Abstract
27-Hydroxycholesterol (27-HC) is the most abundant oxysterol that increases the risk of breast cancer progression. However, little is known about epigenetic regulation of 27-HC metabolism and its role in breast tumor initiation. Using genetic mouse mammary tumor and human breast cancer models, we showed here that the histone reader ZMYND8 was selectively expressed in breast cancer stem cells (BCSCs) and promoted epithelial-mesenchymal transition (EMT), BCSC maintenance and self-renewal, and oncogenic transformation through its epigenetic functions, leading to breast tumor initiation. Mechanistically, ZMYND8 was a master transcriptional regulator of 27-HC metabolism. It increased cholesterol biosynthesis and oxidation but blocked cholesterol efflux and 27-HC catabolism, leading to accumulation of 27-HC in BCSCs. Consequently, 27-HC promoted EMT, oncogenic transformation, and tumor initiation through activation of liver X receptor. These findings reveal that ZMYND8 is an epigenetic booster that drives breast tumor initiation through metabolic reprogramming.
An epigenetic mechanism of metabolic signaling identifies the cause of breast cancer.
INTRODUCTION
Breast cancer originates from a small population of tumor-initiating cells, also known as breast cancer stem cells (BCSCs), whose ability to drive breast tumorigenesis is controlled by the intrinsic factors (1). Cholesterol controls mammalian cell membrane integrity and lipid metabolism to support cell survival and proliferation (2). Intracellular cholesterol is synthesized through the de novo biosynthesis pathway and also supplemented from the diet and is further metabolized into oxysterols, of which 27-hydroxycholesterol (27-HC) is the most abundant in human blood (2). 27-HC is an endogenous selective estrogen receptor (ER) modulator and promotes ER-positive breast cancer progression (3–5). Recent studies have revealed elevated cholesterol biosynthesis in BCSCs (6, 7) and also link 27-HC with a breast cancer risk factor hypercholesterolemia (3). However, the significance of 27-HC in BCSCs and breast tumor initiation remains unknown.
Metabolic alterations are largely influenced by epigenetic regulators (8). While the interplay between epigenetics and metabolism is known to regulate tumor progression (8, 9), little is known about the role of epigenetic regulation of cellular metabolism in BCSC plasticity and breast tumorigenesis. ZMYND8 belongs to the bromodomain superfamily and functions as a histone reader recognizing acetyl lysine 16 of histone H4 (H4K16ac), acetyl lysine 14 of histone H3, monomethyl lysine 4 of histone H3, di- or trimethyl lysine 36 of histone H3, and histone H3.3G34R via a reader cassette containing plant homeodomain, bromodomain, and Pro-Trp-Trp-Pro domain at its N terminus (10, 11). Accumulating studies have shown that ZMYND8 is a dual regulator of gene activation and repression in cancer cells (12–15). ZMYND8 coactivates thousands of genes by interacting with hypoxia-inducible factor-1/2α, BRD4, and P-TEFb complex (12, 16), whereas it binds to gene repressors including KDM5C, NuRD complex, and EZH2 to repress gene transcription (10, 14, 15). In addition, ZMYND8 blocks the cGAS-STING–nuclear factor κB signaling pathway by maintaining genome stability, which leads to inhibition of interferon-β and interferon-stimulated genes in breast cancer cells (13). ZMYND8 is highly expressed in human breast tumors and promotes breast tumor progression in vitro and in xenograft mouse models (12, 13, 15). Its tumor growth function has been recently characterized in other human cancers, including acute myeloid leukemia, colorectal cancer, glioblastoma, clear cell renal cell carcinoma, bladder cancer, and liver cancer (11, 14, 17–20).
In the present study, we identified a previously unknown role of ZMYND8 in BCSC plasticity and tumor initiation. ZMYND8 was selectively expressed in BCSCs in vitro and in vivo. ZMYND8 increased 27-HC biosynthesis but blocked 27-HC catabolism, leading to 27-HC accumulation, which further activated liver X receptor (LXR) to induce epithelial-mesenchymal transition (EMT) and maintain survival and self-renewal of BCSCs, thereby promoting breast tumor initiation. These findings define a previously unappreciated signaling pathway from epigenetics to metabolism in BCSCs that control breast tumorigenesis.
RESULTS
ZMYND8 is required for mammary tumor initiation and progression in vivo
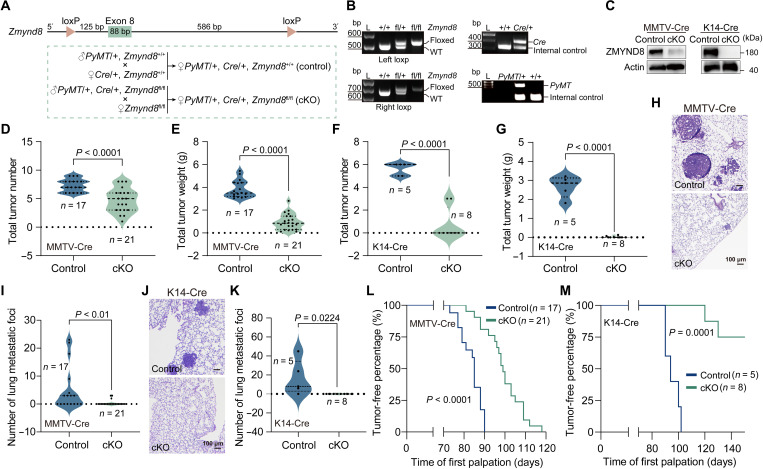
To determine the precise role of ZMYND8 in breast tumor development, we generated Zmynd8-floxed (Zmynd8fl/fl) mice by the CRISPR-Cas9 technique and crossed them with MMTV-PyMT and MMTV-Cre transgenic mice (Fig. 1, A and B). ZMYND8 protein was conditionally depleted in mammary glands isolated from MMTV-PyMT/MMTV-Cre/Zmynd8fl/fl (Zmynd8MMTV-cKO) mice (Fig. 1C, left). At postnatal day 145, we found that both mammary tumor number and weight were significantly reduced in Zmynd8MMTV-cKO mice compared with control mice MMTV-PyMT/MMTV-Cre/Zmynd8+/+ (Fig. 1, D and E). Much more robust inhibition of mammary tumor growth by ZMYND8 conditional knockout (cKO) was observed in another MMTV-PyMT/K14-Cre/Zmynd8fl/fl (Zmynd8K14-cKO) mouse model (Fig. 1, C, right, F and G). Spontaneous lung metastasis was significantly eliminated in Zmynd8MMTV-cKO and Zmynd8K14-cKO mice (Fig. 1, H to K). These results strongly support our previous findings in xenograft models that ZMYND8 promotes breast cancer progression in mice (12, 13). Notably, we found that mammary tumor initiation was significantly inhibited in both Zmynd8MMTV-cKO and Zmynd8K14-cKO mouse mammary tumor models as these mice experienced the longer tumor-free period than their respective control mice (Fig. 1, L and M). Collectively, the results from two genetically engineered mouse models indicate that ZMYND8 is required for mammary tumor initiation and progression in mice.
Fig. 1. Loss of Zmynd8 suppresses mouse mammary tumor initiation and progression.
(A) Scheme of Zmynd8 cKO strategy in mice. (B) Genotyping of loxP, MMTV-PyMT, MMTV-Cre, and K14-Cre. WT, wild type. L, ladder. (C) ZMYND8 protein knockout (KO) in MMTV-PyMT tumors. (D and E) Mammary tumor number (D) and weight (E) in control (MMTV-PyMT/+, MMTV-Cre/+, and Zmynd8+/+) and Zmynd8MMTV-cKO (MMTV-PyMT/+, MMTV-Cre/+, and Zmynd8fl/fl) mice. (F and G) Mammary tumor number (F) and weight (G) in control (MMTV-PyMT/+, K14-Cre/+, and Zmynd8+/+) and Zmynd8K14-cKO (MMTV-PyMT/+, K14-Cre/+, and Zmynd8fl/fl) mice. (H and I) Representative lung hematoxylin and eosin (H&E) images (H) and quantification of lung metastatic foci (I) in control and Zmynd8MMTV-cKO mice. (J and K) Representative lung H&E images (J) and quantification of lung metastatic foci (K) in control and Zmynd8K14-cKO mice. (L and M) Tumor-free period of control mice and Zmynd8MMTV-cKO (L) or Zmynd8K14-cKO (M) mice. Biological replicates (n) are shown on each figure panel. P value was determined by two-tailed Student’s t test (D to G, I, and K) and the log-rank (Mantel-Cox) test (L and M).
ZMYND8 induces EMT and oncogenic transformation
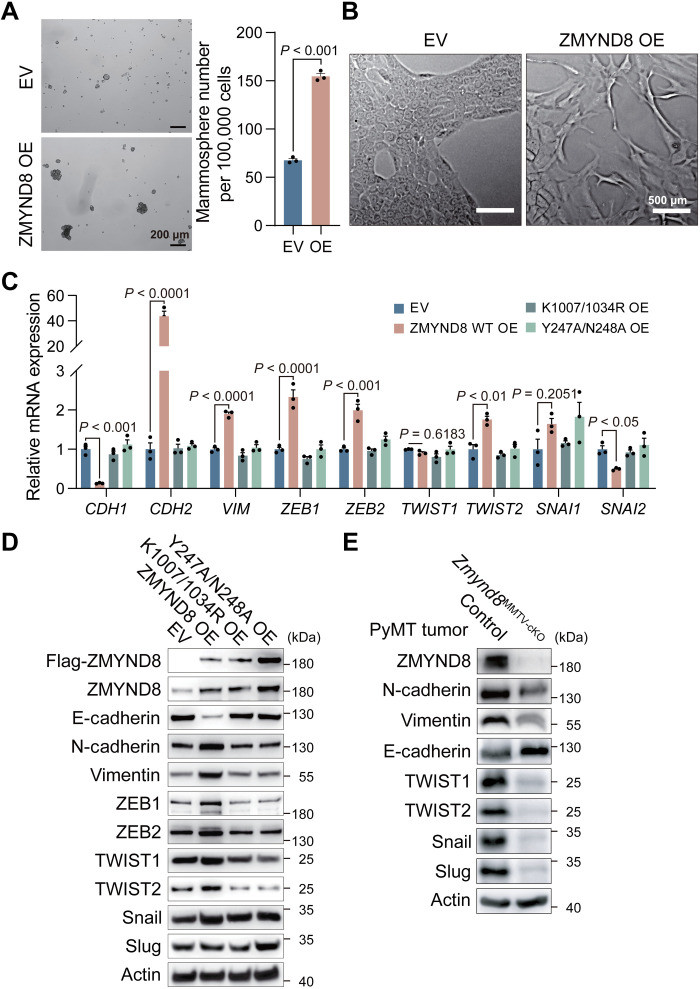
To determine the role of ZMYND8 in oncogenic transformation, we transduced nonmalignant normal mammary epithelial MCF-10A cells with lentivirus carrying ZMYND8 complementary DNA or empty vector (EV) and assessed the ability of ZMYND8 to facilitate MCF-10A cells to form spheres. As expected, few and tiny spheres were found in MCF-10A-EV cells (Fig. 2A). In contrast, overexpression (OE) of ZMYND8 induced more and bigger MCF-10A spheres (Fig. 2A), suggesting that ZMYND8 expression is sufficient to induce stemness and oncogenic transformation of normal breast epithelial cells in vitro. A subpopulation of MCF-10A monolayers exhibited a spindle-like cell morphology when ZMYND8 was overexpressed (Fig. 2B). Moreover, the epithelial marker E-cadherin (encoded by CDH1 gene) was repressed, but the mesenchymal markers N-cadherin and vimentin (encoded by CDH2 and VIM genes, respectively) as well as EMT transcription factors including ZEB1, ZEB2, and TWIST2 all were up-regulated in MCF-10A-ZMYND8 cells as shown by quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay (Fig. 2C), indicating that ZMYND8 drives the EMT phenotype. To determine the requirement of ZMYND8 epigenetic functions for these gene expression, we generated MCF-10A cells expressing ZMYND8 Y247A/N248A mutant [that prevents ZMYND8 binding to H4K16ac (21)] or ZMYND8 K1007/1034R mutant [that blocks BRD4 binding (12)]. Neither Y247A/N248A nor K1007/1034R mutant affected EMT markers in MCF-10A cells (Fig. 2C). ZMYND8-induced mRNA changes of EMT markers were confirmed at their protein levels in MCF-10A cells (Fig. 2D). In contrast, we found that ZMYND8 cKO robustly increased E-cadherin but decreased N-cadherin, vimentin, TWIST1, TWIST2, Snail, and Slug in Zmynd8MMTV-cKO tumors (Fig. 2E). Collectively, these findings indicate that ZMYND8 is necessary and sufficient to induce EMT and also promotes oncogenic transformation of normal mammary epithelial cells.
Fig. 2. ZMYND8 promotes EMT and oncogenic transformation.
(A) Representative of mammosphere images (left) and quantification (right; means ± SEM, n = 3 biological replicates) of mammospheres derived from MCF-10A cells expressing empty vector (EV) or ZMYND8. OE, overexpression. (B) Morphology of MCF-10A-EV and ZMYND8 OE cells (n = 3 biological replicates). (C and D) Analysis of mRNA (C) and protein (D) expression of EMT markers in MCF-10A cells expressing EV, WT ZMYND8, ZMYND8 K1007/1034R mutant, or ZMYND8 Y247A/N248A mutant (means ± SEM, n = 3 biological replicates). (E) Analysis of protein expression of EMT markers in control (MMTV-PyMT/+, MMTV-Cre/+, and Zmynd8+/+) and Zmynd8MMTV-cKO (MMTV-PyMT/+, MMTV-Cre/+, and Zmynd8fl/fl) tumors (n = 3 biological replicates). P value was determined by two-tailed Student’s t test (A) and one-way ANOVA corrected with Tukey’s test (C).
ZMYND8 increases BCSC activity through its epigenetic functions, leading to tumor initiation
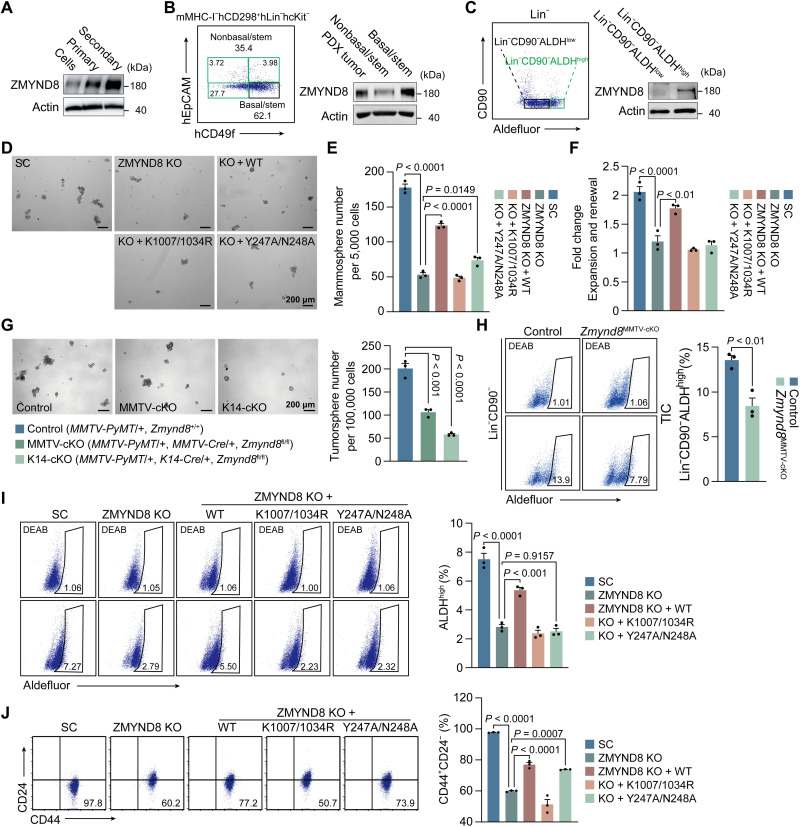
We next studied whether ZMYND8 regulates BCSC activity to promote breast tumor initiation. By comparing breast cancer cell monolayer cultures with BCSC-enriched mammosphere cultures, we found that ZMYND8 protein was up-regulated in primary MDA-MB-231 spheres and further increased in secondary MDA-MB-231 spheres (Fig. 3A). Similar results were observed in BCSC-enriched primary and secondary MCF-7 spheres (fig. S1A). To assess ZMYND8 expression in BCSCs in vivo, we generated a human triple-negative breast cancer patient-derived xenograft (PDX) model and isolated mMHC-I−hCD298+hLin−hcKit−hCD49fhighhEpCAM− BCSCs from PDX tumors by flow cytometry. These BCSCs highly expressed ZMYND8 protein compared with non-BCSCs (Fig. 3B). We also sorted out tumor-initiating cells (Lin−CD90−ALDHhigh) and non–tumor-initiating cells (Lin−CD90−ALDHlow) from MMTV-PyMT mammary tumors by flow cytometry and found that ZMYND8 was selectively expressed in tumor-initiating cells in vivo (Fig. 3C). Together, these in vitro and in vivo findings revealed that ZMYND8 is preferentially expressed in BCSCs.
Fig. 3. ZMYND8 is expressed in BCSCs and required for BCSC activity.
(A) ZMYND8 protein expression in MDA-MB-231 monolayers and mammospheres. Cells, monolayers. Primary, primary mammoshperes. Secondary, secondary mammospheres. n = 3. (B) Flow cytometry sorting of basal/stem cells (mMHC-I−hCD298+hLin−hcKit−hCD49fhighhEpCAM−) and nonbasal/stem cells (mMHC-I−hCD298+hLin−hcKit−hCD49flowhEpCAM− and mMHC-I−hCD298+hLin−hcKit−hEpCAM+) from a triple-negative breast cancer PDX tumor (left). m, mouse; h, human. Analysis of ZMYND8 protein expression in basal/stem cells compared with whole PDX tumor and nonbasal/stem cells (right). n = 3 biological replicates. (C) Flow cytometry sorting of tumor-initiating cells (Lin−CD90−ALDHhigh) and non–tumor-initiating cells (Lin−CD90−ALDHlow) from MMTV-PyMT tumors (left). Analysis of ZMYND8 protein expression in tumor-initiating cells and non–tumor-initiating cells (right). n = 3 biological replicates. (D to F) Mammosphere formation assay of MDA-MB-231-SC, ZMYND8 KO, ZMYND8 KO rescued with WT ZMYND8, K1007/1034R mutant, or Y247A/N248A mutant. Representative mammosphere images (D). Quantification of primary mammosphere numbers (E) and mammosphere expansion and self-renewal (F; means ± SEM, n = 3 biological replicates). (G) Tumorsphere formation assay of control, Zmynd8MMTV-cKO, and Zmynd8K14-cKO tumors. Representative tumorsphere images (left). Quantification of tumorsphere numbers (right; means ± SEM, n = 3 biological replicates). (H) Flow cytometry analysis (left) and quantification (right; means ± SEM, n = 3 biological replicates) of tumor-initiating cells (TIC) in control and Zmynd8MMTV-cKO tumors. DEAB, N,N-diethylaminobenzaldehyde. (I and J) Aldefluor (I) and CD44+CD24− (J) assays of MDA-MB-231-SC, ZMYND8 KO, ZMYND8 KO rescued with WT, K1007/1034R mutant, or Y247A/N248A mutant cells. Representative gates of cell populations (left) and quantification of ALDHhigh or CD44+CD24− cells (right; means ± SEM, n = 3 biological replicates). P value was determined by one-way ANOVA corrected with Tukey’s test (E, F, I, and J) and Dunnett’s test (G), and two-tailed Student’s t test (H).
To determine whether ZMYND8 regulates BCSC activity, we first performed mammosphere formation assay. Primary sphere formation of both MDA-MB-231 and MCF-7 cells was significantly inhibited by ZMYND8 KO (Fig. 3, D and E, and fig. S1, B and C). Reexpression of wild-type (WT) but not K1007/1034R ZMYND8 restored MDA-MB-231 sphere formation in vitro (Fig. 3, D and E). Y247A/N248A mutant marginally rescued MDA-MB-231 sphere formation in vitro (Fig. 3, D and E). The secondary mammosphere assay showed that ZMYND8 KO significantly reduced self-renewal of BCSCs in vitro, which was rescued by WT ZMYND8 but not K1007/1034R or Y247A/N248A mutant (Fig. 3F). Similar results were also found in MCF-7 spheres (fig. S1D). We further found that ZMYND8 cKO by either MMTV-Cre or K14-Cre reduced MMTV-PyMT tumorsphere formation ex vivo (Fig. 3G). Last, we studied the effect of ZMYND8 cKO on tumor-initiating cells in mammary tumors by flow cytometry and found that ZMYND8 cKO significantly decreased Lin−CD90−ALDHhigh tumor-initiating cells in Zmynd8MMTV-cKO tumors in vivo (Fig. 3H). These results indicate that ZMYND8 is required for maintenance and self-renewal of BCSCs in vitro and in vivo.
Both aldehyde dehydrogenase (ALDH) and CD44+CD24− are classic BCSC markers, which represent the proliferative epithelial-like and the invasive mesenchymal-like BCSCs, respectively (22). Flow cytometry analysis showed that ZMYND8 KO significantly decreased the percentage of ALDHhigh or CD44+CD24− BCSCs in both MDA-MB-231 and MCF-7 cells (Fig. 3, I and J, and fig. S1, E and F). WT but not K1007/1034R or Y247A/N248A ZMYND8 rescued ALDHhigh BCSCs in MDA-MB-231 cells (Fig. 3I). However, CD44+CD24− BCSCs were restored by WT and Y247A/N248A but not K1007/1034R ZMYND8 in MDA-MB-231 cells (Fig. 3J). Our findings indicate that ZMYND8 controls the mesenchymal-like BCSC subpopulation through BRD4 and the epithelial-like BCSC subpopulation through both BRD4 and H4K16ac.
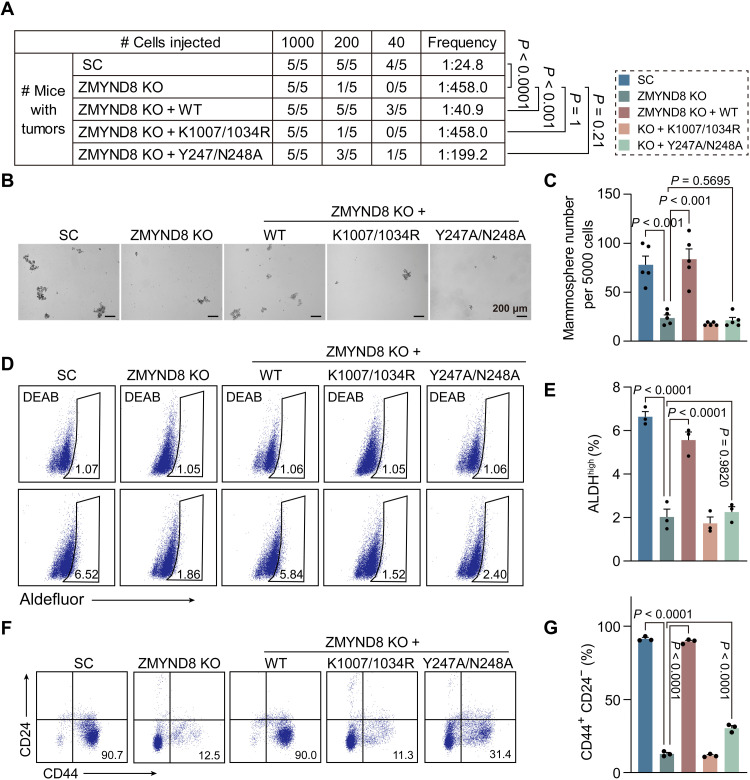
To determine whether ZMYND8 regulates BCSC activity to promote tumor initiation in vivo, we performed limiting dilution assay in NOD-scid IL2Rgammanull (NSG) mice. Scrambled control (SC), ZMYND8 KO, and ZMYND8 rescue MDA-MB-231 cells with three serial dilutions were orthotopically implanted into the mammary fat pad of female NSG mice, respectively. ZMYND8 KO significantly decreased the tumor incidence in mice, which was rescued by WT but not K1007/1034R or Y247A/N248A ZMYND8 (Fig. 4A). In line with our in vitro results (Fig. 3, D to F, I, and J), ZMYND8 KO significantly inhibited MDA-MB-231 tumorsphere formation ex vivo and decreased the number of ALDHhigh and CD44+CD24− BCSC subpopulations in MDA-MB-231 tumors in vivo (Fig. 4, B to G). Expression of WT ZMYND8 but not K1007/1034R mutant in KO tumors rescued BCSC activity ex vivo (Fig. 4, B to G). Y247A/N248A mutant had a marginal rescued effect on CD44+CD24− subpopulation but not ALDHhigh BCSCs and tumorsphere formation (Fig. 4, B to G). Likewise, ZMYND8 KO significantly inhibited MCF-7 BCSC activity in vivo and tumor initiation in mice (fig. S2, A to C). Collectively, these findings indicate that ZMYND8 increases BCSC activity through its epigenetic functions to promote tumor initiation.
Fig. 4. ZMYND8 promotes breast tumor-initiating potential through its epigenetic functions.
(A) Limiting dilution assay of MDA-MB-231-SC, ZMYND8 KO, ZMYND8 KO rescued with WT, K1007/1034R mutant, or Y247A/N248A mutant cells in NSG mice. (B and C) Tumorsphere formation assay in MDA-MB-231-SC, ZMYND8 KO, ZMYND8 KO + WT, ZMYND8 KO + K1007/1034R, and ZMYND8 KO + Y247A/N248A tumors. Representative tumorsphere images (B). Quantification of tumorsphere numbers (C; means ± SEM, n = 5 biological replicates). (D and E) Aldefluor assay in MDA-MB-231-SC, ZMYND8 KO, ZMYND8 KO + WT, ZMYND8 KO + K1007/1034R, and ZMYND8 KO + Y247A/N248A tumors. Representative gates of cell populations (D). Quantification of ALDHhigh cells (E; means ± SEM, n = 3 biological replicates). (F and G) CD44+CD24− assay in MDA-MB-231-SC, ZMYND8 KO, ZMYND8 KO + WT, ZMYND8 KO + K1007/1034R, and ZMYND8 KO + Y247A/N248A tumors. Representative gates of cell populations (F). Quantification of CD44+CD24− cells (G; means ± SEM, n = 3 biological replicates). P value was determined by chi-square test (A) and one-way ANOVA corrected with Tukey’s test (C, E, and G).
ZMYND8 controls 27-HC metabolism in BCSCs
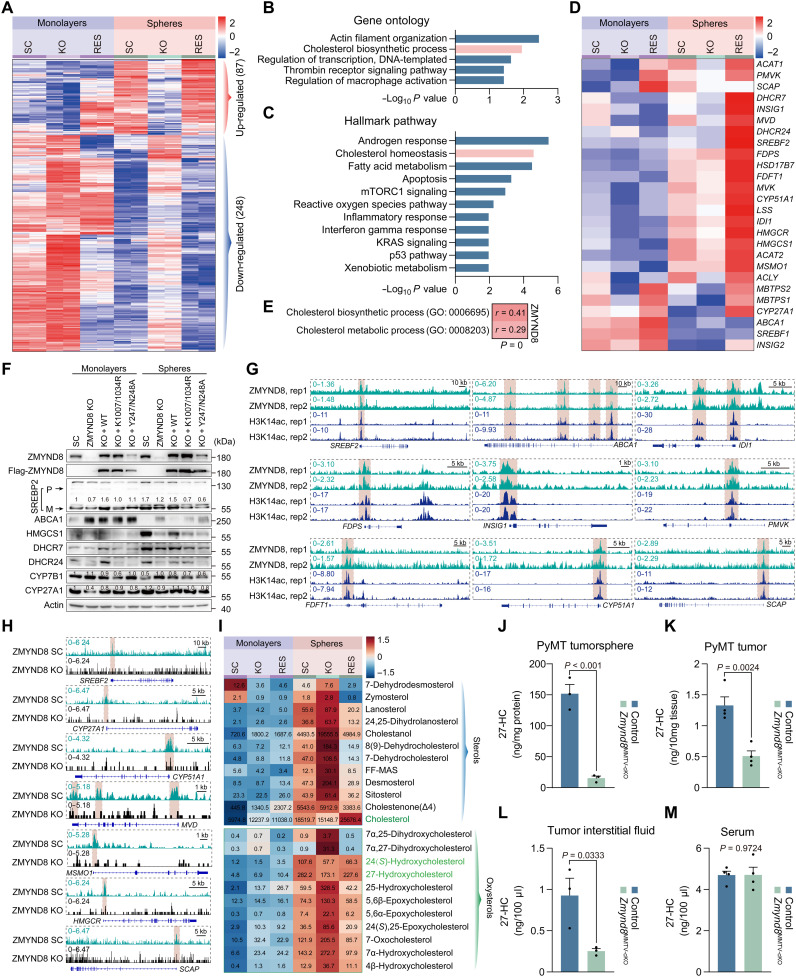
To define the molecular mechanism underlying ZMYND8-induced BCSC activity and tumor initiation, we compared the whole transcriptome by RNA sequencing (RNA-seq) in SC, ZMYND8 KO, and ZMYND8 rescue MDA-MB-231 cells under monolayer and BCSC-enriched sphere culture conditions. Eighty-seven genes were up-regulated, while 248 genes were down-regulated in MDA-MB-231 spheres in a ZMYND8-dependent manner (false discovery rate < 0.05, logCPM > 0, fold change > 1.5; Fig. 5A). Analyses of both gene ontology and hallmark pathway revealed that genes in the cholesterol biosynthetic pathways were significantly enriched in the up-regulation group (Fig. 5, B and C). To validate RNA-seq results, we performed qRT-PCR in the same experimental settings, and mRNA expression of almost all cholesterol biosynthesis genes, including ACLY, ACAT1, ACAT2, HMGCS1, HMGCR, MVK, PMVK, MVD, IDI1, FDFT1, LSS, CYP51A1, HSD17B7, DHCR7, and DHCR24, was up-regulated in MDA-MB-231-SC mammospheres compared with its monolayers, which was blocked by ZMYND8 KO (Fig. 5D). Down-regulation of cholesterol biosynthesis genes in KO spheres was fully reversed by ZMYND8 reexpression (Fig. 5D), suggesting the specific induction of cholesterol biosynthesis genes by ZMYND8. ZMYND8 also enhanced the mRNA expression of the regulatory machinery of cholesterol biosynthesis including SREBF2, MBTPS1, MBTPS2, and INSIG1 in MDA-MB-231 spheres (Fig. 5D). Most of these cholesterol regulating genes were similarly up-regulated in MCF-7 spheres but repressed by ZMYND8 KO1 or KO2 (fig. S3A). In contrast, the cholesterol efflux transporters ABCA1 and ABCG1 were down-regulated in MDA-MB-231 and MCF-7 spheres, respectively, which was reversed by ZMYND8 KO (Fig. 5D and fig. S3B). Again, reexpression of ZMYND8 restored ABCA1 down-regulation in MDA-MB-231-ZMYND8 KO spheres (Fig. 5D). ZMYND8 mRNA was significantly correlated with the cholesterol biosynthesis/metabolism gene signatures in human breast tumors from The Cancer Genome Atlas (TCGA) cohort (Fig. 5E), which further supports a role of ZMYND8 in the expression of cholesterol-regulating genes in breast tumors in vivo. In line with mRNA regulation, the protein levels of SREBP2, HMGCS1, DHCR7, and DHCR24 were reduced in MDA-MB-231-ZMYND8 KO spheres, which were restored by WT ZMYND8 but not K1007/1034R or Y247A/N248A mutant (Fig. 5F). CYP27A1, a metabolic enzyme that oxidizes cholesterol into 27-HC, was also reduced in MDA-MB-231-ZMYND8 KO spheres (Fig. 5F). In contrast, ZMYND8 inhibited the expression of ABCA1 and CYP7B1 proteins in MDA-MB-231 spheres (Fig. 5F). We further found that both ZMYND8 and CYP27A1 were up-regulated in human breast tumors, whereas CYP7B1 was down-regulated in human breast tumors (fig. S3, C to E). Collectively, these gene expression results indicate that ZMYND8 induces enzymes and transcriptional regulators of cholesterol biosynthesis and 27-HC production but represses cholesterol efflux transporters ABCA1 and ABCG1 and a 27-HC downstream catabolic enzyme CYP7B1 in BCSC-enriched mammospheres.
Fig. 5. ZMYND8 induces 27-HC accumulation in BCSCs.
(A) Heatmap of dysregulated genes identified with RNA-seq in MDA-MB-231-SC, ZMYND8 KO, and ZMYND8 rescue (RES) cells (monolayers) and their mammospheres (spheres). n = 2 biological replicates. (B and C) Gene ontology (B) and hallmark pathway enrichment (C) analyses of 87 ZMYND8–up-regulated genes. (D) RT-qPCR analysis of mRNA expression of cholesterol metabolic genes in MDA-MB-231-SC, ZMYND8 KO, and ZMYND8 rescue cells and their mammospheres. n = 3 biological replicates. (E) Correlation analysis between ZMYND8 mRNA expression and cholesterol biosynthesis/metabolism gene signatures in human breast tumors from TCGA cohort. (F) Immunoblot analysis of proteins involved in cholesterol/27-HC metabolism in MDA-MB-231-SC, ZMYND8 KO, and ZMYND8 rescue cells and their mammospheres. n = 3 biological replicates. P, precursor form. M, mature form. (G) Enrichment of ZMYND8 and H3K14ac (highlighted in badshahi brown) on 27-HC metabolic genes in MDA-MB-231 monolayers. (H) Enrichment of ZMYND8 on 27-HC metabolic genes in parental and ZMYND8 KO MDA-MB-231 mammospheres. (I) Mass spectrometry analysis of sterols and oxysterols in MDA-MB-231-SC, ZMYND8 KO, and ZMYND8 rescue cells and their mammospheres (mean, n = 4). (J to M) Mass spectrometry analysis of 27-HC in control (MMTV-PyMT/+ and Zmynd8+/+) and Zmynd8MMTV-cKO (MMTV-PyMT/+, MMTV-Cre/+, and Zmynd8fl/fl) tumorspheres (J; means ± SEM, n = 3 biological replicates), PyMT tumors (K; means ± SEM, n = 4), tumor interstitial fluid (L; means ± SEM, n = 3), and serum (M; means ± SEM, n = 4 biological replicates). P value was determined by two-tailed Student’s t test.
Next, we studied whether ZMYND8 directly controls the expression of cholesterol-regulating genes. Chromatin immunoprecipitation sequencing (ChIP-seq) identified the binding of ZMYND8 on SREBF2, ABCA1, INSIG1, SCAP, MVD, IDI1, FDPS, FDFT1, CYP51A1, and PMVK genes, which was well correlated with H3K14ac enrichment in MDA-MB-231 cells (Fig. 5G and fig. S3F). In contrast, no robust ZMYND8 binding was detected on other cholesterol-regulating genes (fig. S3F). We further confirmed ZMYND8 binding to these cholesterol regulating genes in MDA-MB-231-SC spheres but not ZMYND8 KO spheres (Fig. 5H and fig. S3G), suggesting the specificity of our assay. Notably, ZMYND8 binding to CYP27A1, HMGCR, ACLY, ACAT1, and ACAT2 became substantial in MDA-MB-231 spheres (Fig. 5H and fig. S3G). SREBP2 is known to bind and activate cholesterol biosynthesis genes (23). Coimmunoprecipitation (IP) assay showed that Flag-ZMYND8 did not physically interact with endogenous mature SREBP2 in MDA-MB-231 monolayers or mammospheres (fig. S3H), excluding a role of direct SREBP2 coactivation by ZMYND8 in cholesterol-regulating gene expression. Collectively, our results indicate that ZMYND8 controls these cholesterol-regulating genes via its direct binding and indirectly through induction of SREBP2 expression.
Last, we measured cholesterol, sterols, and oxysterols by mass spectrometry to further determine whether ZMYND8 regulates cholesterol biosynthesis and oxidation in BCSCs. Cholesterol and most of its upstream and downstream metabolites sterols and oxysterols were robustly elevated in MDA-MB-231-SC and MCF-7 spheres compared with their respective monolayers, albeit their levels were relatively low in MCF-7 cells (Fig. 5I and fig. S3I). ZMYND8 KO significantly decreased levels of total cholesterol, 27-HC, and 24(S)-hydroxycholesterol [24(S)-HC], which were reversed by reexpression of ZMYND8 in MDA-MB-231 spheres (Fig. 5I). Neither K1007/1034R nor Y247A/N248A mutant had a rescued effect on 27-HC levels in MDA-MB-231 spheres (fig. S3J). We also detected marked reduction of 27-HC in Zmynd8MMTV-cKO tumorspheres ex vivo (Fig. 5J). Likewise, ZMYND8 cKO significantly reduced the intracellular and tumor interstitial levels of 27-HC in MMTV-PyMT tumors (Fig. 5, K and L). 27-HC dysregulation by ZMYND8 cKO occurred locally within the tumor as circulating 27-HC levels in blood serum were not altered by ZMYND8 cKO in tumor-bearing mice (Fig. 5M). In contrast, ZMYND8 OE increased 27-HC in MCF-7 spheres and most of the sterols and oxysterols in MCF-7 monolayers (fig. S3I), although ZMYND8 KO had minimal effects on levels of cholesterol, sterols, and oxysterols in MDA-MB-231 monolayers (Fig. 5I). In line with altered CYP7B1 (Fig. 5F), 27-HC downstream metabolite 7α,27-dihydroxycholesterol was robustly increased in MDA-MB-231-ZMYND8 KO spheres but decreased in MCF-7-ZMYND8 OE spheres as compared with their respective control spheres (Fig. 5I and fig. S3I). Other sterols and oxysterols were regulated by ZMYND8 in a cell type–specific manner (Fig. 5I and fig. S3I), which requires future investigations to understand the underlying mechanism. By integrating the expression of cholesterol-regulating genes and cholesterol metabolic profiling, we conclude that ZMYND8 enhances cholesterol biosynthesis and oxidation but inhibits cholesterol efflux and 27-HC catabolism, leading to accumulation of 27-HC in BCSC-enriched mammospheres.
27-HC accumulation by ZMYND8 promotes BCSC activity and tumor initiation through activation of LXR
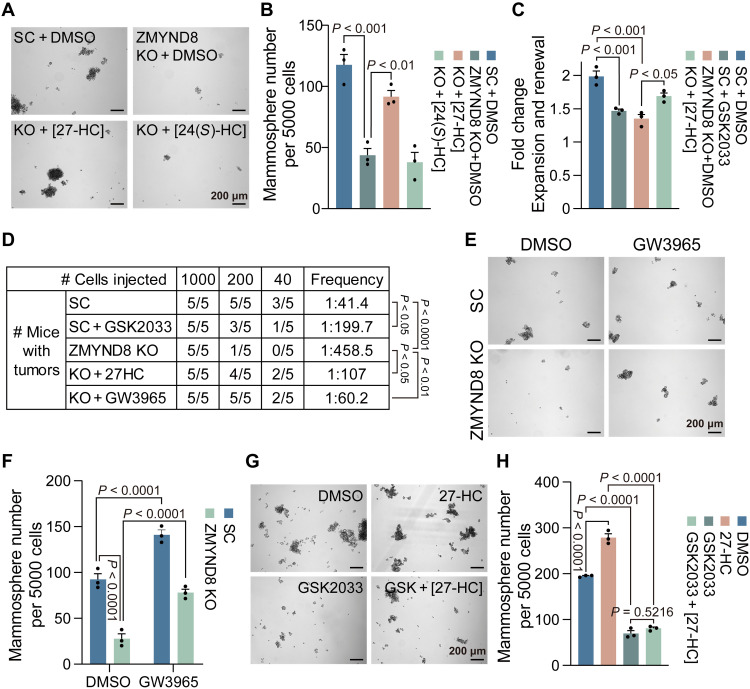
To determine the role of cholesterol metabolism in ZMYND8-induced BCSC activity, we generated MDA-MB-231 cells expressing Flag-tagged ZMYND8 or EV by lentiviral transduction (fig. S4A) and assessed whether blockade of cholesterol metabolism by simvastatin prevents ZMYND8-induced mammosphere formation. ZMYND8 OE robustly increased MDA-MB-231 mammosphere formation, which was blocked by the treatment of 10 μM simvastatin (fig. S4B). We next treated MDA-MB-231-SC and MDA-MB-231-ZMYND8 KO spheres with water-soluble cholesterol (5 μg/ml) to determine whether cholesterol can rescue mammosphere formation. Unfortunately, cholesterol treatment failed to influence formation of both MDA-MB-231-SC and MDA-MB-231-ZMYND8 KO spheres (fig. S4C), indicating that there may be downstream rate-limiting enzymes. Likewise, 24(S)-HC (0.5 μM) had no effect on ZMYND8 KO sphere formation (Fig. 6, A and B, and fig. S4, D and E). However, the treatment of 27-HC at the physiological concentration of 0.5 μM robustly rescued maintenance and self-renewal of MDA-MB-231-ZMYND8 KO spheres (Fig. 6, A to C). We also observed a similar rescued effect of 27-HC on formation of MCF-7-ZMYND8 KO spheres (fig. S4, D and E). In line with these in vitro results (Fig. 6, A to C), 27-HC administration (40 mg/kg per day) significantly prevented inhibition of tumor initiation conferred by ZMYND8 KO in mice (Fig. 6D). These results indicate that ZMYND8 induces BCSC activity and tumor initiation through 27-HC.
Fig. 6. 27-HC–LXR axis mediates ZMYND8-induced breast tumor initiation.
(A and B) Mammosphere formation assay of MDA-MB-231-SC and ZMYND8 KO cells treated with DMSO, 27-HC, or 24(S)-HC. Representative mammosphere images (A). Quantification of mammosphere numbers (B; means ± SEM, n = 3 biological replicates). (C) Quantification of mammosphere expansion and self-renewal of MDA-MB-231-SC cells treated with DMSO or GSK2033 and ZMYND8 KO cells treated with DMSO or 27-HC (means ± SEM, n = 3 biological replicates). (D) Limiting dilution assay of MDA-MB-231-SC cells treated with DMSO or GSK2033 and ZMYND8 KO cells treated with DMSO, 27-HC, or GW3965 in NSG mice. (E and F) Mammosphere formation assay of MDA-MB-231-SC and ZMYND8 KO cells treated with DMSO or GW3965. Representative mammosphere images (E). Quantification of mammosphere numbers (F; means ± SEM, n = 3 biological replicates). (G and H) Mammosphere formation assay of MDA-MB-231 cells treated with DMSO, GSK2033, 27-HC, or both. Representative mammosphere images (G). Quantification of mammosphere numbers (H; means ± SEM, n = 3 biological replicates). P value was determined by two-way ANOVA corrected with Tukey’s test (B, C, and F), chi-square test (D), or one-way ANOVA corrected with Tukey’s test (H).
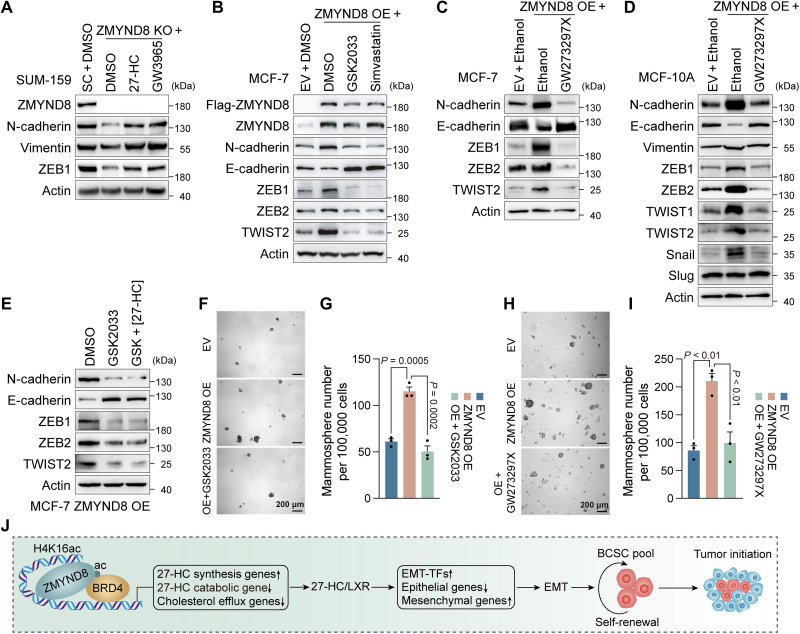
27-HC is a known agonist of the nuclear receptor LXR (24). To determine whether 27-HC activates LXR to mediate ZMYND8-induced BCSC activity and tumor initiation, we treated MDA-MB-231-SC and MDA-MB-231-ZMYND8 KO spheres with a synthetic LXR agonist GW3965 (1 μM). Activation of LXR by GW3965 significantly increased formation of MDA-MB-231-SC spheres and rescued ZMYND8 KO spheres in vitro (Fig. 6, E and F), which phenocopied 27-HC (Fig. 6A). A similar rescued effect of GW3965 was also observed in MCF-7-ZMYND8 KO spheres (fig. S4, D and E). Administration of GW3965 robustly restored ZMYND8 KO tumor initiation in mice (Fig. 6D). In contrast, a selective LXR antagonist GSK2033 (1 μM) inhibited MDA-MB-231-SC sphere formation and expansion in vitro and tumor initiation in mice (Fig. 6, C, D, G, and H). GSK2033 treatment also abolished the 27-HC effect on MDA-MB-231 sphere formation (Fig. 6, G and H). Next, we assessed whether 27-HC and LXR are responsible for ZMYND8-mediated EMT. The levels of endogenous 27-HC in MCF-7 and MDA-MB-231 cells were well correlated with their respective epithelial and mesenchymal states (Fig. 5I and fig. S3I). ZMYND8 KO inhibited N-cadherin, vimentin, and ZEB1 in mesenchymal SUM-159 cells, which were fully restored by the treatment of 27-HC or GW3965 (Fig. 7A). In contrast, ZMYND8 OE inhibited E-cadherin but increased N-cadherin, ZEB1, ZEB2, and TWIST2 in epithelial MCF-7 cells, which was reversed by the treatment of GSK2033 (Fig. 7B). Treatment of CYP27A1 inhibitor GW273297X (5 nM) similarly reversed ZMYND8-induced expression of EMT markers in MCF-7 and MCF-10A cells (Fig. 7, C and D). 27-HC treatment failed to antagonize the GSK2033 effect on EMT in MCF-7-ZMYND8 OE cells (Fig. 7E). Likewise, simvastatin treatment also abolished ZMYND8-mediated EMT in MCF-7 cells (Fig. 7B). Last, we found that the treatment of either GSK2033 or GW273297X blocked ZMYND8-induced stemness and oncogenic transformation of normal mammary epithelial MCF-10A cells (Fig. 7, F to I). Together, these results indicate that 27-HC stimulates LXR activation to mediate ZMYND8-induced EMT, oncogenic transformation, and maintenance and self-renewal of BCSCs, leading to tumor initiation.
Fig. 7. 27-HC–LXR axis mediates ZMYND8-induced EMT and oncogenic transformation.
(A to E) Immunoblot analysis of EMT markers in SUM-159-SC and ZMYND8 KO cells treated for 48 hours with DMSO, 27-HC, or GW3965 (A); MCF-7-EV and ZMYND8 OE cells treated for 48 hours with DMSO, GSK2033, or simvastatin (B); MCF-7-EV and ZMYND8 OE cells treated for 48 hours with ethanol or GW273297X (C); MCF-10A-EV and ZMYND8 OE cells treated for 48 hours with ethanol or GW273297X (D); and MCF-7 ZMYND8 OE cells treated for 48 hours with DMSO, GSK2033, or GSK2033 + 27-HC (E). n = 3 biological replicates. (F to I) Mammosphere formation assay of MCF-10A-EV and ZMYND8 OE cells treated with vehicle (DMSO or ethanol), GSK2033 (F and G), or GW273297X (H and I). Representative mammosphere images (G and H). Quantification of mammosphere numbers (G and I; means ± SEM, n = 3 biological replicates). (J) A proposed model of ZMYND8-induced breast cancer stemness and tumor initiation via activation of 27-HC–LXR axis. TFs, transcription factors. P value was determined by two-way ANOVA corrected with Tukey’s test (G and I).
DISCUSSION
Recent studies from mouse xenograft models showed the inconsistent oncogenic and tumor suppressor roles of ZMYND8 (12, 14, 15, 25, 26). In the present study, we used the genetically modified mouse models to confirm the oncogenic role of ZMYND8 in breast cancer progression and to further characterize an unforeseen function of ZMYND8 in BCSC plasticity and tumor initiation (Fig. 7J), which greatly expands our understanding of ZMYND8 pathobiology in cancer. We showed that ZMYND8 induces EMT to promote stemness and oncogenic transformation of normal mammary epithelial cells. Epigenetic priming is a critical step for the maintenance of tumor-initiating cells and boosts tumorigenesis caused by genetic alterations (27). Our findings identified ZMYND8 as an important epigenetic booster for oncogenic transformation. ZMYND8 is selectively expressed in BCSCs and required for maintenance and self-renewal of BCSCs, leading to breast tumor initiation. EMT is well known to induce stemness in development and cancer (28). ZMYND8 induces EMT through metabolic regulation of 27-HC production and subsequent LXR activation. Previous studies reported that 27-HC is a selective ER modulator in ER-positive breast cancer cells and a selective LXR modulator in myeloid immune cells (4, 29). Our studies from both ER-positive and triple-negative breast cancer cells revealed the ER-independent but LXR-dependent function of 27-HC in EMT, breast cancer stemness, and tumor initiation. Collectively, the ZMYND8–27-HC–LXR axis represents a previously unappreciated signaling pathway from epigenetics to metabolism contributing to breast tumor initiation (Fig. 7J).
Our present studies identified that ZMYND8 is a master transcriptional regulator of 27-HC metabolism in BCSCs, which provides new evidence of ZMYND8’s dual gene regulation functions in metabolism and cancer development. While ZMYND8 does not directly coactivate SREBP2, a master transcription factor that regulates cholesterol biosynthesis (23), it induces the expression of SREBP2 and other genes involved in 27-HC metabolism but inhibits cholesterol efflux transporters and 27-HC catabolic enzyme in BCSCs through BRD4. Thus, both gene coactivator and corepressor functions of ZMYND8 are coordinated to trigger multiple layers of direct and indirect mechanisms underlying 27-HC accumulation in BCSCs. We and others have previously demonstrated a critical role of ZMYND8 in gene activation and repression in non-BCSCs (12–15). The distinct gene signatures controlled by ZMYND8 in non-BCSCs and BCSCs indicate that ZMYND8-dependent gene regulation is cell state dependent.
Previous studies primarily focused on cholesterol biosynthesis in CSCs (6, 17). Our studies here reveal that altered cholesterol metabolism in BCSCs also attributes to decreased cholesterol efflux, in addition to increased cholesterol biosynthesis. Moreover, BCSCs exhibit enhanced cholesterol oxidation but decreased 27-HC catabolism. As a result, 27-HC levels are accumulated in BCSCs. We found that 27-HC but not cholesterol per se is a key factor that contributes to maintenance and self-renewal of BCSCs and tumor initiation. 27-HC has also been shown to play critical roles in myeloid cell biology and subsequent effects on cancer progression (30–32). Whether ZMYND8 is involved in this cell type will be important to explore. These findings provide new insights into the deep mechanism of aberrant cholesterol metabolism–caused breast cancer plasticity. We found that ZMYND8 is a crucial regulator of reprogramming of 27-HC metabolism in BCSCs through controlling enzymes and transporters in the 27-HC metabolic pathways. Early clinical studies showed the benefits of statin in reducing breast cancer recurrence and mortality (33). However, a window-of-opportunity trial showed increased 3-hydroxy-3-methylglutaryl–coenzyme A reductase expression in breast tumors from patients treated with atorvastatin (34), suggesting that these tumors had evolved to make their own local cholesterol. Because statins do not have a posthepatic circulation of very high concentrations, it is unlikely that they have systemic effects on tissues/tumors, making their own cholesterol/27-HC. In addition to de novo biosynthesis, intracellular cholesterol is also supplemented from the diet, particularly in patients with obesity (35). Therefore, blocking de novo cholesterol biosynthesis alone is unlikely sufficient to reduce 27-HC in the cell. Together with recent studies about 27-HC on breast tumor growth and metastasis (3, 5, 30), our findings provide the strong rationale for 27-HC–lowering drugs such as ZMYND8 inhibitor in the prevention and treatment of breast cancer.
In conclusion, ZMYND8 is a master epigenetic regulator of 27-HC metabolism in BCSCs, which increases BCSC plasticity to promote tumor initiation and progression (Fig. 7J). Thus, ZMYND8 is not only an important therapeutic target for the treatment of breast cancer but also a valuable biomarker that can predict breast cancer risk in patients.
MATERIALS AND METHODS
Animal studies
Animal experiments were approved by the Animal Care and Use Committee at UT Southwestern Medical Center. Zmynd8fl/fl mice were generated by Transgenic Core at UT Southwestern Medical Center. Fertilized C57BL/6N oocytes were microinjected with cocktail containing Cas9 protein, Zmynd8 crRNAs were annealed with tracrRNAs, and single-stranded Zmynd8 donor oligonucleotides were inserted with loxP sequences (Fig. 1A and table S1) and implanted into pseudopregnant female Institute of Cancer Research (ICR) mice. F0 founders were crossed with C57BL/6J (the Jackson Laboratory, stock no. 000664) to generate Zmynd8fl/fl mice. Zmynd8fl/fl mice were genotyped and confirmed by Sanger sequencing. To study the roles of ZMYND8 in mammary tumor initiation and progression, Zmynd8fl/fl mice were crossed with MMTV-PyMT mice [B6.FVB-Tg(MMTV-PyVT)634Mul/LellJ; the Jackson Laboratory, stock no. 022974] and MMTV-Cre mice [Tg(MMTV-cre)4Man/J; the Jackson Laboratory, stock no. 003553] or K14-Cre mice [a gift from L. Le laboratory; Tg(KRT14-cre)1Amc/J mice from the Jackson Laboratory, stock no. 004782; backcrossed with C57BL/6J for years]. Tumor initiation time was determined with palpation and measurement (diameter ≥ 2 mm). All tumors were harvested, counted, and weighted at postnatal day 145. Lungs were perfused with phosphate-buffered saline (PBS), inflated with 0.5% agarose, fixed in formalin, embedded in paraffin, and analyzed with hematoxylin and eosin staining.
For limiting dilution assay, different numbers of cells suspended in 100 μl of PBS/Matrigel (1:1, Corning) were implanted into the second left mammary fat pad of female NSG mice (6 to 8 weeks old; the Jackson Laboratory). Mice were euthanized to confirm tumor initiation at day 45 or 50 after cell injection. A slow-release 17β-estradiol pellet (0.72 mg/60-day release/pellet, Innovative Research of America) was implanted subcutaneously into mice the day before MCF-7 cell injection. Mice were administrated for 45 days subcutaneously with 27-HC (40 mg/kg per day; MedChemExpress) in 30% 2-hydroxypropyl-β-cyclodextrin (Sigma-Aldrich), intraperitoneally with GSK2033 (30 mg/kg per day; MedChemExpress) in 10% dimethyl sulfoxide (DMSO)/90% corn oil (Sigma-Aldrich) or with GW3965 (30 mg/liter; MedChemExpress) in drinking water.
For the PDX model, a triple-negative breast tumor collected from a patient during surgical resection (UT Southwestern Medical Center, demographics were not collected) was cut into 8-mm3 fragments and implanted into the right fourth mammary fat pad of female NSG mice (6 to 8 weeks old; the Jackson Laboratory). Tumors were harvested when the tumor volume reached 500 mm3. The study was approved by the Institutional Review Board at UT Southwestern Medical Center with informed consent.
Co-IP and immunoblot assays
Homogenized tissues or cells were lysed in NETN lysis buffer [150 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl (pH 8.0), 0.5% Igepal, and protease inhibitor cocktail] for 30 min on ice and followed by sonication for 15 s. The supernatant was collected for IP or immunoblot after centrifugation. Protein concentration was determined with the Bradford assay (Bio-Rad). For co-IP, the supernatant was incubated with Flag (Sigma-Aldrich) or immunoglobulin G antibody overnight in the presence of protein A/G magnetic beads (Bio-Rad) at 4°C. Next day, after washing four times with NETN lysis buffer, the bound proteins were boiled in 1× Laemmli buffer for 5 min and fractionated by SDS–polyacrylamide gel electrophoresis, followed by immunoblot assay using the antibodies listed in table S2.
Cell culture and lentivirus
MDA-MB-231 (a gift from R. Brekken, UT Southwestern, Dallas, TX, USA), SUM-159 (a gift from G. L. Semenza, Johns Hopkins School of Medicine, Baltimore, MD, USA), MCF-7 (American Type Culture Collection), and human embryonic kidney (HEK) 293FT (Invitrogen) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) or DMEM/Ham’s F-12 medium (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich) at 37°C in a 5% CO2/95% air incubator. MCF-10A cells (a gift from G. L. Semenza, Johns Hopkins School of Medicine, Baltimore, MD, USA) were cultured in DMEM/Ham’s F-12 medium with 5% horse serum (Sigma-Aldrich), epidermal growth factor (EGF) (20 ng/ml; Sigma-Aldrich), hydrocortisone (0.5 μg/ml; Sigma-Aldrich), cholera toxin (100 ng/ml; Sigma-Aldrich), and insulin (10 μg/ml; Sigma-Aldrich) at 37°C in a 5% CO2/95% air incubator. CRISPR-Cas9 KO cell lines have been described previously (12). Lentivirus was generated in HEK293FT cells as described previously (12). All cell lines were mycoplasma free and authenticated by short tandem repeat DNA profiling analysis.
Plasmid constructs
Y247A/N248A mutant was generated by the site-directed mutagenesis PCR and cloned into pcFUGW-3XFLAG vector. Other plasmids have been described previously (12). All plasmids were confirmed by nucleotide sequence analysis.
RT-qPCR assay
Total RNA was extracted from cultured cells with TRIzol reagent (Thermo Fisher Scientific), treated with deoxyribonuclease (DNase) I (Ambion), and reverse transcribed using the iScript Reverse Transcription Supermix (Bio-Rad). qPCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) with primers listed in table S3. The mRNA fold change was calculated on the basis of the threshold cycle (Ct) as 2−Δ(ΔCt), where ΔCt = Cttarget − Ct18S rRNA and Δ(ΔCt) = ΔCtZMYND8 KO, WT, or mutant − ΔCtEV or SC.
Flow cytometry
Human PDX tumors, mouse MMTV-PyMT tumors, or MDA-MB-231 and MCF-7 xenograft tumors were crosscut into small pieces in Hanks’ balanced salt solution (HBSS; Sigma-Aldrich), washed, and digested for 45 min with collagenase/hyaluronidase/DNase (Sigma-Aldrich) in a 37°C shaker. Cultured cells were trypsinized and dissociated into single-cell suspensions. Cells were stained for 30 min with fluorescent-conjugated antibodies listed in table S2 on ice, followed by washing with HBSS and centrifugation at 500g for 5 min. Cells were examined on a FACSCanto RUO Cell Analyzer (Becton Dickinson) or a CytoFLEX Flow Cytometer (Beckman Coulter) or sorted on a FACSAria II SORP Cell Sorter (Becton Dickinson). Aldefluor assay was performed with an ALDEFLUOR kit according to the manufacturer’s protocol (STEMCELL Technologies).
Mammosphere formation and self-renewal assays
MMTV-PyMT tumors were harvested, crosscut into small pieces in HBSS, washed, and digested for 45 min with gentle collagenase/hyaluronidase (STEMCELL Technologies) in a 37°C shaker. After filtering with a 40-μm cell strainer (Thermo Fisher Scientific) and centrifugation, single cell was resuspended in DMEM/Ham’s F-12 medium with B-27 Supplement (Thermo Fisher Scientific), EGF (20 ng/ml; Sigma-Aldrich), basic fibroblast growth factor (20 ng/ml; ProteinTech), heparin (4 μg/ml; STEMCELL Technologies), and 1% penicillin/streptomycin/neomycin (Sigma-Aldrich) and plated overnight on a collagen I–coated plate (Thermo Fisher Scientific) at 37°C in a 5% CO2/95% air incubator. Next day, cells were trypsinized and reseeded on an ultralow attachment six-well plate or dish (Corning) at 37°C in a 5% CO2/95% air incubator for continuous incubation for 7 days. Mammospheres were imaged and harvested for mRNA and protein analysis or 27-HC measurement.
MDA-MB-231, MCF-7, or MCF-10A cells were trypsinized to single-cell suspensions; washed with serum containing medium and then with HBSS; resuspended in MammoCult Medium (STEMCELL Technologies) with or without DMSO (Sigma-Aldrich), 10 μM simvastatin (Sigma-Aldrich), water-soluble cholesterol (5 μg/ml; Sigma-Aldrich), 0.5 μM 27-HC (Sigma-Aldrich), 0.5 μM 24(S)-HC (Sigma-Aldrich), 1 μM GW3965 (MedChemExpress), 1 μM GSK2033 (MedChemExpress), or 5 nM GW273297X (Sai Life); and seeded for 7 days on an ultralow attachment six-well plate at 37°C in a 5% CO2/95% air incubator. Mammospheres were imaged under a Zeiss Axio Observer Z1 microscope.
MDA-MB-231 and MCF-7 xenograft tumors were dissociated into single-cell suspensions as described in the flow cytometry method. Cells were seeded for 7 days on an ultralow attachment six-well plate at 37°C in a 5% CO2/95% air incubator. Mammospheres were imaged under a Zeiss Axio Observer Z1 microscope.
For self-renewal assay, all primary mammospheres were harvested and trypsinized to single-cell suspensions. The resulting cell suspensions were seeded on an ultralow attachment six-well plates for secondary mammosphere culture. The fold change of expansion and self-renewal was calculated as following: (secondary − primary mammosphere number)/primary mammosphere number.
RNA-seq assay
Total RNA was isolated using an RNeasy mini kit (QIAGEN) and treated with DNase (QIAGEN). DNA-free RNA was used for library preparation with the KAPA mRNA HyperPrep Kit (Roche) and sequenced on the Illumina NextSeq 500 with the read configuration as 76–base pair (bp) single end. Bioinformatics were performed as described previously (12).
ChIP-seq assay
Cells were cross-linked with 1% formaldehyde and quenched in 125 mM glycine. Chromatin was isolated using a SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology), sonicated to 200 to 300 bp in length, and subjected to IP with ZMYND8 (Bethyl Laboratories) or H3K14ac (Abcam) antibody (table S2). ChIP DNA libraries were prepared with the NEBNext Ultra II DNA Library Prep Kit for Illumina and sequenced on the Illumina NextSeq 2 K. Bioinformatics were performed as described previously (12).
Sterols and oxysterols measurement
Monolayers were collected with a cell scraper and washed with PBS. Mammospheres were centrifugated at 500g for 5 min followed by PBS washing. Whole blood was collected from ZMYND8 WT or cKO PyMT tumor-bearing mice (5 months old) into a Vacutainer blood collection tube (Becton Dickinson) and left undisturbed at room temperature until its clotting. After removing the clot by centrifuging at 1000g to 2000g for 10 min at 4°C, serum was transferred into a clean polypropylene tube and stored at −80°C immediately. To isolate tumor interstitial fluid, fresh ZMYND8 WT and cKO PyMT tumors were wrapped with nylon mesh with pores of ∼15 × 20–μm, placed at the bottom of the centrifuge tube, and centrifuged at 500g for 10 min at 4°C. The isolated tumor interstitial fluid was collected and transferred into a clean tube on ice. Tumors were repeatedly centrifuged until enough fluids were collected. After centrifugation, tumor interstitial fluid was stored at −80°C immediately. Sterols and oxysterols were extracted from cell pellets, serum, tumor interstitial fluid, and tumors and determined by high-performance liquid chromatography (HPLC)–mass spectrometry (MS)–atmospheric pressure chemical ionization (AB Sciex 4000 QTrap) and HPLC-MS–electrospray ionization (AB Sciex API-5000), respectively (36). Sterols and oxysterols were quantitated by isotope dilution and normalized to protein concentration.
TCGA breast cancer data analysis
RNA-seq data from TCGA human breast cancer cohort were downloaded from the UCSC Cancer Browser (https://genome-cancer.ucsc.edu). ZMYND8, CYP27A1, and CYP7B1 mRNA expression was queried in adjacent normal breast tissues and breast tumors.
Statistical analysis
Statistical analysis was performed by two-tailed Student’s t test between two groups, and one-way or two-way analysis of variance (ANOVA) with multiple testing correction within multiple groups. ELDA software was used to calculate BCSC frequency and statistical significance (http://bioinf.wehi.edu.au/software/elda/) (37). Tumor initiation assay was analyzed with log-rank (Mantel-Cox) test. Experiments except for RNA-seq and ChIP-seq that were the duplicates were repeated at least three times and expressed as means ± SEM. P < 0.05 is considered significant.
Acknowledgments
We thank D. W. Russell and D. J. Mangelsdorf for discussion and L. Le for K14-Cre mice. We also thank the UTSW Transgenic Core for generation of Zmynd8-floxed mice and the UTSW Cancer Center Tissue Resource for assistance in immunohistochemistry, which was supported by NCI Cancer Center Grant P30CA142543. W.L. is a CPRIT Scholar in Cancer Research.
Funding: American Cancer Society–Lisa Dean Moseley Foundation RSG-19-229-01-DMC (W.L.); Cancer Prevention and Research Institute of Texas grants RR140036 and RP190358 (W.L.); National Institutes of Health grants R01CA222393 (W.L.), R35GM124693 and R01AG066166 (Y.W.), P01HL020948 (J.G.M.), and R01CA234025 (E.R.N.); Mary Kay Foundation 08-19 (W.L.); and Department of Defense Breast Cancer Research Program Era of Hope Scholar Award W81XWH-20-BCRP-EOHS/BC200206 (E.R.N.).
Author contributions: Conceptualization: W.L. and Yingfei Wang. Methodology: M.L., L.B., J.G.M., C.X., Yingfei Wang, and W.L. Investigation: M.L., L.B., Y.C., Y.X., Yong Wang, B.Z., C.W., C.D.C., J.G.M., A.K., C.X., and J.E.W. Visualization: M.L., Yingfei Wang, and W.L. Resources: Y.F. and E.R.N. Funding acquisition: W.L., Yingfei Wang, E.R.N., and J.G.M. Project administration: W.L. and Yingfei Wang. Supervision: W.L. and Yingfei Wang. Writing—original draft: W.L., Yingfei Wang, and M.L. Writing—review and editing: M.L., L.B., Y.C., Y.X., Yong Wang, B.Z., C.W., C.D.C., J.G.M., A.K., C.X., Y.F., E.R.N., J.E.W., Yingfei Wang, and W.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The RNA-seq and ChIP-seq data were deposited at the GEO database with accession numbers GSE186543 and GSE203054. The Zmynd8-floxed mice and TNBC PDX tumor can be provided by W.L.’s pending scientific review and a completed material transfer agreement. Requests for the Zmynd8-floxed mice or TNBC PDX tumor should be submitted to W.L. (Weibo.Luo@UTSouthwestern.edu).
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Tables S1 to S3
REFERENCES AND NOTES
- 1.Polyak K., Breast cancer: Origins and evolution. J. Clin. Invest. 117, 3155–3163 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang B., Song B. L., Xu C., Cholesterol metabolism in cancer: Mechanisms and therapeutic opportunities. Nat. Metab. 2, 132–141 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Nelson E. R., Wardell S. E., Jasper J. S., Park S., Suchindran S., Howe M. K., Carver N. J., Pillai R. V., Sullivan P. M., Sondhi V., Umetani M., Geradts J., McDonnell D. P., 27–Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science 342, 1094–1098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DuSell C. D., Umetani M., Shaul P. W., Mangelsdorf D. J., McDonnell D. P., 27-Hydroxycholesterol is an endogenous selective estrogen receptor modulator. Mol. Endocrinol. 22, 65–77 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Q., Ishikawa T., Sirianni R., Tang H., McDonald J. G., Yuhanna I. S., Thompson B., Girard L., Mineo C., Brekken R. A., Umetani M., Euhus D. M., Xie Y., Shaul P. W., 27–Hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep. 5, 637–645 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehmsen S., Pedersen M. H., Wang G., Terp M. G., Arslanagic A., Hood B. L., Conrads T. P., Leth-Larsen R., Ditzel H. J., Increased cholesterol biosynthesis is a key characteristic of breast cancer stem cells influencing patient outcome. Cell Rep. 27, 3927–3938.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Ginestier C., Monville F., Wicinski J., Cabaud O., Cervera N., Josselin E., Finetti P., Guille A., Larderet G., Viens P., Sebti S., Bertucci F., Birnbaum D., Charafe-Jauffret E., Mevalonate metabolism regulates basal breast cancer stem cells and is a potential therapeutic target. Stem Cells 30, 1327–1337 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Grunt T. W., Interacting cancer machineries: Cell signaling, lipid metabolism, and epigenetics. Trends Endocrinol. Metab. 29, 86–98 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Jiang P., Du W., Mancuso A., Wellen K. E., Yang X., Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 493, 689–693 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong F., Chiu L. Y., Cox B., Aymard F., Clouaire T., Leung J. W., Cammarata M., Perez M., Agarwal P., Brodbelt J. S., Legube G., Miller K. M., Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 29, 197–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiao F., Li Z., He C., Xu W., Yang G., Liu T., Shen H., Cai J., Anastas J. N., Mao Y., Yu Y., Lan F., Shi Y. G., Jones C., Xu Y., Baker S. J., Shi Y., Guo R., RACK7 recognizes H3.3G34R mutation to suppress expression of MHC class II complex components and their delivery pathway in pediatric glioblastoma. Sci. Adv. 6, eaba2113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y., Zhang B., Bao L., Jin L., Yang M., Peng Y., Kumar A., Wang J. E., Wang C., Zou X., Xing C., Wang Y., Luo W., ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J. Clin. Invest. 128, 1937–1955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Luo M., Chen Y., Wang Y., Zhang B., Ren Z., Bao L., Wang Y., Wang J. E., Fu Y. X., Luo W., Wang Y., ZMYND8 expression in breast cancer cells blocks T-lymphocyte surveillance to promote tumor growth. Cancer Res. 81, 174–186 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang B., Sun R., Wang D., Sheng H., Wei T., Wang L., Zhang J., Ho T. H., Yang L., Wei Q., Huang H., ZMYND8 preferentially binds phosphorylated EZH2 to promote a PRC2-dependent to -independent function switch in hypoxia-inducible factor-activated cancer. Proc. Natl. Acad. Sci. U.S.A. 118, e2019052118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen H. F., Zhang W. J., Huang Y., He Y. H., Hu G. S., Wang L., Peng B. L., Yi J., Li T. T., Rong R., Chen X. Y., Liu J. Y., Li W. J., Ohgi K., Li S. W., Rosenfeld M. G., Liu W., The dual function of KDM5C in both gene transcriptional activation and repression promotes breast cancer cell growth and tumorigenesis. Adv. Sci. (Weinh) 8, 2004635 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh K., Tang M., Kumari N., Nandy A., Basu S., Mall D. P., Rai K., Biswas D., Positive regulation of transcription by human ZMYND8 through its association with P-TEFb complex. Cell Rep. 24, 2141–2154.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Q., Zhong S., Wang H., Wang X., Li N., Li Y., Zhang G., Yuan H., Lian Y., Chen Q., Han Y., Guo J., Liu Q., Qiu T., Jiang J., Li Q., Tan M., Yin H., Peng J., Xiao Y., Qin J., The ZMYND8-regulated mevalonate pathway endows YAP-high intestinal cancer with metabolic vulnerability. Mol. Cell 81, 2736–2751.e8 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Dou C., Mo H., Chen T., Liu J., Zeng Y., Li S., Guo C., Zhang C., ZMYND8 promotes the growth and metastasis of hepatocellular carcinoma by promoting HK2-mediated glycolysis. Pathol. Res. Pract. 219, 153345 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Qiu F., Jin Y., Pu J., Huang Y., Hou J., Zhao X., Lu Y., Aberrant FBXW7-mediated ubiquitination and degradation of ZMYND8 enhances tumor progression and stemness in bladder cancer. Exp. Cell Res. 407, 112807 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Cao Z., Budinich K. A., Huang H., Ren D., Lu B., Zhang Z., Chen Q., Zhou Y., Huang Y. H., Alikarami F., Kingsley M. C., Lenard A. K., Wakabayashi A., Khandros E., Bailis W., Qi J., Carroll M. P., Blobel G. A., Faryabi R. B., Bernt K. M., Berger S. L., Shi J., ZMYND8-regulated IRF8 transcription axis is an acute myeloid leukemia dependency. Mol. Cell 81, 3604–3622.e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adhikary S., Sanyal S., Basu M., Sengupta I., Sen S., Srivastava D. K., Roy S., Das C., Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of all-trans-retinoic acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J. Biol. Chem. 291, 2664–2681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colacino J. A., Azizi E., Brooks M. D., Harouaka R., Fouladdel S., McDermott S. P., Lee M., Hill D., Madden J., Boerner J., Cote M. L., Sartor M. A., Rozek L. S., Wicha M. S., Heterogeneity of human breast stem and progenitor cells as revealed by transcriptional profiling. Stem. Cell Rep. 10, 1596–1609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hua X., Yokoyama C., Wu J., Briggs M. R., Brown M. S., Goldstein J. L., Wang X., SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. U.S. A. 90, 11603–11607 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu X., Menke J. G., Chen Y., Zhou G., MacNaul K. L., Wright S. D., Sparrow C. P., Lund E. G., 27-hydroxycholesterol is an endogenous ligand for liver X receptor in cholesterol-loaded cells. J. Biol. Chem. 276, 38378–38387 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Shen H., Xu W., Guo R., Rong B., Gu L., Wang Z., He C., Zheng L., Hu X., Hu Z., Shao Z. M., Yang P., Wu F., Shi Y. G., Shi Y., Lan F., Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell 165, 331–342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N., Li Y., Lv J., Zheng X., Wen H., Shen H., Zhu G., Chen T. Y., Dhar S. S., Kan P. Y., Wang Z., Shiekhattar R., Shi X., Lan F., Chen K., Li W., Li H., Lee M. G., ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol. Cell 63, 470–484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vicente-Duenas C., Hauer J., Cobaleda C., Borkhardt A., Sanchez-Garcia I., Epigenetic priming in cancer initiation. Trends Cancer 4, 408–417 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Wilson M. M., Weinberg R. A., Lees J. A., Guen V. J., Emerging mechanisms by which EMT programs control stemness. Trends Cancer 6, 775–780 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Ma L., Vidana Gamage H. E., Tiwari S., Han C., Henn M. A., Krawczynska N., Dibaeinia P., Koelwyn G. J., Das Gupta A., Bautista Rivas R. O., Wright C. L., Xu F., Moore K. J., Sinha S., Nelson E. R., The liver x receptor is selectively modulated to differentially alter female mammary metastasis-associated myeloid cells. Endocrinology, bqac072 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baek A. E., Yu Y. A., He S., Wardell S. E., Chang C. Y., Kwon S., Pillai R. V., McDowell H. B., Thompson J. W., Dubois L. G., Sullivan P. M., Kemper J. K., Gunn M. D., McDonnell D. P., Nelson E. R., The cholesterol metabolite 27 hydroxycholesterol facilitates breast cancer metastasis through its actions on immune cells. Nat. Commun. 8, 864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He S., Ma L., Baek A. E., Vardanyan A., Vembar V., Chen J. J., Nelson A. T., Burdette J. E., Nelson E. R., Host CYP27A1 expression is essential for ovarian cancer progression. Endocr. Relat. Cancer 26, 659–675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma L., Wang L., Nelson A. T., Han C., He S., Henn M. A., Menon K., Chen J. J., Baek A. E., Vardanyan A., Shahoei S. H., Park S., Shapiro D. J., Nanjappa S. G., Nelson E. R., 27-Hydroxycholesterol acts on myeloid immune cells to induce T cell dysfunction, promoting breast cancer progression. Cancer Lett. 493, 266–283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cauley J. A., Zmuda J. M., Lui L. Y., Hillier T. A., Ness R. B., Stone K. L., Cummings S. R., Bauer D. C., Lipid-lowering drug use and breast cancer in older women: A prospective study. J Womens Health (Larchmt) 12, 749–756 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Bjarnadottir O., Romero Q., Bendahl P. O., Jirstrom K., Ryden L., Loman N., Uhlen M., Johannesson H., Rose C., Grabau D., Borgquist S., Targeting HMG-CoA reductase with statins in a window-of-opportunity breast cancer trial. Breast Cancer Res. Treat. 138, 499–508 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Mc Auley M. T., Effects of obesity on cholesterol metabolism and its implications for healthy ageing. Nutr. Res. Rev. 33, 121–133 (2020). [DOI] [PubMed] [Google Scholar]
- 36.McDonald J. G., Smith D. D., Stiles A. R., Russell D. W., A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J. Lipid Res. 53, 1399–1409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y., Smyth G. K., ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1 to S3