Abstract
Purpose
Our aim was to investigate the age‐associated plasma protein profiles in pneumonia‐derived sepsis between infants and toddlers and identify potential age‐adapted prognostic markers for poor outcome of pneumonia‐derived pediatric sepsis.
Experimental Design
A nested case‐control study strategy was applied. The plasma proteomes of pneumonia‐derived pediatric septic patients with different outcomes between infants and toddlers were respectively analysed compared to their age‐matched controls.
Results
Compared to toddlers, pneumonia‐derived sepsis in infants was characterized by increased upregulation of protein processing in the ER, proteasome and antigen processing and presentation; and reduced downregulation in complement and coagulation cascades and cholesterol metabolism. Among them, the pentose phosphate pathway as well as the complement and coagulation cascades were possibly associated with poor outcome of pneumonia‐derived sepsis. Furthermore, we confirmed that HP, THBS1, and SAA1/2 were potential prognostic markers for poor outcome of pneumonia‐derived sepsis in infant patient groups.
Conclusions and Clinical Relevance
Age‐associated plasma protein profiles of pneumonia‐derived pediatric septic patients provided potential age‐adapted biomarkers for a more precise prognosis of poor outcome in pneumonia‐derived pediatric sepsis and helped to improve the survival of septic children.
Keywords: development, paediatric intensive care unit, prognosis, proteomics, sepsis
Abbreviations
- PICU
paediatric intensive care unit;
- PELOD
paediatric logistic organ dysfunction score
- SI
symptom improvement within 7 days after admission
- NI
no symptom improvement within 7 days after admission
- NS
nonsurvivors within 14 days after admission
1. INTRODUCTION
Pneumonia‐derived pediatric sepsis is a leading cause of mortality in children worldwide. Despite advances in diagnosis, treatment and support care, the mortality rate of pneumonia‐derived pediatric sepsis remains 10%–20% [1]. In general, sepsis serves as the final common pathway for children suffering from various primary and secondary infections and presents a dysregulated host response to infectious agents. From birth into teenage years, children undergo not only rapid physiological maturation but also significant changes in their immune systems that affect how they respond to microbes and especially respiratory pathogens. The respiratory system is the most common primary site of infection in several epidemiological investigations [2, 3, 4]. In particular, pediatric septic patients span a long period of developmental ages. In the Pediatric Sepsis Consensus of 2005, six clinically and physiologically meaningful age groups were delineated for age‐specific vital signs and laboratory variables to meet SIRS criteria: new born, neonate, infant, toddler and preschool, school‐aged child, adolescent, and young adult. Within pneumonia‐derived pediatric sepsis, how developmental age‐specific host response impacts the occurrence and development of sepsis remains unclear, especially between infants and toddlers, which were the major susceptible age groups in sepsis. Additionally, the molecular mechanism associated with this age‐related risk is still not completely understood.
In this report, we applied a nested case‐control study strategy. Using a relative quantitative LC–MS/MS method with TMT labelling, we analysed the plasma proteomes of pneumonia‐derived pediatric septic patients with different outcomes between infants (age 1) and toddlers (age 2) with their age‐matched controls. Thus, developmental age‐specific and poor prognosis‐associated plasma proteome profiling would identify the differences in host response to sepsis between infants and toddlers and provide potential novel markers or strategies for a more precise age‐adapted prognosis in pneumonia‐derived pediatric sepsis.
2. MATERIALS AND METHODS
2.1. Study design and patients
Between 2018 and 2019, 294 pneumonia‐derived pediatric septic patients admitted to the pediatric intensive care unit (PICU) of Hunan Children's Hospital were recruited for enrolment in the pneumonia‐derived pediatric sepsis cohort of Hunan Children's Hospital. A total of 90 children aged in infants (7–12 months of age) or toddlers (13–36 months of age) who met the criteria of pneumonia‐derived pediatric sepsis were selected for this proteomic study. The Demographics and clinical characteristics of the patient groups were summarized in supplement 2‐Table S1. Within each age group, patients were sorted into three subgroups based on their prognosis: SI, symptom improvement within 7 days after admission; NI, no symptom improvement within 7 days after admission; NS, nonsurvivors within 14 days after admission. Symptom improvement was evaluated based on the Paediatric Logistic Organ Dysfunction score (PELOD‐2) and clinical symptoms (supplement 2‐Table S2) [5]. Besides, the SI and NI groups at each age were composed of three pooling samples, respectively, namely mild, moderate and severe septic patients. The definitions of sepsis severity were shown in supplement 2‐Table S3. The plasma samples were prepared within 24 h after collection and stored at −80°C. In addition, 20 age‐matched controls were recruited from the child health care department of Hunan Children's Hospital, excluding children with infection, innutrition or any underlying diseases. The study protocol was reviewed and approved by the Medical Ethics Committee of Hunan Children's Hospital (approval No. HCHLL‐2018−28). Informed consent from parents or legal guardians was obtained for sample collection.
Clinical Relevance
Pneumonia‐derived pediatric sepsis is a leading cause of mortality in children worldwide. A poor outcome in septic patients is typically associated with a complicated treatment period. Age is a known risk factor for pediatric sepsis. In cases of pediatric sepsis, how the developmental age‐specific host response impacts the occurrence and development of pneumonia‐derived pediatric sepsis remains unknown, especially between infants and toddlers, which represent the major age groups susceptible to sepsis. Our plasma proteomic analysis of pneumonia‐derived sepsis in infants and toddlers revealed increased upregulation of the pentose phosphate pathway, protein processing in the ER, proteasome and antigen processing and presentation but less downregulation of complement and coagulation cascades and cholesterol metabolism in infants with poor outcomes compared with toddlers. Moreover, the combination of HP and THBS1 expression was demonstrated to be potential infant‐adapted prognostic markers for poor outcomes in infants with sepsis. The identification of these biomarkers promotes the application of age‐adapted precision medicine in pneumonia‐derived pediatric sepsis and helps improve the survival of septic children.
2.2. Plasma sample processing
In total, 110 plasma samples were pooled into 16 samples for assessment using LC–MS/MS as described in Table S2. The highly abundant proteins of plasma samples were removed using a Pierce Top 12 Abundant Protein Depletion Spin Columns Kit (Thermo Fisher). Then the protein samples were processed by reduction, alkylation, trysin digestion, TMT‐16plex labelling and peptide fractionation. The details were described in supplement 1‐Methods.
2.3. LC–MS/MS analysis
The tryptic peptides were dissolved in 0.1% (v/v) formic acid in H2O and directly loaded onto a homemade reversed‐phase analytical column (15 cm length, 75 μm i.d.). The details is found in supplement 1. The peptides were subjected to an NSI source followed by tandem mass spectrometry (MS/MS) in a Q Exactive Plus (Thermo) coupled online to UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350–1800 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were then selected for MS/MS using the NCE setting of 28, and the fragments were detected in the Orbitrap at a resolution of 17,500. A data‐dependent procedure that alternated between one MS scan followed by 20 MS/MS scans with 15.0 s dynamic exclusion. Automatic gain control (AGC) was set at 5E4. Fixed first mass was set as 100 m/z.
The resulting MS/MS data were processed using the Proteome Discoverer 2.4. Tandem mass spectra were searched against the human UniProt database concatenated with the reverse decoy database. Trypsin/P was specified as a cleavage enzyme allowing up to two missing cleavages. The mass tolerance for precursor ions was set as 20 ppm in the first search and 5 ppm in the main search, and the mass tolerance for fragment ions was set as 0.02 Da. Carbamidomethyl on Cys was specified as a fixed modification, and acetylation modification and oxidation on Met were specified as variable modifications. FDR was adjusted to < 1%, and the minimum score for modified peptides was set as > 40.
2.4. Data analysis
Enrichment of Gene Ontology analysis: For each category, a two‐tailed Fisher's exact test was employed to test the enrichment of the differentially expressed protein against all identified proteins.
For further hierarchical clustering based on their functional classification of differentially expressed proteins, those categories, at least enriched in one of the clusters with a p value < 0.05, were selected.
Enrichment of KEGG pathway map: The differentially expressed proteins were annotated by KEGG online service tools KAAS and mapped onto the KEGG pathways by the KEGG online service tools KEGG mapper.
Mfuzz analysis was applied for gene expression pattern analysis. All the differential proteins of each patient group related to the age‐matched control were selected.
The details of data analysis were described in supplement 1‐Methods.
2.5. ELISA
The concentrations of haptoglobin (HP), Thrombospondin 1 (THBS1) and SAA1/2 in patient plasma were analysed in 45 randomly chosen sepsis plasma samples from the cohort of this study using the Human Haptoglobin ELISA Kit (ab108856), Quantikine ELISA Human Thrombospondin‐1 (R&D Systems DTSP10), and Human SAA ELISA Kit (ab100635). Measurements were performed in accordance with the manufacturer's instructions.
3. RESULTS
3.1. Comparison of proteome profiles across the six patient groups
In order to analyse the age‐associated proteome profile of paediatric sepsis, we excluded some major factors which may also affect the outcome of sepsis, such as the weight, nutritional status as well as sepsis severity of the patients. It was found that their no significant differences in the distribution of weight‐for‐age, length‐for‐age and sepsis severity between SI and NI groups at both ages, respectively. The details were shown in supplement 2‐Table S4 ‐S5.
In this study, 16 plasma protein samples were labelled with TMT and quantitatively identified by LC–MS/MS. In total, 667 proteins were identified and quantified. Compared to their age‐matched healthy controls, proteins in any patient group with a fold change ≥ 1.3 were defined as significantly differentially expressed proteins (p < 0.05). Principal Component Analysis (PCA) of patient samples on protein level were presented in supplement 4‐Figure S1.
3.2. Comparison of enriched functions of the differentially regulated proteins across the six patient groups
To investigate the age‐associated and outcome‐associated enriched functions, the differentially expressed proteins in each patient group compared to their age‐matched controls were first categorized based on KEGG pathway analysis of biological functions, molecular functions and protein domains and then presented in clusters in the heatmaps (Figure 1 and Figure S2‐S4). Carbon metabolism was significantly upregulated in pneumonia‐derived pediatric septic patients with poor outcomes at both ages. In particular, the pentose phosphate pathway (PPP) was strongly upregulated with a worse prognosis at both ages and was more activated in the infant group compared with the toddler group. However, cholesterol metabolism was generally repressed during pneumonia‐derived sepsis and was more downregulated in toddlers compared with infant patient groups (Figure 1). Comparisons between infant and toddler groups revealed that younger patients exhibited increased upregulation of antigen processing and presentation (Figure 1). In addition, the IL‐17 signalling pathway was considerably more activated in patients with symptom improvement (SI) compared with those with poor outcomes (NI and NS) in both age groups.
FIGURE 1.
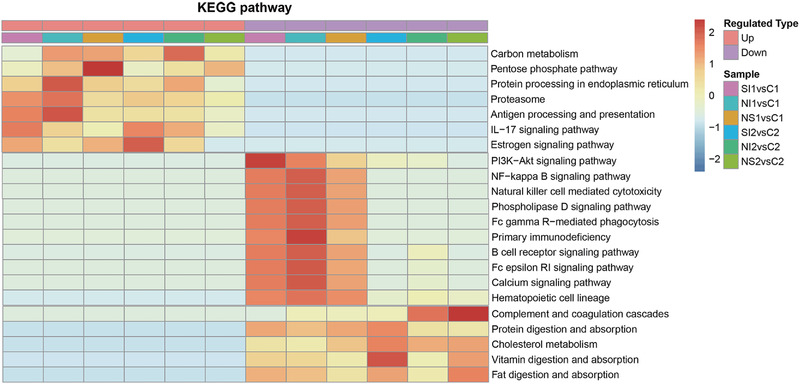
Comparison of the differentially expressed proteins across the six patient groups. KEGG pathway‐based functional enrichment clustering of the differentially expressed proteins in each patient group. SI1 and SI2 refer to subgroups of septic patients with symptom improvement in infants and toddlers, respectively; NI1 and NI2 refer to subgroups of patients without symptom improvement within 7 days in infants and toddlers, respectively; NS1 and NS2 refer to nonsurvivors within 14 days in infants and toddlers, respectively
In this study, we observed that complement and coagulation cascades was decreased more significantly in toddler patient groups with poor outcomes compared with infants (Figure 1). Moreover, Figure 2 illustrated that both complement pathways and coagulation cascades were downregulated in NI2 due to the depletion of complement factors (C3, C5, C6, C7, C8, C9, and C1qrs) and reduced coagulation cascade components, such as coagulation factor IX (F9) and protein C (PC). At the same time, negative regulation of both cascades were weakened, such as complement factor H (FH), heparin cofactor II (HCII), protein C inhibitor (PCI), alpha‐2‐macroglobulin (A2M). In addition, our clinical laboratory test data also showed significantly reduced C3 concentration (p = 0.0276), fibrinogen concentration (p = 0.0068) in NI2 compared to SI2, which also indicated the depletion of complement and coagulation components and confirmed our proteomic results (supplement 4‐Figure S5).
FIGURE 2.
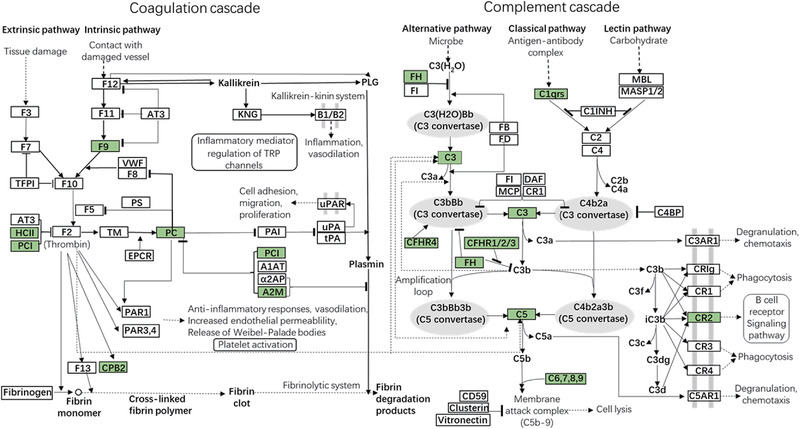
Map of enriched KEGG pathway complement and coagulation cascades in NI2/SI2. Proteins colored with green mean their protein level was downregulated significantly in NI2/SI2. Enriched KEGG pathway map was performed using KEGG online service tools KAAS for protein annotation and visualized using KEGG online service tools KEGG mapper
3.3. Age‐specific gene expression pattern analysis across the infant and toddler septic patient groups
To identify age‐specific potential prognostic biomarkers of pneumonia‐derived sepsis with poor outcome, we investigated the gene expression pattern across septic patient groups with different outcomes at each age. The differentially regulated genes involved in these two age groups were separately clustered based on their expression levels in SI, NI, and NS groups by applying the R package Mfuzz. Here, each age group showed six major clusters (Figure 3).
FIGURE 3.
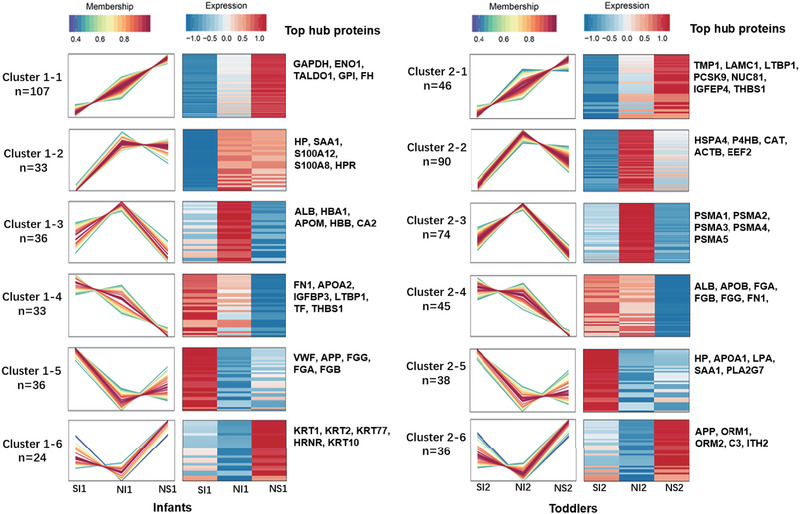
Clustering map of Mfuzz expression patterns. Each cluster corresponds to a line graph, a heatmap and involved core proteins. Line chart: the horizontal axis coordinates are the samples, the vertical axis coordinates are the relative protein expression, a broken line represents a protein, and the line colour represents the membership of the protein in the current class. Heat map: the horizontal axis coordinates are samples, the vertical axis coordinates are different proteins, and the colour of heat map indicates the relative expression of proteins in samples. Top hub proteins: the top seven hub proteins of each cluster based on PPI analysis are shown. In this figure, SI1 and SI2 refer to subgroups of septic patients with symptom improvement in infants and toddlers, respectively; NI1 and NI2 refer to subgroups of patients without symptom improvement within 7 days in infants and toddlers, respectively; NS1 and NS2 refer to nonsurvivors within 14 days in infants and toddlers, respectively
Based on the protein and protein interaction (PPI) analysis on the web‐based platform of the STRING database, node proteins of each cluster were identified. In combination with Cytoscape software, the top hub genes of each cluster were identified by applying cytoHubba based on the local‐based method of maximal clique centrality (MCC) (Figure 3–4 and Figure S6). Haptoglobin (HP), thrombospondin 1 (THBS1), serum amyloid A (SAA1/2), and latent transforming growth factor beta binding protein 1 (LTBP1) showed completely opposite expression patterns between the infant and toddler groups. The protein expression levels of HP and SAA1 decreased in infants with poor outcomes, whereas these levels increased in toddlers with poor outcomes. In addition, THBS1, and LTBP1 expression levels were positively associated with poor outcome in infants but negatively associated with poor outcome in toddlers (Figures 3). This finding implied that these four proteins were possibly age associated and outcome associated. These proteins may be potential age‐associated prognostic biomarkers.
FIGURE 4.
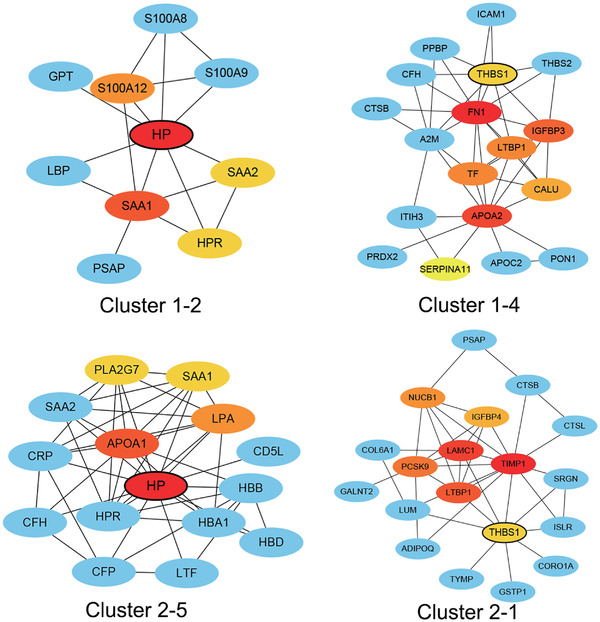
PPI analysis of HP and THBS1 in the Mfuzz clusters of the infant and toddler groups. The top hub genes of each cluster were identified by applying cytoHubba based on the local‐based method of maximal clique centrality (MCC). PPI analysis was based on the string database
3.4. Identification and validation of age‐associated prognostic biomarker candidates for poor outcomes in pneumonia‐derived pediatric sepsis
HP, THBS1, and SAA1/2 proteins were selected to further investigate whether their expression levels were age associated and correlated with poor prognosis in septic patients. Here, HP, THBS1, and SAA1/2 protein levels in 35 plasma samples from septic patients were determined by ELISA. The plasma samples were obtained from the same sample collection and the same grouping criteria for proteomic analysis. The ELISA results showed that the haptoglobin level in infant patients increased with worsened prognosis (p = 0.016), whereas HP levels decreased in toddlers. However, the differences among toddler patient groups SI2, NI2, and NS2 were unfortunately not significant (p = 0.4118) (Figure 5A). The THBS1 expression levels in infant groups significantly decreased with worsened prognosis (p = 0.0171), whereas changes in expression among the three toddler groups were not significant (p = 0.8761). Therefore, HP and THBS1 were possibly correlated with poor prognosis of sepsis exclusively in infants and not in toddlers (Figure 5A). In addition, SAA1/2 levels were not significantly different among infant patient groups as well as toddler groups (Figure 5A).
FIGURE 5.
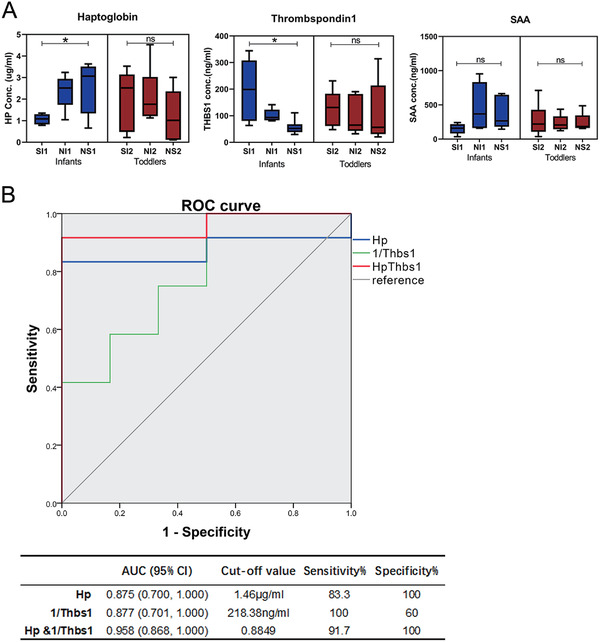
Correlation analysis between biomarkers and sepsis outcome in infants and toddlers. (A) Determination of HP, THBS1, and SAA concentrations by ELISA. (B) ROC curve analysis of infant‐specific prognostic biomarkers of sepsis: HP and THBS1. * indicates a significant difference among the patient groups with p<0.05; ns indicates a difference between the patient groups without statistical significance, p>0.05. THBS1 was evaluated with 1/THBS1 in the ROC curve analysis
To evaluate the prognostic capacity of HP and THBS1 in infant sepsis, their specificity and sensitivity as prognostic biomarkers were determined by ROC curve analysis (Figure 5B). The sensitivity and specificity of HP for poor prognosis in infant sepsis were 83.3% and 100%, respectively, and the AUC value was 0.875 (95% CI: 0.700–1.000). Given that the cut‐off value was 218.38 ng/mL for THBS1, its sensitivity and specificity were 100% and 60%, respectively, and the AUC value was 0.877 (95% CI: 0.701–1.000). Pairwise analysis found that the combination of HP and THBS1 had the best AUC of 0.958 (95% CI: 0.868–1.000) and a sensitivity and specificity of 91.7% and 100%, respectively.
4. DISCUSSION
4.1. Complement and coagulation cascades
In this study, we observed that complement and coagulation cascades were decreased more significantly in toddler patient groups with poor outcomes compared with infants, and presented a depleted and disturbed status of complement system and haemostasis. The explanation for this age‐specific protein expression change may be that the complement and haemostatic systems evolve during childhood, particularly in neonates and infants. On the one hand, complement system in neonates and infants show lower levels of most individual complement proteins as well as lower complement activity, compared to adults [6]. The serum levels of most complement components reached adult levels at 6 months after birth [7, 8]. In the meantime, infant B cells express low levels of the receptor for complement C3d fragment (CD21), which impedes responses to polysaccharide‐complement complexes and reduces antibody response towards polysaccharides [8, 9]. Thus, the contribution of complement system to the infection is relatively limited in infants, while toddlers have more mature complement system, which is more involved in host defense. It may even lead to over activation and disturbed status during severe infection. On the other hand, compared to adults, children exhibit decreased thrombin product in proportion to available prothrombin [10]. Instead of antithrombin (AT), alpha‐2‐macroglobulin is the dominant circulating thrombin inhibitor in young infants [11]. Moreover, infants and children had more time for fibrin formation, slower fibrin formation velocity, less clot formation, larger pore size in the fibrin clot structure and less resistance to fibrinolysis than adults [12]. Therefore, the younger populations had a relatively lower incidence of thrombosis, while older populations were more likely to fall into a reduced and disturbed coagulation status during severe sepsis with poor outcome.
4.2. Diagnosis and therapy
HP is a highly abundant acute‐phase glycoprotein in plasma. HP plays an important role in protecting the foetus and neonates from inflammation‐derived injury [13]. HP scavenges toxic haemoglobin, dampens neutrophilic oxidative bursts, inhibits endotoxin‐induced inflammation, suppresses monocytic TNFα and IL‐12 production, and inhibits B and T lymphocyte proliferation [14, 15, 16, 17, 18]. Generally, HP plasma levels increase during infections, inflammation, and various malignant diseases and decrease with malnutrition, haemolysis, hepatic disease, allergic reactions and seizure disorders [19]. In our study, HP showed increased levels in infant patient groups with poor prognosis but decreased levels in toddler groups with poor prognosis compared to their matched patients with good prognosis. PPI analysis showed that HP interacted mainly with acute inflammatory proteins, such as SAA1/2, S100A8/9, and S100A12, in infant groups (Figure 4). However, in toddlers, HP was coexpressed with haemoglobin proteins (HBA1, HBB, and HBD), lipoproteins (APOA1, LPA, and PLA2G7) and inflammatory proteins (SAA1/2). This result suggested that infant septic patients with poor prognosis were possibly associated with inflammation, whereas toddler patients with poor prognosis were possibly associated with multiple factors, such as haemolysis, malnutrition, and inflammation. This notion was consistent with the literature [19]. Altogether, developmental age affected HP biological function and expression level during sepsis.
Another biomarker candidate is THBS1, which is a glycoprotein that is mainly found in the alpha granules of platelets and secreted by numerous other cells, such as endothelial cells, leukocytes, smooth muscle cells, monocytes and macrophages, under conditions of stimulation [20, 21, 22]. THBS1 interacts with proteins on the cell membrane and extracellular matrix and contributes to platelet aggregation, angiogenesis, wound healing, and the immune response [23, 24]. THBS1 was reported as an element of a multiple biomarker‐based risk model for pediatric septic shock stratification [25]. However, one single‐centre cohort study in the ICU showed that baseline thrombospondin‐1 concentrations were not associated with mortality in adult septic patients [26]. Intriguingly, we found that THBS1 was a prognostic biomarker candidate of poor outcome only in septic infants but not toddler patients. PPI analysis indicated that THBS1 is mainly coexpressed with different proteins involved in platelet degranulation between the infant and toddler groups (Figure 4). This finding suggested that developmental age may affected the biological function of THBS1 during sepsis.
In this study, the response to steroid hormone was significantly downregulated in the group of infants with sepsis but upregulated in nonsurviving toddlers, suggesting that toddlers with extremely poor outcomes may positively respond to steroid hormone therapy and that infants were probably less responsive to steroids (supplement 4‐Figure S2). This finding is consistent with the literatures. In patients with endotype B, older children, and patients with intermediate to high PERSEVERE‐based mortality, the use of corticosteroids was independently associated with a greater than 10‐fold reduction in the risk of a complicated course [27, 28]. Another study demonstrated that adjunctive corticosteroids were independently associated with an increased risk of mortality in the subjects of subclass A (smaller median age), but not in the subjects in subclass B (greater median age) [29]. When testing the interaction between subclass and adjunctive corticosteroids, the effect of corticosteroids on the mortality odds ratio was increased approximately four‐fold in subclass A patients compared with subclass B [30].
4.3. Concluding remarks
Our proteomic analysis combined with clinical laboratory data provided comprehensive insight into the complex, heterogeneous host response to pneumonia‐derived pediatric sepsis and allowed identification of critical protein differences between the two developmental age groups. Moreover, based on a single centre cohort, combined HP and THBS1 was confirmed as a potential infant‐specific prognostic marker of sepsis with an AUC value of 0.958 (95% CI: 0.868–1.000), suggesting that a more accurate prognosis should be age adapted. Therefore, a multicentre cohort study with a large sample size is urgently needed for developmental age‐adapted prognosis of and therapies for pneumonia‐derived pediatric sepsis and improved survival of septic children.
4.4. Associated data
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD026656 [31].
CONFLICT OF INTEREST
The authors have declared no conflicts of interest.
Supporting information
Supporting Information
Table S1. Demographics and clinical characteristics of the patient groups
Table S2. Description of the plasma samples for LC ms/ms in this study
Table S3 Criterion for cliassification of sepsis severity
Table S4 The distribution of weight‐for‐age between SI and NI at both ages
Table S5 The distribution of Length/height‐for‐age between SI and NI at both ages
Supporting Information
Figure S1: Principal Component Analysis (PCA) of patient samples on protein level
Figure S2: Biological function enrichment based clustering of the differentially expressed proteins in each group
Figure S3: Molecular function enrichment based clustering of the differentially expressed proteins in each group
Figure S4: Protein domain enrichment‐based clustering of the differentially expressed proteins in each group
Figure S5: Clinical laboratory tests of coagulation function and complement components
Figure S6: Identification of hub proteins of each cluster by PPI analysis with cytoHubba calculation
ACKNOWLEDGMENTS
The authors thank the Hunan Provincial Natural Science Foundation of China (NO. 2020SK2114, Xiulann Lu), Hunan Provincial Natural Science Foundation of China (NO. 2021JJ70080, Ting Luo), the Key Laboratory of Emergency Medicine for Children, Hunan Provincial Natural Science Foundation of China (NO. 2020SK1014‐3, Zhenghui Xiao), and the Ministry of Science and Technology in China (NO. 2012BAI04B01, Yimin Zhu) for financially supporting of this work.
Luo, T. , Yan, H. , Li, X. , Deng, Y. , Huang, J. , Li, L. , Xiao, Z. , & Lu, X. (2022). Proteomic analysis identified potential age‐associated prognostic biomarkers in pneumonia‐derived pediatric sepsis. Proteomics – Clinical Applications, 16, e2100036. 10.1002/prca.202100036
Ting Luo and Haipeng Yan contributed equally to this work.
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- 1. Black, R. E. , Cousens, S. , Johnson, H. L. , Lawn, J. E. , Rudan, I. , Bassani, D. G. , Jha, P. , Campbell, H. , Walker, C. F. , Cibulskis, R. , Eisele, T. , Liu, L. , & Mathers, C. (2010). Global, regional, and national causes of child mortality in 2008: A systematic analysis. Lancet, 375(9730), 1969–1987. 10.1016/S0140-6736(10)60549-1 [DOI] [PubMed] [Google Scholar]
- 2. Xiao, C. , Wang, S. , Fang, F. , Xu, F. , Xiao, S. , Li, B. , Zhang, G. , Luo, X. , Jiang, J. , Huang, B. , Chen, Y. , Chen, J. , Wang, H. , Yu, J. , Ren, D. , Ren, X. , & Tang, C. (2019). Epidemiology of pediatric severe sepsis in main PICU centers in southwest china. Pediatric Critical Care Medicine, 20(12), 1118–1125. 10.1097/PCC.0000000000002079 [DOI] [PubMed] [Google Scholar]
- 3. Hartman, M. E. , Saeed, M. J. , Powell, K. N. , & Olsen, M. A. (2019). The comparative epidemiology of pediatric severe sepsis. Journal of Intensive Care Medicine, 34(6), 472–479. 10.1177/0885066617735783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Souza, D. C. , Barreira, E. R. , & Faria, L. S. (2017). The epidemiology of sepsis in childhood. Shock (Augusta, Ga.), 47(1S Suppl 1), 2–5. 10.1097/SHK.0000000000000699 [DOI] [PubMed] [Google Scholar]
- 5. Leteurtre, S. , Duhamel, A. , Salleron, J. , Grandbastien, B. , Lacroix, J. , & Leclerc, F. (2013). PELOD‐2. Critical Care Medicine, 41(7), 1761–1773. 10.1097/CCM.0b013e31828a2bbd [DOI] [PubMed] [Google Scholar]
- 6. Kollmann, T. R. , Kampmann, B. , Mazmanian, S. K. , Marchant, A. , & Levy, O. (2017). Protecting the newborn and young infant from infectious diseases: Lessons from immune ontogeny. Immunity, 46(3), 350–363. 10.1016/j.immuni.2017.03.009 [DOI] [PubMed] [Google Scholar]
- 7. Mcgreal, E. P. , Hearne, K. , & Spiller, O. B. (2012). Off to a slow start: Under‐development of the complement system in term newborns is more substantial following premature birth. Immunobiology, 217(2), 176–186. 10.1016/j.imbio.2011.07.027 [DOI] [PubMed] [Google Scholar]
- 8. Simon, A. K. , Hollander, G. A. , & Mcmichael, A. (2015). Evolution of the immune system in humans from infancy to old age. Proceedings. Biological sciences / The Royal Society, 282(1821), 20143085. 10.1098/rspb.2014.3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Griffioen, A. W. , Rijkers, G. T. , Janssens‐Korpela, P. , & Zegers, B. J. (1991). Pneumococcal polysaccharides complexed with C3d bind to human B lymphocytes via complement receptor type 2. Infection and Immunity, 59(5), 1839–1845. 10.1128/iai.59.5.1839-1845.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andrew, M. , Schmidt, B. , Mitchell, L. , Paes, B. , & Ofosu, F. (1990). Thrombin generation in newborn plasma is critically dependent on the concentration of prothrombin. Thrombosis and Haemostasis, 63(1), 27–30. [PubMed] [Google Scholar]
- 11. Schmidt, B. , Mitchell, L. , Ofosu, F. A. , & Andrew, M. (1989). Alpha‐2‐macroglobulin is an important progressive inhibitor of thrombin in neonatal and infant plasma. Thrombosis and Haemostasis, 62(4), 1074–1077. [PubMed] [Google Scholar]
- 12. Ignjatovic, V. , Pelkmans, L. , Kelchtermans, H. , Al Dieri, R. , Hemker, C. , Kremers, R. , Bloemen, S. , Karlaftis, V. , Attard, C. , De Laat, B. , & Monagle, P. (2015). Differences in the mechanism of blood clot formation and nanostructure in infants and children compared with adults. Thrombosis Research, 136(6), 1303–1309. 10.1016/j.thromres.2015.10.034 [DOI] [PubMed] [Google Scholar]
- 13. Buhimschi, C. S. , Bhandari, V. , Dulay, A. T. , Nayeri, U. A. , Abdel‐Razeq, S. S. , Pettker, C. M. , Thung, S. , Zhao, G. , Han, Y. W. , Bizzarro, M. , & Buhimschi, I. A. (2011). Proteomics mapping of cord blood identifies haptoglobin “switch‐on” pattern as biomarker of early‐onset neonatal sepsis in preterm newborns. Plos One, 6(10), e26111. 10.1371/journal.pone.0026111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oliviero, S. , & Cortese, R. (1989). The human haptoglobin gene promoter: Interleukin‐6‐responsive elements interact with a DNA‐binding protein induced by interleukin‐6. Embo Journal, 8(4), 1145–1151. 10.1002/j.1460-2075.1989.tb03485.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oh, S. ‐ K. , Pavlotsky, N. , & Tauber, A. I. (1990). Specific binding of haptoglobin to human neutrophils and its functional consequences. Journal of Leukocyte Biology, 47(2), 142–148. 10.1002/jlb.47.2.142 [DOI] [PubMed] [Google Scholar]
- 16. Arredouani, M. S. , Kasran, A. , Vanoirbeek, J. A. , Berger, F. G. , Baumann, H. , & Ceuppens, J. L. (2005). Haptoglobin dampens endotoxin‐induced inflammatory effects both in vitro and in vivo. Immunology, 114(2), 263–271. 10.1111/j.1365-2567.2004.02071.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baseler, M. W. , & Burrell, R. (1983). Purification of haptoglobin and its effects on lymphocyte and alveolar macrophage responses. Inflammation, 7(4), 387–400. 10.1007/BF00916303 [DOI] [PubMed] [Google Scholar]
- 18. Quaye, I. K. (2008). Haptoglobin, inflammation and disease. Transactions of the Royal Society of Tropical Medicine and Hygiene, 102(8), 735–742. 10.1016/j.trstmh.2008.04.010 [DOI] [PubMed] [Google Scholar]
- 19. Di Masi, A. , De Simone, G. , Ciaccio, C. , D'orso, S. , Coletta, M. , & Ascenzi, P. (2020). Haptoglobin: From hemoglobin scavenging to human health. Molecular Aspects of Medicine, 73, 100851. 10.1016/j.mam.2020.100851 [DOI] [PubMed] [Google Scholar]
- 20. Mcpherson, J. , Sage, H. , & Bornstein, P. (1981). Isolation and characterization of a glycoprotein secreted by aortic endothelial cells in culture. Apparent identity with platelet thrombospondin. Journal of Biological Chemistry, 256(21), 11330–11336. 10.1016/S0021-9258(19)68595-9 [DOI] [PubMed] [Google Scholar]
- 21. Isenberg, J. S. , Martin‐Manso, G. , Maxhimer, J. B. , & Roberts, D. D. (2009). Regulation of nitric oxide signalling by thrombospondin 1: Implications for anti‐angiogenic therapies. Nature Reviews Cancer, 9(3), 182–194. 10.1038/nrc2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lopez‐Dee, Z. , Pidcock, K. , & Gutierrez, L. S. (2011). Thrombospondin‐1: Multiple paths to inflammation. Mediators of Inflammation, 2011, 296069. 10.1155/2011/296069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baenziger, N. L. , Brodie, G. N. , & Majerus, P. W. (1971). A thrombin‐sensitive protein of human platelet membranes. Proceedings of the National Academy of Sciences of the United States of America, 68(1), 240–243. 10.1073/pnas.68.1.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baenziger, N. L. , Brodie, G. N. , & Majerus, P. W. (1972). Isolation and properties of a thrombin‐sensitive protein of human platelets. Journal of Biological Chemistry, 247(9), 2723–2731. 10.1016/S0021-9258(19)45271-X [DOI] [PubMed] [Google Scholar]
- 25. Standage, S. W. , & Wong, H. R. (2011). Biomarkers for pediatric sepsis and septic shock. Expert Review of Anti‐Infective Therapy, 9(1), 71–79. 10.1586/eri.10.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Der Wekken, R. J. , Kemperman, H. , Roest, M. , & De Lange, D. W. (2017). Baseline thrombospondin‐1 concentrations are not associated with mortality in septic patients: A single‐center cohort study on the intensive care unit. Intensive Care Med Exp, 5(1), 7. 10.1186/s40635-017-0120-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wong, H. R. , Atkinson, S. J. , Cvijanovich, N. Z. , Anas, N. , Allen, G. L. , Thomas, N. J. , Bigham, M. T. , Weiss, S. L. , Fitzgerald, J. C. , Checchia, P. A. , Meyer, K. , Quasney, M. , Hall, M. , Gedeit, R. , Freishtat, R. J. , Nowak, J. , Raj, S. S. , Gertz, S. , & Lindsell, C. J. (2016). Combining prognostic and predictive enrichment strategies to identify children with septic shock responsive to corticosteroids*. Critical Care Medicine, 44(10), e1000–e1003. 10.1097/CCM.0000000000001833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Atkinson, S. J. , Cvijanovich, N. Z. , Thomas, N. J. , Allen, G. L. , Anas, N. , Bigham, M. T. , Hall, M. , Freishtat, R. J. , Sen, A. , Meyer, K. , Checchia, P. A. , Shanley, T. P. , Nowak, J. , Quasney, M. , Weiss, S. L. , Banschbach, S. , Beckman, E. , Howard, K. , Frank, E. , …, Wong, H. R. (2014). Corticosteroids and pediatric septic shock outcomes: A risk stratified analysis. Plos One, 9(11), e112702. 10.1371/journal.pone.0112702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wong, H. R. , Cvijanovich, N. , Lin, R. , Allen, G. L. , Thomas, N. J. , Willson, D. F. , Freishtat, R. J. , Anas, N. , Meyer, K. , Checchia, P. A. , Monaco, M. , Odom, K. , & Shanley, T. P. (2009). Identification of pediatric septic shock subclasses based on genome‐wide expression profiling. Bmc Medicine [Electronic Resource], 7, 34. 10.1186/1741-7015-7-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wong, H. R. , Cvijanovich, N. Z. , Anas, N. , Allen, G. L. , Thomas, N. J. , Bigham, M. T. , Weiss, S. L. , Fitzgerald, J. , Checchia, P. A. , Meyer, K. , Shanley, T. P. , Quasney, M. , Hall, M. , Gedeit, R. , Freishtat, R. J. , Nowak, J. , Shekhar, R. S. , Gertz, S. , Dawson, E. , …, Lindsell, C. J. (2015). Developing a clinically feasible personalized medicine approach to pediatric septic shock. American Journal of Respiratory and Critical Care Medicine, 191(3), 309–315. 10.1164/rccm.201410-1864OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perez‐Riverol, Y. , Csordas, A. , Bai, J. , Bernal‐Llinares, M. , Hewapathirana, S. , Kundu, D. J. , Inuganti, A. , Griss, J. , Mayer, G. , Eisenacher, M. , Pérez, E. , Uszkoreit, J. , Pfeuffer, J. , Sachsenberg, T. , Yılmaz, Ş. , Tiwary, S. , Cox, J. , Audain, E. , Walzer, M. , …, Vizcaíno, J. A. (2019). The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Research, 47(D1), D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Table S1. Demographics and clinical characteristics of the patient groups
Table S2. Description of the plasma samples for LC ms/ms in this study
Table S3 Criterion for cliassification of sepsis severity
Table S4 The distribution of weight‐for‐age between SI and NI at both ages
Table S5 The distribution of Length/height‐for‐age between SI and NI at both ages
Supporting Information
Figure S1: Principal Component Analysis (PCA) of patient samples on protein level
Figure S2: Biological function enrichment based clustering of the differentially expressed proteins in each group
Figure S3: Molecular function enrichment based clustering of the differentially expressed proteins in each group
Figure S4: Protein domain enrichment‐based clustering of the differentially expressed proteins in each group
Figure S5: Clinical laboratory tests of coagulation function and complement components
Figure S6: Identification of hub proteins of each cluster by PPI analysis with cytoHubba calculation
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


