Abstract
Climate change is gradual, but it can also cause brief extreme heat waves that can exceed the upper thermal limit of any one organism. To study the evolutionary potential of upper thermal tolerance, we evolved the cold-adapted Antarctic bacterium Pseudoalteromonas haloplanktis to survive at 30°C, beyond its ancestral thermal limit. This high-temperature adaptation occurred rapidly and in multiple populations. It involved genomic changes that occurred in a highly parallel fashion and mitigated the effects of protein misfolding. However, it also confronted a physiological limit, because populations failed to grow beyond 30°C. Our experiments aimed to facilitate evolutionary rescue by using a small organism with large populations living at temperatures several degrees below their upper thermal limit. Larger organisms with smaller populations and living at temperatures closer to their upper thermal tolerances are even more likely to go extinct during extreme heat waves.
The evolutionary potential of upper thermal tolerance encounters a physiological limit mediated by protein misfolding.
INTRODUCTION
Organismal populations experience incessant environmental changes, especially in light of ongoing global warming. Average global temperatures have already increased by 1°C since the preindustrial era (1) and are predicted to increase an additional 0.4° to 4.8°C in the 21st century (2). Moreover, climate change is associated with an increase in the frequency and length of heat waves (3, 4), including marine heat waves. The most important marine heat waves up to date lasted between 10 and 380 days and showed maximum intensities between 3.5 and 9.5°C above sea surface temperatures (5). The frequency of marine heat waves has already increased more than 20 times because of ongoing climate change. These heat waves are predicted to increase even more in frequency, length, and intensity if warming continues, especially if global temperatures rise by 3°C or more (6). As a result, the upper thermal limit of organismal growth is expected to experience increased selective pressure (7). It is not known whether adaptive evolution can rapidly extend upper thermal limits (7, 8) to respond to heat waves, but hard physiological boundaries may limit their extension (9, 10). These boundaries may be caused by the effect that temperature has on organisms, especially on protein denaturation and membrane fluidity, which single-step adaptive mutations may not be able to overcome.
When exposed to severe environmental stress, populations can avoid extinction through evolutionary adaptation, a phenomenon known as evolutionary rescue (11). The success of evolutionary rescue depends on the severity of the stressor, on the speed of its onset, and on population size (11–17). Successful rescue is more likely in large populations facing gradual change (13–16). Temperature changes are important stressors, because temperature affects how organisms grow and reproduce, how fast their biochemical reactions proceed, and how well their proteins function (18). For example, above a temperature threshold called the melting temperature, proteins lose their tertiary structure and their ability to function, which is why the biophysics of protein folding limits the temperature range in which organisms can survive (19). Thermostable proteins tend to be enriched in charged and hydrophobic residues, the latter of which preferentially occur in protein cores to reduce solvent exposure (20). Adaptation to high temperatures has been studied in many organisms, both in the wild, for example, in water fleas and cornflowers, and in the laboratory, for example, by experimentally evolving fruit flies, fish, and the bacterium Escherichia coli (1, 10, 18, 21–29). Bacteria have large population sizes, short generation times, and small genomes, which make them ideal candidates to study rapid adaptation to high temperature and its genomic basis. Adaptive evolution allows E. coli to improve survival at temperatures that are above its optimal growth temperature (28, 29). It adapts rapidly to high temperatures (27), but the genomic basis of adaptations to temperatures that exceed its ancestral upper thermal limit—and their eventual failure—is unknown (30).
To study the evolutionary potential of upper thermal tolerance, identify its limits, and characterize their genomic basis, we used a cold-adapted (psychrophilic) bacterium. Organisms from cold-adapted environments are understudied but ecologically important. They produce the largest fraction of biomass on Earth and are thus central to global biogeochemical cycles (31). Cold-adapted bacteria have a larger thermal safety margin than mesophiles, that is, they live at temperatures that are several degrees below their upper thermal tolerance. In addition, while the fastest growth of mesophiles is close to their environmental temperature, cold-adapted bacteria grow faster at temperatures that are far beyond the temperature in their natural habitat (32). Cold-adapted bacteria might thus adapt more rapidly to high temperatures than mesophiles.
RESULTS
Adapting a cold-adapted bacterium to high temperatures
The Antarctic γ-proteobacterium Pseudoalteormonas haloplanktis strain TAC125 is one of the best-studied cold-adapted bacteria (33). Its genome consists of two chromosomes and two plasmids (34–36). It is able to grow between −2.5° and 29°C, but it already shows signs of heat stress above 20°C (37, 38). Because P. haloplanktis is a natural isolate from Antarctic coastal seas with limited exposure to laboratory conditions (34), we first adapted it to these conditions for 145 generations at 15°C. We then started all further experiments from the preadapted clone with the highest growth increase relative to the wild-type P. haloplanktis TAC125 strain. The likely reason for the clone’s growth increase is a duplication of its entire chromosome 2, which encodes a gluconate transporter and a gluconokinase, both involved in the transport and metabolism of d-gluconic acid, the sole carbon source in our growth medium (table S1).
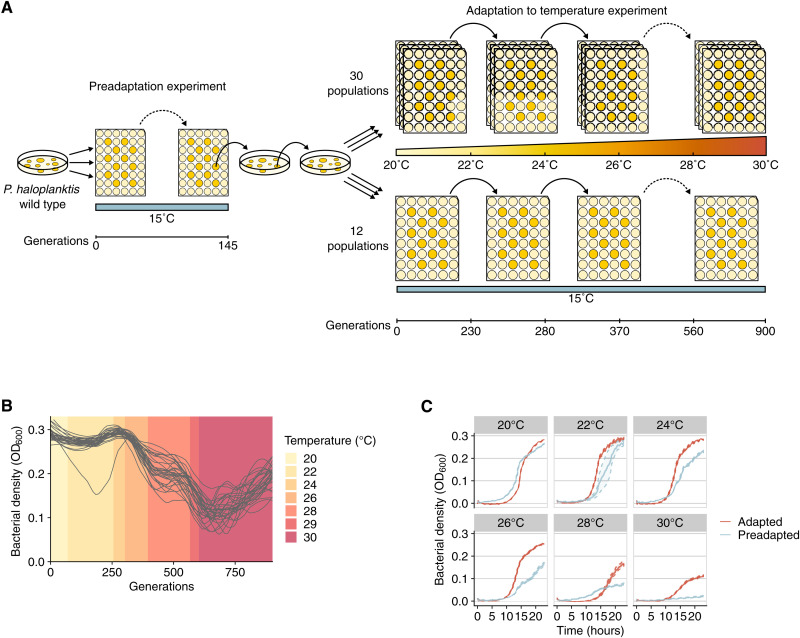
Starting from this preadapted clone, we challenged populations of P. haloplanktis to adapt to gradually increasing temperatures that eventually surpassed the upper thermal limit of our wild-type strain TAC125, resembling a heat wave. Briefly, we evolved 30 replicate populations by serial transfer in batch culture, gradually increasing the temperature from 20° to 22°, 24°, 26°, 28°, 29°, and 30°C. In parallel, we evolved 12 replicate control populations at a constant temperature of 15°C, the wild-type optimal growth temperature (Fig. 1A). At 30°C, i.e., beyond the temperature limit of the wild type and the preadapted clone (fig. S1), population size decreased considerably at first (by 26% on average compared to growth at 29°C and by 37% compared to growth at 28°C). However, population size recovered in all populations. Recovery showed the typical U-shaped curve of evolutionary rescue, where a dip in population size is followed by an increase (Fig. 1B). This rescue was very rapid. Depending on the population, it required only between 70 and 270 generations (fig. S2). In summary, after 900 generations of evolution at gradually increasing temperatures, all 30 replicate populations were able to grow at a temperature beyond the ancestor’s thermal limit (Fig. 1C and fig. S3). Adaptation to 30°C did not involve, in general, trade-offs with growth at lower temperatures (fig. S4 and Supplementary Text), but trade-offs at the level of individual mutations might exist.
Fig. 1. Adaptation to gradual temperature increases.
(A) Experimental design. We evolved 30 replicate populations started from a single preadapted clone during 900 generations at gradually increasing temperatures. To determine the genetic basis of adaptation to rising temperatures, we sequenced the genomes of clones isolated throughout the experiment. (B) Bacterial density, quantified by the optical density of a bacterial culture at 600 nm (OD600) as a function of time, for each of 30 evolving populations. Each line corresponds to data from one population. Background color (see legend) indicates the temperature at which we measured bacterial density. (C) Fitness gains increase with temperature. Growth curves measured during 23 hours at different temperatures (see panel labels) using the initial preadapted clone (blue line) or populations adapted to each temperature (red line). The growth curve for the preadapted clone is the average of 12 biological replicates, and the growth curve for the evolved populations is the average of 90 biological replicates (three replicate population samples for each of 30 evolving populations). Shaded areas and dashed lines indicate 95% confidence intervals.
Fitness increased throughout the experiment, but the highest relative increase occurred at the highest temperatures (Fig. 1C), which is consistent with previous experiments (27). However, even after 300 generations of evolution at 30°C, our populations failed to grow at higher temperatures. Less than half of the populations grew to very low optical density at 31°C, and no population could grow at 32°C (fig. S5). To confirm these observations, we revived end point populations and, after an acclimation period at 30°C, grew them at 31°C for two serial transfers. In these experiments, end point populations could not grow at 31°C, even if we performed serial transfer every 48 hours instead of every 24 hours. While populations incubated (and nongrowing) at 31°C resumed growth after incubation at 15°C, populations incubated at 32°C did not resume growth at 15°C. These results suggest that while the upper thermal limit for growth lies somewhere between 30° and 31°C, the upper thermal limit for survival lies between 31° and 32°C. In summary, we uncovered a limit, within the context of our experiments, for evolutionary rescue in this bacterium.
Genetic changes implicated in temperature adaptation
To identify mutations associated with thermal adaptation, we sequenced 192 single clones isolated from populations adapted to different temperatures, that is, from different time points during the experiment (22°, 26°, 28°, and 30°C), as well as from the control populations evolved at 15°C. For intermediate time points and control populations, we sequenced one clone per population, and for end point populations adapted to 30°C, we sequenced three clones per population. We note one inherent limitation of our experimental design, namely, that clones isolated from higher temperatures have been evolving for longer and in populations with a higher background genetic variation than clones isolated from lower temperatures.
We identified 940 mutations in the sequenced genomes (data file S1), all of which must have occurred de novo, because we started the experiment from a single clone with known genomic sequence. Genetic drift is negligible on the time scale of our experiment, because our populations comprised at least ~108 individuals. Most observed mutations are thus beneficial or hitchhike to high frequency with beneficial mutations. The most frequent mutations are point mutations (62%), followed by indels (32%) (table S2 and data file S2). Of the 585 point mutations that we identified, 498 (85.1%) are missense variants, and only 20 (3.4%) are synonymous point mutations. Half of the indels cause in-frame insertions and deletions, and only 10% of all mutations cause frameshifts and stop codon gains that might abolish protein function. Only 12% of the mutations fall outside coding regions (table S3).
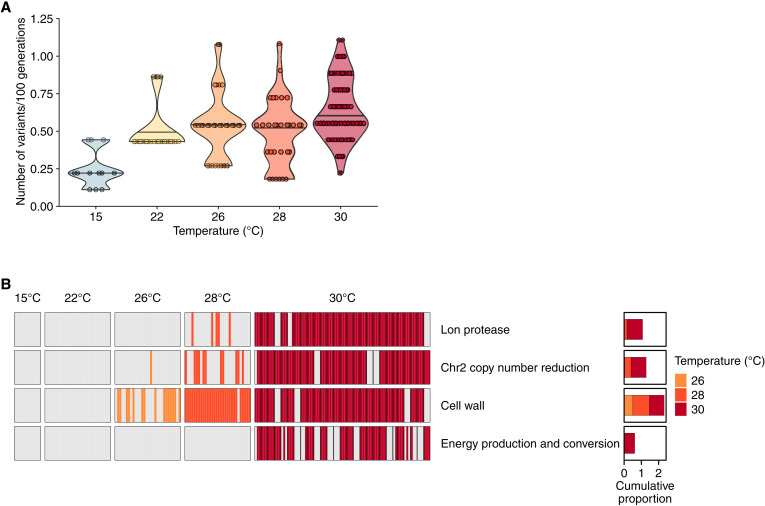
At 30°C, we detected a higher number of new variants per clone than at all other temperatures [Wilcoxon rank sum test, two-tailed false discovery rate (FDR)–adjusted P value of <0.005 in all pairwise comparisons] (Fig. 2A), possibly because selection intensifies as temperature increases. These variants preferentially occur in genes involved in protein turnover and chaperone functions, in cell wall/membrane biogenesis, as well as in energy production and conversion (Fisher test, one-tailed FDR-corrected P value of ≤0.05) (fig. S6, table S4, and Supplementary Text). The two main effects of temperature increases on cell physiology are protein denaturation and the disruption of membrane integrity caused by increased fluidity (39). Mutations in protein turnover and chaperone functions, as well as in cell wall and membrane biogenesis might help to overcome both.
Fig. 2. The number of genetic variants, as well as the incidence of parallel evolution, increases with temperature, and variation accumulates preferentially in some classes of genes.
(A) Ratio of the number of variants identified at each temperature, normalized by the number of generations (×100) elapsed since the preceding temperature. This number of generations equaled 232 generations from 20° to 22°C, 139 generations from 22°to 26°C, 182 generations from 26° to 28°C, and 350 generations from 28° to 30°C. Each data point represents a clone. (B) We clustered mutated genes into functional categories (data file S3), and we indicate for each sequenced clone at each temperature in which gene category mutations occur. The temperature is displayed on the top horizontal axis and the functional gene categories on the right vertical axis. Small colored vertical bars denote each of the sequenced clones. Clones adapted to 30°C are grouped by population. Next to each functional category, a bar plot indicates the proportion of clones that harbor a mutation falling in that category at a given temperature. Only some functional categories are displayed; the full list is displayed in fig. S9.
In addition, at high temperatures, we preferentially observe amino acid substitutions that tend to increase protein stability. These substitutions reduce the content of glycine and alanine, which increase protein flexibility, and they increase the frequency of leucine and isoleucine (both hydrophobic), which help to stabilize protein cores (tables S5 and S6 and Supplementary Text) (40).
Highly parallel mutations are concentrated in few genes and genomic regions
A new genetic variant is especially likely to be adaptive if it occurs multiple times in populations that evolve in parallel. To identify the variants primarily responsible for evolutionary rescue, we next examined the parallel mutations occurring at 30°C in greater detail. The main target of these mutations was the Lon protease: 90% of clones adapted to 30°C harbor mutations in the lon gene. In addition, 87.5% of the clones harbor mutations that decrease the number of copies of chromosome 2 (via two genetic routes; figs. S7 and S8, table S7, and Supplementary Text), and 85% carry mutations in one or several genes involved in cell wall biosynthesis (among others mipA, lapB, bamA, and lpxC). Taken together, all clones adapted to 30°C display at least one change in one of these three categories of variants (Fig. 2B, fig. S9, and Supplementary Text).
Both mutations in lon and mutations that decrease the number of chromosome 2 copies appear at sublethal temperatures (28° and 26°C, respectively) and increase with temperature (Fig. 2B, fig. S9, table S8, and Supplementary Text). Moreover, growth curves measured at different temperatures reveal that the chromosome 2 duplication favors growth below 26°C (fig. S1), most probably because it increases the transport and metabolism of d-gluconic acid (chromosome 2 harbors a gluconate transporter), which is the only carbon source present in our minimal medium. However, at temperatures above 26°C, the duplication reduces growth (fig. S1).
In summary, our analysis strongly suggests that the two most frequent mutations at 30°C, mutations in lon and decreasing chromosome 2 copy number, are driving adaptation to high temperatures and are crucial to rescue populations from extinction. Moreover, our observations validate a prediction from evolutionary rescue theory, namely, that variants favored during sublethal conditions facilitate adaptation to lethal conditions (11).
Protein misfolding limits the extension of upper thermal tolerance
The Lon protease is an ATP (adenosine 5’-triphosphate)-dependent protease that is found in bacteria, Archaea, and Eukaryota (41). It degrades misfolded and mutant proteins, including many regulatory proteins (41). The Lon protease recognizes hydrophobic motifs, which are typically located in a protein’s core and only exposed when the protein is misfolded (41).
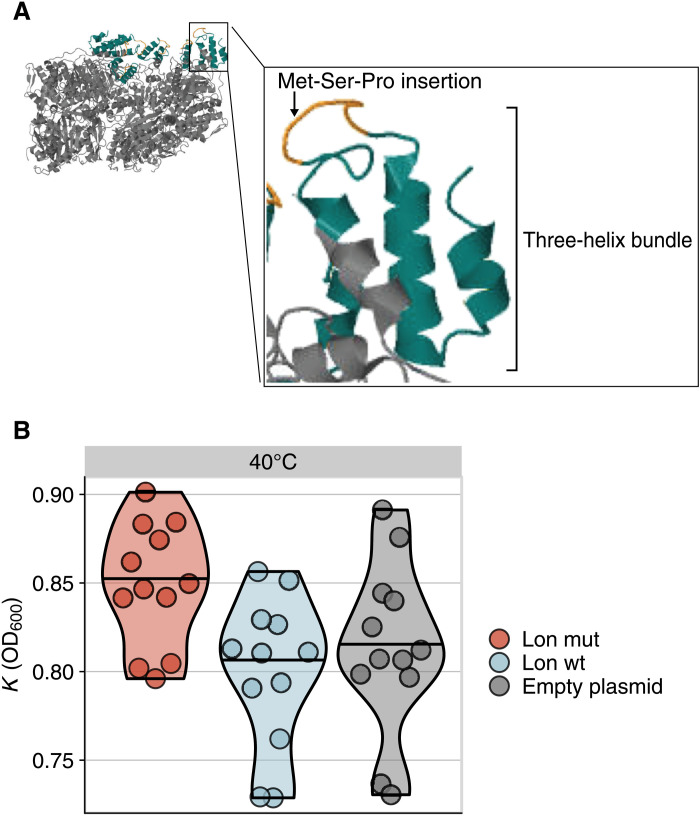
It is remarkable that 90% of clones surviving at 30°C harbor Lon protease mutations but even more remarkable that 72.5% of these clones (58 clones from 20 different populations) share the exact same mutation, a three–amino acid insertion (table S9). We can exclude cross-contamination as a possible origin for this extreme parallelism because we find the insertion in clones isolated from populations located in noncontinuous wells and even on different plates. Furthermore, the amino acid insertion is the only mutation shared among clones. Because our ancestor did not harbor the mutation, these parallel mutations must have occurred independently in our evolving populations. The most plausible explanation is that the insertion, which falls inside a repetitive region (TGC CGA CAT TGG CGA CAT), originated by DNA replication slippage, which led to the addition of a third repeat unit (TGG CGA CAT). An insertion like this might reduce protein expression, but Western blot experiments reveal that this is not the case (fig. S10). The insertion is also not likely to be beneficial by stabilizing the Lon protease, because it localizes to the protein surface. Stability assessments using the FoldX (42) software suggest that the mutation is destabilizing (ΔΔG = 5.523 kcal/mol). The insertion occurs in the N-terminal domain (Fig. 3A and Supplementary Text), which is essential for the binding of Lon to misfolded protein substrates (41).
Fig. 3. Mutations in Lon protease are key for adaptation to high temperature.
(A) Homology modeling of the three–amino acid (Met-Ser-Pro) insertion mutation (orange) of P. haloplanktis Lon protease (amino acids 253 to 775) using SWISS-MODEL and PDB structure 6v11 (https://www.rcsb.org/structure/6V11) as a template. The structure is a homohexamer in which four of the six monomers are bound to ADP (adenosine 5’-diphosphate). The insertion occurs in a region known as the three-helix bundle in the N-terminal domain, which is colored in turquoise. (B) Maximum population density (K) in E. coli BL21 strain carrying an empty plasmid (gray), a plasmid expressing the wild-type Lon protease from P. haloplanktis (blue), or a plasmid expressing the mutated (Met-Ser-Pro insertion) Lon protease from P. haloplanktis (red). We performed 12 biological replicate measurements for each E. coli and plasmid combination (circles). mut, mutant; wt, wild type.
Because P. haloplanktis is not a model organism, tools to engineer specific genes in its genome are limited. To confirm that the Lon protease insertion mutant confers adaptation to high temperature, we thus expressed both the wild-type Lon protease and the insertion mutant in the E. coli strain BL21, which is deficient for Lon protease. Because our experimental results predict that the Lon protease mutant should improve growth, but only at high temperatures, we measured its effect on growth at 30°, 37°, and 40°C. At 37°C, the mutant conferred no advantage, and at 30°C, overexpression of wild-type and mutated Lon protease was disadvantageous (Kneg = 0.740, KLonwt = 0.658, KLonmut = 0.638, Wilcoxon rank sum test, one-tailed P value of <0.01 in all pairwise comparisons). Crucially, at 40°C E. coli grew to significantly higher densities when expressing mutant Lon protease (KLonwt = 0.800, KLonmut = 0.850, one-sided Wilcoxon rank sum test, P = 0.006) (Fig. 3B, figs. S11 to S13, and table S10).
DISCUSSION
Although evolutionary rescue is key to avoid the extinction of populations exposed to environmental stress, it is not well known whether it can rescue populations exposed to temperatures beyond their upper thermal tolerance. Our experiments encountered a limit on evolutionary rescue that exists for our experimental conditions. Specifically, P. haloplanktis populations adapted to grow at 30°C, a temperature that exceeds the upper thermal limit by 1°C, but they could not adapt to survive at higher temperatures.
The mutations that allow P. haloplanktis to survive at 30°C can also help us understand what prevents the further extension of this temperature limit. The most prevalent mutation occurs in the Lon protease, a protein found in all domains of life. The Lon protease is the main protease from the protein quality control mechanism, a mechanism that avoids the accumulation of misfolded proteins by expressing chaperones and proteases (41). The second-most abundant mutation reduces the copy number of chromosome 2. The fitness costs caused by protein misfolding increase with protein abundance (43), thus the reduction in chromosome 2 copy number may provide a fitness benefit because it can help reduce the expression of hundreds of proteins (13.5% of the organism’s genes). Notably, chromosome 2 duplication enhances fitness at low temperatures and reduces fitness only at the highest temperatures. A chromosome 2 duplication has not been reported in natural populations, and its reversion may not be relevant to them, but it is consistent with a key role for protein misfolding in our experiment.
The connection between the thermal limit and protein misfolding is further strengthened by mutations that increase protein stability, such as a gain of leucine and isoleucine residues, and a loss of glycine and alanine residues at the highest temperatures. Considering all the evidence, we hypothesize that adaptation to high temperature reduces the burden of misfolded proteins through mutations in Lon protease, which might increase the degradation of misfolded proteins, or by avoiding protein denaturation through stabilizing mutations. Because this mitigation strategy breaks down beyond 30°C, we conclude that the limit on evolutionary rescue detected within the context of our experiment is imposed by protein misfolding. However, one limitation of our study is that genomic data from clones adapted to 30°C only provide indirect evidence of what limits evolutionary rescue.
What causes the limit on evolutionary rescue remains unknown. One possibility is that the mutations that extend the upper thermal limit are rare or were outcompeted by clones carrying fitter adaptive mutations that do not extend the limit. Another possibility is that there are no single-step adaptive mutations able to overcome the physiological boundaries caused by high temperature. The later would suggest the existence of a hard limit (9, 10). Even hard limits can be overcome with enough time, but this amount of time depends on the necessary number of genetic changes. For example, temperature-induced cell death in E. coli is driven by the denaturation of 83 proteins, 14 of them essential (44). With a genomic mutation rate of 10−3 nucleotide mutations per generation (45), independent mutations in 14 to 83 genes would require very long waiting times, even in large populations. By comparison, during 900 generations of our experiments, P. haloplanktis displayed on average only nine new genetic variants per genome. Even very long waiting times are not prohibitive, however, when environmental change is gradual enough.
Evolutionary rescue is easiest in large populations that become only gradually stressed (13–16). However, even large populations are not immune to extinction (46). In addition, although global climate change is a gradual process, it also leads to abrupt and extreme climatic events, such as heat waves, hurricanes, and droughts (3, 4, 47). Events like this can affect wild populations markedly and have already led to the extinction of some local populations (e.g., bumblebees, corals, flying foxes, and kelp forests) (46–51). For example, a marine heat wave affecting Australia’s Great Southern Reef increased the temperature above the thermal tolerance of kelp (≈2.5°C above maximum sea temperature), causing the loss of kelp forest and its replacement by seaweed turfs (49).
Models predict that if global warming reaches 3.5°C by the end of 21st century, then marine heat waves will increase in length, intensity, and frequency, reaching an average duration of 112 days and 2.5°C over the maximum sea surface temperature (52). The aim of our experiments was to evaluate the evolutionary potential of populations to quickly modify their upper thermal limit, such as in the context of marine heat waves. The upper thermal limit here refers to a thermal reproductive limit, which is expected to be lower than the critical thermal limit, which is a lethality threshold (53). Our experiments were designed to make evolutionary rescue as easy as possible, by using an environment that changes gradually, large populations of a small organism, and an organism that lives at temperatures several degrees below its upper thermal tolerance, which a priori should facilitate adaptation. Even under these favorable conditions, however, we could only extend the upper thermal limit by 1°C, which might not be sufficient according to predictions if global warming reaches 3.5°C. Our observations suggest a low evolvability of upper thermal limits in our study organism, which is consistent with recent results from tropical fish (10). Protein misfolding is a major cause of this limit to evolutionary rescue. Because evolutionary rescue is more difficult in large macroscopic organisms with smaller populations, limits on rescue will be more frequent for these organisms, especially if they live at temperatures close to their thermal tolerance and are affected by extreme climatic events, which can cause extreme temperatures that exceed their thermal tolerance.
MATERIALS AND METHODS
Bacterial strains and growth conditions
We obtained P. haloplanktis strain TAC125 from the Centre de Ressources Biologiques de l’Institute Pasteur (CIP108707). We cultured the strain in minimal marine seawater medium [KH2PO4 (1 g/liter), NH4NO3 (1 g/liter), NaCl (10 g/liter), MgSO4 · 7H2O (0.2 g/liter), FeSO4 · 7H2O (0.01 g/liter), and CaCl2 · 2H2O (0.01 g/liter) (pH = 7)] (37) supplemented with d-gluconic acid (0.1%), at 15°C and 225 rpm of shaking on an incubating shaker (INFORS HT Ecotron). We used E. coli DH5α and E. coli TOP10 for cloning and amplification purposes, growing E. coli cultures in LB medium at 37°C. We cultured recombinant strains in the presence of kanamycin (30 μg/ml).
Preadaptation experiment
P. haloplanktis TAC125 is a natural isolate, and to adapt it to laboratory conditions and to the minimal seawater medium, we performed a short preadaptation experiment. Briefly, we evolved 12 replicate populations starting from a single clone in 48-well plates (P-5ML-48-C, Axygen) in 2 ml of minimal marine seawater medium supplemented with d-gluconic acid (0.1%) at 15°C with shaking at 400 rpm (VWR microplate shaker) in an incubating shaker (INFORS HT multitron). We diluted each population 100-fold every 48 hours for 15 days and then daily for 15 days, which constitutes approximately 145 generations of evolution.
At the end of the preadaptation experiment, we isolated several single clones and assessed their adaptation to minimal medium measuring growth curves recorded during 48 hours. To do so, we cultured freezer stocks of the preadapted clones overnight, in minimal medium in 48-well plates, at 15°C with shaking at 400 rpm. We then diluted the overnight cultures 100-fold into 200-μl final volume of minimal medium in 96-well plates (TPP 92096) and measured absorbance at 600 nm every 10 min during 48 hours at 20°C using a plate reader (Tecan Infinite Pro F200). We used an in-house made external cooler to cool down the plate reader to 20°C. For each clone, we used the Growthcurver R package (54) to estimate growth parameters, and we determined its relative fitness as the difference in maximum population density between the preadapted clone and the wild-type clone. We chose the clone showing the highest relative fitness for further experiments. We henceforth refer to this clone as the preadapted clone.
Experimental evolution
We evolved 30 replicate populations. We initiated each of these populations from a single colony isolated from the preadapted clone, in 48-well plates in 2 ml of minimal marine seawater medium supplemented with d-gluconic acid (0.1%) and shaking at 400 rpm and 20°C. To reduce the possibility of contamination across wells of the same plate, we arranged the replicate populations in a checkerboard pattern, alternating wells containing bacterial cultures with wells containing only growth medium (marine broth 2216, BD Difco). In addition, we did not use the outer wells of each plate, because we had found that medium evaporation is higher in these wells. As described in detail below, we propagated the 30 replicate populations by daily transfer to a new plate and increased the temperature gradually, starting the experiment at 20°C and ending it at 30°C after 900 generations of evolution. To monitor adaptation to temperature, we assessed the relative fitness of evolving populations every 7 days by measuring growth curves during 23 hours using a plate reader (Tecan Infinte Pro F200). Specifically, on the first day of growth at a given temperature, we diluted each of the evolving populations 100-fold in a final volume of 200 μl into 96-well plates (TPP 92096), in three replicates per evolving population, and we recorded the growth curve of each replicate. After 1 week of daily transfer, we repeated this procedure, but this time, we standardized the cell density of cultures to start the growth curves with the same density as on the first day of growth at the focal temperature. We used the Growthcurver R package (54) to fit the logistic equation to the data and to estimate growth parameters of each population. We calculated relative fitness as the difference in maximum population density between evolving populations and populations on the first day of growth at a given temperature. If more than half of the evolving populations had increased their relative fitness, we increased the temperature by an additional 2°C. If fewer than half of the populations had increased fitness, we maintained the current temperature for one additional week and repeated the whole process thereafter. We could not use competitive fitness assays to estimate fitness because our ancestral clone does not grow at temperatures higher than 29°C.
For daily transfer of cultures during experimental evolution, we used a dilution factor that allowed us to maintain a stable population size for at least half of the replicate populations. Specifically, to avoid diluting populations at a rate that would entail their eventual extinction, we required that populations reach stationary phase close to the transfer time, which we determined by measuring the previously mentioned weekly growth curves. In the course of the experiment, we increased the temperature six times (22°, 24°, 26°, 28°, 29°, and 30°C). Because the populations grew successively more slowly and achieved lower population density at higher temperature, we had to adjust the dilution factor over time. Specifically, we used a dilution factor of 1:100 between 20° and 28°C, 1:50 at 28°C and 29°C, and 1:25 at 30°C. Populations spent approximately the following numbers of generations between transfers: 46 at 20°C, 186 at 22°C, 46 at 24°C, 93 at 26°C, 182 at 28°C, 39 at 29°C, and 311 at 30°C.
In parallel to our 30 main populations, we evolved 12 control populations at a constant temperature of 15°C, in a volume of 2 ml, and at a daily 100-fold dilution. We chose the temperature of 15°C because it is the optimal temperature for P. haloplanktis (34). We archived all evolving populations once per week by preparing glycerol stocks. Specifically, we added 1 ml of cell culture to 500 ml of sterile 60% glycerol and stored the resulting suspension at −80°C.
We used several approaches to control for contamination. First, if we observed growth in a well supposed to contain only medium, we restarted the experiment from glycerol stocks. Second, to control for possible contaminations with E. coli (routinely used in the laboratory), we performed a weekly 100-fold dilution of the evolving populations into 48-well plates containing LB medium (Difco 244620). We incubated the plates at 37°C in a microtiter plate shaker (Stuart Microtiter 51505) at 400 rpm for 24 hours and visually monitored any observable growth. We did not find any instance of such growth, suggesting that contamination with E. coli did not occur. Third, once per month, we performed a polymerase chain reaction (PCR) using 20 μl of culture of each of the evolving population. We used four sets of primers: two primers specific for PSHA_RS07527 and PSHA_RS01895 P. haloplanktis TAC125 genes and two set of primers specific for EG11498 and EG11973 E. coli genes (table S11). In addition, we randomly selected five populations and sequenced the 16S ribosomal RNA gene to confirm that the evolving populations harbored only P. haloplanktis. Fourth, we carefully inspected genome sequence data for evidence of cross-contamination but did not find any such evidence. To do so, we computed a heatmap for each time point (22°, 26°, 28°, and 30°C) using all identified mutations and examined this heatmap for clusters of clones. For any clones that clustered in this heatmap, we identified the mutations they shared. We found that all clusters were caused by single mutations in genes that evolved in a highly parallel fashion (e.g., Lon protease and repA).
Estimating the costs of adaptation to 30°C
To study whether adaptation to 30°C involves trade-offs at 20°C, the temperature we used to start the evolution experiment, we grew clones adapted to 30°C at 20°C. To do so, we started overnight cultures from glycerol stocks for 80 clones adapted to 30°C. We grew these clones overnight in 2 ml of minimal marine seawater medium supplemented with d-gluconic acid (0.1%) and incubated them at 20°C. We then diluted the overnight cultures 100-fold into 200 μl of minimal medium and measured optical density at 600 nm (OD600) every 10 min with a plate reader for 23 hours. We performed five biological replicates per clone. We used an external cooler to cool down the plate reader to 20°C for the purpose of these measurements.
Whole-genome sequencing
To study the genetic basis of adaptation to temperature, we isolated clones from populations adapted to different temperatures (i.e., from different time points during the experiment). Specifically, we selected clones from the last day of growth at 22°, 26°, 28°, and 30°C; these are the four temperatures at which the populations spent the largest number of generations. We used minimal marine seawater agar supplemented with 0.1% d-gluconic acid to isolate single clones from freezer stocks and prepared new frozen stocks for each isolated clone. To confirm a clone’s adaptation to a given temperature, we measured growth curves for each isolated clone. Specifically, we prepared overnight cultures from a glycerol stock of the clone in 2 ml of minimal marine seawater medium supplemented with d-gluconic acid and incubated the culture at the corresponding temperature with shaking. We then diluted the overnight culture into 200-μl final volume of medium in a 96-well plate, following the same protocol as during experimental evolution, and measured its growth curve during 23 hours in a plate reader.
We extracted genomic DNA (gDNA) from overnight cultures of the isolated clones (2 × 109 cells) grown in minimal marine seawater medium supplemented with 0.1% d-gluconic acid. To this end, we used the DNeasy Blood & Tissue Kit (QIAGEN). Briefly, we incubated lysates at 56°C for 1 hour after addition of Proteinase K. Then, we added buffer AL to the lysates and incubated at 70°C for 10 min before adding ethanol. We eluted DNA in 100 μl of EB buffer (QIAGEN). To assess the amount and quality of the extracted gDNA, we used a Qubit Fluorometer dsDNA Broad Range assay (Life Technologies) and a NanoDrop (NanoDrop ND-1000, Thermo Fisher Scientific) spectrophotometer and checked the integrity of DNA on 0.7% agarose gels. We used the same procedure to extract gDNA of E. coli MG1655 for cloning purposes.
Library preparation and sequencing [HiSeq 4000, 150–base pair (bp) paired-end reads] was conducted at the Oxford Genomics Centre. We sequenced one clone per population for populations adapted to 22°, 26°, and 28°C, and three clones per population for populations adapted to 30°C, except for population 13 (from which no clones grew on agar plates) and 25 (where we obtained only two clones due to technical errors). In total, we sequenced 192 clones: 1 P. haloplanktis TAC125 wild-type clone, 1 P. haloplanktis TAC125 preadapted clone, 12 control clones, 30 clones adapted to 22°C, 30 clones adapted to 26°C, 30 clones adapted to 28°C, and 86 clones adapted to 30°C. To analyze the sequencing data, we updated a pipeline that we had previously developed (55). Briefly, in this pipeline, we used FastQC (v.0.11.9) (RRID:SCR_014583) for initial sequence quality control. We then trimmed the reads using Trim Galore (v0.4.5) (RRID:SCR_011847) and used BWA-MEM (0.7.17) (56) to map the filtered reads to the P. haloplanktis TAC125 genome (NC_007481.1, NC_007482.1 (34)), including its two described plasmids pMtBL (AJ224742.1 (35)) and pMEGA (MN400773.1 (36)). Then, we used Picard (v.2.17.11) (RRID:SCR_006525) to remove duplicate reads and GATK (v3.8) (57) for indel realignment. We used Samtools (v1.7) (58) and GATK (v3.8) (57) for variant calling and Pindel (v0.2.5b8) (59) and BreakDancer (v1.1) (60) for structural variant calling. We annotated the identified variants and predicted their functional effect using SnpEff (v4.3T) (61). Last, we used QualiMap (v2.2.1) (62) to assess the quality of the sequencing data. The mean coverage for our samples is 114 reads per genomic site. In parallel, we also used the pipeline breseq (v0.32.1a) (63) to identify the mutations that occurred during our experiment, obtaining essentially the same results as using our in-house pipeline. We identified six hypermutator clones in populations adapted to 30°C. They comprise three clones isolated from population 1 (mutation in mutL gene) and three clones isolated from population 11 (mutation in mutS gene). We discarded these clones from all further analyses. We classified a mutation as intergenic if it is located more than 150 bp upstream the start of a gene.
For some analyses, we only considered mutations that originated independently from each other. For these analyses, we only counted each mutation once, i.e., at the first temperature at which we had identified it. For clones adapted to 30°C, if a mutation occurred in more than one of the three clones isolated from the same population, we only counted it once.
Functional enrichment analysis
To investigate the functional implications of the mutations observed in our experiment, we used the COG [Clusters of Orthologous Groups (64)] categories assigned to P. haloplanktis TAC125 genes, which we downloaded from MicroScope (https://mage.genoscope.cns.fr/microscope/home/index.php). Specifically, for each temperature and COG category, we compared the proportion of mutations assigned to a given COG category at a given temperature to the corresponding proportion of mutations but for all other temperatures taken together. To do so, we built a 2 × 2 contingency table using (i) the number of mutations assigned to the query COG category at the selected temperature, (ii) the number of mutations assigned to other COG categories at the selected temperature, (iii) the number of mutations assigned to the query COG category at all other temperatures, and (iv) the number of mutations assigned to other COG categories at all other temperature. We used this table to perform a one-tailed Fisher test (65) to assess statistical significance. Our null hypothesis was that there were no temperature-dependent differences in the proportion of mutations assigned to a given COG category. We corrected for multiple testing with the Benjamini-Hochberg method (66).
Structural analysis of Lon protease
Because a tertiary structure for P. haloplanktis Lon protease does not exist, we used a homology modeling approach to obtain a tertiary structure for P. haloplanktis wild-type Lon protease, as well as for Lon protease carrying the three–amino acid insertion we had identified. We modeled the tertiary protein structure with two software packages, SWISS-MODEL (67) and I-TASSER (68). In SWISS-MODEL, we used the PDB structure 6v11 of Lon protease from Yersinia pestis (79.12% sequence identity to Lon protease of P. haloplanktis) as a template for homology modeling. We used Jmol (69) to visualize tertiary structures. To assess whether the three–amino acid insertion has an effect on protein stability, we used the FoldX algorithm (42) and the tertiary structure obtained using I-TASSER (68). To this end, we first repaired the tertiary structure using the RepairPDB command in FoldX (42). We then optimized the repaired tertiary structure using the command Optimize. Last, we computed stability using the Stability command. We compared the stability of the Lon wild type to the Lon mutant and computed ΔΔG from this comparison.
Western blots to assess Lon protease expression
To assess protein expression levels of the wild-type and different mutated versions of Lon protease, we carried out Western blot experiments. We harvested 1 ml of bacterial culture by centrifugation (5 min, 5000 rpm at room temperature) and resuspended the pellet in 4× Laemmli sample buffer (LDS) to a final ratio of 100 μl per OD600 of culture. We separated the protein fraction of these cell extracts on an SDS–polyacrylamide gel electrophoresis gel (TruPAGE 4 to 12% precat gels, Sigma-Aldrich) and transferred the proteins to a polyvinylidene fluoride membrane using a Bio-Rad Mini-PROTEAN kit, according to the manufacturer’s protocol. After incubation with 3% nonfat milk in phosphate-buffered saline (PBS) and 0.1% Tween 20 for 1 hour, we washed the membrane once with PBS and 0.1% Tween 20 and incubated overnight at 4°C with antibodies against Lon protease (Biorbyt, 1:5000 dilution). We washed membranes three times with PBS and 0.1% Tween 20 for 10 min each and incubated with a 1:2500 dilution of horseradish peroxidase–conjugated anti-rabbit antibodies (GE Healthcare) for 2 hours at room temperature. Last, we washed blots three times with PBS and 0.1% Tween 20 and developed them with the ECL system (GE Healthcare), according to the manufacturer’s protocols on an LAS 4000 Imager (Fujifilm).
Expression of Lon protease in E. coli
To confirm the key role of mutations in Lon protease for adaptation to high temperatures, we expressed wild-type Lon protease and Lon protease with the most common mutation (three–amino acid insertion) in E. coli. To this end, we used plasmid pUCNOmpA-EYFP-1 (fig. S14), which contains the constitutive promoter ompA for protein expression, a pUC origin, a kanamycin resistance gene, and an EYFP (enhanced yellow fluorescent protein)–coding gene (table S12). Briefly, to construct this plasmid, we replaced the pSC101 origin of plasmid pmss201_ompA (70) with a pUC origin obtained from pBAD202/D-TOPO (Invitrogen, K420201). From the plasmid thus modified, we eliminated the EYFP-coding gene to obtain an empty control plasmid using whole-plasmid PCR (see table S11 for primers). Next, we amplified the Lon-Phwt (wild-type Lon protease) and Lon-Ph14.1 (Lon protease carrying the three–amino acid insertion) genes from gDNA of P.haloplanktis TAC125 wild-type (CIP108707) and P. haloplanktis TAC125 30°C-14.1 clone (clone 1 isolated from population 14 adapted to 30°C), respectively. We then used Gibson PCR (71) to insert these genes into pUCNOmpA under the constitutive promoter OmpA, using the NEBuilderHiFi DNA Assembly Master Mix (New England Biolabs), which resulted in the two plasmids pUCNOmpA-LonPhwt and pUCNOmpA-LonPh14.1 (table S12).
We transformed the three plasmids we had engineered (pUCNOmpA-LonPhwt carrying wild-type Lon protease, pUCNOmpA-LonPh14.1 carrying mutated Lon protease, and the empty plasmid pUCNOmpA) into E. coli BL21 (DE3) electrocompetent cells (Sigma-Aldrich). This E. coli strain lacks the endogenous Lon protease of E. coli. We inoculated a single colony of the transformed bacteria into LB medium supplemented with kanamycin (30 μg/ml) and grew the culture overnight at 37°C and with shaking at 220 rpm. We diluted the culture to an OD600 of 0.005 in a 96-well plate (TPP 92096), with 12 replicates per culture, and monitored bacterial growth by measuring OD600 on a plate reader (Tecan Infinite F200 Pro) for 48 hours. We measured growth curves at 30°, 37°, and 40°C, to investigate how the expression of the different Lon proteases affects the growth of E. coli at different temperatures. We used the Growthcurver R package (54) to estimate the maximum population density for each of the growth curves.
Statistical analyses
We performed all statistical analyses and produced all graphics using R (72).
Acknowledgments
We would like to thank J. van Gestel, J. Payne, Á. San Millán, K. Sprouffske, and J. Zheng for discussions and suggestions on the experimental design. We thank J. Zheng for providing us with the plasmid pUCNOmpA-EYFP-1. We thank L. Mohn for quantitative PCR guidance and Y. Choffat for technical support. We thank the Oxford Genomics Centre at the Wellcome Centre for Human Genetics (funded by Wellcome Trust grant reference 203141/Z/16 /Z) for the generation and initial processing of the sequencing data.
Funding: This work was supported by the Swiss National Science Foundation, Ambizione grant PZ00P3_161545 (to M.T.-R.); the Swiss National Science Foundation, PRIMA grant PR00P3_185899 (to M.T.-R.); the European Research Council under grant agreement no. 739874 (to A.W.); the Swiss National Science Foundation, grant 31003A_172887 (to A.W.); and the University Priority Research Program in Evolutionary Biology at the University of Zurich (to A.W.).
Author contributions: Conceptualization: M.T.-R. and A.W. Experiment design: M.T.-R. Experiment performance: M.T.-R. and M.O. Data analysis: M.T.-R., F.C.-G., and A.W. Visualization: M.T.-R. and F.C.-G. Supervision: M.T.-R. and A.W. Writing—original draft: M.T.-R. and A.W. Writing—review and editing: M.T.-R., M.O., F.C.-G., and A.W.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Sequence reads are available in the European Nucleotide Archive database under the accession code PRJEB50060. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Correspondence and requests for all materials should be addressed to M.T.-R. (mtollriera@gmail.com) or A.W. (andreas.wagner@ieu.uzh.ch).
Supplementary Materials
This PDF file includes:
Supplementary Text
Supplementary Methods
Figs. S1 to S15
Tables S1 to S13
References
Other Supplementary Material for this manuscript includes the following:
Data S1 to S3
REFERENCES AND NOTES
- 1.Scheffers B. R., De Meester L., Bridge T. C. L., Hoffmann A. A., Pandolfi J. M., Corlett R. T., Butchart S. H. M., Pearce-Kelly P., Kovacs K. M., Dudgeon D., Pacifici M., Rondinini C., Foden W. B., Martin T. G., Mora C., Bickford D., Watson J. E. M., The broad footprint of climate change from genes to biomes to people. Science 354, aaf7671 (2016). [DOI] [PubMed] [Google Scholar]
- 2.IPPC, Climate Change 2013: The Physical Science Basis Working Group I Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge Univ. Press, 2013). [Google Scholar]
- 3.Drijfhout S., Bathiany S., Beaulieu C., Brovkin V., Claussen M., Huntingford C., Scheffer M., Sgubin G., Swingedouw D., Catalogue of abrupt shifts in Intergovernmental Panel on Climate Change climate models. Proc. Natl. Acad. Sci. U.S.A. 112, E5777–E5786 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Pol M., Jenouvrier S., Cornelissen J. H. C., Visser M. E., Behavioural, ecological and evolutionary responses to extreme climatic events: Challenges and directions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smale D. A., Wernberg T., Oliver E. C. J., Thomsen M., Harvey B. P., Straub S. C., Burrows M. T., Alexander L. V., Benthuysen J. A., Donat M. G., Feng M., Hobday A. J., Holbrook N. J., Perkins-Kirkpatrick S. E., Scannell H. A., Sen Gupta A., Payne B. L., Moore P. J., Marine heatwaves threaten global biodiversity and the provision of ecosystem services. Nat. Clim. Chang. 9, 306–312 (2019). [Google Scholar]
- 6.Laufkötter C., Zscheischler J., Frölicher T. L., High-impact marine heatwaves attributable to human-induced global warming. Science 369, 1621–1625 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Blackburn S., Van Heerwaarden B., Kellermann V., Sgro C. M., Evolutionary capacity of upper thermal limits: Beyond single trait assessments. J. Exp. Biol. 217, 1918–1924 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Bradshaw W. E., Holzapfel C. M., Evolutionary response to rapid climate change. Science 312, 1477–1478 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Araújo M. B., Ferri-Yáñez F., Bozinovic F., Marquet P. A., Valladares F., Chown S. L., Heat freezes niche evolution. Ecol. Lett. 16, 1206–1219 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Morgan R., Finnøen M. H., Jensen H., Pélabon C., Jutfelt F., Low potential for evolutionary rescue from climate change in a tropical fish. Proc. Natl. Acad. Sci. U.S.A. 117, 33365–33372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell G., Evolutionary rescue and the limits of adaptation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120080 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez A., Bell G., Evolutionary rescue and adaptation to abrupt environmental change depends upon the history of stress. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120079 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell G., Evolutionary rescue. Annu. Rev. Ecol. Evol. Syst. 48, 605–627 (2017). [Google Scholar]
- 14.Bell G., Gonzalez A., Evolutionary rescue can prevent extinction following environmental change. Ecol. Lett. 12, 942–948 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Ramsayer J., Kaltz O., Hochberg M. E., Evolutionary rescue in populations of Pseudomonas fluorescens across an antibiotic gradient. Evol. Appl. 6, 608–616 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samani P., Bell G., Adaptation of experimental yeast populations to stressful conditions in relation to population size. J. Evol. Biol. 23, 791–796 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Lindsey H. A., Gallie J., Taylor S., Kerr B., Evolutionary rescue from extinction is contingent on a lower rate of environmental change. Nature 494, 463–467 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Tenaillon O., Rodríguez-Verdugo A., Gaut R. L., McDonald P., Bennett A. F., Long A. D., Gaut B. S., The molecular diversity of adaptive convergence. Science 335, 457–461 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Elias M., Wieczorek G., Rosenne S., Tawfik D. S., The universality of enzymatic rate-temperature dependency. Trends Biochem. Sci. 39, 1–7 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Rigoldi F., Donini S., Redaelli A., Parisini E., Gautieri A., Review: Engineering of thermostable enzymes for industrial applications. APL Bioeng. 2, 011501 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandberg T. E., Pedersen M., Lacroix R. A., Ebrahim A., Bonde M., Herrgard M. J., Palsson B. O., Sommer M., Feist A. M., Evolution of Escherichia coli to 42°C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol. Biol. Evol. 31, 2647–2662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deatherage D. E., Kepner J. L., Bennett A. F., Lenski R. E., Barrick J. E., Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc. Natl. Acad. Sci. U.S.A. 114, E1904–E1912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann A. A., Sgró C. M., Climate change and evolutionary adaptation. Nature 470, 479–485 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Geerts A. N., Vanoverbeke J., Vanschoenwinkel B., Van Doorslaer W., Feuchtmayr H., Atkinson D., Moss B., Davidson T. A., Sayer C. D., De Meester L., Rapid evolution of thermal tolerance in the water flea Daphnia. Nat. Clim. Chang. 5, 665–668 (2015). [Google Scholar]
- 25.Thomann M., Imbert E., Engstrand R. C., Cheptou P. O., Contemporary evolution of plant reproductive strategies under global change is revealed by stored seeds. J. Evol. Biol. 28, 766–778 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Mallard F., Nolte V., Tobler R., Kapun M., Schlötterer C., A simple genetic basis of adaptation to a novel thermal environment results in complex metabolic rewiring in Drosophila. Genome Biol. 19, 119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett A. F., Dao K. M., Lenski R. E., Rapid evolution in response to high-temperature selection. Nature 346, 79–81 (1990). [DOI] [PubMed] [Google Scholar]
- 28.Bennett A. F., Lenski R. E., Mittler J. E., Evolutionary adaptation to temperature. I. Fitness responses of Escherichia coli to changes in its thermal environment. Evolution 46, 16–30 (1992). [DOI] [PubMed] [Google Scholar]
- 29.Mongold J. A., Bennett A. F., Lenski R. E., Evolutionary adaptation to temperature. IV. Adaptation of Escherichia coli at a niche boundary. Evolution 50, 35–43 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Rudolph B., Gebendorfer K. M., Buchner J., Winter J., Evolution of Escherichia coli for growth at high temperatures. J. Biol. Chem. 285, 19029–19034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siddiqui K. S., Williams T. J., Wilkins D., Yau S., Allen M. A., Brown M. V., Lauro F. M., Cavicchioli R., Psychrophiles. Annu. Rev. Earth Planet. Sci. 41, 87–115 (2013). [Google Scholar]
- 32.Williams T. J., Lauro F. M., Ertan H., Burg D. W., Poljak A., Raftery M. J., Cavicchioli R., Defining the response of a microorganism to temperatures that span its complete growth temperature range (−2°C to 28°C) using multiplex quantitative proteomics. Environ. Microbiol. 13, 2186–2203 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Parrilli E., Tedesco P., Fondi M., Tutino M. L., Lo Giudice A., de Pascale D., Fani R., The art of adapting to extreme environments: The model system Pseudoalteromonas. Phys. Life Rev. 137–161 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Médigue C., Krin E., Pascal G., Barbe V., Bernsel A., Bertin P. N., Cheung F., Cruveiller S., D’Amico S., Duilio A., Fang G., Feller G., Ho C., Mangenot S., Marino G., Nilsson J., Parrilli E., Rocha E. P. C., Rouy Z., Sekowska A., Tutino M. L., Vallenet D., von Heijne G., Danchin A., Coping with cold: The genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15, 1325–1335 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tutino M. L., Duilio A., Parrilli R., Remaut E., Sannia G., Marino G., A novel replication element from an Antarctic plasmid as a tool for the expression of proteins at low temperature. Extremophiles 5, 257–264 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Qi W., Colarusso A., Olombrada M., Parrilli E., Patrignani A., Tutino M. L., Toll-Riera M., New insights on Pseudoalteromonas haloplanktis TAC125 genome organization and benchmarks of genome assembly applications using next and third generation sequencing technologies. Sci. Rep. 9, 16444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sannino F., Giuliani M., Salvatore U., Apuzzo G. A., de Pascale D., Fani R., Fondi M., Marino G., Tutino M. L., Parrilli E., A novel synthetic medium and expression system for subzero growth and recombinant protein production in Pseudoalteromonas haloplanktis TAC125. Appl. Microbiol. Biotechnol. 101, 725–734 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Piette F., D’Amico S., Mazzucchelli G., Danchin A., Leprince P., Feller G., Life in the cold: A proteomic study of cold-repressed proteins in the antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Appl. Environ. Microbiol. 77, 3881–3883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. J. J. Angilletta, Thermal Adaptation: A Theoretical and Empirical Synthesis (Oxford Univ. Press, 2009). [Google Scholar]
- 40.Metpally R. P. R., Reddy B. V. B., Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: Insights into the molecular basis of cold adaptation of proteins. BMC Genomics 10, 11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gur E., The Lon AAA+ protease. Subcell. Biochem. 66, 35–51 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L., The FoldX web server: An online force field. Nucleic Acids Res. 33, W382–W388 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geiler-Samerotte K. A., Dion M. F., Budnik B. A., Wang S. M., Hartl D. L., Drummond D. A., Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 680–685 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leuenberger P., Ganscha S., Kahraman A., Cappelletti V., Boersema P. J., Von Mering C., Claassen M., Picotti P., Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science 355, eaai7825 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Lee H., Popodi E., Tang H., Foster P. L., Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, E2774–E2783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bell G., Collins S., Adaptation, extinction and global change. Evol. Appl. 1, 3–16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grant P. R., Grant B. R., Huey R. B., Johnson M. T. J., Knoll A. H., Schmitt J., Evolution caused by extreme events. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welbergen J. A., Klose S. M., Markus N., Eby P., Climate change and the effects of temperature extremes on Australian flying-foxes. Proc. R. Soc. B Biol. Sci. 275, 419–425 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wernberg T., Bennett S., Babcock R. C., De Bettignies T., Cure K., Depczynski M., Dufois F., Fromont J., Fulton C. J., Hovey R. K., Harvey E. S., Holmes T. H., Kendrick G. A., Radford B., Santana-Garcon J., Saunders B. J., Smale D. A., Thomsen M. S., Tuckett C. A., Tuya F., Vanderklift M. A., Wilson S., Climate-driven regime shift of a temperate marine ecosystem. Science 353, 169–172 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Hughes T. P., Kerry J. T., Baird A. H., Connolly S. R., Dietzel A., Eakin C. M., Heron S. F., Hoey A. S., Hoogenboom M. O., Liu G., McWilliam M. J., Pears R. J., Pratchett M. S., Skirving W. J., Stella J. S., Torda G., Global warming transforms coral reef assemblages. Nature 556, 492–496 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Soroye P., Newbold T., Kerr J., Climate change contributes to widespread declines among bumble bees across continents. Science 367, 685–688 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Frölicher T. L., Fischer E. M., Gruber N., Marine heatwaves under global warming. Nature 560, 360–364 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Walsh B. S., Parratt S. R., Hoffmann A. A., Atkinson D., Snook R. R., Bretman A., Price T. A. R., The impact of climate change on fertility. Trends Ecol. Evol. 34, 249–259 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Sprouffske K., Wagner A., Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17, 172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.San Millan A., Peña-Miller R., Toll-Riera M., Halbert Z. V., McLean A. R., Cooper B. S., MacLean R. C., Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 5, 5208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Depristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., Philippakis A. A., Del Angel G., Rivas M. A., Hanna M., McKenna A., Fennell T. J., Kernytsky A. M., Sivachenko A. Y., Cibulskis K., Gabriel S. B., Altshuler D., Daly M. J., A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye K., Schulz M. H., Long Q., Apweiler R., Ning Z., Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fan X., Abbott T. E., Larson D., Chen K., BreakDancer: Identification of genomic structural variation from paired-end read mapping. Curr. Protoc. Bioinformatics 45, 15.6.1–15.6.11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., Wang L., Land S. J., Lu X., Ruden D. M., A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okonechnikov K., Conesa A., García-Alcalde F., Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32, 292–294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deatherage D. E., Barrick J. E., Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V., The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fisher R. A., On the Interpretation of χ2 from contingency tables, and the calculation of P. J. R. Stat. Soc. 85, 87 (1922). [Google Scholar]
- 66.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 67.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., De Beer T. A. P., Rempfer C., Bordoli L., Lepore R., Schwede T., SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jmol: An open-source Java viewer for chemical structures in 3D; http://jmol.sourceforge.net/.
- 70.Zaslaver A., Bren A., Ronen M., Itzkovitz S., Kikoin I., Shavit S., Liebermeister W., Surette M. G., Alon U., A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3, 623–628 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. III, Smith H. O., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 72.R Core Team, R: A Language and Environment for Statistical Computing (2020); www.r-project.org.
- 73.Feller G., Gerday C., Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 1, 200–208 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Vollmer W., Von Rechenberg M., Höltje J. V., Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274, 6726–6734 (1999). [DOI] [PubMed] [Google Scholar]
- 75.Höltje J.-V., Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62, 181–203 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klein G., Kobylak N., Lindner B., Stupak A., Raina S., Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J. Biol. Chem. 289, 14829–14853 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Corsaro M. M., Lanzetta R., Parrilli E., Parrilli M., Tutino M. L., Ummarino S., Influence of growth temperature on lipid and phosphate contents of surface polysaccharides from the antarctic bacterium Pseudoalteromonas haloplanktis TAC 125. J. Bacteriol. 186, 29–34 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo H., Suzuki T., Rubinstein J. L., Structure of a bacterial atp synthase. eLife 8, e43128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Russell J. B., Cook G. M., Energetics of bacterial growth: Balance of anabolic and catabolic reactions. Microbiol. Rev. 59, 48–62 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharma S. K., De Los Rios P., Christen P., Lustig A., Goloubinoff P., The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat. Chem. Biol. 6, 914–920 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Menon A. S., Waxman L., Goldberg A. L., The energy utilized in protein breakdown by the ATP-dependent protease (La) from Escherichia coli*. J. Biol. Chem. 262, 722–726 (1987). [PubMed] [Google Scholar]
- 82.Lin C. C., Su S. C., Su M. Y., Liang P. H., Feng C. C., Wu S. H., Chang C. I., Structural insights into the allosteric operation of the Lon AAA+ protease. Structure 24, 667–675 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Cooper V. S., Bennett A. F., Lenski R. E., Evolution of thermal dependence of growth rate of Escherichia coli populations during 20,000 generations in a constant environmENT. Evolution 55, 889–896 (2001). [DOI] [PubMed] [Google Scholar]
- 84.Rodríguez-Verdugo A., Carrillo-Cisneros D., Gonzaĺez-González A., Gaut B. S., Bennett A. F., Different tradeoffs result from alternate genetic adaptations to a common environment. Proc. Natl. Acad. Sci. U.S.A. 111, 12121–12126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lenski R. E., Bennett A. F., Evolutionary response of Escherichia coli to thermal stress. Am. Nat. 142, S47–S64 (1993). [DOI] [PubMed] [Google Scholar]
- 86.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J. A. M., Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J., qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, R19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F., Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Supplementary Methods
Figs. S1 to S15
Tables S1 to S13
References
Data S1 to S3