Abstract
The N6‐methyladenosine (m6A) RNA methyltransferase METTL16 is an emerging player in the RNA modification landscape of the human cell. Originally thought to be a ribosomal RNA methyltransferase, it has now been shown to bind and methylate the MAT2A messenger RNA (mRNA) and U6 small nuclear RNA (snRNA). It has also been shown to bind the MALAT1 long noncoding RNA and several other RNAs. METTL16's methyltransferase domain contains the Rossmann‐like fold of class I methyltransferases and uses S‐adenosylmethionine (SAM) as the methyl donor. It has an RNA methylation consensus sequence of UACAGARAA (modified A underlined), and structural requirements for its known RNA interactors. In addition to the methyltransferase domain, METTL16 protein has two other RNA binding domains, one of which resides in a vertebrate conserved region, and a putative nuclear localization signal. The role of METTL16 in the cell is still being explored, however evidence suggests it is essential for most cells. This is currently hypothesized to be due to its role in regulating the splicing of MAT2A mRNA in response to cellular SAM levels. However, one of the more pressing questions remaining is what role METTL16's methylation of U6 snRNA plays in splicing and potentially cellular survival. METTL16 also has several other putative coding and noncoding RNA interactors but the definitive methylation status of those RNAs and the role METTL16 plays in their life cycle is yet to be determined. Overall, METTL16 is an intriguing RNA binding protein and methyltransferase whose important functions in the cell are just beginning to be understood.
This article is categorized under:
RNA Processing > RNA Editing and Modification
RNA Interactions with Proteins and Other Molecules > RNA‐Protein Complexes
Keywords: epitranscriptomics, METTL16, N6‐methyladenosine, RNA binding, RNA methyltransferase
METTL16 is an N6‐methyladenosine RNA methyltransferase with a variety of targets including the U6 small nuclear RNA, the MALAT1 long noncoding RNA, and the MAT2A messenger RNA. The role of METTL16 in the cell is still being explored as are the functions and importance of its various domains (RBD, N terminal RNA binding domain; VCR, vertebrate conserved region).
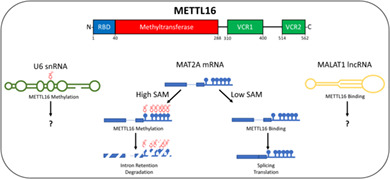
1. INTRODUCTION
RNA modifications of messenger RNA (mRNA) play an important role in posttranscriptional and translational regulation (Boo & Kim, 2020; Peer et al., 2019; Roundtree et al., 2017; Roundtree & He, 2016; Zhao et al., 2017). As such, interest in RNA‐modifying enzymes such as METTL16 has dramatically increased in recent years. There are over 100 known RNA modifications, and it is well established that ribosomal RNA (rRNA) and transfer RNA (tRNA) are rife with modifications (Boccaletto et al., 2018; Boccaletto & Baginski, 2021; Cantara et al., 2011). In fact, the most prevalent RNA modification, pseudouridine, is found mostly in rRNA and tRNA (Ge & Yu, 2013; Wiener & Schwartz, 2021). It is for this reason that modifications in these RNAs have been well studied. Many of these studied modifications in tRNA and rRNA have been revealed to fine‐tune their functions in translational efficiency (Chujo & Tomizawa, 2021; Grosjean, 2005; Sloan et al., 2017; Suzuki, 2021). To date, eukaryotic mRNA has been found to contain at least 13 modifications, including N6‐methyladenosine (m6A), 5‐methylcytidine (m5C), and 7‐methylguanosine, but the functions of these modifications are just beginning to be elucidated (Jonkhout et al., 2017; Nachtergaele & He, 2017).
Interest in understanding the role of RNA modification in regulating mRNA has exploded over the last few years, fueled largely by the identification of the RNA binding proteins responsible for writing, reading, and erasing some of these modifications (Jiang et al., 2021; Shi et al., 2019; Zaccara et al., 2019). Of these, methylation of the nitrogen at the sixth carbon residue on adenosine, otherwise known as m6A has arguably received the most attention to date (He & He, 2021). One reason for this is that both m6A “writers” (including METTL16 and METTL3/METTL14) and m6A “erasers” (FTO and ALKBH5) acting on mRNA have been identified, suggesting that the presence of the modification (and hence the fate of the affected mRNA) may be regulated in some manner (Shi et al., 2019; Zaccara et al., 2019).
2. m6A MODIFICATION
The m6A modification, found in most eukaryotes, is the most abundant modification in mRNA (Desrosiers et al., 1974), with estimates of approximately three m6As per mRNA (Yue et al., 2015). This modification has been shown to be important for the stability and translational efficiency of a number of mRNAs (Du et al., 2016; Fry et al., 2017; A. Li et al., 2017; Shi et al., 2017; X. Wang, Lu, et al., 2014; X. Wang, Zhao, et al., 2015), and is involved in many different physiological processes including embryonic development (Geula et al., 2015; Y. Wang, Li, et al., 2014; Zhao & He, 2015), circadian rhythm (Fustin et al., 2013; Shi et al., 2018), and many types of cancer (Jiang et al., 2021; Lan et al., 2019). In addition to METTL16, which is the focus of this review, there are a number of other m6A methyltransferases that have been identified (Table 1).
TABLE 1.
Human m6A RNA methyltransferases and their identified methylated RNAs
| m6A methyltransferase | Validated RNA methylome |
|---|---|
| METTL3/METTL14 | mRNAs, ncRNAs, microRNAs, circRNAs |
| METTL16 | U6 snRNA, MAT2A pre‐mRNA |
| METTL5 | 18S rRNA |
| ZCCHC4 | 28S rRNA |
2.1. METTL3/METTL14/WTAP complex
Interest in the m6A modification has recently grown in part due to the identification of a methyltransferase complex responsible for m6A‐methylating nascent pre‐mRNA within the nucleus. The core complex consists of METTL3 and METTL14, as well as Wilms' tumor associating protein (Agarwala et al., 2012; Ke et al., 2017; Lee et al., 2014; J. Liu et al., 2014; Ping et al., 2014). Other proteins including KIAA1429, RBM15, and RBM15B have also been shown to function with the core methyltransferase complex as loss of these proteins decreases cellular m6A levels (Patil et al., 2016; Schwartz et al., 2014). METTL3 contains a Rossmann‐like fold domain typical of other class I methyltransferases, and utilizes S‐adenosylmethionine (SAM) as a substrate to methylate adenosines within a DRACH consensus sequence (modified adenosine underlined), often found in 3′ UTR's and around both start and stop codons of mRNA (Chen et al., 2015; Y. Li et al., 2014; J. Liu et al., 2014; Luo et al., 2014; Meyer et al., 2012). METTL3 also contains two CCCH‐type zinc fingers outside of the methyltransferase domain that are required for RNA binding and hence methylation (Huang et al., 2019; P. Wang, Doxtader, & Nam, 2016). METTL14 lacks a SAM binding site and catalytic activity but does participate in mRNA binding/targeting (Sledz & Jinek, 2016; P. Wang, Doxtader, & Nam, 2016; X. Wang, Feng, et al., 2016). Although predominantly found in the nucleus, the methyltransferase complex and its activity has been found within the cytoplasm as well and is thought to be responsible for the bulk of cellular m6A mRNA modification (Bokar et al., 1997; Gokhale et al., 2016; Harper et al., 1990; Lin et al., 2016; Ping et al., 2014).
2.2. METTL16
More recently, another m6A methyltransferase, METTL16, has been identified although its targets appear to be more limited in number (Aoyama et al., 2020; Brown et al., 2016; Nance et al., 2020; Pendleton et al., 2017; Ruszkowska, 2021; Warda et al., 2017). Like METTL3, METTL16 contains the Rossmann‐like fold of class I methyltransferases and uses SAM as the methyl donor but appears to have additional regulatory and RNA binding domains which we will discuss below. The two verified targets of METTL16 share a consensus sequence (UACAGARAA) that is different from METTL3/14's consensus and the RNA methylation sites of the known METTL16 targets also harbor structural elements shown to be important for methylation (Doxtader et al., 2018; Mendel et al., 2018; Pendleton et al., 2017). Like METTL3, METTL16 contains additional regions outside of its methyltransferase domain that also interact with RNA substrates and presumably provide specificity (Mendel et al., 2018; Ruszkowska et al., 2018). Interestingly, in global mRNA binding protein identification studies in which polyadenylated mRNA is isolated along with endogenous interacting proteins, METTL16 is frequently captured, while components of the METTL3/14 complex are not (Baltz et al., 2012; Brannan et al., 2016; Castello et al., 2012; Conrad et al., 2016; Kwon et al., 2013). In addition, while many METTL16 RNA interactors have been identified, only two have been verified as methylation substrates, perhaps suggesting additional roles for METTL16 beyond its catalytic activity.
2.3. rRNA m6A methyltransferases
Even though m6A is prevalent in mRNA, it is also found in other types of RNAs including rRNA, long noncoding RNA (lncRNA), microRNA, and small nuclear RNA (snRNA) (Berulava et al., 2015; Iwanami & Brown, 1968; Reddy et al., 1972; Zhou et al., 2016). Due to its homology with the Escherichia coli 23S rRNA m6A methyltransferase, ybiN/rlmF, METTL16 was originally predicted to mediate eukaryotic rRNA m6A methylations. However, METTL5 has recently been shown to methylate 18S rRNA and ZCCHC4 has been identified as the eukaryotic 28S m6A methyltransferase (Ma et al., 2019; Pinto et al., 2020; Ren et al., 2019; van Tran et al., 2019). Interestingly, both enzymes appear to be very specific to their rRNA substrates although other RNA interactors have been preliminarily identified. While the function of these rRNA modifications is not yet known, structure predictions suggest that the m6A sites are located in areas important for translational fidelity (Dinman & Wickner, 1995; Piekna‐Przybylska et al., 2008; Qin et al., 2012). Thus, while METTL16's role in rRNA methylation is still unclear, substantial progress into its structure, function, and preferred RNA substrates have been made in recent years.
3. METTL16 DOMAINS AND THEIR FUNCTIONS
There are two known m6A RNA methyltransferases with verified mRNA targets in humans: the METTL3/14 complex, and METTL16. While there is a large overlap of structural similarity between these two enzymes (Doxtader et al., 2018; Ruszkowska et al., 2018) there are differences that suggest they have unique responsibilities in the cell. METTL3/14 has been extensively researched and many RNA interactors and methylation targets have been identified (a review of METTL3/14 can be found in S. Liu et al., 2020). METTL16 has been investigated much less and therefore has only a few known interactors.
METTL16 is a conserved protein, with homologs found from vertebrates to yeast and bacteria (Aoyama et al., 2020; Mendel et al., 2018; Pendleton et al., 2017). A phylogenetic tree showing a few of these homologs with respective protein diagrams is shown in Figure 1. From this analysis, it is clear the protein has evolved, adding an additional domain in vertebrates. METTL16's protein domains have been investigated and, for the human protein, are categorized as an N‐terminal RNA‐binding, methyltransferase, and vertebrate conserved region (VCR) domains (Figure 2). Although METTL16 has never been crystallized in its full form, four groups have published fragment structures of METTL16 (Aoyama et al., 2020; Doxtader et al., 2018; Mendel et al., 2018; Ruszkowska et al., 2018); there is also one unpublished structure listed as 2h00 in PDB (an excellent review of METTL16 structure can be found in Ruszkowska, 2021). Here we briefly discuss each domain, their responsibilities, and the implications in RNA interaction. We also discuss the region predicted to contain a nuclear localization sequence.
FIGURE 1.
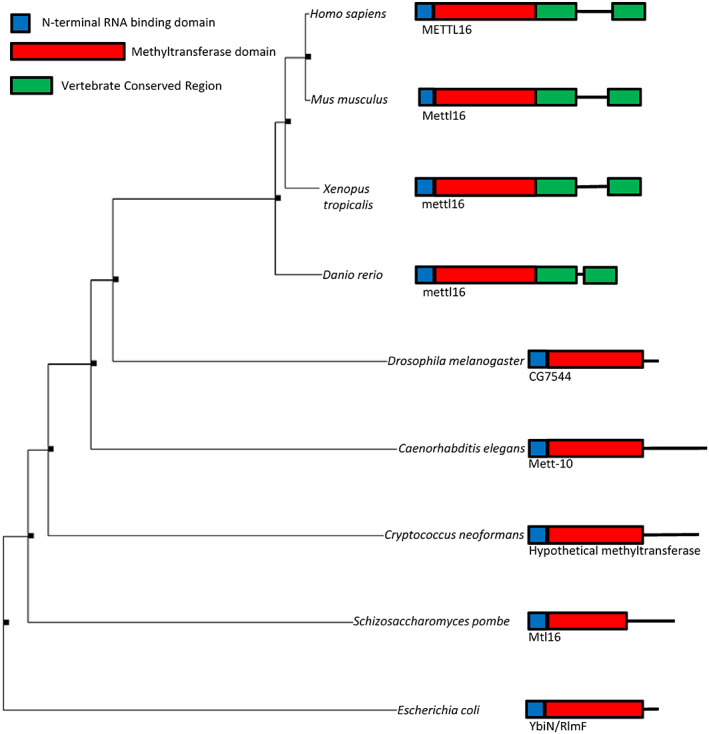
Phylogenetic tree of METTL16 and homologs. Simple protein diagram of each organism's homolog and the protein name is located next to each respective species. Tree was generated using Seaview using distance methods under standard parameters (Gouy et al., 2010). Sizes of protein diagrams are approximate and not to scale
FIGURE 2.

Schematic of human METTL16 protein. Numbers indicate amino acids from N to C terminals. RBD, N‐terminal RNA binding domain; VCR1, vertebrate conserved region 1; VCR2, vertebrate conserved region 2
3.1. N‐terminal RNA binding domain
The N‐terminus of METTL16 contains a group of polar and positive‐charged amino acids that form a groove large enough to accommodate double‐stranded RNA (Mendel et al., 2018; Ruszkowska et al., 2018) (see Figure 2). Among vertebrates it is highly conserved (Mendel et al., 2018; Ruszkowska et al., 2018) with lesser but still significant conservation in lower organisms (see Figure 1). This region is sometimes grouped with the methyltransferase domain as it was not distinguished from the methyltransferase domain until 2018 by Ruszkowska et al. who identified this region as one unique from other RNA methyltransferases (Ruszkowska et al., 2018). A few months later, Mendel et al. showed that mutations to charged residues (the first five arginines and lysines) in this region abolished RNA binding, and subsequently, methylation of RNAs (Mendel et al., 2018). Therefore, while the methyltransferase domain forms a groove to accommodate RNA during methylation, it is unable to secure the RNA for catalysis without the N‐terminal binding domain. While only five amino acids in this region have been shown to be responsible for RNA binding, the first 40 N‐terminal amino acids have been classified as an RNA binding domain, even though it does not follow “canonical” RNA‐binding motifs. This domain forms a projection from the methyltransferase domain of the protein and is thought to act as a “claw” to further stabilize the RNA in the methyltransferase RNA groove (Ruszkowska et al., 2018). Interestingly, in the co‐crystal structure, these residues do not appear to contact RNA (Doxtader et al., 2018), however, functionally they clearly play a role in RNA binding (Mendel et al., 2018). This domain is one of those unique to METTL16, potentially dictating the group of RNAs with which it can interact.
3.2. Methyltransferase domain
The methyltransferase domain of METTL16 is highly conserved through vertebrates (Aoyama et al., 2020; Ruszkowska et al., 2018), with lower conservation still identified through E. coli (Sergiev et al., 2008; Shima et al., 2017) (see Figure 1). As mentioned above, approximately the first half of the protein was termed the methyltransferase domain until 2018, whereas it is now considered to be comprised of amino acids 40–288 (see Figure 2). The methyltransferase domain has the conserved Rossmann‐like fold of class I methyltransferases containing the amino acids needed for several actions which are almost identical to METTL3 (Ruszkowska et al., 2018). A hydrophobic pocket and several hydrogen bonds are required for the proper placement of the adenosine portion of SAM while the canonical “NPPF” amino acid sequence stabilizes the methionine region in SAM (Doxtader et al., 2018). This “NPPF” region is also needed for destabilization of the sixth nitrogen on the RNA adenosine for transfer of the methyl group (Doxtader et al., 2018). The methyltransferase domain also provides additional support to the N‐terminal RNA‐binding domain with positive‐charged amino acids that line the internal groove (Mendel et al., 2018; Ruszkowska et al., 2018). As mentioned above, ridding METTL16 of the N‐terminal RNA binding domain abrogates RNA binding (Mendel et al., 2018). Even though the methyltransferase domain contains a groove lined with amino acids that can attract the negative‐charged RNA backbone, it alone does not appear to be strong enough to retain the RNA in the groove. Intriguingly, this RNA accommodating region is not an open‐ended groove like METTL14, which is needed to bind single‐stranded RNA at an internal site, rather it is a closed groove (Mendel et al., 2018; X. Wang, Feng, et al., 2016). It is therefore suggested that the RNA interaction here must either be at the end of the strand or in a hairpin secondary structure.
One unique aspect of METTL16's methyltransferase domain is the “auto‐regulatory K‐loop” (amino acids 163–167) which lowers methylation efficiency by obstructing the SAM binding pocket (Doxtader et al., 2018). Lowered methylation efficiency is suggested to attenuate the overall activity of METTL16 in the cell (Doxtader et al., 2018) and could perhaps contribute to its role as a methyl sensor. It is therefore plausible that mutations in this region, which have been shown to produce a hyperactive methyltransferase, could result in dysregulation (Doxtader et al., 2018). A more detailed account of the biochemical mechanics of METTL16's RNA methylation can be found here (Lence et al., 2019).
3.3. VCR domain
The VCR domain is located at the C‐terminus of METTL16 from amino acids 310–562 (see Figure 2). The domain contains two regions (VCRs) conserved among vertebrates (Aoyama et al., 2020; Pendleton et al., 2017; Ruszkowska et al., 2018) (see Figure 1). Amino acids 289–400 are classified as VCR1, 514–562 are classified as VCR2, and 401–513 are considered a linker region between these two (Aoyama et al., 2020). The domain was first discussed in 2017 as a potential splicing director for MAT2A RNA when cellular SAM levels were below a critical level (Pendleton et al., 2017). Due to its highly dynamic nature, the crystal structure of this region was not determined until 2020 (Aoyama et al., 2020). Crystallization was achieved by removing the unstructured linker region between the two VCRs (Aoyama et al., 2020). Because of the presumed motion of this domain, it is believed to bind near the target adenosine (in a double‐stranded region) to bend the RNA and allow better access of the adenosine to the core methyltransferase domain (Aoyama et al., 2020).
Interestingly, the VCR domain is structurally homologous (although not in sequence) with the kinase‐associated 1 (KA1) domain of TUT1, a U6 snRNA‐specific terminal uridylyl transferase (Aoyama et al., 2020). As in TUT1's KA1 domain, the VCR domain of METTL16 acts as a clamp that binds double‐stranded RNA. The KA1 domain in TUT1 is known to aid U6 snRNA binding for polyuridylation (Trippe et al., 1998), however the KA1 domain is also found in other proteins such as serine/threonine kinases (Paung & Seeliger, 2018). The proposed functions of the domain in these kinases include autoinhibition and targeting the protein for anionic membranes (Marx et al., 2010; Moravcevic et al., 2010). It has been noted that these domains are observed to show low sequence conservation while maintaining high structural homology (Paung & Seeliger, 2018). With these similarities between the VCR and KA1 domains, it is possible that METTL16 has roles outside those currently known and solidifies U6 snRNA as one of the main METTL16 interactors. Because the VCR domain of METTL16 has only recently been identified, the next few years of investigation into this region will be exciting.
3.4. Potential nuclear localization sequence
Located in the region between the end of the methyltransferase domain and the beginning of the VCR domain, a vertebrate‐conserved sequence of amino acids has a computationally predicted (yet unverified) nuclear localization sequence (SKRRKLEKPRK, amino acids 300–310; Nguyen Ba et al., 2009). The Caenorhabditis elegans homolog of METTL16, METT‐10, which contains a verified nuclear localization sequence in a non‐conserved region, is transported to the nucleus by dynein light chain during mitosis (Dorsett & Schedl, 2009). Originally thought to be predominantly nuclear in human cells as well, our recent study using biochemical fractionation of several cell types suggests METTL16 is present in both the nucleus and cytoplasm (Nance et al., 2020). In addition, when METTL16 was overexpressed, more was observed in the cytoplasmic fraction. Therefore, it may be that METTL16's main cellular compartment is the nucleus, with accumulated excess located in the cytoplasm.
4. METTL16 RNA INTERACTORS
To date, three RNAs have been established as bona fide interactors of METTL16: MAT2A mRNA, U6 snRNA, and MALAT1 lncRNA. While MALAT1's methylation via METTL16 has not been proven, both MAT2A and U6 are definitively m6A methylated by METTL16. Similarity in the methylated region of these two RNAs led to the identification of a consensus nonamer sequence “UACAGARAA,” where the fourth base (underlined) is modified. While it is unlikely this consensus is a mere coincidence, it does not exclude the possibility of additional sequence determinants. For example, studies have found that this nonamer sequence, which is found in thousands of other human mRNAs, is not widely methylated (Pendleton et al., 2017). Furthermore, in the METTL16 knockout mouse embryo, expression level of RNAs with the consensus nonamer sequence was not affected by decreased METTL16 levels (Mendel et al., 2018). These and other observations suggest that while potentially necessary for methylation, perhaps this nonamer sequence alone is not sufficient for METTL16 methylation. Indeed, current data suggests that the nonamer sequence needs to be in a specific secondary structure for methylation to occur (Doxtader et al., 2018). However, since the consensus sequence is based on only a few known interactors, this may also suggest the need to expand the RNAs considered for METTL16 binding and/or methylation. In this section, we review the three well‐established RNAs and their interactions with METTL16. We also discuss other potential RNA interactors and the resulting implications.
4.1. MALAT1 lncRNA
Metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1), also known as NEAT2 (nuclear‐enriched abundant transcript 2), is a common and conserved mammalian lncRNA known for its widespread role in transcriptional regulation, alternative splicing, and sequestering miRNAs (Amodio et al., 2018; Hutchinson et al., 2007). MALAT1 lncRNA is ~8000 bases, with the last ~100 bases folded in a triple helix structure, as shown in Figure 3. Most notably, MALAT1 expression has multiple implications in cancer progression (reviews of MALAT1 can be found in Amodio et al., 2018; Arun et al., 2020). It was also the first RNA discovered to associate with METTL16 through the use of electrophoretic mobility assays and UV crosslinking in HeLa cells (Brown et al., 2016). Up until this time, METTL16 was predicted to be a putative rRNA methyltransferase. The specific interaction region of MALAT1 with METTL16 was identified using RNA‐proximity ligation assays (Brown et al., 2016) and shown to include the element for nuclear expression. Moreover, the ligation did not occur without the VCR domain of METTL16, providing evidence that this was the region of MALAT1 binding. Interestingly, an m6A‐modified adenosine has been identified in MALAT1 at adenosine 8290 near the location of METTL16‐MALAT1 interaction site (Jin et al., 2019; Linder et al., 2015), but it does not appear to be deposited by METTL16. Instead, the sequence with the m6A modification overlaps with the “RRACH” consensus sequence favored by the METTL3/14 complex (R = purine and H = A, C, or U) (J. Liu et al., 2014) and has been shown to increase with METTL3 overexpression, suggesting that it is deposited by METTL3/14 (Jin et al., 2019). The purpose of METTL16 binding to MALAT1 is currently unknown. If methylation does not occur, it may a way to sequester MALAT1 or target it for interaction with an unidentified METTL16 protein cofactor. The triple helix of MALAT1 has been shown to stabilize and delay degradation of the RNA (Brown et al., 2012) but the role of METTL16 in this process has not been explored. This RNA–protein interaction is intriguing not only because methylation does not seem to occur, but also because METTL16 does not have any other identified RNA interactors in the triple helical form. Whether MALAT1 is unique in its status of being bound but not methylated by METTL16 remains to be determined. It is possible that this is another way METTL16 regulates its RNA interactors.
FIGURE 3.
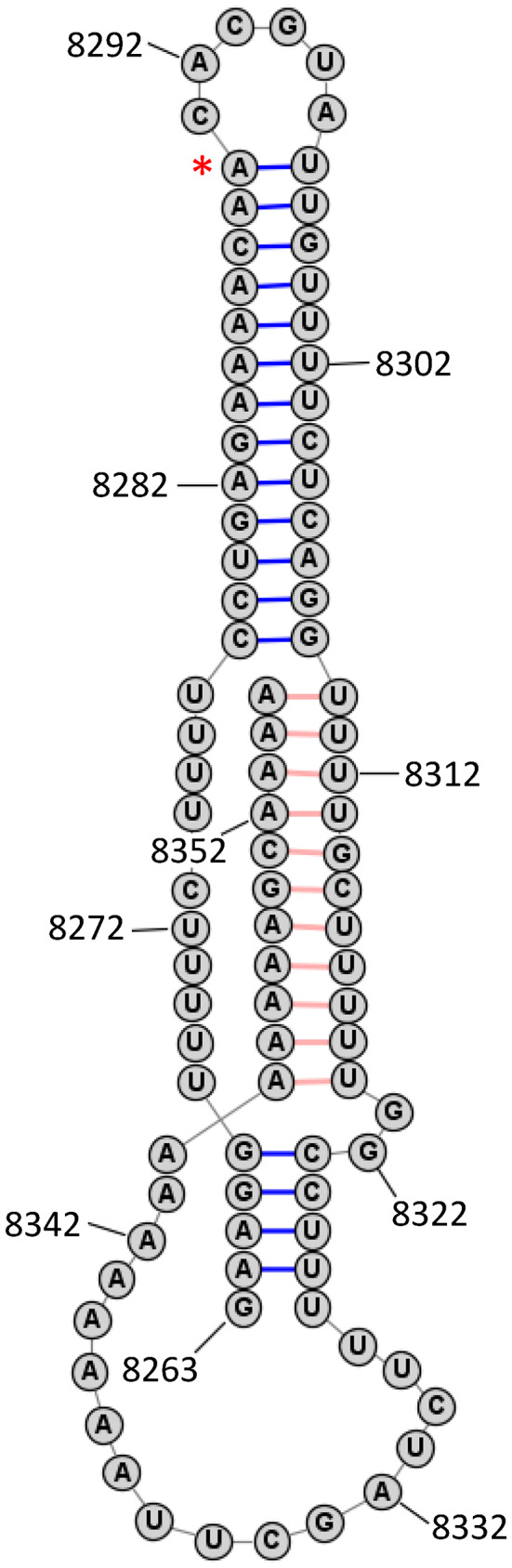
Partial RNA structure of human lncRNA MALAT1 predicted to bind METTL16. Structure shows triple helix region where METTL16 is reported to interact. Asterisk indicates predicted m6A site. Adapted from Brown et al. (2012)
4.2. MAT2A mRNA
MAT2A is an mRNA that encodes the “A” (or alpha) subunit of methionine adenosyltransferase 2 (MAT2), which is the catalytic subunit of this enzyme. MAT2, as the name suggests, is responsible for fusing a methionine to adenosine to produce S‐adenosyl methionine (Cantoni & Durell, 1957). This resulting molecule, known also as AdoMet or SAM, is responsible for most methyl donation reactions in the cell, whether it be for DNA, RNA, protein, or other molecules (Halim et al., 1999). This reaction is energetically expensive, costing the cell one ATP molecule per SAM molecule produced (Cantoni & Durell, 1957). As such, production of SAM is highly regulated to control the amount of methylation reactions that occur.
SAM regulation varies among kingdoms of life and it seems mammals have evolved post‐transcriptional regulation of MAT2A as one way to achieve this (there is also evidence of two miRNAs targeting both MAT2A and MAT2B; Lo et al., 2013; Simile et al., 2019, not discussed here). A 2017 study demonstrated METTL16‐regulated differential splicing of MAT2A RNA in response to SAM levels in human embryonic kidney cells (Pendleton et al., 2017). When SAM levels are adequate, METTL16 binds and methylates adenosines in several hairpin loops located in the MAT2A 3′ UTR (Figure 4). This impairs splicing of the terminal intron, leading to intron retention and degradation of the mRNA. However, when methionine levels in the media drop below a certain level (11–33 μM), METTL16 binds but is unable to complete the methylation reaction and remains bound to the hairpins. METTL16 then recruits splicing machinery to the last intron allowing for expression of a full‐length mature mRNA and proper translation of the MAT2A protein (Scarborough et al., 2021). Thus, through the sensing of SAM levels via its catalytic activity, METTL16 is able to maintain adequate SAM levels for all cellular methylation reactions. It should be noted that the MAT2A hairpins in Figure 4 are displayed as their stable structure when unbound by METTL16 (Parker et al., 2011). It has been shown that when bound to METTL16 (at least for hairpins 1 and 6), the structure changes (this structure can be found here: Doxtader et al., 2018).
FIGURE 4.
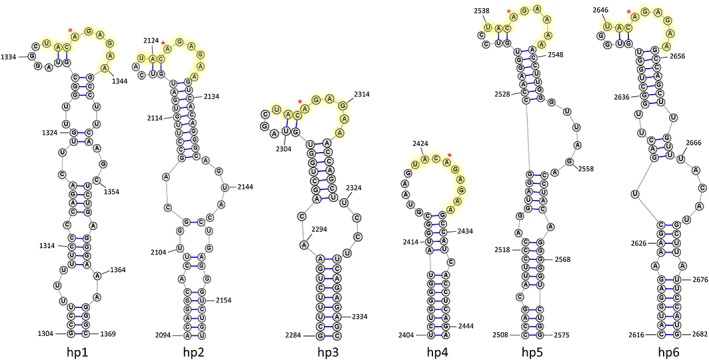
Partial RNA structures of human mRNA MAT2A 3′ UTR. The six hairpins are labeled relative to 5′ to 3′ direction. Nonamer sequence is highlighted, and asterisk indicates predicted m6A sites. Structures shown are when unbound to METTL16. Adapted from Parker et al. (2011)
Previous studies have also shown that MAT2A's mRNA stability is also dependent on SAM levels (Martinez‐Chantar et al., 2003) and recently METTL16 has been confirmed to bind several regions of the 3′ UTR of MAT2A mRNA (Warda et al., 2017). Furthermore, upon m6A methylation of the 3′ UTR hairpins, MAT2A reporter mRNA is bound by the m6A reader YTHDC1 and targeted for degradation in HeLa cells (Shima et al., 2017). All six MAT2A 3′ UTR hairpins were found to be m6A modified (and also had lower levels of modification after METTL16 knockdown), a finding supported by a single‐base‐resolution m6A study as well (Koh et al., 2019). Ridding the MAT2A mRNA reporter of only the fifth and sixth hairpins impaired the mRNA stability response to low SAM levels, however, to fully interrupt this response, at least four hairpins had to be mutated. Interestingly, both studies (Pendleton et al., 2017; Shima et al., 2017) showed increased MAT2A mRNA in response to treatment with cycloleucine, a competitive inhibitor of the MAT2A enzyme, although Pendleton et al. showed higher levels of the intron‐retained nuclear isoform compared to Shima et al. Thus while there may be small differences due to cell type, growth conditions, media, detection methods, and so on, in both cases MAT2A mRNA was posttranscriptionally responsive to SAM levels in a METTL16‐dependent manner.
Shima et al. noted two further observations. Downregulation of FTO, a m6A demethylase, resulted in lower expression of MAT2A reporter mRNA suggesting that demethylation may play a role in maintaining basal MAT2A mRNA levels even in the presence of sufficient SAM. Also, despite showing clear regulation of a MAT2A reporter RNA, YTHDC1 knockdown showed no effect on endogenous MAT2A RNA expression (Shima et al., 2017). It may be that the 5′ UTR is regulated in a way that is redundant with YTHDC1 interaction (since this would be present on the endogenous mRNA but not included in the reporter RNA), however this is speculative. It is also possible that this is an artifact of using a reporter RNA, but again that possibility would need to be explored further. Still, it is clear that methylation via METTL16 regulates MAT2A mRNA stability.
Overall, these two studies consistently demonstrate that when cellular SAM levels are adequate, MAT2A mRNA is targeted for degradation both in the nucleus (through aberrant splicing and subsequent degradation) and the cytoplasm (though increased degradation) via METTL16 activity. Conversely, when cellular SAM levels decrease and MAT2A is needed, increased splicing, along with increased mRNA half‐life ensure the cell can increase its SAM synthesis to meet demands. This redundancy may be a fail‐safe mechanism because if METTL16 levels were depleted for some reason, intron retention would reduce the production of new mature mRNA, however reduced m6A methylation would also lead to increased half‐life of the mature mRNA. It is very possible that all mechanisms mentioned, and potentially others not yet known, can contribute to an elaborate posttranscriptional regulation process of intracellular SAM levels.
Currently, only MAT2A hairpins 1 and 6 have been crystallized in complex with the N‐terminal and methyltransferase domains of METTL16 (amino acids 1–291) (Doxtader et al., 2018). These structures were acquired through stabilization of the RNA by removing the mismatches normally found in the wild‐type MAT2A hairpin stems. Using hairpin 1, Aoyama et al. determined methylation by full‐length METTL16 occurred at K m = 0.027 μM and had a K d = 0.042 μM (Aoyama et al., 2020). Interestingly, when using only the first two domains of METTL16 (amino acids 1–300, lacking the VCR domain), methylation was much lower at K m = 0.76 μM. Of note, several previous studies have used this truncated version of the METTL16 protein to obtain their results as well (Doxtader et al., 2018; Pendleton et al., 2017; Ruszkowska et al., 2018) so variations between publications are likely dependent on the exact nature of the METTL16 construct used.
Because of the numerous implications of SAM regulation through MAT2A levels, other studies have investigated METTL16's role in this process. An attempt to knockout METTL16 in mice revealed viable embryos up to the implantation stage where it is known a large DNA methylation event (following massive demethylation) occurs to radically change RNA expression (Mendel et al., 2018). Even though this group did not observe a large change in the 3′ intronic region of MAT2A (the region of METTL16‐directed alternative splicing), aberrant splicing was reported, and it was concluded the embryo cannot recover from the demethylation event without METTL16's involvement.
There are several papers that focus on correlating RNA expression with survival rates of specific cancers that have shown anomalous METTL16 levels to be consistently related to poor outlook (Hou et al., 2020; K. Li et al., 2020; X. Liu et al., 2019; P. Wang et al., 2020; S. Wang et al., 2021; Yeon et al., 2018; Zhang et al., 2020). Although it has not been proven, it may be that dysregulation of MAT2A is the main cause for this relation, given that unregulated methylation events in the cell can lead to a plethora of overexpressed protooncogenes/oncogenes or under‐expressed tumor suppressors through epigenetic mechanisms (Kalev et al., 2021; K. Li et al., 2020; S. Liu et al., 2020). This is speculative since it has not been shown that METTL16 is directly responsible. There is even a study relating the lack of a typical microbiome with MAT2A hypomethylation and subsequent lower expression (due to downregulation of METTL16) in mice intestine, colon, and brain cells (Jabs et al., 2020). Whether METTL16's influence on MAT2A is the sole cause of these observations or there are influences left to be discovered, it is certain that METTL16 is a key component in overall cellular processes.
4.3. U6 snRNA
snRNA U6 directs intron splicing by associating with and destabilizing the 5′ splice site, as well as serving as the catalytic subunit of the splicing machinery. About 40 years ago, human U6 snRNA was determined to have an m6A at A43 (Epstein et al., 1980), located in a stem bulge of the major hairpin structure (Figure 5). However, it was not until a few years ago that METTL16 was identified as the responsible m6A methyltransferase in vitro for A43 (Pendleton et al., 2017). Interestingly, while METTL16 homologs have been identified in most species, the m6A status of U6 snRNA in species other than human and yeast has not been investigated. More research is needed to determine the methylation status of these snRNA homologs and, if present, whether it is due to the respective METTL16 homolog.
FIGURE 5.
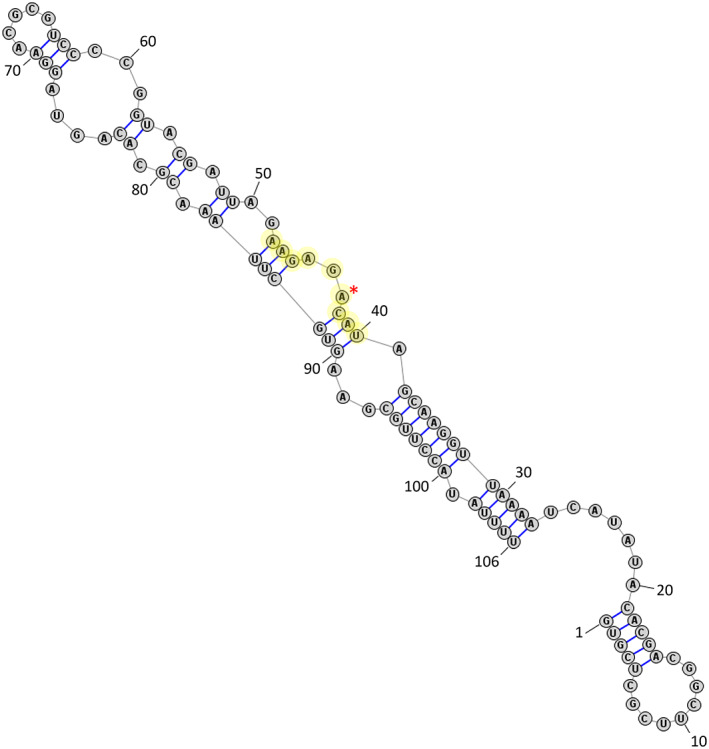
Full RNA structure of human snRNA U6 predicted to bind METTL16. Numbered bases indicate 5′ to 3′ direction. Nonamer sequence is highlighted, and asterisk indicates m6A site (A43). Adapted from Aoyama et al. (2020)
Human U6 is methylated by METTL16 with a K m = 0.025 μM and a K d = 0.016 μM when using full length METTL16 (Aoyama et al., 2020). These values are comparable to those of MAT2A methylation and binding, but surprisingly, removing the METTL16 VCR domain increases the K m to more than 4 μM for U6. For comparison, the K m for MAT2A using the same truncated METTL16 was 0.76 μM. The authors suggest the structural differences surrounding the methylation sites in each RNA explain this, with U6 snRNA showing more flexibility than the MAT2A mRNA (Aoyama et al., 2020). Thus, it is proposed that without the stabilization of the RNA via the VCR domain, METTL16 cannot easily keep U6 in the binding pocket long enough for methylation to occur. Furthermore, it is suggested that the VCR domain binds and bends U6 so the adenosine to be modified is more accessible to the pocket where methyl transfer occurs. Thus, through U6‐METTL16 kinetics, the importance of the VCR domain is highlighted.
It should be noted that while the region surrounding the m6A methylation site of U6 is necessary for properly directing splicing, it has not been proven (although it has been speculated) that an unmethylated adenosine in this position could not perform the same function. Warda et al. deeply explored the U6‐METTL16 interaction and determined modification of A43 occurs after the 5′ capping of U6 but before association of U6 with the rest of the spliceosome complex (Warda et al., 2017). They speculated that, because U6 associates with pre‐mRNA via weak interactions, the m6A thermodynamically “fine‐tunes” this interaction. It is known that double‐stranded RNA is stabilized by an m6A near but not in the duplex, whereas it is destabilizing if located in the duplex (Roost et al., 2015). Therefore, the m6A at A43 in U6 may further weaken binding with the pre‐mRNA for proper splicing and dissociation after splicing is complete. Interestingly, knockdown of METTL16 in HEK293 cells resulted in no change in overall U6 levels but ~50% reduction in U6 containing an m6A (Warda et al., 2017). However, because there is an additional m6A at A76 in U6 RNA, this may be expected if the second m6A is not deposited by METTL16 (Warda et al., 2017). Embryonic knockout of METTL16 in mice showed no global changes in splicing in the transcriptome, suggesting this U6 methylation may not be necessary for splicing in mammals (Mendel et al., 2018). Regardless, U6 is a reliable RNA of the human METTL16 interactome and methylome, although the exact function of the m6A modification remains a mystery.
4.4. Other RNA Interactors
4.4.1. 18S and 28S rRNA
MAT2A, U6, and MALAT1 are the most widely accepted RNA interactors of METTL16. However, several other types of RNAs have been implicated as potential interactors. It is well established that rRNAs contain modified nucleotides including m6A (Boccaletto et al., 2018; Cantara et al., 2011). Immunoprecipitations of both endogenous and exogenous METTL16 and identification of associated RNAs have revealed several rRNAs as potential interactors (Brown et al., 2016; Nance et al., 2020; Warda et al., 2017), although this could be attributed to non‐specific crosslinking (due to proximity) or interactions (due to the large amount of rRNAs in total cellular RNA). The closest related METTL16 homolog in E. coli, ybiN/rlmF (~31% sequence homology), is the methyltransferase responsible for depositing an m6A on the 23S rRNA (Sergiev et al., 2008). Additionally, a probable 28S rRNA m5C methyltransferase, NOP2, has been identified in complex with METTL16 (Mendel et al., 2018). Therefore, it is plausible that METTL16 binds and/or methylates rRNA. While rRNA m6A levels have not been investigated, knockdown of METTL16 does not result in differences in overall expression of rRNAs but this is not surprising due to the shear amount and half‐life of this class of RNA. However, as mentioned previously, there are two other identified rRNA m6A methyltransferases in vertebrates (METTL5, ZCCHC4), so it is possible that through evolution one or both of those methyltransferases took on this responsibility and METTL16 no longer methylates rRNA in human cells.
4.4.2. DNA damage associated small RNAs
Recently, a study investigating DNA damage via ultraviolet radiation observed the recruitment of METTL16 to the region of damage and association with small RNAs in the vicinity (Svobodova Kovarikova et al., 2020). While recruitment of m6A‐modified RNAs to the region of UV‐induced DNA damage has been shown, a previous study identified recruitment of METTL3/14 to the damaged region (Y. Xiang et al., 2017). The more recent study showed a small percentage of irradiated cells demonstrating higher localized METTL16 levels during subsequent repair. While this finding awaits further verification, it is an interesting development that expands upon the RNAs expected to interact with METTL16.
4.4.3. Other mRNAs
Identifying additional METTL16 RNA interactors has been more difficult. There are multiple deep sequencing datasets of RNA resulting from immunoprecipitations of exogenous METTL16 and knockdown studies of endogenous METTL16. However, there is little consensus among these sets other than the three established RNA interactors (MAT2A, U6, and MALAT1) already described. We caution that some that the immunoprecipitations and RNA expressions could result from non‐specific pulldowns or secondary effects due to dysregulation of MAT2A (and therefore cellular SAM and m6A levels; (Koh et al., 2019; Mikutis et al., 2020). Many of the potential RNA interactors were identified using an RNA–protein crosslinking method. Because METTL16 has multiple RNA binding domains (one of which seems highly dynamic and could potentially bind generic double‐stranded RNA), it is possible many of these “interactors” are simply RNAs that were briefly in contact with METTL16 but not truly bound by the protein (Ascano et al., 2012; Hafner et al., 2021; Mukherjee et al., 2011; Wheeler et al., 2018). One must also consider the proteins found to interact with METTL16: if these proteins bring RNA near METTL16, it could again become crosslinked with METTL16 without being a true interactor. It is therefore imperative to use other methods and evidence to verify these as bona fide METTL16 interactors.
Using m6A immunoprecipitation datasets published by Pendleton et al. and others, one group produced a bioinformatics‐based approach to determine the overall cellular processes affected by several m6A methyltransferases (Song et al., 2019). These datasets revealed the RNAs that were hypomethylated after METTL16 knockdown in HEK293 cells (Pendleton et al., 2017). They categorized the resulting high‐scoring RNAs and highlighted the top 10 processes with the most RNAs in each group. The highest scored groups were: “endoplasmic reticulum‐associated misfolded protein catabolic process,” “regulation of cell cycle,” “actin cytoskeleton organization,” “positive regulation of apoptotic process,” and “protein ubiquitination.” To avoid potential overlap, they excluded genes that were shown to be reliant on more than one methyltransferase. This may have eliminated processes that are affected by multiple methyltransferases from the higher scoring processes. They also warned that, as only one dataset was available for the METTL16 methylome at the time of publication, future datasets are needed to solidify these findings, but will also likely skew the current readout. While they provided an in‐depth library of genomic features to determine likely types of and regions on methylated RNAs, they warn it is not completely extensive. Even given these caveats, this is still one of the publications that fills a hole in the much‐needed knowledge of METTL16's responsibilities, and the addition of more datasets is eagerly awaited.
A more recent publication using synthetic reactive SAM, which is added to RNA by its corresponding methyltransferase, revealed ~70% overlap with previous antibody‐based datasets for METTL16 interactors (Mikutis et al., 2020). The overlap was identified in MOLM‐13 cells which stably expressed a short hairpin RNA targeting METTL16 and confirmed with m6A IP. The identified RNAs strongly aligned with a “UACAG” consensus, shortened from the “UACAGARAA” consensus sequence found in MAT2A and U6. This is an exciting addition to METTL16 investigation by giving verification to the somewhat fickle datasets produced through immunoprecipitation alone. Within this group of RNAs, ~7600 are mRNA and ~900 are lncRNA. However, as with other studies, there was concern about the potential of a secondary effect due to METTL16's influence on MAT2A and hence cellular SAM levels. As a solution, RNA from the METTL16 set was compared to RNA identified as METTL3‐methylated. If an RNA showed dependence on both methyltransferases, it was assumed to most likely be a METTL3 target with a secondary effect from METTL16. Similarly, a study from 2019 used an approach similar to an m6A IP (m6A‐crosslinking‐exonuclease‐sequencing [m6ACE‐seq]) coupled with knockdown of the respective methyltransferase in HEK293T cells to identify METTL3 and METTL16 RNA interactors. They suggested that a large portion of RNAs identified as METTL16‐dependent were actually the result of indirect dependency due to SAM regulation (Koh et al., 2019). The Mikutis study also discovered a striking similarity between the METTL16‐dependent lncRNAs m6A region and intronic polyadenylated sites suggesting that METTL16 may have a role in intronic polyadenylation which could direct splicing and contribute to cancer progression (Mikutis et al., 2020). As mentioned previously, the VCR domain of METTL16 is structurally homologous to the KA1 domain of TUT1 (Aoyama et al., 2020). In TUT1, the KA1 domain binds double‐stranded RNA for polyuridylation to occur (Trippe et al., 1998). This suggests the VCR domain could act in a similar fashion and therefore adds to the theory of METTL16's involvement in intronic polyadenylation.
Our recent publication using endogenous METTL16 immunoprecipitation without crosslinking identified several RNAs bound to METTL16: NT5DC2, MYC, HIF1A, β2M, and STUB1 (Nance et al., 2020). Knockdown of METTL16 in vitro shows differential expression in some but not all these identified RNAs. Interestingly, these RNAs have been identified in some of the previously published exogenous METTL16 immunoprecipitation and endogenous knockdown datasets as well (Koh et al., 2019; Pendleton et al., 2017; Warda et al., 2017). Reoccurring identification of these RNAs in multiple datasets obtained via different methods argues against non‐specific pulldowns but could suggest they may simply be weaker interactors with METTL16 which will require more evidence to confirm binding and methylation and determine the role of each.
5. METTL16 IN MODEL ORGANISMS
5.1. C. elegans
There are two groups who have studied the C. elegans METTL16 homolog, METT‐10. The first group published two papers in 2009 after METT‐10 was identified in a screen for effectors of cell proliferation and differentiation (Dorsett et al., 2009; Dorsett & Schedl, 2009). Worm lines with genetic variations in METT‐10 showed that it promotes cell cycle progression and potentially meiotic entry. Nuclear accumulation of METT‐10 was observed during meiosis, whereas METT‐10 knockout promoted mitotic entry. METT‐10 had previously been identified as a dynein light chain 1 (DLC‐1) interactor from a genome‐wide yeast two‐hybrid study (S. Li et al., 2004). Interestingly, knockdown of dlc‐1 caused a decrease in nuclear METT‐10 protein in both germ and somatic cells using fluorescent microscopy. However, overall METT‐10 protein levels were also reduced, prompting further investigation. With the dlc‐1 RNA knockdown, they showed mett‐10 RNA was also reduced, although the mechanism was not investigated. It was also determined that METT‐10 exists as an oligomer. With mutated METT‐10 studies, they determined that METT‐10's catalytic activity is essential for proper cell cycle progression, although it seems when nuclear localization was disrupted, cytoplasmic METT‐10 could still modify RNAs before they entered the nucleus and not disrupt cell cycle progression.
More recently, another group produced a METT‐10 knockout line of C. elegans and report, with both m6A‐RIP‐Seq and SCARLET methods, that m6A was undetectable in both U6 snRNA and sams‐3, sams‐4, and sams‐5 (the SAM synthetase mRNAs) (Mendel et al., 2021). The SAM synthetase RNAs' splicing was inhibited upon m6A modification by METT‐10, leading to RNA degradation and no functional protein produced. To investigate the similarity of METT‐10 function to human METTL16, a sams‐3 mRNA reporter was introduced into human HeLa cell extracts. Splicing of the reporter RNA was inhibited in the same manner as in worm extracts suggesting human METTL16 could carry out the reaction. Furthermore, the U2 auxiliary factor 35 (U2AF35) was unable to bind to the m6A‐modified sams fragment, which ultimately blocked splicing in this area. Interestingly, U2AF35 was not needed if the pre‐mRNA had a strong polypyrimidine tract adjacent to the 3′ splice site, in which case only U2AF65 was needed. While C. elegans do not show these strong polypyrimidine tracts, it may be that other organisms use this method to regulate splicing only in certain RNAs. Interestingly, no global splicing changes were observed upon loss of U6 methylation. METT‐10 knockout in C. elegans did show a strong fertility defect suggesting METT‐10 may contribute to fertility in ways other than those discussed. These results suggest that METT‐10 can inhibit splicing through 3′ splice site m6A modification, however most predicted METTL16 m6A sites in human cells are not at 3′ splice sites. Furthermore, there is currently no evidence that the m6A deposited by METT‐10 in human extracts specifically inhibits splicing or activates alternative splicing.
5.2. Mus musculus
To date, the only in vivo knockout of METTL16 in mice was accomplished by insertion of a triple‐stop codon into the Mettl16 gene (Mendel et al., 2018). Unable to obtain viable homozygous mice, heterozygous mice were used to produce knockout mouse zygotes and embryonic development tracked. At embryonic day 2.5, the METTL16 null mice were present in normal Mendelian ratios and showed only ~20 RNAs differentially expressed, including MAT2A, compared to the heterozygous and wild‐type Mettl16 mice embryos. At embryonic day 3.5, normal Mendelian ratios were also obtained, however there were ~5000 differentially expressed RNAs. By embryonic day 6.5, the METTL16 knockout mice were only 2% of the population and at embryonic day 8.5, no knockout embryos were detected. It was theorized that the large DNA methylation and demethylation that occurs around this time of development was unable to occur without proper MAT2A regulation by Mettl16 leading to loss of viability. While the RNAs identified in this paper are not human, they can be compared to the human homologs to determine the RNAs most likely regulated by Mettl16 during development. This group has also produced a conditional knockout of Mettl16, which was induced once the mice were adults (Mendel et al., 2021). The males, which show the highest level of Mettl16 tissue expression in the testes, were infertile due to lack of germ cell development. While they did not investigate this further, they determined that Mettl16 has important roles beyond that in embryonic development.
6. CONCLUDING PERSPECTIVES
Here we have discussed the individual domains of the METTL16 protein, their proposed functions, and known and predicted RNA interactors of the enzyme. The amount of published work on METTL16 is impressive, given that the first study citing function was only published in 2016. Even so, there is quite a bit still unknown. METTL16 has been shown by several CRISPR screens to be essential for life, and all reported attempts to remove METTL16 via CRISPR‐Cas9 from normal and cancerous cells have been unsuccessful (Barbieri et al., 2017; Guzzardo et al., 2017; Mendel et al., 2018; T. Wang, Birsoy, et al., 2015). While there has been speculation, it is not yet proven what METTL16 does that makes it indispensable. Two main theories are currently discussed: regulation of MAT2A mRNA or regulation of U6 snRNA. It should be noted that neither has been proven and there is still the possibility that one, neither, or both are the reason for METTL16's essentiality.
MAT2A transcription and mRNA half‐life are regulated by multiple pathways in addition to METTL16 (Halim et al., 1999, 2001; Ramani & Tomasi, 2012; Vazquez‐Chantada et al., 2010). As previously mentioned, MAT2A is the enzymatic subunit of methionine adenosyltransferase 2, responsible for producing SAM, which acts as a methyl group donor for most cellular needs. This is just one part of the methionine cycle in the cell, where several enzymes work together to keep methionine readily available for methylation and other reactions, such as protein synthesis and the glutathione pathway. Because humans do not synthesize methionine, it is critical for the cell to tightly regulate its usage. Moreover, excessive methyl‐donating molecules in the cell can lead to hypermethylation of chromatin, RNA, and others, resulting in aberrant cell functions. At the same time, under‐production of the methyl‐donators would lead to hypomethylation, again skewing cell functions. Therefore, it is plausible that loss of interaction with METTL16 would be enough to permanently injure SAM production and cause lethality.
In our previous studies, transient knockdown of METTL16 did not result in a change in MAT2A protein expression (Nance et al., 2020). However, upon both METTL16 knockdown and cycloleucine treatment, others have reported decreased MAT2A protein levels (Pendleton et al., 2017; Shima et al., 2017). A potential solution to understanding the role of SAM in METTL16's essentiality would be to overexpress MAT2A before removing METTL16 from the genome. Supplementing the culture media with extra methionine could ensure enough substrate for optimal methionine adenosyltransferase performance, avoiding a potential complication. In addition, deeper investigation into the specific in vivo interaction between METTL16 and MAT2A could be accomplished by inactivating a particular METTL16 functional domain (methyltransferase, N‐terminal RNA binding, etc.) and observe the effect, if any, on MAT2A mRNA stability and modification.
The other major METTL16 interactor is U6 snRNA, a spliceosome RNA. This RNA binds a pre‐mRNA at the 5′ splice site, helping to destabilize this region, so when the 3′ splice site is brought into proximity, transesterification can occur. It is unknown whether the m6A modification is needed for proper function of U6. The modification of U6 was shown to be inefficient without the VCR domain of METTL16 (Aoyama et al., 2020). As mentioned earlier, the VCR domain is unique to vertebrates. However, because the m6A in U6 is highly conserved from vertebrates to yeast (Brow & Guthrie, 1988), either the m6A is deposited another way outside of the vertebrate classification, or the methylation may not be necessary for proper function. It could also be that methylation is necessary for some but not all species. A METTL16 homolog has not been reported in Saccharomyces cerevisiae, however, fission yeast such as Schizosaccharomyces pombe do have a METTL16 homolog (Mtl16; see Figure 1) and their U6 snRNA is m6A modified. Removal of the Mtl16 homolog is not lethal, although it does result in slower growth rates (Watts et al., 2018). A more recent study in S. pombe found that without U6 snRNA m6A modification, splicing of some RNAs were affected. Pre‐mRNAs that contained an AAG sequence at the 5′ splice site were unaffected by the m6A status of U6, however those that contain a BBH sequence required the m6A in U6 for proper splicing to occur (Ishigami et al., 2021).
One of the largest obstacles in the RNA modification field today is detection. Several studies have attempted to produce new protocols to isolate and correctly identify m6A in modified RNAs (Koh et al., 2019; N. Liu et al., 2013; Mikutis et al., 2020; Oerum et al., 2019; Ovcharenko et al., 2021; S. Xiang et al., 2021). Because METTL3/14 has many known targets, it is often used in these studies with METTL16 often being included experimentally (as there are few verified interactors). This does come with some issues. First, it is generally regarded that METTL3/14 and METTL16 have different consensus sequences (Doxtader et al., 2018; Mendel et al., 2018; Pendleton et al., 2017; Shima et al., 2017). Therefore, some m6As that are in close proximity may be assigned to only one methyltransferase. Also, these studies tend to use polyadenylated RNA selection for more efficient sequencing, however this is very likely to exclude some interactors, since it is known that METTL16 interacts with RNAs other than mRNA. Furthermore, some studies show results as dependence of methylation on a certain methyltransferase. If an RNA shows dependence on both METTL3/14 and METTL16, it seems it is considered a “true” METTL3/14 target while showing secondary dependence on METTL16 due to the indirect effect of MAT2A (Koh et al., 2019; Song et al., 2019). Looking to refine a consensus motif among these results will exclude potential interactors and could be why there are differences among published results. Lastly, if MAT2A function is crucially dependent on METTL16, the knockdown of METTL16 (which is used to compare RNAs subsequently hypomethylated) could lower the cellular concentration of SAM. This is the basis of the argument of secondary dependence mentioned before. However, demethylases could also be responsive to the induced lower level of m6A‐modified RNA and demethylate further for SAM recycling, which could result in RNAs appearing dependent on the methyltransferase knocked‐down. These speculations are not verifiable until more information is known about the innerworkings of methyltransferases, demethylases, and their regulation on/by the methionine cycle.
In this review, we have discussed the current state of METTL16 understanding, predictions, and speculations. Even though there is little information that is considered to be confirmed about this protein, it is evident that it has a significant and intriguing influence in humans. The goal of this publication is to combine the results and implications of studies on this protein to educate and guide the m6A community to what is and is not yet known about METTL16. It is our hope to open up the possibilities of METTL16's function based on past publications to further productive research into this exciting enzyme.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
AUTHOR CONTRIBUTIONS
Emily Satterwhite: Conceptualization; writing‐original draft; writing‐review & editing. Kyle Mansfield: Conceptualization; funding acquisition; writing‐original draft; writing‐review & editing.
RELATED WIREs ARTICLES
RNA methylation in nuclear pre‐mRNA processing
Dynamic and reversible RNA N6‐methyladenosine methylation
A birds'‐eye view of the activity and specificity of the mRNA m6A methyltransferase complex
ACKNOWLEDGMENTS
We would like to thank members of the lab including Kristen Carraway and Mohammed Dorgham for helpful discussions and advice during the writing of this manuscript. This work was supported by a Research Scholar Grant, RSG‐19‐044‐01‐RMC, from the American Cancer Society (KDM). The funders had no role in the preparation of the manuscript.
Satterwhite, E. R. , & Mansfield, K. D. (2022). RNA methyltransferase METTL16: Targets and function. Wiley Interdisciplinary Reviews: RNA, 13(2), e1681. 10.1002/wrna.1681
Edited by: Carol Lutz, Associate Editor and Jeff Wilusz, Editor‐in‐Chief
Funding information American Cancer Society (KDM), Grant/Award Number: RSG‐19‐044‐01‐RMC
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Agarwala, S. D. , Blitzblau, H. G. , Hochwagen, A. , & Fink, G. R. (2012). RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genetics, 8(6), e1002732. 10.1371/journal.pgen.1002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio, N. , Raimondi, L. , Juli, G. , Stamato, M. A. , Caracciolo, D. , Tagliaferri, P. , & Tassone, P. (2018). MALAT1: A druggable long non‐coding RNA for targeted anti‐cancer approaches. Journal of Hematology & Oncology, 11(1), 63. 10.1186/s13045-018-0606-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama, T. , Yamashita, S. , & Tomita, K. (2020). Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Research, 48(9), 5157–5168. 10.1093/nar/gkaa227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun, G. , Aggarwal, D. , & Spector, D. L. (2020). MALAT1 long non‐coding RNA: Functional implications. Noncoding RNA, 6(2), 1–17. 10.3390/ncrna6020022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano, M. , Hafner, M. , Cekan, P. , Gerstberger, S. , & Tuschl, T. (2012). Identification of RNA‐protein interaction networks using PAR‐CLIP. WIREs RNA, 3(2), 159–177. 10.1002/wrna.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz, A. G. , Munschauer, M. , Schwanhausser, B. , Vasile, A. , Murakawa, Y. , Schueler, M. , Youngs, N. , Penfold‐Brown, D. , Drew, K. , Milek, M. , Wyler, E. , Bonneau, R. , Selbach, M. , Dieterich, C. , & Landthaler, M. (2012). The mRNA‐bound proteome and its global occupancy profile on protein‐coding transcripts. Molecular Cell, 46(5), 674–690. 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Barbieri, I. , Tzelepis, K. , Pandolfini, L. , Shi, J. , Millan‐Zambrano, G. , Robson, S. C. , Aspris, D. , Migliori, V. , Bannister, A. J. , Han, N. , De Braekeleer, E. , Ponstingl, H. , Hendrick, A. , Vakoc, C. R. , Vassiliou, G. S. , & Kouzarides, T. (2017). Promoter‐bound METTL3 maintains myeloid leukaemia by m(6)A‐dependent translation control. Nature, 552(7683), 126–131. 10.1038/nature24678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berulava, T. , Rahmann, S. , Rademacher, K. , Klein‐Hitpass, L. , & Horsthemke, B. (2015). N6‐adenosine methylation in MiRNAs. PLoS One, 10(2), e0118438. 10.1371/journal.pone.0118438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto, P. , & Baginski, B. (2021). MODOMICS: An operational guide to the use of the RNA modification pathways database. Methods in Molecular Biology, 2284, 481–505. 10.1007/978-1-0716-1307-8_26 [DOI] [PubMed] [Google Scholar]
- Boccaletto, P. , Machnicka, M. A. , Purta, E. , Piatkowski, P. , Baginski, B. , Wirecki, T. K. , de Crécy‐Lagard, V. , Ross, R. , Limbach, P. A. , Kotter, A. , Helm, M. , & Bujnicki, J. M. (2018). MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Research, 46(D1), D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar, J. A. , Shambaugh, M. E. , Polayes, D. , Matera, A. G. , & Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet‐binding subunit of the human mRNA (N6‐adenosine)‐methyltransferase. RNA, 3(11), 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Boo, S. H. , & Kim, Y. K. (2020). The emerging role of RNA modifications in the regulation of mRNA stability. Experimental & Molecular Medicine, 52(3), 400–408. 10.1038/s12276-020-0407-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan, K. W. , Jin, W. , Huelga, S. C. , Banks, C. A. , Gilmore, J. M. , Florens, L. , Washburn, M. P. , Van Nostrand, E. L. , Pratt, G. A. , Schwinn, M. K. , Daniels, D. L. , & Yeo, G. W. (2016). SONAR discovers RNA‐binding proteins from analysis of large‐scale protein‐protein interactomes. Molecular Cell, 64(2), 282–293. 10.1016/j.molcel.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow, D. A. , & Guthrie, C. (1988). Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature, 334(6179), 213–218. 10.1038/334213a0 [DOI] [PubMed] [Google Scholar]
- Brown, J. A. , Kinzig, C. G. , DeGregorio, S. J. , & Steitz, J. A. (2016). Methyltransferase‐like protein 16 binds the 3′‐terminal triple helix of MALAT1 long noncoding RNA. Proceedings of the National Academy of Sciences of the United States of America, 113(49), 14013–14018. 10.1073/pnas.1614759113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. A. , Valenstein, M. L. , Yario, T. A. , Tycowski, K. T. , & Steitz, J. A. (2012). Formation of triple‐helical structures by the 3′‐end sequences of MALAT1 and MENbeta noncoding RNAs. Proceedings of the National Academy of Sciences of the United States of America, 109(47), 19202–19207. 10.1073/pnas.1217338109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantara, W. A. , Crain, P. F. , Rozenski, J. , McCloskey, J. A. , Harris, K. A. , Zhang, X. , Vendeix, F. A. P. , Fabris, D. , & Agris, P. F. (2011). The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Research, 39(Database issue), D195–D201. 10.1093/nar/gkq1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni, G. L. , & Durell, J. (1957). Activation of methionine for transmethylation. II. The methionine‐activating enzyme; studies on the mechanism of the reaction. The Journal of Biological Chemistry, 225(2), 1033–1048. 10.1016/s0021-9258(18)64899-9 [DOI] [PubMed] [Google Scholar]
- Castello, A. , Fischer, B. , Eichelbaum, K. , Horos, R. , Beckmann, B. M. , Strein, C. , Davey, N. E. , Humphreys, D. T. , Preiss, T. , Steinmetz, L. M. , Krijgsveld, J. , & Hentze, M. W. (2012). Insights into RNA biology from an atlas of mammalian mRNA‐binding proteins. Cell, 149(6), 1393–1406. 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Chen, K. , Lu, Z. , Wang, X. , Fu, Y. , Luo, G. Z. , Liu, N. , Han, D. , Dominissini, D. , Dai, Q. , Pan, T. , & He, C. (2015). High‐resolution N(6) ‐methyladenosine (m(6) A) map using photo‐crosslinking‐assisted m(6) A sequencing. Angewandte Chemie (International Ed. in English), 54(5), 1587–1590. 10.1002/anie.201410647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo, T. , & Tomizawa, K. (2021). Human transfer RNA modopathies: Diseases caused by aberrations in transfer RNA modifications. The FEBS Journal. 10.1111/febs.15736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, T. , Albrecht, A. S. , de Melo Costa, V. R. , Sauer, S. , Meierhofer, D. , & Orom, U. A. (2016). Serial interactome capture of the human cell nucleus. Nature Communications, 7, 11212. 10.1038/ncomms11212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers, R. , Friderici, K. , & Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America, 71(10), 3971–3975. 10.1073/pnas.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman, J. D. , & Wickner, R. B. (1995). 5 S rRNA is involved in fidelity of translational reading frame. Genetics, 141(1), 95–105. 10.1093/genetics/141.1.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett, M. , & Schedl, T. (2009). A role for dynein in the inhibition of germ cell proliferative fate. Molecular and Cellular Biology, 29(22), 6128–6139. 10.1128/MCB.00815-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett, M. , Westlund, B. , & Schedl, T. (2009). METT‐10, a putative methyltransferase, inhibits germ cell proliferative fate in Caenorhabditis elegans . Genetics, 183(1), 233–247. 10.1534/genetics.109.105270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxtader, K. A. , Wang, P. , Scarborough, A. M. , Seo, D. , Conrad, N. K. , & Nam, Y. (2018). Structural basis for regulation of METTL16, an S‐adenosylmethionine homeostasis factor. Molecular Cell, 71(6), 1001–1011.e1004. 10.1016/j.molcel.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H. , Zhao, Y. , He, J. , Zhang, Y. , Xi, H. , Liu, M. , Ma, J. , & Wu, L. (2016). YTHDF2 destabilizes m(6)A‐containing RNA through direct recruitment of the CCR4‐NOT deadenylase complex. Nature Communications, 7, 12626. 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, P. , Reddy, R. , Henning, D. , & Busch, H. (1980). The nucleotide sequence of nuclear U6 (4.7 S) RNA. The Journal of Biological Chemistry, 255(18), 8901–8906. 10.1016/s0021-9258(18)43587-9 [DOI] [PubMed] [Google Scholar]
- Fry, N. J. , Law, B. A. , Ilkayeva, O. R. , Holley, C. L. , & Mansfield, K. D. (2017). N(6)‐methyladenosine is required for the hypoxic stabilization of specific mRNAs. RNA, 23(9), 1444–1455. 10.1261/rna.061044.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin, J. M. , Doi, M. , Yamaguchi, Y. , Hida, H. , Nishimura, S. , Yoshida, M. , Isagawa, T. , Morioka, M. S. , Kakeya, H. , Manabe, I. , & Okamura, H. (2013). RNA‐methylation‐dependent RNA processing controls the speed of the circadian clock. Cell, 155(4), 793–806. 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- Ge, J. , & Yu, Y. T. (2013). RNA pseudouridylation: New insights into an old modification. Trends in Biochemical Sciences, 38(4), 210–218. 10.1016/j.tibs.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula, S. , Moshitch‐Moshkovitz, S. , Dominissini, D. , Mansour, A. A. , Kol, N. , Salmon‐Divon, M. , Hershkovitz, V. , Peer, E. , Mor, N. , Manor, Y. S. , Ben‐Haim, M. S. , Eyal, E. , Yunger, S. , Pinto, Y. , Jaitin, D. A. , Viukov, S. , Rais, Y. , Krupalnik, V. , Chomsky, E. , … Hanna, J. H. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science, 347(6225), 1002–1006. 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- Gokhale, N. S. , McIntyre, A. B. R. , McFadden, M. J. , Roder, A. E. , Kennedy, E. M. , Gandara, J. A. , Hopcraft, S. E. , Quicke, K. M. , Vazquez, C. , Willer, J. , Ilkayeva, O. R. , Law, B. A. , Holley, C. L. , Garcia‐Blanco, M. A. , Evans, M. J. , Suthar, M. S. , Bradrick, S. S. , Mason, C. E. , & Horner, S. M. (2016). N6‐methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host & Microbe, 20(5), 654–665. 10.1016/j.chom.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy, M. , Guindon, S. , & Gascuel, O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution, 27(2), 221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Grosjean, H. (2005). Fine‐tuning of RNA functions by modification and editing. Springer. [Google Scholar]
- Guzzardo, P. M. , Rashkova, C. , Dos Santos, R. L. , Tehrani, R. , Collin, P. , & Burckstummer, T. (2017). A small cassette enables conditional gene inactivation by CRISPR/Cas9. Scientific Reports, 7(1), 16770. 10.1038/s41598-017-16931-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner, M. , Katsantoni, M. , Köster, T. , Marks, J. , Mukherjee, J. , Staiger, D. , Ule, J. , & Zavolan, M. (2021). CLIP and complementary methods. Nature Reviews Methods Primers, 1(1), 20. 10.1038/s43586-021-00018-1 [DOI] [Google Scholar]
- Halim, A. B. , LeGros, L. , Chamberlin, M. E. , Geller, A. , & Kotb, M. (2001). Regulation of the human MAT2A gene encoding the catalytic alpha 2 subunit of methionine adenosyltransferase, MAT II: Gene organization, promoter characterization, and identification of a site in the proximal promoter that is essential for its activity. The Journal of Biological Chemistry, 276(13), 9784–9791. 10.1074/jbc.M002347200 [DOI] [PubMed] [Google Scholar]
- Halim, A. B. , LeGros, L. , Geller, A. , & Kotb, M. (1999). Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. The Journal of Biological Chemistry, 274(42), 29720–29725. 10.1074/jbc.274.42.29720 [DOI] [PubMed] [Google Scholar]
- Harper, J. E. , Miceli, S. M. , Roberts, R. J. , & Manley, J. L. (1990). Sequence specificity of the human mRNA N6‐adenosine methylase in vitro. Nucleic Acids Research, 18(19), 5735–5741. 10.1093/nar/18.19.5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, P. C. , & He, C. (2021). m(6) A RNA methylation: From mechanisms to therapeutic potential. EMBO Journal, 40(3), e105977. 10.15252/embj.2020105977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, M. , Guo, X. , Chen, Y. , Cong, L. , & Pan, C. (2020). A prognostic molecular signature of N(6)‐methyladenosine methylation regulators for soft‐tissue sarcoma from the cancer genome atlas database. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research, 26, e928400. 10.12659/MSM.928400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Dong, X. , Gong, Z. , Qin, L.‐Y. , Yang, S. , Zhu, Y.‐L. , Wang, X. , Zhang, D. , Zou, T. , Yin, P. , & Tang, C. (2019). Solution structure of the RNA recognition domain of METTL3‐METTL14 N6‐methyladenosine methyltransferase. Protein & Cell, 10(4), 272–284. 10.1007/s13238-018-0518-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, J. N. , Ensminger, A. W. , Clemson, C. M. , Lynch, C. R. , Lawrence, J. B. , & Chess, A. (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics, 8, 39. 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigami, Y. , Ohira, T. , Isokawa, Y. , Suzuki, Y. , & Suzuki, T. (2021). A single m(6)A modification in U6 snRNA diversifies exon sequence at the 5′ splice site. Nature Communications, 12(1), 3244. 10.1038/s41467-021-23457-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanami, Y. , & Brown, G. M. (1968). Methylated bases of ribosomal ribonucleic acid from HeLa cells. Archives of Biochemistry and Biophysics, 126(1), 8–15. 10.1016/0003-9861(68)90553-5 [DOI] [PubMed] [Google Scholar]
- Jabs, S. , Biton, A. , Becavin, C. , Nahori, M. A. , Ghozlane, A. , Pagliuso, A. , Spanò, G. , Guérineau, V. , Touboul, D. , Gianetto, Q. G. , Chaze, T. , Matondo, M. , Dillies, M.‐A. , & Cossart, P. (2020). Impact of the gut microbiota on the m(6)A epitranscriptome of mouse cecum and liver. Nature Communications, 11(1), 1344. 10.1038/s41467-020-15126-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X. , Liu, B. , Nie, Z. , Duan, L. , Xiong, Q. , Jin, Z. , Yang, C. , & Chen, Y. (2021). The role of m6A modification in the biological functions and diseases. Signal Transduction and Targeted Therapy, 6(1), 74. 10.1038/s41392-020-00450-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, D. , Guo, J. , Wu, Y. , Du, J. , Yang, L. , Wang, X. , Di, W. , Hu, B. , An, J. , Kong, L. , Pan, L. , & Su, G. (2019). m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1‐miR‐1914‐3p‐YAP axis to induce NSCLC drug resistance and metastasis. Journal of Hematology & Oncology, 12(1), 135. 10.1186/s13045-019-0830-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jonkhout, N. , Tran, J. , Smith, M. A. , Schonrock, N. , Mattick, J. S. , & Novoa, E. M. (2017). The RNA modification landscape in human disease. RNA, 23(12), 1754–1769. 10.1261/rna.063503.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalev, P. , Hyer, M. L. , Gross, S. , Konteatis, Z. , Chen, C. C. , Fletcher, M. , Lein, M. , Aguado‐Fraile, E. , Frank, V. , Barnett, A. , Mandley, E. , Goldford, J. , Chen, Y. , Sellers, K. , Hayes, S. , Lizotte, K. , Quang, P. , Tuncay, Y. , Clasquin, M. , … Marjon, K. (2021). MAT2A inhibition blocks the growth of MTAP‐deleted cancer cells by reducing PRMT5‐dependent mRNA splicing and inducing DNA damage. Cancer Cell, 39(2), 209–224 e211. 10.1016/j.ccell.2020.12.010 [DOI] [PubMed] [Google Scholar]
- Ke, S. , Pandya‐Jones, A. , Saito, Y. , Fak, J. J. , Vagbo, C. B. , Geula, S. , Hanna, J. H. , Black, D. L. , Darnell, J. E., Jr. , & Darnell, R. B. (2017). m(6)A mRNA modifications are deposited in nascent pre‐mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes & Development, 31(10), 990–1006. 10.1101/gad.301036.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, C. W. Q. , Goh, Y. T. , & Goh, W. S. S. (2019). Atlas of quantitative single‐base‐resolution N(6)‐methyl‐adenine methylomes. Nature Communications, 10(1), 5636. 10.1038/s41467-019-13561-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S. C. , Yi, H. , Eichelbaum, K. , Fohr, S. , Fischer, B. , You, K. T. , Castello, A. , Krijgsveld, J. , Hentze, M. W. , & Kim, V. N. (2013). The RNA‐binding protein repertoire of embryonic stem cells. Nature Structural & Molecular Biology, 20(9), 1122–1130. 10.1038/nsmb.2638 [DOI] [PubMed] [Google Scholar]
- Lan, Q. , Liu, P. Y. , Haase, J. , Bell, J. L. , Huttelmaier, S. , & Liu, T. (2019). The critical role of RNA m(6)A methylation in cancer. Cancer Research, 79(7), 1285–1292. 10.1158/0008-5472.CAN-18-2965 [DOI] [PubMed] [Google Scholar]
- Lee, M. , Kim, B. , & Kim, V. N. (2014). Emerging roles of RNA modification: m(6)A and U‐tail. Cell, 158(5), 980–987. 10.1016/j.cell.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Lence, T. , Paolantoni, C. , Worpenberg, L. , & Roignant, J. Y. (2019). Mechanistic insights into m(6)A RNA enzymes. Biochimica et Biophysica Acta – Gene Regulatory Mechanisms, 1862(3), 222–229. 10.1016/j.bbagrm.2018.10.014 [DOI] [PubMed] [Google Scholar]
- Li, A. , Chen, Y. S. , Ping, X. L. , Yang, X. , Xiao, W. , Yang, Y. , Sun, H. Y. , Zhu, Q. , Baidya, P. , Wang, X. , Bhattarai, D. P. , Zhao, Y. L. , Sun, B. F. , & Yang, Y. G. (2017). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Research, 27(3), 444–447. 10.1038/cr.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K. , Luo, H. , Luo, H. , & Zhu, X. (2020). Clinical and prognostic pan‐cancer analysis of m6A RNA methylation regulators in four types of endocrine system tumors. Aging, 12(23), 23931–23944. 10.18632/aging.104064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Armstrong, C. M. , Bertin, N. , Ge, H. , Milstein, S. , Boxem, M. , Vidalain, P. O. , Han, J. D. , Chesneau, A. , Hao, T. , Goldberg, D. S. , Li, N. , Martinez, M. , Rual, J. F. , Lamesch, P. , Xu, L. , Tewari, M. , Wong, S. L. , Zhang, L. V. , … Vidal, M. (2004). A map of the interactome network of the metazoan C. elegans . Science, 303(5657), 540–543. 10.1126/science.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Wang, X. , Li, C. , Hu, S. , Yu, J. , & Song, S. (2014). Transcriptome‐wide N(6)‐methyladenosine profiling of rice callus and leaf reveals the presence of tissue‐specific competitors involved in selective mRNA modification. RNA Biology, 11(9), 1180–1188. 10.4161/rna.36281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. , Choe, J. , Du, P. , Triboulet, R. , & Gregory, R. I. (2016). The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Molecular Cell, 62(3), 335–345. 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder, B. , Grozhik, A. V. , Olarerin‐George, A. O. , Meydan, C. , Mason, C. E. , & Jaffrey, S. R. (2015). Single‐nucleotide‐resolution mapping of m6A and m6Am throughout the transcriptome. Nature Methods, 12(8), 767–772. 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Yue, Y. , Han, D. , Wang, X. , Fu, Y. , Zhang, L. , Jia, G. , Yu, M. , Lu, Z. , Deng, X. , Dai, Q. , Chen, W. , & He, C. (2014). A METTL3‐METTL14 complex mediates mammalian nuclear RNA N6‐adenosine methylation. Nature Chemical Biology, 10(2), 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N. , Parisien, M. , Dai, Q. , Zheng, G. , He, C. , & Pan, T. (2013). Probing N6‐methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA, 19(12), 1848–1856. 10.1261/rna.041178.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Zhuo, L. , Wang, J. , Zhang, Q. , Li, Q. , Li, G. , Yan, L. , Jin, T. , Pan, T. , Sui, X. , Lv, Q. , & Xie, T. (2020). METTL3 plays multiple functions in biological processes. American Journal of Cancer Research, 10(6), 1631–1646. [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Liu, L. , Dong, Z. , Li, J. , Yu, Y. , Chen, X. , Ren, F. , Cui, G. , & Sun, R. (2019). Expression patterns and prognostic value of m(6)A‐related genes in colorectal cancer. American Journal of Translational Research, 11(7), 3972–3991. [PMC free article] [PubMed] [Google Scholar]
- Lo, T. F. , Tsai, W. C. , & Chen, S. T. (2013). MicroRNA‐21‐3p, a berberine‐induced miRNA, directly down‐regulates human methionine adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS One, 8(9), e75628. 10.1371/journal.pone.0075628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, G. Z. , MacQueen, A. , Zheng, G. , Duan, H. , Dore, L. C. , Lu, Z. , Liu, J. , Chen, K. , Jia, G. , Bergelson, J. , & He, C. (2014). Unique features of the m6A methylome in Arabidopsis thaliana . Nature Communications, 5, 5630. 10.1038/ncomms6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H. , Wang, X. , Cai, J. , Dai, Q. , Natchiar, S. K. , Lv, R. , Chen, K. , Lu, Z. , Chen, H. , Shi, Y. G. , Lan, F. , Fan, J. , Klaholz, B. P. , Pan, T. , Shi, Y. , & He, C. (2019). N(6‐)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nature Chemical Biology, 15(1), 88–94. 10.1038/s41589-018-0184-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Chantar, M. L. , Latasa, M. U. , Varela‐Rey, M. , Lu, S. C. , Garcia‐Trevijano, E. R. , Mato, J. M. , & Avila, M. A. (2003). l‐Methionine availability regulates expression of the methionine adenosyltransferase 2A gene in human hepatocarcinoma cells: Role of S‐adenosylmethionine. The Journal of Biological Chemistry, 278(22), 19885–19890. 10.1074/jbc.M211554200 [DOI] [PubMed] [Google Scholar]
- Marx, A. , Nugoor, C. , Panneerselvam, S. , & Mandelkow, E. (2010). Structure and function of polarity‐inducing kinase family MARK/Par‐1 within the branch of AMPK/Snf1‐related kinases. The FASEB Journal, 24(6), 1637–1648. 10.1096/fj.09-148064 [DOI] [PubMed] [Google Scholar]
- Mendel, M. , Chen, K. M. , Homolka, D. , Gos, P. , Pandey, R. R. , McCarthy, A. A. , & Pillai, R. S. (2018). Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Molecular Cell, 71(6), 986–1000.e1011. 10.1016/j.molcel.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel, M. , Delaney, K. , Pandey, R. R. , Chen, K. M. , Wenda, J. M. , Vagbo, C. B. , Steiner, F. A. , Homolka, D. , & Pillai, R. S. (2021). Splice site m(6)A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell, 184, 3125–3142.e25. 10.1016/j.cell.2021.03.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, K. D. , Saletore, Y. , Zumbo, P. , Elemento, O. , Mason, C. E. , & Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell, 149(7), 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikutis, S. , Gu, M. , Sendinc, E. , Hazemi, M. E. , Kiely‐Collins, H. , Aspris, D. , Vassiliou, G. S. , Shi, Y. , Tzelepis, K. , & Bernardes, G. J. L. (2020). meCLICK‐Seq, a substrate‐hijacking and RNA degradation strategy for the study of RNA methylation. ACS Central Science, 6(12), 2196–2208. 10.1021/acscentsci.0c01094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic, K. , Mendrola, J. M. , Schmitz, K. R. , Wang, Y. H. , Slochower, D. , Janmey, P. A. , & Lemmon, M. A. (2010). Kinase associated‐1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell, 143(6), 966–977. 10.1016/j.cell.2010.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, N. , Corcoran, D. L. , Nusbaum, J. D. , Reid, D. W. , Georgiev, S. , Hafner, M. , Ascano, M., Jr. , Tuschl, T. , Ohler, U. , & Keene, J. D. (2011). Integrative regulatory mapping indicates that the RNA‐binding protein HuR couples pre‐mRNA processing and mRNA stability. Molecular Cell, 43(3), 327–339. 10.1016/j.molcel.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele, S. , & He, C. (2017). The emerging biology of RNA post‐transcriptional modifications. RNA Biology, 14(2), 156–163. 10.1080/15476286.2016.1267096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance, D. J. , Satterwhite, E. R. , Bhaskar, B. , Misra, S. , Carraway, K. R. , & Mansfield, K. D. (2020). Characterization of METTL16 as a cytoplasmic RNA binding protein. PLoS One, 15(1), e0227647. 10.1371/journal.pone.0227647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba, A. N. , Pogoutse, A. , Provart, N. , & Moses, A. M. (2009). NLStradamus: A simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics, 10, 202. 10.1186/1471-2105-10-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerum, S. , Catala, M. , Atdjian, C. , Brachet, F. , Ponchon, L. , Barraud, P. , Iannazzo, L. , Droogmans, L. , Braud, E. , Ethève‐Quelquejeu, M. , & Tisne, C. (2019). Bisubstrate analogues as structural tools to investigate m(6)A methyltransferase active sites. RNA Biology, 16(6), 798–808. 10.1080/15476286.2019.1589360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko, A. , Weissenboeck, F. P. , & Rentmeister, A. (2021). Tag‐free internal RNA labeling and photocaging based on mRNA methyltransferases. Angewandte Chemie (International Ed. in English), 60(8), 4098–4103. 10.1002/anie.202013936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, B. J. , Moltke, I. , Roth, A. , Washietl, S. , Wen, J. , Kellis, M. , Breaker, R. , & Pedersen, J. S. (2011). New families of human regulatory RNA structures identified by comparative analysis of vertebrate genomes. Genome Research, 21(11), 1929–1943. 10.1101/gr.112516.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, D. P. , Chen, C. K. , Pickering, B. F. , Chow, A. , Jackson, C. , Guttman, M. , & Jaffrey, S. R. (2016). m(6)A RNA methylation promotes XIST‐mediated transcriptional repression. Nature, 537(7620), 369–373. 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paung, Y. , & Seeliger, M. A. (2018). KA1 domains: Unity in mechanistic diversity. Structure, 26(8), 1045–1047. 10.1016/j.str.2018.07.002 [DOI] [PubMed] [Google Scholar]
- Peer, E. , Moshitch‐Moshkovitz, S. , Rechavi, G. , & Dominissini, D. (2019). The epitranscriptome in translation regulation. Cold Spring Harbor Perspectives in Biology, 11(8), 1–13. 10.1101/cshperspect.a032623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton, K. E. , Chen, B. , Liu, K. , Hunter, O. V. , Xie, Y. , Tu, B. P. , & Conrad, N. K. (2017). The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell, 169(5), 824–835.e814. 10.1016/j.cell.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna‐Przybylska, D. , Decatur, W. A. , & Fournier, M. J. (2008). The 3D rRNA modification maps database: With interactive tools for ribosome analysis. Nucleic Acids Research, 36(Database issue), D178–D183. 10.1093/nar/gkm855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping, X. L. , Sun, B. F. , Wang, L. , Xiao, W. , Yang, X. , Wang, W. J. , Adhikari, S. , Shi, Y. , Lv, Y. , Chen, Y. S. , Zhao, X. , Li, A. , Yang, Y. , Dahal, U. , Lou, X. M. , Liu, X. , Huang, J. , Yuan, W. P. , Zhu, X. F. , … Yang, Y. G. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6‐methyladenosine methyltransferase. Cell Research, 24(2), 177–189. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, R. , Vagbo, C. B. , Jakobsson, M. E. , Kim, Y. , Baltissen, M. P. , O'Donohue, M. F. , Guzmán, U. H. , Małecki, J. M. , Wu, J. , Kirpekar, F. , Olsen, J. V. , Gleizes, P.‐E. , Vermeulen, M. , Leidel, S. A. , Slupphaug, G. , & Falnes, P. O. (2020). The human methyltransferase ZCCHC4 catalyses N6‐methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Research, 48(2), 830–846. 10.1093/nar/gkz1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, D. , Liu, Q. , Devaraj, A. , & Fredrick, K. (2012). Role of helix 44 of 16S rRNA in the fidelity of translation initiation. RNA, 18(3), 485–495. 10.1261/rna.031203.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani, K. , & Tomasi, M. L. (2012). Transcriptional regulation of methionine adenosyltransferase 2A by peroxisome proliferator‐activated receptors in rat hepatic stellate cells. Hepatology, 55(6), 1942–1953. 10.1002/hep.25594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, R. , Ro‐Choi, T. S. , Henning, D. , Shibata, H. , Choi, Y. C. , & Busch, H. (1972). MOdified nucleosides of nuclear and nucleolar low molecular weight ribonucleic acid. The Journal of Biological Chemistry, 247(22), 7245–7250. 10.1016/s0021-9258(19)44620-6 [DOI] [PubMed] [Google Scholar]
- Ren, W. , Lu, J. , Huang, M. , Gao, L. , Li, D. , Wang, G. G. , & Song, J. (2019). Structure and regulation of ZCCHC4 in m(6)A‐methylation of 28S rRNA. Nature Communications, 10(1), 5042. 10.1038/s41467-019-12923-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost, C. , Lynch, S. R. , Batista, P. J. , Qu, K. , Chang, H. Y. , & Kool, E. T. (2015). Structure and thermodynamics of N6‐methyladenosine in RNA: A spring‐loaded base modification. Journal of the American Chemical Society, 137(5), 2107–2115. 10.1021/ja513080v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree, I. A. , Evans, M. E. , Pan, T. , & He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell, 169(7), 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree, I. A. , & He, C. (2016). RNA epigenetics – chemical messages for posttranscriptional gene regulation. Current Opinion in Chemical Biology, 30, 46–51. 10.1016/j.cbpa.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszkowska, A. (2021). METTL16, methyltransferase‐like protein 16: Current insights into structure and function. International Journal of Molecular Sciences, 22(4), 2176–2196. 10.3390/ijms22042176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszkowska, A. , Ruszkowski, M. , Dauter, Z. , & Brown, J. A. (2018). Structural insights into the RNA methyltransferase domain of METTL16. Scientific Reports, 8(1), 5311. 10.1038/s41598-018-23608-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarborough, A. M. , Flaherty, J. N. , Hunter, O. V. , Liu, K. , Kumar, A. , Xing, C. , Tu, B. P. , & Conrad, N. K. (2021). SAM homeostasis is regulated by CFIm‐mediated splicing of MAT2A. eLife, 10. 10.7554/eLife.64930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S. , Mumbach, M. R. , Jovanovic, M. , Wang, T. , Maciag, K. , Bushkin, G. G. , Mertins, P. , ter‐Ovanesyan, D. , Habib, N. , Cacchiarelli, D. , Sanjana, N. E. , Freinkman, E. , Pacold, M. E. , Satija, R. , Mikkelsen, T. S. , Hacohen, N. , Zhang, F. , Carr, S. A. , Lander, E. S. , & Regev, A. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Reports, 8(1), 284–296. 10.1016/j.celrep.2014.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergiev, P. V. , Serebryakova, M. V. , Bogdanov, A. A. , & Dontsova, O. A. (2008). The ybiN gene of Escherichia coli encodes adenine‐N6 methyltransferase specific for modification of A1618 of 23 S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. Journal of Molecular Biology, 375(1), 291–300. 10.1016/j.jmb.2007.10.051 [DOI] [PubMed] [Google Scholar]
- Shi, H. , Wang, X. , Lu, Z. , Zhao, B. S. , Ma, H. , Hsu, P. J. , Liu, C. , & He, C. (2017). YTHDF3 facilitates translation and decay of N(6)‐methyladenosine‐modified RNA. Cell Research, 27(3), 315–328. 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Wei, J. , & He, C. (2019). Where, when, and how: Context‐dependent functions of RNA methylation writers, readers, and erasers. Molecular Cell, 74(4), 640–650. 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Zhang, X. , Weng, Y. L. , Lu, Z. , Liu, Y. , Lu, Z. , Li, J. , Hao, P. , Zhang, Y. , Zhang, F. , Wu, Y. , Delgado, J. Y. , Su, Y. , Patel, M. J. , Cao, X. , Shen, B. , Huang, X. , Ming, G. L. , Zhuang, X. , … Zhou, T. (2018). m(6)A facilitates hippocampus‐dependent learning and memory through YTHDF1. Nature, 563(7730), 249–253. 10.1038/s41586-018-0666-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima, H. , Matsumoto, M. , Ishigami, Y. , Ebina, M. , Muto, A. , Sato, Y. , Kumagai, S. , Ochiai, K. , Suzuki, T. , & Igarashi, K. (2017). S‐Adenosylmethionine synthesis is regulated by selective N(6)‐adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Reports, 21(12), 3354–3363. 10.1016/j.celrep.2017.11.092 [DOI] [PubMed] [Google Scholar]
- Simile, M. M. , Peitta, G. , Tomasi, M. L. , Brozzetti, S. , Feo, C. F. , Porcu, A. , Cigliano, A. , Calvisi, D. F. , Feo, F. , & Pascale, R. M. (2019). MicroRNA‐203 impacts on the growth, aggressiveness and prognosis of hepatocellular carcinoma by targeting MAT2A and MAT2B genes. Oncotarget, 10(29), 2835–2854. 10.18632/oncotarget.26838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz, P. , & Jinek, M. (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. eLife, 5, e18434. 10.7554/eLife.18434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan, K. E. , Warda, A. S. , Sharma, S. , Entian, K. D. , Lafontaine, D. L. J. , & Bohnsack, M. T. (2017). Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biology, 14(9), 1138–1152. 10.1080/15476286.2016.1259781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. , Xu, Q. , Wei, Z. , Zhen, D. , Su, J. , Chen, K. , & Meng, J. (2019). Predict epitranscriptome targets and regulatory functions of N(6)‐methyladenosine (m(6)A) writers and erasers. Evolutionary Bioinformatics Online, 15, 1176934319871290. 10.1177/1176934319871290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T. (2021). The expanding world of tRNA modifications and their disease relevance. Nature Reviews. Molecular Cell Biology, 22, 375–392. 10.1038/s41580-021-00342-0 [DOI] [PubMed] [Google Scholar]
- Svobodova Kovarikova, A. , Stixova, L. , Kovarik, A. , Komurkova, D. , Legartova, S. , Fagherazzi, P. , & Bartova, E. (2020). N(6)‐adenosine methylation in RNA and a reduced m3G/TMG level in non‐coding RNAs appear at microirradiation‐induced DNA lesions. Cell, 9(2), 360. 10.3390/cells9020360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe, R. , Sandrock, B. , & Benecke, B. J. (1998). A highly specific terminal uridylyl transferase modifies the 3′‐end of U6 small nuclear RNA. Nucleic Acids Research, 26(13), 3119–3126. 10.1093/nar/26.13.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tran, N. , Ernst, F. G. M. , Hawley, B. R. , Zorbas, C. , Ulryck, N. , Hackert, P. , Bohnsack, K. E. , Bohnsack, M. T. , Jaffrey, S. R. , Graille, M. , & Lafontaine, D. L. J. (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Research, 47(15), 7719–7733. 10.1093/nar/gkz619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez‐Chantada, M. , Fernandez‐Ramos, D. , Embade, N. , Martinez‐Lopez, N. , Varela‐Rey, M. , Woodhoo, A. , Luka, Z. , Wagner, C. , Anglim, P. P. , Finnell, R. H. , Caballería, J. , Laird‐Offringa, I. A. , Gorospe, M. , Lu, S. C. , Mato, J. M. , & Martinez‐Chantar, M. L. (2010). HuR/methyl‐HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology, 138(5), 1943–1953. 10.1053/j.gastro.2010.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Doxtader, K. A. , & Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Molecular Cell, 63(2), 306–317. 10.1016/j.molcel.2016.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Wang, X. , Zheng, L. , & Zhuang, C. (2020). Gene signatures and prognostic values of m6A regulators in hepatocellular carcinoma. Frontiers in Genetics, 11, 540186. 10.3389/fgene.2020.540186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Fan, X. , Zhu, J. , Xu, D. , Li, R. , Chen, R. , Hu, J. , Shen, Y. , Hao, J. , Wang, K. , Jiang, X. , Wang, Y. , Jiang, Y. , Li, J. , & Zhang, J. (2021). The differentiation of colorectal cancer is closely relevant to m6A modification. Biochemical and Biophysical Research Communications, 546, 65–73. 10.1016/j.bbrc.2021.02.001 [DOI] [PubMed] [Google Scholar]
- Wang, T. , Birsoy, K. , Hughes, N. W. , Krupczak, K. M. , Post, Y. , Wei, J. J. , Lander, E. S. , & Sabatini, D. M. (2015). Identification and characterization of essential genes in the human genome. Science, 350(6264), 1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Feng, J. , Xue, Y. , Guan, Z. , Zhang, D. , Liu, Z. , Gong, Z. , Wang, Q. , Huang, J. , Tang, C. , Zou, T. , & Yin, P. (2016). Structural basis of N(6)‐adenosine methylation by the METTL3‐METTL14 complex. Nature, 534(7608), 575–578. 10.1038/nature18298 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Lu, Z. , Gomez, A. , Hon, G. C. , Yue, Y. , Han, D. , Fu, Y. , Parisien, M. , Dai, Q. , Jia, G. , Ren, B. , Pan, T. , & He, C. (2014). N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature, 505(7481), 117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Zhao, B. S. , Roundtree, I. A. , Lu, Z. , Han, D. , Ma, H. , Weng, X. , Chen, K. , Shi, H. , & He, C. (2015). N(6)‐methyladenosine modulates messenger RNA translation efficiency. Cell, 161(6), 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Li, Y. , Toth, J. I. , Petroski, M. D. , Zhang, Z. , & Zhao, J. C. (2014). N6‐methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature Cell Biology, 16(2), 191–198. 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warda, A. S. , Kretschmer, J. , Hackert, P. , Lenz, C. , Urlaub, H. , Hobartner, C. , Sloan, K. E. , & Bohnsack, M. T. (2017). Human METTL16 is a N(6)‐methyladenosine (m(6)A) methyltransferase that targets pre‐mRNAs and various non‐coding RNAs. EMBO Reports, 18(11), 2004–2014. 10.15252/embr.201744940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, B. R. , Wittmann, S. , Wery, M. , Gautier, C. , Kus, K. , Birot, A. , Heo, D. H. , Kilchert, C. , Morillon, A. , & Vasiljeva, L. (2018). Histone deacetylation promotes transcriptional silencing at facultative heterochromatin. Nucleic Acids Research, 46(11), 5426–5440. 10.1093/nar/gky232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, E. C. , Van Nostrand, E. L. , & Yeo, G. W. (2018). Advances and challenges in the detection of transcriptome‐wide protein‐RNA interactions. WIREs RNA, 9(1), e1436. 10.1002/wrna.1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener, D. , & Schwartz, S. (2021). The epitranscriptome beyond m(6)A. Nature Reviews. Genetics, 22(2), 119–131. 10.1038/s41576-020-00295-8 [DOI] [PubMed] [Google Scholar]
- Xiang, S. , Gao, M. , Cao, J. , Shu, X. , Cheng, M. , Wang, F. , Deng, T. , & Liu, J. (2021). Precise identification of an RNA methyltransferase's substrate modification site. Chemical Communications (Cambridge), 57(20), 2499–2502. 10.1039/d0cc08260k [DOI] [PubMed] [Google Scholar]
- Xiang, Y. , Laurent, B. , Hsu, C. H. , Nachtergaele, S. , Lu, Z. , Sheng, W. , Xu, C. , Chen, H. , Ouyang, J. , Wang, S. , Ling, D. , Hsu, P. H. , Zou, L. , Jambhekar, A. , He, C. , & Shi, Y. (2017). RNA m(6)A methylation regulates the ultraviolet‐induced DNA damage response. Nature, 543(7646), 573–576. 10.1038/nature21671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeon, S. Y. , Jo, Y. S. , Choi, E. J. , Kim, M. S. , Yoo, N. J. , & Lee, S. H. (2018). Frameshift mutations in repeat sequences of ANK3, HACD4, TCP10L, TP53BP1, MFN1, LCMT2, RNMT, TRMT6, METTL8 and METTL16 genes in colon cancers. Pathology Oncology Research, 24(3), 617–622. 10.1007/s12253-017-0287-2 [DOI] [PubMed] [Google Scholar]
- Yue, Y. , Liu, J. , & He, C. (2015). RNA N6‐methyladenosine methylation in post‐transcriptional gene expression regulation. Genes & Development, 29(13), 1343–1355. 10.1101/gad.262766.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara, S. , Ries, R. J. , & Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nature Reviews. Molecular Cell Biology, 20(10), 608–624. 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Gu, Y. , & Jiang, G. (2020). Expression and prognostic characteristics of m(6) A RNA methylation regulators in breast cancer. Frontiers in Genetics, 11, 604597. 10.3389/fgene.2020.604597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. S. , & He, C. (2015). Fate by RNA methylation: m6A steers stem cell pluripotency. Genome Biology, 16(1), 43. 10.1186/s13059-015-0609-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. S. , Roundtree, I. A. , & He, C. (2017). Post‐transcriptional gene regulation by mRNA modifications. Nature Reviews. Molecular Cell Biology, 18(1), 31–42. 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, K. I. , Parisien, M. , Dai, Q. , Liu, N. , Diatchenko, L. , Sachleben, J. R. , & Pan, T. (2016). N(6)‐methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. Journal of Molecular Biology, 428(5 Pt A), 822–833. 10.1016/j.jmb.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


