Abstract
The similarities and differences in the structures of the nifH gene pools of six different soils (Montrond, LCSA-p, Vernon, Dombes, LCSA-c, and Thysse Kaymor) and five soil fractions extracted from LCSA-c were studied. Bacterial DNA was directly extracted from the soils, and a region of the nifH gene was amplified by PCR and analyzed by restriction. Soils were selected on the basis of differences in soil management, plant cover, and major physicochemical properties. Microenvironments differed on the basis of the sizes of the constituent particles and the organic carbon and clay contents. Restriction profiles were subjected to principal-component analysis. We showed that the composition of the diazotrophic communities varied both on a large scale (among soils) and on a microscale (among microenvironments in LCSA-c soil). Soil management seemed to be the major parameter influencing differences in the nifH gene pool structure among soils by controlling inorganic nitrogen content and its variation. However, physicochemical parameters (texture and total C and N contents) were found to correlate with differences among nifH gene pools on a microscale. We hypothesize that the observed nifH genetic structures resulted from the adaptation to fluctuating conditions (cultivated soil, forest soil, coarse fractions) or constant conditions (permanent pasture soil, fine fractions). We attempted to identify a specific band within the profile of the clay fraction by cloning and sequencing it and comparing it with the gene databases. Unexpectedly, the nifH sequences of the dominant bacteria were most similar to sequences of unidentified marine eubacteria.
Soil diazotrophs are the main source of the nitrogen input in primary-production ecosystems. In the biosphere, except for anthropic nitrogen inputs, nitrogen fixation is the principal way in which the nitrogen supply is maintained and increased. Nitrogen fixation occurs in a wide range of bacterial phyla, from Archaebacteria to Eubacteria (54). All N2 fixers carry a nifH gene, which encodes the Fe protein of the nitrogenase.
This nifH gene has been largely studied by culture-independent approaches. These approaches provide a more complete picture of the diazotrophic community than culture-based approaches. Various techniques, such as PCR cloning (55, 56), denaturing gradient gel electrophoresis (36, 37), PCR-restriction fragment length polymorphism (RFLP), and fluorescently labeled terminal (FLT)-RFLP (10, 31, 32, 48, 53), have been used to analyze the composition of nifH gene pools in various environments. These studies found that the nifH gene is present in diverse environments: forest soil (48, 53), the rhizosphere of native wetland species, such as Spartina (10, 36, 37), or of crop species, such as rice (52), aquatic (7, 55, 56) or polar (34) cyanobacteria, and the bacteria found in termite guts (31, 32, 33). All these studies described a large number of unknown sequences which correspond to diverse unidentified diazotrophs. Some nifH genes are characteristic of an ecological niche (10, 48). Shaffer et al. (48) evoked the possible relationship between the habitats of soil nitrogen-fixing bacteria and the structure of nifH gene pools.
Environmental parameters affecting the activity of soil bacteria, especially N2 fixation, have been detailed over many years (3, 13). In grasslands, plant species may affect microbial biomass and activity (5). Riffkin et al. (42) showed that N2 fixation is influenced by different soil factors, including soil texture. Cejudo and Paneque (9) and Limmer and Drake (29) suggested that the nitrogen status of the soil may also influence N2 fixation by diazotrophs. The role of inorganic nitrogen, such as ammonium and nitrate, in preventing N2 fixation may be related to the limitation of gene expression and to the inactivation of the nitrogenase enzyme in some bacteria (45).
We aimed to investigate the nifH gene pools in soils in relation to differences in their texture, plant cover, and management to determine whether similarities among pools exist and which common environmental factor(s) could explain such similarities. Contrasting microenvironments within the soil were also studied, because they overwhelm the global factors (plant cover, soil management) and may reveal the specific influence of local factors (organic matter, clay minerals, contact with soil solution, etc.). The structure of the nifH gene pool was investigated by RFLP analysis of the nifH gene, which had been amplified from DNA directly extracted from soil samples. Restriction patterns were compared using a principal-component analysis (PCA) to estimate the relatedness of nifH gene pools and to identify some of the soil characteristics involved in these relationships. We attempted to identify the diazotrophs by cloning and sequencing a specific band within the profile of the microenvironment and comparing the sequences obtained with a gene data bank.
MATERIALS AND METHODS
Soil samples.
Samples were collected from the upper layer (0 to 20 cm) of the studied soils. Five soils from France and one tropical ultisol (Senegal) were sampled. The main characteristics of the soils, the dominant plant species, soil management, and location are given in Table 1.
TABLE 1.
Characteristics of the studied soils
| Soila | Locationb | Soil management | Soil type | Dominant plant species | Total N (‰) | CECc (cmol/kg) | pH (H2O) |
|---|---|---|---|---|---|---|---|
| LCSA-p | SE France | Permanent pasture | Loam | Graminae sp. | 2.80 | 108 | 5.77 |
| Montrond | SE France | Permanent pasture | Clay loam | Graminae sp. | 3.60 | 243 | 6.06 |
| Vernon | NW France | Permanent pasture | Silt loam | Agrostis vulgaris | 1.53 | 88 | 6.27 |
| LCSA-c | SE France | Cultivation | Loam | Zea mays | 1.03 | 64 | 6.98 |
| Thysse K. | Senegal | Cultivation | Sandy loam | Eleusine coracana | 0.70 | 31 | 7.20 |
| Dombes | SE France | Forest | Silt loam | Alnus glutinosa | 2.25 | 94 | 3.90 |
LCSA-p, La Côte Saint André soil under pasture; LCSA-c, La Côte Saint André soil under cultivation.
SE, southeast; NW, northwest.
CEC, cation exchange capacity.
The cultivated LCSA-c soil sample was separated into five fractions, corresponding to various sizes of particles and aggregates, by a size fractionation procedure (25). The 250- to 2,000-μm and 50- to 250-μm fractions were coarse and fine sands, respectively, with associated macroaggregates. The 20- to 50-μm and 2- to 20-μm size fractions were microaggregates with particles of silt and loam, respectively, and the <2-μm fraction consisted of dispersible clays and organic colloids. The fractionation procedure was carried out in duplicate on subsamples (30 g equivalent dry weight) from field samples sieved through a 2-mm mesh. The proportions of the various fractions and their characteristics are presented in Table 2.
TABLE 2.
Physicochemical characteristics of LCSA-c soil fractionsa
| Fraction (μm) | Weight distribution (%)b | Proportion (‰) of:
|
|||||
|---|---|---|---|---|---|---|---|
| Sands | Silts | Clays | Organic matter | Total N | C/N | ||
| >250 | 29 | 901 | 0* | 91 | 7.6 | 0.9 | 5* |
| 250–50 | 20.7 | 812* | 79* | 100 | 8.8 | 0.1 | 51* |
| 50–20 | 19.5 | 0* | 878 | 112 | 10 | 0.1 | 58* |
| 20–2 | 14.7 | 0* | 617 | 344 | 38.9 | 2.6 | 9* |
| <2 | 8.2 | 0* | 0* | 941* | 58.6 | 1.8 | 19* |
*, calculated value.
Percentage of unfractionated soil by mass.
Extraction and purification of DNA from soil and fraction samples.
Bacterial DNA was directly extracted from soil samples and from soil microenvironments by a direct-lysis method (39). DNA was extracted from each replicate of the fractionation process and in duplicate on unfractionated soils. DNA was purified and quantified as described previously by Ranjard et al. (39).
PCR amplification of the nifH gene fragment.
One hundred nanograms of DNA was used as template in PCR. Selected primers PolF and PolR (5′ TGC GAY CCS AAR GCB GAC TC 3′ and 5′ ATS GCC ATC ATY TCR CCG GA 3′, respectively) (38) were used to amplify a 360-bp region between sequence positions 115 and 476 (referring to the Azotobacter vinelandii nifH coding sequence [M20568]). PCR amplification was carried out as described by Poly et al. (38).
RFLP analysis.
Ten microliters of each PCR product was directly used for restriction enzyme cleavage. The reaction enzyme mixture contained 1× restriction enzyme buffer and 1.25 U of restriction endonuclease. MnlI, HaeIII, and NdeII (Biolabs) were selected for their specificity for the amplified region of nifH (38) and were used as specified by the manufacturer. The PCR products were digested overnight. Digested DNA samples were analyzed by electrophoresis in a 5% polyacrylamide gel (19:1) (Bio-Rad). The electrophoresis conditions were 15 h at 35 V in 1× Tris-borate-EDTA buffer, followed by 30 min of staining in 1× SYBR Green I (FMC BioProducts). This procedure was repeated at least two times for each sample to verify the consistency of the patterns. To assess the possible influence of the time of sampling, RFLP analysis was carried out four times over a 90-day period between April and July on Vernon soil.
Analysis of restriction profiles and statistical analysis of data.
The band intensity and band running times of each fragment were automatically integrated with Molecular Analyst software (Bio-Rad). A matrix was built using the relative intensity of each band compared to the total intensity of the profile.
PCA on the covariance data matrix was performed with soils (or soil microenvironments) as the rows and the relative intensities of the bands from the three restriction enzymes as the columns. This provided an ordering of nifH gene pools, which were plotted on two-dimensional maps. PCA on the correlation data matrix obtained from physicochemical characteristics (as columns) (Table 2) of each fraction (as rows) was performed. The Monte Carlo test was carried out with 10,000 random permutations to test the significance of the PCA results.
PCA analysis and the permutation test were carried out using the ADE-4 software (49).
Characterization of a MnlI nifH restriction fragment.
A band of approximately 250 bp, specific to the <2-μm fraction of LCSA-c soil, was isolated from the MnlI restriction profile. The fragment was excised from the polyacrylamide gel (19:1) and purified by electroelution with a Mini-Protean II apparatus (Bio-Rad), and the purified nifH fragment was recovered in 20 μl of ultrapure water.
A clone library was constructed with the SureClone ligation kit (Pharmacia, Orsay, France). The restriction fragment resulting from MnlI digestion was ligated to pUC18 (Promega, Charbonnieres, France) and transformed into competent Escherichia coli DH5α (Life Biotechnologies, Cergy Pontoise, France) in accordance with the manufacturer's instructions. Cells were grown in Luria-Bertani medium at 37°C for 24 h. Fifty clones with the insert (white colonies) were sampled, suspended in 100 μl of ultrapure water, lysed by being boiled for 3 min in a bath, and then frozen for 5 min in liquid nitrogen. Cell residues were pelleted by centrifugation for 3 min at 3,000 × g.
Plasmid inserts were collected from each clone by amplifying 1 μl of the supernatant lysate with primers M13R and M13F, which annealed to the polylinker of pUC18 (Promega). The amplicon was run in a 2% agarose gel to determine the size of the insert. Only inserts of 250 ± 30 bp were screened for insert diversity. The amplified inserts were digested separately with NdeII, HaeIII, and TaqI (Biolabs) from 8 μl of the PCR product. The resulting fragments were separated by gel electrophoresis in 4% Metaphor agarose (FMC BioProducts). The electrophoretic patterns of restriction fragments were analyzed. Individual clones were grouped into restriction groups or phylotypes based on a 100% identity threshold of the restriction patterns for the three enzymes used.
Determination of nucleotide sequences and phylogenetic analysis of clones.
The fluorescence DiDeoxy termination method was used to sequence both strands of the plasmid inserts in an automated fluorescence sequencing system (Genome Express, Grenoble, France). In phylotypes 1 to 5 we sequenced 4, 2, 1, 1, and 1 clones, respectively.
Sequences were aligned with the Clustal W package (50) and then corrected by manual inspection. A phylogenetic tree was constructed using the neighbor-joining method (46) on sequence fragments (260 bp in positions 214 to 476 of the Azotobacter vinelandii nifH coding sequence [M20568]). The topology of this distance tree was tested by resampling data with 1,000 bootstraps (15) to provide confidence estimates for tree topologies. Parsimony and maximum-likelihood analysis was done using the Phylo-Win program (16).
Nucleotide sequence accession numbers.
DNA sequences were deposited in GenBank with the following accession numbers: AF312941, AF312942, AF312943, AF312944, AF312945, AF312946, AF312947, AF312948, and AF312949.
RESULTS
Soil and microenvironment properties.
Soils were compared on the basis of their physicochemical characteristics (Table 1), resulting in four soil types. Physicochemical properties grouped LCSA-c and LCSA-p soils, loam soils which were sampled at the same location and which differed mainly in their soil management (crop cultivation versus permanent pasture). Vernon and Dombes soils, two soils which were a long way apart geographically, were both silt loam. Montrond soil, which has high organic matter and clay contents, represented a third type, clay loam soil, and Thysse Kaymor (Thysse K.), which has a high fine sand content and a low organic matter content, represented the fourth type, sandy loam soil.
The physicochemical characteristics of soil fractions from LCSA-c (Table 2) were compared by PCA (data not shown). PCA classified fractions into size categories correlated with their organic matter, nitrogen, and clay contents.
RFLP analysis of nifH gene pools from soils.
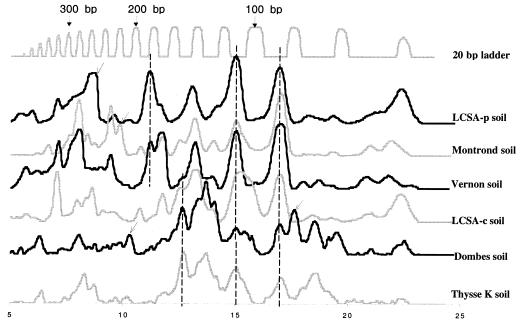
Amplification of nifH with degenerate primers yielded a single band of the expected size (approximately 360 bp) (data not shown). Reproducible restriction profiles were obtained for duplicate soil samples and also for samples collected at various times from the same field (Fig. 1). Different soils gave contrasting patterns (Fig. 2 for MnlI; data not shown for NdeII and HaeIII), with differences in the presence or absence of fragments and in the relative intensities of fragments. A different number of fragments was observed depending on the restriction endonuclease used. For example, HaeIII provided 19 different bands from the six unfractionated soils, whereas MnlI and NdeII resulted in 31 and 33 bands, respectively. Some fragments were found in all soils, such as the 110- and 85-bp MnlI bands (Fig. 2) and the 300- and 280-bp NdeII bands (data not shown). Other bands were found to be characteristic of one soil: MnlI 245-bp (Fig. 2), NdeII 120-bp, and HaeIII 170-bp bands for LCSA-p; MnlI 220-bp band for Montrond; HaeIII 355-bp band for Vernon; MnlI 320-bp band for LCSA-c; and MnlI 210-bp, MnlI 80-bp, and NdeII 140-bp bands for Dombes. Bands common to the three nonpasture soils included the MnlI 160-bp, NdeII 165-bp, and HaeIII 110- and 75-bp bands; the MnlI 180-bp band (Fig. 2) was specific to pasture soils.
FIG. 1.
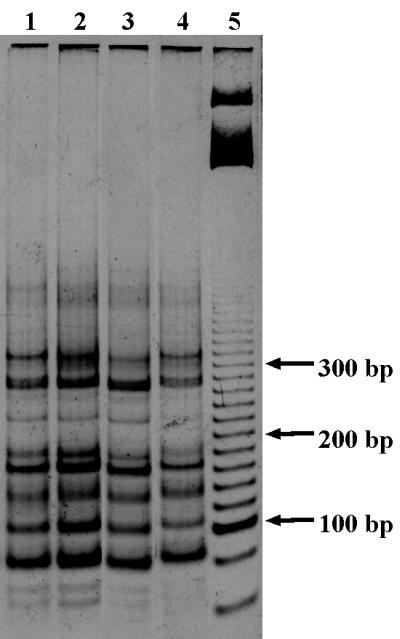
MnlI RFLPs of nifH PCR products obtained from Vernon soil on four sampling dates. Lane 1, 6 April 1998; lane 2, 4 May 1998; lane 3, 10 June 1998; lane 4, 17 July 1998. Migration was performed on a 5% polyacrylamide (19:1) gel, and the molecular size marker (lane 5) was 20 bp.
FIG. 2.
Electrophoretogram of MnlI RFLPs of nifH PCR products obtained from the six studied soils. Dashed lines, peaks common to several soils; light arrows, characteristic fragments (see text).
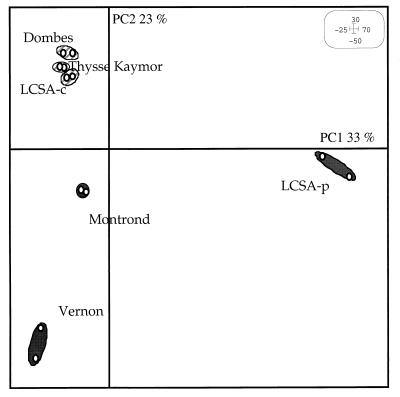
Pairwise analysis of nifH gene profiles by PCA allowed the ordering of nifH gene pools. The first principal component, PC1, and the second principal component, PC2, explained 33 and 23% of the variance of the data, respectively (Fig. 3). The factorial map (Fig. 3) showed that three of the studied soils (LCSA-c, Dombes, and Thysse K.) were grouped, whereas the other soils were not. PCA indicated that there was a large variability in nifH pools in permanent pasture soils (LCSA-p, Montrond, and Vernon) and a low variability in nonpasture soils. The significance of the separation of nonpasture soils from pasture soils was tested with a Monte Carlo test. Results revealed a significant difference (P = 0.0017) between pasture and nonpasture soils.
FIG. 3.
PCA generated from soil nifH restriction profiles by HaeIII, NdeII, and MnlI. Dark spots, pasture soils; hatched spots, nonpasture soils.
RFLP analysis of nifH gene pools from LCSA-c soil fractions.
Some differences among the patterns obtained from the different fractions of LCSA-c soil occurred (Fig. 4). Differences were mainly due to differences in the relative intensities of common bands among profiles. The numbers of different bands with HaeIII, MnlI, and NdeII were 15, 19, and 27, respectively. The number of restriction bands classified enzymes in the same order (HaeIII < MnlI < NdeII) as the soil study.
FIG. 4.
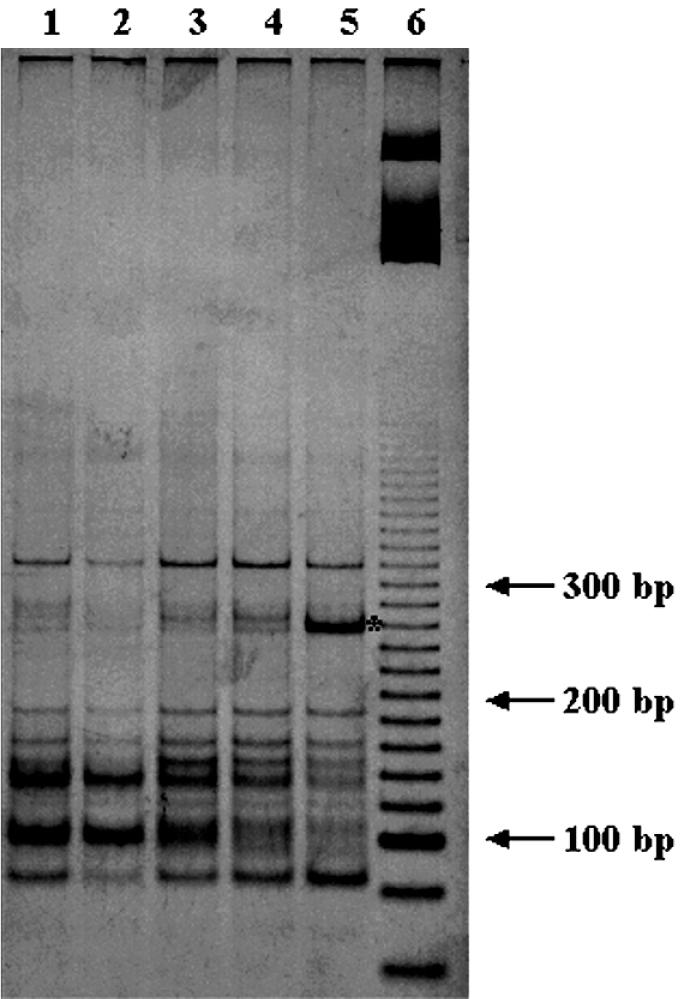
Polyacrylamide gel electrophoresis of MnlI RFLPs from nifH PCR products obtained from LCSA-c soil fractions. Lane 1, >250-μm fraction; lane 2, 250- to 50-μm fraction; lane 3, 50- to 20-μm fraction; lane 4, 20- to 2-μm fraction; lane 5, <2-μm fraction; lane 6, 20-bp molecular size marker. Asterisk, 250-bp fragment characteristic of the <2-μm fraction.
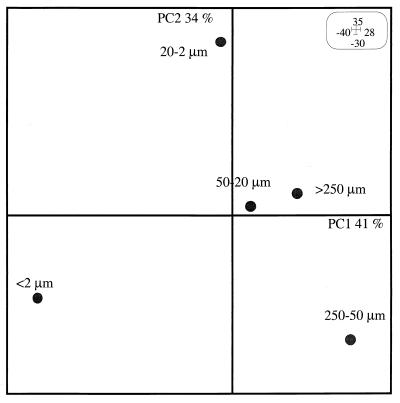
The first and the second principal components, PC1 and PC2, explained 41 and 34% of the variance of the data, respectively (Fig. 5). PCA on the three enzyme patterns (Fig. 5) showed that ordering on PC1 mostly corresponded to sizes of the fractions and showed that the finest-size fraction (<2 μm) and the sand fractions (>50 μm) were at opposite ends on the PC1 axis (Fig. 5). PC2 differentiated the 50- to 250-μm fraction from the >250-μm fraction and the 2- to 20-μm fraction from the 20- to 50-μm fraction. Some bands were associated with certain microenvironments; for example, the nifH gene MnlI RFLP profile from the DNA of the <2-μm fraction exhibited one dominant band at 250 bp (Fig. 4).
FIG. 5.
PCA generated from nifH restriction profiles from LCSA-c soil microenvironments by HaeIII, NdeII, and MnlI.
Cloning and sequencing of the MnlI 250-bp band.
This band was chosen for further characterization to study the diversity of the nifH sequences associated with this fragment. Fifty clones were screened for the nifH insert, and 45 clones (90%) had an insert of the expected size (250 bp). Restriction analysis with TaqI, NdeII, and HaeIII resulted in division of the clones into 16 phylotypes. Phylotypes 1 to 5 accounted for 33, 29, 6.6, 4.5, and 4.5% of the clones, respectively. Each of the additional 11 phylotypes were represented by a single clone.
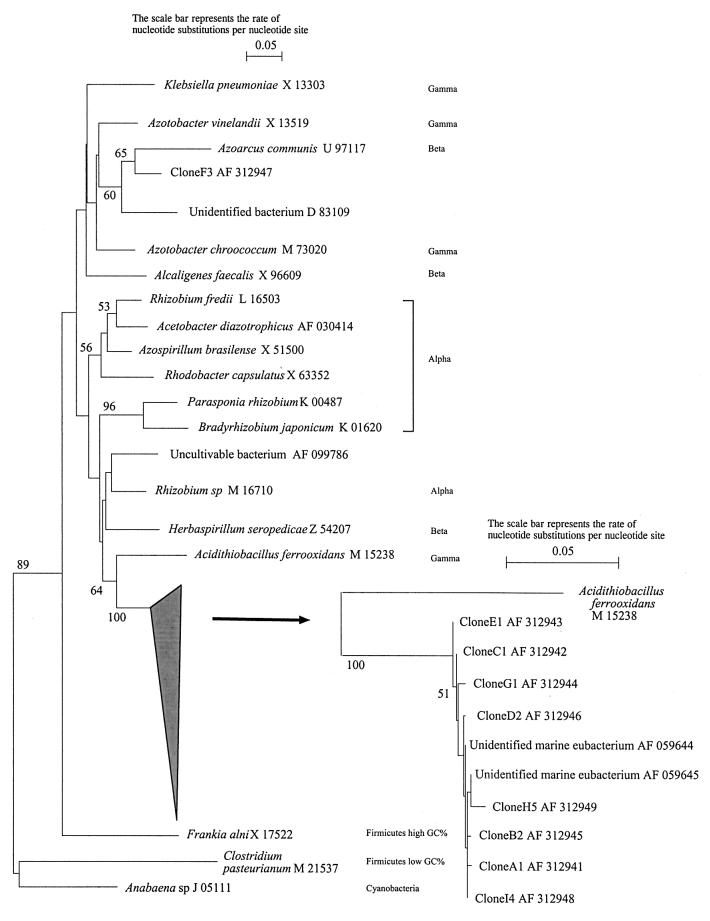
The five phylotypes that contained more than one clone were sequenced. The nucleotide sequences of the nifH insert were aligned and compared to nifH sequences (Fig. 6) from databases. All the clones sequenced were located at the 3′ end of nifH PCR products, at positions 214 to 476 of the A. vinelandii nifH coding sequence (GenBank accession no. M20568). The sequences of clones from phylotypes 1, 2, 4, and 5 were very similar (Fig. 6). Phylotype 2 nifH sequences had one change compared to those of phylotype 1; the C residue at position 474 was replaced by a G residue (with reference to A. vinelandii M20568), which removed one of the HaeIII restriction sites. Phylotype 4 (4.5%) lacked a 33-bp region at the 3′ end. Phylotype 5 (4.5%) had the largest sequence (260 bp) and differed from the others by four nucleotides in positions 244, 462, 465, and 474. The nifH sequences of phylotypes 1, 2, 4, and 5 were all similar to the sequence of an unidentified marine eubacterium (AF059644, AF059645) (56) and clustered to Acidithiobacillus ferrooxidans (M15238). Clones from phylotype 3 (6.6% of the selected clones) harbored a 244-bp fragment in which 20% of the nucleotides did not match those in any of the other phylotypes. The sequence of phylotype 3 was similar to the sequence of a β-proteobacterium, identified as Azoarcus communis (U97117).
FIG. 6.
Phylogeny of nifH nucleotide sequences using 21 partial nifH gene sequences from the GenBank database and 9 sequences obtained from the cloning of the 250-bp MnlI band from the LCSA-c clay fraction. GenBank database accession numbers are indicated next to the bacterial names. Locations of the nifH fragments used for the analysis correspond to a sequence fragment of ≈250 bp in positions 214 to 476 (referring to the A. vinelandii nifH coding sequence [M20568]). The tree was constructed by the neighbor-joining method, and bootstrap values above 50 from 1,000 resamplings are shown for each node.
DISCUSSION
We used RFLP-PCR on nifH gene pools to investigate the genetic structure of the diazotrophic communities associated with various soils and microenvironments. Considering the taxonomy of diazotrophs, Young (54) reported that the phylogeny of the nifH gene is broadly consistent with that based on 16S rRNA, showing that nifH could be considered a good marker of diazotrophic community structure. Other studies (33, 52, 55) reported that the analysis of partial nifH gene sequences provided information on the phylogeny and composition of diazotroph natural communities.
PCA ordering of soil and microenvironment gene pools was compared to the ordering of soil based on soil properties to identify the environmental factors controlling the observed structure of the diazotroph communities. An attempt to identify the diazotrophic pool carrying a nifH gene fragment specific to the clay fraction of the LCSA-c soil was also made. This fragment was cloned, sequenced, and aligned with known nifH sequences published in GenBank.
Comparison of the structure of the nifH gene pool among soils.
PCA ordering (Fig. 3) revealed two soil groups: the first group included the two cultivated soils (LCSA-c and Thysse K.) and the alder forest soil (Dombes). The nifH gene pool of the second group, consisting of the three soils under permanent pasture (LCSA-p, Montrond, and Vernon), exhibited a more distinctive composition than that of the gene pool of the first group of soils. The Monte Carlo test confirmed that two groups were significantly separated (P = 0.0017), suggesting that the structure of the nifH gene pool is not controlled by the geographical location of the soils. The time stability of the RFLP profiles was revealed by comparing the Vernon soil profiles derived from samples collected at four sampling times over a 90-day period (from April to July). No differences among profiles could be detected (Fig. 1), suggesting that the nifH pool structure of a soil remains stable over several months. Similarly, Shaffer et al. (48) showed that the nifH gene profiles of a forest soil were similar over a 16-month period and Piceno and Lovell (36, 37) showed that even dramatic modifications in nutrient availability (nitrogen, carbon, and phosphorus) did not affect the diazotroph pool in the rhizosphere of Spartina alterniflora in the short term.
Most studies usually report the influence of soil physics (42) and chemical properties (13, 18) on diazotrophic activity. Our results revealed that the observed differences in nifH gene pool structure among various soils cannot be explained by the measured physicochemical characteristics (Table 1). This discrepancy highlights the finding that diazotrophic activity and diazotrophic community structure are not similarly affected by soil properties. The structure of the nifH gene pool might not be related to gene expression or to nitrogenase activity. A study by Alexander (2) showed that the presence or absence of particular culturable bacterial genera may depend on soil parameters. We studied bacteria without regard for their ability to grown on synthetic media. Nonculturable bacteria represent a large part of soil diazotrophs (52, 53), and this may explain why the influence of soil parameters observed by Alexander on culturable bacteria only (2) was not predominant for all diazotrophs.
The lack of relationships between nifH gene pools and the considered physicochemical characteristics suggested that other soil properties are responsible for the observed nifH gene pool ordering. Bardgett et al. (5) suggested that plant species affect the soil microbial community more than the physical or chemical properties of the soil. Our results did not support this suggestion: the tightly clustered group of nonpasture soils contained distinct plant species (maize, millet, and alder); contrastingly the soils from the three pastures, characterized by similar complex gramineous associations (data not shown), were completely disjointed. These results suggest that plant species are not the main factor that influences the nifH gene pool.
Another parameter that could influence the diazotrophic community structure is the amount and quality of organic matter, especially nitrogen. The total amounts (inorganic and organic) of nitrogen in all soils were measured (Table 1) and were not found to be correlated to the observed nifH gene pool differentiation. Various studies have indicated that the activity (9, 29) and abundance of total diazotrophs or of specific populations can be influenced by the amounts of the inorganic nitrogenous forms. For example, ammonium and nitrate inhibit the nitrogenase enzyme even at low concentrations (35), and the nitrate content was reported to be negatively correlated with the number of diazotrophs (22), such as azospirilla on maize roots (26) or Acetobacter diazotrophicus in sugar cane fields (14). In our study, the differences among the nifH gene pools in the various studied soils may result from selection or the adaptation of diazotrophs to distinct inorganic nitrogen environmental conditions. Although we did not identify and quantify the different nitrogen forms, it can be supposed that the studied pasture soils and the nonpasture soils offered these contrasting conditions, which influence nitrogen mineralization and consequently the balance between organic and inorganic forms. Soils under permanent pasture are characterized by a lower nitrogen mineralization than forest or cultivated soils (47). Denitrification (28) and plant nutrition processes lower the nitrate content in pastures. Furthermore, the amount of inorganic nitrogen in cultivated soils and in forest soil can be increased by processes such as fertilizer application and the rapid degradation of organic matter. The application of inorganic fertilizer (21) and tillage (4, 8) stimulate the mineralization of native soil organic matter. A high nitrogen content and a low lignin content have been observed in the litter of alder (12); these lead to a rapid degradation of organic matter (21, 51) and consequently to the production of inorganic nitrogen (17). Fertilization and degradation of organic matter are discontinuous processes (44) which temporarily alter the amount of bioavailable inorganic nitrogen. Consequently, the structure of the nifH pools analyzed in our study might result from the adaptation to different amounts of inorganic nitrogen forms and also from the rhythm of inorganic nitrogen production (constant in pastures and fluctuating in nonpasture soils). The inorganic nitrogen status of soils is in turn influenced by interactions among soil chemical properties, plant species, and soil management.
Comparison of nifH gene pools among LCSA-c soil factions.
Restriction profiles from the various microenvironments of LCSA-c soil were found to be different from the profile of the unfractionated soil and from each other. Ordering on PC1 (Fig. 5) revealed that most differences in genetic structure occurred between the coarse fractions (>250 μm and 50 to 250 μm) and the clay fraction (<2 μm). Various studies, such as whole-cell counting (24, 40) and biomass measurements (23), as well as specific bacterial enumerations (25) and determinations of the genetic structures (40) and activities (6, 28, 30) of bacterial subcommunities, have shown that soil microenvironments differ from each other. Ordering on PC1 grouped nifH gene pools located in microenvironments with similar granulometric characteristics: the two coarse fractions (>250 μm and 50 to 250 μm) were closely related, as were the two medium fractions (20 to 50 μm and 2 to 20 μm). The <2-μm fraction was distinct from the others. PCA on the physicochemical characteristics of the fractions resulted in an ordering of size fractions on PC1 (data not shown) that was similar to the ordering based on nifH patterns. Therefore, the structure of nifH gene pools in fractions is probably correlated to the main characteristics of these fractions (clay, organic matter, and nitrogen contents). On a similar microscale, bacterial activities, such as the mineralization of organic matter (11), respiration, and denitrification (28), and the structure of bacterial populations associated with the size fractions (40) have also been reported to be influenced by the same parameters (clay, organic matter, and nitrogen contents).
The amount of the available inorganic nitrogen may vary among microenvironments as well as among different soils. Several studies have shown that the amount of mineralized nitrogen was greater in macroaggregates than in microaggregates and clay fractions (19, 47). Similarly, the finest fractions have a higher denitrifying activity and a lower inorganic nitrogen content (28). Furthermore, microorganisms associated with coarse fractions are probably in close contact with the soil solution and are probably subjected to greater fluctuations in conditions (water, nutrients, aeration status, fertilizer input, etc.) than microorganisms associated with microaggregates and clay fractions (20, 24, 43). The structure of nifH gene pools in the microenvironments might also result from a specific adaptation of diazotrophs to fluctuating environmental conditions (such as inorganic nitrogen release) in coarse fractions, whereas the more-constant conditions encountered in the microaggregates and the clay fractions favor other nifH genes and other diazotrophs.
Identification of diazotrophs.
We attempted to identify the diazotrophs by use of a specific nifH gene band within a profile. The presence of numerous nifH gene sequences in databases and the similar phylogenetic trees derived from both the 16S rRNA genes and the nifH genes should facilitate the identification of diazotrophs. However, the amplified fragment (360 bp) and the small restriction fragments derived from this amplicon restricted identification. A characteristic dominant 250-bp band (the main MnlI nifH restriction fragment in the clay fraction profile) (Fig. 4) was cloned and sequenced.
The RFLP profiles of the cloned fragments led to the assignment of 15 phylotypes, of which four phylotypes (1, 2, 4, and 5) represented 71% of the selected clones. These phylotypes have very similar sequences, revealing the low diversity of nifH sequences in this band. It is probably not due to a discriminative amplification by the primers used because Poly et al. (38) showed that these primers are effective on most of the bacteria belonging to the cluster I branch of nifH phylogeny (7). Other explanations include the high sensitivity of the method, because a band can discriminate strains from the same species (38), or the specificity of the clay environment, which reduces the diversity of the associated diazotrophs.
Although the bootstrap values were low and mainly nonsignificant, the phylogenetic tree obtained from the small nifH sequence was consistent with the phylogenetic tree (1, 52, 54) deduced from the comparison with the larger nifH sequence. Young (54), Ueda et al. (52), and Achouak et al. (1) also found that the nifH sequence from Acidithiobacillus ferrooxidans, a γ-proteobacterium (27), grouped with those from some α-proteobacteria. The same unexpected presence of a β-proteobacterium in the α-proteobacterium cluster was found for Herbaspirillum seropedicae.
BLAST homologies and the positioning of the clones in the nifH partial sequence-derived tree showed that the four dominant phylotypes grouped with two sequences described by Zehr et al. (56) from Pacific Ocean diatom samples. This relationship between nifH sequences from marine and soil environments is surprising due to the different environmental conditions encountered. However, previous studies have mentioned similarities between nifH genes from bacteria associated with zooplankton or marine microbial mats and from bacteria living in termite guts (7). Similarly, the latter were found to be similar to bacteria associated with rice rhizospheres (33). Phylotype 3, which represented 6.6% of the clones, was found to be similar to the Azoarcus genus. This was less surprising as this genus is commonly found in soil and can colonize the roots of many gramineous plants (41). The next step of this study will be to isolate the bacteria carrying these genes to evaluate how they are adapted to the environments they originated from.
Conclusion.
This study showed that the composition of the nifH gene pool varies both on a large scale (among soils) and on a microscale (among microenvironments isolated from one soil). Soil management seemed to be the dominant parameter influencing the genetic structure in the unfractionated soils studied by controlling inorganic nitrogen content and its fluctuation. On a microscale, physical and chemical properties (texture and total C and N contents) were correlated with differences among nifH gene pools. We hypothesize that the observed nifH genetic structure resulted from adaptation to fluctuating conditions (cultivated soil, forest soil, coarse fractions) compared to constant conditions (permanent pasture soil, fine fractions). The diazotroph that is specific to the clay environment in LCSA-c soil was identified by cloning, sequencing, and comparing new sequences with those of known nifH genes. This strategy proved to be successful even on short DNA fragments. A further step would be to isolate and identify diazotrophs that are adapted to fluctuating inorganic nitrogen and to constant and low inorganic nitrogen content.
ACKNOWLEDGMENTS
We express our gratitude to J. Thioulouse (UMR-CNRS 5558, Biométrie et Biologie Evolutive) for his help with the multivariate analysis of data.
This investigation was supported by the Ecocompatibility of Solid Wastes program (grant 9674056/DIMT/mfb) by the Agence de l'Environnement et de la Maîtrise de l'Energie (ADEME).
REFERENCES
- 1.Achouak W, Normand P, Heulin T. Comparative phylogeny of rrs and nifH genes in the Bacillaceae. Int J Syst Bacteriol. 1999;49:961–967. doi: 10.1099/00207713-49-3-961. [DOI] [PubMed] [Google Scholar]
- 2.Alexander M, editor. Microbial ecology. New York, N.Y: John Wiley & Sons, Inc.; 1971. [Google Scholar]
- 3.Atlas R M, Bartha R. Microbial ecology. Fundamentals and applications. Reading, Mass: Addison-Wesley Publishing Co.; 1981. [Google Scholar]
- 4.Balesdent J, Mariotti A, Boisgontier D. Effect of tillage on soil organic carbon mineralization estimated from 13C abundance in maize fields. J Soil Sci. 1990;41:587–596. [Google Scholar]
- 5.Bardgett R D, Mawdsley J L, Edwards S, Hobbs P J, Rodwell J S, Davies W J. Plant species and nitrogen effects on soil biological properties of template upland grasslands. Funct Ecol. 1999;13:650–660. [Google Scholar]
- 6.Beauchamp E G, Seech A G. Denitrification with different sizes of soil aggregates obtained from dry-sieving and from sieving with water. Biol Fertil Soils. 1990;10:188–193. [Google Scholar]
- 7.Braun S T, Proctor L M, Zani S, Mellon M T, Zehr J P. Molecular evidence for zooplankton-associated nitrogen-fixing anaerobes based on amplification of nifH gene. FEMS Microbiol Ecol. 1999;28:273–279. [Google Scholar]
- 8.Cambardella C A, Eliott E T. Particulate soil organic-matter changes across a grassland cultivation sequence. Soil Sci Soc Am J. 1992;56:777–783. [Google Scholar]
- 9.Cejudo F J, Paneque A. Short-term nitrate (nitrite) inhibition of nitrogen fixation in Azotobacter chroococcum. J Bacteriol. 1986;165:240–243. doi: 10.1128/jb.165.1.240-243.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chelius M K, Lepo J E. Restriction fragment length polymorphism analysis of PCR-amplified nifH sequences from wetland plant rhizosphere communities. Environ Technol. 1999;20:883–889. [Google Scholar]
- 11.Christensen B T. Physical fractionation of soil and organic matter in primary particle size and density separates. Adv Soil Sci. 1992;20:1–89. [Google Scholar]
- 12.Domenach A M, Moiroud A, Jocteur Monrozier L. Leaf carbon and nitrogen constituents of some actinorhizal tree species. Soil Biol Biochem. 1994;26:649–653. [Google Scholar]
- 13.Dommergues Y, Mangenot F, editors. Soil microbial ecology. Paris, France: Masson; 1970. [Google Scholar]
- 14.Dos Reis F B, Jr, Reis V M, Urquiaga S, Dobereiner J. Influence of nitrogen fertilisation on the population of diazotrophic bacteria Herbaspirillum spp. and Acetobacter diazotrophicus in sugar cane (Saccharum spp.) Plant Soil. 2000;219:153–159. [Google Scholar]
- 15.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 16.Galtier N, Gouy M, Gautier C. Sea View and Phylo-Win: two graphic molecular tools for sequence alignment and molecular phylogeny. Comput Appl Biol Sci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 17.George T, Ladha J K, Buresh R J, Garrily D P. Managing native and legume-fixed nitrogen in lowland rice-based cropping systems, p 69–92. In: Ladha J K, George T, Bohlool B B, editors. Biological nitrogen fixation for sustainable agriculture. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. [Google Scholar]
- 18.Giller K E, Witter E, McGrath S P. Toxicity of heavy metals to microorganisms and microbial process in agricultural soils: a review. Soil Biol Biochem. 1998;30:1389–1414. [Google Scholar]
- 19.Gupta V V S R, Germida J J. Distribution of microbial biomass and its activity in different soil aggregate size classes as affected by cultivation. Soil Biol Biochem. 1988;20:777–786. [Google Scholar]
- 20.Hattori T. Soil aggregates as microhabitats of microorganisms. Biol Fertil Soils. 1988;6:189–203. [Google Scholar]
- 21.Haynes R J. The decomposition process: mineralization, immobilization, humus formation, and degradation. In: Haynes R J, editor. Mineral nitrogen in plant-soil system. Physiological ecology. London, United Kingdom: Academic Press Inc.; 1986. pp. 52–126. [Google Scholar]
- 22.Herridge D F, Brockwell J. Contributions of fixed nitrogen and soil nitrate to the nitrogen economy of irrigated soybean. Soil Biol Biochem. 1988;20:711–717. [Google Scholar]
- 23.Jocteur Monrozier L, Ladd J N, Fitzpatrick R W, Foster R C, Maupach M. Components and microbial biomass content of size fractions in soils of contrasting aggregation. Geoderma. 1991;49:37–62. [Google Scholar]
- 24.Jocteur Monrozier L, Guez P, Chalamet A, Bardin R, Martins J, Gaudet J P. Distribution of microorganisms and fate of xenobiotic molecules in insaturated soil environments. Sci Tot Environ. 1993;136:121–133. [Google Scholar]
- 25.Kabir M, Chotte J L, Rahman M, Bally R, Jocteur Monrozier L. Distribution of soil fractions and location of soil bacteria in a vertisol under cultivation and perennial grass. Plant Soil. 1994;163:243–255. [Google Scholar]
- 26.Kalininskaya T A. The influence of different forms of combined nitrogen on nitrogen-fixing activity of Azospirilla in the rhizosphere of rice plants. In: Vancura V, Kunc F, editors. Proceedings of the International Symposium on Interrelationships between Microorganisms and Plants in Soil. New York, N.Y: Elsevier Science Publishing, Inc.; 1989. pp. 283–286. [Google Scholar]
- 27.Kelly D P, Wood A P. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int J Syst Evol Microbiol. 2000;50:511–516. doi: 10.1099/00207713-50-2-511. [DOI] [PubMed] [Google Scholar]
- 28.Lensi R, Clays-Josserand A, Jocteur Monrozier L. Denitrifiers and denitrifying activity in size fractions of a mollisol under permanent pasture and continuous cultivation. Soil Biol Biochem. 1995;27:61–69. [Google Scholar]
- 29.Limmer C, Drake H L. Effect of carbon, nitrogen, and electron acceptor availability on anaerobic N2 fixation in beech forest soil. Soil Biol Biochem. 1998;30:153–158. [Google Scholar]
- 30.Nacro H, Benest D, Abbadie L. Distribution of microbial activities and organic matter according to particle size in a humid savanna soil (Lamto, Côte d'Ivoire) Soil Biol Biochem. 1996;28:1687–1697. [Google Scholar]
- 31.Noda S, Ohkuma M, Usami R, Horikoshi K, Kudo T. Culture-independent characterization of gene responsible for nitrogen fixation in the symbiotic microbial community in the gut of the termite Neotermes koshunensis. Appl Environ Microbiol. 1999;65:4935–4942. doi: 10.1128/aem.65.11.4935-4942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohkuma M, Noda S, Kudo T. Phylogenetic diversity of nitrogen fixation genes in the symbiotic microbial community in the gut of diverse termites. Appl Environ Microbiol. 1999;65:4926–4934. doi: 10.1128/aem.65.11.4926-4934.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohkuma M, Noda S, Usami R, Horikoshi K, Kudo T. Diversity of nitrogen fixation genes in the symbiotic intestinal microflora of the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:2747–2752. doi: 10.1128/aem.62.8.2747-2752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson J B, Steppe T F, Litaker R W, Pearl H W. N2-fixing microbial consortia associated with the ice cover of Lake Bonney, Antarctica. Microb Ecol. 1998;36:231–238. doi: 10.1007/s002489900110. [DOI] [PubMed] [Google Scholar]
- 35.Peoples M B, Crasswell E T. Biological nitrogen fixation: investments, expectations and actual contributions to agriculture. Plant Soil. 1992;141:13–39. [Google Scholar]
- 36.Piceno Y M, Lovell C R. Stability in natural bacterial communities. I. Nutrient addition effects on rhizosphere diazotroph assemblage composition. Microb Ecol. 2000;39:32–40. doi: 10.1007/s002489900192. [DOI] [PubMed] [Google Scholar]
- 37.Piceno Y M, Lovell C R. Stability in natural bacterial communities. II. Plant resource allocation effects on rhizosphere diazotroph assemblage composition. Microb Ecol. 2000;39:41–48. doi: 10.1007/s002489900191. [DOI] [PubMed] [Google Scholar]
- 38.Poly F, Jocteur Monrozier L, Bally R. Improvement in RFLP procedure to study the community of nitrogen fixers in soil through the diversity of nifH gene. Res Microbiol. 2001;152:95–103. doi: 10.1016/s0923-2508(00)01172-4. [DOI] [PubMed] [Google Scholar]
- 39.Ranjard L, Poly F, Combrisson J, Richaume A, Nazaret S. A single procedure to recover DNA from the surface or inside aggregates and in various size fractions of soil suitable for PCR based assays of bacteria. Eur J Soil Biol. 1998;34:89–97. [Google Scholar]
- 40.Ranjard L, Poly F, Combrisson J, Richaume A, Gourbière F, Thioulouse J, Nazaret S. Heterogeneous cell density and genetic structure of bacterial pools associated with various soil microenvironments as determined by enumeration and DNA fingerprinting approach (RISA) Microb Ecol. 2000;39:263–272. [PubMed] [Google Scholar]
- 41.Reinhold Hurek B, Hurek T. Interaction of gramineous plants with Azoarcus spp and other diazotrophs: identification, localization, and perspectives to study their function. Crit Rev Plant Sci. 1998;17:29–39. [Google Scholar]
- 42.Riffkin P A, Quigley P E, Kearney G A, Cameron F J, Gault R R, Peoples M B, Thies J E. Factors associated with biological nitrogen fixation in dairy pastures in south-western Victoria. Aust J Agric Res. 1999;50:261–272. [Google Scholar]
- 43.Robert M, Chenu C. Interactions between soil minerals and microorganisms. In: Stotzky G, Bollag J M, editors. Soil biochemistry. New York, N.Y: Marcel Dekker Inc.; 1992. pp. 307–404. [Google Scholar]
- 44.Robertson P G, Vitousek P M. Nitrification potentials in primary and secondary succession. Ecology. 1981;62:376–386. [Google Scholar]
- 45.Rudnick P, Meletzus D, Green A, He L, Kennedy C. Regulation of nitrogen fixation by ammonium in diazotrophic species of proteobacteria. Soil Biol Biochem. 1997;29:831–841. [Google Scholar]
- 46.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 47.Scott N A. Soil aggregation and organic matter mineralization in forest and grasslands: plant species effect. Soil Sci Soc Am J. 1998;62:1081–1089. [Google Scholar]
- 48.Shaffer B T, Widmer F, Porteous L A, Seidler R J. Temporal and spatial distribution of the nifH gene of N2-fixing bacteria in forests and clearcuts in western Oregon. Microb Ecol. 2000;39:12–21. doi: 10.1007/s002489900183. [DOI] [PubMed] [Google Scholar]
- 49.Thioulouse J, Chessel D, Dolédec S, Olivier J M. ADE-4: a multivariate analysis and graphical display software. Stat Comput. 1997;7:75–83. [Google Scholar]
- 50.Thompson J D, Higgins D G, Gibson T J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trinsoutrot I, Recous S, Bentz B, Linères M, Chèneby D, Nicolardot B. Biochemical quality of crop residues and carbon and nitrogen mineralization kinetics under nonlimiting nitrogen conditions. Soil Sci Soc Am J. 2000;64:918–926. [Google Scholar]
- 52.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Widmer F, Shaffer B T, Porteous L A, Seidler R J. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade mountain range. Appl Environ Microbiol. 1999;65:374–380. doi: 10.1128/aem.65.2.374-380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young J P W. Phylogenetic classification of nitrogen-fixing organisms. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 43–86. [Google Scholar]
- 55.Zehr J P, Mellon M, Braun S, Litaker W, Steppe T, Paerl H W. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl Environ Microbiol. 1995;61:2527–2532. doi: 10.1128/aem.61.7.2527-2532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zehr J P, Mellon M T, Zani S. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl Environ Microbiol. 1998;64:3444–3450. doi: 10.1128/aem.64.9.3444-3450.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]