Abstract
Alveolar macrophages (AMs) are critical mediators of pulmonary inflammation. Given the unique lung tissue environment, whether there exist AM-specific mechanisms that control inflammation is not known. Here, we found that among various tissue-resident macrophage populations, AMs specifically expressed Lepr, encoding receptor for a key metabolic hormone leptin. AM-intrinsic Lepr signaling attenuated pulmonary inflammation in vivo, manifested as subdued acute lung injury yet compromised host defense against Streptococcus pneumoniae infection. Lepr signaling protected AMs from necroptosis and thus constrained neutrophil recruitment and tissue damage secondary to release of proinflammatory cytokine interleukin-1α. Mechanistically, Lepr signaling sustained activation of adenosine monophosphate–activated protein kinase in a Ca2+ influx–dependent manner and rewired cellular metabolism, thus preventing excessive lipid droplet formation and overloaded metabolic stress in a lipid-rich alveolar microenvironment. In conclusion, our results defined AM-expressed Lepr as a metabolic checkpoint of pulmonary inflammation and exemplified a macrophage tissue adaptation strategy for maintenance of immune homeostasis.
Lepr is specifically expressed in alveolar macrophages and signals to dampen pulmonary inflammation.
INTRODUCTION
To fulfil tissue-specific functions, tissue-resident macrophages (TRMs) in various microenvironments exhibit unique gene expression profiles and cellular features (1–3). Alveolar macrophages (AMs) represent highly specialized macrophage populations located in the lung alveoli and constitute the first line of immune defense in the respiratory tract (4). AMs rapidly respond to inhaled pathogenic microbes and initiate pulmonary inflammatory responses via releasing factors such as interleukin-1α (IL-1α) that mediates recruitment of neutrophils to sites of infection (5, 6). These inflammatory responses aim to clear pathogens yet often inevitably lead to tissue damage. Thus, like other TRMs, AM-mediated inflammation needs to strike a fine balance between eliciting sufficient protective immunity and preventing excessive damage to lung tissues beyond the extent of repair (7). Unbalanced AM activation and function may result in dysregulated immune responses and contribute to pathogenesis of pulmonary diseases including bacterial pneumonia and acute lung injury (8–12).
In addition to the common dilemma between host defense and tissue damage, the unique pulmonary microenvironment imposes additional challenges for AM tissue adaptation. Residing in alveoli where lipid species are highly enriched in the form of surfactants, AMs are metabolically and transcriptionally distinct from other macrophage populations in the body (13). AMs heavily rely on active lipid metabolism to maintain pulmonary surfactant homeostasis and to stabilize the local microenvironment (14). Relative to other TRMs, AMs contain large quantities of lipid droplets and highly express lipid metabolism–associated genes such as peroxisome proliferator–activated receptor gamma (Pparg) (15, 16). Situated in unique tissue niches, it is conceivable that AMs have metabolic features that deviate from the canonical paradigms (17, 18). Recent reports imply that instead of turning to glycolysis, activated AMs resort to mitochondrial respiration for cellular energy demand (19). However, how AMs develop a distinct cellular program to adapt to the microenvironment of alveoli and to coordinate the pulmonary inflammatory responses is not known.
Leptin is produced and secreted into circulation from white adipose tissues (20–22). After reaching the brain, leptin binds to leptin receptor (Lepr) on neurons in hypothalamus, which, in turn, controls the appetite and body energy expenditure (23–25). Encoded by one Lepr gene, Lepr protein can be presented as the long or short isoform, with the long isoform being the major signaling receptor in hypothalamus (26). Engagement of the long isoform by leptin triggers the canonical Janus kinase–signal transducers and activators of transcription (Jak-STAT) pathway and STAT3-coordinated transcriptional program along with activation of additional signaling molecules such as AKT serine/threonine kinase 1 (Akt) (22, 27). Recent evidence suggests that the effects of leptin are not restricted to the central nervous system and may be extended to other systems as well (28). In the immune system, a regulatory role of leptin has been implicated in processes such as CD4+ T cell differentiation (29, 30). However, the ob/ob (Lep mutant) and db/db mice (Lepr mutant) that are morbidly obese are commonly used to study the function of leptin or Lepr in immune regulation. Thus, the direct effects of Lepr signaling on the immune cells are inevitably confounded by the markedly altered nutritional and overall physiological status of these animals. As a result, whether leptin exerts direct regulatory function in an immune cell–intrinsic manner remains elusive.
In the search of key “signature genes” of AMs, we found that Lepr was highly and specifically expressed in AMs among TRMs, which prompted us to investigate the role of the leptin-Lepr axis in AMs. Using myeloid lineage–specific deletion of Lepr without affecting global metabolism and physiology, we found that intrinsic Lepr signaling in AMs imposed a metabolic checkpoint on pulmonary inflammation. Lepr signaling orchestrated an adenosine monophosphate–activated protein kinase (AMPK)–centric network to maintain the metabolic fitness of AMs and restricted necroptosis-associated release of inflammatory mediators upon microbial challenges. Deficiency of Lepr signaling exacerbated pulmonary inflammation in vivo as manifested by worsened acute lung injury and heightened immune resistance to Streptococcus pneumoniae (Spn) infection. Together, our results identified Lepr expression as a tissue adaptation feature of AMs and revealed the direct and specific effects of Lepr signaling on immune cells with critical functional consequences.
RESULTS
Lepr is highly and specifically expressed in AMs
To profile the expression of Lepr in various TRMs, Ai14 mice were crossed with Lepr-Cre mice to generate the Lepr reporter mice (referred to as tdTomato/+, Lepr-Cre), in which Lepr-expressing cells were labeled as tdTomato+. AMs (CD11c+ Siglec-F+) distinguished by flow cytometry showed tdTomato positivity in reporter mice, indicating robust Lepr expression (Fig. 1, A and B). In contrast to the positive signals in AMs, Lepr was minimally expressed in other in vivo TRMs and commonly studied bone marrow–derived macrophages (BMDMs) (Fig. 1, A and B). Moreover, Lepr was not detected in various immune cell populations in peripheral blood and spleen (fig. S1, A and B), neither in lung interstitial macrophages (fig. S1C), further signifying the tissue and cell type specificity of its expression pattern. Protein expression, as reported by tdTomato, was corroborated by measurement of transcripts, showing robust Lepr mRNA expression in AMs with hypothalamus, known for high Lepr levels (31), serving as a positive control (Fig. 1C). In contrast, Lepr mRNA was barely detectable in other primary TRMs and several macrophage cell lines including MH-S, a murine AM cell line (Fig. 1C and fig. S1D). In lung cryosections of Lepr reporter mice, tdTomato+ cells were located adjacent to alveolar lining cells (Fig. 1D), which resembled the tissue distribution of AMs and further confirmed Lepr expression. In line with the murine study, analyses of a recently reported healthy human bronchoalveolar lavage fluid (BALF) single-cell RNA sequencing (scRNA-seq) dataset showed strong expression of LEPR in human AMs relative to other cell types (Fig. 1E). Together, these results demonstrated uniquely high expression of Lepr in AMs, which prompted us to investigate its immune cell–specific function.
Fig. 1. Lepr is highly and specifically expressed in AMs.
(A) Flow cytometry analysis of different macrophage populations from littermate control (tdTomato/+, +/+) and Lepr-reporter mice (tdTomato/+, Lepr-Cre). WAT, white adipose tissue. (B) Mean fluorescence intensity (MFI) of cells from reporter mice (tdTomato/+, Lepr-Cre) relative to control mice (tdTomato/+, +/+), calculated from (A). Int. mac, interstitial macrophage; NK, natural killer. (n = 2 to 8). (C) Relative mRNA level of Lepr in AMs, peritoneal macrophages, BMDMs, and hypothalamus in wild-type (WT) C57BL/6 mice. Representative results from three independent experiments. (D) Visualization of tdTomato+ cells in lung frozen sections from Lepr-reporter mice. Scale bars, 100 μm. DAPI, 4′,6-diamidino-2-phenylindole. (E) Expression level of LEPR in cells from healthy human BALF. Data are shown as means ± SEM.
Loss of Lepr signaling in AMs protects host from S. pneumoniae infection
To investigate the role of Lepr signaling in AMs, we crossed Lepr flox mice (Leprfl/fl) with myeloid lineage–specific Lyz2-Cre mice to generate Lepr conditional knockout mice (Leprfl/fl Lyz2-Cre, referred to as Lepr cKO mice hereafter). The gene deletion efficiency was verified in AMs by quantitative polymerase chain reaction with Lyz2-Cre mice as controls (fig. S1E). We also validated the deletion specificity by examining the expression of Lepr in several peripheral tissues, including mammary gland, intestine, and spleen, as reported in an earlier study (32). Notably, the expression level of Lepr was relatively low in those tissues in comparison with AMs (fig. S1F), and Lyz2-Cre did not induce detectable deletion of Lepr (fig. S1G). In the follow-up characterization, we observed that in contrast to db/db mice that are morbidly obese because of global Lepr mutation, Lepr cKO mice showed normal body weight compared to control mice regardless of gender (fig. S2A), indicating normal energy expenditure. At the resting state, Lepr cKO mice exhibited normal AM frequency and number (fig. S2, B to D) as well as normal expression of anti-inflammatory cytokines by AMs such as Il10 and Tgfb (fig. S2E), which encoded cytokines essential for maintenance of airway tolerance. Moreover, the gross lung architecture appeared normal in Lepr cKO mice (fig. S2F). These results implicated that myeloid-derived Lepr signaling was dispensable for maintaining systemic energy homeostasis and lung function at the steady state. Nevertheless, leptin protein was readily detectable in BALF (fig. S2G) at the levels sufficient for engaging receptor signaling (27), suggesting that the leptin-Lepr axis may impose important functions on AMs, potentially under nonsteady state conditions.
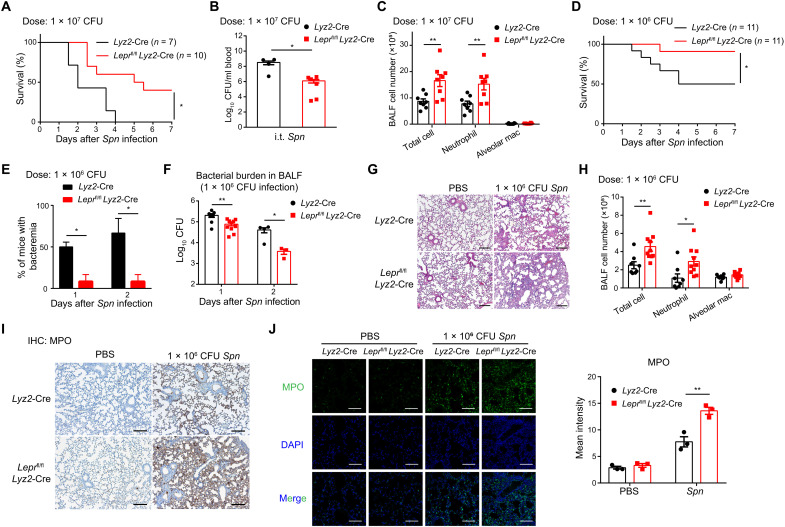
Next, we sought to investigate the role of Lepr signaling in pulmonary inflammation. We adopted a lung infection model of Spn, the leading cause of community-acquired pneumonia worldwide (10, 12). Upon intratracheal (i.t.) infection of strain D39 at a lethal dose of 1 × 107 colony forming units (CFU) (33), while all control animals succumbed to bacterial challenges, Lepr cKO mice showed significantly improved survival (Fig. 2A). Accordingly, bacteremia, as indicated by bacterial burdens in blood circulation, was significantly alleviated in Lepr cKO mice (Fig. 2B). At the early stage of Spn infection, neutrophils are rapidly recruited to sites of infection and become a predominant effector population in BALF for pathogen clearance (9), a notion confirmed by our results that CD11b+ Ly-6G+ neutrophils constituted most BALF immune cells at day 1 after lethal infection (fig. S3, A and B). We found that the cell numbers of neutrophils were significantly increased in Lepr cKO mice, resulting in a proportional increase of total BALF cells, while AMs appeared as a minor population, and their numbers did not change significantly after infection (Fig. 2C). As all control mice died rapidly in responses to the lethal dose of infection, which hampered subsequent phenotypical analyses, we switched to a model of sublethal dose (1 × 106 CFU) infection for the following experiments. Similar to the lethal challenge, Lepr signaling deficiency significantly protected animals from sublethal Spn infection manifested as improved survival, alleviated bacteremia, and reduced BALF bacterial burdens (Fig. 2, D to F). In line with enhanced host defense, histological assessments of infected lungs revealed increased inflammatory cell infiltration and severe hemorrhage in Lepr cKO mice (Fig. 2G). Given that AM numbers did not differ between control and Lepr cKO animals, increased total BALF cells in Lepr cKO mice were largely due to the increase in neutrophil percentages and numbers (Fig. 2H and fig. S3C). Flow cytometry analyses were further corroborated by in situ visualizations of neutrophils in lung sections using myeloperoxidase (MPO) staining, which showed markedly increased lung-infiltrating neutrophils in Lepr cKO mice after infection (Fig. 2, I and J). Nonetheless, the increased degree of pulmonary inflammation in Lepr cKO mice led to more efficient control of bacterial infection within the lung and reduced dissemination of bacteria into blood, which therefore conferred protection from detrimental effects of the pathogen. Together, these results implicated that myeloid-intrinsic Lepr signaling dampened excessive neutrophil-mediated inflammation at the cost of compromised host defense against bacterial infection.
Fig. 2. Loss of Lepr signaling in AMs protects host from S. pneumoniae infection.
Lyz2-Cre and Leprfl/fl Lyz2-Cre mice were intratracheally (i.t.) infected with Spn at the dose of 1 × 107 CFU per mouse. (A) Survival of mice was monitored (n = 7 to 10). (B) Bacterial burden in blood at day 3 after infection was measured (n = 5 to 8). (C) Flow cytometry analysis of cell population in BALF at day 1 after infection (n = 8). Lyz2-Cre and Leprfl/fl Lyz2-Cre mice were intratracheally infected with Spn at the dose of 1 × 106 CFU per mouse. (D) Survival of mice was monitored (n = 11). (E) Percentage of mice with bacteremia at days 1 and 2 after infection was calculated (n = 3). (F) Bacterial burden in BALF at days 1 and 2 after infection (n = 3 to 10). (G) Hematoxylin and eosin (H&E) staining of lung sections from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice at day 1 after intratracheal instillation of phosphate-buffered saline (PBS) or Spn. Scale bar, 200 μm. (H) Flow cytometry analysis of cell populations in BALF at day 1 after infection (n = 8 to 10). (I) Immunohistochemistry (IHC) of MPO in lung sections from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice at day 1 after intratracheal instillation of PBS or Spn (n = 3). Scale bars, 200 μm. (J) Immunofluorescence staining and mean intensity of MPO in lung sections from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice at day 1 after intratracheal instillation of PBS or Spn (n = 3). Scale bars, 100 μm. Data are shown as means ± SEM. *P < 0.05 and **P < 0.01, Mantel-Cox test (A and D) or unpaired t test (other panels).
Lepr signaling in AMs plays a protective role in acute lung injury
As outcomes of pulmonary inflammation could be manifested as host defense and tissue damage, having examined the host defense aspect, we next assessed the role of Lepr signaling with intratracheal instillation of lipopolysaccharide (LPS), a widely used model for acute lung injury (8). After LPS instillation, the neutrophil numbers in Lepr cKO mice were significantly higher than those in control mice, leading to a parallel increase of total BALF cell numbers (Fig. 3, A and B). The AM numbers did not change significantly (Fig. 3C). Increased neutrophil infiltration in Lepr cKO mice was confirmed by immunofluorescence staining of MPO (Fig. 3D). As neutrophils could cause damage to the host tissue, we further assessed the extent of lung injury and found increased lung vascular permeability in Lepr cKO mice compared with control animals during acute lung injury (Fig. 3E). Excessive inflammatory cell infiltration in alveoli and severe lung injury assessed by hematoxylin and eosin (H&E) staining of tissue sections were observed in Lepr cKO mice after LPS instillation (Fig. 3, F and G). Furthermore, severe bleeding in BALF was observed in Lepr cKO mice (Fig. 3H). These results demonstrated exaggerated inflammation in Lepr cKO mice during LPS-induced acute lung injury. In conclusion, these results indicated that Lepr signaling negatively regulated the magnitude of pulmonary inflammation to avoid severe tissue damage.
Fig. 3. Lepr signaling in AMs plays a protective role in acute lung injury.
LPS was intratracheally administrated (0.5 mg/kg) in Lyz2-Cre and Leprfl/fl Lyz2-Cre mice. (A) Total cell numbers in BALF were quantified. Numbers of neutrophils (B) and AMs (C) were analyzed by flow cytometry. n = 8 to 14. (D) Immunofluorescence staining and mean intensity of MPO in lung sections from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice at 6 hours after intratracheal instillation of PBS or LPS (n = 2 to 4). Scale bars, 100 μm. (E) Measurement of lung vascular permeability in lungs from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice at 6 hours after intratracheal instillation of PBS or LPS (n = 4). H&E staining (F) and lung injury score (G) of lung sections from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice at 6 hours after intratracheal instillation of PBS or LPS (n = 5). Scale bars, 200 μm. (H) Bleeding in BALF from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice 4 hours after LPS administration (n = 4). Data are shown as means ± SEM. *P < 0.05; **P < 0.01; and not significant (ns), P > 0.05, unpaired t test.
Lepr signaling maintains plasma membrane integrity and restrains IL-1α release in inflammatory AMs
Next, we wished to explore the underlying mechanisms of leptin-mediated effects in pulmonary inflammation. Myeloid-specific Lyz2-Cre may drive target gene deletion in neutrophils in addition to macrophages, and thus, we examined Lepr expression in lung-infiltrating neutrophils to determine whether Lepr signaling in neutrophils could potentially contribute to the observed phenotypes. In Spn or LPS-treated Lepr reporter mice, minimal expression of Lepr was detected in neutrophils (fig. S3, D to G), excluding the possibility of neutrophil-intrinsic leptin effects and suggesting that enhanced neutrophil-mediated inflammation associated with Lepr deficiency was likely secondary to alterations of macrophages.
We first analyzed prototypical inflammatory cytokines and chemokines IL-6, tumor necrosis factor–α (TNFα), and CXCL1 in Spn infection and LPS-induced acute lung injury models but found comparable production in BALF between control and Lepr cKO mice (fig. S4, A and B). Leptin levels in BALF were also comparable (fig. S4, C and D). Consistent with the above in vivo results, deficiency of Lepr signaling in AMs did not significantly alter Il6, Tnf, and Cxcl1 expression upon in vitro LPS stimulation (fig. S4E). Furthermore, RNA-seq analyses of control and Lepr KO AMs under resting and LPS-activated conditions did not yield obvious candidates that may underscore the phenotypical differences (fig. S4F). Therefore, we postulated that leptin might regulate AM inflammatory function at the posttranscriptional level. It has been reported that necrosis of AMs contributes to pathogenesis of LPS-induced acute lung injury (34). When the viability of AMs was assessed by staining with either 7-aminoactinomycin D (7-AAD) or Fixable Viability Dye eFluor 506, we unexpectedly observed that the mean fluorescence intensity (MFI) of both 7-AAD and Fixable Viability Dye eFluor 506 signals were increased in Lepr-deficient AMs after LPS administration and Spn infection (Fig. 4, A to D, and fig. S4G to J), indicating enhanced plasma membrane permeability. High-resolution visualization of cell surfaces by scanning electron microscope revealed increased pore formation in Lepr-deficient AMs after live Spn infection (Fig. 4, E and F). In addition, the release of lactate dehydrogenase (LDH), an indicator of cell death, was also found to be elevated in BALF from Lepr cKO mice (Fig. 4G). Thus, these results suggested that Lepr signaling sustained AM plasma membrane integrity during pulmonary inflammation.
Fig. 4. Lepr signaling maintains plasma membrane integrity and restrains IL-1α release in inflammatory AMs.
(A) Flow cytometry analysis of 7-AAD in AMs (gated on CD45+ CD11c+ Siglec-F+) from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice with intratracheal instillation of PBS or LPS. (B) MFI was calculated from (A). n = 3 to 7. (C) Flow cytometry analysis of 7-AAD in AMs (gated on CD45+ CD11c+ Siglec-F+) from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice at day 1 after intratracheal instillation of Spn. (D) MFI was calculated from (C). n = 7 to 10. (E) Scanning electron microscopy of Lyz2-Cre and Leprfl/fl Lyz2-Cre AMs at resting state and after live Spn infection [multiplicity of infection (MOI) = 50] in vitro for 4 hours. Scale bars, 5 μm. (F) Quantification of pores (diameter < 50 nm) on plasma membrane from (E). n = 10 to 14. (G) Measurement of LDH release in BALF from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice at day 1 after Spn infection (n = 7 to 8). OD490, optical density at 490 nm. (H) Measurement of IL-1α level by enzyme-linked immunosorbent assay in BALF from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice at day 1 after Spn infection (n = 11 to 13). (I and J) Lyz2-Cre and Leprfl/fl Lyz2-Cre mice were intratracheally infected with Spn plus control immunoglobulin G1 (IgG1)/anti–IL-1α IgG, and cells in BALF (I) were analyzed by flow cytometry (n = 6 to 7). 7-AAD staining (J) was analyzed in AMs (n = 6 to 7). Data are shown as means ± SEM. *P < 0.05; **P < 0.01; ****P < 0.0001; and ns, P > 0.05, unpaired t test.
Compromised plasma membrane integrity could lead to release of inflammatory cell contents that further intensifies inflammation. Among the key factors affected by cellular permeability, we detected increased IL-1α protein levels in BALF of Lepr cKO mice after Spn infection (Fig. 4H) despite comparable mRNA expression in AMs (fig. S4K), implying that Lepr signaling likely regulated release but not transcription of IL-1α. To investigate the causal relationship between IL-1α release and pulmonary phenotypes, an IL-1α–neutralizing antibody was administrated in vivo to Spn-infected animals. Augmentation of total cell number and neutrophil infiltration in Lepr cKO mice was nearly completely abrogated after IL-1α blockade (Fig. 4I), indicating that increased neutrophil infiltration was attributed to elevated IL-1α release. Neutralization of IL-1α had minimal impact on AM plasma membrane permeability as measured by 7-AAD staining in both control and Lepr cKO mice (Fig. 4J), suggesting that IL-1α was a downstream effector of AM cell death. In summary, these results implicated that Lepr signaling maintained AM plasma membrane integrity and thus restricted release of inflammatory cell contents, such as IL-1α, to attenuate pulmonary inflammation.
Lipid droplets accumulate in Lepr-deficient AMs
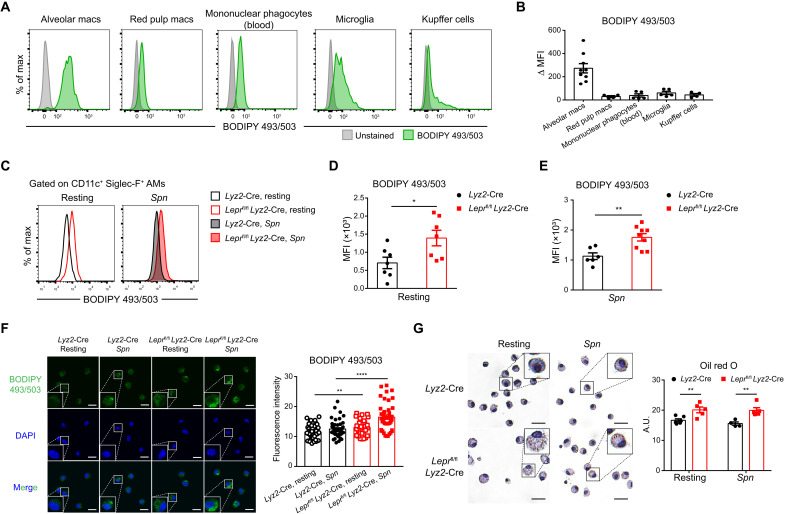
Among various TRMs, AMs show active lipid metabolism that is endowed by the lung microenvironment (14). We first used a fat-soluble dye, 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene (BODIPY 493/503), to quantitate lipid droplets and found that AMs exhibited the highest lipid droplet contents among the TRM populations examined (Fig. 5, A and B). Given the notable parallel patterns of Lepr expression and lipid abundances, we attempted to determine whether Lepr signaling might control lipid storage in AMs. The lipid droplet contents were significantly enriched in Lepr-deficient AMs relative to control cells at resting state and after Spn infection (Fig. 5, C to E). In line with flow cytometry analyses, direct visualization of lipid droplets with BODIPY 493/503 and Oil red O staining showed consistent enrichment in Lepr KO AMs at resting state and after Spn stimulation (Fig. 5, F and G). Thus, these results indicated that Lepr signaling constrained lipid droplet formation and maintained lipid homeostasis in AMs.
Fig. 5. Lipid droplets accumulate in Lepr-deficient AMs.
(A and B) Lipid droplet in different TRMs of WT mice was measured by flow cytometry analysis of BODIPY 493/503. MFI relative to unstained control was calculated. n = 4 to 9. (C to E) Representative histograms (C) and MFI (D and E) showing BODIPY 493/503 staining in AMs from Lyz2-Cre and Leprfl/fl Lyz2-Cre mice at resting state and at day 1 after Spn infection. n = 6 to 9. (F) Confocal microscopy imaging of BODIPY 493/503 staining in Lyz2-Cre and Leprfl/fl Lyz2-Cre AMs at resting state and after heat-inactivated Spn stimulation (MOI = 20) in vitro for 4 hours. Fluorescence intensity of BODIPY 493/503 in each cell was calculated. Scale bars, 10 μm. n = 29 to 47. (G) Representative sections of oil red O staining in Lyz2-Cre and Leprfl/fl Lyz2-Cre AMs at resting state and after heat-inactivated Spn stimulation (MOI = 20) in vitro for 4 hours. Oil red O staining was quantified. Scale bars, 20 μm. A.U., arbitrary unit. Data are shown as means ± SEM. *P < 0.05, **P < 0.01, and ****P < 0.0001, unpaired t test.
Lepr signaling orchestrates AM cellular metabolism to maintain a balanced energy status
Lipid overload could result in metabolic stress and compromised cellular functionality (35, 36). We then examined the metabolic alterations associated with lipid overload in Lepr-deficient AMs. Seahorse analysis showed that oxygen consumption rate (OCR) in Lepr-deficient AMs was higher than that in control AMs upon Spn stimulation (Fig. 6, A and B), suggesting that oxidative phosphorylation (OXPHOS) might be up-regulated to meet the catabolic demands of excessive lipid species. In contrast, extracellular acidification rate (ECAR), an indicator of glycolytic activities, was reduced in Lepr KO AMs (Fig. 6, A and B), implicating that Lepr signaling rewired metabolic programs in AMs by favoring glycolysis over OXPHOS. To further profile cellular metabolism in an unbiased manner, resting and Spn-challenged AMs were subjected to targeted metabolomics analysis focusing on essential metabolic modules including tricarboxylic acid (TCA) cycle and glycolysis pathway. Principal components analysis (PCA) showed that, under the resting condition, the overall metabolic state of AMs was indistinguishable on the basis of Lepr signaling competency or deficiency (Fig. 6C). However, upon Spn stimulation, the metabolic programs in Lepr-deficient AMs markedly deviated from those in control AMs (Fig. 6C), implying tight coupling of metabolic changes with macrophage activation. In Spn-activated AMs, deficiency of Lepr signaling led to increased levels of TCA cycle intermediates isocitrate/citrate and oxoglutarate yet decreased levels of glucose and pyruvate (Fig. 6D), consistent with the above Seahorse analysis. Moreover, the level of AMP and ratio of [AMP] × [ADP] to [ATP], key indicators of cellular energy status, were reduced in Lepr KO AMs (Fig. 6E). Together, Lepr signaling plays a key role in orchestrating metabolism to maintain energy balance in AMs (Fig. 6F).
Fig. 6. Lepr signaling orchestrates AM cellular metabolism to maintain a balanced energy status.
(A) OCR and ECAR of Lyz2-Cre and Leprfl/fl Lyz2-Cre AMs at resting state and after heat-inactivated Spn stimulation (MOI = 10) were measured by Seahorse XFe96 Analyzer. Representative results from three independent experiments. A/R, antimycin A/rotenone. (B) Statistical analysis of maximal respiration and glycolytic capacity from (A). n = 3 to 6. (C) PCA of targeted metabolomics results of Lyz2-Cre and Leprfl/fl Lyz2-Cre AMs at resting state and after Spn stimulation for 4 hours. (D) Fold change (Leprfl/fl Lyz2-Cre versus Lyz2-Cre) of top five increased and decreased metabolites from metabolomics results after Spn stimulation for 4 hours. TCA cycle– and glycolysis-related metabolites were shown in red and blue, respectively. (E) Fold change (Leprfl/fl Lyz2-Cre versus Lyz2-Cre) of AMP and ratio of [AMP] × [ADP] to [ATP] from metabolomics results after Spn stimulation. “[]” indicated abundance of each adenine nucleotide. (F) Metabolic pathway map depicting alterations of metabolites at regulatory steps in glycolysis and TCA cycle from Lyz2-Cre and Leprfl/fl Lyz2-Cre AMs. Data are shown as means ± SEM. *P < 0.05 and ns, P > 0.05, paired t test.
Lepr signaling maintains AM metabolic fitness through AMPK
The ratio of adenine nucleotides is one of the determinants that control activities of AMPK, a central sensor and orchestrator of cellular energy status (37). Given the altered ratio of adenine nucleotides in Lepr-deficient AMs, we postulated that Lepr may influence AMPK signaling. In wild-type (WT) AMs, leptin treatment rapidly induced phosphorylation of AMPK on threonine 172 [pAMPKα (T172)] (Fig. 7A), which positively indicated AMPK activities. Moreover, loss of function of Lepr in activated AMs led to consistent reduction of AMPK phosphorylation (Fig. 7B), suggesting that Lepr signaling contributed to full-fledged AMPK activities in AMs. To mechanistically connect Lepr with AMPK, we examined pathways and factors known to activate AMPK (37) and zoomed our attention on Ca2+ signaling. Upon Spn stimulation, Lepr-competent AMs exhibited marked Ca2+ influx, which was impaired in Lepr-deficient AMs (Fig. 7C). Furthermore, treatment of Spn-stimulated AMs with STO-609, a calmodulin–dependent protein kinase kinase β (CaMKKβ) inhibitor, consistently resulted in decreased AMPK activation (Fig. 7D), implying that Lepr signaling sustained AMPK via the Ca2+-CaMKKβ axis. In addition to AMPK, various signaling pathways downstream of Lepr have been reported, the most prominent being the canonical Jak-STAT pathway (22, 23). Nevertheless, in AMs, we did not detect leptin-induced activation of Jak2-STAT3 signaling nor of other pathways including Akt (fig. S5, A to D). Lepr can be present in one of two isoforms encoded by the same gene, the short (Lepr-a) and long (Lepr-b) isoforms that differ in intracellular domain and thus in their capacity to engage Jak-STAT signaling (fig. S5E). Compared with hypothalamus known to predominantly express the long isoform Lepr-b (23), AMs expressed relatively low level of Lepr-b but high level of Lepr-a (fig. S5F), plausibly explaining the inability of AMs to engage canonical signaling in response to leptin. Together, these results indicated that Lepr in AMs primarily functioned through Ca2+-AMPK signaling pathway.
Fig. 7. Lepr signaling maintains AM metabolic fitness through AMPK.
(A) WT AMs were incubated in FBS-free medium for 2 hours and were treated with leptin (1 ng/ml). pAMPKα (T172) was analyzed by immunoblotting, n = 4. (B) Immunoblotting analysis of pAMPKα (T172) in AMs after live Spn infection (MOI = 50) in vitro for 4 hours, n = 3. (C) AMs were loaded with 1 μM Fluo-4 AM in Hanks’ balanced salt solution (2 mM Ca2+). Ca2+ oscillation before and after heat-inactivated Spn stimulation (MOI = 20) was analyzed. Representative results of three independent experiments. N = 53 or 25. N, number of cells that were analyzed. (D) WT AMs were stimulated by heat-inactivated Spn (MOI = 20) for 4 hours with/without STO-609 pretreatment (2 hours, 15 μM). pAMPKα (T172) was analyzed by immunoblotting. n = 4. (E and F) Lyz2-Cre and Leprfl/fl Lyz2-Cre AMs were pretreated with vehicle/AICAR (0.5 mM) overnight (14 hours) and were stimulated with heat-inactivated Spn for 4 hours (MOI = 20). Representative histograms (E) and MFI (F) were shown for BODIPY 493/503 staining. n = 4. (G) Immunoblotting analysis of pMLKL (S345) in AMs after live Spn infection (MOI = 50) in vitro for 4 hours. n = 3. (H) Lyz2-Cre and Leprfl/fl Lyz2-Cre mice were intranasally pretreated with AICAR (500 μg per mouse/day) for 3 consecutive days and were then intratracheal instilled with Spn + vehicle/AICAR. At day 1 after infection, 7-AAD staining was analyzed in AMs by flow cytometry. n = 5 to 7. Data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; and ns, P > 0.05, paired (A, B, and G) or unpaired t test (D, F, and H).
AMPK activation skews cellular metabolism toward lipid catabolism (38, 39), and the reduced AMPK activation in Lepr KO AMs correlated with lipid overload. To probe the causal relationship between AMPK and lipid metabolism in AMs, we reactivated AMPK in Lepr KO AMs by an AMPK activator, 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR), and found that AICAR treatment abrogated increased lipid droplet formation in Lepr KO AMs (Fig. 7, E and F), suggesting that defective AMPK activation in Lepr-deficient AMs causally led to the accumulation of lipid droplets. AMPK has been reported to suppress inflammatory necroptosis (40), and we then analyzed activation of necroptosis executor, mixed-lineage kinase domain–like protein (MLKL). Lepr-null AMs exhibited enhanced MLKL phosphorylation upon Spn infection (Fig. 7G), consistent with compromised plasma membrane integrity associated with Lepr deficiency. Last, the increase of 7-AAD staining in Lepr KO AMs was abrogated with in vivo AICAR administration (Fig. 7H), further substantiating the suppressive effect of AMPK on AM necroptosis. In contrast to MLKL, cleaved Caspase-1 was not detected in Spn-infected AMs (fig. S5G), suggesting minimal involvement of inflammasome in the process. Therefore, Lepr sustains AMPK signaling in AMs to inhibit necroptosis and subsequently attenuate pulmonary inflammation.
DISCUSSION
AMs are uniquely located in a tissue compartment where inhaled air enters the body and thus are frequently exposed to airborne innocuous particles and harmful pathogens. As the first line of pulmonary defense, AMs hold the trigger of initiating a cascade of subsequent immune and inflammatory responses, signified by rapid and massive recruitment of neutrophils to sites of infection, a process that is typically irreversible and often leads to tissue damage. As a result, the threshold for AM-initiated pulmonary inflammation needs to be precisely set to ensure sufficient protective immunity yet avoid unnecessary tissue damage. Another critical role of AMs is to phagocytose and recycle excessive pulmonary surfactants, and thus, AMs are also burdened with the task of constantly degrading numerous lipid species (4, 16, 41). Therefore, it is highly intriguing that the leptin-Lepr axis, a potent regulator of global lipid mass, is operational in an AM-specific manner as a metabolic checkpoint to set the threshold of pulmonary inflammation. In Lepr-sufficient AMs, AMPK activation is sustained to constrain lipid overload, and thus, AMs appear “lean” on the cellular level (fig. S6). The resulting bioenergetic patterns further maintain AMPK activities to reinforce a positive feedforward loop that renders AMs metabolically fit and less prone to inflammation. On the contrary, lipid droplets accumulate in Lepr-deficient AMs, and the cells become “obese,” accompanied by rewired energetic metabolism and markedly reduced threshold for inflammatory responses (fig. S6). Therefore, Lepr signaling sustains the metabolic fitness of AMs to control pulmonary inflammation, which represents a prominent tissue adaptation identity of AMs.
Leptin and Lepr were originally identified as obesity-associated genes (20). Leptin is present in circulation of lean mice at the range of 5 to 15 ng/ml (42, 43). Leptin can be detected in BALF at the picogram range (figs. S2G and S4, C and D), which is likely derived from circulation. As leptin is present in alveoli not only under resting state but also after LPS instillation and Spn infection with relatively constant levels, it may provide tonic signaling to establish functional identity of AMs. In AMs, leptin-activated AMPK was Ca2+ influx dependent. The effect of Lepr signaling on Ca2+ influx has been reported in pancreatic β cell (44) and pericytes (45), implicating that engagement of Ca2+ signaling might represent one specific mode of leptin action on nonneuronal cell types. The inability of Lepr to activate Jak2-STAT3 in AMs might be explained by the dominance of the short isoform that lacks the portion of intracellular domain containing STAT docking sites: The homodimer of the short isoform is incapable of mediating the Jak2-STAT3 signal; the heterodimer of Lepr-a–Lepr-b failed to activate STAT3 signaling shown in an earlier study (46), which could also occur in AMs; and the short isoform might as well act as a decoy receptor to suppress Jak2-STAT3 signaling initiated by long isoform. Previous studies have shown that expression of Lepr short isoform, Lepr-a, is widely distributed in peripheral tissues and that expression level of Lepr-a is specifically high in the lung (32, 47). When Lepr-a–deficient mice were generated (48), in contrast to db/db mice, they showed normal body weight on regular chow diet and only modestly increased body weight on high-fat diet. Therefore, the precise physiological function of Lepr-a remains elusive. Our results with AMs exemplify that the leptin to Lepr-a signal may represent a tonic pathway that primarily functions in distinct cell types in a peripheral organ-specific manner.
The unique expression pattern of Lepr ensures the AM-specific regulation among TRM populations. Although different TRMs share several core macrophage programs, TRMs develop distinct mechanisms as they face diverse challenges in local tissues. Investigation on tissue-specific mechanisms of TRMs may provide insights into understanding physiological significance of TRMs. Accumulating works have noted that macrophages are highly plastic, and nutrient availability within different tissue niches varies markedly, which may differentially reprogram the metabolism of TRMs (14, 18). Our results showed that AMs down-regulated glycolysis after bacterial infection (Fig. 6A), which was unexpected, given the prevailing paradigm of macrophage metabolism supporting the up-regulation of glycolytic pathway in inflammatorily activated macrophages (49). This unique metabolic reprogramming may be explained by special nutrient availability in alveoli, where glucose concentration is relatively low, yet oxygen is particularly enriched. To survive and effectively mount immune response in such a microenvironment, AMs are likely to adopt OXPHOS instead of glycolysis as the preferential machinery for energy generation. Consistent with our results, a recent report also demonstrates that AMs do not rely on glycolysis under the LPS-stimulated condition (19). Together, these results suggest that metabolic adaptation may constitute an important functional parameter to establish TRM identity and diversify macrophage responses most appropriate to local microenvironments.
Spn infection is still a leading cause of community-acquired pneumonia worldwide in recent years, with especially high incidence in infants and elderly people (9, 12). As antibiotic resistance and evasion of vaccination are emerging, in-depth understanding of host-pathogen interaction and subsequent conceptualization of immune-infection therapeutic targets are urgently needed. Our study provides new insights into understanding innate immune responses against this pathogen and implies manipulation of AMs as a potential tool for drug discovery and vaccine optimization. Pulmonary bacterial infections often cause tissue damage, which is manifested as acute lung injury and may develop into acute respiratory distress syndrome (ARDS), a lethal or disabling condition (8, 11). ARDS is also commonly associated with pulmonary viral infections such as coronavirus disease 2019 (50). Thus, further work is needed to fully understand function of Lepr in other lung injury–associated pulmonary diseases, such as viral infection and pulmonary fibrosis. Given the unique expression of Lepr in AMs, targeted intervention of Lepr signaling through respiratory tract may represent a promising therapeutic approach of pulmonary inflammatory conditions such as ARDS.
MATERIALS AND METHODS
Experimental design
The goal of this study was to investigate lung-specific role of Lepr in AMs during pulmonary inflammation. Lepr-reporter mice were used to profile the expression pattern of Lepr among various TRMs. Lepr cKO mice, in which Lepr was specifically deleted in myeloid lineage cells, were intratracheally infected with Spn or administrated with LPS. Survival of mice, bacterial burden, inflammatory cell infiltration, and pathological changes in the lung were analyzed to assess the role of Lepr during pulmonary inflammation. Changes of cellular responses and signaling pathways in AMs after Spn infection were analyzed to study the underlying mechanism. Sample sizes and replicates were specified in figure legends.
Mice
All mice were at the Laboratory Animal Resources Center of Tsinghua University. The mice were maintained on a 12-hour light/12-hour light/dark cycle (0700 to 1900), 22° to 26°C, 40 to 70% humidity, with sterile pellet food and water ad libitum, and under specific pathogen-free condition. Mice with myeloid lineage–specific deletion of Lepr (Leprfl/fl Lyz2-Cre) were generated by crossing Leprfl/fl mice with Lyz2-Cre mice (the Jackson Laboratory). Lyz2-Cre mice were used as controls. Lepr-reporter (tdTomato/+, Lepr-Cre) mice were generated by crossing Ai14 mice [Gt(ROSA)26Sortm14(CAG-tdTomato)Hze] and Lepr-Cre mice [Leprtm3(cre)Mgmj] (the Jackson Laboratory). Littermate (tdTomato/+, +/+) mice, in which tdTomato was heterozygous and Cre recombinase was not expressed, were used as controls. Six- to 12-week-old male and female mice were used for experiments. Control and KO/reporter mice were age- and gender-matched. The laboratory animal facility has been accredited by Association for Assessment and Accreditation of Laboratory Animal Care International. The Institutional Animal Care and Use Committee (IACUC) of Tsinghua University approved all animal protocols used in this study.
Cell culture and reagents
Murine AMs were obtained by collecting BALF from mice and were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Gibco). Murine BMDMs were obtained as previously described (51) and were maintained in DMEM supplemented with 10% FBS and 10% L929 cell supernatant as conditioned medium providing macrophage colony-stimulating factor. Cell culture–grade LPS was purchased from Sigma-Aldrich, and cells were stimulated at a concentration of 10 ng/ml.
Collection of immune cells from tissue
Tissues were harvested from euthanized mice. Lung was cut into small pieces and was digested in digestion buffer [RPMI 1640 medium (Macgene) containing collagenase IV (2 mg/ml; Sigma-Aldrich) and deoxyribonuclease (DNase I; 50 μg/ml; Sigma-Aldrich)] at 37°C and 200 rpm for 1 hour. Brain and white adipose tissues were cut into small pieces and were digested in digestion buffer [phosphate-buffered saline (PBS) containing collagenase IV (4 mg/ml) and DNase I (50 μg/ml)] at 37°C and 200 rpm for 45 min. Cells from the spleen and liver were harvested by homogenization. Peritoneal macrophages were harvested by collecting lavage of peritoneal cavity. Colonic macrophages were isolated as previously described (52). Peripheral blood was collected, and red blood cells were lysed by ammonium-chloride-potassium lysis buffer. If necessary, cells were passed through 70-μm cell strainer and were collected by 1500 rpm centrifugation for 5 min at 4°C.
AMs were identified as CD45+ CD11c+ Siglec-F+ cells. Peritoneal macrophages, white adipose tissue macrophages, and splenic red pulp macrophages were identified as CD45+ CD11b+ F4/80+ cells. Blood monocytes were identified as CD45+ Ly-6G− CD115+ cells. Colonic macrophages were identified as CD45+ CD11b+ CD11c+ Ly-6Clo CD64+ cells. Kupffer cells were identified as CD45+ Ly-6C− F4/80+ CD11b+ cells. Microglia were identified as CD45+ CD11b+ F4/80+ CX3CR1+ cells. Natural killer (NK) cells were identified as CD45+ NKp46+ NK1.1+ cells.
scRNA-seq data analysis
ScRNA-seq data of cells from healthy human control BALFs were acquired from Gene Expression Omnibus (GEO) database by accession no. GSE145926 (53). Data analysis was conducted in R v4.0 by packages from Seurat v3.1 (54). In total, 26,185 cells were analyzed. The data were first preprocessed as the following criteria: filter cells that have unique feature counts >5000 or < 200 and filter cells that have >25% mitochondrial counts. Then, the data were normalized by “LogNormalize” with a scale factor of 10,000. By “vst” method, 2000 features were identified for downstream analysis. After scaling data and performing PCA, top 12 principal components were used for further analysis. The cells were clustered at the resolution of 0.4, and nonlinear dimensional reduction was run using Uniform Manifold Approximation and Projection. Last, expression of LEPR in each cluster was visualized by violin plot.
Bacterial cultivation and infection
Spn D39 (serotype 2) was cultured in Todd-Hewitt broth (OXOID) supplemented with 0.5% yeast extract (THY medium) or on trypticase soy agar (BD Biosciences) supplemented with 5% defibrinated sheep blood (SolarBio Life Sciences) at 37°C with 5% CO2 as described previously (55). Lung infection was performed in age- and gender-matched mice according to the animal protocols approved by the IACUC in Tsinghua University, as described in (56). Briefly, Spn frozen stocks in 15% glycerol were thawed, washed by PBS once, and diluted to a desirable density in PBS before intratracheal administration. For in vitro live bacterial infection, the bacteria were diluted to a desirable density in cell culture medium (penicillin-streptomycin free). Bacterial attachment to host cells was facilitated by centrifugation of 500g at room temperature (RT) for 5 min. Two hours after infection, gentamicin (SolarBio Life Sciences) was added to a final concentration of 100 μg/ml to avoid bacterial overgrowth. Two hours later, cells were lysed for further experiments. In vitro infection of cells with heat-inactivated bacteria was carried out in a similar manner, except for resuspension of live Spn in cell culture medium containing penicillin-streptomycin, and inactivated at 60°C for 10 min before mixing with host cells.
Targeted metabolomics and data analysis
After heat-inactivated Spn stimulation [multiplicity of infection (MOI = 10)] for 4 hours, the culture medium was aspirated, and cells were washed with PBS three times. Next, 1 ml 80% (v/v) methanol (prechilled to −80°C) was added to cells, and the plate was incubated at −80°C overnight. The cells were scraped, and the lysates were transferred to a 1.5-ml tube. Tubes were centrifuged at 14,000g at 4°C for 20 min, and the supernatant was transferred to a new tube. Samples were dried for targeted metabolomics analysis.
Targeted metabolomics experiment was performed on TSQ Quantiva (Thermo Fisher Scientific, CA). This analysis mainly measured metabolites in the TCA cycle, glycolysis, pentose phosphate pathway, amino acids, and purine metabolism. For C18-based reverse phase chromatography, 10 mM tributylamine and 15 mM acetate (diluted in water) served as mobile phase A and 100% methanol as mobile phase B. A 25-min gradient from 5 to 90% mobile B was used, and data were acquired in positive-negative ion switching mode. The resolution for both Q1 and Q3 was 0.7 full width at half maximum. The source voltage was 3500 V for positive and 2500 V for negative ion mode. Source parameters were as follows: spray voltage of 3000 V, capillary temperature of 320°C, heater temperature of 300°C, sheath gas flow rate of 35, and auxiliary gas flow rate of 10. Metabolites were identified on the basis of TraceFinder search with home-built database containing about 300 compounds. Data analysis was performed on MetaboAnalyst 5.0 (https://metaboanalyst.ca/) for PCA (57).
Measurements of ECAR and OCR
Cells were seeded in an XFe96 Cell Culture Microplate at the density of 2 × 104 cells per well. Cells were attached to the plate overnight and were stimulated with heat-inactivated Spn for 4 hours as previously described. Before the assay, the culture medium was removed, and assay medium was added in each well. For ECAR measurement, the assay medium was XF base medium supplemented with 4 mM l-glutamine (pH = 7.4 ± 0.05). For OCR measurement, the assay medium was XF base medium supplemented with 25 mM glucose, 4 mM l-glutamine, and 1 mM sodium pyruvate (pH = 7.4 ± 0.05). Cells were incubated in assay medium for 1 hour at 37°C (without CO2). Sensor cartridge was pretreated with XF calibrant at 37°C (without CO2) for >12 hours. Then, compounds were added to the sensor cartridge. For ECAR, the compounds were added in the order of glucose (80 mM, 8×), oligomycin (18 μM, 9×), and 2-deoxyglucose (1 M, 10×). For OCR, the compounds were added in the order of oligomycin (16 μM, 8×), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (18 μM, 9×), and antimycin A/rotenone (2.5 mM, 10×). The sensor cartridge was calibrated, and the assay was performed on a Seahorse XFe96 Analyzer according to the manufacturer’s instructions. The results were analyzed on a Seahorse Wave Desktop 2.6.1. All reagents, plates, and machines were purchased from Agilent.
Lipid droplet staining
BODIPY 493/503 (Invitrogen) was used for lipid droplet staining. For confocal imaging, cells were grown on a coverslip. After treatment, cells were fixed with 4% paraformaldehyde at RT for 15 min. Then, cells were washed with PBS for three times and were permeabilized with 0.2% Triton X-100 (Sigma-Aldrich) at RT for 10 min. After washing with PBS, lipid droplet was stained with BODIPY 493/503 (1 μg/ml) in PBS at RT for 15 min. Next, cells were stained with 4′,6-diamidino-2-phenylindole for 5 min and washed with double-distilled H2O. The coverslip was mounted with Fluoroshield mounting medium (Abcam). Images were acquired on Olympus FV3000 confocal laser scanning microscope. For flow cytometry analysis, after treatment, cells were washed with PBS and were stained with BODIPY 493/503 (1 μg/ml in PBS) in cell culture incubator for 20 min. After washing with PBS for twice, cells were resuspended in PBS for flow cytometry analysis.
In in vitro staining of lipid droplet, cholesterol (100 or 200 μM; Sigma-Aldrich) was added into cell culture and was incubated with cells overnight (for ~14 hours). In in vitro AICAR treatment experiments, BALF was collected with culture medium. The cells were incubated with BALF medium overnight for following assay. AICAR (Selleck) was added to a concentration of 0.5 mM.
Bulk RNA-seq
Total RNA was extracted by TRIzol (Life Technologies). Library construction and sequencing were conducted on Illumina HiSeq platform by Beijing Genomics Institute (BGI). Data processing was also performed by BGI. Total reads were cleaned and were mapped to mm10 reference genome. Differentially expressed genes were identified as expression of KO/WT fold change of ≥2 (up-regulated in KO) or ≤0.5 (down-regulated in KO). False discovery rate was set as ≤0.001.
Statistical analysis
Statistical analysis was performed in GraphPad Prism 7. Two-tailed paired t test or unpaired t test was indicated in figure legends, and P value was calculated. Data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; and not significant, P > 0.05.
Acknowledgments
We thank H. An and C. Qian from J.-R.Z.’s laboratory (Tsinghua University) for assistance in infection experiments. We thank X. Wang from Y. Shi’s laboratory (Tsinghua University) for expertise and providing equipment in Ca2+ imaging experiments.
Funding: This work was funded by the National Natural Science Foundation of China grant nos. 31725010, 31821003, 31991174, 32030037, and 82150105 (to X.H.), Minister of Science and Technology of China grants 2020YFA0509100 (to X.H.) and 2017YFA0505800 (to W.Z.), Tsinghua University COVID-19 Scientific Research Program 2020Z99CFZ024 (to X.H.), and funds from Tsinghua-Peking Center for Life Sciences and Institute for Immunology at Tsinghua University (X.H. and W.Z.).
Author contributions: Conceptualization: X.H. and W.Z. Data curation: Z.G. Formal analysis: Z.G. Funding acquisition: X.H. and W.Z. Investigation: Z.G. and H.Y. Methodology: Z.G., X.H., W.Z., and J.-R.Z. Project administration: Z.G. and H.Y. Resources: X.H., W.Z., and J.Z. Supervision: X.H. and W.Z. Visualization: Z.G. Writing—original draft: Z.G. Writing—review and editing: Z.G., X.H., W.Z., and J.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All of RNA-seq datasets were deposited in the National Center for Biotechnology Information GEO database (https://ncbi.nlm.nih.gov/geo/). ScRNA-seq datasets can be accessed through GEO no. GSE145926. Bulk RNA-seq datasets can be accessed through GEO no. GSE185218.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S6
Table S1
REFERENCES AND NOTES
- 1.Okabe Y., Medzhitov R., Tissue biology perspective on macrophages. Nat. Immunol. 17, 9–17 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Bleriot C., Chakarov S., Ginhoux F., Determinants of resident tissue macrophage identity and function. Immunity 52, 957–970 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Guilliams M., Thierry G. R., Bonnardel J., Bajenoff M., Establishment and maintenance of the macrophage niche. Immunity 52, 434–451 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Hussell T., Bell T. J., Alveolar macrophages: Plasticity in a tissue-specific context. Nat. Rev. Immunol. 14, 81–93 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki A., Foxman E. F., Molony R. D., Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 17, 7–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nobs S. P., Kopf M., Tissue-resident macrophages: Guardians of organ homeostasis. Trends Immunol. 42, 495–507 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Allard B., Panariti A., Martin J. G., Alveolar macrophages in the resolution of inflammation, tissue repair, and tolerance to infection. Front. Immunol. 9, 1777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson B. T., Chambers R. C., Liu K. D., Acute respiratory distress syndrome. N. Engl. J. Med. 377, 562–572 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Brooks L. R. K., Mias G. I., Streptococcus pneumoniae’s virulence and host immunity: Aging, diagnostics, and prevention. Front. Immunol. 9, 1366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiser J. N., Ferreira D. M., Paton J. C., Streptococcus pneumoniae: Transmission, colonization and invasion. Nat. Rev. Microbiol. 16, 355–367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthay M. A., Zemans R. L., Zimmerman G. A., Arabi Y. M., Beitler J. R., Mercat A., Herridge M., Randolph A. G., Calfee C. S., Acute respiratory distress syndrome. Nat. Rev. Dis. Primers 5, 18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torres A., Cilloniz C., Niederman M. S., Menendez R., Chalmers J. D., Wunderink R. G., van der Poll T., Pneumonia. Nat. Rev. Dis. Primers 7, 25 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Amit I., Winter D. R., Jung S., The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat. Immunol. 17, 18–25 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Caputa G., Castoldi A., Pearce E. J., Metabolic adaptations of tissue-resident immune cells. Nat. Immunol. 20, 793–801 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Kopf M., Schneider C., Nobs S. P., The development and function of lung-resident macrophages and dendritic cells. Nat. Immunol. 16, 36–44 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Lavin Y., Mortha A., Rahman A., Merad M., Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 15, 731–744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill L. A., Kishton R. J., Rathmell J., A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zago G., Saavedra P. H. V., Keshari K. R., Perry J. S. A., Immunometabolism of Tissue-resident macrophages - an appraisal of the current knowledge and cutting-edge methods and technologies. Front. Immunol. 12, 665782 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woods P. S., Kimmig L. M., Meliton A. Y., Sun K. A., Tian Y., O’Leary E. M., Gokalp G. A., Hamanaka R. B., Mutlu G. M., Tissue-resident alveolar macrophages do not rely on glycolysis for LPS-induced inflammation. Am. J. Respir. Cell Mol. Biol. 62, 243–255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Proenca R., Maffei M., Barone M., Leopold L., Friedman J. M., Positional cloning of the mouse obese gene and its human homologue. Nature 372, 425–432 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Zhu Q., Scherer P. E., Immunologic and endocrine functions of adipose tissue: Implications for kidney disease. Nat. Rev. Nephrol. 14, 105–120 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Friedman J. M., Leptin and the endocrine control of energy balance. Nat. Metab. 1, 754–764 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Pan W. W., Myers M. G. Jr., Leptin and the maintenance of elevated body weight. Nat. Rev. Neurosci. 19, 95–105 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Zeng W., Lu Y. H., Lee J., Friedman J. M., Reanalysis of parabiosis of obesity mutants in the age of leptin. Proc. Natl. Acad. Sci. U.S.A. 112, E3874–E3882 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng W., Pirzgalska R. M., Pereira M. M., Kubasova N., Barateiro A., Seixas E., Lu Y. H., Kozlova A., Voss H., Martins G. G., Friedman J. M., Domingos A. I., Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell 163, 84–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman J. M., Halaas J. L., Leptin and the regulation of body weight in mammals. Nature 395, 763–770 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Wauman J., Zabeau L., Tavernier J., The leptin receptor complex: Heavier than expected? Front. Endocrinol. (Lausanne) 8, 30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francisco V., Pino J., Campos-Cabaleiro V., Ruiz-Fernandez C., Mera A., Gonzalez-Gay M. A., Gomez R., Gualillo O., Obesity, fat mass and immune system: Role for leptin. Front. Physiol. 9, 640 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naylor C., Petri W. A. Jr., Leptin regulation of immune responses. Trends Mol. Med. 22, 88–98 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Kiernan K., MacIver N. J., The role of the adipokine leptin in immune cell function in health and disease. Front. Immunol. 11, 622468 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tartaglia L. A., Dembski M., Weng X., Deng N., Culpepper J., Devos R., Richards G. J., Campfield L. A., Clark F. T., Deeds J., Muir C., Sanker S., Moriarty A., Moore K. J., Smutko J. S., Mays G. G., Wool E. A., Monroe C. A., Tepper R. I., Identification and expression cloning of a leptin receptor, OB-R. Cell 83, 1263–1271 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Ghilardi N., Ziegler S., Wiestner A., Stoffel R., Heim M. H., Skoda R. C., Defective STAT signaling by the leptin receptor in diabetic mice. Proc. Natl. Acad. Sci. U.S.A. 93, 6231–6235 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C., An H., Hu J., Li J., Zhang W., Lan X., Deng H., Zhang J. R., MetR is a molecular adaptor for pneumococcal carriage in the healthy upper airway. Mol. Microbiol. 116, 438–458 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Dagvadorj J., Shimada K., Chen S., Jones H. D., Tumurkhuu G., Zhang W., Wawrowsky K. A., Crother T. R., Arditi M., Lipopolysaccharide induces alveolar macrophage necrosis via CD14 and the P2X7 receptor leading to interleukin-1α release. Immunity 42, 640–653 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon H., Shaw J. L., Haigis M. C., Greka A., Lipid metabolism in sickness and in health: Emerging regulators of lipotoxicity. Mol. Cell 81, 3708–3730 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz A. L. S., Barreto E. A., Fazolini N. P. B., Viola J. P. B., Bozza P. T., Lipid droplets: Platforms with multiple functions in cancer hallmarks. Cell Death Dis. 11, 105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez A., Hall M. N., Lin S. C., Hardie D. G., AMPK and TOR: The Yin and Yang of cellular nutrient sensing and growth control. Cell Metab. 31, 472–492 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Herzig S., Shaw R. J., AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 19, 121–135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinberg G. R., Carling D., AMP-activated protein kinase: The current landscape for drug development. Nat. Rev. Drug Discov. 18, 527–551 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Lee S. B., Kim J. J., Han S. A., Fan Y., Guo L. S., Aziz K., Nowsheen S., Kim S. S., Park S. Y., Luo Q., Chung J. O., Choi S. I., Aziz A., Yin P., Tong S. Y., Fiesel F. C., Springer W., Zhang J. S., Lou Z., The AMPK-Parkin axis negatively regulates necroptosis and tumorigenesis by inhibiting the necrosome. Nat. Cell Biol. 21, 940–951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies L. C., Jenkins S. J., Allen J. E., Taylor P. R., Tissue-resident macrophages. Nat. Immunol. 14, 986–995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maffei M., Halaas J., Ravussin E., Pratley R. E., Lee G. H., Zhang Y., Fei H., Kim S., Lallone R., Ranganathan S., Kern P. A., Friedman J. M., Leptin levels in human and rodent: Measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat. Med. 1, 1155–1161 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Ahren B., Mansson S., Gingerich R. L., Havel P. J., Regulation of plasma leptin in mice: Influence of age, high-fat diet, and fasting. Am. J. Physiol. 273, R113–R120 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Park S. H., Ryu S. Y., Yu W. J., Han Y. E., Ji Y. S., Oh K., Sohn J. W., Lim A., Jeon J. P., Lee H., Lee K. H., Lee S. H., Berggren P. O., Jeon J. H., Ho W. K., Leptin promotes KATP channel trafficking by AMPK signaling in pancreatic β-cells. Proc. Natl. Acad. Sci. U.S.A. 110, 12673–12678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butiaeva L. I., Slutzki T., Swick H. E., Bourguignon C., Robins S. C., Liu X., Storch K. F., Kokoeva M. V., Leptin receptor-expressing pericytes mediate access of hypothalamic feeding centers to circulating leptin. Cell Metab. 33, 1433–1448.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Bahrenberg G., Behrmann I., Barthel A., Hekerman P., Heinrich P. C., Joost H. G., Becker W., Identification of the critical sequence elements in the cytoplasmic domain of leptin receptor isoforms required for Janus kinase/signal transducer and activator of transcription activation by receptor heterodimers. Mol. Endocrinol. 16, 859–872 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Fei H., Okano H. J., Li C., Lee G. H., Zhao C., Darnell R., Friedman J. M., Anatomic localization of alternatively spliced leptin receptors (Ob-R) in mouse brain and other tissues. Proc. Natl. Acad. Sci. U.S. A. 94, 7001–7005 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z., Ceccarini G., Eisenstein M., Tan K., Friedman J. M., Phenotypic effects of an induced mutation of the ObRa isoform of the leptin receptor. Mol. Metab. 2, 364–375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung J., Zeng H., Horng T., Metabolism as a guiding force for immunity. Nat. Cell Biol. 21, 85–93 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Mangalmurti N., Hunter C. A., Cytokine storms: Understanding COVID-19. Immunity 53, 19–25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu H., Zhu J., Smith S., Foldi J., Zhao B., Chung A. Y., Outtz H., Kitajewski J., Shi C., Weber S., Saftig P., Li Y., Ozato K., Blobel C. P., Ivashkiv L. B., Hu X., Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat. Immunol. 13, 642–650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang L., Zhang X., Ji L., Kou T., Smith S. M., Zhao B., Guo X., Pineda-Torra I., Wu L., Hu X., The colonic macrophage transcription factor RBP-J orchestrates intestinal immunity against bacterial pathogens. J. Exp. Med. 217, e20190762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z., Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu L., Ma Y., Zhang J. R., Streptococcus pneumoniae recruits complement factor H through the amino terminus of CbpA. J. Biol. Chem. 281, 15464–15474 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Wang Y., Wen Z., Pan X., Briles D. E., He Y., Zhang J. R., Novel immunoprotective proteins of streptococcus pneumoniae identified by opsonophagocytosis killing screen. Infect. Immun. 86, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pang Z., Chong J., Zhou G., de Lima Morais D. A., Chang L., Barrette M., Gauthier C., Jacques P. E., Li S., Xia J., MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 49, W388–W396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S6
Table S1