Abstract
This review summarizes the current understanding of the development of the neuroendocrine gonadotropin‐releasing hormone (GnRH) system, including discussion on open questions regarding (1) transcriptional regulation of the Gnrh1 gene; (2) prenatal development of the GnRH1 system in rodents and humans; and (3) paracrine and synaptic communication during migration of the GnRH cells.
Keywords: development, GnRH, guidance cues, migratory pathway, olfactory ensheathing cells, olfactory placode, synaptic and paracrine signals, terminal nerve, transcription factors
The development of the neuroendocrine gonadotropin‐releasing hormone (GnRH) system is a conserved event in vertebrates. These cells originate in the nasal placode (A, gray area, asterisk) yet end of distributed in the forebrain (B, mouse parasagittal section depicted. Dots = GnRH cells). This journey is depicted in C‐F. C. Mouse, E11.5 stage, GnRH cells (brown) can are found within the nasal placode (boxed area). D. Mouse, E14.5 stage, GnRH cells (green, arrows) migrate along Contactin‐2 positive fibers (magenta) toward the developing forebrain. E. Schematic depicting the multiple interactions, paracrine and synaptic, that occur between GnRH, olfactory ensheathing cells, as well as non‐GnRH cells during this journey. F. Migration of GnRH cells (green) along peripherin axons (red) visualized using 3DISCO technique on human fetus, stage CS19. Abbr. TV= telencephalic vesicle, IV= fourth ventricle, aVNO= anlage of the vomeronasal organ (VNO), OB= olfactory bulb, ON = olfactory nerve, NFJ = nasal forebrain junction, FB= forebrain.

1. INTRODUCTION
Sequencing of the gonadotropin‐releasing hormone (GnRH) peptide 1 , 2 revolutionized GnRH research because antibodies were produced that allowed (1) the anatomical location of GnRH cells to be characterized in a variety of species; (2) secretion studies to be performed; and (3) electrophysiological properties of GnRH neurons to be examined, as verified by post‐hoc immunodetection of recorded cells. The majority of these studies concentrated on the GnRH system postnatally as a function of reproductive status. However, between 1984 and 1991, 3 , 4 , 5 , 6 , 7 , 8 , 9 a series of landmark papers transformed our knowledge of the developmental, anatomical, and physiological features of the GnRH. These papers initiated a ‘new’ look at the neuroendocrine GnRH system and subsequently a wealth of information has been published, 10 yet many important questions remain for future GnRH researchers.
Postnatally, in all vertebrates, the neuroendocrine gonadotropin‐releasing hormone cells (GnRH, GnRH1 in mammals and GnRH3 in zebrafish) are not confined to classic anatomical nuclei, and are often distributed rostrally from the olfactory bulbs to the caudal hypothalamus (Figure 1A), with the exact distribution being species dependent. This posed an intriguing question: where were the GnRH progenitors and how did the system develop? Thirty‐two years ago, two groups independently presented work addressing this question. These studies demonstrated that the GnRH neuroendocrine cells in mouse arise outside the central nervous system (CNS), in the developing olfactory pit, and migrate into the forebrain during embryonic development 4 , 5 , 6 (Figure 1B, C), and that disruption of GnRH migration in humans is associated with Kallmann syndrome. 7 Subsequent to these initial reports, the migration of GnRH cells from the bilateral developing olfactory pits has been documented in all vertebrates, 10 helping to explain the symmetry of the neuroendocrine GnRH cells around the midline brain axis (Figure 1A), with the final location of (the majority of) neuroendocrine GnRH cells in each species related to guidance cues, as well as the brain development of that particular species.
FIGURE 1.
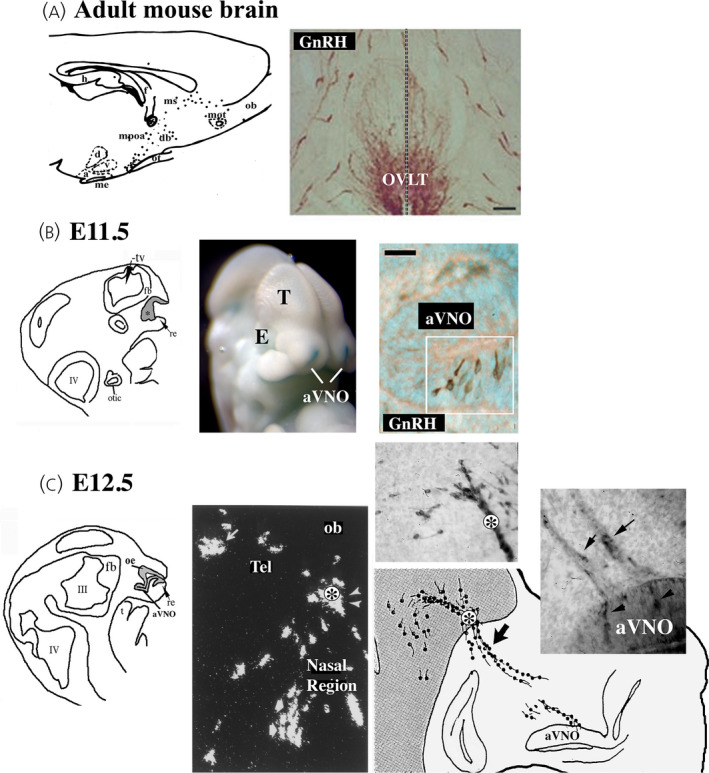
Overview of the development of the neuroendocrine gonadotropin‐releasing hormone (GnRH) system. (A) Adult distribution in the mouse. Left: Schematic of GnRH cells (black dots) in a parasagittal adult mouse brain section showing that GnRH cells are distributed from the olfactory bulbs to the caudal hypothalamus. Ob, olfactory bulbs; mot, medialolfactory tract; ms, medial septum; dbb, diagonal band of broca; mpoa, medial preoptic area; ot, optic tract; h, hippocampus; d, dorsomedial hypothalamic nucleus; v, ventromedial hypothalamic nucles; a, arcuate nucles; me, median eminence. Right: Coronal section at the level of the organum vasculosum lamina terminalis (OVLT) immunostained for GnRH showing bilateral distribution across midline (dotted line) and dispersed distribution of cells. (B) Left: Schematic illustrating structure of embryonic day (E)11.5 mouse embryo. Grey areas represent the developing olfactory pit (*, olfactory epithelium and anlage of the vomeronasal organ [aVNO]). Tv, telencephalic vesicle; re, respiratory epithelium; t, tongue; fb, forebrain. Middle: Image showing whole mount E11.5 Fgf8 lacZ/+ knock‐in mouse embryo 16 with olfactory pits (blue reaction, white arrows), eye (E) and telencephalon (T). Right: E11.5, immunostaining against GnRH detects immunopositive cells (brown, cells in boxed region) in the ventral portion of the anlage of the VNO. (C) Schematic of E12.5 mouse embryonic section through nasal region, olfactory epithelium (OE) and anlage of the VNO are visible. Middle: in situ hybridization histochemistry included in the original 1989 paper 5 showing cells expressing GnRH mRNA in the nasal region, at the nasal forebrain junction (asterisk) and within the forebrain (Tel). Right: Schematic of E12.5 head showing location of two upper panels that are photomicrographs of sections immunostained for GnRH showing positive cells emerging from the VNO area, as well as crossing the nasal forebrain junction (asterisk)
In addition to disruption of GnRH neuron migration resulting in reproductive dysfunction, the association of GnRH progenitors with a placodal structure has implications for transcriptional regulation and subsequently GnRH expression. Similar to CNS structures, placodes are initially patterned by gradients of growth and guidance factors. These factors, in turn, induce the expression of transcription factors in a concentration‐dependent manner. These broadly expressed transcription factors then act in a combinatorial fashion to specify subregions prior to the onset of neurogenesis and subsequently can have additional roles in controlling the specification and differentiation of individual neuronal subtypes. As such, perturbation of expression of growth and guidance factors in the developing nasal region can have profound effects on GnRH neuron development and/or GnRH expression.
2. THE GNRH GENE: STRUCTURE AND EXPRESSION PATTERN
The Gnrh (or Gnrh1) gene sequence was first characterized in 1984, encoding the entire 92‐amino acid GnRH peptide precursor, prepro‐GnRH. 3 This precursor contains a signal peptide located at the N‐terminus, GnRH, a cleavage signal, and the GnRH‐associated peptide (GAP) located at the C‐terminus. Removing the signaling peptide from prepro‐GnRH gives rise to the pro‐GnRH peptide, which, after being processed by an endopeptidase, produces GnRH and GAP. The final maturation step for GnRH involves removing the basic amino acids at the C‐terminus by a carboxypeptidase and conversion of the N‐terminal glutamine by a glutaminyl cyclase, resulting in the bioactive GnRH decapeptide. 1 , 2 In the mouse, these events occur early in development, indicating that the factors needed for initiating GnRH gene expression and peptide processing of GnRH are present within the nasal compartment.
The Gnrh gene contains four exons and three introns. The processing mechanism resulting in Gnrh mRNA and its ultimate translation to the GnRH decapeptide is complex. Exonic splicing enhancers located in exon 3 and 4 of the Gnrh gene are required for splicing of exon 1 from its preceding intron. 11 In the absence of exons 3 and 4, as is the case in the hypogonadal hpg mouse, splicing of exon 1 from the preceding intron is unsuccessful, leading to defective translation and ultimately resulting in the lack of the GnRH peptide. 12 , 13 Few mutations in the GNRH gene itself, as a primary cause of reproductive dysfunction in human patients, have been found (see below).
Many of the studies on transcriptional regulation of Gnrh discussed below would not have been possible without the creation of two GnRH‐secreting cell lines. In 1990, Mellon et al. 8 generated the GT1‐7 cell line that models mature GnRH neurons, whereas the GN11 cell line, generated by Radovick et al. 9 in 1991, models the migratory nature of less mature GnRH neurons. Both cell lines were created by introducing the oncogene SV40 T‐antigen driven by the GnRH promoter into transgenic mice and culturing the cells from the resultant tumors. Together, these cell lines have advanced our understanding of the mechanistic processes that involve the signaling and transcription factors needed for the fine‐tuned transcriptional control of Gnrh.
3. TRANSCRIPTION FACTORS REGULATING GNRH1
3.1. Developmental transcription factors: Otx2 and Vax1
Otx2 and Vax1 are important transcription factors for forebrain development. In addition, both play an important role in GnRH neuronal development, with a deficiency of either perturbing the development of GnRH neurons in nasal regions. Otx2 is a homeodomain protein that is expressed in GnRH neurons, binds to the proximal promoter of GnRH (Figure 2A), and is upregulated during GnRH neuronal development. Mutations in the Otx2 gene are associated with hypogonadotropic hypogonadism, although a mechanistic understanding of the function of Otx2 in development was limited because the complete loss of Otx2 is embryonically lethal. Thus, transgenic animal models that targeted Otx2 deletion in a cell‐specific manner were developed. Using a GnRH‐Cre/Otx2 knockout (KO) mouse model, Diaczok et al. 14 showed that loss of Otx2 in GnRH neurons resulted in a significant decrease in the number of GnRH neurons, which led to a suite of reproductive impairments such as delayed pubertal onset, abnormal estrous cycles, and infertility associated with hypogonadotropic hypogonadism. To determine whether the decreased number of GnRH neurons was a result of cell death, a lineage tracing strategy was used that crossed a Rosa26‐EYFP reporter mouse with the GnRH‐Cre/Otx2 KO mice. This approach allowed the investigators to track GnRH neurons that had expressed Gnrh during any time during developmental, including cells that terminally cease to express Gnrh. Decreased numbers of EYFP‐labeled GnRH neurons were found in the KO mice, demonstrating that GnRH neurons underwent cell death. This finding was supported by cleaved caspase‐1 staining along the GnRH migratory route in KO mice. Thus, Otx2 is critical for GnRH expression and survival during embryonic development prior to the cells entering the forebrain.
FIGURE 2.

Key transcriptional sites and regulatory elements in the mouse and human Gnrh 5′‐region. (A) The mouse gonadotropin‐releasing hormone (GnRH) gene (mGnRH) contains a GnRH neuron‐specific element (NSE) in the proximal promoter, which extends to −1005 bp. In addition, a GnRH neuronal enhancer region is present that also serves as an ovarian GnRH repressor region. The GnRH enhancer contains a kisspeptin‐response element (KsRE) with an Otx2 binding site important for GnRH transcriptional activation. Otx2 binding sites are also present in the enhancer, outside of the KsRE, and in the proximal promoter. (B) The human GnRH gene (hGnRH) has a promoter region extending to −551 bp and contains an AP‐1 site important for IGF‐1 responsiveness. There is also a GnRH NSE upstream of the promoter that contains binding sites for Brn‐2, a POU homeodomain factor. ERK, extracellular signal regulated kinase; PI3K, phosphoinositide 3‐kinase; pTN, putative terminal nerve. Created with bioRENDER (biorender.com)
Vax1, similar to Otx2, is crucial for GnRH neuronal function. This was initially demonstrated in studies of Vax1 null murine models that do not survive long after birth. Hoffman et al. 15 investigated the heterozygous Vax1 KO mouse, which is viable and lives to adulthood. It was observed that both male and female Vax1 heterozygotes were subfertile, with a reduction in GnRH neurons of more than 50%. 15 Later, this group 16 clarified the role of VAX1 by deleting Vax1 specifically in GnRH neurons using an established GnRH‐Cre mouse line. 17 The GnRH‐Cre/Vax1 KO mouse model had a complete loss of GnRH neuronal immunostaining, which was associated with a hypogonadal, infertile state. 16 However, when crossing this KO mouse with a LacZ reporter strain, they observed that GnRH neurons were still present but not actively expressing Gnrh. To address how Vax1 regulates Gnrh expression, they used luciferase and electrophoretic mobility shift assays in GN11 and GT1‐7 cells and determined that Vax1 (which was only highly expressed in GT1‐7 cells) acts through the GnRH proximal and enhancer 1 regions to regulate Gnrh.
4. TRANSCRIPTIONAL DIRECTION OF GNRH NEURON‐SPECIFIC EXPRESSION
Notably the GnRH gene is also expressed in other reproductive tissues, such as the mammary gland, ovaries, and placenta. 18 , 19 Although the central function of GnRH neurons in the regulation of reproduction and fertility was established soon after its discovery, the mechanisms underlying the cell‐specific expression of the GnRH gene and protein in neuroendocrine GnRH cells, as well as the role of GnRH in the extrahypothalamic tissues, remained a mystery. In addition, during development, GnRH is expressed transiently in other neuronal cell types. In 2002, Wolfe et al. 20 sought to investigate the cis‐regulatory elements that specifically target GnRH transcription to hypothalamic neurons. By generating transgenic mice with a luciferase reporter construct bound to the human GnRH promoter (hGnRH), they identified two binding sites for a POU homeodomain transcription factor known as Brn‐2 (Figure 2B) as an essential transcriptional target for directing hypothalamic‐specific GnRH expression. 20 Brn‐2 is characterized by its binding to an octameric DNA sequence (ATGCAAAT), which is common among this family of POU transcription factors. Not only was Brn‐2 mRNA determined to be expressed in GnRH neurons in mice, but also Brn‐2 was shown to stimulate mouse GnRH (mGnRH) gene expression by transfecting GN11 cells with a Brn‐2 expression vector. In another study, the same study group identified specific promoter regions that directed GnRH expression to either the hypothalamus or the ovary (Figure 2A) using different GnRH promoter deletion fragments fused to a luciferase reporter. These experiments showed that the DNA sequence located −1005 bp from the mGnRH promoter was sufficient to direct both hypothalamic and ovarian GnRH expression. 21 Additionally, an enhancer region between −3446 and −2078 bp was identified that (1) specified neuronal GnRH expression and (2) contained an ovarian repressor element (Figure 2A). To determine the physiological significance of this GnRH regulatory element, this neuron‐specific enhancer region was further characterized by generating a mouse with a deletion of GnRH promoter fragment between −2806 and −2078 bp. In addition to decreased hypothalamic GnRH gene expression and increased ovarian GnRH immunohistochemical staining, this GnRH regulatory element KO mouse also exhibited delayed pubertal onset and abnormal estrous cyclicity. 22 Together, these studies advanced our understanding of the specific elements needed to direct GnRH expression to neurons in critical tissues (i.e., hypothalamus) and repress its expression in other tissues, which may negatively impact the function of the reproductive axis. Clearly, these are interesting regions of the GnRH promoter, for which further research is needed to identify additional proteins that might bind to these sequences, forming complexes that may differentially act on GnRH transcription.
5. KISSPEPTIN AS THE MAJOR NEUROPEPTIDE REGULATOR OF GNRH EXPRESSION
The hypothalamic neuropeptide kisspeptin (Kiss1) is now considered to be the major secretagogue for GnRH and activates signaling cascades through its G‐protein coupled receptor Kiss1R, which is highly expressed in GnRH neurons. In vitro studies using the GnRH‐secreting cell lines, GN11 and GT1‐7, demonstrated how Kiss1 upregulates Gnrh gene expression and how active transcriptional regulation of Gnrh requires dynamic chromatin conformational changes and accessibility to key enhancer regions. 23 , 24 Novaira et al. 24 used chromatin immunoprecipitation assays to determine the mechanisms by which Kiss1 mediates the regulation of the GnRH gene. They discovered a kisspeptin‐response element (KsRE) between −3446 and −2806 bp of the mGnRH gene using luciferase reporter gene deletions. Furthermore, their study also determined that the KsRE in enhancer region 2 was a binding site for Otx2 and that Kiss1 treatment increased Otx2 gene and protein expression and its binding to the KsRE (Figure 3).
FIGURE 3.

Kisspeptin induction of gonadotropin‐releasing hormone (GnRH) gene expression. Kisspeptin induces signaling cascades via activation of its G‐protein coupled receptor, Kiss1R. These signaling cascades activate chromatin conformational changes by recruiting the transcription factor Otx2 at specific binding sites in the kisspeptin response element (KsRE) of the GnRH enhancer region and the neuron‐specific element (NSE) in the proximal promoter region. Created with bioRENDER (biorender.com)
Prior to identifying the KsRE and its role in Otx2 recruitment, another Otx2 binding site had been located at −356 and −249 bp in the regulatory region of the mGnRH promoter 25 as a neuron‐specific element (NSE) (Figure 2A) which directed the expression of the mGnRH gene specifically to neurons. The 2009 study by Novaira et al. 24 also showed that Kiss1 treatment increased the interaction between this Otx2 NSE and the KsRE, providing a model of how chromatin loop formation regulates mGnRH gene transcription. Whether other GPCRs can act on Gnrh transcription in a manner similar to that outlined for the signaling through the Kiss1R (i.e., recruit Otx2 and mediate chromatin modifications) remains to be determined.
6. GNRH TRANSCRIPTION AND METABOLIC/GROWTH FACTOR SIGNALS: INSULIN AND INSULIN‐LIKE GROWTH FACTOR‐1
The hypthalamic–gonadal axis is sensitive to environmental factors such as nutrition, with feedback from peripheral metabolic signals, such as insulin, affecting Gnrh gene expression. Both animal studies and cell culture models have been used to explore the regulatory role of insulin on the reproductive axis, and specifically on GnRH function. It has been reported by several groups that a reduction in insulin levels results in decreased GnRH pulsatile release, whereas administration of insulin stimulates GnRH neurons in vivo and in vitro. To investigate the intracellular mechanisms that mediate GnRH regulation by insulin, studies were performed in the GnRH cell lines GT1‐7 and GN11, which express the insulin receptor. Kim et al. 26 used a luciferase reporter transfection assay to show that insulin treatment increased mGnRH promoter activity in a region located between −1250 and −587 bp from the transcriptional start site and that mitogen‐activated protein kinase (MAPK) signaling mediated the insulin response because this effect was blocked by a MEK inhibitor. Early growth response‐1 (Egr‐1) had been shown to be a transcription factor regulated by insulin in non‐reproductive tissues and was activated through MAPK signaling. Experiments using GN11 cells showed that Egr‐1 binds to the proximal promoter region (−67 and −76 bp) of the mGnRH (Figure 2A) and mutations of this region demonstrated its role in mediating the insulin‐induced increase in GnRH. 27
Insulin and insulin‐like growth factor 1 (IGF‐1) are members of the insulin‐like family that overlap in activating their cognate receptors, insulin receptor and IGF‐1 receptor. Insulin and IGF‐1 share structural homology suggesting the existence of signaling crosstalk in the hypothalamic reproductive axis. Similar to the insulin studies above, IGF‐1 has also been shown to stimulate GnRH secretion in vivo, as well as induce GnRH gene transcription. Using the GnRH‐secreting NLT cell line. 9 , 28 Zhen et al. 29 demonstrated that IGF‐1 induced GnRH transcription by activating an AP‐1 binding site in the promoter region of the human GnRH (hGnRH) (Figure 2B) gene that was transfected into the NLT cells. The AP‐1 site, which has a consensus sequence of (TGACTCA), is located between −402 and −396 bp in the 5′ flanking promoter region of the hGnRH gene. Although a classic AP‐1 binding site was not found in the mGnRH promoter, Fos/Jun heterodimers can produce an AP‐1 transcriptional factor to mediate the effects of IGF‐1 on GnRH transcription. Thus, the obstacles presented by the divergence of cis‐regulatory elements in the promoter regions of human and mouse GnRH can potentially be overcome by targeting similar signaling pathways such as MAPK signaling, as suggested previously. 30 How such regulatory elements might change in accordance with the species is an interesting question, the answer to which may help unravel unique reproductive strategies used by species to cope with environmental challenges.
7. MICRORNA REGULATION OF GNRH
MicroRNAs (miRNAs) are single‐stranded non‐coding RNAs that have short sequences (approximately 22 nucleotides). Despite their small size, miRNAs play very important regulatory roles at the post‐transcriptional level. Recent advances have identified specific miRNAs that enhance GnRH transcription, especially prior to pubertal onset, when the establishment of tonic GnRH peptide secretion is critical. Messina et al. 31 identified two important miRNAs that regulate GnRH in the infantile period: miR‐200 and miR‐155. MiR‐200 regulates the GnRH gene directly and through the GnRH repressor Zeb1, whereas miR‐155 regulates GnRH through Cebpb, a nitric oxide‐mediated repressor of GnRH. More recently, miR‐375 was shown to indirectly regulate GnRH expression in GT1‐7 cells by inhibiting Sp1, resulting in the activation of the GnRH repressor, Cebpb. 32 Another miRNA that was shown to be important in regulating pubertal onset is miR‐29. This miRNA inhibits GnRH expression by targeting a transcriptional activator, Tbx21. In studies using GT1‐7 cells, it was shown that blocking miR‐29 increased GnRH gene expression. In mice, the loss of brain‐specific miR‐29 resulted in earlier puberty onset and increases in luteinizing hormone secretion. 33
8. MUTATIONS IN THE GNRH GENE
Isolated or idiopathic hypogonadotropic hypogonadism (IHH), either normosmic (nIHH) or accompanied by anosmia (KS), is a rare disorder characterized by reproductive impairments such as pubertal delays/absence, hypogonadism, and infertility. Mutations in many genes involved in either the migration or development of GnRH neurons (e.g., FGF8, KAL1, PROK2, and CHD7) or the synthesis and release of GnRH (e.g., KISS1, KISS1R, and TAC3) have been described. Often, nIHH or KS are associated with variants in two interacting genes, which makes identification of the etiologies difficult. However, mutations in the GNRH gene itself as a primary cause of nIHH so far account for < 2% of cases. Although the characterization of the hpg mouse, harboring a deletion mutation in the Gnrh gene that truncates protein synthesis, set a precedent for mutations of GNRH, it was almost three decades before new mutations in the Gnrh gene were discovered. In 2009, Chan et al. 34 reported a homozygous frameshift mutation of GNRH in a male patient with severe congenital nIHH. This mutation (p.G29GfsX12) changes the codon that encodes the sixth amino acid of the GnRH decapeptide and prematurely introduces a stop codon. In addition to this homozygous mutation, Chan et al. 34 discovered rare heterozygous sequence variants of GNRH in four different patients with nIHH. A different homozygous frameshift mutation in siblings with nHH has also been reported. These subjects were found to have an insertion of an adenine (c.18‐19insA) in the N‐terminal region of the signaling sequence of the precursor peptide prepro‐GnRH. This mutation results in the aberrant production of a peptide that lacks the functional GnRH decapeptide sequence. 35 In these studies, the affected patients had a parent and/or siblings with heterozygous mutations, but no nIHH presentation. However, one heterozygous variant discovered in the study by Chan et al., 34 p.R31C, was directly implicated in GnRH deficiency and predicted to be the primary cause of nIHH in that individual. 34 The R31C variant is a missense mutation that changes an arginine in codon 31 to a cysteine in the conserved GnRH decapeptide sequence and results in a mutant GnRH peptide that has a much lower binding affinity for the GnRH receptor (GnRH‐R). Subsequent to its discovery in 2009, the R31C variant has been found in other unrelated individuals with nIHH. 36
It is worth noting that the lack of GnRH itself, such as in the hpg mouse, does not perturb migration of ‘GnRH’ cells into the forebrain, 37 whereas disruption of the region of the olfactory pit where GnRH cells are first detected, such as mutations in fibroblast growth factor 8 (FGF8)/fibroblast growth factor receptor 1, results in disruption of GnRH cell development. 38 In addition, during development, GnRH cells express many of the receptors identified in postnatal GnRH cells. 10 Thus, transcriptional regulators, such as kisspeptin, 39 may act either developmentally and/or postnatally to alter GnRH expression.
9. THE GNRH SYSTEM IN RODENTS AND HUMANS
To understand the beauty and challenges of the neuroendocrine GnRH system, one most delve into the development of craniofacial regions and, specifically, the olfactory pit. The olfactory pit, where early GnRH cells were detected in mice, develops from the olfactory placode, a structure which gives rise to both the main olfactory epithelium (olfactory sensory neurons), as well as the vomeronasal organ (VNO, pheromone sensory neurons). GnRH cells are found within the boundaries of the developing VNO. Based on location, it was proposed that, in mammals, GnRH cells originate in the olfactory placode, a structure that starts out as a thickening of ectoderm cells at the tip of the nose. Studies in other species, including human (see below), have reported similar results. Subsequent studies indicated that disrupting genes involved in olfactory placode development or olfactory axon targeting to the olfactory bulb often disrupted the GnRH system and consequently function, 10 reinforcing the association of GnRH neurons with the olfactory placode. Certainly, the identification of the prenatal journey of GnRH cells led to an explosion in research and an understanding of KS. Together, the data on the development of the GnRH neuroendocrine cells have conclusively shown that the development of the GnRH system and the olfactory system are entwined. However, the developing olfactory system is composed of many cell types that include (1) the neural crest derived olfactory ensheathing cells (OECs, see last section) and (2) sensory neurons that send axons to the brain which detect chemical (main olfactory epithelium) or pheromones (sensory cells in the VNO), as well as (3) an ill‐defined structure known as the terminal ganglion, the axons of which also course through nasal areas but then turn toward the rostral forebrain at the nasal/forebrain junction (NFJ). Components of these cell types form a migratory mass as they cross the nasal region. Under certain circumstances, perturbations of each of these other cell types can alter the migration of GnRH cells into the brain.
9.1. Olfactory placode and GnRH ontogenesis
In mice, between embryonic days (E)10.5 and 11.5, the olfactory placode undergoes a first neurogenic wave that gives rise to several migratory neuronal populations, including the GnRH neurons. 40 , 41 Recent data indicate that the GnRH neurons originate from Achaete‐scute homolog 1 (Ascl‐1+) progenitors, likely located in the ventral portion of the area between the prospective respiratory epithelium and the vomeronasal organ primordium, 42 (Figure 4A). The anlage of the vomeronasal organ of mice starts to form during the secondary invagination of the olfactory pit between E10.5 and 11.5. At E11.5 the non‐neurogenic prospective respiratory epithelium, positive for transcription factor AP‐2 alpha acts as a source of FGF8, which is crucial for GnRH neurons, as well as vomeronasal neurogenesis and craniofacial development, 43 (Figure 4A). In adult humans, the VNO is a vestigial structure for which the possible function is still debated. 44 , 45 However, early work 7 showed GnRH cells in embryonic human nasal regions and recent work documented the association of GnRH cells with the embryonic human vomeronasal organ. 46 In humans, the olfactory placodes invaginate at Carnegie stage (CS) 16 (approximately day 39 of gestation) (Figure 4D). After invagination occurs, similar to the mouse, 47 a gradient expression of the transcription factors AP‐2α, which peaks in the nasal mesenchyme surrounding the respiratory epithelium, and PAX6, for which expression peaks in the presumptive olfactory/vomeronasal epithelia, was documented 46 (Figure 4D).
FIGURE 4.

Ontogenesis and molecular signatures of olfactory placode and gonadotropin‐releasing hormone (GnRH)‐1 neurons. (A) Cartoon illustrating the olfactory pit of mouse at embryonic day (E)11.5. The respiratory epithelium (blue, RE) acts as source of fibroblast growth factor 8 (FGF8), whereas the ventral portions of the nasal mesenchyme release BMP4 (proximal to the respiratory epithelium) and Noggin (proximal to the neurogenic vomeronasal organ [VNO]). GnRH‐1 neurons (Cyan) form in the ventral regions of the VNO anlage and migrate out of the developing VNO along axons of the putative terminal nerve (pTN) (gold). The TN neurons express transient axonal glycoprotein‐1 (TAG‐1)/contactin‐2 (Cntn‐2); peripherin; roundabout‐3 (Robo)3 and N‐CAM. (B) E14.5 Immunolabelling against GnRH‐1 Cntn‐2/TAG‐1: GnRH‐1 neurons (white) migrate out of the VNO along Cntn‐2/TAG‐1 positive axons. (C) E14.5 GnRH‐1 neurons (white) migrating from the VNO to the preoptic area (POA) along Robo3 immunoreactive (magenta) axons of the terminal nerve (TN). Cell bodies of the terminal nerve ganglion (TNg) can be detected proximal to the VNO. (D) AP‐2α (green) and PAX6 (red) expression in a sagittal section of a Carnegie stage (CS) 16 embryo head. AP‐2α is expressed by rostro‐ and ventromedial nasal mesenchyme, as well as by a few ectodermal cells of the dorsal and ventral portion of the respiratory epithelium (RE). Pax6 is expressed in the olfactory epithelium (OE) with an opposite gradient to AP‐2α. (E) GnRH‐1 (green) and ßIII‐tubulin (red) expression in a coronal section of a CS 18 embryo. ßIII‐tubulin‐positive VNN axons emerge from the developing VNO and form the axonal scaffold for the GnRH‐1 migratory process. (F, G) Expression of Cntn‐2/TAG‐1 (F, arrows) and GnRH‐1 (G) in a CS 19 embryo shows that GnRH‐1 cells use Cntn‐2/TAG‐1 positive fibers as scaffold for migration (G, merge). (H) LV, lateral ventricle; CX, cortex; HIP, hippocampus; THAL, thalamus; SEP, septum; CP, caudate putamen; HYP, hypothalamus; TN, terminal nerve; OB, olfactory bulb; VNN, vomeronasal nerve; VNO, vomeronasal organ; OE, olfactory epithelium. Left: Schematic of a sagittal section of a gestational week (GW)9 human fetus. Red fiber tracts indicate the VNN and grey fiber tracts represent the projections of the TN. Right: Sagittal section through the nose of a GW 9 foetus immunostained for neuropilin‐2 (Npn‐2; red; molecular marker of the developing VNO) and GnRH‐1 (green). Npn‐2 is expressed by axons emerging from the VNO (arrows). GnRH‐1 neurons migrate toward the forebrain along the Npn‐2‐immunoreactive VNO axons. Scale bars: (B, C) 100 µm; (D) 200 µm; (E) 500 µm; (F) 60 µm; (G) 20 µm; and (H) 100 µm. (A–C) Previously unpublished images. 64 , 67 (D–H) Adapted with permission 46
In rodents, the neurogenesis of the vomeronasal sensory neurons is established after the ontogenesis of the migratory neurons, between E11.5 and E12.5. 45 , 48 , 49 Notably, GLI family zinc finger 3 (Gli3) loss‐of‐function negatively affects the ontogeny of vomeronasal sensory neurons but not GnRH neurogenesis. 42 These data suggest that the GnRH neurons form from a distinct site of neurogenic progenitors compared to those giving rise to vomeronasal neurons. Consistent with this, formation of a proliferative vomeronasal primordium has been observed during embryonic development in many animal species, such as birds, which do not form a functional sensory vomeronasal neuroepithelium at later stages but have been shown to have migrating GnRH cells in nasal areas during development. In addition, during human fetal development, the VNO also retains proliferative potential at least until week 14 of gestation. 46 These data suggest that the human embryonic VNO anlage contains actively proliferating progenitors comprising potential precursors of the cellular types belonging to the migratory mass that coalesce during the GnRH migratory process, and which most likely contribute to the proper migration and differentiation of GnRH neurons. 45
9.2. Olfactory region and GnRH migration
‘Pioneer’ olfactory axons have been documented in mouse crossing the nasal region and targeting the base of the developing telencephalon. 50 Documentation of such axons, as well as the source of these axons (olfactory sensory cells, pheromone sensory cells, terminal ganglion cells), is needed in other species. Such data will help to unravel the cues that direct this group of axons and determine whether these pioneer axons are a prerequisite for the guidance of all future olfactory/vomeronasal/terminal nerve axons and subsequently appropriate migration of GnRH cells to the developing forebrain. In addition, migratory neuronal progenitor cells and neuronal precursors have been identified leaving the nasal placode prior to our ability to mark pioneer axonal outgrowth in nasal regions. 40 , 41 Identification of the early guidance cues that participate in these processes and whether they are the same as directing pioneer axons remains to be addressed. Subsequently, the GnRH cells migrate in association with a subset of intermingled ‘olfactory‐derived’ axons, together with OECs and blood vessels in nasal regions. All of these components migrate through the cribriform plate and then most GnRH cells turn caudally on a olfactory‐derived pathway (often termed the terminal nerve, the nervus terminalis or the caudal branch of the vomeronasal nerve) into the developing forebrain, in contrast to olfactory and vomeronasal sensory axons and OECs, which enter the olfactory bulb. Thus, specific cues, guidance factors, and cell adhesion molecules enable GnRH cells to differentiate between axons ending within olfactory regions and those reaching the forebrain. In the embryonic mouse, a few markers have highlighted this pathway and include peripherin 42 , 50 (a sensory intermediate filament), TAG‐1 51 also known as Cntnt2 (transient axonal glycoprotein‐1/contactin‐2), roundabout‐3 (Robo)3 and DCC 52 (deleted in colorectal cancer; receptor for the guidance molecule netrin‐1). In DCC KO mice, this caudal branch aberrantly turns towards the cerebral cortex instead of the hypothalamus, and so too do the GnRH cells, 52 indicating the importance of this caudal axonal pathway for the establishment of the adult‐like GnRH cell distribution in mouse. However, within the brain of several species, GnRH cells had been documented in areas outside the ‘continuum’, such as the cortex and hippocampus in non‐mutant animals. As visualization techniques have improved, the distribution of GnRH cells appears to be much more diverse than previously assumed (see below) (Figure 5). How GnRH cells migrate to areas outside the ‘normal neuroendocrine’ domain, and whether these are neuroendocrine cells in nature, is unclear and requires further research. In general, however, with respect to movement of the neuroendocrine GnRH cells in either nasal or forebrain regions, to our knowledge, no report has documented a true uncoupling of GnRH cells from an axonal pathway they migrate on.
FIGURE 5.
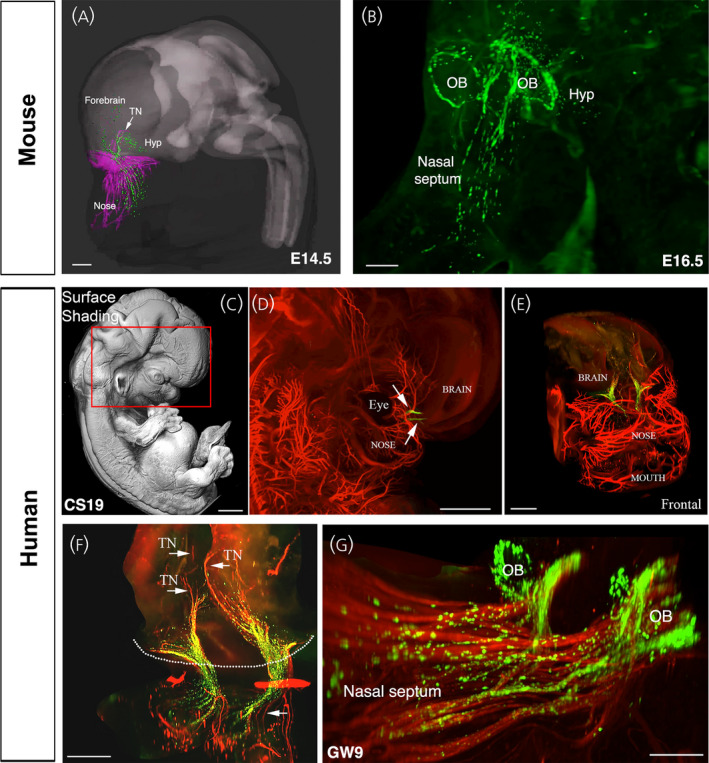
The gonadotropin‐releasing hormone (GnRH)‐1 migratory pathway in mice and humans. (A) 3D‐rendering depicting whole‐body iDISCO experiments in embryonic day (E)14.5 mouse embryos. E14.5 embryos were immunolabelled for peripherin (magenta) and GnRH‐1 (green), cleared using iDISCO and imaged with a light‐sheet microscope (LSM) and finally 3D‐rendered using Imaris (Oxford Instruments); Hyp, hypothalamus; TN, terminal nerve. (B) Lateral projection of the whole head of an E16.5 mouse embryo immunostained for GnRH‐1 and cleared using 3DISCO. At this stage, GnRH‐1 neurons enter the brain, progress dorsally and then turn ventrally toward the hypothalamus. GnRH‐1 neurons are also found in the neocortex and septal/diagonal band of Broca, and they organize in a ring‐like structure around the olfactory bulbs (OB). (C) Surface shading image and (D, E) peripherin labelling (red) of peripheral nerves (D, E) and GnRH‐1 neurons (green; D, E) at CS 19 immunolabed using 3DISCO. (E, F) Different views and magnifications of the entire transparent head depicting the GnRH‐1 migratory neurons (green) and the migratory scaffold (red, peripherin‐immunoreactive). (G) Whole‐mount immunolabeling (GnRH‐1, green; TAG‐1, red) and 3DISCO clearing of a gestational week (GW)9 foetus. In human foetuses, as in mouse embryos, GnRH‐1 neurons form a ring‐like structure around the olfactory bulbs (OB). Scale bars: (A) 400 µm; (B) 200 µm; (C, D), 1000 µm; (E), 800 µm; (F), 300 µm; and (G), 200 µm. Adapted with permission. 46113
9.3. The pathway
In mouse, at around E11.5 days of development, GnRH neurons can be immunodetected in the anlage of the prospective VNO. 4 , 5 , 6 From this area, the GnRH neurons migrate along TAG‐1/ Cntn‐2; peripherin; Robo3 positive axonal projections, towards the NFJ 50 , 51 , 52 (Figure 4A–C). In human fetuses, peripherin (Figure 4E) and Cntn‐2/TAG‐1 (Figure 4F–H) are also expressed by the putative vomeronasal/terminal axons, which form the scaffold for the GnRH migration. 46 In the past, as a result of the high density and often intermingled neuronal fibers in the developing nasal area and the broad immunoreactivity of peripheral nerves against peripherin and Cntn‐2/TAG‐1, it has been difficult to discriminate between axons of distinct neuronal populations present in the nasal area that GnRH cells associate with. Over the years, researchers proposed that the GnRH neurons reach the hypothalamus migrating along a subset of vomeronasal nerve fibers. 10 , 51 Other studies, based on the fact that birds and other vertebrates do not have an evident VNO, proposed that the GnRH neurons migrate to the hypothalamus along the axons of the olfactory sensory neurons, whereas others suggested that the GnRH neurons migrate on the axons of ganglionic neurons of the terminal nerve. 7
The terminal nerve, also known as cranial nerve XIII, nervus terminalis or cranial nerve zero “0”, is an enigmatic ganglionic structure that projects to the chemosensory mucosa and into the brain, and in some animal species also to the retina. 53 The terminal nerve was first described in 1895 54 as a supernumerary nerve in sharks. Subsequently, the terminal nerve has been described in multiple animal species, including cetaceans, which do not have an olfactory epithelium or olfactory bulbs, and humans, which do not have a functional VNO in adulthood. 55 , 56 , 57 , 58 In humans, the nervus terminalis has been defined as a microscopic plexus of unmyelinated peripheral nerve fascicles in the subarachnoid space. 59 , 60 The terminal nerve invades the brain as a small bundle of fibers in the region caudal to the olfactory bulbs. Thus, because of the small size of the terminal nerve plexus and partial antigenic overlaps with other peripheral nerves a distinction between other nasal sensory axons and the terminal nerve projection has been incomplete. The unclear definition of what neurons form the GnRH migratory track, together with the documented olfactory defects in patients with KS, suggested a direct developmental relationship between aberrant olfactory development and reproductive defects. However, anosmia and olfactory bulb agenesis do not necessarily translate into IHH in humans. 61 Moreover, despite many genetic and phenotypical correlations, definitive experimental evidence demonstrating connectivity of the olfactory and/or vomeronasal sensory neurons to the brain is necessary for GnRH neuronal migration into the brain.
If the terminal nerve is distinct from olfactory sensory neurons, can we dissociate the terminal nerve and olfactory development? Arx‐1 is an X‐linked homeobox gene and the null animals have defective proliferation and migration of olfactory bulb interneurons. Such developmental impairment translates into absence of protruding olfactory bulbs together with abnormal axonal termination of olfactory sensory neurons. 62 Characterization of the GnRH neuronal migration in Arx‐1 deficient mice revealed, quite unexpectedly, that lack of connection of olfactory/vomeronasal sensory neurons to the forebrain, secondary to olfactory agenesis, does not prevent the GnRH neurons from invading the brain. Indeed, in Arx‐1 null mutants, the GnRH neurons cross tangles of olfactory fibers and OECs forming the fibrocellular mass and invade the brain along the projections of the putative terminal nerve. In line with this, delayed, but successful, GnRH neuronal migration to the hypothalamus has been previously described in Gli3pdn hypomorph mice, another mouse mutant with defective olfactory bulb development. 63 These data indicate that the GnRH neurons can invade the brain migrating along terminal nerve axons and that the projections of the terminal nerve follow different guidance cues from those guiding olfactory and vomeronasal sensory neurons to either the main or the accessory olfactory bulbs. 64
Coupling whole‐mount immunofluorescence with tissue‐clearing techniques in both intact mouse and human fetuses provided evidence that GnRH neurons migrate into several hypothalamic and extrahypothalamic brain regions in tight association with peripherin‐positive vomeronasal nerve/terminal nerve fibers 46 (Figure 5), supporting the notion that the VNO and the terminal ganglion may play key roles in the ontogenesis and migration of GnRH neurons in humans, similar to other mammals. Based on those data, it is tempting to speculate that some IHH could be the result of defective central projections of the terminal nerve rather than defective projections of the main olfactory axons to the olfactory bulb, thus leading to insufficient/aberrant GnRH migration. In agreement with this hypothesis, previous studies in mice have shown that intracranial projections of the vomeronasal nerve/terminal nerve, expressing neuropilin 1 (NRP1), the receptor of semaphorin 3A, fail to enter the brain in mice lacking a functional semaphorin‐binding domain in NRP1 (Nrp1sema/sema mice) or semaphorin 3A (Sema3a−/− mice) 65 Indeed, the terminal nerve and GnRH neurons invade the brain in areas highly enriched in guidance cues Sema3A and slit guidance ligand 1 (Slit1). Notably Sema3A‐ and Slit1‐mediated repulsion prevent the olfactory and vomeronasal fibers from connecting to wrong parts of the olfactory bulb or invading the brain. 66 However, the terminal nerve does not appear to be repelled by these two guidance cues. 64 GnRH neurons invade the brain in areas rich of the repulsive cue Slit1 and fail to invade the brain in Sem3A null mutants, suggesting that Sema3A could function as an attractive, rather than repulsive, cue for the terminal nerve. 64 Without question, this proposal still needs further refinement because single and double KO mice in Robo3, a key player in silencing Slit mediated repulsion, and neural EGFL like 2 (NELL2), a guidance cue for Robo3, do not display major defects in GnRH neuronal migration. 67 Several mouse models carrying mutations in guidance cues have shown defects in GnRH neuronal migration. 10 Whether some of these molecules are specifically expressed on the terminal nerve awaits further investigation. Thus, the mechanisms that control GnRH and terminal nerve trajectories to the basal forebrain are still unclear. Understanding what the terminal nerve is, what genes control its development, and its physiological role are long lasting open questions that need to be answered to decipher the cellular and molecular mechanisms underlying congenital forms of anosmia, KS and nIHH.
9.4. Extra‐hypothalamic GnRH cells
As noted above, GnRH cells had been documented in areas outside the ‘continuum’, such as the cortex and hippocampus in several species, but largely ignored. Recent work in mouse and humans using iDISCO (i.e., immunolabeling‐enabled three‐dimensional imaging of solvent‐cleared organs) has revitalized this issue, with the number and distribution of GnRH cells being larger and more diverse than previously assumed. Until recently, data in humans were relatively scarce. 7 , 68 , 69 , 70 However, the use of histological clearing techniques coupled to light‐sheet microscopy on human fetuses, from gestational week (GW)5 to GW14, has provided the first chronological and quantitative analysis of GnRH neuron ontogenesis, differentiation, and migration, as well as a 3D atlas of their distribution in intact fetuses 46 (Figure 5). GnRH cell quantification established that approximately 10,000 GnRH‐immunoreactive neurons are present in the human fetal brain with approximately 2000 GnRH neurons scattered along the ventral forebrain, corresponding to the hypothalamic area, whereas approximately 8000 GnRH neurons migrate towards pallial and/or subpallial structures. These observations agree with other anatomical studies on primates, which identified GnRH mRNA and protein expression in similar extrahypothalamic regions unrelated to reproduction. 71 , 72 , 73 Moreover, a recent study has shown that, in adult human post‐mortem brains, between 150,000 and 200,000 GnRH synthesizing cells are in the basal ganglia and basal forebrain. 74 RNA‐sequencing analysis of cholinergic interneurons and medium spiny projection neurons microdissected from the human putamen showed selective expression of GNRH1 and GNRHR‐1 autoreceptors in the cholinergic cell population. 74 However, whether non‐reproductive functions regulated by GnRH in the human putamen exist, similar to the previously proposed regulation of systemic aging, 75 requires further investigations. Identification of the GnRH population that migrates into the forebrain, as well as their location and function, in a variety of species is needed.
10. EMBRYONIC PARACRINE AND SYNAPTIC COMMUNICATION DURING GNRH NEURONAL DEVELOPMENT (Figure 6)
In addition to adhesion and guidance molecules that are critical for leading GnRH neurons on the correct migration path, cell–cell interactions along the migration route also affect the behavior of cells through both paracrine and synaptic communication. These interactions change the electrical activity of the cells, modulate their calcium homeostasis, and activate a variety of signaling pathways that ultimately affect the motility of the cells, their passage through the different migration compartments, and their molecular and physiological maturation.
11. ELECTRICAL ACTIVITY AND CALCIUM CONTROL MIGRATION
Electrical activity of neurons affects their migration directly by affecting cytoskeleton dynamics, or indirectly through modulating gene expression patterns in the cells. In both processes, calcium acts as the prime second messenger, which conveys the information encoded by neuron excitability to the cytoskeleton or to the nucleus. The direct pathway in which calcium affects neuronal motility has been well studied in many types of migrating neurons, 76 as well as in GnRH cells. 77 , 78 Membrane depolarization causes the opening of voltage‐gated calcium channels and the ensuing intracellular calcium rise (from both extracellular sources as well as from intracellular stores) relays the information to the cytoskeleton of cells, thereby affecting their motility. The direct evidence that electrical activity leads to short‐lived increases in cytosolic calcium has recently been confirmed in migrating fish GnRH neurons recorded in vivo. 79 In mammalian GnRH neurons, calcium release from intracellular stores was also shown to cause actin flow into the leading process of the migrating neurons, thereby stimulating their forward movement. 78
There is ample evidence for the role of electrical activity on GnRH neuron migration, although there are species differences about the nature of transmitters involved. In zebrafish, glutamatergic inputs through NMDA receptors were shown to be the main excitatory neurotransmitter for GnRH cells during development, 79 although GABAergic inputs were also reported. 80 In mammals, GABA, through its excitatory GABAA receptors, is considered to be the dominant neurotransmitter affecting GnRH migration, 81 , 82 , 83 although glutamate probably also plays a role in the control of migration. 84 The electrical activity and migration of GnRH neurons can also be modulated by other signaling molecules, such as cholecystokinin (CCK), anti‐Mullerian hormone (AMH) and stromal derived growth factor‐1. 81 , 85 , 86 , 87
In addition to its direct role on cytoskeleton remodeling, electrical activity can also play an indirect role on neuronal migration by causing wide scale shifts in gene expression patterns. Such changes not only affect the speed of migration, but also cause a switch in migration mode and affect the repertoire of guidance molecules that the cells use to direct their migration. 88 , 89 , 90 , 91 This pathway is of particular interest in the case of GnRH neurons as they cross different extracellular environments, requiring them to switch migration modes along their journey from the nasal placode into the brain. The crossing of the cribriform plate, the NFJ, that separates the nasal compartment from the forebrain is one such instance.
At the NFJ, multiple tissues are juxtaposed to each other, including midline nasal cartilage, nasal mesenchyme, and the cribriform plate, as well as meninges and developing CNS tissue. Chemokines, growth factors, and neurotransmitters localized to, or forming gradients at, the NFJ, have been shown to alter movement of GnRH cells into the forebrain/out of nasal regions. Notably, GnRH cells have been shown to pause in chicken, 92 mammals, 46 , 82 and fish 79 at the NFJ (Figure 6, top). The reason for this pause is just beginning to be addressed, but it may ensure (1) maturation of GnRH1 neurons; (2) establishment/targeting of migratory pathway to appropriate brain regions; and/or (3) necessary changes in the cellular and/or extracellular matrix composition both within the nasal and/or brain regions. Thus, the pause of GnRH cells at the NFJ is considered to be an adaptation period in which the cells mature and gain the ability to cross from one environment to another, 79 , 82 although the underlying mechanisms causing the maturation were unclear. A decrease of GnRH cells as they enter forebrain, or shortly thereafter, has been documented in GAD67 mutant mice, Necdin mutant mice, AP‐2 mutant mice and Nhlh2 mutant mice, 10 consistent with premature entrance of GnRH neurons into CNS regions being detrimental to the cells. Recently, work in zebrafish has revealed that spontaneous coordinated electrical activity within the GnRH population induces a phenotypic switch in neurons that allows them to exit the pause and cross the NFJ to enter the brain. In rodents, electrical activity may play a similar role because GABAergic excitatory input onto GnRH neurons paused at the NFJ was suggested to be a possible driver of cell maturation and NFJ crossing. 82 Although the molecular details of the switch remain to be determined, single‐cell transcriptomic studies of migrating cells in the different compartments are sure to provide valuable insight into the identity of the genes that drive the process.
FIGURE 6.
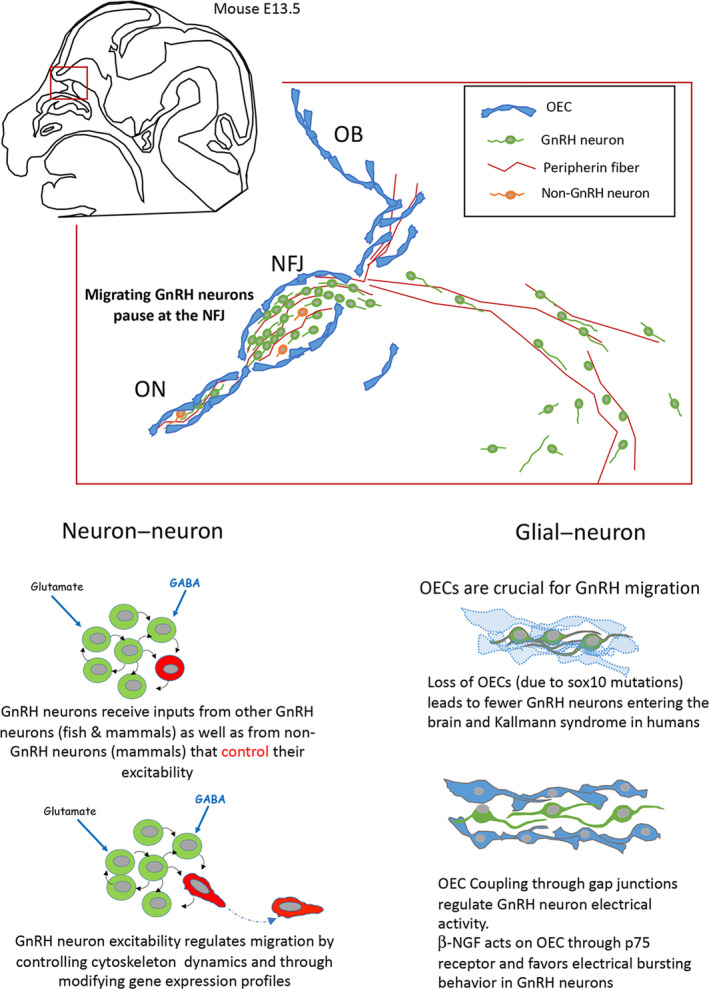
Schematic depicting paracrine and synaptic communication during gonadotropin‐releasing hormone (GnRH) neuronal development. (A) Schematic representation of a parasagittal section of an embryonic day (E)13.5 mouse embryo head and the nasal/forebrain junction (NFJ). Peripherin expressing vomeronasal (VN) and terminal nerves (TN) axons are the substrate for neuronal migration. Olfactory ensheathing cells (OECs) form a cellular sheath around neurons and VN and TN fibers. All together, they form the migratory mass. At the NFJ, the migratory mass stops and as in a rail yard, axons of the VNO and OECs turn dorsally towards olfactory bulbs (OB), whereas TN axons and GnRH neurons turn ventro‐caudally. (B) At the NFJ, GnRH neurons receive inputs from other GnRH neurons and non‐GnRH neurons. This synaptic activity controls GnRH neurons excitability and regulates gene expression as well as cell motility through intracellular calcium signaling. (C) OECs are glial cells and, as such, they can play an active role in regulating GnRH neurons excitability. For example, beta nerve growth factor (β‐NGF) regulates electrical activity by favoring bursting behavior, which may result in an increase in Ca2+ transients and an increase in motility. Sox10 codes for an important transcription factor involved in the development of neural crest and in the migratory mass and is restricted to OECs. Sox10 disruption and loss of function result in the loss of OECs and the blockade of GnRH neurons at the NFJ, suggesting that OECs play a major role in re‐activating GnRH neuronal migration towards the central nervous system
12. DRIVERS OF ACTIVITY
Despite our increasing appreciation for the importance of electrical activity in the migration of GnRH neurons, the cellular identity of the activity drivers is less understood. Indeed, the GnRH migratory environment is effectively isolated by a special type of glial cells, OECs, making it difficult for external inputs to reach the migrating cells and affect their migration. Therefore, the prime candidates for driving electrical activity of migrating GnRH neurons are cells of the migratory cell mass and OECs.
12.1. Neuron–neuron interactions (Figure 6B)
Within the migratory mass, direct interactions between GnRH neurons may be an important route by which the GnRH population controls the fate of its individual units. In fish, migratory GnRH neurons form an isolated, synaptically‐wired glutamatergic network with emergent electrical activity. 80 In mammals, where the excitatory effects of GABA on GnRH neuronal activity and migration are well established, approximately 30% of the migrating GnRH neurons are GABAergic, 93 suggesting direct communication between migrating GnRH cells. Direct GnRH‐GnRH signaling can be inferred from synchronized activity of migrating GnRH neurons 86 and the combined expression of excitatory ligand–receptor pairs such as glutamate, 84 , 94 AMH, 87 and GnRH 95 , 96 in migratory GnRH neurons. Apart from direct interaction between GnRH cells, other neurons within the migratory cell mass may also contribute to the regulation of electrical activity in GnRH neurons. Non‐GnRH GABAergic cells that are present along the migration pathway 93 and CCK‐expressing sensory cells of the developing olfactory epithelium and vomeronasal organ 85 are possible candidates for such a role. Taken together, these findings suggest that direct synaptic and paracrine interactions between neurons within the migratory cell mass play an important role in regulating the electrical activity of GnRH neurons and therefore in controlling their migration.
12.2. Glial–neuron interactions (Figure 6C)
OECs are glial cells that accompany GnRH neurons and olfactory axons in their migration from the nasal cavity to the olfactory bulbs. They form a sheath around GnRH neurons and axonal fibers and are a component of the migratory mass between nasal cavities and the cribriform plate of the ethmoid bone. 97 Because OECs form a close association with the migratory cell mass, these glial cells may be an important source of non‐synaptic input onto migrating GnRH neurons. They display both astrocytic‐ (GFAP, S100b, NCAM, Cx43, Aldh1L1, and Ptprz1) and Schwann cell‐ (vimentin, O4, P75, Mpz, and Galc) markers. 98 Notably, they can display myelinization properties when they are transplanted along damaged nerves, but they do not form myelin sheath in their natural location (nasal septum, external nerve layer of the olfactory bulb). OECs are known to improve neuronal survival, axon regeneration, and neurite outgrowth. 99 , 100 They modulate the immune response and release anti‐inflammatory proteins via exosomes and display phagocytic properties. 101 OECs cannot be unambiguously identified by a specific marker, which has hampered their specific targeting using genetic tools. Moreover, OECs display remarkable phenotypic variability. Whether this reflects genuinely different populations, different maturation status or the influence of microenvironmental factors remains unclear. OECs are neural crest‐derived, 47 , 102 migrating into the olfactory placode and then leaving together with olfactory neuronal axons, GnRH neurons, and other neurons through the nasal septum to the NFJ, together forming the migratory mass. OECs have been shown to leave the embryonic nasal placode at E10 along olfactory axons, before the onset of GnRH neuronal migration. At later stages, OECs are found in the vicinity of GnRH neurons up to the NFJ. 100
The direct role of OECs on the migration of GnRH neurons was clearly demonstrated in Sox10LacZ mutant mice where Sox10 expression was perturbed by the insertion of the LacZ reporter gene. Sox10 codes for an important transcription factor involved in the development of neural crest and, in the migratory mass, is restricted to OECs. In Sox10LacZ mice, OEC differentiation failed, and only a few LacZ‐positive cells were detected in the proximal part of the olfactory nerve and within the olfactory nerve layer. These cells did not express classical markers of OECs in the olfactory nerve layer, but did express BLBP and NPY and were probably undifferentiated neural crest cells. 103 Sox10 deletion seriously compromised olfactory axon targeting, reduced the number of olfactory neurons in the olfactory epithelium, and strongly affected migration of GnRH neurons towards the anterior part of the brain. The number of GnRH neurons detected in the preoptic area of Sox10 mutants at E16.5 embryos was reduced to one‐quarter of that observed in wild‐type embryos, whereas the proportion of GnRH neurons in the nasal parenchyma increased, indicating a critical role of OECs in GnRH neuronal migration. This role was further validated in humans in which Sox10 loss‐of‐function mutations have been found in patients with KS associated with deafness. 104 OECs express receptors that are involved in GnRH neuronal migration. GPR37 is a Parkin‐associated endothelial‐like G‐protein coupled receptor displaying a strong expression during development along the migratory route of GnRH neurons. Gpr37 and GPR37‐ligand prosaposin expression were detected in both OECs and GnRH neurons. In Gpr37 KO mice, the migration of GnRH neurons was delayed at embryonic stages, and adults displayed more GnRH in the rostral regions than wild‐type mice. 105 In vitro, the inhibition of GPR37, slows GnRH neuronal migration.
OECs can also play an indirect role on GnRH neuronal migration by regulating the electrical activity of GnRH neurons. Using embryonic mouse nasal explant cultures that recapitulate many aspects of nasal development in vivo, 48 , 106 , 107 the presence of p75 nerve growth factor receptor (NGFR) was demonstrated on OECs closely associated with GnRH neurons. The chronic application of a functional blocking p75NGFR antibody in explants induced a decrease in olfactory axon fasciculation, a reduced number of GnRH neurons, and a change in the morphology of OECs. 106 Acute beta nerve growth factor (β‐NGF) application induced a dose‐dependent increase in spontaneous electrical activity and a remodeling of PSA‐NCAM membrane immunoreactivity on GnRH neurons that suggested changes in GnRH neuron–OEC membrane–membrane interactions. 108 Whether the action of b‐NGF is direct or not remains unclear because a reverse transcriptase‐polymerase chain reaction showed the expression of Ngfr in embryonic GnRH neurons. Therefore, b‐NGF/p75NGFR can modulate the rate of GnRH neuron migration through a possible mechanism involving OECs. To date, no hypogonadotropic hypogonadism involving β‐NGF or p75NGFR has been reported.
Clearly, OECs are an important part of the developing GnRH neuron environment. OECs relay metabolic information and contribute to the regulation of olfactory function. Recent work in Drosophila showed that gut‐derived inflammatory cytokines can modulate the metabolic coupling between OECs and neurons, leading to a temporary change in olfaction. 109 Whether such mechanisms exist at embryonic stages and affect GnRH neurons is not known, although it opens the field of nutritional or metabolic programming of puberty in utero or in ovo. These initial findings in model organisms are now being complemented by studies in human fetuses, 46 accumulating genetic data in human patients suffering from reproductive disorders, 110 and manipulations of human induced pluripotent stem cells 111 that can be differentiated into functional organoids. 112 These new insights may be used to diagnose and treat neurodevelopmental reproductive disorders such as hypogonadotropic hypogonadism and central precocious puberty. In addition, these insights provide valuable tools for the manipulation and control of reproduction in commercially important animal species in which sexual maturation inhibits the desired somatic growth.
13. CONCLUSIONS
Many receptors and molecules are expressed by GnRH neurons during their migration into the forebrain. 10 However, where they might influence GnRH neuron migration in vivo may require further evaluation. It should be remembered that GnRH cells travel spatially and temporally across 3 main areas (nasal region, NFJ, forebrain), with each region having an unique composition of guidance molecules and factors. Thus, even if the GnRH cells express a specific receptor throughout their journey, it may be activated once in a specific region or multiple times in different regions. In addition, the GnRH population is already highly heterogeneous during embryonic development with different receptors (of guidance molecules) differentially expressed by the GnRH neuronal population, perhaps protecting the system from one single gene mutation disrupting it altogether. Indeed, KS and other forms of GnRH deficiency are usually polygenic in nature. In addition, many molecules (e.g., chemokines) are not confined to anatomical boundaries and may produce different responses depending on the concentration that the GnRH cells are exposed to. In addition, the GnRH neurons migrate to the brain along the terminal nerve fibers, and their migration depends on correct olfactory ensheathing cells development. As a community, we should make an effort to elucidate a deeper understanding of the cellular and molecular mechanisms underlying formation and development of both the terminal nerve and olfactory ensheathing cells. Clearly, to fully comprehend the migration of GnRH cells, we must put all the pieces in their appropriate sequence, and this is what makes this system both challenging and exciting.
With its multifactorial control of migration and the heterogeneity and isolation of its migration environment, the GnRH system provides a particularly attractive opportunity for studying neural migration. In addition to numerous extracellular cues, direct cell–cell interactions add a novel regulatory layer to the complex array of mechanisms that control GnRH neuronal development and migration. These discoveries assign a functional role for OECs that affect GnRH migration and reveal the importance of the dynamic interactions taking place between the neurons of the migratory cell mass. According to this emerging view, GnRH neurons not only follow guidance cues provided by the environment, but also form an inter‐hemispheric circuit that serves to relay information and shapes the behavior of the individual units, thus affecting the assembly of the adult circuit.
The journey of the GnRH cells is long in distance and one that is continually expanding as the embryo develops (i.e., in mice, the first GnRH1 cells leave the nasal placode around E11 with cells continuing to migrate through the cribriform at E16.5, at which time the embryo has approximately tripled in size). During migration of the GnRH cells, the milieu through which GnRH cells migrate changes as well. Much is known about the neurogenesis GnRH cells in the forming placode, as well as the pathway, cell interactions, and signals/cues that they use to cross the nasal region. New data confirms the importance of the NFJ region as a dynamic point for GnRH cell maturation, and indicates that, in mammals, non‐neuroendocrine GnRH cells enter the brain, and populate diverse regions.
Once within the brain, independent of final location, the majority of neuroendocrine GnRH axons project to the median eminence where they access (via fenestrated capillaries in the median eminence) the pituitary portal capillary system. Little is known about the molecules used to guide GnRH axons (or other neuroendocrine axons for that matter) to the median eminence. Certainly, however, it is one of the crucial final steps in assuring a functional reproductive axis. Continued research on the development of GnRH neuron is a prerequisite to understanding GnRH neuronal populations postnally, as well as pathophysiologic conditions that disrupt reproductive function. Thus, unraveling the cues/processes underlying the development of the GnRH system, both neuroendocrine as well as non‐neuroendocrine, will yield a wealth of information relevant to neuronal development of the brain and reproductive function.
CONFLICT OF INTERESTS
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
Anne Hélène Duittoz: Writing – original draft. Paolo E. E. Forni: Writing – original draft. Paolo Giacobini: Writing – original draft. Matan Golan: Writing – original draft. Patrice Mollard: Writing – original draft. Ariel L Negron: Writing – original draft. Sally Radovick: Writing – original draft. Susan Wray: Writing – original draft, editing final manuscript.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/jne.13087.
ACKNOWLEDGMENTS
The research reported in this publication was supported by the French National Research Agency (ANR) – Project NEED (CES 2008‐011); by the National Institute for Research in Alimentation and Environment (INRAE) – Department of Animal Physiology and Breeding Systems (PHASE) – Project – Playing Pools (2012‐2014) (AD); by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under the Awards R15‐HD096411 (PEF); R01‐HD097331/HD/NICHD (PEF); by the National Institute of Deafness and other Communication Disorders of the National Institutes of Health under the Award R01‐DC017149 (PEF); by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (ERC‐2016‐CoG, grant agreement no. 725149/REPRODAMH) (PG); by the Institut National de la Santé et de la Recherche Médicale (INSERM), France (grant number U1172) (PG); by the Agence Nationale de la Recherche (ANR‐15‐CE14‐0012, ANR‐18‐CE14‐0017), Leducq Foundation ERPT grant and FranceBioImaging ANR‐10‐INSB‐04 (PM).; by H2020‐MSCA‐IF‐2014 grant number 656763‐GTHREG (MG); by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under the Award R01 HD068777 (SR); and by the Intramural Research Program of the National Institutes of Health, National Institute of Neurological Disorders and Stroke (SW, ZIA NS002824).
Duittoz AH, Forni PE, Giacobini P, et al. Development of the gonadotropin‐releasing hormone system. J Neuroendocrinol. 2022;34:e13087. doi: 10.1111/jne.13087
Ariel L. Negrón, Sally Radovick contributed to Sections 2‐8.
Paolo E. Forni, Paolo Giacobini, Susan Wray contributed to Sections 1, 9 and 13.
Anne H. Duittoz, Matan Golan, Patrice Mollard contributed to Sections 10‐12.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this review because no new data were created or analyzed.
REFERENCES
- 1. Burgus R, Butcher M, Amoss M, et al. Primary structure of the ovine hypothalamic luteinizing hormone‐releasing factor (LRF) (LH‐hypothalamus‐LRF‐gas chromatography‐mass spectrometry‐decapeptide‐Edman degradation). Proc Natl Acad Sci USA. 1972;69(1):278‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matsuo H, Baba Y, Nair RM, Arimura A, Schally AV. Structure of the porcine LH‐ and FSH‐releasing hormone. I. The proposed amino acid sequence. Biochem Biophys Res Commun. 1971;43(6):1334‐1339. [DOI] [PubMed] [Google Scholar]
- 3. Seeburg PH, Adelman JP. Characterization of cDNA for precursor of human luteinizing hormone releasing hormone. Nature. 1984;311(5987):666‐668. [DOI] [PubMed] [Google Scholar]
- 4. Wray S, Nieburgs A, Elkabes S. Spatiotemporal cell expression of luteinizing hormone‐releasing hormone in the prenatal mouse: evidence for an embryonic origin in the olfactory placode. Brain Res Dev Brain Res. 1989;46(2):309‐318. [DOI] [PubMed] [Google Scholar]
- 5. Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone‐releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA. 1989;86(20):8132‐8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwanzel‐Fukuda M, Pfaff DW. Origin of luteinizing hormone‐releasing hormone neurons. Nature. 1989;338(6211):161‐164. [DOI] [PubMed] [Google Scholar]
- 7. Schwanzel‐Fukuda M, Bick D, Pfaff DW. Luteinizing hormone‐releasing hormone (LHRH)‐expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res Mol Brain Res. 1989;6(4):311‐326. [DOI] [PubMed] [Google Scholar]
- 8. Radovick S, Wray S, Lee E, et al. Migratory arrest of gonadotropin‐releasing hormone neurons in transgenic mice. Proc Natl Acad Sci USA. 1991;88(8):3402‐3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mellon PL, Windle JJ, Goldsmith PC, Padula CA, Roberts JL, Weiner RI. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron. 1990;5(1):1‐10. [DOI] [PubMed] [Google Scholar]
- 10. Cho HJ, Shan Y, Whittington NC, Wray S. Nasal placode development, GnRH neuronal migration and kallmann syndrome. Front Cell Dev Biol. 2019;7:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seong JY, Park S, Kim K. Enhanced splicing of the first intron from the gonadotropin‐releasing hormone (GnRH) primary transcript is a prerequisite for mature GnRH messenger RNA: presence of GnRH neuron‐specific splicing factors. Mol Endocrinol. 1999;13(11):1882‐1895. [DOI] [PubMed] [Google Scholar]
- 12. Cattanach BM, Iddon CA, Charlton HM, Chiappa SA, Fink G. Gonadotrophin‐releasing hormone deficiency in a mutant mouse with hypogonadism. Nature. 1977;269(5626):338‐340. [DOI] [PubMed] [Google Scholar]
- 13. Mason AJ, Hayflick JS, Zoeller RT, et al. A deletion truncating the gonadotropin‐releasing hormone gene is responsible for hypogonadism in the hpg mouse. Science. 1986;234(4782):1366‐1371. [DOI] [PubMed] [Google Scholar]
- 14. Diaczok D, DiVall S, Matsuo I, Wondisford FE, Wolfe AM, Radovick S. Deletion of Otx2 in GnRH neurons results in a mouse model of hypogonadotropic hypogonadism. Mol Endocrinol. 2011;25(5):833‐846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoffmann U, Dirisamer A, Heher S, Kostner K, Widhalm K, Neunteufl T. Relation of peripheral flow‐mediated vasodilatation and coronary arterial calcium in young patients with heterozygous familial hypercholesterolemia. Am J Cardiol. 2002;90(1):70‐73. [DOI] [PubMed] [Google Scholar]
- 16. Hoffmann HM, Trang C, Gong P, Kimura I, Pandolfi EC, Mellon PL. Deletion of Vax1 from gonadotropin‐releasing hormone (GnRH) neurons abolishes GnRH expression and leads to hypogonadism and infertility. J Neurosci. 2016;36(12):3506‐3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolfe A, Divall S, Singh SP, et al. Temporal and spatial regulation of CRE recombinase expression in gonadotrophin‐releasing hormone neurones in the mouse. J Neuroendocrinol. 2008;20(7):909‐916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dong KW, Yu KL, Roberts JL. Identification of a major up‐stream transcription start site for the human progonadotropin‐releasing hormone gene used in reproductive tissues and cell lines. Mol Endocrinol. 1993;7(12):1654‐1666. [DOI] [PubMed] [Google Scholar]
- 19. Radovick S, Wondisford FE, Nakayama Y, Yamada M, Cutler GB Jr, Weintraub BD. Isolation and characterization of the human gonadotropin‐releasing hormone gene in the hypothalamus and placenta. Mol Endocrinol. 1990;4(3):476‐480. [DOI] [PubMed] [Google Scholar]
- 20. Wolfe A, Kim HH, Tobet S, Stafford DE, Radovick S. Identification of a discrete promoter region of the human GnRH gene that is sufficient for directing neuron‐specific expression: a role for POU homeodomain transcription factors. Mol Endocrinol. 2002;16(3):435‐449. [DOI] [PubMed] [Google Scholar]
- 21. Kim HH, Wolfe A, Smith GR, Tobet SA, Radovick S. Promoter sequences targeting tissue‐specific gene expression of hypothalamic and ovarian gonadotropin‐releasing hormone in vivo. J Biol Chem. 2002;277(7):5194‐5202. [DOI] [PubMed] [Google Scholar]
- 22. Novaira HJ, Yates M, Diaczok D, Kim H, Wolfe A, Radovick S. The gonadotropin‐releasing hormone cell‐specific element is required for normal puberty and estrous cyclicity. J Neurosci. 2011;31(9):3336‐3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Novaira HJ, Fadoju D, Diaczok D, Radovick S. Genetic mechanisms mediating kisspeptin regulation of GnRH gene expression. J Neurosci. 2012;32(48):17391‐17400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Novaira HJ, Ng Y, Wolfe A, Radovick S. Kisspeptin increases GnRH mRNA expression and secretion in GnRH secreting neuronal cell lines. Mol Cell Endocrinol. 2009;311(1–2):126‐134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim HH, Wolfe A, Cohen RN, et al. In vivo identification of a 107‐base pair promoter element mediating neuron‐specific expression of mouse gonadotropin‐releasing hormone. Mol Endocrinol. 2007;21(2):457‐471. [DOI] [PubMed] [Google Scholar]
- 26. Kim HH, DiVall SA, Deneau RM, Wolfe A. Insulin regulation of GnRH gene expression through MAP kinase signaling pathways. Mol Cell Endocrinol. 2005;242(1–2):42‐49. [DOI] [PubMed] [Google Scholar]
- 27. DiVall SA, Radovick S, Wolfe A. Egr‐1 binds the GnRH promoter to mediate the increase in gene expression by insulin. Mol Cell Endocrinol. 2007;270(1–2):64‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhen S, Dunn IC, Wray S, et al. An alternative gonadotropin‐releasing hormone (GnRH) RNA splicing product found in cultured GnRH neurons and mouse hypothalamus. J Biol Chem. 1997;272(19):12620‐12625. [DOI] [PubMed] [Google Scholar]
- 29. Zhen S, Zakaria M, Wolfe A, Radovick S. Regulation of gonadotropin‐releasing hormone (GnRH) gene expression by insulin‐like growth factor I in a cultured GnRH‐expressing neuronal cell line. Mol Endocrinol. 1997;11(8):1145‐1155. [DOI] [PubMed] [Google Scholar]
- 30. Wolfe A, Divall S, Wu S. The regulation of reproductive neuroendocrine function by insulin and insulin‐like growth factor‐1 (IGF‐1). Front Neuroendocrinol. 2014;35(4):558‐572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Messina A, Langlet F, Chachlaki K, et al. A microRNA switch regulates the rise in hypothalamic GnRH production before puberty. Nat Neurosci. 2016;19(6):835‐844. [DOI] [PubMed] [Google Scholar]
- 32. Li H, Li X, Zhang D, Li J, Cui S. MiR‐375 potentially enhances GnRH expression by targeting Sp1 in GT1‐7 cells. In Vitro Cell Dev Biol Anim. 2021;57(4):438‐447. [DOI] [PubMed] [Google Scholar]
- 33. Li X, Xiao J, Fan Y, et al. miR‐29 family regulates the puberty onset mediated by a novel Gnrh1 transcription factor TBX21. J Endocrinol. 2019;242(3):185‐197. [DOI] [PubMed] [Google Scholar]
- 34. Chan YM, de Guillebon A, Lang‐Muritano M, et al. GNRH1 mutations in patients with idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2009;106(28):11703‐11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bouligand J, Ghervan C, Tello JA, et al. Isolated familial hypogonadotropic hypogonadism and a GNRH1 mutation. N Engl J Med. 2009;360(26):2742‐2748. [DOI] [PubMed] [Google Scholar]
- 36. Maione L, Albarel F, Bouchard P, et al. R31C GNRH1 mutation and congenital hypogonadotropic hypogonadism. PLoS One. 2013;8(7):e69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gill JC, Wadas B, Chen P, et al. The gonadotropin‐releasing hormone (GnRH) neuronal population is normal in size and distribution in GnRH‐deficient and GnRH receptor‐mutant hypogonadal mice. Endocrinology. 2008;149(9):4596‐4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chung WCJ, Tsai PS. Role of fibroblast growth factor signaling in gonadotropin‐releasing hormone neuronal system development. Front Horm Res. 2010;3937–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar D, Periasamy V, Freese M, Voigt A, Boehm U. In utero development of kisspeptin/GnRH neural circuitry in male mice. Endocrinology. 2015;156(9):3084‐3090. [DOI] [PubMed] [Google Scholar]
- 40. Fornaro M, Geuna S, Fasolo A, Giacobini‐Robecchi MG. HuC/D confocal imaging points to olfactory migratory cells as the first cell population that expresses a post‐mitotic neuronal phenotype in the chick embryo. Neuroscience. 2003;122(1):123‐128. [DOI] [PubMed] [Google Scholar]
- 41. Miller AM, Treloar HB, Greer CA. Composition of the migratory mass during development of the olfactory nerve. J Comp Neurol. 2010;518(24):4825‐4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taroc EZM, Naik AS, Lin JM, et al. Gli3 regulates vomeronasal neurogenesis, olfactory ensheathing cell formation, and GnRH‐1 neuronal migration. J Neurosci. 2020;40(2):311‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Forni PE, Bharti K, Flannery EM, Shimogori T, Wray S. The indirect role of fibroblast growth factor‐8 in defining neurogenic niches of the olfactory/GnRH systems. J Neurosci. 2013;33(50):19620‐19634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ortiz‐Leal I, Torres MV, Villamayor PR, Lopez‐Beceiro A, Sanchez‐Quinteiro P. The vomeronasal organ of wild canids: the fox (Vulpes vulpes) as a model. J Anat. 2020;237(5):890‐906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Katreddi RR, Forni PE. Mechanisms underlying pre‐ and postnatal development of the vomeronasal organ. Cell Mol Life Sci. 2021;78(12):5069‐5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Casoni F, Malone SA, Belle M, et al. Development of the neurons controlling fertility in humans: new insights from 3D imaging and transparent fetal brains. Development. 2016;143(21):3969‐3981. [DOI] [PubMed] [Google Scholar]
- 47. Forni PE, Taylor‐Burds C, Melvin VS, Williams T, Wray S. Neural crest and ectodermal cells intermix in the nasal placode to give rise to GnRH‐1 neurons, sensory neurons, and olfactory ensheathing cells. J Neurosci. 2011;31(18):6915‐6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wray S. From nose to brain: development of gonadotrophin‐releasing hormone‐1 neurones. J Neuroendocrinol. 2010;22(7):743‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Taroc EZM, Katreddi RR, Forni PE. Identifying Isl1 genetic lineage in the developing olfactory system and in GnRH‐1 neurons. Front Physiol. 2020;11:601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wray S, Key S, Qualls R, Fueshko SM. A subset of peripherin positive olfactory axons delineates the luteinizing hormone releasing hormone neuronal migratory pathway in developing mouse. Dev Biol. 1994;166(1):349‐354. [DOI] [PubMed] [Google Scholar]
- 51. Yoshida K, Tobet SA, Crandall JE, Jimenez TP, Schwarting GA. The migration of luteinizing hormone‐releasing hormone neurons in the developing rat is associated with a transient, caudal projection of the vomeronasal nerve. J Neurosci. 1995;15(12):7769‐7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deiner MS, Sretavan DW. Altered midline axon pathways and ectopic neurons in the developing hypothalamus of netrin‐1‐ and DCC‐deficient mice. J Neurosci. 1999;19(22):9900‐9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Demski LS, Schwanzel‐Fukuda M. The terminal nerve (nervus terminalis): structure, function, and evolution. Introduction. Ann N Y Acad Sci. 1987;519:ix‐xi. [DOI] [PubMed] [Google Scholar]
- 54. Kingsbury AC. Danazol and fetal masculinization: a warning. Med J Aust. 1985;143(9):410‐411. [DOI] [PubMed] [Google Scholar]
- 55. Demski LS. Phylogeny of luteinizing hormone‐releasing hormone systems in protochordates and vertebrates. Ann N Y Acad Sci. 1987;519:1‐14. [DOI] [PubMed] [Google Scholar]
- 56. Ridgway SH, Demski LS, Bullock TH, Schwanzel‐Fukuda M. The terminal nerve in odontocete cetaceans. Ann N Y Acad Sci. 1987;519:201‐212. [DOI] [PubMed] [Google Scholar]
- 57. Wirsig‐Wiechmann CR. Introduction to the anatomy and function of the nervus terminalis. Microsc Res Tech. 2004;65(1–2):1. [DOI] [PubMed] [Google Scholar]
- 58. Pena‐Melian A, Cabello‐de la Rosa JP, Gallardo‐Alcaniz MJ, et al. Cranial pair 0: the nervus terminalis. Anat Rec. 2019;302(3):394‐404. [DOI] [PubMed] [Google Scholar]
- 59. Jin ZW, Cho KH, Shibata S, Yamamoto M, Murakami G, Rodriguez‐Vazquez JF. Nervus terminalis and nerves to the vomeronasal organ: a study using human fetal specimens. Anat Cell Biol. 2019;52(3):278‐285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fuller GN, Burger PC. Nervus terminalis (cranial nerve zero) in the adult human. Clin Neuropathol. 1990;9(6):279‐283. [PubMed] [Google Scholar]
- 61. Tunsuriyawong P, Pongpirul K, Chaisam T, Prajuabpansri P. Olfactory bulb agenesis with normal sexual hormones. BMJ Case Rep. 2017;2017:bcr‐2017‐221899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yoshihara S, Omichi K, Yanazawa M, Kitamura K, Yoshihara Y. Arx homeobox gene is essential for development of mouse olfactory system. Development. 2005;132(4):751‐762. [DOI] [PubMed] [Google Scholar]
- 63. Naruse I, Fukui Y, Keino H, Taniguchi M. The arrest of luteinizing hormone‐releasing hormone neuronal migration in the genetic arhinencephalic mouse embryo (Pdn/Pdn). Brain Res Dev Brain Res. 1994;81(2):178‐184. [DOI] [PubMed] [Google Scholar]
- 64. Taroc EZM, Prasad A, Lin JM, Forni PE. The terminal nerve plays a prominent role in GnRH‐1 neuronal migration independent from proper olfactory and vomeronasal connections to the olfactory bulbs. Biol Open. 2017;6(10):1552‐1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cariboni A, Davidson K, Rakic S, Maggi R, Parnavelas JG, Ruhrberg C. Defective gonadotropin‐releasing hormone neuron migration in mice lacking SEMA3A signalling through NRP1 and NRP2: implications for the aetiology of hypogonadotropic hypogonadism. Hum Mol Genet. 2011;20(2):336‐344. [DOI] [PubMed] [Google Scholar]
- 66. Nguyen‐Ba‐Charvet KT, Di Meglio T, Fouquet C, Chedotal A. Robos and slits control the pathfinding and targeting of mouse olfactory sensory axons. J Neurosci. 2008;28(16):4244‐4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Taroc EZM, Lin JM, Tulloch AJ, Jaworski A, Forni PE. GnRH‐1 neural migration from the nose to the brain is independent from Slit2, Robo3 and NELL2 signaling. Front Cell Neurosci. 2019;13:fncel.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Quinton R, Hasan W, Grant W, et al. Gonadotropin‐releasing hormone immunoreactivity in the nasal epithelia of adults with Kallmann's syndrome and isolated hypogonadotropic hypogonadism and in the early midtrimester human fetus. J Clin Endocrinol Metab. 1997;82(1):309‐314. [DOI] [PubMed] [Google Scholar]
- 69. Schwanzel‐Fukuda M, Crossin KL, Pfaff DW, Bouloux PM, Hardelin JP, Petit C. Migration of luteinizing hormone‐releasing hormone (LHRH) neurons in early human embryos. J Comp Neurol. 1996;366(3):547‐557. [DOI] [PubMed] [Google Scholar]
- 70. Kim KH, Patel L, Tobet SA, King JC, Rubin BS, Stopa EG. Gonadotropin‐releasing hormone immunoreactivity in the adult and fetal human olfactory system. Brain Res. 1999;826(2):220‐229. [DOI] [PubMed] [Google Scholar]
- 71. Quanbeck C, Sherwood NM, Millar RP, Terasawa E. Two populations of luteinizing hormone‐releasing hormone neurons in the forebrain of the rhesus macaque during embryonic development. J Comp Neurol. 1997;380(3):293‐309. [PubMed] [Google Scholar]
- 72. Rance NE, Young WS 3rd, McMullen NT. Topography of neurons expressing luteinizing hormone‐releasing hormone gene transcripts in the human hypothalamus and basal forebrain. J Comp Neurol. 1994;339(4):573‐586. [DOI] [PubMed] [Google Scholar]
- 73. Terasawa E, Busser BW, Luchansky LL, et al. Presence of luteinizing hormone‐releasing hormone fragments in the rhesus monkey forebrain. J Comp Neurol. 2001;439(4):491‐504. [DOI] [PubMed] [Google Scholar]
- 74. Skrapits K, Sarvari M, Farkas I, et al. The cryptic gonadotropin‐releasing hormone neuronal system of human basal ganglia. Elife. 2021;10:e67714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang G, Li J, Purkayastha S, et al. Hypothalamic programming of systemic ageing involving IKK‐beta, NF‐kappaB and GnRH. Nature. 2013;497(7448):211‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zheng JQ, Poo MM. Calcium signaling in neuronal motility. Annu Rev Cell Dev Biol. 2007;23(1):375‐404. [DOI] [PubMed] [Google Scholar]
- 77. Toba Y, Pakiam JG, Wray S. Voltage‐gated calcium channels in developing GnRH‐1 neuronal system in the mouse. Eur J Neurosci. 2005;22(1):79‐92. [DOI] [PubMed] [Google Scholar]
- 78. Hutchins BI, Klenke U, Wray S. Calcium release‐dependent actin flow in the leading process mediates axophilic migration. J Neurosci. 2013;33(28):11361‐11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Golan M, Boulanger‐Weill J, Pinot A, et al. Synaptic communication mediates the assembly of a self‐organizing circuit that controls reproduction. Sci Adv. 2021;7:eabc8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Song Y, Tao B, Chen J, et al. GABAergic neurons and their modulatory effects on GnRH3 in zebrafish. Endocrinology. 2017;158(4):874‐886. [DOI] [PubMed] [Google Scholar]
- 81. Casoni F, Hutchins BI, Donohue D, Fornaro M, Condie BG, Wray S. SDF and GABA interact to regulate axophilic migration of GnRH neurons. J Cell Sci. 2012;125(Pt 21):5015‐5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fueshko SM, Key S, Wray S. GABA inhibits migration of luteinizing hormone‐releasing hormone neurons in embryonic olfactory explants. J Neurosci. 1998;18(7):2560‐2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Moore JP Jr, Shang E, Wray S. In situ GABAergic modulation of synchronous gonadotropin releasing hormone‐1 neuronal activity. J Neurosci. 2002;22(20):8932‐8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Simonian SX, Herbison AE. Differing, spatially restricted roles of ionotropic glutamate receptors in regulating the migration of gnrh neurons during embryogenesis. J Neurosci. 2001;21(3):934‐943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Giacobini P, Kopin AS, Beart PM, Mercer LD, Fasolo A, Wray S. Cholecystokinin modulates migration of gonadotropin‐releasing hormone‐1 neurons. J Neurosci. 2004;24(20):4737‐4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Giacobini P, Wray S. Cholecystokinin directly inhibits neuronal activity of primary gonadotropin‐releasing hormone cells through cholecystokinin‐1 receptor. Endocrinology. 2007;148(1):63‐71. [DOI] [PubMed] [Google Scholar]
- 87. Malone SA, Papadakis GE, Messina A, et al. Defective AMH signaling disrupts GnRH neuron development and function and contributes to hypogonadotropic hypogonadism. Elife. 2019;8:e47198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bando Y, Irie K, Shimomura T, et al. Control of spontaneous Ca2+ transients is critical for neuronal maturation in the developing neocortex. Cereb Cortex. 2016;26(1):106‐117. [DOI] [PubMed] [Google Scholar]
- 89. Hurni N, Kolodziejczak M, Tomasello U, et al. Transient cell‐intrinsic activity regulates the migration and laminar positioning of cortical projection neurons. Cereb Cortex. 2017;27(5):3052‐3063. [DOI] [PubMed] [Google Scholar]
- 90. Kitazawa A, Kubo K, Hayashi K, Matsunaga Y, Ishii K, Nakajima K. Hippocampal pyramidal neurons switch from a multipolar migration mode to a novel "climbing" migration mode during development. J Neurosci. 2014;34(4):1115‐1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ohtaka‐Maruyama C, Okamoto M, Endo K, et al. Synaptic transmission from subplate neurons controls radial migration of neocortical neurons. Science. 2018;360(6386):313‐317. [DOI] [PubMed] [Google Scholar]
- 92. Mulrenin EM, Witkin JW, Silverman AJ. Embryonic development of the gonadotropin‐releasing hormone (GnRH) system in the chick: a spatio‐temporal analysis of GnRH neuronal generation, site of origin, and migration. Endocrinology. 1999;140(1):422‐433. [DOI] [PubMed] [Google Scholar]
- 93. Tobet SA, Chickering TW, King JC, et al. Expression of gamma‐aminobutyric acid and gonadotropin‐releasing hormone during neuronal migration through the olfactory system. Endocrinology. 1996;137(12):5415‐5420. [DOI] [PubMed] [Google Scholar]
- 94. Iremonger KJ, Constantin S, Liu X, Herbison AE. Glutamate regulation of GnRH neuron excitability. Brain Res. 2010;1364:35‐43. [DOI] [PubMed] [Google Scholar]
- 95. Abraham E, Palevitch O, Ijiri S, Du SJ, Gothilf Y, Zohar Y. Early development of forebrain gonadotrophin‐releasing hormone (GnRH) neurones and the role of GnRH as an autocrine migration factor. J Neuroendocrinol. 2008;20(3):394‐405. [DOI] [PubMed] [Google Scholar]
- 96. Romanelli RG, Barni T, Maggi M, et al. Expression and function of gonadotropin‐releasing hormone (GnRH) receptor in human olfactory GnRH‐secreting neurons: an autocrine GnRH loop underlies neuronal migration. J Biol Chem. 2004;279(1):117‐126. [DOI] [PubMed] [Google Scholar]
- 97. Ramon‐Cueto A, Valverde F. Olfactory bulb ensheathing glia: a unique cell type with axonal growth‐promoting properties. Glia. 1995;14(3):163‐173. [DOI] [PubMed] [Google Scholar]
- 98. Franceschini IA, Barnett SC. Low‐affinity NGF‐receptor and E‐N‐CAM expression define two types of olfactory nerve ensheathing cells that share a common lineage. Dev Biol. 1996;173(1):327‐343. [DOI] [PubMed] [Google Scholar]
- 99. Roet KC, Verhaagen J. Understanding the neural repair‐promoting properties of olfactory ensheathing cells. Exp Neurol. 2014;261:594‐609. [DOI] [PubMed] [Google Scholar]
- 100. Geller S, Lomet D, Caraty A, Tillet Y, Duittoz A, Vaudin P. Rostro‐caudal maturation of glial cells in the accessory olfactory system during development: involvement in outgrowth of GnRH neurites. Eur J Neurosci. 2017;46(10):2596‐2607. [DOI] [PubMed] [Google Scholar]
- 101. Saglam A, Calof AL, Wray S. Novel factor in olfactory ensheathing cell‐astrocyte crosstalk: anti‐inflammatory protein alpha‐crystallin B. Glia. 2021;69(4):1022‐1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Barraud P, Seferiadis AA, Tyson LD, et al. Neural crest origin of olfactory ensheathing glia. Proc Natl Acad Sci USA. 2010;107(49):21040‐21045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Barraud P, St John JA, Stolt CC, Wegner M, Baker CV. Olfactory ensheathing glia are required for embryonic olfactory axon targeting and the migration of gonadotropin‐releasing hormone neurons. Biol Open. 2013;2(7):750‐759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Pingault V, Bodereau V, Baral V, et al. Loss‐of‐function mutations in SOX10 cause Kallmann syndrome with deafness. Am J Hum Genet. 2013;92(5):707‐724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Saadi H, Shan Y, Marazziti D, Wray S. GPR37 signaling modulates migration of olfactory ensheathing cells and gonadotropin releasing hormone cells in mice. Front Cell Neurosci. 2019;13:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Raucci F, Tiong JD, Wray S. P75 nerve growth factor receptors modulate development of GnRH neurons and olfactory ensheating cells. Front Neurosci. 2013;72:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Geller S, Kolasa E, Tillet Y, Duittoz A, Vaudin P. Olfactory ensheathing cells form the microenvironment of migrating GnRH‐1 neurons during mouse development. Glia. 2013;61(4):550‐566. [DOI] [PubMed] [Google Scholar]
- 108. Pinet‐Charvet C, Fleurot R, Derouin‐Tochon F, et al. Beta‐nerve growth factor stimulates spontaneous electrical activity of in vitro embryonic mouse GnRH neurons through a P75 mediated‐mechanism. Sci Rep. 2020;10(1):10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cai XT, Li H, Borch Jensen M, et al. Gut cytokines modulate olfaction through metabolic reprogramming of glia. Nature. 2021;596(7870):97‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cangiano B, Swee DS, Quinton R, Bonomi M. Genetics of congenital hypogonadotropic hypogonadism: peculiarities and phenotype of an oligogenic disease. Hum Genet. 2021;140(1):77‐111. [DOI] [PubMed] [Google Scholar]
- 111. Lund C, Yellapragada V, Vuoristo S, et al. Characterization of the human GnRH neuron developmental transcriptome using a GNRH1‐TdTomato reporter line in human pluripotent stem cells. Dis Model Mech. 2020;13:dmm040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Huang WK, Wong SZH, Pather SR, et al. Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell. 2021;28(9):1657‐70 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Belle M, Godefroy D, Couly G, et al. Tridimensional visualization and analysis of early human development. Cell. 2017;169(1):161‐73 e12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this review because no new data were created or analyzed.


