 | 0.218 | F | 0.373 | 0.168 |
| T2T2+T1T2 vs.T1T1 | 2568/2828 | 1.27[1.03,1.58] | 0.023* | 43 | 0.035 | R | 0.373 | 0.201 | |
| T2T2 vs.T1T2+T1T1 | 2568/2828 | 1.18[0.55,2.55] | 0.670 | 52.1 | 0.015 | R | 0.360 | 0.469 | |
| T2T2 vs.T1T1 | 2046/2322 | 1.23[0.57,2.65] | 0.605 | 52.2 | 0.014 | R | 0.428 | 0.455 | |
| T1T2 vs.T1T1 | 2509/2772 | 1.19[1.03,1.38] | 0.019* | 39 | 0.058 | F | 0.428 | 0.263 | |
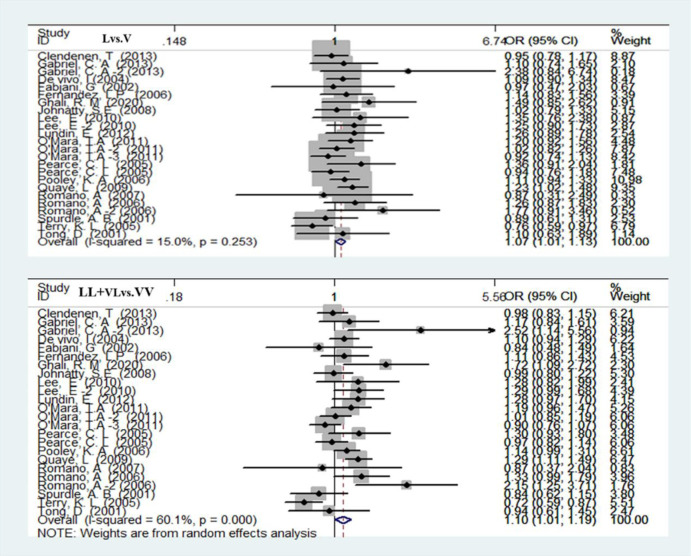
| V660L | L vs. V | 16685/22577 | 1.07[1.00,1.13] | 0.031* | 15 | 0.253 | F | 0.130 | 0.081 |
| LL+VL vs. VV | 16685/22577 | 1.10[1.01,1.19] | 0.027* | 60.1 | 0.000 | R | 0.503 | 0.265 | |
| LL vs. VL+VV | 16685/22577 | 1.13[0.99,1.29] | 0.075 | 0 | 0.476 | F | 0.413 | 0.244 | |
| LL vs. VV | 12481/17361 | 1.07[0.93,1.23] | 0.325 | 5 | 0.392 | F | 0.673 | 0.219 | |
| VL vs. VV | 16257/22084 | 1.09[1.00,1.18] | 0.056 | 60.7 | 0.000 | R | 0.385 | 0.204 | |
F: Fixed model, R: Random model.
Fig 2. Forest plot of overall cancer risk associated with Alu-insertion PgR polymorphism (T2 vs. T1 and T2T2+T1T2vs.T1T1).
Fig 3. Forest plot of overall cancer risk associated with V660L PgR polymorphism (L vs. V and LL+VL vs. VV).
Table 4. Pooled odds ratios (ORs) in subgroups.
| SNP/subgroups | No. of study | Allele model | Dominant model | Recessive model | Homozygous model | Heterozygous model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | ||
| Alu insertion | ||||||||||||||||
| OC | 5 | 1.20 | 0.95–1.51 | 0.125 | 1.23 | 0.96–1.56 | 0.100 | 1.59 | 0.56–4.49 | 0.382 | 1.67 | 0.60–4.69 | 0.330 | 1.13 | 0.93–1.37 | 0.232 |
| BC | 7 | 1.28 | 0.95–1.72 | 0.101 | 1.30 | 0.86–1.97 | 0.219 | 1.14 | 0.32–4.04 | 0.836 | 1.17 | 0.33–4.29 | 0.818 | 1.23 | 0.97–1.55 | 0.086 |
| EC | 1 | 1.08 | 0.53–2.19 | 0.828 | 1.31 | 0.76–2.28 | 0.170 | 0.06 | 0.00–1.17 | 0.063 | 0.06 | 0.00–1.27 | 0.071 | 1.53 | 0.86–2.73 | 0.146 |
| white | 9 | 1.21 | 1.00–1.45 | 0.046* | 1.23 | 0.99–1.54 | 0.066 | 1.40 | 0.62–3.14 | 0.413 | 1.44 | 0.84–3.28 | 0.377 | 1.14 | 0.98–1.33 | 0.099 |
| Asian | 1 | 2.67 | 0.81–8.81 | 0.108 | 3.12 | 1.25–7.79 | 0.015* | 1.00 | 0.06–16.21 | 1 | 1.15 | 0.07–18.65 | 0.923 | 3.44 | 1.30–9.09 | 0.013* |
| African | 1 | 1.00 | 0.21–4.62 | 0.996 | 0.81 | 0.25–2.63 | 0.731 | —— | —— | —— | —— | —— | —— | 0.81 | 0.25–2.63 | 0.731 |
| Mix | 1 | 1.08 | 0.53–2.19 | 0.828 | 1.31 | 0.75–2.28 | 0.329 | 0.06 | 0.00–1.17 | 0.063 | 0.06 | 0.00–1.27 | 0.071 | 1.53 | 0.86–2.73 | 0.019 |
| HWE > 0.05 | 8 | 1.21 | 0.98–1.50 | 0.076 | 1.27 | 0.98–1.65 | 0.066 | 0.85 | 0.38–1.91 | 0.043 | 1.12 | 0.48–2.61 | 0.797 | 1.22 | 1.02–1.45 | 0.026 |
| HWE < 0.05 | 4 | 1.23 | 0.90–1.68 | 0.188 | 1.30 | 0.85–1.98 | 0.230 | 1.85 | 0.22–15.66 | 0.004 | 1.88 | 0.23–15.50 | 0.556 | 1.13 | 0.86–1.48 | 0.379 |
| V660L | ||||||||||||||||
| OC | 6 | 1.06 | 0.94–1.20 | 0.334 | 1.09 | 0.82–1.45 | 0.566 | 1.24 | 0.94–1.64 | 0.126 | 1.19 | 0.89–1.59 | 0.230 | 1.05 | 0.78–1.43 | 0.733 |
| BC | 10 | 1.06 | 0.98–1.15 | 0.139 | 1.09 | 1.00–1.19 | 0.052 | 1.07 | 0.88–1.29 | 0.518 | 1.00 | 0.82–1.22 | 0.979 | 1.09 | 0.99–1.20 | 0.064 |
| EC | 3 | 1.07 | 0.96–1.20 | 0.215 | 1.10 | 0.97–1.26 | 0.137 | 1.16 | 0.90–1.50 | 0.256 | 1.11 | 0.85–1.44 | 0.461 | 1.09 | 0.96–1.23 | 0.195 |
| white | 17 | 1.06 | 0.99–1.12 | 0.081 | 1.07 | 0.98–1.17 | 0.134 | 1.10 | 0.95–1.27 | 0.189 | 1.06 | 0.91–1.22 | 0.471 | 1.06 | 0.97–1.16 | 0.209 |
| Asian | 1 | 1.20 | 0.92–1.56 | 0.186 | 1.19 | 0.96–1.47 | 0.109 | 1.51 | 0.83–2.76 | 0.179 | 1.35 | 0.73–2.53 | 0.341 | 1.16 | 0.93–1.45 | 0.190 |
| African | 1 | 2.38 | 0.84–6.74 | 0.103 | 2.52 | 1.14–5.56 | 0.022* | 0.75 | 0.04–15.82 | 0.855 | 0.29 | 0.02–6.55 | 0.434 | 2.83 | 1.26–6.35 | 0.012* |
| Mix | 2 | 1.35 | 0.76–2.38 | 0.305 | 1.28 | 1.01–1.62 | 0.041* | 1.49 | 0.80–2.79 | 0.212 | 1.23 | 0.65–2.42 | 0.499 | 1.25 | 0.97–1.60 | 0.081 |
| HWE > 0.05 | 15 | 1.04 | 0.97–1.11 | 0.229 | 1.03 | 0.93–1.14 | 0.003 | 1.17 | 1.00–1.37 | 0.46 | 1.15 | 0.98–1.35 | 0.084 | 1.03 | 0.95–1.13 | 0.452 |
| HWE < 0.05 | 6 | 1.16 | 1.02–1.31 | 0.020 | 1.34 | 1.10–1.64 | 0.003 | 1.01 | 0.76–1.29 | 0.975 | 0.85 | 0.64–1.14 | 0.277 | 1.29 | 1.06–1.57 | 0.009 |
Val 660 Leu polymorphism and the risk of female reproductive cancer
A total of 16685 cancer patients and 22577 healthy women in 18 studies were used to assess the relationship between the V660 locus and female reproductive cancer risk using the allele genetic model (L vs. V), homozygous genetic model (LL vs. VV), heterozygous genetic model (VL vs. VV), dominant genetic model (VL+LL vs. VV) and recessive genetic models (LL vs. VL+VV). The V660L mutation increased the risk of female reproductive cancer in the allele genetic model (OR = 1.02 95% CI = 1.00–1.13) and dominant genetic model (OR = 1.10 95% CI = 1.01–1.19). The heterozygote genetic model confirmed (OR = 1.09 95% CI = 1.00–1.18) that the V660L mutation increased the risk of female reproductive cancer (Fig 3 and Table 3).
The subgroup analysis found a significant association under the dominant genetic model (OR = 1.21 95% CI = 1.00–1.45) of the breast cancer group (Table 4).
Publication bias
Begg’s and Egger’s analyses showed that no publication bias in the Alu insertion or V660L (Table 3).
Sensitivity and heterogeneity
A sensitivity analysis was performed to determine whether changes in the inclusion criteria for meta-analysis affected the final results. The author deleted individual studies involved in each meta-analysis to reflect the impact of a single dataset on the merged ORs. Most of the corresponding merged ORs did not change substantially (data not shown). We also changed the effect model to test the impact on the results, and no substantial changes were found on the combined OR, which showed that our results were statistically robust. I2 statistics were used to test the heterogeneity (Table 3), and no heterogeneity was observed in any of the genetic models.
Discussion
Current evidence indicates that progesterone plays a vital role in regulating female reproduction. The physiological role of progesterone is mediated by the progesterone receptor (PgR), which includes a total of 8 exons and 7 introns [48]. PgR-A and PgR-B are the two subtypes of PgR. The co-expression levels in most normal progesterone-targeted cells are similar. The balance between subtypes regulates the expression of many other genes. Abnormal expression of PgR-A or PgR-B causes a significant change in the ratio between subtypes, which leads to changes in the transmission of progesterone information, and these changes affect physiological functions and trigger a series of serious physiological consequences. The Alu insertion together with V660L and H770H is called PROGINS, which is an important polymorphism of the PgR gene. The Alu insertion affects the binding properties of receptors and hormones and induces amino acid changes, which cause female reproductive cancer.
Our meta-analysis included 30 studies with 25405 controls and 19253 female reproductive cancer cases. These studies examined the relationship between PgR gene PROGINS polymorphisms (Alu insertion and V660L) and female reproductive tract cancer. Our meta-analysis results demonstrated a significant association between PROGINS and female reproductive cancer, and PROGINS mutations increased the risk of female reproductive cancer. We also performed a sensitivity analysis to test the validity of the results, and the results of the meta-analysis were stable. The association between PgR mutations and female reproductive cancer varies between races. The meta-analysis of the dominant genetic model of the Alu-insertion polymorphism showed that women with T2 mutations had a significantly higher risk of developing female reproductive tract cancer than women with T1T1 genotypes in the general population. There was a significant association between Alu insertion and female reproductive cancer in whites (OR = 1.25, 95% CI = 1.01–1.56), but this association disappeared in Asians and Africans. The difference in correlation may be caused by several factors. First, the frequency of Alu insertion is different due to different ethnic groups, different ethnic groups of genetic backgrounds, different lifestyles, and different environmental factors. Second, there are few reports of the locus in Asians and Africans.
Although some studies showed linkage disequilibrium reactions between Alu insertions and V660L, V660L cannot replace Alu insertions in the analysis of genetic polymorphisms based on these meta-analysis data. The disease correlation between the two polymorphisms was different between ethnicities. Alu insertion was associated with female reproductive cancer incidence in white (Allele model: OR = 1.21, 95% CI = 1.00–1.45; Heterozygous model: OR = 3.44, 95% CI = 1.30–9.09) and Asian populations (Dominant model: OR = 3.12, 95% CI = 1.25–7.79), but the association disappeared for African and mixed racial groups.
Our results showed a significant relationship between V660L and the susceptibility to female reproductive cancer in the allele genetic model, dominant genetic model and heterozygous genetic model.
However, our research has some potential limitations. First, studies that met the inclusion criteria or were unpublished may have been missed. Second, although the control group was primarily selected from healthy people, some people did not mention their physiological status or whether they had benign disease. Finally, 26 studies included whites in the ethnic subgroup analysis, but few studies included Asians and Africans. Therefore, the differences in the associations between different ethnic subgroups should be carefully interpreted. In conclusion, although there are limitations, the results in this article provide significant evidence that PROGINS increases the risk of female reproductive cancer.
Supporting information
(DOCX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by the Research Fund Project of the Education Bureau of Hunan Province, China (Grant No.19A419) with a grant of CNY 80,000. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hoffjan S. Dissecting the genetic background of multifactorial diseases and traits—A major challenge for genetic research. Molecular and cellular probes. 2016;30(6):345. doi: 10.1016/j.mcp.2016.11.003 . [DOI] [PubMed] [Google Scholar]
- 2.Schrodi SJ, Mukherjee S, Shan Y, Tromp G, Sninsky JJ, Callear AP, et al. Genetic-based prediction of disease traits: prediction is very difficult, especially about the future. Frontiers in genetics. 2014;5:162. doi: 10.3389/fgene.2014.00162 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: a cancer journal for clinicians. 2022;72(1):7–33. Epub 2022/01/13. doi: 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 4.Garg D, Ng SSM, Baig KM, Driggers P, Segars J. Progesterone-Mediated Non-Classical Signaling. Trends in endocrinology and metabolism: TEM. 2017;28(9):656–68. Epub 2017/06/28. doi: 10.1016/j.tem.2017.05.006 . [DOI] [PubMed] [Google Scholar]
- 5.Kim JJ, Kurita T, Bulun SE. Progesterone action in endometrial cancer, endometriosis, uterine fibroids, and breast cancer. Endocrine reviews. 2013;34(1):130–62. Epub 2013/01/11. doi: 10.1210/er.2012-1043 ; PubMed Central PMCID: PMC3565104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilton HN, Clarke CL, Graham JD. Estrogen and progesterone signalling in the normal breast and its implications for cancer development. Molecular and cellular endocrinology. 2018;466:2–14. Epub 2017/08/31. doi: 10.1016/j.mce.2017.08.011 . [DOI] [PubMed] [Google Scholar]
- 7.Gompel A. Progesterone and endometrial cancer. Best practice & research Clinical obstetrics & gynaecology. 2020;69:95–107. Epub 2020/08/01. doi: 10.1016/j.bpobgyn.2020.05.003 . [DOI] [PubMed] [Google Scholar]
- 8.Terry KL, De Vivo I, Titus-Ernstoff L, Sluss PM, Cramer DW. Genetic variation in the progesterone receptor gene and ovarian cancer risk. American journal of epidemiology. 2005;161(5):442–51. Epub 2005/02/19. doi: 10.1093/aje/kwi064 ; PubMed Central PMCID: PMC1380205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulac-Jericevic B, Mullinax RA, DeMayo FJ, Lydon JP, Conneely OM. Subgroup of reproductive functions of progesterone mediated by progesterone receptor-B isoform. Science. 2000;289(5485):1751–4. Epub 2000/09/08. doi: 10.1126/science.289.5485.1751 . [DOI] [PubMed] [Google Scholar]
- 10.Stenzig J, Schweikert A, Piasecki A, Höppner G, Eschenhagen T, Rau T. Progesterone receptor variants associated with the PROGINS haplotype exhibit functional properties similar to those of wild-type progesterone receptor. Pharmacogenetics and genomics. 2012;22(8):629–41. doi: 10.1097/FPC.0b013e3283558256 . [DOI] [PubMed] [Google Scholar]
- 11.Payer LM, Steranka JP, Kryatova MS, Grillo G, Lupien M, Rocha PP, et al. Alu insertion variants alter gene transcript levels. Genome research. 2021;31(12):2236–48. Epub 2021/11/21. doi: 10.1101/gr.261305.120 ; PubMed Central PMCID: PMC8647820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romano A, Delvoux B, Fischer DC, Groothuis P. The PROGINS polymorphism of the human progesterone receptor diminishes the response to progesterone. Journal of molecular endocrinology. 2007;38:331–50. doi: 10.1677/jme.1.02170 . [DOI] [PubMed] [Google Scholar]
- 13.Mir R, Altayar MA, Hamadi A, Tayeb FJ, Saeedi NH, Jalal MM, et al. Molecular determination of progesterone receptor’s PROGINS allele (Alu insertion) and its association with the predisposition and susceptibility to polycystic ovary syndrome (PCOS). Mammalian genome: official journal of the International Mammalian Genome Society. 2022. Epub 2022/01/09. doi: 10.1007/s00335-021-09941-w . [DOI] [PubMed] [Google Scholar]
- 14.Wang-Gohrke S, Chang-Claude J, Becher H, Kieback DG, Runnebaum IB. Progesterone receptor gene polymorphism is associated with decreased risk for breast cancer by age 50. Cancer research. 2000;60(9):2348–50. Epub 2000/05/16. . [PubMed] [Google Scholar]
- 15.Dunning AM, Healey CS, Pharoah PD, Teare MD, Ponder BA, Easton DF. A systematic review of genetic polymorphisms and breast cancer risk. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1999;8(10):843–54. Epub 1999/11/05. . [PubMed] [Google Scholar]
- 16.Rowe S, Coughlan S, McKenna N, Garrett E, Kieback D, Carney D, et al. Ovarian carcinoma-associated TaqI restriction fragment length polymorphism in intron G of the progesterone receptor gene is due to an Alu sequence insertion. Cancer research. 1995;55(13):2743–5. . [PubMed] [Google Scholar]
- 17.De Vivo I, Huggins GS, Hankinson SE, Lescault PJ, Boezen M, Colditz GA, et al. A functional polymorphism in the promoter of the progesterone receptor gene associated with endometrial cancer risk. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(19):12263–8. Epub 2002/09/10. doi: 10.1073/pnas.192172299 ; PubMed Central PMCID: PMC129433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Junqueira MG, da Silva ID, Nogueira-de-Souza NC, Carvalho CV, Leite DB, Gomes MT, et al. Progesterone receptor (PROGINS) polymorphism and the risk of endometrial cancer development. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2007;17(1):229–32. Epub 2007/02/13. doi: 10.1111/j.1525-1438.2006.00767.x . [DOI] [PubMed] [Google Scholar]
- 19.De Vivo I, Hankinson SE, Colditz GA, Hunter DJ. The progesterone receptor Val660—>Leu polymorphism and breast cancer risk. Breast cancer research: BCR. 2004;6(6):R636–9. Epub 2004/11/13. doi: 10.1186/bcr928 ; PubMed Central PMCID: PMC1064075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spurdle AB, Hopper JL, Chen X, McCredie MR, Giles GG, Venter DJ, et al. The progesterone receptor exon 4 Val660Leu G/T polymorphism and risk of breast cancer in Australian women. ?. 2002;11(5):439–43. Epub 2002/05/16. . [PubMed] [Google Scholar]
- 21.Spurdle AB, Webb PM, Purdie DM, Chen X, Green A, Chenevix-Trench G. No significant association between progesterone receptor exon 4 Val660Leu G/T polymorphism and risk of ovarian cancer. Carcinogenesis. 2001;22(5):717–21. Epub 2001/04/27. doi: 10.1093/carcin/22.5.717 . [DOI] [PubMed] [Google Scholar]
- 22.Pearce CL, Hirschhorn JN, Wu AH, Burtt NP, Stram DO, Young S, et al. Clarifying the PROGINS allele association in ovarian and breast cancer risk: a haplotype-based analysis. Journal of the National Cancer Institute. 2005;97(1):51–9. Epub 2005/01/06. doi: 10.1093/jnci/dji007 . [DOI] [PubMed] [Google Scholar]
- 23.McKenna NJ, Kieback DG, Carney DN, Fanning M, McLinden J, Headon DR. A germline TaqI restriction fragment length polymorphism in the progesterone receptor gene in ovarian carcinoma. British journal of cancer. 1995;71(3):451–5. Epub 1995/03/01. doi: 10.1038/bjc.1995.92 ; PubMed Central PMCID: PMC2033643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manolitsas TP, Englefield P, Eccles DM, Campbell IG. No association of a 306-bp insertion polymorphism in the progesterone receptor gene with ovarian and breast cancer. British journal of cancer. 1997;75(9):1398–9. Epub 1997/01/01. doi: 10.1038/bjc.1997.238 ; PubMed Central PMCID: PMC2228221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lancaster JM, Berchuck A, Carney ME, Wiseman R, Taylor JA. Progesterone receptor gene polymorphism and risk for breast and ovarian cancer. British journal of cancer. 1998;78(2):277. Epub 1998/07/31. doi: 10.1038/bjc.1998.480 ; PubMed Central PMCID: PMC2062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Runnebaum IB, Wang-Gohrke S, Vesprini D, Kreienberg R, Lynch H, Moslehi R, et al. Progesterone receptor variant increases ovarian cancer risk in BRCA1 and BRCA2 mutation carriers who were never exposed to oral contraceptives. Pharmacogenetics. 2001;11(7):635–8. Epub 2001/10/23. doi: 10.1097/00008571-200110000-00010 . [DOI] [PubMed] [Google Scholar]
- 27.Tong D, Fabjani G, Heinze G, Obermair A, Leodolter S, Zeillinger R. Analysis of the human progesterone receptor gene polymorphism progins in Austrian ovarian carcinoma patients. International journal of cancer. 2001;95(6):394–7. Epub 2001/10/23. doi: . [DOI] [PubMed] [Google Scholar]
- 28.Donaldson CJ, Crapanzano JP, Watson JC, Levine EA, Batzer MA. PROGINS Alu insertion and human genomic diversity. Mutation research. 2002;501(1–2):137–41. Epub 2002/04/06. doi: 10.1016/s0027-5107(02)00015-5 . [DOI] [PubMed] [Google Scholar]
- 29.Fabjani G, Tong D, Czerwenka K, Schuster E, Speiser P, Leodolter S, et al. Human progesterone receptor gene polymorphism PROGINS and risk for breast cancer in Austrian women. Breast cancer research and treatment. 2002;72(2):131–7. Epub 2002/06/01. doi: 10.1023/a:1014813931765 . [DOI] [PubMed] [Google Scholar]
- 30.Lancaster JM, Wenham RM, Halabi S, Calingaert B, Marks JR, Moorman PG, et al. No relationship between ovarian cancer risk and progesterone receptor gene polymorphism in a population-based, case-control study in North Carolina. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2003;12(3):226–7. MEDLINE:12646513. [PubMed] [Google Scholar]
- 31.Fernandez LR, Milne RL, Barroso E, Cuadros M, Arias JI, Ruibal A, et al. Estrogen and progesterone receptor gene polymorphisms and sporadic breast cancer risk: a Spanish case-control study. International journal of cancer. 2006;119(2):467–71. doi: 10.1002/ijc.21847 WOS:000238267300029. [DOI] [PubMed] [Google Scholar]
- 32.Pooley KA, Healey CS, Smith PL, Pharoah PD, Thompson D, Tee L, et al. Association of the progesterone receptor gene with breast cancer risk: a single-nucleotide polymorphism tagging approach. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2006;15(4):675–82. Epub 2006/04/15. doi: 10.1158/1055-9965.EPI-05-0679 . [DOI] [PubMed] [Google Scholar]
- 33.Romano A, Lindsey PJ, Fischer DC, Delvoux B, Paulussen AD, Janssen RG, et al. Two functionally relevant polymorphisms in the human progesterone receptor gene (+331 G/A and progins) and the predisposition for breast and/or ovarian cancer. Gynecologic oncology. 2006;101(2):287–95. Epub 2005/12/20. doi: 10.1016/j.ygyno.2005.10.040 . [DOI] [PubMed] [Google Scholar]
- 34.Govindan S, Ahmad SN, Vedicherla B, Kodati V, Jahan P, Rao KP, et al. Association of progesterone receptor gene polymorphism (PROGINS) with endometriosis, uterine fibroids and breast cancer. Cancer biomarkers: section A of Disease markers. 2007;3(2):73–8. Epub 2007/05/25. doi: 10.3233/cbm-2007-3201 . [DOI] [PubMed] [Google Scholar]
- 35.Romano A, Baars M, Martens H, Brandao R, Detisch Y, Jongen E, et al. Impact of Two Functional Progesterone Receptor Polymorphisms (PRP): +331G/A and PROGINS on the Cancer Risks in Familial Breast/Ovarian Cancer. The Open Cancer Journal. 2007;1(1):1–8. doi: 10.2174/1874079000701010001 [DOI] [Google Scholar]
- 36.Johnatty SE, Spurdle AB, Beesley J, Chen X, Hopper JL, Duffy DL, et al. Progesterone receptor polymorphisms and risk of breast cancer: results from two Australian breast cancer studies. Breast cancer research and treatment. 2008;109(1):91–9. doi: 10.1007/s10549-007-9627-3 WOS:000255031000010. [DOI] [PubMed] [Google Scholar]
- 37.Leite DB, Junqueira MG, de Carvalho CV, Massad-Costa AM, Goncalves WJ, Nicolau SM, et al. Progesterone receptor (PROGINS) polymorphism and the risk of ovarian cancer. Steroids. 2008;73(6):676–80. Epub 2008/04/04. doi: 10.1016/j.steroids.2008.02.005 . [DOI] [PubMed] [Google Scholar]
- 38.Quaye L, Tyrer J, Ramus SJ, Song H, Wozniak E, DiCioccio RA, et al. Association between common germline genetic variation in 94 candidate genes or regions and risks of invasive epithelial ovarian cancer. PloS one. 2009;4(6):e5983. doi: 10.1371/journal.pone.0005983 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Surekha S, Nageswararao R. LACK OF INFLUENCE OF PROGIN POLYMORPHISM IN BREAST CANCER DEVELOPMENT AND PROGRESSION. Cell Tissue Res. 2009. [Google Scholar]
- 40.Lee E, Hsu C, Haiman CA, Razavi P, Horn-Ross PL, Van Den Berg D, et al. Genetic variation in the progesterone receptor gene and risk of endometrial cancer: a haplotype-based approach. Carcinogenesis. 2010;31(8):1392–9. Epub 2010/06/16. doi: 10.1093/carcin/bgq113 ; PubMed Central PMCID: PMC2915632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Mara TA, Fahey P, Ferguson K, Marquart L, Lambrechts D, Despierre E, et al. Progesterone receptor gene variants and risk of endometrial cancer. Carcinogenesis. 2011;32(3):331–5. Epub 2010/12/15. doi: 10.1093/carcin/bgq263 ; PubMed Central PMCID: PMC3105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundin E, Wirgin I, Lukanova A, Afanasyeva Y, Krogh V, Axelsson T, et al. Selected polymorphisms in sex hormone-related genes, circulating sex hormones and risk of endometrial cancer. Cancer Epidemiology. 2012;36(5):445–52. doi: 10.1016/j.canep.2012.04.006 WOS:000309818500017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clendenen T, Zeleniuch-Jacquotte A, Wirgin I, Koenig KL, Afanasyeva Y, Lundin E, et al. Genetic Variants in Hormone-Related Genes and Risk of Breast Cancer. PloS one. 2013;8(7). doi: 10.1371/journal.pone.0069367 WOS:000325211000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gabriel CA, Mitra N, Demichele A, Rebbeck T. Association of progesterone receptor gene (PGR) variants and breast cancer risk in African American women. Breast cancer research and treatment. 2013;139(3):833–43. Epub 2013/06/15. doi: 10.1007/s10549-013-2592-0 ; PubMed Central PMCID: PMC3810299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patricia GAM, Figuera LE, Gomez Flores-Ramos L, Maria Puebla-Perez A, Moises Zuniga-Gonzalez G. Association of the Alu insertion polymorphism in the progesterone receptor gene with breast cancer in a Mexican population. Archives of Medical Science. 2015;11(3):551–60. doi: 10.5114/aoms.2015.52357 WOS:000357049400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghali RM, Al-Mutawa MA, Ebrahim BH, Jrah HH, Zaied S, Bhiri H, et al. Progesterone Receptor (PGR) Gene Variants Associated with Breast Cancer and Associated Features: a Case-Control Study. Pathology oncology research: POR. 2018;26(1):141–7. doi: 10.1007/s12253-017-0379-z . [DOI] [PubMed] [Google Scholar]
- 47.Albalawi IA, Mir R, Abu-Duhier FM. Molecular evaluation of PROGINS mutation in Progesterone Receptor gene and determination of its frequency, distribution pattern and association with Breast Cancer susceptibility in Saudi Arabia. Endocrine, metabolic & immune disorders drug targets. 2019. doi: 10.2174/1871530319666191125153050 . [DOI] [PubMed] [Google Scholar]
- 48.DV I., Huggins GS, Hankinson SE, Lescault PJ, Boezen M, Colditz GA, et al. A functional polymorphism in the promoter of the progesterone receptor gene associated with endometrial cancer risk. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(19):12263–8. doi: 10.1073/pnas.192172299 . [DOI] [PMC free article] [PubMed] [Google Scholar]