Abstract
Extrapolation strategies from adult data for designing pediatric drug development programs are explored using the quantitative systems pharmacology (QSP) modeling approach, a mechanistic drug and disease modeling framework that can predict clinical response and guide pediatric drug development in general. This innovative model‐informed drug discovery and development approach can leverage adult‐pediatric pharmacology and disease similarity metrics to validate extrapolation assumptions. We describe the QSP model strategy and framework for extrapolation to design pediatric drug development programs by leveraging adult data across a wide range of therapeutic areas and illustrating stage‐gate decisions informed by such an approach.
UNDERSTANDING THE CONTEXT FOR EXTRAPOLATION OF ADULT DATA TO SUPPORT PEDIATRIC DRUG DEVELOPMENT
Evidence‐based extrapolation approaches from well‐controlled adult studies for drugs for which effectiveness and safety have already been established form a cornerstone for authorization of such drug(s) in pediatric use. Supported by legislation in the United States and the European Union, such an extrapolation step could mitigate the off‐label drug use in the pediatric population due to a lack of or delay in using well‐tested medications in children. Nevertheless, the prevalent off‐label use of the drug(s) in the pediatric population hinders clinical investigations of drugs required to obtain sufficient data in such populations. 1 Modeling and simulating data from adult clinical data, disease registries, and other sources in a pediatric population has been useful in determining the extent of similarity in the dose exposure‐response relationship between adult and pediatric populations. 2
Modeling approaches utilizing systems biology and systems pharmacology are applied extensively to interpret human biology, 3 disease progression, 4 drug effectiveness, 5 and safety. 6 Implementing model‐informed drug development (MIDD) principles to de‐risk development is one of the latest evolutions of clinical pharmacology explored by several pharmaceutical and biotechnology companies. The approach is also endorsed by the global regulatory community, including the European Medical Association (EMA), Pharmaceuticals and Medical Devices Agency (PMDA), and the US Food and Drug Administration (FDA). 7 , 8 , 9 Using modeling and simulation approaches to de‐risk decision making in the drug development process is not new. However, the systematic integration of the unique model assets 10 in an evolving computing environment that expands with knowledge on the drug candidate is still a work in progress for many pharmaceutical sponsors. Feedback from early adopters suggests that the MIDD approach can reduce both time and cost in drug development 11 when conducted appropriately.
In the context of pediatric drug development, the MIDD approach is beneficial to leverage the adult drug development experience, especially when knowledge of the adult therapeutic window is considered portable to the pediatric target populations. However, there are situations when the confidence of the adult therapeutic window cannot be conferred to the pediatric target population, especially when there are potential differences in the underlying disease process or the response to treatment between the two populations. In such a situation, significant effort from several stakeholders is required to assess the extent to which the adult clinical experience can be used as a guide for pediatric drug development. Developing a robust pediatric extrapolation strategy must describe the underlying assumptions, expose relevant data sources, and examine methods and approaches to extrapolate the adult experience. There is a need to consider the validity of assumptions and develop plans to generate and/or collect relevant pediatric data to reduce uncertainty and inform planning. In this endeavor, multiple approaches are often useful to illustrate the fidelity of the strategy. Of these computational approaches, the quantitative systems pharmacology (QSP) modeling approach offers unique insight not obtained from other solutions, particularly ones unable to quantifiably address the disease biology or pathophysiology.
QSP AS A DISCIPLINE
The QSP model, in a quantitative and mechanistic manner, aims to integrate and inform biological or toxicological disease processes and drug action complexity in response to therapeutic modulation. 12 As part of the overarching MIDD approach, QSP informs critical decision making via quantitative analysis focused on de‐risking underlying assumptions and data collected at the time of stage‐gate decision and the uncertainty assessment. 8 Unfortunately, the prevailing opinion regarding the scope of influence of these models is still centered on early stage drug development. As a result, the implementation of QSP efforts still tends to be drug discovery‐focused in scope. There exists an overlap and distinct advantages of QSP compared to more traditional, less complex or mechanistic methods, but the overall value to late‐stage development is underappreciated. Complexity still represents too high a hurdle for some stakeholders to embrace fully, and the discipline has often been panned as an “academic exercise” given the presumed investment and lack of appreciated return on investment. QSP is inherently appropriate for extrapolation purposes, and the appropriateness of bridging populations is well within the QSP framework. The model structure can accommodate disease progression, potential pharmacokinetic/pharmacodynamic (PK/PD) differences, and their interdependencies or potential confounding features. 10 , 13 , 14 As these features (disease progression and PK/PD) are more often viewed as value‐added, it may simply be an issue of messaging, but certainly, there is a need for further education.
PURPOSE AND INTENTION OF QSP
QSP is a mechanistic modeling tool that links disease and drug’s molecular mechanisms to biomarkers and clinical end points used to assess the disease and therapeutic effect. It is suited to understanding the system‐level response to treatment across multiple PD markers and clinical end points and assessing patient variability on a mechanistic basis. Given QSP’s strength in integrating biomarker and end point data to elucidate the mechanistic basis of pharmacological proof of concept (POC), it has found a natural home within translational medicine development. 15 This allows QSP modelers to leverage the wealth of information that is part of the discovery and nonclinical investigation of novel therapeutics and act as a quantitative translational bridge to clinical POC and proof of mechanism. As the QSP models transition from preclinical to clinical applications, a more fit‐for‐purpose approach allows the tailoring of these models to represent clinical disease characteristics pertinent to evaluating POC. A significant opportunity for QSP is to assess the translatability and similarity of adult clinical disease to pediatric disease, thereby providing a vital capability for pediatric drug development. The similarity of disease is a crucial component of the assessment of the pediatric extrapolation approach. QSP modeling can make disease similarity and treatment response assessments more model and data‐driven and thus provide tools for further exploration even in a postmarketing setting.
The QSP platform is an innovative tool for extending the value of clinical data and disease knowledge and filling the gap through simulation. By integrating molecular, cellular, and tissue level models to local therapeutic effect, the QSP model is well‐positioned to elucidate and evaluate disease similarity and treatment response between adult and pediatric populations. These mechanistic‐based models can quantitatively translate human physiology, cellular dynamics, and pharmacology mechanisms into disease‐specific parameters to replicate trial outcomes and generating virtual patient cohorts. 16 , 17 , 18 , 19 , 20 Thereby, we highlight the steps and considerations a product development team can take when developing a pediatric extrapolation plan and leverage QSP modeling for disease similarity assessment, supporting a robust pediatric extrapolation approach.
STRENGTH, WEAKNESSES, OPPORTUNITIES, AND THREATS ANALYSIS AND USE CASES
We have attempted to create a strength, weaknesses, opportunities, and threats analysis grid around the utility of using the QSP model as an extrapolation strategy (Figure 1). Although there may not be a universal agreement with this initial assessment, it provides a baseline for future discussion.
FIGURE 1.
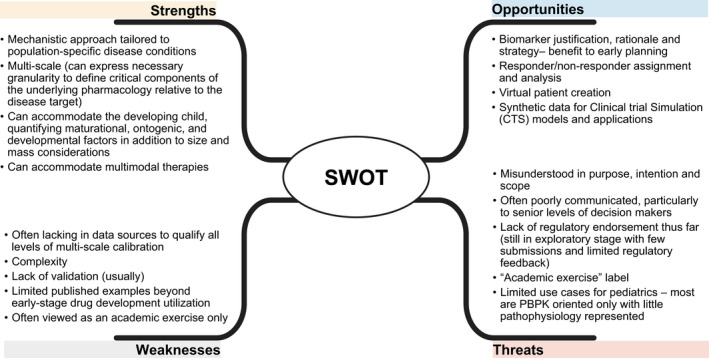
Strength, weaknesses, opportunities, and threat analysis supporting the use of quantitative systems pharmacology methodologies and approaches towards the support of pediatric extrapolation strategies. PBPK, physiologically‐based pharmacokinetic
A critical component of the pediatric extrapolation plan is assessing disease similarity, which incorporates two key components─underlying disease mechanisms and physiology and disease progression. 21 , 22 Breaking down underlying disease mechanisms and physiological processes into a multiscale framework and capturing the known mechanisms at the pathway, cellular, and target organ levels allows integration of existing knowledge and data to evaluate any gaps in making a case for pediatric extrapolation. This multiscale approach facilitates a translational strategy that links data‐driven understanding of disease mechanisms to differences across the two populations via a biomarker strategy. The biomarker strategy captures a quantitative representation of disease severity and links pharmacologically induced changes in disease biomarkers to relevant clinical end points. This biomarker strategy and multiscale disease knowledge forms the basis for disease similarity assessment between adult and pediatric populations. Through available and historical data registries and clinical trial data, one can assemble the biomarker axes of comparison between the two populations, guided by clinical input. Moreover, changes in the chosen biomarkers over time will provide the basis for considering disease progression into the QSP modeling framework for disease similarity. This approach sets up the primary components necessary for a model‐driven assessment of disease similarity. Moreover, it paves the way to use the same QSP model for treatment response similarity—another critical assessment for pediatric extrapolation.
QSP MODELING STRATEGY AND FRAMEWORK
The first step in developing the QSP modeling strategy and building the model is the knowledge discovery phase. Here, we utilize knowledge discovery platforms (e.g., text mining and machine learning capabilities), as well as current and historical data along the biological axes relevant to the disease (Figure 2). Through this process, one identifies the relevant genotype and phenotype mappings associated with existing patient segments, potentially representing the patient population into groups with differentiated disease severity levels, as defined by a biomarker panel and disease end points. Example information at this stage of the process includes identified genetic mutations that may be prevalent by geography/location, demographic, or other intrinsic/extrinsic factors, that may bear downstream effects on underlying disease pathophysiology leading to differences in disease progression rates or severity levels (disease registries). Following this chain of data down a mechanistic path, systems biology databases can provide data to support the implications of these genetic changes at the cellular and metabolic scales. Finally, how these changes at a cellular level map to specific organs of interest that results in clinical manifestation and changes in disease end points; incorporation of this continuum of disease mechanism data into a systems biology approach identifies, supplements, or confirms a biomarker panel that may be used to represent disease severity and progression characteristics and can be statistically linked to the clinical end point. Two important aspects of this bottom‐up representation are capturing variability in underlying biomarker and end point distributions across pediatric age groups and adult population, as well as recognizing and representing the uncertainties associated with quantitative measures of the disease mechanisms. These uncertainties can be propagated forward into uncertainty in the biomarker distribution or defined data gaps that would be learned as part of the biomarker strategy for the adult or pediatric trial. Table 1 shows the snapshot of information assembled during this process stage, defining the population’s disease severity levels and prevalence, biomarkers used to track disease improvements, and link to clinical end points. A further breakdown of the identified mechanisms and commensurate biomarkers from the adult stage to adolescents and younger age groups is critical, especially as it relates to capturing disease processes that may be pediatric age group specific, or to establish key differentiated disease pathways across pediatric age groups and the adult population. This catalogue of biomarkers and associated biological mechanisms, and end point distributions across defined pediatric age groups leading into the adult population becomes critical to define the commensurate virtual populations in the QSP model, establishing disease similarity criteria and building a pediatric investigation plan and study designs informed by a data and model driven age de‐escalation strategy. Finally, this strategy may surface gaps in knowledge or uncertainties that would need to be informed through carefully designed clinical trials.
FIGURE 2.
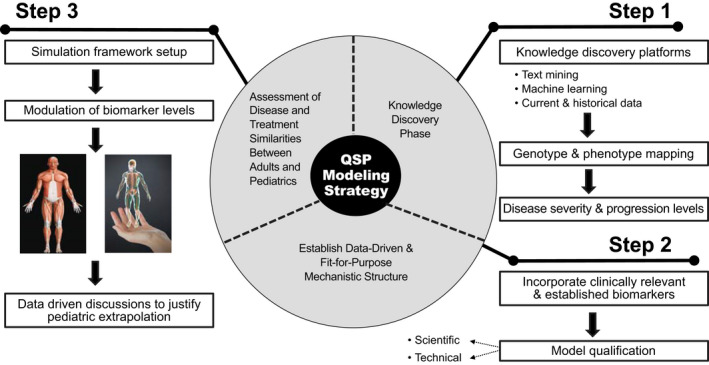
Quantitative systems pharmacology modeling strategy and framework
TABLE 1.
Sample data collected during the knowledge discovery phase of quantitative systems pharmacology model development of Gaucher’s disease type 1
| Disease severity | Genotype | Biomarker | Clinical end point | Prevalence (%) |
|---|---|---|---|---|
| Mild | N370S/N370S |
GL‐1 Ceraminde GM‐3 |
Spleen volume | 46.5 |
| Moderate |
Other/other (neither unspecified allele is categorized as severe) N370S/Other (unspecified allele may be categorized as severe) |
GL‐1 Ceraminde GM‐3 |
Spleen volume | 49.3 |
| Severe | G202R, L444P, D409H, 84GG, and R463C alleles categorized as severe mutation |
GL‐1 Ceraminde GM‐3 |
Spleen volume | 4.2 |
Note: Example data collected during the knowledge discovery phase of the QSP model was designed to predict treatment response via different modalities within the heterogeneous Gaucher’s Disease Type 1 patient population. 26
The second step in building the QSP modeling framework is establishing the mechanistic structure and level of detail in the model that will be fit‐for‐purpose and data‐driven. However, this framework should have sufficient mechanisms to differentiate the identified disease severity and progression levels using the scope of knowledge and data discovered in the knowledge discovery phase, and that captures spectrum of mechanisms across pediatric age groups and the adult population. This is an iterative process, with the QSP modelers working with relevant internal and external biology and clinical experts, including pediatricians, to ensure the model incorporates the clinically relevant biomarkers used in adult clinical trials to assess treatment response across as broad as possible of a pediatric age group. Model qualification is a vital prerequisite before finalizing this process and using the model to determine disease similarity and treatment response. 23 , 24 Thus, we lay out the over‐arching principles of QSP model qualification, namely scientific and technical (Table 2), but this will likely require additional scientific and regulatory dialogue and input to refine to a more structured approach to QSP model qualification for extrapolation purposes.
TABLE 2.
Various aspects of quantitative systems pharmacology model qualification in the context of an extrapolation strategy
| Type of qualification | Definition used | Context |
|---|---|---|
| Scientific | The process of identifying and describing the key questions and the model concepts, assumptions, and limitations. | The model concepts refer to the representation of the scientific components and their interactions. The assumptions and limitations are specific to answering the critical questions with sufficient reliability (e.g., accuracy and precision) and interpreting modeling results. |
| Technical | The process of ensuring proper model implementation represents the intended model concepts and assumptions. | This requires checking the fidelity of the mathematical methodology to the model concepts and assumptions, the computer code to the mathematical representation of the model, and the code results to those anticipated for the intended model. |
The final step is the use of the model for the assessment of disease and treatment similarities. Setting up a QSP simulation framework requires the team‐level agreement of the biomarkers that will be measured to assess disease and treatment levels, and the pediatric age de‐escalation approach. Because the biomarkers have been established at this point, this step pertains to underlying variables in the QSP model that will contribute to the modulation of the biomarker levels and evaluation of how variability in underlying disease segments and age groups can be wired in the model and captured at the biomarker level, to represent biomarker distributions from existing or historical datasets. This process enables the generation of virtual patients within pediatric and adult populations that represent respective severity and progression characteristics reflective of the clinical populations. Figure 3 shows a hypothetical simulation of a pediatric population based on a model fitted to adult data. The QSP simulations set the stage for informing data‐driven discussions on disease and treatment response similarity. One of the advantages of having a QSP model anchor clinical trial simulations used to support the proposed clinical development plan is that such a model can explore previously untested biomarkers (including pediatric specific biomarkers) in addition to those used to support adult registration as Figure 3 alludes. With this approach, sponsors and regulators can evaluate the generalizability of adult PK/PD relationships to pediatric target populations as well as the information value that a new or pediatric‐specific biomarker can bring to the study design, inclusion/exclusion criteria, and dose recommendation. Clinical end points and sampling scheme can be similarly explored with this approach.
FIGURE 3.
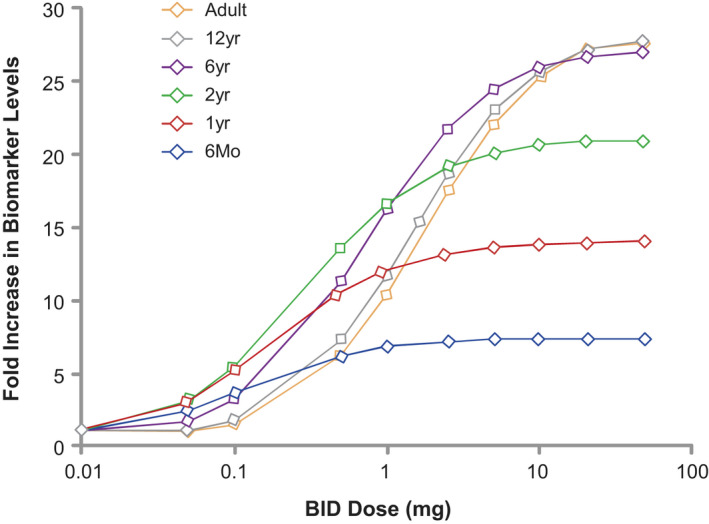
Hypothetical simulation of biomarker response in pediatric population based on a model fitted to adult data. Drug A is an oral hypothetical competitive inhibitor of an enzyme involved in a disease that affects adults and children. The graph denotes a hypothetical increase in biomarker levels in pediatric patients (categorized per age) based on adult parameters/information entered in the quantitative systems pharmacology model
The second essential part of this process is establishing and agreeing with the team and regulators about the quantitative differences or overlap in the biomarkers identified for similarity assessment that would dictate the similarity of the two populations to justify pediatric extrapolation. The team would then need to develop data‐driven scenarios based on the biomarkers used as to how much overlap and what minimal set of biomarkers would justify disease similarity. As previously mentioned, such QSP‐informed scenarios can be used to verify that disease similarity is consistent across age strata in addition to the typical pairwise comparisons made using traditional PK/PD approaches. Figure 4 shows how, while building the QSP model, multiple parameters are utilized to determine the variability/similarity between adult and pediatric populations and taken into account for extrapolation of data. Coupling this variability assessment with the predefined range for the treatment response and diseases to be considered similar in the pediatric versus adult population sets the stage for a model‐informed discussion with the team and regulators regarding the level of evidence to support pediatric extrapolation.
FIGURE 4.
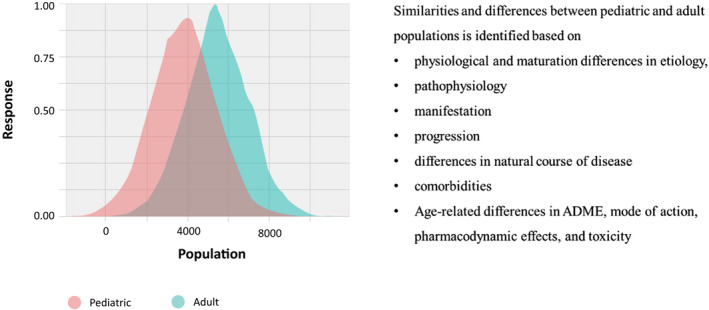
Assessment of disease and treatment similarity/difference between pediatric and adult population. ADME, Absorption, Distribution, Metabolism, and Excretion
PUTTING IT ALL TOGETHER
QSP framework can bring forward important capability towards drug development, not only for the pediatric population but also for rare and complex diseases. It could reduce the number, size, or duration of pediatric studies by leveraging adult‐pediatric drug pharmacology and disease similarity metrics and evaluating different therapeutic strategies bridging across rare disease segments (e.g., along a common pathway), reducing the need for additional trials and accelerating the development of novel therapies for pediatric diseases. This model‐informed pediatric extrapolation approach benefits from incorporation of differences in disease pathophysiology at defined pediatric age groups to inform biomarker and clinical end point simulations, leading to an optimized strategy for pediatric drug development, and specifically age de‐escalation and clinical trial design. Likewise, simulations constructed with various de‐escalation approaches can be used as the basis for meaningful labeling statements especially when they are verified via clinical evaluation or real‐world evidence postapproval. Tools for advancing the science and technology of QSP modeling and its application to extrapolation strategies are evolving but require further investment. Integrating diverse data sources and modeling methodologies, such as data science, systems biology, bioinformatics, and pharmacology, are necessary to build and harness the mechanistic continuum needed to formulate disease severity and treatment response models. The feasibility of operationalizing QSP within drug development timelines and cost constraints necessitates commensurate investment in resources, data lakes and infrastructure, scalable computing environments, and technical advances to enable rapid prototyping, model development, and simulation studies. It would help if there were a common platform where QSP models could be collaboratively co‐developed and shared among the community of scientists and stakeholders. Similarly, the International Council for Harmonization E11 guidance suggests incorporating appropriate modeling and simulation techniques into pediatric drug development as part of an overall strategy recognized via multidisciplinary discussions on assumptions, objectives, methods, deliverables, and timelines of the program. 25 A mechanism to export models to various platforms would also benefit pharmaceutical and clinical end‐users.
Although this QSP model‐driven approach to enabling extrapolation to pediatrics brings important opportunities to advance the science of pediatric drug development and accelerate pediatric drug approvals, there are a number of challenges ahead that can be overcome via further investment and additional real‐world examples. One challenge is setting and defining the bar for appropriate qualification of the QSP model (both structure and parameter values) for the purpose of extrapolation, in coordination with regulatory authorities, and setting acceptable criteria for model‐based extrapolation. Linking specific mechanisms of disease progression to biomarkers and ultimately end points necessitate additional experimental data to be collected for informing these mechanistic links, as well as assumptions that need to be continually evaluated as new scientific and clinical information emerges. Facilitating appropriate access and data platforms to leverage patient registry and historical clinical trial data is invaluable for developing and calibrating the QSP model to match progression characteristics and focusing on the patient relevant biomarker and end points of disease. Finally, advancing technical methodologies for parameter estimation in more complex model structures, and advancing tools to quantify and propagate model uncertainty and capture patient variability will translate into higher confidence in simulation results and broader adoption of model simulation outputs for high impact decisions and applications.
Nevertheless, the opportunity of applying QSP models to efficiently inform a pediatric investigation and extrapolation plan, and in turn facilitate disease and treatment response similarity is significant. This allows drug developers to accelerate pediatric drug development not only by leveraging adult data but also from one pediatric age group to another, and maximize the information value of pediatric trials through optimized pediatric trial design using this model‐based approach. In addition, this model‐based investigation expands the evidence and knowledge base for internal communication and with regulators on the overall therapeutic effect on disease burden and extension to the pediatric population, filling in gaps through simulation that would otherwise necessitate further clinical investigations, especially in pediatrics.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Yeruk Mulugeta and Dr. Lynne P. Yao for reviewing and providing valuable content and perspective towards improving the manuscript. The authors would also like to thank Md. Najeeb Ashraf, SciVoc Consulting Inc., for his medical writing and editing support toward the development of this manuscript.
Azer K, Barrett JS. Quantitative system pharmacology as a legitimate approach to examine extrapolation strategies used to support pediatric drug development. CPT Pharmacometrics Syst Pharmacol. 2022;11:797‐804. doi: 10.1002/psp4.12801
Funding information
No funding was received for this work.
REFERENCES
- 1. Sun H, Temeck J, Chambers W, Perkins G, Bonnel R, Murphy D. Extrapolation of efficacy in pediatric drug development and evidence‐based medicine: progress and lessons learned. Ther Innov Regul Sci. 2017;52:199‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van den Anker J. Paediatric extrapolation: the panacea for paediatric drug development? Br J Clin Pharmacol. 2019;85:672‐674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nyman E, Brannmark C, Palmer R, et al. A hierarchical whole‐body modeling approach elucidates the link between in vitro insulin signaling and in vivo glucose homeostasis. J Biol Chem. 2011;286(29):26028‐26041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hardy T, Abu‐Raddad E, Porksen N, De Gaetano A. Evaluation of a mathematical model of diabetes progression against observations in the Diabetes Prevention Program. Am J Physiol Endocrinol Metab. 2012;303(2):E200‐E212. [DOI] [PubMed] [Google Scholar]
- 5. Liu J, Ogden A, Comery TA, Spiros A, Roberts P, Geerts H. Prediction of efficacy of vabicaserin, a 5‐HT2C agonist, for the treatment of schizophrenia using a quantitative systems pharmacology model. CPT Pharmacometrics Syst Pharmacol. 2014;3:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Messinis DE, Melas IN, Hur J, Varshney N, Alexopoulos LG, Bai JPF. Translational systems pharmacology‐based predictive assessment of drug‐induced cardiomyopathy. CPT Pharmacometrics Syst Pharmacol. 2018;7(3):166‐174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhattaram VA, Booth BP, Ramchandani RP, et al. Impact of pharmacometrics on drug approval and labeling decisions: a survey of 42 new drug applications. AAPS J. 2005;7:E503‐E512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Madabushi R, Wang Y, Zineh I. A holistic and integrative approach for advancing model‐informed drug development. CPT Pharmacometrics Syst Pharmacol. 2019;8(1):9‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. EFPIA MID3 Workgroup , Marshall SF, Burghaus R, et al. Good practices in model‐informed drug discovery and development: practice, application, and documentation. CPT Pharmacometrics Syst Pharmacol. 2016;5(3):93‐122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Polak S, Tylutki Z, Holbrook M, Wisniowska B. Better prediction of the local concentration–effect relationship: the role of physiologically based pharmacokinetics and quantitative. Drug Dis Today. 2019;24:7. [DOI] [PubMed] [Google Scholar]
- 11. Lalonde RL, Kowalski KG, Hutmacher MM, et al. Model‐based drug development. Clin Pharmacol Ther. 2007;82:21‐32. [DOI] [PubMed] [Google Scholar]
- 12. Bradshaw EL, Spilker ME, Zang R, et al. Applications of quantitative systems pharmacology in model‐informed drug discovery: perspective on impact and opportunities. CPT Pharmacometrics Syst Pharmacol. 2019;8:777‐791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Musuamba FT, Manolis E, Holford N, et al. Advanced methods for dose and regimen finding during drug development: summary of the EMA/EFPIA workshop on dose finding (London 4–5 December 2014). CPT Pharmacometrics Syst Pharmacol. 2017;6:418‐429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Musante CJ, Ramanujan S, Schmidt BJ, Ghobrial OG, Lu J, Heatherington AC. Quantitative systems pharmacology: a case for disease models. Clin Pharmacol Ther. 2017;101(1):24‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gadkar K, Kirouac D, Parrott N, Ramanujan S. Quantitative systems pharmacology: a promising approach for translational pharmacology. Drug Discov Today Technol. 2016;21‐22:57‐65. [DOI] [PubMed] [Google Scholar]
- 16. Nijsen MJMA, Wu F, Bansal L, et al. Preclinical QSP modeling in the pharmaceutical industry: an IQ consortium survey examining the current landscape. CPT Pharmacometrics Syst Pharmacol. 2018;7(3):135‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Visser SA, de Alwis DP, Kerbusch T, Stone JA, Allerheiligen SR. Implementation of quantitative and systems pharmacology in large pharma. CPT Pharmacometrics Syst Pharmacol. 2014;3(10):e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rockne RC, Trister AD, Jacobs J, et al. A patient‐specific computational model of hypoxia‐modulated radiation resistance in glioblastoma using 18F‐FMISO‐PET [published correction appears in J R Soc Interface. 2015 Nov 6;12(112). pii: 20150927.]. J R Soc Interface. 2015;12(103):20141174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Helmlinger G, Sokolov V, Peskov K, et al. Quantitative systems pharmacology: an exemplar model‐building workflow with applications in cardiovascular, metabolic, and oncology drug development. CPT Pharmacometrics Syst Pharmacol. 2019;8(6):380‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jarrett AM, Shah A, Bloom MJ, et al. Experimentally‐driven mathematical modeling to improve combination targeted and cytotoxic therapy for HER2+ breast cancer. Sci Rep. 2019;9(1):12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. FDA . Guidance for industry providing clinical evidence of effectiveness for human drugs and biological products. 1998. https://www.fda.gov/media/71655/download
- 22. European Medicines Agency . Reflection paper on the use of extrapolation in the development of medicines for pediatrics. 2018. https://www.ema.europa.eu/en/documents/scientific‐guideline/adopted‐reflection‐paper‐use‐extrapolation‐development‐medicines‐paediatrics‐revision‐1_en.pdf
- 23. Hasegawa C, Duffull SB. Selection and qualification of simplified qsp models when using model order reduction techniques. AAPS J. 2017;20(1):2. [DOI] [PubMed] [Google Scholar]
- 24. Friedrich CM. A model qualification method for mechanistic physiological QSP models to support model‐informed drug development. CPT Pharmacometrics Syst Pharmacol. 2016;5(2):43‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. ICH E11(R1) Addendum: Clinical Investigation of Medicinal Products in the Pediatric Population. Guidance for Industry. 2008;1‐11. https://www.fda.gov/media/101398/download [Google Scholar]
- 26. Abrams R, Kaddi CD, Tao M, et al. A quantitative systems pharmacology model of gaucher disease type 1 provides mechanistic insight into the response to substrate reduction therapy with eliglustat. CPT Pharmacometrics Syst Pharmacol. 2020;9:374‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]


