Abstract
Arthrobacter globiformis D47 was shown to degrade a range of substituted phenylurea herbicides in soil. This strain contained two plasmids of approximately 47 kb (pHRIM620) and 34 kb (pHRIM621). Plasmid-curing experiments produced plasmid-free strains as well as strains containing either the 47- or the 34-kb plasmid. The strains were tested for their ability to degrade diuron, which demonstrated that the degradative genes were located on the 47-kb plasmid. Studies on the growth of these strains indicated that the ability to degrade diuron did not offer a selective advantage to A. globiformis D47 on minimal medium designed to contain the herbicide as a sole carbon source. The location of the genes on a plasmid and a lack of selection would explain why the degradative phenotype, as with many other pesticide-degrading bacteria, can be lost on subculture. A 22-kb EcoRI fragment of plasmid pHRIM620 was expressed in Escherichia coli and enabled cells to degrade diuron. Transposon mutagenesis of this fragment identified one open reading frame that was essential for enzyme activity. A smaller subclone of this gene (2.5 kb) expressed in E. coli coded for the protein that degraded diuron. This gene and its predicted protein sequence showed only a low level of protein identity (25% over ca. 440 amino acids) to other database sequences and was named after the enzyme it encoded, phenylurea hydrolase (puhA gene).
Microbial degradation is the main process affecting the environmental persistence of pesticides (1). Removal of unwanted residues, as well as pesticide efficacy, is ultimately dependent on the presence, numbers, and enzymatic capability of soil microorganisms. However, bioavailability and physical parameters such as pH and soil type strongly influence the rate at which degradation occurs (1). Substituted phenylurea herbicides are used to control weeds in a wide range of crops and in amenity horticulture, but detection of these compounds in drinking water supplies has led to restrictions in their use. Many bacterial and fungal isolates that are able to break down some of these compounds have been reported, but most strains have not been well characterized (34). Cullington and Walker (7) isolated a bacterial strain, named D47, from a soil that had developed the ability to rapidly degrade the herbicide diuron, which is usually relatively persistent in soil. In minimal medium this strain was also able to degrade the herbicides isoproturon, chlortoluron, linuron, and monolinuron. Degradation of all of these compounds was shown to occur through hydrolysis of the urea carbonyl group rather than by the usual route of successive demethylation (7, 36). For diuron an increase in 3,4-dichloroaniline (DCA) concentration equivalent to the loss of parent herbicide was found (7). Therefore, the term degradation in this paper refers only to a partial breakdown of this molecule. Subsequent characterization of strain D47 using 16S rRNA sequence analysis and biochemical tests indicated that it belonged within the Arthrobacter globiformis group (36). Arthrobacter species form part of the gram-positive coryneform bacteria and are considered one of the major groups of aerobic soil bacteria. A number of these isolates have been reported to utilize a wide variety of organic chemicals, including carbamate herbicides and chlorinated biphenyls (29).
Many pesticide degradation genes present in soil bacteria have been shown to reside on plasmids, a common location for other degradation genes (2, 6, 12, 15, 16, 23, 28, 29). Degradation genes have been identified for the pesticides carbofuran (35), atrazine (3, 8), 2,4-dichlorophenoxyacetic acid (2,4-D) (9, 31), and parathion (17, 31, 35). However, no genes involved in the breakdown of substituted phenylurea herbicides have been described. Strain D47 was found to have a broad range of degradative ability within the urea-based herbicides which was stable on subculture and storage at −70°C. This paper reports the isolation and growth characteristics of degradative and nondegradative mutants of A. globiformis D47, the location of the degradative gene(s) on a single plasmid, and the expression and characterization of a single gene capable of degrading diuron.
MATERIALS AND METHODS
Bacterial strains and media.
A. globiformis D47 was isolated from Deep Slade field (Horticulture Research International, Wellesbourne, United Kingdom) using three successive treatments of a soil subsample with diuron and four successive rounds of enrichment in minimal medium containing diuron (7). A. globiformis D47 and mutants derived from it were routinely grown on Luria-Bertani (LB) agar (Merck, Poole, United Kingdom) at 30°C for 2 days and kept at 4°C for up to 1 month. Fresh cultures from a −70°C stock were regularly obtained to ensure maintenance of the degradative phenotype. Diuron was added to medium and buffers using the method of Cullington and Walker (7). Diuron (20-mg ml−1 stock solution prepared in 100% methanol) was added to a sterile Schott bottle, and the methanol was left to evaporate in a laminar flow bench. Minimal salts medium [MSM] (7) or phosphate-buffered saline (PBS) was added aseptically, and the bottle was shaken for 30 min using a wrist action shaker. All Escherichia coli (DH5α or DH10B) clones were routinely grown on LB agar or in LB broth at 37°C with ampicillin (100 μg ml−1), kanamycin (50 μg ml−1), or trimethoprim (25 μg ml−1) to select for cosmid and plasmid vectors, as required.
Degradation studies in soil.
A. globiformis D47 was grown on nutrient agar containing diuron (20 μg ml−1) at 25°C for 3 days. The cells were suspended in minimal salts solution, and 5 ml was added to 250 g (wet weight) of soil to yield an inoculation density of 1.2 × 107 CFU g of soil−1. The soil used was from Little Cherry field (Horticulture Research International) and was a sandy-loam soil of the Wick series containing 16% (wt/wt) clay, 70% (wt/wt) sand, and 1.9% (wt/wt) organic matter. The soil (matric potential, −33 kPa) was sieved (2 mm), air dried overnight, and, after inoculation and herbicide addition, rewetted back to its original moisture content (13%). The urea-based herbicides were applied to soil samples as manufacturer's formulations to mimic how they would be applied when used commercially. These were chlorotoluron (water solubility, 74 mg liter−1 at 25°C) (formulation dicuran with 50% active ingredient from Novartis); diuron (water solubility, 36 mg liter−1 at 25°C) (formulation karmex with 80% active ingredient from DuPont); isoproturon (water solubility, 55 mg liter−1 at 22°C) (formulation arelon with 48% active ingredient from AgrEvo); linuron (water solubility, 64 mg liter−1 at 20°C) (formulation linuron-50 with 50% active ingredient from DuPont); monolinuron (water solubility, 735 mg liter−1 at 25°C) (formulation aresin with 50% active ingredient from AgrEvo); and monuron (water solubility, 230 mg liter−1 at 25°C) (formulation telvar with 80% active ingredient from DuPont). Stocks of these were added to duplicate soil samples, which were then well mixed to provide a final concentration of 20 μg g of soil−1. Over a 10-day period at 20°C, 10 g (wet weight) of soil was removed and extracted with 15 ml of methanol, except for monuron, where 15 ml of 90% (vol/vol) acetonitrile was used. Samples were analyzed by high-pressure liquid chromatography (HPLC) using acetonitrile-water-phosphoric acid (75:25:0.25 by volume) as the mobile-phase solvent and a Lichrosorb RP18 column (250 by 4 mm; Merck). The mobile-phase flow rate was 1 ml min−1, and detection was by UV absorbance at 240 nm. A one-way model, analysis of variance, was used to estimate the least significant difference for the pattern of decline for each herbicide, which was plotted on the graph as a bar (P < 0.05). Controls included noninoculated soil and soil with no herbicide added.
Diuron degradation assay.
For A. globiformis D47 and derived strains, single colonies were inoculated into sterile glass vials (ca. 3-ml volume) containing 0.5 ml of mineral salts medium and diuron (20 μg ml−1). The samples were incubated at 30°C for 3 days. To each sample 1 ml of the mobile phase solvent, acetonitrile-water-phosphoric acid (75:25:0.25 by volume), was added. The presence of diuron and its major degradation product (DCA) was determined by HPLC using a Lichrosorb RP18 column (250 by 4 mm; Merck) with a mobile-phase flow rate of 1 ml min−1 and detection by UV absorbance at 240 nm. For E. coli clones, each strain was grown in 5 ml of LB medium containing the appropriate antibiotic at 37°C for 24 h. The cells were collected by centrifugation at 6,000 × g for 5 min and washed twice with PBS (0.1 M phosphate [pH 7.2], 0.85% [wt/vol] NaCl). The cell pellet was resuspended in 100 μl of PBS and subjected to three rounds of freeze-thaw treatment (−70°C for 30 min and 30°C for 30 min). To this lysate, 10 μg of diuron ml−1 was added in PBS, and the sample was incubated at 30°C for 3 days. Samples were analyzed by HPLC to determine the levels of diuron and DCA. Controls included E. coli, E. coli (pUC18), and A. globiformis D47 grown in LB medium.
Plasmid curing of strain D47.
Using 1 ml of an 18-h starter culture of A. globiformis D47 (50 ml of LB medium, 30°C, 150 rpm), 50 ml of fresh LB medium was inoculated and incubated at 30 and 35°C for 24 h. Every 24 h for 10 days successive transfers were made (1 to 50 ml) from each culture. On transfer, samples (0.1 ml) were taken, diluted, plated onto nutrient agar, and incubated at 30°C for 2 days. One hundred fifty individual colonies were selected and checked for their ability to degrade diuron using the MSM-based degradation assay. A selection (approximately 50) of degradative and nondegradative strains were screened to determine their plasmid composition.
Plasmid DNA profiling and purification.
A small-scale plasmid preparation method for profiling strain D47 and its derivatives was used. A single colony was used to inoculate 5 ml of LB medium that was incubated at 30°C for 18 h. Cells were collected by centrifugation at 8,000 × g for 10 min and resuspended in 300 μl of buffer P1 (Qiagen, Crawley, United Kingdom) containing 1 mg of lysozyme ml−1. After 10 min at room temperature, 300 μl of buffer P2 was added and held on ice for 5 min. To this, 300 μl of P3 was added, and the sample was centrifuged at 13,000 × g for 15 min to remove the debris. Isopropanol (0.8 volume) was added, and the sample was recentrifuged at 13,000 × g for 30 min. The pellet was washed with 70% (vol/vol) ethanol and resuspended in 50 μl of 1 mM Tris-HCl (pH 8.0). Samples (usually 20 μl) were analyzed by gel electrophoresis (0.7% [wt/vol] agarose). Larger-scale purified plasmid preparations were obtained from 500-ml cultures in a similar way. The cell pellet was resuspended in 3 ml of P1 containing lysozyme, held at room temperature for 15 min, and then washed three times with P1 buffer to remove the lysozyme. After this, 3-ml volumes of P2 and P3 were added as described above. A Qiagen tip 100 column was used to purify the DNA, using the manufacturer's instructions, and the eluate from the column was isopropanol precipitated, washed with 70% (vol/vol) ethanol, and resuspended in a final volume of 100 μl.
DNA cloning.
Plasmid DNA from A. globiformis D47 was digested, individually, with the restriction enzymes EcoRI and SstI. Cut DNA was ligated to the vectors Supercos (Stratagene, Amsterdam, The Netherlands), which was cut with EcoRI, and pUC18, which was cut with SstI, using standard procedures. The ligated DNA was heat treated at 65°C for 5 min, dialyzed, and electroporated (12.5 kV cm2) using a Bio-Rad system into E. coli cells. Colonies were selected on LB agar supplemented with kanamycin (Supercos) or ampicillin (pUC18). Over 100 of these colonies were screened using the small-scale plasmid preparation method described above without the addition of lysozyme. The plasmid DNA was digested with either EcoRI or SstI to confirm the presence of inserts and to determine the insert size. Clones with similar-sized inserts were grouped, and restriction digestion was used to identify clones that gave identical patterns. One representative of each plasmid clone with fragment sizes that matched those seen for A. globiformis D47 plasmid DNA was selected. These plasmids were labeled using a digoxigenin kit (Boehringer-Roche, Lewis, United Kingdom) and used as hybridization probes. Target DNA was prepared from A. globiformis D47 and degradative, nondegradative, and plasmid-free strains. Equal quantities (1 μg) of target DNAs were boiled for 5 min, placed on ice for 10 min, and spotted onto a nylon membrane (5 μl). The DNA was fixed to the membrane by UV treatment. For each probe a standard hybridization was carried out. The filter was prehybridized in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.5% (wt/vol) blocking reagent at 68°C for 6 h. The probe was added, and the hybridization was continued for 18 h. The membrane was successively washed with 2×, 1×, and 0.2× SSC containing 0.1% sodium dodecyl sulfate at 68°C for 20 min. The filter was developed using the reagent disodium 2-chloro-5-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7] decan}4-yl)-1-phenyl phosphate according to the instructions of the manufacturer (Boehringer-Roche).
DNA sequencing.
A system of transposon mutagenesis, subcloning from larger fragments, and designing of sequencing primers to walk out from known sequences was used to obtain the sequence of the degradative gene. All sequencing reactions were performed using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Warrington, United Kingdom) and analyzed on an automated DNA sequencer (Applied Biosystems). Sequences were edited and assembled using DNA* (DNAStar Inc., Madison, Wis.) software, and sequence analysis was performed using the packages FASTA (6.0) and the EBI internet system. DNA mapping and translation were performed using the programs Clone Manager and Enhance (Scientific & Educational Software, Durham, N.C.).
Transposon mutagenesis.
Transposon mutagenesis using the artificial transposon AT2 was carried out using a modification of the manufacturer's protocol (Primer Island kit; Applied Biosystems). E. coli plasmid DNA (1 μg in 1 μl) was mixed with the transposon AT2 in 1× buffer (final volume, 20 μl) and incubated at 30°C for 1 h. The reaction was stopped by the addition of EDTA (final concentration, 10 mM) and sodium dodecyl sulfate (final concentration, 0.05% [wt/vol]) and heat treatment (65°C for 15 min). The sample was dialyzed by placing a 10-μl droplet on a 0.025-μm-pore-size filter (Millipore, Watford, United Kingdom) floating on H2O. After 20 min, the sample was electroporated into E. coli cells. Transposon mutants were selected on LB medium containing 50 μg of trimethoprim ml−1. Mutants were tested for their ability to degrade diuron. To locate transposon insertion positions in selected mutants, plasmid DNAs from mutated clones were purified and the Primer Island ± primers (Applied Biosystems) were used to sequence outward from the transposon ends.
RESULTS AND DISCUSSION
Degradation of urea-based herbicides.
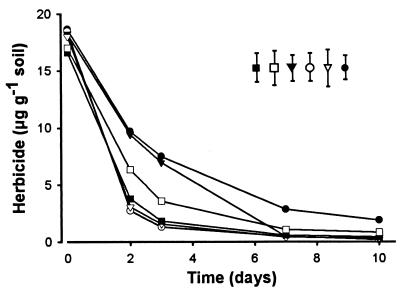
When A. globiformis D47 was added to soil samples, the strain degraded chlorotoluron, diuron, isoproturon, linuron, monolinuron, and monuron (Fig. 1). The different herbicides were introduced at 20 μg g of soil−1 and declined to below 2 μg g of soil−1 over 10 days. Herbicide concentrations in noninoculated controls dropped to 13.8 μg g of soil−1 for chlorotoluron, 15.0 μg g−1 for diuron, 14.3 μg g−1 for isoproturon, 15.8 μg g−1 for linuron, 19.3 μg g−1 for monolinuron, and 14.5 μg g−1 for monuron after 10 days. Although it was known that A. globiformis D47 could degrade some of these chemicals in pure culture (7), this is the first report of their degradation in soil. These results illustrate the potential use of this bacterium to degrade many urea-based herbicides in soil. With such a broad spectrum of activity, the gene(s) and enzyme(s) involved in urea-based herbicide breakdown are of particular interest.
FIG. 1.
Degradation of substituted urea-based herbicides in soil by A. globiformis D47. Chlorotoluron (○), diuron (□), isoproturon (▾), linuron (▪), monolinuron (▿), and monuron (●) were tested. Data are the means from two replicate experiments. LSD (P < 0.05) is indicated.
Although this strain cannot completely mineralize these chemicals, it could provide a key step in pesticide degradation when part of a microbial consortium. In addition, a microbial strain or plant could also be modified to contain a number of degradative genes for herbicide breakdown.
Plasmid profiles of A. globiformis D47 and mutants.
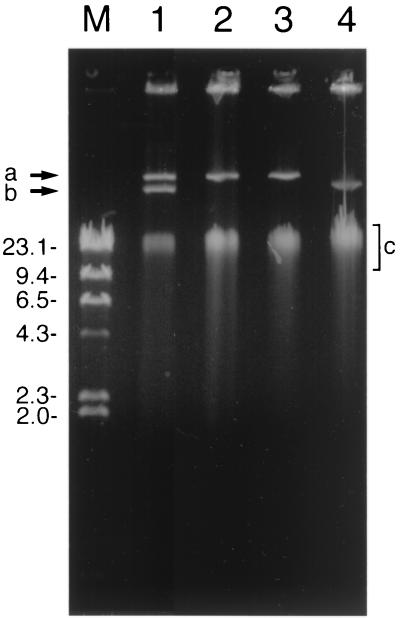
Two plasmids were observed in A. globiformis D47 (Fig. 2). In a screen of single colonies at the end of repeated subculture of the wild-type strain, those that maintained the ability to degrade diuron had either both plasmids or the larger of the two plasmids, pHRIM620. Isolates that could no longer degrade diuron had either no detectable plasmids or the smaller of the two plasmids, pHRIM621. These results strongly indicated that the larger plasmid gave the wild-type strain the ability to degrade diuron. Restriction analysis of plasmid DNAs of strains with both plasmids and ones with either of the two plasmids indicated that pHRIM620 and pHRIM621 were approximately 47 and 35 kb, respectively. Many of the degradative functions in Arthrobacter species have been attributed to plasmid-associated genes, including those that degrade the herbicide S-ethyl-N,N-dipropylthiocarbamate (33) and the insecticide carbaryl (1-naphthyl-N-methylcarbamate) (11). The degradative plasmids described in these studies have ranged in size from 2.5 MDa to 160 kb, and the strains were shown to have between one and four plasmids. The location of the degradative genes on plasmids can aid or promote transfer to other strains and lead to an increase in the metabolic diversity of the soil microbial population. New gene combinations could also allow the degradation of related compounds, degradation via different pathways, or recombination between related genes to generate even greater metabolic diversity (14).
FIG. 2.
Agarose gel illustrating the plasmid profiles of A. globiformis D47 and mutants. Lane 1, strain D47; lanes 2 and 3, degrading mutants; lane 4, nondegrading mutant; lane M, markers (lambda DNA cut with HindIII), in kilobases. Positions of the two plasmids (arrows a and b) and chromosomal DNA (area c) are indicated.
Growth of mutants on media with diuron.
Isolation and maintenance of strains that utilize a pesticide as a sole carbon source can often be difficult, as many strains grown on minimal medium cannot degrade the pesticide or have lost the ability to degrade it. The selective nature of the isolation media originally used to obtain the wild-type degradative strain from soil (7) was investigated using selected strains that differed only in their plasmid composition. The growth of a nondegrading strain was compared to that of the degrading strain on LB agar and MSM agar containing diuron. On LB agar, diuron inhibited the growth of the wild-type strain at concentrations in excess of 20 μg ml−1. The nondegrader was also inhibited at this concentration; therefore, the ability to degrade diuron did not offer the wild-type strain a selective advantage. On minimal medium both strains grew equally, but their growth was limited. Since the nondegrader cannot utilize diuron, this indicated that other carbon sources within the medium (such as traces of methanol used to introduce diuron or contaminants in the agar) had been metabolized for primary growth. Since a key step in isolation of degradative strains is the formation of colonies on minimal medium, only bacteria capable of degrading the herbicide should grow. However, out of 32 colonies selected on MSM containing diuron, only 5 degraded diuron, and they were subsequently characterized as the same organism (7). We therefore do not know why the selective enrichment was successful. Others have reported the loss of degradative ability on subculture of strains, even when minimal medium is used (23, 24). If the degradative phenotype is unstable, constant selection is required to maintain it in the population, which again relies on the medium allowing only the growth of isolates with degradative ability. During enrichment culture, the transfer of soil into MSM would render the medium nonminimal, and subculture would also transfer dead bacteria that could act as an alternative carbon source. In addition, impurities in the agar may have provided alternatives to diuron as an energy source, which may explain why only 16% of the isolates showed diuron degradation (7). There have been reports of the isolation of nondegrading bacteria on media specifically designed to select only for isolates capable of degradation, and in these cases the pesticide is probably not acting as the sole carbon source (23, 24). In most studies, broth culture appears to provide the most stable conditions for maintaining degradation, which suggests that the addition of agar to the medium may introduce additional nutrient sources. A. globiformis D47 was unable to grow in liquid MSM with diuron, although degradation of diuron could be detected, indicating that the enzyme within the cells was active. However, the liquid enrichment culture must offer some selective advantage for the growth of strains to have enabled the original isolation of A. globiformis D47 from the complex microbial population in soil.
Cloning of the degradation gene.
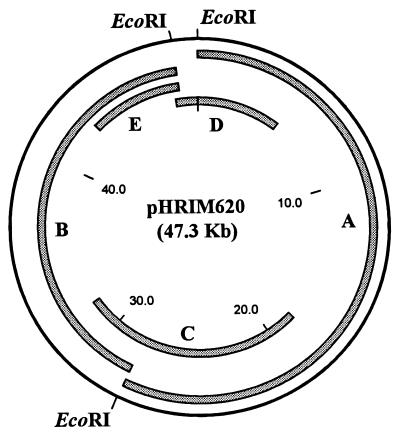
Digestion of pHRIM620 with EcoRI gave two large bands (17 and 22 kb) and one smaller band (2.5 kb), while digestion with SstI gave numerous smaller bands. Plasmid DNA from A. globiformis D47 was cloned into Supercos and pUC18. To select clones with inserts from pHRIM620, a series of unique clones were used as hybridization probes and screened against strains with both plasmids, pHRIM620 alone, or pHRIM621 alone and DNA from a plasmid-free strain. Clones that hybridized to all samples were believed to have inserts that originated from chromosomal DNA. Clones that hybridized with the strains containing both plasmids and just pHRIM620 were selected. Plasmids were cut with EcoRI, BamHI, and SalI to determine the locations of subclones within pHRIM620. Sequence information generated from primer sites present on the cloning vector were used to design reverse primers to PCR amplify and sequence back across the EcoRI restriction sites on pHRIM620. This provided a simple EcoRI plasmid map. Using DNA hybridization, individual clones were tested for hybridization to each other, which identified overlapping fragments. Using this information, inserts that provided complete coverage of pHRIM620 with a good overlap at each EcoRI restriction site were selected (Fig. 3). These clones were tested for their ability to degrade diuron after growth in minimal medium (M9) containing diuron or in LB medium at 25, 30, and 37°C for 8, 24, and 48 h. Cells were disrupted by sonication or freeze-thaw treatment. One of the clones, E. coli(pHRIM624) with a large insert (22 kb), was found to degrade diuron. With this clone, the greatest degradation of diuron was observed after growth in LB medium for 24 h and freeze-thaw lysis treatment of the cells. These results indicated that the degradative gene(s) on pHRIM624 was expressed from its own promoter(s) in E. coli. A. globiformis D47 is a member of the gram-positive coryneform bacteria, and therefore it is surprising that a promoter from this strain can function in E. coli. However a few A. globiformis genes have been expressed in E. coli (2, 4, 5, 10, 20, 22, 27), and some are also known to be expressed from their own promoters (27).
FIG. 3.
Map of pHRIM620 indicating the positions of the EcoRI restriction enzyme sites and the subclones (A to E) used. Sizes are in kilobases.
Identification of the degradative gene(s) and sequence analysis.
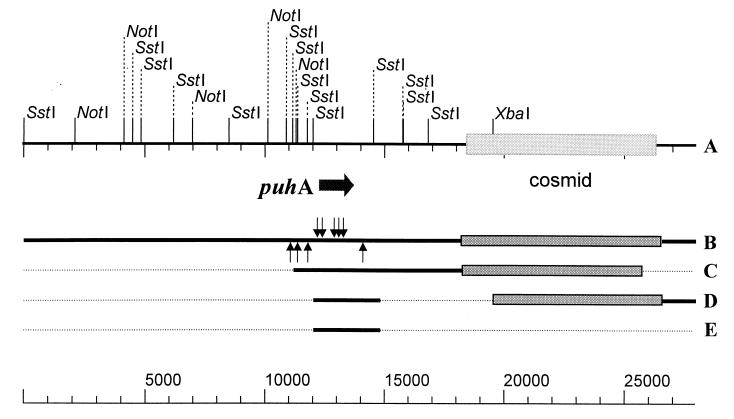
Over 150 transposon mutants of pHRIM624 were tested for their ability to degrade diuron. A series of mutants were shown to be unable to degrade diuron, and the locations of these mutations were determined (Fig. 4). All eight of these insertions were located in one open reading frame within a 2.5-kb SstI fragment; other insertions in front of and behind the gene did not disrupt enzyme activity. Digestion of pHRIM624 with NotI, followed by religation, deleted a 12-kb region from one end of the insert. This clone, when expressed in E. coli, retained the ability to degrade diuron. Digestion of pHRIM624 with SstI and religation provided a clone with just the 2.5-kb SstI fragment and a small fragment next to the cosmid. This clone, when expressed, was able to degrade diuron. Since the same region adjacent to the cosmid was deleted in the pHRIM624-NotI construct, this suggested that the small SstI fragment encoded activity. The 2.5-kb SstI fragment was cut out from pHRIM624-SstI and inserted into the SstI site in pUC19. When expressed in E. coli, this clone was able to degrade diuron. DNA sequence analysis indicated that a 1,368-bp open reading frame encoded this enzyme, and it showed little similarity to any other sequence in the EMBL database. The gene was predicted to code for a 456-amino-acid protein with an estimated size of 48.9 kDa and a pI of 5.2. The protein sequence, when compared to those in the EMBL database, showed a low level of sequence similarity (ca. 25% over 200 amino acids) to sequenced organophosphorus hydrolases, as outlined in Table 1.
FIG. 4.
Map of plasmid pHRIM622 (A) indicating the positions of transposon insertions (B) that inactivated the degradative ability (arrows above the line) or had no effect (arrows below the line). The position of gene puhA is indicated. The maps of active subclones (C, D, and E) obtained using the enzymes NotI (C) and SstI (D and E) are also presented. The fragment in line E was cloned into the plasmid pUC18. Sizes are in base pairs.
TABLE 1.
Sequence similarity of phenylurea hydrolase (PuhA) to proteins in the SWISSPROT databasea
| % Identity | Overlap (amino acids) | Accession no. (reference) | Organism | Enzyme |
|---|---|---|---|---|
| 23.4 | 435 | Q9S1C6 (22) | Arthrobacter sp. | Organophosphorus hydrolase |
| 26.0 | 460 | Q50432 (19) | Mycobacterium sp. | Organophosphate acid anhydrase |
| 25.7 | 458 | CAC04032 (25) | Streptomyces sp. | Organophosphate acid anhydrase |
| 24.0 | 425 | BAB06654 (32) | Bacillus sp. | Aryldialkylphosphatase |
| 24.6 | 425 | O27577 (30) | Methanobacterium | Aryldialkylphosphatase |
Percent identity and the length of comparison (overlap) are presented. Sequences are ordered as they are listed in the FASTA 6.0 table, which takes into account other similarity factors not presented here.
The other pesticide degradation genes that have been characterized are involved in the breakdown of 2,4-D, atrazine, carbofuran, and parathion (3, 8, 9, 17, 31, 35). These can be complex degradative systems, involving many genes and gene clusters and located on the chromosome as well as on plasmids. For example, the pathway for 2,4-D degradation is highly complex (9, 13). The degradation gene reported in our study bears greater resemblance to simpler systems such as the single genes which encode carbofuran hydrolase (mcd) (35) and parathion hydrolase (pah) (17). In a similar way, these single-gene systems all have a broad substrate specificity. We have named the gene identified in this study puhA, as it encodes a phenylurea hydrolase and cleaves the carbonyl bond in this group of herbicides. The puhA gene could be expressed in an appropriate microorganism and used as part of a strategy to clean up a contaminated site (18), or it could act as a biocatalyst for pesticide detoxification (21, 26). The sequence information could also be used to provide a DNA probe to study the prevalence and diversity of this gene in soil microbial populations. This will provide a useful tool with which to study changes in bacterial populations during the development of enhanced phenylurea herbicide degradation in soils.
ACKNOWLEDGMENTS
We thank Suzanne Lincoln at Horticulture Research International for technical help with the DNA cloning work.
We acknowledge the financial support of the Biotechnology and Biological Sciences Research Council (BBSRC) and the Ministry for Agriculture Fisheries and Food (MAFF), United Kingdom.
REFERENCES
- 1.Aislabie J, Lloyd-Jones G. A review of bacterial degradation of pesticides. Aust J Soil Res. 1995;33:925–942. [Google Scholar]
- 2.Bernauer H, Mauch L, Brandsch R. Interaction of the regulatory protein NicR1 with the promoter region of the pAO1-encoded 6-hydroxy-d-nicotine oxidase gene of Arthrobacter oxidans. Mol Microbiol. 1992;6:1809–1820. doi: 10.1111/j.1365-2958.1992.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 3.Boundy-Mills K L, De Souza M L, Mandelbaum R T, Wackett L P, Sadowsky M J. The atzB gene of Pseudomonassp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl Environ Microbiol. 1997;63:916–923. doi: 10.1128/aem.63.3.916-923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandsch R, Hakkanen A E, Mauch L, Nagursky H, Decker K. 6-Hydroxy-d-nicotine oxidase of Arthrobacter oxidans. Gene structure of the flavoenzyme and its relationship to 6-hydroy-d-nicotine oxidase. Eur J Biochem. 1987;167:315–320. doi: 10.1111/j.1432-1033.1987.tb13338.x. [DOI] [PubMed] [Google Scholar]
- 5.Brandsch R, Faller W, Schneider K. Plasmid pAO1 of Arthrobacter oxidans encodes 6-hydroxy-d-nicotine oxidase: cloning and expression of the gene in Escherichia coli. Mol Gen Genet. 1986;202:96–101. doi: 10.1007/BF00330523. [DOI] [PubMed] [Google Scholar]
- 6.Chung J M, Ka J O. Isolation and characterization of 2,4-dichlorophenoxyacetic acid-degrading bacteria from paddy soils. J Microbiol. 1998;36:256–261. [Google Scholar]
- 7.Cullington J E, Walker A. Rapid biodegradation of diuron and other phenylurea herbicides by a soil bacterium. Soil Biol Biochem. 1999;31:677–686. [Google Scholar]
- 8.De Souza L M, Seffernick J, Martinez B, Sadowsky M J, Wackett L P. The atrazine catabolism genes atzABCare widespread and highly conserved. J Bacteriol. 1998;180:1951–1954. doi: 10.1128/jb.180.7.1951-1954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Don R H, Weightman A J, Knackmuss H-J, Timmis K N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophusJMP134(pJP4) J Bacteriol. 1985;161:85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haraguchi K, Mori S, Hayashi K. Cloning of inulin fructotransferase (DFA III-producing) gene from Arthrobacter globiformisC11–1. J Biosci Bioeng. 2000;89:590–595. doi: 10.1016/s1389-1723(00)80062-6. [DOI] [PubMed] [Google Scholar]
- 11.Hayatsu M, Hirano M, Tadahiro N. Involvement of two plasmids in the degradation of carbaryl by Arthrobactersp. strain RC100. Appl Environ Microbiol. 1999;65:1015–1019. doi: 10.1128/aem.65.3.1015-1019.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holben W E, Schroeter B M, Calabrese G M, Olsen R H, Kukor J K, Biederbeck V O, Smith A E, Tiedje J M. Gene probe analysis of soil microbial populations selected by amendment with 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol. 1992;58:3941–3948. doi: 10.1128/aem.58.12.3941-3948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ka J O, Holben W E, Tiedje J M. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl Environ Microbiol. 1994;60:1106–1115. doi: 10.1128/aem.60.4.1106-1115.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karns J S. Molecular genetics of pesticide degradation by soil bacteria. ACS Symp Ser. 1990;426:141–152. [Google Scholar]
- 15.Laemmli C M, Leveau J H J, Zehnder A J B, van der Meer J R. Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutrophaJMP134(pJP4) J Bacteriol. 2000;182:4165–4172. doi: 10.1128/jb.182.15.4165-4172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mae A A, Marits R O, Ausmees N R, Koiv V M, Heinaru A L. Characterization of a new 2,4-dichlorophenoxyacetic acid degrading plasmid pEST4011—physical map and localization of catabolic genes. J Gen Microbiol. 1993;139:3165–3170. [Google Scholar]
- 17.Mulbury W M, Karns J S, Kearney P C, Nelson J O, McDaniel C S, Wild J R. Identification of a plasmid-borne parathion hydrolase gene from Flavobacterium sp. by Southern hybridization with opd from Pseudomonas diminuta. Appl Environ Microbiol. 1986;51:926–930. doi: 10.1128/aem.51.5.926-930.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulbury W M, Kearney P C. Degradation of pesticides by micro-organisms and the potential for genetic manipulation. Crop Prot. 1991;10:334–346. [Google Scholar]
- 19.Mulbury W M. The aryldialkylphosphatase-encoding gene ADPB from Nocardia sp. strain-B-1—cloning, sequencing and expression in Escherichia coli. Gene. 1992;121:149–153. doi: 10.1016/0378-1119(92)90174-n. [DOI] [PubMed] [Google Scholar]
- 20.Nishiya Y, Toda A, Imanaka T. Gene cluster for creatinine degradation in Arthrobactersp. TE1826. Mol Gen Genet. 1998;257:581–586. doi: 10.1007/s004380050685. [DOI] [PubMed] [Google Scholar]
- 21.Ohkawa H, Shiota N, Imaishi H, Yamada T, Inui H, Ohkawa Y. Cytochrome P450 monooxygenases metabolizing herbicides. Biotechnol Biotechnol Equip. 1998;12:17–22. [Google Scholar]
- 22.Oshiro K, Kakuta T, Nikaidou N, Watanabe T, Uchiyama T. Molecular cloning and nucleotide sequencing of organophosphorus insecticide hydrolase gene from Arthrobactersp. strain B-5. J Biosci Bioeng. 1999;87:531–534. doi: 10.1016/s1389-1723(99)80105-4. [DOI] [PubMed] [Google Scholar]
- 23.Parekh N R, Hartmann A, Charnay M-P, Fournier J-C. Diversity of carbofuran-degrading soil bacteria and detection of plasmid-encoded sequences homologous to the mcdgene. FEMS Microbiol Ecol. 1995;17:149–160. [Google Scholar]
- 24.Parekh N R, Hartmann A, Marie-Paule C, Fournier J-C. PCR detection of the MCD gene and evidence of sequence homology between the degradative genes and plasmids from diverse carbofuran-degrading bacteria. Soil Biol Biochem. 1996;28:1797–1804. [Google Scholar]
- 25.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolorA3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 26.Richins R D, Kaneva I, Mulchandani A, Chen W. Biodegradation of organophosphorus pesticides by surface-expressed organophosphorus hydrolase. Nat Biotechnol. 1997;15:984–987. doi: 10.1038/nbt1097-984. [DOI] [PubMed] [Google Scholar]
- 27.Sakurai H, Yokota A, Tomita F. Molecular cloning of an inulin fructotransferase (depolymerizing) gene from Arthrobacter sp. H65–7 and its expression in Escherichia coli. Biosci Biotechnol Biochem. 1997;61:87–92. doi: 10.1271/bbb.61.87. [DOI] [PubMed] [Google Scholar]
- 28.Sarman U, Monoj K R, Singh H D. Isolation of plasmid pRLI from Arthrobacter oxydans317 and demonstration of its role in steroid 1 (2)-dehydration. J Basic Microbiol. 1994;34:183–190. [Google Scholar]
- 29.Sayler G S, Hooper S W, Layton A C, King J M H. Catabolic plasmids of environmental and ecological significance. Microb Ecol. 1990;19:1–20. doi: 10.1007/BF02015050. [DOI] [PubMed] [Google Scholar]
- 30.Smith D R, Doucette-Stamm L A, DeLoughery C, Lee H, DuBois J, Aldredge T, Bashirzadeh R, Blakely D, et al. Complete genome sequence of Methanobacterium thermoautotrophicumΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streber W R, Timmis K N, Meinhart H Z. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophusJMP134. J Bacteriol. 1987;169:2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takami H, Nakasone K, Hirama C, Takaki Y, Masui N, Fuji F, Nakamura Y, Inuone A. An improved physical and genetic map of yhe genome of alkaliphilic Bacillussp. C-125. Extremophiles. 1999;3:21–28. doi: 10.1007/s007920050095. [DOI] [PubMed] [Google Scholar]
- 33.Tam A C, Behki R M, Khan S U. Isolation and characterization of an S-ethyl-N,N-dipropylthiocarbamate-degrading Arthrobacter strain and evidence for plasmid-associated s-ethyl-N,N-dipropylthiocarbamate degradation. Appl Environ Microbiol. 1987;53:1088–1093. doi: 10.1128/aem.53.5.1088-1093.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tixier C, Bogaerts P, Sancelme M, Bonnemoy F, Twagilimana L, Cuer A, Bohatier J, Veschambre H. Fungal biodegradation of a phenylurea herbicide, diuron: structure and toxicity of metabolites. Pest Man Sci. 2000;56:455–462. [Google Scholar]
- 35.Tomasek P H, Karns J S. Cloning of a carbofuran hydrolase gene from Achromobacterstrain WM111 and its expression in gram-negative bacteria. J Bacteriol. 1998;171:4038–4044. doi: 10.1128/jb.171.7.4038-4044.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbull, G. A., J. E. Cullington, A. Walker, and J. A. W. Morgan. Identification and characterization of a diuron-degrading bacterium. Biol. Fertil. Soils, in press.