Abstract
Microbial communities are continuously exposed to unpredictable changes in their environment. To thrive in such dynamic habitats, microorganisms have developed the ability to readily switch phenotypes, resulting in a number of differently adapted subpopulations expressing various traits. In evolutionary biology, a particular case of phenotypic heterogeneity that evolved in an unpredictably changing environment has been defined as bet‐hedging. Bet‐hedging is a risk‐spreading strategy where isogenic populations stochastically (randomly) diversify their phenotypes, often resulting in maladapted individuals that suffer lower reproductive success. This fitness trade‐off in a specific environment may have a selective advantage upon the sudden environmental shift. Thus, a bet‐hedging strategy allows populations to persist in very dynamic habitats, but with a particular fitness cost. In recent years, numerous examples of phenotypic heterogeneity in different microorganisms have been observed, some suggesting bet‐hedging. Here, we highlight the latest reports concerning bet‐hedging phenomena in various microorganisms to show how versatile this strategy is within the microbial realms.
This article is categorized under:
Infectious Diseases > Molecular and Cellular Physiology
Keywords: adaptation, bet‐hedging, evolutionary strategy, persisters, phenotypic heterogeneity
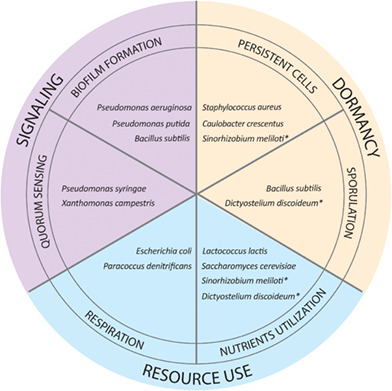
Overview of recent studies on phenotypic heterogeneity and possible employment of bet‐hedging strategies in various microorganisms. In this work, we highlight some recent studies regarding bet‐hedging traits that fall into several categories of microbial lifestyle, including signaling (purple), dormancy (yellow), and resource use (blue). In some cases, the same population manifests different bet‐hedging strategies because of direct or indirect relationships between traits. With an asterisk, we marked examples of studies where a population was shown to employ several bet‐hedging strategies, for example, nutrient utilization is directly involved in spore or persisters formation.
1. INTRODUCTION
In many natural environments, microbial populations are constantly exposed to fluctuations of biotic and abiotic factors. For instance, soil‐inhabiting microorganisms like Bacillus subtilis sense frequent changes of osmolarity caused by interchanging rain and drought periods and accordingly regulate the transport and biosynthesis of osmoprotectants (i.e., proline and glycine betaine) to avoid further cell rupture or desiccation (Bremer & Krämer, 2019). Therefore, microorganisms must evolve various adaptation strategies to sense and process environmental information readily and avoid extinction.
In a direct response to challenging environmental conditions, microorganisms exploit various gene regulatory networks, such as operons and regulons, to modulate their phenotype and/or behavior (Benson & Haldenwang, 1992; Crombach & Hogeweg, 2008; Jacob & Monod, 1961; Krell et al., 2010; Siebring et al., 2012). The capacity to adapt by reversibly switching between different phenotypic states, analogously to ON/OFF switches, is known as phenotypic switching (Henderson et al., 1999; van der Woude & Bäumler, 2004). However, phenotypic switching usually occurs only in a fraction of the population due to the presence of intra‐ and extracellular noise and the topology of regulatory networks involved in the sensing and processing of the environmental signals (i.e., bi‐ or multi‐stable networks; Elowitz et al., 2002; Ozbudak et al., 2002; Paulsson, 2004; Pedraza & van Oudenaarden, 2005; Veening, Smits, et al., 2008). As a result, a nongenetic differentiation within an isogenic population gives rise to several phenotypically distinct subpopulations. This phenomenon is known as phenotypic heterogeneity, and some of its examples include lactose utilization in Escherichia coli (van Hoek & Hogeweg, 2007), cellular differentiation in B. subtilis (Kearns & Losick, 2005; Smits et al., 2005; Veening, Igoshin, et al., 2008; Yüksel et al., 2016), flagellin phase variation in Salmonella enterica (Bonifield & Hughes, 2003), and the development of stress‐resistant yeast (Bishop et al., 2007).
Phenotypic heterogeneity may arise from a responsive event to specific environmental cues (responsive switching), but it can also be the result of random changes in gene expression that are independent of the varying environmental conditions (stochastic switching) (Elowitz et al., 2002; Kussell, 2005; Levine et al., 2013). These strategies have different advantages and disadvantages for populations exposed to certain fluctuating environmental conditions (Figure 1). Since responsive switching strongly depends on the maintenance and the activation of stress‐specific sensory circuits, it causes an adaptive lag that can be critical for survival when the environment fluctuates (Acar et al., 2008; Kaern et al., 2005). In unpredictable environments, stochastic switching can be advantageous over responsive switching by generating a variety of maladapted phenotypes (i.e., phenotypes with reduced fitness), which overall increase the long‐term fitness of the population (Ackermann, 2015; Kussell, 2005; Kussell et al., 2005). This particular form of phenotypic heterogeneity, in which the individuals stochastically express maladapted phenotypes, is known as “bet‐hedging.”
FIGURE 1.
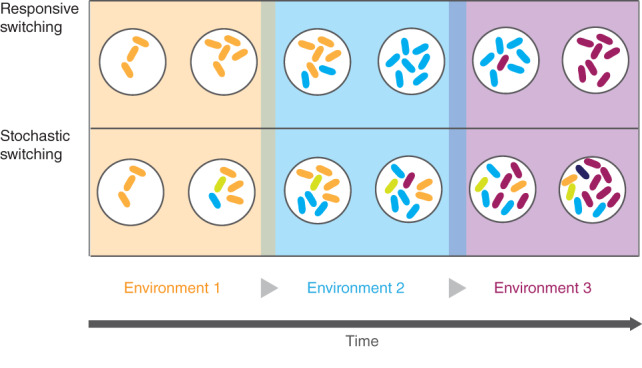
Responsive versus stochastic switching (adapted from Kussell et al., 2005). Isogenic cell populations adapt to changing conditions by switching their phenotype, either responsively (upper panel) or stochastically (lower panel). A schematic representation of the switching strategies is shown, in which the color of the fittest individuals matches the color of the environment. In responsive switching, cells change their phenotype when sensing an environmental change to maximize temporal fitness. The population survives if the majority of the individuals successfully commit to the switch. However, when the environment changes in a stochastic manner, the stochastic switching strategy becomes critical for adaptation. Populations that randomly employ stochastic switching, express a number of maladapted phenotypes of reduced fitness that may suit another environment in the future
In evolutionary biology, bet‐hedging has been described as a risk‐spreading strategy displayed by isogenic populations that explicitly evolved in fluctuating environments (Gillespie, 1974; Kussell, 2005; Philippi & Seger, 1989). Under such conditions, clonal populations stochastically generate phenotypes with different fitness‐related traits, resulting in individuals suffering lower reproductive success. Because the environmental changes favor different phenotypes at different times, the presence of randomly fit individuals may have a selective advantage when the sudden environmental shifts occur. Populations that employ bet‐hedging minimize the temporal fitness variance of surviving offspring and maximize the long‐term geometric mean fitness across generations (de Jong et al., 2011; Grimbergen et al., 2015; Kussell et al., 2005; Seger, 1987; Starrfelt & Kokko, 2012). Importantly, this temporal trade‐off between fitness mean and variance does not occur in any other adaptation strategies.
One of the most prominent and well‐studied cases of how microbes use a bet‐hedging strategy is the formation of persister cells in different bacteria, including E. coli, S. meliloti, or, in more recently reported studies, Staphylococcus aureus and Caulobacter crescentus (Balaban et al., 2004; Huang et al., 2020; Keren et al., 2004; Kussell et al., 2005; Zalis et al., 2019). In this mechanism, under antibiotic stress conditions, a part of the initially isogenic population stochastically and reversibly enters into dormancy and becomes resistant to a killing dose of the antibiotics (Figure 2). Furthermore, once the antibiotic pressure is relieved, the dormant cells can regrow and repopulate the environment. In this regard, the presence of persister cells becomes highly relevant in common antibiotic treatments (Fisher et al., 2017; Lewis, 2007, 2010).
FIGURE 2.
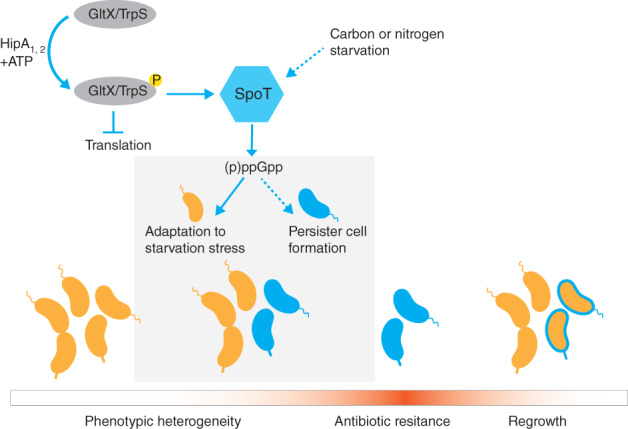
Schematic representation of persister cell formation in Caulobacter crescentus. It has been proposed that antibiotic persistence in C. crescentus is promoted by HipA1 and HipA2 toxins, which are serine/threonine kinases that phosphorylate the aminoacyl‐tRNA synthetases GltX and TrpS, preventing synthesis of charged tRNAs (Huang et al., 2020). Phosphorylation of GltX/TrpS leads to translation arrest and activation of the amino acid starvation‐signaling pathway (SpoT). Activation of SpoT is also stochastically triggered by carbon or nitrogen starvation (indicated by the dashed arrow). Elevated levels of (p)ppGpp, a stringent response alarmone, contribute to translational arrest. Activation of SpoT and further transcriptional changes in the isogenic population of C. crescentus allow most cells to adapt to the starvation conditions, whereas only a fraction of the population becomes dormant (phenotypic heterogeneity). Dormant cells (blue cells) are not metabolically active and can survive high doses of antibiotics. The persister state is reversible; therefore, when the optimal conditions arrive, dormant cells can repopulate the environment (orange cells). The gradient red‐colored bar indicates starvation stress
Theoretical studies have shown that the evolution and success of bet‐hedging strategies depend on many factors, including the environment's reliability, that is, the frequency and magnitude of environmental changes (Gaál et al., 2010; Kussell, 2005); the ability of the population to respond to changes by mutations and rare phenotype selection (King & Masel, 2007; Wolf et al., 2005); the presence of a suitable environmental cue at the time (King & Masel, 2007); and the co‐occurrence of other evolutionary strategies (King & Masel, 2007). Considering all these ecological factors, it is very challenging to provide empirical evidence for true bet‐hedging strategies for several reasons. First, bet‐hedging traits evolve in isogenic populations, and since the microbial genomes are subjected to natural genetic modifications or acquired mutations, the genetic composition of the studied population should be considered in the experimental setup. Second, because adaptive changes usually arise after long periods, and the fitness gains must be quantified across several generations, it is necessary to perform long‐term evolutionary experiments under fluctuating growth conditions.
Consequently, authors of recent reports have discussed six categories of experimental evidence for bet‐hedging strategies (de Jong et al., 2011; Grimbergen et al., 2015; Simons, 2011). Until now, only a few studies have experimentally demonstrated stochastic phenotypic switching in dynamic environments, including the elegant work of Beaumont et al. (2009), which provided the evidence for de novo evolution of bet‐hedging traits in Pseudomonas fluorescens under frequently fluctuating conditions (Beaumont et al., 2009). Here, selection of phenotypes was achieved by repeatedly imposing the exclusion rule and a population bottleneck. Applying both at the point of environmental shift enabled to maintain diversity in the population by excluding the most common phenotype (the exclusion rule) and to select for a random variant among the surviving cells to minimize the cost of bet‐hedging (a bottleneck). Interestingly, the authors showed that in two out of 12 experimental replicates, surviving genotypes persisted due to stochastic phenotype switching. Furthermore, in one of the switching genotypes, they identified a mutation in the carB gene, encoding for a subunit of carbamoylphosphate synthase involved in pyrimidine and arginine biosynthesis. It is speculated that this mutation caused significant changes in the central metabolism of the evolved population that further translated to molecular noise and promoted stochastic phenotype switching.
This experimental example was later revisited by Libby and Rainey (2011). Here, the authors used mathematical models and simulations to investigate the benefits of stochastic switching in populations of P. fluorescens subjected to the exclusion rule and population bottleneck (Libby & Rainey, 2011). Importantly, they observed that switching populations could invade and even replace the nonswitchers despite the high fitness costs and the frequency with which the switching occurred. Besides, the simulations showed that the results are robust to alterations in switching rate, the fidelity of exclusion, bottleneck size, duration of environmental state, and growth rate.
Both studies demonstrated that the exclusion rules and bottlenecks can shape the adaptation of populations responding to fluctuating conditions and that experimental studies, reinforced with theoretical models, can be a superior strategy in proving the evolution of bet‐hedging. Nonetheless, much more remains to be discovered, including the mechanisms of stochastic switching, which is the most challenging to follow.
This mini‐review discusses the most recent empirical studies where authors claim the role of bet‐hedging in observed phenotypic heterogeneity. Specifically, we focus on describing microbial systems that display a bet‐hedging strategy, but without stressing the six categories of experimental evidence proposed by de Jong et al., 2011, Grimbergen et al., 2015 and Simons, 2011. In Figure 3, we compiled all the examples discussed that fall into diverse microbial lifestyle areas, including signaling, resource use and dormancy.
FIGURE 3.
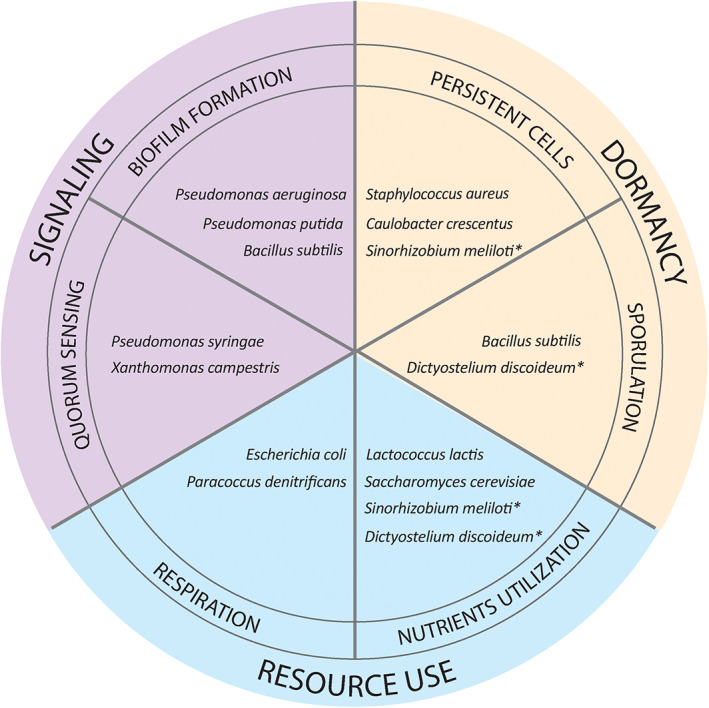
Overview of recent studies on phenotypic heterogeneity and possible employment of bet‐hedging strategies in various microorganisms. In this work, we highlight some recent studies regarding bet‐hedging traits that fall into several categories of microbial lifestyle, including signaling (purple), dormancy (yellow), and resource use (blue). In some cases, the same population manifests different bet‐hedging strategies because of direct or indirect relationships between traits. With an asterisk, we marked examples of studies where a population was shown to employ several bet‐hedging strategies, for example, nutrient utilization is directly involved in spore or persisters formation
2. NOISE IN MICROBIAL CONVERSATIONS: BET‐HEDGING IN QUORUM‐SENSING RESPONSES AND CELL FATE DETERMINATION
It has been long recognized that cells within a microbial community can sense changes in the cell population density and adjust their gene expression accordingly (Nealson et al., 1970). This cell‐to‐cell communication mechanism is known as “quorum sensing” (QS), and is based on the production and sensing of small diffusible molecules known as autoinducers (AIs). AIs trigger cooperative behaviors and the production of “public goods” (Fuqua et al., 1994, 2001; Ng & Bassler, 2009; Williams, 2007). In addition, their chemical nature differs depending on the producing organism. For instance, Gram‐negative bacteria release and sense homoserine lactones (Eberhard et al., 1981), whereas Gram‐positive bacteria produce and sense autoinducing peptides (Kleerebezem et al., 1997; Magnuson et al., 1994). The QS signaling is AI concentration‐dependent, reflecting the number of cells in the population (“quorum”). Accordingly, when the concentration of AIs reaches a threshold, QS‐signaling promotes collective phenotype switching, which is beneficial for the entire population.
Processes reported to be controlled by QS include competence initiation (Håvarstein et al., 1996; Magnuson et al., 1994; Salvadori et al., 2019), endospore formation (Lazazzera et al., 1997; Li et al., 2011; López & Kolter, 2010), bioluminescence (Eberhard et al., 1981; Fuqua et al., 1994; Gray & Garey, 2001), secretion of virulence factors (Ji et al., 1995; Rutherford & Bassler, 2012), bacteriocin production (Brurberg et al., 1997; Kleerebezem, 2004; Kuipers et al., 1998; Shanker & Federle, 2017), biofilm formation (Brindhadevi et al., 2020; Hammer & Bassler, 2003; Kievit et al., 2001; Yarwood et al., 2004), and even changes in the bacteriophage lifecycle (i.e., lysogenic vs. lytic; Harms & Diard, 2019; Høyland‐Kroghsbo et al., 2013; Qin et al., 2017). These experimental works describe QS‐signaling in bulk populations in which the cells at a high density collectively contribute to the common good. Nevertheless, phenotypic heterogeneity is highly present in QS systems, mainly because QS can exploit bistable switches (Bauer et al., 2017). Some examples of phenotypic heterogeneity in QS include the LuxR/LuxI bioluminescence system of Vibrio fischeri and Vibrio harveyi (campbellii) (Anetzberger et al., 2009; Weber & Buceta, 2013), the presence of “social cheaters” in Pseudomonas aeruginosa (Sandoz et al., 2007), and the escape of individual cells from Pseudomonas putida biofilms (Cárcamo‐Oyarce et al., 2015). Other examples of single‐cell studies on QS and phenotypic heterogeneity were collected and neatly presented in the recent reviews by Grote et al. (2015), Mukherjee and Bassler (2019), and Bettenworth et al. (2019).
Interestingly, the occurrence of phenotypic heterogeneity in several cases of QS systems has been suggested to represent a potential bet‐hedging strategy. Pradhan and Chatterjee (2014) used Pseudomonas syringae and Xanthomonas campestris to study bet‐hedging in QS response to AIs (Pradhan & Chatterjee, 2014). To follow the phenotypic variation in response to AIs, they used chromosomally encoded green fluorescent protein reporter fusions, responsive to N‐acyl‐homoserine lactone and diffusible signal factor (cis‐11‐methyl‐2‐dodecenoic acid), respectively. In both experimental conditions, the authors show the coexistence of two subpopulations: QS‐responsive and nonresponsive cells (bright and dark accordingly), at high cell‐density in the presence of external QS signals. Moreover, this phenotypic heterogeneity has been identified as a nonheritable and reversible stochastic event, similar to persister cell formation when antibiotic pressure is removed. Induced and uninduced cells could further differentiate to QS‐responsive or nonresponsive subpopulations, independently of their progeny. Since QS signaling is involved in coordinating multiple social behaviors, like motility or biofilm formation, the authors propose that the production of QS nonresponsive cells would be advantageous under rapidly changing conditions. However, despite the analogy of persister cells' formation, we suggest that in future studies long‐term experiments and fitness estimations are necessary to prove that these mechanisms indeed evolved as a bet‐hedging strategy.
2.1. Spore formation in B. subtilis
In B. subtilis, the production of highly resistant endospores in response to nutritional stress is a prevalent example of an evolved bet‐hedging strategy (Siebring et al., 2014; Veening et al., 2005). Spore formation is regulated by a multicomponent phosphorelay, in which the final threshold concentration of phosphorylated Spo0A, the key sporulation transcription factor, triggers spore‐related gene expression (Fujita et al., 2005; Perego & Hoch, 2014). The Spo0A phosphorylation and thus sporulation initiation depends on several other signaling systems, including RapA‐PhrA, a QS duo that responds to starvation stress (Perego & Hoch, 2014),and the two‐component signal transduction system ComA‐ComP, which indicates the cell density of the population and activates the RapA phosphatase (Lazazzera et al., 1997, 1999; Mueller et al., 1992). Remarkably, noise in the expression of the RapA‐PhrA system has been reported as a source of heterogeneity in sporulation in B. subtilis (Veening et al., 2005). Previous studies demonstrated that cells with a low transcription of rapA sporulate early, whereas cells that strongly upregulate rapA delay their entry to sporulation (Bischofs et al., 2009). Later, Mutlu et al. (2018) showed that RapA‐PhrA determines not only the sporulation timing, but also impacts the spore yield and the spore revival frequency (Mutlu et al., 2018). This phenotypic spore memory that differentiates spores into high and low quality is proposed to be a bet‐hedging strategy to overcome the uncertainty of the environment, where the concentration of nutrients fluctuate. Early spores germinate more efficiently in an environment with low nutrient concentration than late spores, resulting in a fitness benefit under starvation periods. This hypothesis has been further validated theoretically and experimentally by Mutlu et al. (2020). In this work, the authors studied spore revival of B. subtilis strains isolated from gut and soil upon nutrient supply to determine the fitness advantage of different sporulation strategies, emphasizing spore quantity versus quality, and their evolutionary course (Mutlu et al., 2020).
2.2. Sporulation and biofilm formation interplay
Cell fate determination in B. subtilis largely depends on bistable regulatory switches due to their noisy nature, hence a broad range of different phenotypes can be observed within an isogenic population. It has been long recognized that the B. subtilis biofilms are multicellular communities comprising motile, matrix‐producing, and sporulating cells (Vlamakis et al., 2008). Cell differentiation is known to be tightly regulated by the levels of the phosphorylated Spo0A and depending on the intracellular concentration of Spo0A~P, a cell commits either to sporulation (high levels of Spo0A~P) or biofilm matrix formation (intermediate levels of Spo0A~P). In the case of biofilm development, the presence of an adequate concentration of Spo0A~P is essential for the production of SinI. SinI is a direct antagonist of transcription repressor SinR, which allows for transcription of epsA and tasA operons required for polysaccharide synthesis and production of amyloid fibers, by titrating SinR levels. Moreover, the levels of Spo0A~P and cell fate determination can be indirectly affected by other regulatory systems like DegU (responsible for floating biofilm formation) and the QS systems.
In B. subtilis, the ComX signaling peptide, a component of the ComA‐ComP QS system, has been shown to positively regulate transcription of the epsA‐O operon and polysaccharide synthesis by increasing surfactin production. Subsequently, an elevated surfactin concentration activates KinC, a histidine kinase which increases Spo0A~P levels, promoting biofilm formation. In the recent work of Spacapan et al. (2019), the role of QS in cellular development was investigated. The authors examined the role of ComX crosstalk between sporulation and biofilm formation and showed that the ComA‐ComP QS system in floating biofilms reduces the growth rate and increases early stochastic sporulation, which suggests a potential bet‐hedging strategy (Spacapan et al., 2019). The authors used a QS mutant named ∆comQ, which does not produce active signal peptide ComX, and subsequently followed cell differentiation in floating biofilms to prove their hypothesis. By measuring the colony forming units and the number of spores in floating biofilms, the authors showed that floating biofilm of a wild type had a significantly higher amount of spores than the ∆comQ mutant. Moreover, the analysis of activation of the early sporulation promoter PspoIIQ confirmed that the ∆comQ mutation delays entry into sporulation. Interestingly, when the culture was supplemented with external ComX peptide, the promoter's activity resembled the wild type.
Taken all together, ComX was shown to act as a switch that regulates population growth and promotes early sporulation. In this regard, it has been proposed that early sporulation might benefit B. subtilis survival in fluctuating environments where nutrients are often depleted. This benefit is in line with those observed in the spore quantity versus quality trade‐off described in the previous section. Therefore, it is tempting to speculate that this long‐term fitness benefit is the result of a bet‐hedging strategy.
3. BET‐HEDGING IN RESOURCES UPTAKE AND UTILIZATION
3.1. Bet‐hedging in respiratory mechanisms: a safety valve under anoxic spells
Microorganisms inhabiting microenvironments, where oxygen availability is limited (i.e., soil, sediments, biofilms, or marine environments), have developed respiratory mechanisms to utilize alternative oxidizing agents and ultimately generate energy to grow. Prominent examples are denitrifying bacteria, which use nitrogen oxyanions and oxides as electron acceptors during hypoxic/anoxic conditions (Shapleigh, 2006). Denitrifiers reduce nitrate to N2 via nitrite, NO, and N2O stepwise to avoid the entrapment in anoxia for long periods. For each step of reduction: NAR (NO3 − → NO2 −), NIR (NO2 − → NO), NOR (2NO → N2O), and NOS (N2O → N2), they express a set of reductases, which are encoded by nar, nir, nor, and nos gene clusters, respectively. The functionality and cooperation of all reductases prevent the accumulation of toxic intermediates like nitric oxide and conserve energy under frequent fluctuation in oxygen levels (Bergaust et al., 2008; Sullivan et al., 2013).
In the model denitrifier microorganism Paracoccus denitrificans, it has been observed that all cells in an isogenic population synthesize NOS, while only part of the population synthesizes NIR (Lycus et al., 2018). Lycus et al. (2018) described this phenotypic variation based on the NIR system's stochastic initiation, which eventually becomes autocatalytic via NO production. It is suggested that by employing stochastic initiation of NIR, P. denitrificans can hedge its bets and generate different cell variants under imminent anoxia. In this work, the authors propose that the bet‐hedging strategy in NIR initiation is beneficial for conserving energy when the oxygen levels suddenly increase. In this scenario, the cell does not spend its energy synthesizing a denitrifying proteome (Olaya‐Abril et al., 2018); instead, energy is invested in aerobic respiration (a long‐term fitness benefit). However, the NIR expression, in sudden anoxia might cause a severe fitness loss (decreased growth rates caused by graduate NIR expression). Moreover, the authors provide evidence that NIR is localized and stored at the poles of nongrowing cells, ready to use in the case of a sudden switch to anaerobic respiration. Additionally, they discuss the effect of temperature on synthesizing one of the NIR components, NirS. Because the probability of NirS synthesis and expression of the complete denitrification proteome increased with increasing temperatures the authors suggest that the bet‐hedging strategy might have evolved at low temperatures.
This elegant study highlights not only the evolution of a bet‐hedging population. One can speculate that it also suggests a more profound complexity with a mix of survival strategies, where evolved bet‐hedging crosses with the division of labor. The formation of different cell variants in a population of P. denitrificans is thought to contribute to survival via producing common goods (N2O), cross‐feeding interactions, and scavenging by neighboring cells. In this scenario, the division of labor might have evolved from the previous maladaptation that became beneficial on a generation‐wide scale.
It has been recently suggested that not only denitrifying bacteria play the odds in environments when oxygen is scarce. Carey and Goulian (2019) proposed that the aerobically growing population of E. coli hedge its bets to prepare for the future anoxic spells by noisy expression of the torCAD operon (Carey & Goulian, 2019). The signaling two‐component system TorT/TorS/TorR regulates the torCAD operon and allows E. coli cells to sense and utilize trimethylamine oxide (TMAO) as a final electron acceptor instead of oxygen. Previous studies on torCAD have shown that TorT/TorS/TorR complex is expressed as well under aerobic conditions, however with a high cell‐to‐cell variance of the mean expression values (Ansaldi et al., 2007; Carey & Goulian, 2019; Roggiani & Goulian, 2015). Consequently, the noisy expression generates different phenotypic variants resulting in maladapted cells' subpopulations with lower fitness under aerobic conditions. It has been shown that only cells that highly express the torCAD operon upon sudden anoxia continued to grow in anaerobic conditions, suggesting the bet‐hedging strategy (Carey et al., 2018).
3.2. Bet‐hedging in nutrient utilization
Microorganisms require several nutrients to thrive in the niche they occupy, and previous studies illustrate how some hedge their bets in response to specific environmental conditions (Solopova et al., 2014; van Boxtel et al., 2017; Veening, Smits, et al., 2008). For instance, when a population of the amoeba Dictyostelium discoideum encounters nutrient depletion, two subpopulations are distinguished: aggregators (spores) and nonaggregators (vegetative cells; Wonhee & Gomer, 2011). An elegant theoretical work on the study of each subpopulation under short and long starvation conditions provided evidence of the bet‐hedging strategy underlying this case of phenotypic heterogeneity (Martínez‐García & Tarnita, 2017). This work demonstrates that more spores are formed in the long starvation condition, whereas vegetative cells are predominant in the short starvation condition. Therefore, although one phenotype might do better at specific requirements, bet‐hedging ensures the population's survival.
Similarly, a bet‐hedging mechanism in response to starvation in the bacterium S. meliloti has been described (Ratcliff & Denison, 2010, 2011; Zhang & Rainey, 2010). In this work, under starvation conditions, two different cell types emerge after cell division, cells carrying either low or high concentrations of polyhydroxybutyrate (PHB; Ratcliff et al., 2008). The cell having high PHB concentrations can survive for long‐term starvation periods. Besides, the authors of this work correlate this bet‐hedging strategy with bacterial persistence because the high‐PHB‐containing cells show higher tolerance to the antibiotic ampicillin (Ratcliff & Denison, 2011). Importantly, this study provides evidence of the origins of the state of persistence triggered by a nutrient‐stress and bet‐hedging mechanism. In contrast to high‐PHB‐containing cells, the low‐PHB cells are metabolically active, that is, they are nonpersistent cells, and thus this subpopulation of cells can utilize the nutrients that are available in the environment.
3.3. Bet‐hedging for amino acids utilization
Few studies have investigated bet‐hedging strategies on amino acid utilization. We have previously reported a case of phenotypic heterogeneity on the uptake of the amino acid methionine by Lactococcus lactis, where two subpopulations rely each on a methionine transporter with different affinities for the amino acid (Hernandez‐Valdes et al., 2020). Since one of the subpopulations primarily utilizes a high‐affinity transporter, the cells in this subpopulation are likely fitter in environments with minimal amounts of methionine than the subpopulation that relies on a low‐affinity transporter. Notably, this heterogeneity is apparent at the colony level, and therefore it is a remarkable example of phenotypic switching with a low rate. A further study to confirm whether this heterogeneity is a bet‐hedging strategy is envisioned. The low switching rate offers the possibility to subject each subpopulation to evolution and selection experiments.
Based on the seasonal changes (e.g., light and temperature), the presence of nutrients in the environment fluctuates (Kearns et al., 2016; Mello et al., 2016; Paver & Kent, 2017). A study on Saccharomyces cerevisiae shows that the GAP1 gene is responsible for adapting the population to simultaneous changes in the environment (Møller et al., 2013). Due to a hub‐switch in the GAP1 gene, three genotypes are observed: a chromosomal GAP1 gene, an extrachromosomal circle GAP1 gene, and the GAP1 deletion. GAP1 encodes an amino acid transporter for l‐ and d‐amino acids, but participates in the pathway for stress‐protecting sugar trehalose and participates in biofilm formation (Donaton et al., 2003; Torbensen et al., 2012). Therefore, different finesses are observed for each subpopulation in specific growth conditions. For instance, the GAP1 deletion has lower fitness than the wild type when grown in l‐glutamate as a nitrogen source, but it has a higher fitness when grown in either ammonium or allantoin. Interestingly, the presence of GAP1 in extrachromosomal circles resulted in a higher fitness than the wild type in an evolved population with l‐glutamate as a nitrogen source, probably due to an increase in gene number.
3.4. Bet‐hedging for carbohydrate utilization
In contrast to amino acids, the link between carbon source utilization and bet‐hedging has been extensively described in previous studies (Gasperotti et al., 2020; Grimbergen et al., 2015; Siegal, 2015). The carbon catabolite repression in E. coli explains its preference for glucose and how the presence of this sugar prevents the use of other carbon sources so that only when no glucose is present, the other sugars can be utilized (Kremling et al., 2015). Recent work provides extra insight into the mechanism underlying the transition from glucose to other carbon sources in E. coli (Kotte et al., 2014). Kotte et al. (2014) show that during this carbon source transition, characterized by a lag phase, the population of cells is diversified into growing and nongrowing phenotypes. The subpopulation that uses the secondary available carbon source is responsible for the lag phase, whereas the nongrowing subpopulation enters a dormant state. Moreover, these subpopulations are distinguished by different glycolytic rates during their initial growth on glucose, a cell with a low glycolytic rate becomes the subpopulation that enters dormancy. The authors propose that the cells hedge their bets during their growth on glucose, resembling a conditional bet‐hedging mechanism, where it occurs only after glucose is depleted, and the cells with a high glycolytic rate can cope with the change of carbon source.
Likewise, our group reported the diauxic shift from glucose utilization to cellobiose in L. lactis (Solopova et al., 2014). In this case, the transition between both states is determined by catabolite repression and the stringent response. The cells that utilize cellobiose are cells that switch during the release of catabolite repression and activate the stringent response to enable cellobiose utilization.
An interesting bet‐hedging mechanism related to avoiding glucose repression in S. cerevisiae has been described (Garcia et al., 2016; Jarosz et al., 2014). The yeast S. cerevisiae generally consumes glucose, preferably even when other carbon sources are present (Pinu et al., 2014). Previous studies have reported that some strains switch to a metabolic generalistic type despite their characteristic metabolic specialization, allowing them to consume other carbon sources even when glucose is present. Remarkably, this capacity relies on the inheritance of an altered protein conformation—a prion named [GAR+]. The prion is transduced from mother cells to daughter cells at different rates, depending on the nutrients in the environment, suggesting a bet‐hedging mechanism to cope with the fluctuations of carbon sources. Furthermore, bacteria are able to induce [GAR+] through the production of lactic acid in such a way that strong and weak variants of the prion are observed, depending on the different concentrations of lactic acid (Garcia et al., 2016).
4. DISCUSSION AND OUTLOOK
4.1. Does phenotypic heterogeneity always benefit a population?
An environment where resources, especially nutrients, are constantly changing is a scenario in which microorganisms can develop different phenotypes to increase their survival chances (Childs et al., 2010; van Boxtel et al., 2017) (Figure 4). Although many studies have demonstrated how phenotypically variable bacterial strains are able to thrive in a niche, showing successful outcomes of using a bet‐hedging strategy, few studies have studied the adverse effects of heterogeneity in bacterial phenotypes (Levy, 2016). In this regard, do microorganisms always benefit from displaying heterogeneity?
FIGURE 4.
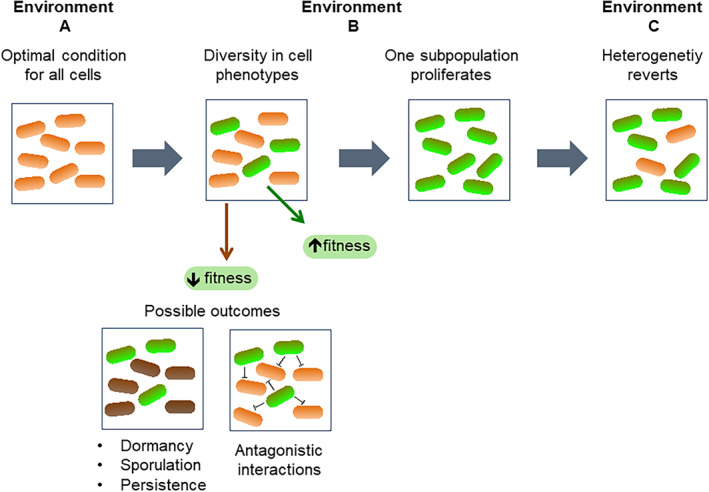
Nutrient fluctuations and cell consequences during a bet‐hedging strategy. Bacteria develop phenotypic heterogeneity during changes in environmental conditions (Environment A to B), and a bet‐hedging strategy results in subpopulations of cells with different fitness. While cells with low fitness are subjected to different outcomes (e.g., sporulation), the fitter cells thrive. Eventually, (change to Environment C) the cells display the diversity in phenotypes by a random switch
Troselj and Wall (2018) investigated the population response of the soil bacterium Myxococcus xanthus when a subpopulation of cells are starving (auxotrophic for amino acids; Troselj et al., 2018; Troselj & Wall, 2018). While no bacterial interaction occurs in a mixed population of prototrophic and auxotrophic cells in rich media, an antagonistic interaction is established in a starvation medium. A toxin produced by the prototrophic cells kills the auxotrophic cells, via the type VI secretion system (T6SS). An analysis of the mechanism underlying this antagonism between sibling cells shows that starving cells have lower amounts of immunity factors than the growing cells. Thus, this observation suggests that growing cells eliminate the less fit cells of the population. Several scenarios have been suggested for the biological implications of this interaction. For instance, it might be that growing cells obtain nutrients from the lysed cells, and therefore, the population becomes fitter without the starving cells, that is, as a homogeneous population. Another possibility is eliminating the nongrowing cells to avoid sporulation, which is a decision at the population level and an energetically expensive process. It would be interesting to explore whether similar antagonistic interactions against the less fit subpopulation are present in some known cases of bet‐hedging strategies by bacteria, where studies have been focused on the exclusive benefits to the subpopulation that is favored under a specific nutritional condition.
Changes in nutrients have revealed bet‐hedging and other phenotypic heterogeneity strategies (Gasperotti et al., 2020; van Boxtel et al., 2017). The importance of this evolutionary adaptation relies on the possibility to persist and thrive in niches where the availability of nutrients fluctuates (Balaban et al., 2004; Martín et al., 2019; Ratcliff & Denison, 2011). Since bacteria live in densely populated environments, and nutrient availability is expected to change, bet‐hedging represents an adaptive evolution strategy that allows bacteria to cope with unstable environmental conditions.
5. CONCLUSIONS
It has been long acknowledged that microbial populations employ bet‐hedging strategies to persist in very dynamic and unpredictable habitats. In the last decades, due to their high relevance in the food industry (e.g., food spoilage caused by germination of persistent spores) and the medical field (e.g., antibiotic‐resistant persister cells), endospore and persister cell formation have been particularly studied in bacteria (Balaban et al., 2004; Lewis, 2007; Veening et al., 2005). Nonetheless, despite the difficulties in identifying bet‐hedging, recent research studies on phenotypic heterogeneity have provided many promising indications of bet‐hedging strategies employed by a plethora of microorganisms. Current studies have shown that bet‐hedging traits may concern many physiological aspects (Figure 3), which provides further evidence of how frequently microorganisms hedge their bets. Yet, more extensive, long‐term evolutionary studies to assess the fitness gains in fluctuating environments are required to confirm the authenticity of bet‐hedging cases described in previous classification reviews (de Jong et al., 2011; Grimbergen et al., 2015; Simons, 2011).
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
AUTHOR CONTRIBUTIONS
Luiza P. Morawska: Conceptualization (equal); investigation (equal); resources (equal); visualization (lead); writing – original draft (lead); writing – review and editing (equal). Jhonatan A. Hernandez‐Valdes: Conceptualization (equal); investigation (equal); resources (equal); visualization (supporting); writing – original draft (supporting); writing – review and editing (equal). Oscar P. Kuipers: Conceptualization (equal); investigation (supporting); supervision (lead); writing – original draft (supporting); writing – review and editing (supporting).
RELATED WIREs ARTICLES
Adaptive resistance to antibiotics in bacteria: a systems biology perspective
ACKNOWLEDGMENT
Luiza P. Morawska was supported by the BE‐Basic RandD Program, which was granted an FES subsidy from the Dutch Ministry of Economic Affairs and Luiza P. Morawska was further supported by a grant from Christian Hansen company, Denmark. Jhonatan A. Hernandez‐Valdes was financially supported by the Netherlands Organization for Scientific Research (NWO), research program TTW (13858).
Morawska, L. P. , Hernandez‐Valdes, J. A. , & Kuipers, O. P. (2022). Diversity of bet‐hedging strategies in microbial communities—Recent cases and insights. WIREs Mechanisms of Disease, 14(2), e1544. 10.1002/wsbm.1544
Edited by: Jessica Lawler, Executive Editor
Funding information BE‐Basic R&D Program; Christian Hansen company, Denmark; the Netherlands Organization for Scientific Research (NWO), Grant/Award Number: 13858
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Acar, M. , Mettetal, J. T. , & van Oudenaarden, A. (2008). Stochastic switching as a survival strategy in fluctuating environments. Nature Genetics, 40(4), 471–475. 10.1038/ng.110 [DOI] [PubMed] [Google Scholar]
- Ackermann, M. (2015). A functional perspective on phenotypic heterogeneity in microorganisms. Nature Reviews Microbiology, 13(8), 497–508. 10.1038/nrmicro3491 [DOI] [PubMed] [Google Scholar]
- Anetzberger, C. , Pirch, T. , & Jung, K. (2009). Heterogeneity in quorum sensing‐regulated bioluminescence of Vibrio harveyi. Molecular Microbiology, 73(2), 267–277. 10.1111/j.1365-2958.2009.06768.x [DOI] [PubMed] [Google Scholar]
- Ansaldi, M. , Théraulaz, L. , Baraquet, C. , Panis, G. , & Méjean, V. (2007). Aerobic TMAO respiration in Escherichia coli . Molecular Microbiology, 66(2), 484–494. 10.1111/j.1365-2958.2007.05936.x [DOI] [PubMed] [Google Scholar]
- Balaban, N. Q. , Merrin, J. , Chait, R. , Kowalik, L. , & Leibler, S. (2004). Bacterial persistence as a phenotypic switch. Science (New York, N.Y.), 305(5690), 1622–1625. 10.1126/science.1099390 [DOI] [PubMed] [Google Scholar]
- Bauer, M. , Knebel, J. , Lechner, M. , Pickl, P. , & Frey, E. (2017). Ecological feedback in quorum‐sensing microbial populations can induce heterogeneous production of autoinducers. eLife, 6, e25773. 10.7554/eLife.25773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont, H. J. E. , Gallie, J. , Kost, C. , Ferguson, G. C. , & Rainey, P. B. (2009). Experimental evolution of bet hedging. Nature, 462(7269), 90–93. 10.1038/nature08504 [DOI] [PubMed] [Google Scholar]
- Benson, A. K. , & Haldenwang, W. G. (1992). Characterization of a regulatory network that controls sigma B expression in Bacillus subtilis . Journal of Bacteriology, 174(3), 749–757. 10.1128/jb.174.3.749-757.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergaust, L. , Shapleigh, J. , Frostegård, Å. , & Bakken, L. (2008). Transcription and activities of NOx reductases in Agrobacterium tumefaciens: The influence of nitrate, nitrite and oxygen availability. Environmental Microbiology, 10(11), 3070–3081. [DOI] [PubMed] [Google Scholar]
- Bettenworth, V., Steinfeld, B., Duin, H., Petersen, K., Streit, W. R., Bischofs, I., & Becker, A. (2019). Phenotypic Heterogeneity in Bacterial Quorum Sensing Systems. Journal of Molecular Biology, 431(23), 4530–4546. 10.1016/j.jmb.2019.04.036 [DOI] [PubMed]
- Bischofs, I. B. , Hug, J. A. , Liu, A. W. , Wolf, D. M. , & Arkin, A. P. (2009). Complexity in bacterial cell‐cell communication: Quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay. Proceedings of the National Academy of Sciences of the United States of America, 106(16), 6459–6464. 10.1073/pnas.0810878106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, A. L. , Rab, F. A. , Sumner, E. R. , & Avery, S. V. (2007). Phenotypic heterogeneity can enhance rare‐cell survival in “stress‐sensitive” yeast populations. Molecular Microbiology, 63(2), 507–520. 10.1111/j.1365-2958.2006.05504.x [DOI] [PubMed] [Google Scholar]
- Bonifield, H. R. , & Hughes, K. T. (2003). Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. Journal of Bacteriology, 185(12), 3567–3574. 10.1128/jb.185.12.3567-3574.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer, E. , & Krämer, R. (2019). Responses of microorganisms to osmotic stress. Annual Review of Microbiology, 73, 313–334. 10.1146/annurev-micro-020518-115504 [DOI] [PubMed] [Google Scholar]
- Brindhadevi, K. , LewisOscar, F. , Mylonakis, E. , Shanmugam, S. , Verma, T. N. , & Pugazhendhi, A. (2020). Biofilm and Quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochemistry, 96, 49–57. 10.1016/j.procbio.2020.06.001 [DOI] [Google Scholar]
- Brurberg, M. B. , Nes, I. F. , & Eijsink, V. G. H. (1997). Pheromone‐induced production of antimicrobial peptides in Lactobacillus. Molecular Microbiology, 26(2), 347–360. 10.1046/j.1365-2958.1997.5821951.x [DOI] [PubMed] [Google Scholar]
- Cárcamo‐Oyarce, G. , Lumjiaktase, P. , Kümmerli, R. , & Eberl, L. (2015). Quorum sensing triggers the stochastic escape of individual cells from Pseudomonas putida biofilms. Nature Communications, 6(1), 5945. 10.1038/ncomms6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey, J. N. , & Goulian, M. (2019). A bacterial signaling system regulates noise to enable bet hedging. Current Genetics, 65(1), 65–70. 10.1007/s00294-018-0856-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey, J. N. , Mettert, E. L. , Roggiani, M. , Myers, K. S. , Kiley, P. J. , & Goulian, M. (2018). Regulated stochasticity in a bacterial signaling network permits tolerance to a rapid environmental change. Cell, 173(1), 196–207. 10.1016/j.cell.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs, D. Z. , Metcalf, C. J. E. , & Rees, M. (2010). Evolutionary bet‐hedging in the real world: Empirical evidence and challenges revealed by plants. Proceedings of the Royal Society B: Biological Sciences, 277(1697), 3055–3064. 10.1098/rspb.2010.0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombach, A. , & Hogeweg, P. (2008). Evolution of evolvability in gene regulatory networks. PLoS Computational Biology, 4(7), e1000112. 10.1371/journal.pcbi.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, I. G. , Haccou, P. , & Kuipers, O. P. (2011). Bet hedging or not? A guide to proper classification of microbial survival strategies. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 33(3), 215–223. 10.1002/bies.201000127 [DOI] [PubMed] [Google Scholar]
- Donaton, M. C. V. , Holsbeeks, I. , Lagatie, O. , Van Zeebroeck, G. , Crauwels, M. , Winderickx, J. , & Thevelein, J. M. (2003). The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Molecular Microbiology, 50(3), 911–929. 10.1046/j.1365-2958.2003.03732.x [DOI] [PubMed] [Google Scholar]
- Eberhard, A. , Burlingame, A. L. , Eberhard, C. , Kenyon, G. L. , Nealson, K. H. , & Oppenheimer, N. J. (1981). Structural identification of autoinducer of photobacterium fischeri luciferase. Biochemistry, 20(9), 2444–2449. 10.1021/bi00512a013 [DOI] [PubMed] [Google Scholar]
- Elowitz, M. B. , Levine, A. J. , Siggia, E. D. , & Swain, P. S. (2002). Stochastic gene expression in a single cell. Science, 297(5584), 1183–1186. 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- Fisher, R. A. , Gollan, B. , & Helaine, S. (2017). Persistent bacterial infections and persister cells. Nature Reviews Microbiology, 15(8), 453–464. 10.1038/nrmicro.2017.42 [DOI] [PubMed] [Google Scholar]
- Fujita, M. , González‐Pastor, J. E. , & Losick, R. (2005). High‐ and low‐threshold genes in the Spo0A regulon of Bacillus subtilis . Journal of Bacteriology, 187(4), 1357–1368. 10.1128/JB.187.4.1357-1368.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua, C. , Parsek, M. R. , & Greenberg, E. P. (2001). Regulation of gene expression by cell‐to‐cell communication: Acyl‐homoserine lactone quorum sensing. Annual Review of Genetics, 35(1), 439–468. 10.1146/annurev.genet.35.102401.090913 [DOI] [PubMed] [Google Scholar]
- Fuqua, W. C. , Winans, S. C. , & Greenberg, E. P. (1994). Quorum sensing in bacteria: The LuxR‐LuxI family of cell density‐responsive transcriptional regulators. Journal of Bacteriology, 176(2), 269–275. 10.1128/jb.176.2.269-275.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaál, B. , Pitchford, J. W. , & Wood, A. J. (2010). Exact results for the evolution of stochastic switching in variable asymmetric environments. Genetics, 184(4), 1113–1119. 10.1534/genetics.109.113431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D. M. , Dietrich, D. , Clardy, J. , & Jarosz, D. F. (2016). A common bacterial metabolite elicits prion‐based bypass of glucose repression. eLife, 5, e17978. 10.7554/eLife.17978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperotti, A. , Brameyer, S. , Fabiani, F. , & Jung, K. (2020). Phenotypic heterogeneity of microbial populations under nutrient limitation. Current Opinion in Biotechnology, 62, 160–167. 10.1016/j.copbio.2019.09.016 [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H. (1974). Natural selection for within‐generation variance in offspring number. Genetics, 76(3), 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, K. M. , & Garey, J. R. (2001). The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology (Reading, England), 147(Pt 8), 2379–2387. 10.1099/00221287-147-8-2379 [DOI] [PubMed] [Google Scholar]
- Grimbergen, A. J. , Siebring, J. , Solopova, A. , & Kuipers, O. P. (2015). Microbial bet‐hedging: The power of being different. Current Opinion in Microbiology, 25, 67–72. 10.1016/j.mib.2015.04.008 [DOI] [PubMed] [Google Scholar]
- Grote, J., Krysciak, D., & Streit, W. R. (2015). Phenotypic Heterogeneity, a Phenomenon That May Explain Why Quorum Sensing Does Not Always Result in Truly Homogenous Cell Behavior. Applied and Environmental Microbiology, 81(16), 5280–5289. 10.1128/AEM.00900-15 [DOI] [PMC free article] [PubMed]
- Hammer, B. K. , & Bassler, B. L. (2003). Quorum sensing controls biofilm formation in Vibrio cholerae. Molecular Microbiology, 50(1), 101–104. 10.1046/j.1365-2958.2003.03688.x [DOI] [PubMed] [Google Scholar]
- Harms, A. , & Diard, M. (2019). Crowd controlled—host quorum sensing drives phage decision. Cell Host & Microbe, 25(2), 179–181. 10.1016/j.chom.2019.01.016 [DOI] [PubMed] [Google Scholar]
- Håvarstein, L. S. , Gaustad, P. , Nes, I. F. , & Morrison, D. A. (1996). Identification of the streptococcal competence‐pheromone receptor. Molecular Microbiology, 21(4), 863–869. 10.1046/j.1365-2958.1996.521416.x [DOI] [PubMed] [Google Scholar]
- Henderson, I. R. , Owen, P. , & Nataro, J. P. (1999). Molecular switches—The ON and OFF of bacterial phase variation. Molecular Microbiology, 33(5), 919–932. 10.1046/j.1365-2958.1999.01555.x [DOI] [PubMed] [Google Scholar]
- Hernandez‐Valdes, J. A. , van Gestel, J. , & Kuipers, O. P. (2020). A riboswitch gives rise to multi‐generational phenotypic heterogeneity in an auxotrophic bacterium. Nature Communications, 11(1), 1203. 10.1038/s41467-020-15017-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyland‐Kroghsbo, N. M. , Mærkedahl, R. B. , & Svenningsen, S. L. (2013). A quorum‐sensing‐induced bacteriophage defense mechanism. MBio, 4(1), e00362–e00312. 10.1128/mBio.00362-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. Y. , Gonzalez‐Lopez, C. , Henry, C. , Mijakovic, I. , & Ryan, K. R. (2020). HipBA toxin‐antitoxin systems mediate persistence in Caulobacter crescentus . Scientific Reports, 10(1), 2865. 10.1038/s41598-020-59283-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, F. , & Monod, J. (1961). Genetic regulatory mechanisms in the synthesis of proteins. Journal of Molecular Biology, 3(3), 318–356. 10.1016/S0022-2836(61)80072-7 [DOI] [PubMed] [Google Scholar]
- Jarosz, D. F. , Lancaster, A. K. , Brown, J. C. S. , & Lindquist, S. (2014). An evolutionarily conserved prion‐like element converts wild fungi from metabolic specialists to generalists. Cell, 158(5), 1072–1082. 10.1016/j.cell.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, G. , Beavis, R. C. , & Novick, R. P. (1995). Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proceedings of the National Academy of Sciences, 92(26), 12055–12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern, M. , Elston, T. C. , Blake, W. J. , & Collins, J. J. (2005). Stochasticity in gene expression: From theories to phenotypes. Nature Reviews. Genetics, 6(6), 451–464. 10.1038/nrg1615 [DOI] [PubMed] [Google Scholar]
- Kearns, D. B. , & Losick, R. (2005). Cell population heterogeneity during growth of Bacillus subtilis . Genes & Development, 19(24), 3083–3094. 10.1101/gad.1373905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, P. J. , Angell, J. H. , Howard, E. M. , Deegan, L. A. , Stanley, R. H. R. , & Bowen, J. L. (2016). Nutrient enrichment induces dormancy and decreases diversity of active bacteria in salt marsh sediments. Nature Communications, 7(1), 12881. 10.1038/ncomms12881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren, I. , Shah, D. , Spoering, A. , Kaldalu, N. , & Lewis, K. (2004). Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli . Journal of Bacteriology, 186(24), 8172–8180. 10.1128/JB.186.24.8172-8180.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kievit, T. R. D. , Gillis, R. , Marx, S. , Brown, C. , & Iglewski, B. H. (2001). Quorum‐sensing genes in pseudomonas aeruginosa biofilms: Their role and expression patterns. Applied and Environmental Microbiology, 67(4), 1865–1873. 10.1128/AEM.67.4.1865-1873.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, O. D. , & Masel, J. (2007). The evolution of bet‐hedging adaptations to rare scenarios. Theoretical Population Biology, 72(4), 560–575. 10.1016/j.tpb.2007.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem, M. (2004). Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides, 25(9), 1405–1414. 10.1016/j.peptides.2003.10.021 [DOI] [PubMed] [Google Scholar]
- Kleerebezem, M. , Quadri, L. E. N. , Kuipers, O. P. , & De Vos, W. M. (1997). Quorum sensing by peptide pheromones and two‐component signal‐transduction systems in Gram‐positive bacteria. Molecular Microbiology, 24(5), 895–904. 10.1046/j.1365-2958.1997.4251782.x [DOI] [PubMed] [Google Scholar]
- Kotte, O. , Volkmer, B. , Radzikowski, J. L. , & Heinemann, M. (2014). Phenotypic bistability in Escherichia coli's central carbon metabolism. Molecular Systems Biology, 10(7), 736. 10.15252/msb.20135022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell, T. , Lacal, J. , Busch, A. , Silva‐Jiménez, H. , Guazzaroni, M.‐E. , & Ramos, J. L. (2010). Bacterial sensor kinases: Diversity in the recognition of environmental signals. Annual Review of Microbiology, 64(1), 539–559. 10.1146/annurev.micro.112408.134054 [DOI] [PubMed] [Google Scholar]
- Kremling, A. , Geiselmann, J. , Ropers, D. , & de Jong, H. (2015). Understanding carbon catabolite repression in Escherichia coli using quantitative models. Trends in Microbiology, 23(2), 99–109. 10.1016/j.tim.2014.11.002 [DOI] [PubMed] [Google Scholar]
- Kuipers, O. P. , de Ruyter, P. G. G. A. , Kleerebezem, M. , & de Vos, W. M. (1998). Quorum sensing‐controlled gene expression in lactic acid bacteria. Journal of Biotechnology, 64(1), 15–21. 10.1016/S0168-1656(98)00100-X [DOI] [Google Scholar]
- Kussell, E. (2005). Phenotypic diversity, population growth, and information in fluctuating environments. Science, 309(5743), 2075–2078. 10.1126/science.1114383 [DOI] [PubMed] [Google Scholar]
- Kussell, E. , Kishony, R. , Balaban, N. Q. , & Leibler, S. (2005). Bacterial persistence: A model of survival in changing environments. Genetics, 169(4), 1807–1814. 10.1534/genetics.104.035352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera, B. A. , Palmer, T. , Quisel, J. , & Grossman, A. D. (1999). Cell density control of gene expression and development in Bacillus subtilis. In G. M. Dunny & S. C. Winans (Eds.), Cell‐cell signaling in bacteria. (pp. 27–46). Washington, DC: American Society for Microbiology Press. [Google Scholar]
- Lazazzera, B. A. , Solomon, J. M. , & Grossman, A. D. (1997). An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis . Cell, 89(6), 917–925. 10.1016/s0092-8674(00)80277-9 [DOI] [PubMed] [Google Scholar]
- Levine, J. H. , Lin, Y. , & Elowitz, M. B. (2013). Functional roles of pulsing in genetic circuits. Science, 342(6163), 1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, S. F. (2016). Cellular heterogeneity: Benefits besides bet‐hedging. Current Biology, 26(9), R355–R357. 10.1016/j.cub.2016.03.034 [DOI] [PubMed] [Google Scholar]
- Lewis, K. (2007). Persister cells, dormancy and infectious disease. Nature Reviews Microbiology, 5(1), 48–56. 10.1038/nrmicro1557 [DOI] [PubMed] [Google Scholar]
- Lewis, K. (2010). Persister cells. Annual Review of Microbiology, 64, 357–372. 10.1146/annurev.micro.112408.134306 [DOI] [PubMed] [Google Scholar]
- Li, J. , Chen, J. , Vidal, J. E. , & McClane, B. A. (2011). The Agr‐like quorum‐sensing system regulates sporulation and production of enterotoxin and beta 2 toxin by clostridium perfringens type A non‐food‐borne human gastrointestinal disease strain F5603. Infection and Immunity, 79(6), 2451–2459. 10.1128/IAI.00169-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby, E. , & Rainey, P. B. (2011). Exclusion rules, bottlenecks and the evolution of stochastic phenotype switching. Proceedings of the Royal Society B: Biological Sciences, 278(1724), 3574–3583. 10.1098/rspb.2011.0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López, D. , & Kolter, R. (2010). Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis . FEMS Microbiology Reviews, 34(2), 134–149. 10.1111/j.1574-6976.2009.00199.x [DOI] [PubMed] [Google Scholar]
- Lycus, P. , Soriano‐Laguna, M. J. , Kjos, M. , Richardson, D. J. , Gates, A. J. , Milligan, D. A. , Frostegård, Å. , Bergaust, L. , & Bakken, L. R. (2018). A bet‐hedging strategy for denitrifying bacteria curtails their release of N2O. Proceedings of the National Academy of Sciences of the United States of America, 115(46), 11820–11825. 10.1073/pnas.1805000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson, R. , Solomon, J. , & Grossman, A. D. (1994). Biochemical and genetic characterization of a competence pheromone from B. subtilis . Cell, 77(2), 207–216. 10.1016/0092-8674(94)90313-1 [DOI] [PubMed] [Google Scholar]
- Martín, P. V. , Muñoz, M. A. , & Pigolotti, S. (2019). Bet‐hedging strategies in expanding populations. PLoS Computational Biology, 15(4), e1006529. 10.1371/journal.pcbi.1006529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐García, R. , & Tarnita, C. E. (2017). Seasonality can induce coexistence of multiple bet‐hedging strategies in dictyostelium discoideum via storage effect. Journal of Theoretical Biology, 426, 104–116. 10.1016/j.jtbi.2017.05.019 [DOI] [PubMed] [Google Scholar]
- Mello, B. L. , Alessi, A. M. , McQueen‐Mason, S. , Bruce, N. C. , & Polikarpov, I. (2016). Nutrient availability shapes the microbial community structure in sugarcane bagasse compost‐derived consortia. Scientific Reports, 6(1), 38781. 10.1038/srep38781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller, H. D. , Andersen, K. S. , & Regenberg, B. (2013). A model for generating several adaptive phenotypes from a single genetic event: Saccharomyces cerevisiae GAP1 as a potential bet‐hedging switch. Communicative & Integrative Biology, 6(3), e23933. 10.4161/cib.23933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. P. , Bukusoglu, G. , & Sonenshein, A. L. (1992). Transcriptional regulation of Bacillus subtilis glucose starvation‐inducible genes: Control of gsiA by the ComP‐ComA signal transduction system. Journal of Bacteriology, 174(13), 4361–4373. 10.1128/jb.174.13.4361-4373.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S., & Bassler, B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nature Reviews Microbiology, 17(6), 371–382. doi: 10.1038/s41579-019-0186-5. [DOI] [PMC free article] [PubMed]
- Mutlu, A. , Kaspar, C. , Becker, N. , & Bischofs, I. B. (2020). A spore quality–quantity tradeoff favors diverse sporulation strategies in Bacillus subtilis . The ISME Journal, 14(11), 2703–2714. 10.1038/s41396-020-0721-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu, A. , Trauth, S. , Ziesack, M. , Nagler, K. , Bergeest, J.‐P. , Rohr, K. , Becker, N. , Höfer, T. , & Bischofs, I. B. (2018). Phenotypic memory in Bacillus subtilis links dormancy entry and exit by a spore quantity‐quality tradeoff. Nature Communications, 9, 69. 10.1038/s41467-017-02477-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson, K. H. , Platt, T. , & Hastings, J. W. (1970). Cellular control of the synthesis and activity of the bacterial luminescent system. Journal of Bacteriology, 104(1), 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, W.‐L. , & Bassler, B. L. (2009). Bacterial quorum‐sensing network architectures. Annual Review of Genetics, 43(1), 197–222. 10.1146/annurev-genet-102108-134304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaya‐Abril, A. , Hidalgo‐Carrillo, J. , Luque‐Almagro, V. M. , Fuentes‐Almagro, C. , Urbano, F. J. , Moreno‐Vivián, C. , Richardson, D. J. , & Roldán, M. D. (2018). Exploring the denitrification proteome of paracoccus denitrificans PD1222. Frontiers in Microbiology, 9, 1137. 10.3389/fmicb.2018.01137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak, E. M. , Thattai, M. , Kurtser, I. , Grossman, A. D. , & van Oudenaarden, A. (2002). Regulation of noise in the expression of a single gene. Nature Genetics, 31(1), 69–73. 10.1038/ng869 [DOI] [PubMed] [Google Scholar]
- Paulsson, J. (2004). Summing up the noise in gene networks. Nature, 427(6973), 415–418. 10.1038/nature02257 [DOI] [PubMed] [Google Scholar]
- Paver, S. F. , & Kent, A. D. (2017). Direct and context‐dependent effects of light, temperature, and phytoplankton shape bacterial community composition. Ecosphere, 8(9), e01948. 10.1002/ecs2.1948 [DOI] [Google Scholar]
- Pedraza, J. M. , & van Oudenaarden, A. (2005). Noise propagation in gene networks. Science, 307(5717), 1965–1969. 10.1126/science.1109090 [DOI] [PubMed] [Google Scholar]
- Perego, M. , & Hoch, J. A. (2014). Two‐component systems, phosphorelays, and regulation of their activities by phosphatases. In Bacillus subtilis and Its Closest Relatives (pp. 473–481). John Wiley & Sons Ltd. 10.1128/9781555817992.ch33 [DOI] [Google Scholar]
- Philippi, T. , & Seger, J. (1989). Hedging one's evolutionary bets, revisited. Trends in Ecology & Evolution, 4(2), 41–44. 10.1016/0169-5347(89)90138-9 [DOI] [PubMed] [Google Scholar]
- Pinu, F. R. , Edwards, P. J. B. , Gardner, R. C. , & Villas‐Boas, S. G. (2014). Nitrogen and carbon assimilation by Saccharomyces cerevisiae during Sauvignon blanc juice fermentation. FEMS Yeast Research, 14(8), 1206–1222. 10.1111/1567-1364.12222 [DOI] [PubMed] [Google Scholar]
- Pradhan, B. B. , & Chatterjee, S. (2014). Reversible non‐genetic phenotypic heterogeneity in bacterial quorum sensing. Molecular Microbiology, 92(3), 557–569. 10.1111/mmi.12575 [DOI] [PubMed] [Google Scholar]
- Qin, X. , Sun, Q. , Yang, B. , Pan, X. , He, Y. , & Yang, H. (2017). Quorum sensing influences phage infection efficiency via affecting cell population and physiological state. Journal of Basic Microbiology, 57(2), 162–170. 10.1002/jobm.201600510 [DOI] [PubMed] [Google Scholar]
- Ratcliff, W. C. , & Denison, R. F. (2010). Individual‐Level Bet Hedging in the Bacterium Sinorhizobium meliloti . Current Biology, 20(19), 1740–1744. 10.1016/j.cub.2010.08.036 [DOI] [PubMed] [Google Scholar]
- Ratcliff, W. C. , & Denison, R. F. (2011). Bacterial persistence and bet hedging in Sinorhizobium meliloti . Communicative & Integrative Biology, 4(1), 98–100. 10.4161/cib.4.1.14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff, W. C. , Kadam, S. V. , & Denison, R. F. (2008). Poly‐3‐hydroxybutyrate (PHB) supports survival and reproduction in starving rhizobia. FEMS Microbiology Ecology, 65(3), 391–399. 10.1111/j.1574-6941.2008.00544.x [DOI] [PubMed] [Google Scholar]
- Roggiani, M. , & Goulian, M. (2015). Oxygen‐dependent cell‐to‐cell variability in the output of the Escherichia coli tor phosphorelay. Journal of Bacteriology, 197(12), 1976–1987. 10.1128/JB.00074-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, S. T. , & Bassler, B. L. (2012). Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harbor Perspectives in Medicine, 2(11), a012427. 10.1101/cshperspect.a012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvadori, G. , Junges, R. , Morrison, D. A. , & Petersen, F. C. (2019). Competence in streptococcus pneumoniae and close commensal relatives: Mechanisms and implications. Frontiers in Cellular and Infection Microbiology, 9, 94. 10.3389/fcimb.2019.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz, K. M. , Mitzimberg, S. M. , & Schuster, M. (2007). Social cheating in Pseudomonas aeruginosa quorum sensing. Proceedings of the National Academy of Sciences, 104(40), 15876–15881. 10.1073/pnas.0705653104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger, J. (1987). What is bet‐hedging? Oxford Surveys in Evolutionary Biology, 4, 182–211. [Google Scholar]
- Shanker, E. , & Federle, M. J. (2017). Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes, 8(1), 15. 10.3390/genes8010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapleigh, J. P. (2006). The denitrifying prokaryotes. In Dworkin M., Falkow S., Rosenberg E., Schleifer K.‐H., & Stackebrandt E. (Eds.), The prokaryotes: Volume 2: Ecophysiology and biochemistry (pp. 769–792). Springer. 10.1007/0-387-30742-7_23 [DOI] [Google Scholar]
- Siebring, J. , Elema, M. J. , Drubi Vega, F. , Kovács, Á. T. , Haccou, P. , & Kuipers, O. P. (2014). Repeated triggering of sporulation in Bacillus subtilis selects against a protein that affects the timing of cell division. The ISME Journal, 8(1), 77–87. 10.1038/ismej.2013.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebring, J. , Sorg, R. A. , Herber, M. , & Kuipers, O. P. (2012). Take it or leave it: Mechanisms underlying bacterial bistable regulatory networks. In Filloux AAM, (ed.), Bacterial Regulatory Networks. (1st ed, pp. 305–332). Caister Academic Press. [Google Scholar]
- Siegal, M. L. (2015). Shifting sugars and shifting paradigms. PLoS Biology, 13(2), e1002068. 10.1371/journal.pbio.1002068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, A. M. (2011). Modes of response to environmental change and the elusive empirical evidence for bet hedging. Proceedings of the Royal Society B: Biological Sciences, 278(1712), 1601–1609. 10.1098/rspb.2011.0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, W. K. , Eschevins, C. C. , Susanna, K. A. , Bron, S. , Kuipers, O. P. , & Hamoen, L. W. (2005). Stripping Bacillus: ComK auto‐stimulation is responsible for the bistable response in competence development: Bistability in competence of B. subtilis . Molecular Microbiology, 56(3), 604–614. 10.1111/j.1365-2958.2005.04488.x [DOI] [PubMed] [Google Scholar]
- Solopova, A. , van Gestel, J. , Weissing, F. J. , Bachmann, H. , Teusink, B. , Kok, J. , & Kuipers, O. P. (2014). Bet‐hedging during bacterial diauxic shift. Proceedings of the National Academy of Sciences, 111(20), 7427–7432. 10.1073/pnas.1320063111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spacapan, M. , Danevčič, T. , Štefanic, P. & Mandic‐Mulec, I. (2019). Quorum sensing in Bacillus subtilis slows down biofilm formation by enabling sporulation bet hedging. BioRxiv, 768671. 10.1101/768671 [DOI]
- Starrfelt, J. , & Kokko, H. (2012). Bet‐hedging‐a triple trade‐off between means, variances and correlations. Biological Reviews, 87(3), 742–755. 10.1111/j.1469-185X.2012.00225.x [DOI] [PubMed] [Google Scholar]
- Sullivan, M. J. , Gates, A. J. , Appia‐Ayme, C. , Rowley, G. , & Richardson, D. J. (2013). Copper control of bacterial nitrous oxide emission and its impact on vitamin B12‐dependent metabolism. Proceedings of the National Academy of Sciences, 110(49), 19926–19931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torbensen, R. , Møller, H. D. , Gresham, D. , Alizadeh, S. , Ochmann, D. , Boles, E. , & Regenberg, B. (2012). Amino acid transporter genes are essential for FLO11‐dependent and FLO11‐independent biofilm formation and invasive growth in Saccharomyces cerevisiae. PLoS One, 7(7), e41272. 10.1371/journal.pone.0041272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troselj, V. , Treuner‐Lange, A. , Søgaard‐Andersen, L. , & Wall, D. (2018). Physiological heterogeneity triggers sibling conflict mediated by the type VI secretion system in an aggregative multicellular bacterium. MBio, 9(1), e01645–e01617. 10.1128/mBio.01645-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troselj, V. , & Wall, D. (2018). Metabolic disharmony and sibling conflict mediated by T6SS. Microbial Cell, 5(5), 256–258. 10.15698/mic2018.05.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boxtel, C. , van Heerden, J. H. , Nordholt, N. , Schmidt, P. , & Bruggeman, F. J. (2017). Taking chances and making mistakes: Non‐genetic phenotypic heterogeneity and its consequences for surviving in dynamic environments. Journal of the Royal Society Interface, 14(132), 20170141. 10.1098/rsif.2017.0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude, M. W. , & Bäumler, A. J. (2004). Phase and antigenic variation in bacteria. Clinical Microbiology Reviews, 17(3), 581–611. 10.1128/CMR.17.3.581-611.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek, M. , & Hogeweg, P. (2007). The effect of stochasticity on the Lac Operon: An evolutionary perspective. PLoS Computational Biology, 3(6), e111. 10.1371/journal.pcbi.0030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening, J.‐W. , Hamoen, L. W. , & Kuipers, O. P. (2005). Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis . Molecular Microbiology, 56(6), 1481–1494. 10.1111/j.1365-2958.2005.04659.x [DOI] [PubMed] [Google Scholar]
- Veening, J.‐W. , Igoshin, O. A. , Eijlander, R. T. , Nijland, R. , Hamoen, L. W. , & Kuipers, O. P. (2008). Transient heterogeneity in extracellular protease production by Bacillus subtilis . Molecular Systems Biology, 4, 184. 10.1038/msb.2008.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening, J.‐W. , Smits, W. K. , & Kuipers, O. P. (2008). Bistability, epigenetics, and bet‐hedging in bacteria. Annual Review of Microbiology, 62(1), 193–210. 10.1146/annurev.micro.62.081307.163002 [DOI] [PubMed] [Google Scholar]
- Vlamakis, H. , Aguilar, C. , Losick, R. , & Kolter, R. (2008). Control of cell fate by the formation of an architecturally complex bacterial community. Genes & Development, 22(7), 945–953. 10.1101/gad.1645008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, M. , & Buceta, J. (2013). Dynamics of the quorum sensing switch: Stochastic and non‐stationary effects. BMC Systems Biology, 7, 6. 10.1186/1752-0509-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, P. (2007). Quorum sensing, communication and cross‐kingdom signalling in the bacterial world. Microbiology (Reading, England), 153(Pt 12), 3923–3938. 10.1099/mic.0.2007/012856-0 [DOI] [PubMed] [Google Scholar]
- Wolf, D. M. , Vazirani, V. V. , & Arkin, A. P. (2005). Diversity in times of adversity: Probabilistic strategies in microbial survival games. Journal of Theoretical Biology, 234(2), 227–253. 10.1016/j.jtbi.2004.11.020 [DOI] [PubMed] [Google Scholar]
- Wonhee, J. , & Gomer, R. H. (2011). Initial cell type choice in dictyostelium. Eukaryotic Cell, 10(2), 150–155. 10.1128/EC.00219-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood, J. M. , Bartels, D. J. , Volper, E. M. , & Greenberg, E. P. (2004). Quorum sensing in Staphylococcus aureus biofilms. Journal of Bacteriology, 186(6), 1838–1850. 10.1128/JB.186.6.1838-1850.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüksel, M. , Power, J. J. , Ribbe, J. , Volkmann, T. , & Maier, B. (2016). Fitness trade‐offs in competence differentiation of Bacillus subtilis . Frontiers in Microbiology, 7, 888. 10.3389/fmicb.2016.00888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalis, E. A. , Nuxoll, A. S. , Manuse, S. , Clair, G. , Radlinski, L. C. , Conlon, B. P. , Adkins, J. , & Lewis, K. (2019). Stochastic variation in expression of the tricarboxylic acid cycle produces persister cells. MBio, 10(5), e01930‐19. 10.1128/mBio.01930-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.‐X. , & Rainey, P. B. (2010). Bet hedging in the underworld. Genome Biology, 11(10), 137. 10.1186/gb-2010-11-10-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


