Abstract
Pluripotent cells are a transient population of the mammalian embryo dependent on transcription factors, such as OCT4 and NANOG, which maintain pluripotency while suppressing lineage specification. However, these factors are also expressed during early phases of differentiation, and their role in the transition from pluripotency to lineage specification is largely unknown. We found that pluripotency factors play a dual role in regulating key lineage specifiers, initially repressing their expression and later being required for their proper activation. We show that Oct4 is necessary for activation of HoxB genes during differentiation of embryonic stem cells and in the embryo. In addition, we show that the HoxB cluster is coordinately regulated by OCT4 binding sites located at the 3′ end of the cluster. Our results show that core pluripotency factors are not limited to maintaining the precommitted epiblast but are also necessary for the proper deployment of subsequent developmental programs.
The role of OCT4 extends beyond pluripotency to regulate in a dual fashion lineage determinants such as the Hox genes.
INTRODUCTION
Pluripotency, the ability of a cell to give rise to derivatives of all embryonic germ layers, occurs in cultured embryonic stem (ES) cells and for a brief period during development of the mammalian embryo. A small group of transcription factors, octamer-binding transcription factor 4 (OCT4), NANOG, and SOX2, controls this state both in vivo and in culture by regulating a large battery of downstream target genes (1). During preimplantation stages of mammalian embryos, these factors are expressed in the epiblast of the blastocyst, which shares various molecular features with ES cells, among them the expression of the core pluripotency factors. Progression from pluripotency toward differentiation requires the dismantling of the pluripotency regulatory network, leading to the expression of lineage determination genes and turning on of specific developmental pathways. However, the expression of pluripotency factors beyond the blastocyst stage suggests roles not directly related to pluripotency maintenance (2). Oct4 (official gene symbol Pou5f1) is continuously expressed up to embryonic day (E) 8.5, initially throughout the epiblast and subsequently showing progressive restriction to the posterior part of the embryo (3). Nanog is reexpressed at E5.5, but only in the posterior-proximal region, where it controls development of the primordial germ cells (4) and is turned off by E7.5 (5). Some studies have suggested roles of these factors beyond pluripotency. For example, Oct4 has been found to promote mesoendodermal differentiation (6, 7) and cardiomyocyte fate (8), while Nanog has been proved to regulate primitive hematopoiesis (9). Nevertheless, loss-of-function approaches to investigating the role of Oct4 and Nanog at these stages have proved difficult because preimplantation lethality precludes analysis of later phenotypes (1). To overcome early lethality, conditional Oct4 mutants have been analyzed at early postimplantation stages. However, loss of Oct4 at these stages leads to tissue disorganization and proliferation defects at gastrulation, which could obscure potential lineage-specific defects (10, 11). Therefore, we still lack a complete understanding of the roles of pluripotency factors during later stages of development, as well as how pluripotency and differentiation programs are coordinated in the embryo.
In this work, we aimed to understand the role of Oct4 and Nanog beyond pluripotency. We have characterized the transcriptional changes caused by gain of function of these factors and determined that they regulate many developmental regulators in a dual fashion, repressing their expression at E7.5 and activating them at E9.5. Among them, we have used as a paradigm the regulation of Hox genes by Oct4. Hox genes are a large conserved family of transcription factors that specify cellular identities along the anterior-posterior axis throughout metazoan evolution (12). They are organized in clusters along the chromosome that determine their temporal and spatial activity (13). By using an inducible Oct4 loss-of-function model, we have determined that it is required for proper activation of the HoxB cluster. We have identified functional OCT4 binding sites in the regulatory regions of the cluster and demonstrated that these regions are essential for proper HoxB gene expression.
RESULTS
Stage-dependent regulation of developmental genes by pluripotency factors
We used doxycycline (dox)–inducible transgenic mouse models providing controlled Oct4 or Nanog expression in postimplantation embryos (14, 15). These mice carry two independent alleles: on the one hand, an insertion of the cDNA for Oct4 or Nanog driven by an rtTA (see below) responsive promoter at the permissive Col1a1 locus and, on the other, the transcriptional transactivator rtTA, which is only active if bound by dox, inserted at the Rosa26 locus. Adding dox to the drinking water of pregnant mice will result in the activation of the transgenes in the embryos in a temporal controlled manner. We chose two different time windows for induction of Oct4 and Nanog: from E4.5 to E7.5 and from E6.5 to E9.5 (Fig. 1A), thus maintaining expression beyond the point when endogenous gene activity is turned off. Robust expression of both transgenes was obtained at E7.5 and E9.5 (fig. S1A), with higher levels in the neural tube and the mesoderm (fig. S1B). Expression levels of Oct4 or Nanog in treated embryos were comparable or even lower than endogenous levels in E14 or R1 (16) ES cells (fig. S1C). We analyzed the transcriptomes of embryos by RNA sequencing (RNA-seq) from untreated and dox-treated females and compared gene expression changes between stages and models (data file S1).
Fig. 1. Oct4 and Nanog coordinate developmental programs in the postgastrulation mouse embryo.
(A) Diagram showing the windows of dox treatment used to induce Oct4 or Nanog expression at postimplantation stages of mouse development. (B) Gene expression changes driven by expression of Oct4 or Nanog for 3 days up to E7.5 or E9.5; unsupervised hierarchical clustering of genes differentially expressed in at least one condition segregated genes into 10 groups according to behavior (clusters #1 to #10). (C) Enrichment in genes located in the vicinity of chromatin immunoprecipitation sequencing (ChIP-seq)–defined OCT4 or NANOG binding peaks, shown as the log10 ratio of each cluster versus genes differentially expressed in at least one of the conditions analyzed. Two-tailed Student’s t test; adj *P < 0.01. (D) Representative examples of developmental genes showing opposite behaviors in response to Oct4 and Nanog; genes are either early epiblast markers that are up-regulated early and down-regulated late (clusters #6 and #10; right) or later lineage specifiers that are down-regulated early and up-regulated late (cluster #8; left). Expression differences between dox-treated and untreated embryos are shown as logFC (fold change) of counts per million from RNA sequencing (RNA-seq) data.
More than 50% of genes differentially expressed upon Oct4 expression up to E7.5 also changed when Nanog was induced in the same time window (fig. S1D) and included major developmental regulators. However, this proportion halved when we compared changes occurring in E9.5 embryos (24%). Similarly, 23% of genes deregulated by Oct4 at E7.5 also changed at E9.5. As for Nanog, 36% of genes changing at E7.5 were shared with Oct4, 16% at E9.5, and only 14% were common at both stages in Nanog-expressing embryos (fig. S1D). Core pluripotency factors activate each other’s expression (1, 17), and we observed positive cross-regulation of Oct4 and Nanog at E7.5 (data file S1), but not at E9.5 (Data file S1; fig. S1A). Furthermore, we did not observe up-regulation of other pluripotency factors, such as Sox2, upon Oct4 or Nanog expression. Therefore, there is not an overall activation of the embryonic pluripotency program in the gastrulating embryo driven by these factors.
We performed unsupervised hierarchical clustering of the data using genes that were differentially expressed in at least one condition (4090 genes; data file S2). Most of the resulting clusters (Fig. 1B) show a stronger tendency for up-regulation or down-regulation in only one condition (e.g., clusters #1 to #5; fig. S2A), confirming the largely independent and stage-specific effects of Oct4 and Nanog expression. Functional annotation of Gene Ontology terms showed that most clusters were enriched for genes involved in development and transcription, with some exceptions such as cluster #5, which is enriched for cell cycle genes, and cluster #7, which includes genes involved in lipid metabolism (fig. S2B and data file S3).
As the majority of changes in gene expression we observed are surely not caused by direct regulation by OCT4 or NANOG, but secondary to other genes or also to changes to cell fate, we aimed to gain insight into putative direct targets among the deregulated genes. We checked for the presence of OCT4 or NANOG binding in the vicinity of differentially expressed genes by examining published chromatin immunoprecipitation sequencing (ChIP-seq) data in ES cells (18, 19). This is not ideal, but as we lack genome-wide binding information for OCT4 and NANOG in early gastrulating mouse embryos, we reasoned that the similarity of ES cells and early embryonic tissues would suffice for an exploratory analysis. We assessed for binding near genes from each cluster compared to genes differentially expressed in at least one condition (Fig. 1C). Unexpectedly, only three clusters (#6, #8, and #10) showed consistent enrichment for genes located near OCT4- or NANOG-bound genomic regions (Fig. 1C). Moreover, these clusters showed an intriguing pattern in response to Oct4 and Nanog: clusters #6 and #10 were up-regulated by both Oct4 and Nanog at E7.5 and down-regulated at E9.5, whereas cluster #8 showed the opposite behavior (fig. S2A). The three clusters are enriched for developmental regulators (fig. S2B and data file S3); however, whereas clusters #6 and #10 include early epiblast genes (e.g., Chordin, Dnmt3a/3b, Eomes, Eras, Esrrb, Etv5, Fgf4/5, Foxa2, Gsc, Lefty2, Mesp1/2, Nodal, Otx2, Snai1, Tdgf1, and Zic5), cluster #8 includes multiple lineage specification genes (e.g., Cdx1/2/4, Dlx5/6, Fgf3, Gata5/6, Gbx2, Gli1/2/3, Hnf1b, Hox genes, Irx3/4/5, Meis1/2/3, Msx1/2, Pitx1/2, Tbx genes, and Wnt3a/5a/5b/6) (Fig. 1D and data file S2).
Widespread regulation of Hox genes by pluripotency factors
The large number of Hox genes in this last group (18 genes; data file S2) prompted us to examine the response of all 39 Hox genes to Oct4 and Nanog (Fig. 2A and data files S1 and S2). Nanog induction up to E7.5 down-regulated the expression of 14 Hox genes, while it up-regulated the expression of 3. Its expression up to E9.5 only up-regulated the expression of 5 Hox genes. On the other hand, Oct4 significantly down-regulated 23 Hox genes when expressed up to E7.5, and up-regulated 24 when expressed up to E9.5 (Fig. 2A). It is noteworthy that in the case of Oct4, no Hox gene was up-regulated at E7.5 or down-regulated at E9.5. Oct4 affects expression of Hox genes from all four clusters (HoxA-D), but excluding most of the posterior Hox genes from paralog groups 10 to 13. Previous work has shown that Oct4 delays the activation of posterior Hox genes in trunk progenitors at later embryonic stages (20), suggesting that Oct4 could have opposite roles in the regulation of anterior and posterior Hox genes during development (21).
Fig. 2. Pluripotency factors mediate a global switch from Hox gene repression to activation.
(A) Schematic representation of the four mouse Hox clusters, indicating gene expression changes induced by Oct4 or Nanog at E7.5 or E9.5, as identified in the RNA-seq analysis. Yellow, down-regulated; blue, up-regulated; gray, unchanged. (B) Diagram depicting the nested windows of 1.5-day dox treatment for Oct4 induction between E5.5 and E9.5. (C) Changes in the expression of selected Hox genes (Hoxa1, Hoxb1, Hoxb4, and Hoxb9) induced by Oct4 during development, shown as log ratios of reverse transcription quantitative polymerase chain reaction (RT-qPCR)–measured relative RNA levels in untreated and dox-treated embryos as in B; *P < 0.05 by two-tailed Student’s t test.
These results indicate that endogenous Oct4 might be regulating Hox genes during postimplantation development, which would require them being coexpressed at these stages. To address whether this was the case, we analyzed published single-cell expression data in gastrulating embryos (E6.5 to E8.5 embryos) from the mouse gastrulation atlas (22). We found that the majority of cells expressing Oct4 also express Hox genes; at E8.0, approximately 80% of Oct4-positive cells also express any of the Hox genes examined (fig. S3A). Pearson correlation of single-cell RNA-seq expression data showed that the highest correlation between Oct4 and Hox genes occurs from E7.75 to E8.25 (fig. S3B). Correlation at these time points is positive, which is coincident with the up-regulation of Hox genes by Oct4 overexpression in the late-time window, but is negative at earlier stages when OCT4 would be repressing Hox gene expression. Analysis of the single-cell RNA-seq data for Oct4 and Hoxb1 expression in different cell types at E8.0, the time point where they show the highest correlation, identified highest levels of both genes in mesodermal derivatives and a near complete overlap between them (fig. S3, C and D). We next analyzed Oct4 mRNA levels in Hox-positive cells and observed that Oct4 is expressed at similar levels in these populations, compared to all Oct4-positive cells from the same stages (fig. S3E). This rules out the possibility that Hox genes are only expressed in cells with low levels of Oct4, which would be turning it off as a prerequisite for proper lineage commitment. The results obtained from the analysis of scRNA-seq data in postimplantation embryos point toward a strong correlation between Oct4 and Hox gene expression throughout the developmental stages that we have examined.
To determine when the switch from Hox gene repression to activation takes place, we induced Oct4 for 1.5-day periods between E5.5 and E8.0, harvesting embryos between E7.0 and E9.5 (Fig. 2B). We examined the expression of Hoxa1 and three HoxB cluster genes (Hoxb1, Hoxb4, and Hoxb9) by reverse transcription quantitative polymerase chain reaction (RT-qPCR) using genotype-matched embryos from non–dox-treated females as controls. Consistent with the transcriptomic data, we observed a switch from repression to activation. However, the switch occurred earlier for Hoxa1 and Hoxb1 than for Hoxb4 and Hoxb9 (Fig. 2C), resembling the collinear activation of the clusters in the early embryo (23).
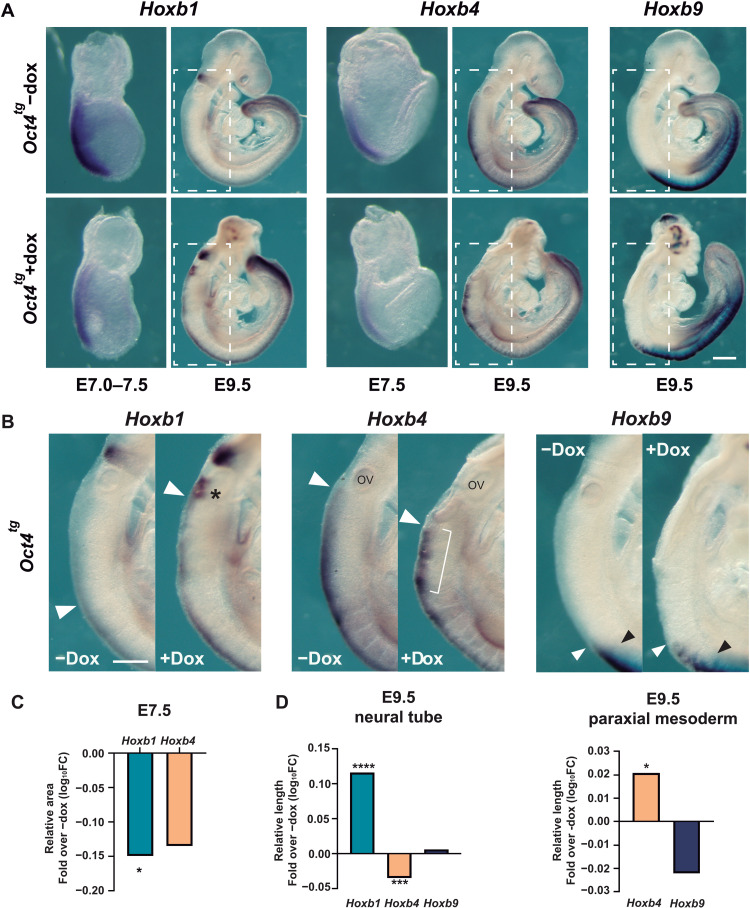
We next examined Oct4-induced changes to HoxB gene expression patterns by in situ hybridization of E7.0 to 7.5 and E9.5 embryos exposed to dox for 3 days. At E7.0 to 7.5, expression of Hoxb1 and Hoxb4 was down-regulated in the posterior region of the embryo, as predicted from the transcriptomic analysis (Fig. 3, A and C), and at E9.5, we observed gain of expression for HoxB genes in several domains of the embryo (Fig. 3, A, B, and D). As by this stage Oct4 is no longer expressed in Hox-positive territories, these results show that Oct4 can regulate multiple Hox genes even if expressed out of its endogenous context. Hoxb1 was the most altered, with a shift along the anterior-posterior axis in the neural tube (Fig. 3, B, arrowhead, and D) together with up-regulation or persistence of expression in presumptive rhombomere 6 territory (Fig. 3B, asterisk). Expression of Hoxb4 was posteriorized in the neural tube (Fig. 3, B, arrowhead, and D), showing an irregular and patchy pattern (Fig. 3B, bracket). However, its expression in the somatic mesoderm shifted anteriorly (Fig. 3D). As for Hoxb9, there is a change in the anterior limit of expression in the neural tube (Fig. 3B, white arrowhead) compared with that in the paraxial mesoderm, which also shows an irregular pattern (Fig. 3B, black arrowhead). Most notably, all three genes examined showed patches of expression in the anterior neural tube (Fig. 3A and fig. S4A), a territory devoid of Hox expression at all developmental stages (23). In situ hybridization in adjacent sections (fig. S4, B and C) revealed that these patches correspond to Oct4-expressing vesicle-like structures, where the anterior marker Otx2 was lost and Hox genes were expressed in various combinations (fig. S4C, arrowheads, and D, arrows). These results suggest a coordinated response of the HoxB cluster to Oct4 gain of function, leading to its activation in Hox-free domains such as the forebrain.
Fig. 3. Coordinated response of HoxB cluster to Oct4 overexpression.
(A) In situ hybridization analysis of HoxB cluster gene expression in E7.5 and E9.5 untreated (control, top row) and dox-treated (bottom row) Oct4tg embryos. n (E7.5, −/+ dox) = 8/12 (Hoxb1), 4 per condition (Hoxb4); n (E9.5) = 3 per condition. Scale bars, 140 μm (E7.0 to 7.5), 170 μm (E7.5), and 380 μm (E9.5). (B) Close-up views of E9.5 embryos shown in (A). The expression of Hoxb1 is expanded anteriorly (white arrowhead) in dox-treated Oct4tg embryos, and higher levels of expression are observed in presumptive rhombomere 6 (asterisk). Neural expression of Hoxb4 shows a patchy and disorganized pattern upon dox treatment (bracket) and is retarded more posterior (white arrowhead). Expression in the somites is shifted to the anterior part of the region. Neural expression of Hoxb9 is shifted anteriorly (white arrowhead) in relation to its limit of expression in the paraxial mesoderm (black arrowhead). n = 3. Scale bar, 250 μm. (C) Quantification of the positive area of in situ experiments in E7.5 embryos shown in (A), normalized by the total area of the embryo. Results are shown as log10; *P < 0.05 by two-tailed Student’s t test. (D) Quantification of the length of the positive area of in situ experiments in the neural tube (left) and paraxial mesoderm (right) of E9.5 embryos shown in (A), normalized by the total length of the embryo measured from the otic vesicle to the end of the tail. Results are shown as log10; analysis was performed by unpaired two-tailed Student’s t test. *P < 0.05; ***P < 0.001; ****P < 0.0001.
On the other hand, Nanog caused no obvious changes in Hox gene expression in E9.5 embryos except for an anterior expansion of the Hoxb1 domain in the neural tube, similar to what we observed for Oct4 (fig. S4, E and F).
Oct4 is necessary for the correct activation of Hox genes
To definitively establish the role of Oct4 in the regulation of Hox genes under endogenous conditions, we analyzed its requirement both in vivo and in ES cell differentiation assays. In the first place, we used a 4-hydroxytamoxifen (4-OHT) inducible Oct4 loss-of-function mouse model. These mice carry a floxed Oct4 allele together with an inducible Cre driven by the ROSA26 promoter (R26CreERT2) (24). To prevent embryo lethality at preimplantation or early postimplantation stages (10, 24), we deleted Oct4 by administering a single dose of 4-OHT at E6.5 (Fig. 4A). Embryos recovered at E7.5 and E8.5 from treated mothers showed no obvious morphological defects, while at E9.5, we observed a partially penetrant phenotype with craniofacial and trunk defects, as has been previously described (11). We subclassified E9.5-treated embryos in those showing a mild (open neural tube but normal posterior trunk) or severe (anterior malformations and posterior truncations with impaired somitogenesis) phenotypes. Quantification of Oct4 expression levels by RT-qPCR at these stages confirms that its expression is gradually lost (Fig. 4B), which suggests that deletion is not complete at the earlier stages examined or that we are observing prolonged stability of Oct4 mRNA because of its intrinsic half-life. However, when we analyzed HoxB genes (Hoxb1, Hoxb4, and Hoxb9), their expression was reduced in Oct4 loss-of-function embryos as compared to controls (non–4-OHT treated) at all stages (Fig. 4B).
Fig. 4. Oct4 is required for correct HoxB expression.
(A) Diagram depicting the experimental procedure followed with the loss-of-function model. (B) Changes in the expression of selected HoxB genes (Hoxb1, Hoxb4, and Hoxb9) induced by Oct4 deletion at E6.5 and analyzed at E7.5, E8.5, and E9.5. Results are shown as log ratios of RT-qPCR relative to untreated embryos. Analysis was performed by unpaired two-tailed Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. n (−/+ tam) = 6/16 (E7.5), 8/14 (E8.5), 7/9 (E9.5 mild), and 10/7 (E9.5 severe). (C) In situ hybridization analysis of HoxB cluster gene expression at E9.5. Images show Oct4fl/fl-untreated embryos (top row) and 4-OH–treated embryos with mild (middle row) and severe phenotype (bottom row). n (−/+ tam) = 5/6 (Hoxb1), 4/8 (Hoxb4), and 7/7 (Hoxb9). Scale bar, 380 μm. (D) Changes in the expression of Hoxb1, Hoxb4, and Hoxb9 in ZHBTc4 ES cells with different levels of Oct4 up to 6 days of differentiation to mesoderm or anterior neural (hindbrain) lineages. mRNA levels were measured by RT-qPCR in ZHBTc4 ES cells treated with tetracycline from day 3 to day 6 (Oct4 WT), from day 0 to day 6 (cells that do not express Oct4 along differentiation; Oct4 LoF), and untreated ZHBTc4 (cells that express constitutively Oct4 along differentiation; Oct4 GoF). Statistical significance was calculated by two-way analysis of variance (ANOVA) followed by Tukey’s test (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We next examined HoxB gene expression in control and Oct4 loss-of-function embryos by whole mount in situ hybridization. We observed a down-regulation of Hoxb1 and Hoxb4 expression in both mild and severe E9.5 Oct4-deleted embryos (Fig. 4C and fig. S5, A to C). However, we did not detect changes in Hoxb9 expression by in situ hybridization.
To determine how changes of Oct4 affect HoxB gene expression during differentiation, we used an ES cell model where we could modulate Oct4 expression in an inducible fashion. The ZHBTc4 ES cell line has both copies of endogenous Oct4 deleted and harbors a tetracycline (tet)–dependent Oct4 transgene (Tet-Off), where addition of the drug leads to the quick down-regulation of Oct4 (6). We differentiated ZHBTc4 cells toward mesodermal-like (via Wnt activation) or anterior neural–like (hindbrain, via retinoic acid induction) fates (25), under three different experimental conditions. In the first case, ZHBTc4 cells were treated with tet 3 days after starting the differentiation process (fig. S5D). This mimics the endogenous dynamics of Oct4 that is strongly down-regulated from day 3 to day 4 in both differentiation protocols (fig. S5E), and thus, we considered this condition equivalent to the wild-type behavior (Oct4 WT). In the second, cells were treated with tet from day 0 and therefore were devoid of Oct4 during the whole differentiation process (Oct4 LoF; fig. S5D), but not during the previous pluripotent phase. Last, we also analyzed untreated ZHBTc4 cells that express Oct4 throughout pluripotency and the differentiation process (Oct4 GoF; fig. S5D).
Under these conditions, we observed the expected up-regulation of Hox genes during differentiation, with Hoxb1 showing an earlier peak in expression than Hoxb4 or Hoxb9 (Fig. 4D). Gain of function of Oct4 leads to increase in Hox gene expression, although not robustly except for the expression of Hoxb4 at day 6 during both mesodermal and neural differentiation. On the other hand, loss of function of Oct4 caused a failure to properly activate Hox genes during differentiation (Fig. 4D). It is noteworthy that we observed these effects during both Wnt-dependent (mesoderm) and Wnt-independent (hindbrain) differentiation protocols. This is relevant because we identified different Wnt genes as putative targets of Oct4 (see above), and a possibility was that the effect we observed of Oct4 on Hox genes was not direct but mediated by Wnts, which are known to activate Hox gene expression in the early embryo (26).
Together, these experiments strongly suggest that Oct4 is required for the correct initiation of the expression of genes from the HoxB cluster, once differentiation has begun starting from the pluripotent epiblast.
Global control of the HoxB cluster by OCT4
To investigate how pluripotency factors regulate Hox genes, we examined previously published ChIP-seq binding profiles of OCT4 and NANOG in mouse ES cells (27). We detected some weak binding of these factors within the HoxA, HoxB, and HoxC clusters, D, but observed very prominent peaks at the anterior ends of the three clusters (Fig. 5A and fig. S6A). As for HoxD cluster, various peaks were found both at the anterior end and within the cluster. In the HoxB cluster, both OCT4 and NANOG bind proximally to the Hoxb1 promoter (P-site) and to a distal region (D-site) approximately 9 kb downstream of its transcriptional start site (Fig. 5A). These sites have been shown to bind OCT4 during ES cell differentiation (28) and are bound both in ES cells and epiblast-like cells in the transition from naïve to primed pluripotency (16).
Fig. 5. Pluripotency factor binding sites establish long-distance contacts in the HoxB cluster.
(A) ChIP-seq binding profiles for OCT4 and NANOG (27) along the HoxB cluster (mm9; chr11:96,029,042-96,307,981; reversed). Distribution of histone marks for active transcription (H3K4m3), repression (H3K27m3), and active regulatory elements (H3K4m1 and H3K27ac) in Bruce4 ES cells (60) is shown below. Red arrowheads indicate two strong binding sites neighboring Hoxb1 at the anterior end of the cluster. (B) ChIP-qPCR of OCT4 binding to the P-site and D-site in ES cells (left, n = 4) and in untreated (−dox) and treated (+dox) E9.5 Oct4tg embryos (right, n = 3). Enrichment is shown as percentage of input for immunoglobulin G (IgG) (negative control) and anti-OCT4 antibody. Horizontal bars indicate means. *P < 0.05 by two-tailed Student’s t test. (C) Chromatin interaction profile established from viewpoints located at the P-site and D-site in a 1-Mb window surrounding the HoxB cluster (mm9; chr11:95,725,993-96,725,993) in ES cells, and untreated (−dox) and treated (+dox) E9.5 Oct4tg embryos. Distribution of normalized reads in two replicates is shown, with significant interactions below. Asterisks indicate novel intracluster interactions established in dox-treated embryos, and boxed regions (dashed line) indicate the interactions established toward the Hoxb13 region.
We confirmed that these sites were occupied by OCT4 in ES cells by ChIP-qPCR (Fig. 5B), using a region from the Nanog promoter shown to be bound by OCT4 (27, 29) as a positive control and other unrelated genomic regions as negative controls (fig. S6B). ChIP in E9.5 embryos showed no binding, but after dox administration, we observed binding at both the P-site and D-site (Fig. 5B), demonstrating that these sites are occupied by OCT4 upon induction of its expression.
The response of multiple Hox genes to Oct4 we observed in the RNA-seq dataset and in whole mounts in situ suggested the existence of global mechanisms of Hox cluster regulation by pluripotency factors. To address whether these regions could be acting as common regulatory elements for the cluster, we examined their interaction profile by circular chromatin conformation capture followed by high-throughput sequencing (4C-seq). We designed viewpoints for both the P-site and D-site and carried out 4C-seq in mouse ES cells, where OCT4 is present and HoxB genes are not expressed, as well as in dox-treated and dox-untreated E9.5 Oct4tg embryos (Fig. 5C). Normalized reads were used to fit a distance-decreasing monotone function, to take into account that nearby fragments will randomly interact more frequently, and contacts that deviated significantly from the normal distribution were identified.
The chromatin structure surrounding the anterior end of the HoxB cluster is relatively stable independently of its expression (Fig. 5C), as has been shown for other viewpoints in the cluster (30). Interaction occurs on both sides of the viewpoints: toward the HoxB cluster itself, with a strong limit near Hoxb13, and outside the cluster toward the telomeric region (Fig. 5C). However, interactions are differently distributed around the viewpoint in ES cells and embryos. In ES cells, there are more interactions toward the Hoxb13 region, possibly reflecting the closed conformation of the cluster mediated by poised promoters (31), whereas in E9.5 embryos, interactions increase toward the telomeric gene desert defined by Skap1 (Fig. 5C), whose expression at E7.0 to E7.75 is limited to embryonic blood progenitors (32). This difference in interactions might reflect the active state of the HoxB cluster in the embryo, where distal regulatory elements located in this region (33) are recruited to define its correct expression, as is also the case for the HoxA cluster (26). Our observations are also in line with recent results that show that the HoxA and HoxD clusters are organized in compact domains in ES cells that open up during differentiation (34). When Oct4 is induced in E9.5 embryos, previously unidentified contacts are established from both the P-site and D-site toward the cluster (Fig. 5C, asterisks) accompanied by a reduction in the interactions with the Hoxb13 domain (Fig. 5C, dashed boxes). We can conclude that these regions at the telomeric end of the HoxB cluster, which are bound by pluripotency factors, establish intracluster interactions in active or inactive states. Furthermore, the presence of OCT4 in E9.5 embryos leads to a reorganization of the local architecture of the HoxB cluster.
Deletion of the distal OCT4 site disrupts the pattern of HoxB expression during differentiation
To complement these observations, we tested the necessity of the OCT4-bound regions described above in the regulation of the HoxB cluster by their deletion in ES cells by CRISPR-Cas9–mediated genome editing. We examined the genomic regions covered by ChIP-seq peaks, finding that the proximal site (P) contains one consensus OCT4 binding site, while the distal site (D) contains at least two (fig. S7A). In the case of the P-site, this consensus lies within the previously described proximal Hoxb1 autoregulatory element (35) and in very close proximity to the promoter. In addition, this region was shown to be bound by SOX/OCT heterodimers being required for the maximal transcriptional activity of Hoxb1 (36). Given the difficulty of deleting this site without compromising other known regulatory inputs on Hoxb1, we decided to only analyze the effect of deleting the D-site. This region does not map to any other known Hoxb1 regulatory elements, such as the two described 3′ retinoic acid response elements (fig. S7A) (37, 38).
We generated two independent ES cell clones deleted for the D-site (clones #30 and #57; fig. S7A) and analyzed changes in HoxB gene expression during their differentiation toward mesoderm-like or anterior neural–like (hindbrain) fates (fig. S7B) (25). Comparison of parental with HoxB D-siteΔ/Δ ES cells along differentiation showed a similar trend for each of the Hox genes examined independently of the differentiation protocol, and a comparable behavior in the two independent clones (Fig. 6A). Hoxb1 is down-regulated as compared to controls at later stages of differentiation. Hoxb4 do not show significant changes along differentiation. Last, Hoxb9 is consistently activated in deleted cell lines throughout the differentiation window analyzed (Fig. 6A). To analyze the effect of the deletion in vivo, we used the HoxB D-siteΔ/Δ ES cells to generate mouse lines. Homozygous mice survive to term, which did not come as a surprise, as deletion of the entire HoxB cluster in homozygosity does not cause embryonic lethality (39). We examined the expression of Hoxb1 and Hoxb4 at E7.5, a time when anterior HoxB genes and Oct4 are still codetected (fig. S3, A and B). RT-qPCR showed down-regulation of Hoxb1 and up-regulation of Hoxb4 in deleted embryos (Fig. 6B). This further confirmed the different roles of Oct4 at this stage, when it would be necessary to achieve proper expression of Hoxb1 but at the same time would be lowering levels of Hoxb4.
Fig. 6. OCT4 D-site is required for correct expression of HoxB genes during differentiation.
(A) Changes in the expression of Hoxb1, Hoxb4, and Hoxb9 as a consequence of the deletion of the distal OCT4 binding site up to 6 days of differentiation (d3 to d6) of ES cells to mesoderm or anterior neural (hindbrain) lineages. RNA expression was measured by RT-qPCR and normalized to Actb in two independent deleted homozygous clones (#30, dark yellow; #57, light yellow). Statistical significance was calculated by two-way ANOVA followed by Dunnett’s test (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (B) Expression of Hoxb1, Hoxb4, and Hoxb9 quantified by RT-qPCR in E7.5 HoxB D-siteΔ/Δ embryos normalized by their expression in control embryos (n = 10). Results are shown as log10; *P < 0.05; **P < 0.01 by two-tailed Student’s t test. (C) Quantification of the positive area of in situ experiments shown in (D), normalized by the total area of the embryo. Results are shown as log10; *P < 0.05 by two-tailed Student’s t test. (D) Expression of Hoxb1 (top) and Hoxb4 (bottom) in E7.5 early head-fold stage HoxB D-siteΔ/Δ (right) embryos compared to controls (left); n (control/HoxB D-siteΔ/Δ) = 4/6 (Hoxb1) and 4/7 (Hoxb4). Scale bar, 200 μm.
Whole mount in situ hybridization at the early head-fold stage showed no clear changes in Hoxb1, but an expansion of the area of expression at the posterior part of the embryo of Hoxb4 (Fig. 6, C and D). We also examined expression of HoxB genes in E9.5 embryos from the HoxB D-siteΔ/Δ line and did not observe major changes except for overall lower levels of Hoxb4 expression (fig. S7, C and D). Therefore, these results suggest that at early stages of expression, OCT4 is necessary to fine-tune expression of HoxB genes in their endogenous domains.
DISCUSSION
It is generally assumed that pluripotency factors act to restrict lineage decisions before gastrulation. However, Oct4 has been shown to participate in several later developmental decisions in the mouse, including primitive endoderm development (40, 41), lineage priming (24, 28, 42), primitive streak proliferation (11), regulation of trunk length (20), or the formation of cranial neural crest (43). Furthermore, recent results have shown how the lack of Oct4 in early gastrulating embryos results in a blockade of epithelial-to-mesenchymal transition in the posterior epiblast, thus disrupting proper axis formation (10). Both single-cell RNA-seq data and in situ detection in mouse show that Oct4 and Hox genes are coexpressed in cells of the gastrulating embryo, from the onset of Hox gene expression and up to E8.5 (3, 32).
We propose that at these early stages, Oct4 plays a dual role, first maintaining Hox genes silent before lineage commitment, and later being required for their proper activation. This behavior is specific for Oct4, as in the case of Nanog, we only observe initial repression of Hox genes, which agrees with the description of the mutual cross-repression of Nanog and Hoxa1 to differentially regulate a common set of downstream target genes involved in early phases of lineage commitment (44).
OCT4 switches from a repressor to an activator at the onset of gastrulation, when Hox genes become activated and when OCT4 is still widely expressed, but the exact timing of this event is yet to be determined. OCT4 associates with both activator and repressor complexes, through the recruitment of different cofactors. For example, OCT4 recruits ERG-associated protein with SET domain (ESET) to silence trophoblast-associated genes (45, 46) but also can bind SALL4 or NURD in a histone deacetylase complex (47). On the other hand, the interaction of OCT4 with WDR5 induces transcription of key self-renewal regulators (48). In addition, other protein partners are key for specific roles of OCT4, such as the recruitment of BRG1 that is essential for its pioneer activity (49) or its cooperation with OTX2 at the transition between naïve and primed states of pluripotency (16). Thus, it is tempting to speculate that an exchange of cofactors would explain the bimodal function of OCT4 in the regulation of Hox genes regulation. In addition, the function of OCT4 on some of its target genes is not a simple on/off binary system, but dependent on its own levels (6). This could also potentially explain our observation that at early developmental stages, OCT4 is repressing genes that will be later up-regulated, when OCT4 levels are lower. The capacity of a single factor of having opposite transcriptional activities allows for a rapid switch in expression of its targets. This is needed for a correct establishment of developmental processes. Also, it is critical immediately after the loss of pluripotency when cells need to take fate decisions in a short time window. Further studies using protein dynamics to complement our studies on gene expression will provide better resolution on OCT4’s role on Hox genes expression.
The concerted response of Hox genes, together with the chromatin interactions established by bound regions we see in the HoxB cluster, suggests that OCT4 regulates globally Hox clusters and forms part of the complex regulatory apparatus that ensures proper Hox gene expression (12, 26). Furthermore, when we examine the expression of anterior, middle, and posterior HoxB genes in gain- and loss-of-function models, we observe a collinear response to Oct4, in line with recent findings of the repression of most posterior 5′ Hox genes by OCT4 (20, 21). It is also interesting to note that pluripotency factors have been involved in the development of the neural crest of amphibians (50) and mammals (43), a multipotent population of cells that are equally patterned by Hox genes. Therefore, we find multiple instances and cell populations during vertebrate development (gastrulation, trunk extension, and the neural crest) where pluripotency factors (and more specifically POU5-like factors such as OCT4) could be regulating Hox genes. Moreover, our expression data indicate that other patterning genes respond in a similar fashion, suggesting that OCT4 and other pluripotency factors mediate a switch from repression to activation of an array of developmental regulators at the time of lineage decisions.
The early activation of Hox genes is dependent on several pathways and factors, such as retinoic acid, fibroblast growth factors (FGFs), Wnts, GDF11, or CDX transcription factors (21). The results we present here suggest that OCT4 is also essential to trigger correct Hox gene activation at a specific time and in cell-specific populations. However, we cannot rule out that OCT4 could be priming or maintaining these regions open for the activation by other transcription factors, as has been described during reprogramming (51). Our data using loss-of-function models are compatible with a pioneering role of OCT4 on Hox early enhancers. On the other hand, when we use gain-of-function models, the results suggest a direct activation of Hox genes by OCT4. These two possibilities are not mutually exclusive and, certainly, OCT4 binding could favor the later recruitment and activity of other factors. For example, the HoxB P-site is embedded within the Hoxb1 autoregulatory element that drives expression in rhombomere 4 of the hindbrain (35). This region binds SOX and OCT proteins that are necessary for the optimal response to the transcriptional activation by HOX/PBX heterodimers (36). Thus, the early binding of OCT4, even before Hoxb1 is expressed, could facilitate later recruitment of HOXB1 to nearby sequences. In agreement with this hypothesis, Hoxb1 expression in rhombomere 4 is greatly diminished in Oct4 mutant embryos.
In summary, initial lineage specification involves not only dismantling of the core pluripotency gene regulatory network (2) but also a switch in function of key factors such as OCT4 from repressors to activators that would supervise the transition from pluripotency to lineage determination. Our results provide new insight into temporally dynamic roles for factors such as OCT4, which beyond their well-described function in pluripotency, are directly responsible for regulating genes associated with differentiation programs.
MATERIALS AND METHODS
Animal models
Mice lines used in this study were housed and maintained in the animal facility at the Centro Nacional de Investigaciones Cardiovasculares (Madrid, Spain) in accordance with national and European legislation. Procedures were approved by the CNIC Animal Welfare Ethics Committee and by the Area of Animal Protection of the Regional Government of Madrid (ref. PROEX 196/14). Double-homozygote transgenic males of the Oct4/rtTA (R26-M2rtTA;Col1a1-tetO-Oct4) (14) or Nanog/rtTA (R26-M2rtTA;Col1a1-tetO-Nanog) (15) mouse lines were mated with CD1 females, which were treated with dox (0.2 or 1 mg/ml) in the drinking water to induce the Oct4 or Nanog transgene, respectively, in embryos. For Oct4 transgene induction in E7.5 embryos to be used for in situ hybridization, a single 100-μl intraperitoneal injection of dox (25 μg/μl) was administered to pregnant females at E5.5, followed by dox administration (0.5 mg/ml) in drinking water. Nontreated mice of the same genotype were used as controls. Double-homozygote R26CreERT2;Oct4LoxP/LoxP (24) mice were mated, and females were treated at E6.5 by administering a single dose of 4-OH (5 mg, at 25 μg/μl) by gavage. Mouse lines deleted for the Oct4 distal site adjacent to Hoxb1 were generated by blastocyst injection of mutated ES cells (see below) following standard procedures (52) and genotyped using the primers specified in data file S4.
RNA sequencing
RNA-seq was performed with three biological replicates, each consisting of pools of 8 to 12 E7.5 embryos or 3 E9.5 embryos obtained from untreated (controls) or dox-treated double heterozygous embryos. Levels of Oct4 or Nanog overexpression were tested by RT-qPCR for each independent litter before RNA-seq. Equally, three biological replicates of E14 ES cells were used for RNA-seq. Single-end sequencing was performed by the CNIC Genomics Unit using a GAIIx sequencer. Adapters were removed with Cutadapt v1.14, and sequences were mapped and quantified using RSEM v1.2.20 to the transcriptome set from Mouse Genome Reference NCBIM37 and Ensembl Gene Build version 67. Differentially expressed genes between groups were identified using the limma bioconductor package. Only P < 0.05 adjusted through the Benjamini-Hochberg procedure was considered as significant. Clustering analysis was conducted for all genes differentially expressed between the induced and control conditions for any of the four conditions. Overrepresented biological categories were identified using DAVID v6.8 (53). Lists of genes located close to OCT4 and NANOG genomic bound regions were generated from published ChIP-seq datasets (18, 19). Each ChIP-seq peak was assigned to the single nearest gene in a 100-kb window. RNA-seq data are available at the NCBI Gene Expression Omnibus (GEO) database under accession number GSE94954.
Single-cell gene expression data from Pijuan-Sala et al. (22) were analyzed to assess coexpression of Oct4 and Hox gene. A gene is considered to be expressed in a cell when its expression level is >0 fragments per million.
RT-qPCR assays
Total RNA from single embryos from E8.0 onward, embryo pools up to E7.5, or ES cells directly lysed in their wells, was extracted with RNeasy kit (Qiagen) and digested with deoxyribonuclease I (Qiagen) to remove genomic DNA. Total RNA (0.5 to 1.0 μg) was reverse transcribed using Quantitech Reverse kit (Applied Biosystems). qPCR was performed with SYBR Green Master Mix (Applied Biosystems) on an AB 7900-Fast-384 machine. qPCR primers are listed in data file S4. Expression values were normalized to the expression of Actb and Ywhaz (whose expression as measured in our RNA-seq data did not change upon Oct4 or Nanog induction) using the comparative CT method (54), and SDs were calculated and plotted using Prism 7.0 software (GraphPad). All assays were performed in triplicate.
In situ hybridization
In situ hybridization in whole mount embryos or sections was performed using digoxigenin-labeled probes as described (55). Probes for Hoxb1, Hoxb4, Hoxb9, and Otx2 were generated by PCR (primers listed in data File S4), and the probe for Oct4 was provided by T. Rodriguez (Imperial College London). Early embryos were staged according to Forlani et al. (56). Quantification of in situ hybridization staining was carried out using Fiji to measure the area and length of the positive domain of Hox gene expression, in both the neural tube and the somites. Values were normalized by the total length or area of the embryo from the otic vesicle to the end of the tail.
4C sequencing
4C was performed as previously described (30, 57) on two replicates of pools of 60 to 70 E9.5 embryos or 1 × 106 to 2 × 106 G4 ES cells. Samples were cross-linked with 2% paraformaldehyde, frozen in liquid nitrogen, and stored at −80°. Chromatin was digested with Dpn II (New England BioLabs) followed by Nla III (New England BioLabs), and ligated with T4 DNA Ligase (Promega). For all experiments, 0.5 to 1 μg of the resulting 4C template was used for the subsequent PCR (primers listed in data file S4). 4C libraries were sequenced (single end) at the CNIC Genomics Unit using an Illumina HiSeq 2500 sequencer. Sequences were mapped and quantified using RSEM v1.2.20 to the Mouse Genome Reference NCBIM37. Reads located in fragments flanked by two restriction sites of the same enzyme, in fragments smaller than 40 base pairs (bp) or within a window of 10 kb around the viewpoint, were filtered out. Mapped reads were converted to reads per first enzyme fragment ends and smoothed using a 30-fragment mean running window algorithm. Smoothed scores from each experiment were then normalized to the total number or reads before the visualization. To calculate the frequency of captured sites per window, Fastq files were demultiplexed using Cutadapt with the viewpoint sequences as indexes. Potential Illumina adaptor contaminants and small chimeric reads were removed. Processed reads were assigned to their corresponding genomic fragment after a virtual digestion of the reference genome with the first and second restriction enzymes. Reads located in fragments 5 kb around the viewpoint were filtered out. Quantification was performed considering each fragment end as one capture site if one or more sequences were mapped to it. The number of capture sites was summarized per 30-fragment window. The frequency of capture sites per window was used to fit a distance-decreasing monotone function, and z scores were calculated from its residuals using a modified version of FourCSeq (58). Significant contacts were considered in cases where the z score was >2 in both replicates and deviated significantly (adjusted P < 0.05) from its normal cumulative distribution in at least one of the replicates. 4C-seq data are available at the NCBI GEO database under accession number GSE94954,
Chromatin immunoprecipitation
ChIP was performed using 10 E9.5 embryos or 1 × 106 ES cells per experiment. After recovery, embryos were treated with collagenase type I (Stemcell Technologies, 07902) at 0.125% for 1 hour at 37°. Then, embryos were desegregated using a pipette and washed with cold phosphate-buffered saline. Samples were fixed, and protein-DNA complexes were cross-linked by treatment with 1% formaldehyde (Pierce, 289069) for 15 min rocking at room temperature. To stop fixation, glycine (Nzytech, MBO1401) was added to a final concentration of 125 mM during 10 min. Next, ChIP was performed using the ChIP-IT High Sensitivity kit (Active Motif, 53040), following the manufacturer’s instructions. DNA was sheared into fragments ranging from 200 to 1000 bp using a sonicator (Diagenode Bioruptor Water Bath Sonicator, 30 s on 30 s off for 30 min). Immunoprecipitations were carried out using rabbit polyclonal anti-OCT4 antibody (Abcam, ab19857), and anti-Rabbit immunoglobulin G polyclonal antibody (Abcam, ab171870) was used as negative control. Enrichment was measured by qPCR. A fragment from the Nanog promoter was used as a positive control (27, 29), and genomic fragments from the loci of Anks1b, Smg6, and Tiam1 were used as negative controls (59) after checking they did not contain OCT4 bound peaks (27). qPCR primers used are listed in data file S4.
ES culture, cell editing, and differentiation
ZHBTc4 cells (6) were maintained in culture on 0.1% gelatin (Sigma-Aldrich) using Corning p24 plates with CellBIND surface. Medium contained inactivated fetal calf serum (HyClone), leukemia inhibitory factor (LIF) (produced in-house), and 2i (PD0325901 and CHIR99021, Sigma-Aldrich). G4 mouse ES cells were maintained in culture on 0.1% gelatin (Sigma-Aldrich) in medium containing inactivated fetal calf serum (HyClone) and LIF (produced in-house). CRISPR-Cas9–mediated deletions in G4 mouse ES cells were generated using two guide RNAs (g1D-g2D; data file S4) together with a plasmid for Cas9. Cells were transfected, selected by sorting, and replated. Individual clones (clone #30 and clone #57) were genotyped using the primers specified in data file S4.
For anterior neural (hindbrain) and paraxial mesoderm differentiation, cells were treated as described (25). Briefly, cells were grown in monolayer using Corning p24 plates with CellBIND surface and with 0.1% gelatin (Sigma-Aldrich) added 30 min before passing in N2B27 media supplemented with basic Fgf (10 ng/ml) (R&D Systems) for 3 days (d1 to d3) and then were transferred into different media depending on the differentiation process. To induce anterior neural identity, 10 nM RA (Sigma-Aldrich) was added from D3 to D5. To induce mesodermal differentiation, the cells were treated with CHIR990215uM from D3 to D5. Cells were collected at each time point by adding lysis buffer directly to the wells. ZHBTc4 cells were treated with tet (1 μg/ml) (Sigma-Aldrich) from D0, from D3, or nontreated to activate Oct4 expression.
Statistical analysis
No blinding or randomization method was used for mouse experiments, and sample size was not predetermined. Statistical tests used are described above where relevant and in the figure legends.
Acknowledgments
We wish to dedicate this work to the memory of J. L. Gomez-Skarmeta, a most dear friend and a passionate scientist. We thank B. Fernandez-Tresguerres for initial input to this work; M. Serrano and K. Hochedlinger for the Nanogtg mouse line; J. Nichols for the Oct4 conditional KO mice and advice; A. Smith for the ZHBTc4 ES cell line; T. Rodriguez for reagents and discussions; T. Rayon, M. Torres, and M. Mallo for comments and suggestions; J. J. Tena for advice; the CNIC Genomics and Transgenesis Units for assistance; S. Bartlett for English editing; and members of Manzanares laboratory for continued support.
Funding: This work was funded by the Spanish government (grants BFU2017-84914-P and PID2020-115755GB-I00 to M.M.; BFU2016-74961-P and BFU2016-81887-REDT to J.L.G.-S.), the Andalusian government (grant BIO-396 to J.L.G.-S.), and the European Research Council (ERC; grant agreement 740041 to J.L.G.-S.). M.T. held Juan de la Cierva fellowships from the Spanish government (FJCI-2017-31791 and IJC2019-038897-I), R.R. and R.D.A. held FPU fellowships from the government, and J.V. was the recipient of a “La Caixa” fellowship. Work in the laboratory of J.L.G.-S. was supported by a María de Maetzu Unit of Excellence Grant (MDM-2016-0687) to the Department of Gene Regulation and Morphogenesis of the CABD. The CBMSO is supported by an institutional grant from the Fundación Ramon Areces, and the CNIC is supported by the Instituto de Salud Carlos III (ISCIII), the Ministerio de Ciencia e Innovación (MCIN), and the Pro CNIC Foundation, and is a Severo Ochoa Center of Excellence (grant CEX2020-001041-S funded by MICIN/AEI/10.13039/501100011033).
Author contributions: M.T., E.L.-J., J.S.d.A., A.B., J.V., C.B.-C., I.R., R.R., E.S., and J.A. performed the experiments. H.S.-I., R.D.A., and C.T. analyzed the data. J.L.G.-S., G.G., F.S.-C., and M.M. supervised the research. M.T., E.L.-J., J.S.d.A. and M.M. wrote the manuscript with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Sequencing data are deposited at the NCBI Gene Expression Omnibus Database (GEO) under accession number GSE94954. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Other Supplementary Material for this manuscript includes the following:
Data files S1 to S4
REFERENCES AND NOTES
- 1.Nichols J., Smith A., Pluripotency in the embryo and in culture. Cold Spring Harb. Perspect. Biol. 4, a008128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osorno R., Tsakiridis A., Wong F., Cambray N., Economou C., Wilkie R., Blin G., Scotting P. J., Chambers I., Wilson V., The developmental dismantling of pluripotency is reversed by ectopic Oct4 expression. Development 139, 2288–2298 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downs K. M., Systematic localization of Oct-3/4 to the gastrulating mouse conceptus suggests manifold roles in mammalian development. Dev. Dyn. 237, 464–475 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Zhang M., Leitch H. G., Tang W. W. C., Festuccia N., Hall-Ponsele E., Nichols J., Surani M. A., Smith A., Chambers I., Esrrb complementation rescues development of Nanog-Null germ cells. Cell Rep. 22, 332–339 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hart A. H., Hartley L., Ibrahim M., Robb L., Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev. Dyn. 230, 187–198 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Niwa H., Miyazaki J., Smith A. G., Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Thomson M., Liu S. J., Zou L. N., Smith Z., Meissner A., Ramanathan S., Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145, 875–889 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeineddine D., Papadimou E., Chebli K., Gineste M., Liu J., Grey C., Thurig S., Behfar A., Wallace V. A., Skerjanc I. S., Pucéat M., Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev. Cell 11, 535–546 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Sainz de Aja J., Menchero S., Rollan I., Barral A., Tiana M., Jawaid W., Cossio I., Alvarez A., Carreño-Tarragona G., Badia-Careaga C., Nichols J., Göttgens B., Isern J., Manzanares M., The pluripotency factor NANOG controls primitive hematopoiesis and directly regulates Tal1. EMBO J. 38, e99122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulas C., Chia G., Jones K. A., Hodgson A. C., Stirparo G. G., Nichols J., Oct4 regulates the embryonic axis and coordinates exit from pluripotency and germ layer specification in the mouse embryo. Development 145, dev159103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeVeale B., Brokhman I., Mohseni P., Babak T., Yoon C., Lin A., Onishi K., Tomilin A., Pevny L., Zandstra P. W., Nagy A., van der Kooy D., Oct4 is required ~E7.5 for proliferation in the primitive streak. PLOS Genet. 9, e1003957 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander T., Nolte C., Krumlauf R., Hox genes and segmentation of the hindbrain and axial skeleton. Annu. Rev. Cell Dev. Biol. 25, 431–456 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Deschamps J., Duboule D., Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 31, 1406–1416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochedlinger K., Yamada Y., Beard C., Jaenisch R., Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121, 465–477 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Piazzolla D., Palla A. R., Pantoja C., Cañamero M., de Castro I. P., Ortega S., Gómez-López G., Dominguez O., Megías D., Roncador G., Luque-Garcia J. L., Fernandez-Tresguerres B., Fernandez A. F., Fraga M. F., Rodriguez-Justo M., Manzanares M., Sánchez-Carbayo M., García-Pedrero J. M., Rodrigo J. P., Malumbres M., Serrano M., Lineage-restricted function of the pluripotency factor NANOG in stratified epithelia. Nat. Commun. 5, 4226 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Buecker C., Srinivasan R., Wu Z., Calo E., Acampora D., Faial T., Simeone A., Tan M., Swigut T., Wysocka J., Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14, 838–853 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K. Y., Sung K. W., Lee C. W. H., Zhao X. D., Chiu K. P., Lipovich L., Kuznetsov V. A., Robson P., Stanton L. W., Wei C. L., Ruan Y., Lim B., Ng H. H., The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H., Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Marson A., Levine S. S., Cole M. F., Frampton G. M., Brambrink T., Johnstone S., Guenther M. G., Johnston W. K., Wernig M., Newman J., Calabrese J. M., Dennis L. M., Volkert T. L., Gupta S., Love J., Hannett N., Sharp P. A., Bartel D. P., Jaenisch R., Young R. A., Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521–533 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aires R., Jurberg A. D., Leal F., Nóvoa A., Cohn M. J., Mallo M., Oct4 is a key regulator of vertebrate trunk length diversity. Dev. Cell 38, 262–274 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Mallo M., Reassessing the role of Hox genes during vertebrate development and evolution. Trends Genet. 34, 209–217 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Pijuan-Sala B., Griffiths J. A., Guibentif C., Hiscock T. W., Jawaid W., Calero-Nieto F. J., Mulas C., Ibarra-Soria X., Tyser R. C. V., Ho D. L. L., Reik W., Srinivas S., Simons B. D., Nichols J., Marioni J. C., Göttgens B., A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deschamps J., van Nes J., Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 132, 2931–2942 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Le Bin G. C., Muñoz-Descalzo S., Kurowski A., Leitch H., Lou X., Mansfield W., Etienne-Dumeau C., Grabole N., Mulas C., Niwa H., Hadjantonakis A.-K., Nichols J., Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development 141, 1001–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouti M., Delile J., Stamataki D., Wymeersch F. J., Huang Y., Kleinjung J., Wilson V., Briscoe J., A gene regulatory network balances neural and mesoderm specification during vertebrate trunk development. Dev. Cell 41, 243–261.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neijts R., Amin S., van Rooijen C., Tan S., Creyghton M. P., de Laat W., Deschamps J., Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev. 30, 1937–1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whyte W. A., Orlando D. A., Hnisz D., Abraham B. J., Lin C. Y., Kagey M. H., Rahl P. B., Lee T. I., Young R. A., Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simandi Z., Horvath A., Wright L. C., Cuaranta-Monroy I., de Luca I., Karolyi K., Sauer S., Deleuze J. F., Gudas L. J., Cowley S. M., Nagy L., OCT4 acts as an integrator of pluripotency and signal-induced differentiation. Mol. Cell 63, 647–661 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P., Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Noordermeer D., Leleu M., Schorderet P., Joye E., Chabaud F., Duboule D., Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. eLife 3, e02557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbieri M., Xie S. Q., Torlai Triglia E., Chiariello A. M., Bianco S., de Santiago I., Branco M. R., Rueda D., Nicodemi M., Pombo A., Active and poised promoter states drive folding of the extended HoxB locus in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 24, 515–524 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Scialdone A., Tanaka Y., Jawaid W., Moignard V., Wilson N. K., Macaulay I. C., Marioni J. C., Göttgens B., Resolving early mesoderm diversification through single-cell expression profiling. Nature 535, 289–293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nolte C., Jinks T., Wang X., Martinez Pastor M. T., Krumlauf R., Shadow enhancers flanking the HoxB cluster direct dynamic Hox expression in early heart and endoderm development. Dev. Biol. 383, 158–173 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Kundu S., Ji F., Sunwoo H., Jain G., Lee J. T., Sadreyev R. I., Dekker J., Kingston R. E., Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol. Cell 65, 432–446.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popperl H., Bienz M., Studer M., Chan S. K., Aparicio S., Brenner S., Mann R. S., Krumlauf R., Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell 81, 1031–1042 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Di Rocco G., Gavalas A., Popperl H., Krumlauf R., Mavilio F., Zappavigna V., The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 auto-regulatory enhancer function. J. Biol. Chem. 276, 20506–20515 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Marshall H., Studer M., Pöpperl H., Aparicio S., Kuroiwa A., Brenner S., Krumlauf R., A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature 370, 567–571 (1994). [DOI] [PubMed] [Google Scholar]
- 38.Langston A. W., Thompson J. R., Gudas L. J., Retinoic acid-responsive enhancers located 3′ of the Hox A and Hox B homeobox gene clusters. Functional analysis. J. Biol. Chem. 272, 2167–2175 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Medina-Martinez O., Bradley A., Ramirez-Solis R., A large targeted deletion of Hoxb1-Hoxb9 produces a series of single-segment anterior homeotic transformations. Dev. Biol. 222, 71–83 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Frum T., Halbisen M. A., Wang C., Amiri H., Robson P., Ralston A., Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev. Cell 25, 610–622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stirparo G. G., Kurowski A., Yanagida A., Bates L. E., Strawbridge S. E., Hladkou S., Stuart H. T., Boroviak T. E., Silva J. C. R., Nichols J., OCT4 induces embryonic pluripotency via STAT3 signaling and metabolic mechanisms. Proc. Natl. Acad. Sci. U.S.A. 118, e2008890118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radzisheuskaya A., le Bin Chia G., dos Santos R. L., Theunissen T. W., Castro L. F. C., Nichols J., Silva J. C. R., A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat. Cell Biol. 15, 579–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zalc A., Sinha R., Gulati G. S., Wesche D. J., Daszczuk P., Swigut T., Weissman I. L., Wysocka J., Reactivation of the pluripotency program precedes formation of the cranial neural crest. Science 371, eabb4776 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Kumar B., Parker H. J., Parrish M. E., Lange J. J., Slaughter B. D., Unruh J. R., Paulson A., Krumlauf R., Dynamic regulation of Nanog and stem cell-signaling pathways by Hoxa1 during early neuro-ectodermal differentiation of ES cells. Proc. Natl. Acad. Sci. U.S.A. 114, 5838–5845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeap L. S., Hayashi K., Surani M. A., ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin 2, 12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan P., Han J., Guo G., Orlov Y. L., Huss M., Loh Y. H., Yaw L. P., Robson P., Lim B., Ng H. H., Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 23, 2507–2520 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M. M., Choudhary J., An expanded Oct4 interaction network: Implications for stem cell biology, development, and disease. Cell Stem Cell 6, 382–395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ang Y. S., Tsai S. Y., Lee D. F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B., Wang J., Rendl M., Bernstein E., Schaniel C., Lemischka I. R., Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183–197 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.King H. W., Klose R. J., The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife 6, e22631 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buitrago-Delgado E., Nordin K., Rao A., Geary L., LaBonne C., Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells. Science 348, 1332–1335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soufi A., Donahue G., Zaret K. S., Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.A. Nagy, M. Gertsensten, K. Vintersten, R. Behringer, Manipulating the Mouse Embryo: A Laboratory Manual (CSHL Press, ed. 3, 2003). [Google Scholar]
- 53.Huangda W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Schmittgen T. D., Livak K. J., Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Ariza-McNaughton L., Krumlauf R., Non-radioactive in situ hybridization: Simplified procedures for use in whole-mounts of mouse and chick embryos. Int. Rev. Neurobiol. 47, 239–250 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Forlani S., Lawson K. A., Deschamps J., Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development 130, 3807–3819 (2003). [DOI] [PubMed] [Google Scholar]
- 57.van de Werken H. J., Landan G., Holwerda S. J. B., Hoichman M., Klous P., Chachik R., Splinter E., Valdes-Quezada C., Öz Y., Bouwman B. A. M., Verstegen M. J. A. M., de Wit E., Tanay A., de Laat W., Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods 9, 969–972 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Klein F. A., Pakozdi T., Anders S., Ghavi-Helm Y., Furlong E. E. M., Huber W., FourCSeq: Analysis of 4C sequencing data. Bioinformatics 31, 3085–3091 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Handoko L., Xu H., Li G., Ngan C. Y., Chew E., Schnapp M., Lee C. W. H., Ye C., Ping J. L. H., Mulawadi F., Wong E., Sheng J., Zhang Y., Poh T., Chan C. S., Kunarso G., Shahab A., Bourque G., Cacheux-Rataboul V., Sung W. K., Ruan Y., Wei C. L., CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V. V., Ren B., A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Data files S1 to S4