Abstract
Objective
Despite the absolute requirement of Delta/Notch signaling to activate lateral inhibition during early blood vessel development, many mechanisms remain unclear about how this system is regulated. Our objective was to determine the involvement of Epsin 15 Homology Domain Containing 2 (EHD2) in delta‐like ligand 4 (Dll4) endocytosis during Notch activation.
Approach and Results
Using both in vivo and in vitro models, we demonstrate that EHD2 is a novel modulator of Notch activation in endothelial cells through controlling endocytosis of Dll4. In vitro, EHD2 localized to plasma membrane‐bound Dll4 and caveolae. Chemical disruption of caveolae complexes resulted in EHD2 failing to organize around Dll4 as well as loss of Dll4 internalization. Reduced Dll4 internalization blunted Notch activation in endothelial cells. In vivo, EHD2 is primarily expressed in the vasculature, colocalizing with junctional marker VE‐cadherin and Dll4. Knockout of EHD2 in zebrafish produced a significant increase in dysmorphic sprouts in zebrafish intersomitic vessels during development and a reduction in downstream Notch signaling.
Conclusions
Overall, we demonstrate that EHD2 is necessary for Dll4 transcytosis and downstream Notch activation.
Keywords: angiogenesis, blood vessel development, delta‐like 4 protein, EHD2, endocytosis, Notch, trafficking, transcytosis, Zebrafish
Abbreviations
- Dll4
Delta‐like ligand 4
- ECs
endothelial cells
- EHD2
epsin 15 Homology Domain Containing 2
- NECD
Notch extracellular domain
1. INTRODUCTION
Angiogenesis is the process in which new blood vessels develop from an existing vascular bed. During this event, in response to a chemotactic gradient, a single endothelial cell (EC) in a group must identify itself as a tip cell. 1 This tip cell will then lead the charge up a growth factor gradient, while stalk cells trail behind. 2 Tip and stalk cells each have distinct morphological and functional identities: the tip defined by its spiny, branching filopodia reaching forward as the cell migrates; the stalk defined by its smoothened appearance and heightened junctional stability. 3 , 4 For proper angiogenic growth to proceed, the maintenance of tip/stalk cell specification is paramount. Central to tip/stalk cell specification is the Notch signaling pathway. Notch is a transmembrane protein composed of an extracellular domain (NECD) and an intracellular domain (NICD). Endothelial cells with high‐Notch activation will adopt a stalk cell identity, whereas an EC deficient in Notch or Notch signaling will adopt a tip cell identity. 5 , 6
Delta‐like proteins are transmembrane Notch ligands. The NECD of a Notch presenting cell will bind Delta on an adjacent cell. This Delta/Notch binding elicits two consecutive cleavage events. First, obscured within two domains (LNR and HD) of Notch is a cleavage site termed S2. When exposed, the S2 site is cleaved by a disintegrin and metalloprotease (ADAM) complex leaving the NECD attached to Delta. 7 This event precedes the second cleavage by γ‐secretase at the S3 site to release the NICD. Once freed, the NICD translocates to the nucleus, binding the transcription factor CSL to upregulate downstream genes that promote lateral inhibition. 8 , 9 , 10
One proposed mechanism for this activation of Notch by Delta is the application of a mechanical force generated by Delta/NECD transcytosis (ie, endocytosis of Delta while bound to NECD) to expose the extracellular S2 domain. This pulling force has been shown to be on the order of 19 pN per single bond 11 and is necessary to force apart the LNR/HD interaction, thus exposing the S2 and subsequent S3 site for cleavage. To date, studies on the endocytic mechanisms that underlie Delta/Notch transcytosis have only focused on Delta‐like ligand 1 (Dll1) in non‐endothelial tissue. 11 , 12 , 13 Despite the absolute requirement of Delta‐like ligand 4 (Dll4) for Notch signaling in vascular tissue, the mechanisms of Dll4 transcytosis remain unknown.
In mammals, there are four Epsin15 homology domain (EHD) proteins, EHD1‐4 are each involved in endocytic processes, although EHD2 stands alone from this group in being the only one with a solved crystal structure and the only to interact with caveolae. 14 EHD2 multimerizes through an interaction between the G‐domain and the EH domain. 14 , 15 The multimerization of EHD2 allows it to form a circular ring around an endocytic vesicle to mediate invaginating pit stability. 14 This complex localizes to caveolae, assisting in caveolin‐mediated endocytosis through stabilization of the pit structure formed by hairpin shaped proteins in the membrane. 16 , 17 The breadth of EHD2 function in ECs remains poorly understood.
In this article, we identify EHD2 as a novel regulator of Notch activation in ECs through controlling endocytosis of Dll4. In vitro, EHD2 localized to plasma membrane‐bound Dll4 and caveolae independently of clathrin. Disruption of caveolin endocytosis resulted in EHD2 failing to organize around Dll4 as well as loss of Dll4 internalization in ECs. In vivo, knockout of EHD2 produced a significant increase in dysmorphic sprouts in zebrafish intersomitic vessels (ISVs) during development and a reduction in Notch signaling. Overall, we demonstrate that EHD2 impacts Dll4 endocytosis and downstream Notch signaling important for blood vessel development.
2. MATERIALS AND METHODS
2.1. Zebrafish studies
All zebrafish used in this study were the AB strain. Zebrafish housing and protocols were approved by the University of Denver Institutional Animal Care and Use Committee. Zebrafish embryos were raised in a 28℃ incubator in embryo buffer for 2 days. Tg(kdrl:GFP) strain was previously published by Choi et al. 18 Tg(fli:LifeAct‐GFP) strain was previously published by Hen et al. 2015. 19 Tg(kdrl:mCherry) strain was previously published by Proulx et al. 2010. 20 Tg(cdh5:gal4ff) strain was previously published by Bussmann et al. 2011. 21
Tol2 transposase RNA was synthesized from pT3TS‐Tol2 (Addgene, #31831) using the MEGAscript™ T3 Transcription Kit (Thermo Fisher Scientific, AM1338) and stored at −80°C at a dilution of 100 ng/µl. Injection mixture was prepared on ice containing 300 ng Tol2 transposase RNA and 500 ng recombinant plasmid and was brought to 10 µl total volume with 0.1% phenol‐red (VWR, 470301–974) in water. 1–4 cell embryos were injected directly into cell with 2 pl injection mixture.
4‐guide CRISPR/Cas9 targeted gene KO was performed as outlined by Wu et al. 2018. 22 In brief, 4 single guide RNA templates fused to a scaffold were synthesized for each target gene using HiScribe™ SP6 RNA Synthesis Kit (New England BioLabs, E2070S). Injection mixture was prepared on ice containing 5 µM Cas9 (PNA Bio, CP02), 1 µg/µl sgRNA, and brought to 6 µl with 0.1% phenol‐red in water. Cas9 and sgRNA guides were pre‐complexed at 37℃ for 5 min. 1–2 cell embryos were injected directly into yolk with 2 pl injection mixture. Validation of guide knock outs was carried out by amplification of a DNA flanking targets sites and then cloning into a pME backbone (see Table S1) followed by Sanger sequencing.
2.2. In situ hybridization
In situ hybridization was performed as outlined by Thisse et al. 2007. 23 DNA was primed from a zebrafish cDNA library (supplemental data) and inserted into a pME backbone containing both T7 and SP6 promoters via Gibson reaction. Antisense probes were converted to RNA from this template using the HiScribe™ SP6 RNA synthesis Kit (New England BioLabs, E2040S). Sense probes were converted to RNA from this template using the HiScribe™ T7 RNA synthesis Kit. In each RNA synthesis reaction, UTP was substituted for DIG RNA labeling mix (UTP) (Sigma Aldrich, 11277073910). Probes were designed to be roughly 800 bp in size, which has shown to produce the most efficient labeling in zebrafish. Antisense probes were used to detect the transcript of interest, sense probes were used as a control to monitor over‐development of staining solution (225 µl Nitro Blue Tetrazolium [50 mg dissolved in 0.7 ml N,N‐dimethylformamide anhydrous and 0.3 ml water], 50 ml Alkaline Tris Buffer [100 mM 1 M Tris‐HCl pH 9.5, 50 mM MgCl2, 100 mM 5 M NaCl, 0.1% Tween 20 20%], 175 µl 5‐Bromo 4‐Chloro 3‐indolyl Phosphate [50 mg dissolved in 1 ml N,N‐dimethylformamide anhydrous]).
2.3. Plasmids
The follow plasmids were used in the current study: pShuttle‐CMV was a gift from Bert Vogelstein (Addgene plasmid # 16403): AdEasier‐1 cells (strain) was a gift from Bert Vogelstein (Addgene plasmid # 16399); and mEmerald‐Clathrin‐15 was a gift from Michael Davidson (Addgene plasmid # 54040). EHD2 was purchased from Origene (MR220542). Dll4 was purchased from Origene (MR212151).
2.4. Cell culture
Human umbilical vein endothelial cells were purchased from Sigma Aldrich and cultured in proprietary media (Promocell) at 37°C at 5% CO2. For imaging experiments glass‐bottomed imaging dishes were exposed to UV light for 6 min and then coated with 15 µg/ml laminin mouse protein (Thermo Fisher Scientific, 23017015) overnight at 37°C. Cells were plated onto laminin coated dishes for 4–6 h prior to imaging or fixation. 0.9 µM siRNA (Thermo Fisher Scientific, s225944, s26959, am4611) was introduced to primary human umbilical vein endothelial cells (HUVEC) using the Neon® transfection system (Thermo Fisher Scientific).
Adenovirus constructs (tagRFP‐EHD2 and Emerald‐Clathrin) were created as previously described. 24 In brief, constructs were introduced via Gibson Assembly into pShuttle‐CMV. PShuttle‐CMV plasmids were then digested overnight with MssI (Thermo Fisher Scientific, IVGN0244) and purified via gel extraction. Linearized pShuttle‐CMV plasmids were transformed into the final viral backbone using electrocompetent AdEasier‐1 cells. Successful incorporation of pShuttle‐CMV construct into AdEasier‐1 cells confirmed via digestion with PacI (Thermo Fisher Scientific, IVGN0184). 5000 ng plasmid was then digested at 37°C overnight, then 85°C for 10 min and transfected in a 3:1 polyethylenimine (PEI, Sigma Aldrich, 408747):DNA ratio into 70% confluent HEK 293A cells (Thermo Fisher Scientific, R70507).
Over the course of approximately 2–4 weeks, fluorescent cells became swollen and burst or budded‐off the plate. Once approximately 50% of the cells had lifted off of the plate, cells were removed and centrifuged at 500x g for 5 min in a 15 ml conical tube. The cells were resuspended in 1 ml DPBS (Genesee Scientific, 25‐508B). Cells were then lysed by 3 consecutive quick freeze‐thaw cycles in liquid nitrogen, spun down for 5 min at 500x g, and supernatant was added to two 70% confluent T‐75 flasks. Propagation continued and collection repeated for infection of 10 qty, 15 cm dishes. After collection, 8 ml viral supernatant was collected and combined with 4.4 g CsCl (Sigma Aldrich, 289329) in 10 ml DPBS. Solution was overlaid with mineral oil and spun at 100 000x g at 100°C for 18 h. Viral fraction was collected with a syringe and stored in a 1:1 ratio with a storage buffer containing 10 mM Tris, pH 8.0, 100 mM NaCl, 0.1 percent BSA, and 50% glycerol.
2.5. Sprouting angiogenesis assay
Fibrin‐bead assay was performed as reported by Nakatsu et al. 2007. 25 Briefly, HUVECs were coated onto microcarrier beads (Amersham) and plated overnight. The following day, the EC‐covered microbeads were embedded in a fibrin matrix. Once the clot was formed media was overlaid along with 100 000 NHLFs. Media was changed daily along with monitoring of sprout development. For imaging the fibrin‐bead assay, first fibroblasts were removed from the clot with a 1‐min trypsin incubation. Following incubation, the trypsin was neutralized with DMEM containing 10% bovine serum albumin, washed 3 times with PBS, and fixed using 4% paraformaldehyde for 40 min. After fixation, the clot was washed 3 times with PBS, permeabilized with 0.5% Triton‐X for 2 h and then blocked with 2% BSA for 1 h prior to overnight incubation with primary antibodies. The following day, primary antibodies were removed and the clot was washed 5 times with PBS and secondary antibody was added with 2% BSA and incubated overnight. Prior to imaging the clot was washed 5 times with PBS. All primary and secondary antibodies are listed in the Data Supplement.
2.6. Proximity ligation assay
The proximity ligation assay was purchased from Sigma (DUO92101) and carried out as previously described. 26
2.7. pHrodo internalization assay
PHrodo™ iFL Red STP Ester (Thermo Fisher Scientific, P36010) was resuspended to 10 mM with DMSO. On the day of antibody labeling, pHrodo™ iFL Red STP Ester was diluted to 2 mM in DMSO (VWR Life Science, 97063–136). Antibody was brought up in DPBS (2 mg/ml final concentration for Dll4 polyclonal antibody [Thermo Fisher Scientific, PA5‐46974]; 1 mg/ml final concentration for recombinant human Notch‐1 Fc Chimera [R&D Systems, P46531]) and added to 1/10 volume 1 M sodium bicarbonate, pH 8.5. 3.3 µl of 2 mM pHrodo™ dye was added to the antibody and allowed to react in the dark for 1 h with gentle flicking every 15 min. While this reaction is occurring, Zeba™ Spin Desalting Column (Thermo Fisher Scientific, 89882) was washed 3 times with 300 µl DPBS at 1500x g for 1 min. After 1 h, the labeled antibody solution was loaded into the desalting column and allowed to absorb. DPBS was overlaid on top of labeled antibody to bring total column load volume to 70 µl and spun at 1500x g for 2 min. Flow‐through was stored at 4°C.
For bead tethered pHrodo, 6 µl of pHrodo‐conjugated antibody brought to 200 µl TBST and added to 10 µl of either Dynabeads™ Protein G (Thermo Fisher Scientific, 10004D) or Protein A Agarose Resin (Gold Biotechnology, P‐400–5). Beads and antibody were incubated and rotated at room temperature for 10 min. Conjugated beads were washed 3 times with 200 µl TBST then stored in a final volume of 10 µl TBST at 4°C.
Three microliters pHrodo‐labeled antibody and 1 µl Hoechst 33342 trihydrochloride, trihydrate (Thermo Fisher Scientific, H3570) was added to 70%–80% confluent HUVECs plated on laminin coated dishes in 1 ml media and incubated at 37°C for 10 min. After 10 min, the cells were washed 3 times with 1 ml DPBS and then placed in 2 ml media. 10 z‐stack images were taken for each condition, this marks time point 0 min. After images were taken of each group, cells were returned to 37°C for 10 min and imaged again.
2.8. Microscopy
Images were taken on a Nikon Eclipse Ti inverted microscope equipped with a CSU‐X1 Yokogawa spinning disk field scanning confocal system and a Hamamatusu EM‐CCD digital camera. Cell culture images were captured using a Nikon Plan Apo 60x NA 1.40 oil objective using Olympus type F immersion oil NA 1.518 (ThorLabs, MOIL‐30). Fish images were taken using either Nikon Apo LWD 20x NA 0.95 or Nikon Apo LWD 40x NA 1.15 water objectives. For transmission electron microscopy images, primary HUVEC of specified treatment were fixed in 2% glutaraldehyde, 2% PFA, 0.2 M Cacodylate buffer and imaged at the University of Colorado Anschutz Medical Campus.
2.9. Immunohistochemistry
2D cell culture was fixed in 4% paraformaldehyde in DPBS for 10 min. Cells were then washed 3 times for 5 min in TBST and permeabilized in 0.1% Triton‐X 100 for 10 min. Cells were then washed 3 times for 5 min and blocked for 1 h in 2% bovine serum albumin. Primary antibodies were applied at specified dilutions in Key Resources Table (Appendix S1) overnight. Cells were washed 3 times for 10 min in TBST and then moved to secondary for 2 h at specified dilutions in Key Resources Table. Cells were washed again 3 times for 15 min in TBST before imaging.
2.10. Western blot
Primary HUVEC culture was trypsinized and lysed using Ripa buffer (20 mM Tris‐HCl [pH 7.5], 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP‐40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β‐glycerophosphate, 1 mM Na3VO4, 1 µg/ml leupeptin) containing 1x ProBlock™ Protease Inhibitor Cocktail −50, Plus EDTA (GoldBio, GB‐334–20). Total concentration of protein in lysate was quantified using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, 23225) measured at 562 nm and compared to a standard curve. 20–50 µg protein was prepared in 0.52 M SDS, 1.2 mM bromothymol blue, 58.6% glycerol, 75 mM Tris pH 6.8, and 0.17 M DTT. Samples were boiled for 10 min, then 35 µl was loaded in a 7%–12% SDS gel and run at 170 V. Protein was then transferred to Immun‐Blot PVDF Membrane (BioRad, 1620177) at 4°C, 100 V for 1 h 10 min. Blots were blocked in 2% milk for 1 h, then put in primary antibody at specified concentrations overnight. After 3 10‐min washes with TBST, secondary antibodies at specified concentrations were applied for 4 h. After 3 additional TBST washes, blots were developed with ProSignal® Pico ECL Spray (Genesee Scientific, 20‐300S). All images were processed using ImageJ (Fiji).
2.11. Pharmacological treatment
DAPT (Sigma Aldrich, D5942) was applied to cells for 3 days at a final concentration of 5 µM. Dynasore hydrate (Sigma Aldrich, D7693) was applied to cells for 30 min at a final concentration of 100 µM. LY‐411575 (Sigma Aldrich, SML0506) was diluted in egg water to a final dilution of 2 µM from 30–48 hpf. Latrunculin A (Sigma Aldrich, 428021‐100UG) was applied to cells for 1 h at a final concentration of 5 µM. Methyl‐β‐Cyclodextrin (Sigma Aldrich, M7439‐1G) was applied to cell for 10 min at a final concentration of 10 mM.
2.12. Quantification and statistical analysis
Dysmorphic vessels were defined by a sprout emerging out of or separate from the defined ISV and quantified in 48 hpf embryos expressing tg(kdrl:GFP). RT‐PCR was quantified using the gel analysis function in Fiji image analysis software. 27 In sum, rectangular sections were drawn around individual lanes in gray‐scale, high‐quality gel image using the pathway Analyze > Gel > Select First Lane, Analyze > Gel > Select Next Lane. After all lanes are selected, the pathway Analyze > Gel > Plot Lanes was used. The peaks of each lane were then segmented using the Straight‐Line selection tool and highlighted with the Wand tool. Selection of the area inside the peak generates a Results window with the area and percent of each peak. The percent value of each sample was divided by the percent value of the control to obtain a relative density. Relative densities of the gene of interest (eg, Hey2) were divided by the relative density of the housekeeping gene (GAPDH) to obtain a final adjusted density value.
Cellular uptake of pHrodo‐labeled antibody was also quantified using Fiji image analysis software. Stack files were z‐stacked at maximum intensity, and each color channel was adjusted so that the background was zero. Each individual cell was outlined with the Freehand Selections tool. The color channels were then separated, and any background fluorescence (488 channel) was subtracted from the pHrodo fluorescent intensity (561 channel) using pathway Process > Image Calculator. The Integrated Density of pHrodo fluorescent intensity within each cell boundary was then recorded for every cell at each time point. Each experiment was performed in triplicate.
Pearson's coefficient was calculated using the Image J Plugin Just Another Colocalization Plugin (JACoP). 28 All statistical analysis was performed in GraphPad Prism8. Comparisons between two conditions were made using a t‐test, comparisons between multiple conditions were made using a One‐Way ANOVA. Zebrafish sex was not accounted for as sex‐determination did not occur in the developmental window in which the zebrafish larvae were assayed.
3. RESULTS
3.1. EHD2 is enriched in blood vessels
In a screen for vascular specific proteins, we observed that EHD2 expression was largely isolated to the vasculature via in situ hybridization in E9.5 embryos (Figure 1A). To confirm this we also assayed for EHD2 transcript in P7 retinas 29 and observed EHD2 was largely restricted to blood vessels with a noticeable increase in expression at the vascular front (Figure 1B). These results demonstrate that EHD2 expression is highly enriched in blood vessels.
FIGURE 1.
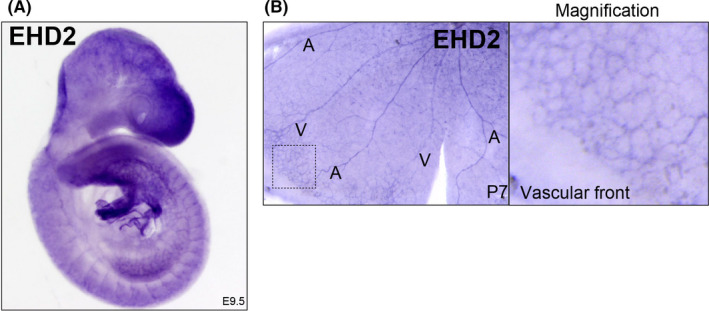
EHD2 expression is localized to blood vessels. (A) In situ hybridization of EHD2 in mouse embryo (E9.5). (B) In situ hybridization in P7 mouse retina. Box marks magnification area of vascular front. A, artery and V, vein
3.2. EHD2 and Caveolae localize to membranous Dll4
As EHD2 expression was more robust in the vascular front we hypothesized that EHD2 may play a role in Notch signaling. 30 More specifically, given EHD2’s biological function has previously been shown to be involved in caveolae stabilization, 14 we hypothesized that EHD2 may be interacting with Dll4 during Dll4/NECD transcytosis. To determine the potential endocytic route EHD2 employed, we moved to an in vitro culture‐based model using primary ECs (Human umbilical vein ECs). We first tested if Dll4 and EHD2 were in close proximity using a proximity ligation assay. 31 Using an antibody directed at a myc‐tagged Dll4 we demonstrated that EHD2 and Dll4 were indeed in close contact (<10 nm; Figure 2A,B). As a control, we also overexpressed an EHD2‐related protein EHBP1 32 with Dll4‐myc and did not detect a significant increase in proximity labeling events (Figure 2B).
FIGURE 2.
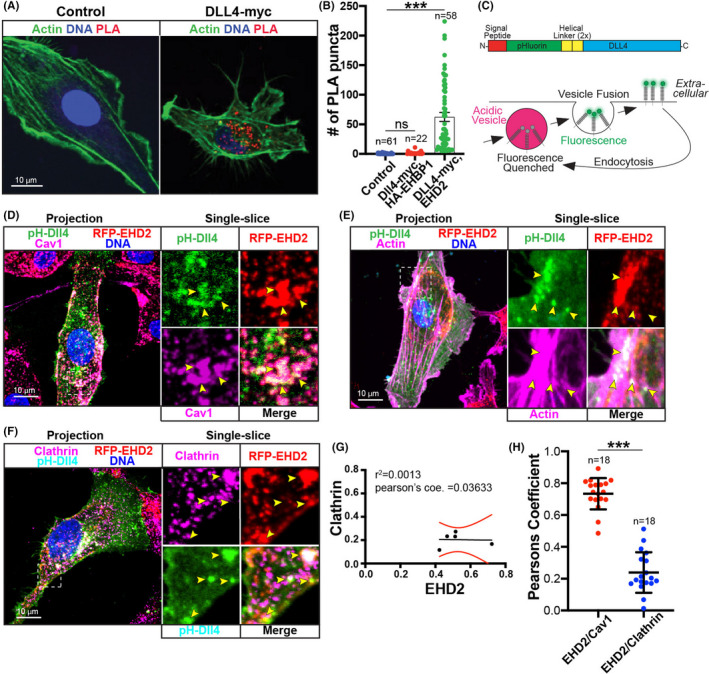
Membranous Dll4 localizes with EHD2 and caveolin‐1. (A) Representative image of proximity ligation assay (PLA). Cells were stained as indicated and PLA reaction is marked by red puncta. Control condition was not transfected. Dll4‐myc condition was only transfected with Dll4‐myc and stained for endogenous EHD2. (B) Graph of number of PLA puncta by condition. Control condition was not transfected. EHBP1 condition was transfected with EHBP1‐HA and Dll4‐myc. Dll4‐myc condition was only transfected with Dll4‐myc and stained for endogenous EHD2. (C) Schematic of engineered Dll4 (top). Bottom, cartoon of pH‐dependent function of GFP variant pHluorin tag. PHluorin fluoresces on the membrane at neutral pH but is quenched when internalized into acidic endosomes allowing for visualization of only membranous Dll4. (D) Representative image of endothelial cell (HUVEC) stained for caveolin‐1 (Cav1) expressing pHluorin‐Dll4 (pH‐Dll4) and RFP‐EHD2. (E) Representative image of cell stained for actin expressing pH‐Dll4 and RFP‐EHD2. (F) Representative image of cell stained for clathrin expressing pH‐Dll4 and RFP‐EHD2. (G) Proportion of coincidence of clathrin (y‐axis) and EHD2 (x‐axis) around Dll4 puncta. (H) Pearson's correlation between indicated proteins. N, number of cells. Boxes denote magnified images on right. Yellow arrowheads show areas of pH‐Dll4 puncta. ***p < .001. Error bars are 95% confidence intervals. All experiments were done at minimum in triplicate
To focus only on the extracellular, membrane‐inserted pool of Dll4 that would be available for Notch binding, we constructed a pHluorin‐tagged Dll4 (pH‐Dll4) vector. PHluorin is a GFP variant that fluoresces at neutral pH and is quenched when internalized into low pH vesicles allowing for visualization of the extracellular, membrane‐bound Dll4 population 33 (Figure 2C). Co‐expression of pH‐Dll4 and red fluorescence protein (RFP)‐EHD2 demonstrated strong colocalization with EHD2 surrounding Dll4 puncta (Figure 2D). Staining for caveolin‐1 confirmed caveolae structures also heavily colocalized with EHD2 and Dll4 (Figure 2D). Next, we used the same overexpression approach and stained for actin as endocytic caveolae have been reported to be in close association with filamentous actin. 34 We observed several instances where membranous pH‐Dll4 and EHD2 were in close proximity to actin fibers (Figure 2E). These data suggest that EHD2 associates with membrane‐bound Dll4 in caveolar structures.
How Dll4, per se, is transcytosed has not been reported; however, Dll1 has been shown to be internalized via a clathrin‐dependent route. 11 , 13 , 35 , 36 To determine if clathrin‐mediated endocytosis was also involved in Dll4 endocytosis, we stained for endogenous clathrin while expressing pH‐Dll4 and RFP‐EHD2. Clathrin showed less localization to pH‐Dll4 puncta compared with EHD2 (Figure 2F). Here, large Dll4 puncta showed elevated colocalization with clathrin, while small Dll4 puncta was largely devoid of clathrin. Localization analysis between clathrin and EHD2 showed no significant correlation to Dll4 position (Figure 2G), suggesting these proteins are likely not associated. Pearson's colocalization analysis of EHD2 and caveolin‐1 demonstrated a strong correlation, while EHD2 and clathrin showed a weak Pearson's correlation index (Figure 2H). To further confirm clathrin was not associated with Dll4, we siRNA knocked down either caveolin‐1 or clathrin light chain beta (CLTB, integral clathrin protein) and probed for the association between EHD2 and pH‐Dll4. Knock down of caveolin‐1 ablated EHD2’s localization, or any detectable levels of EHD2, with pH‐Dll4, while loss of CLTB did not impact EHD2 and Dll4s localization (Figure S1A‐C). Overall, these results indicate that Dll4 and EHD2 are rarely associated with clathrin endocytic machinery.
3.3. EHD2 and caveolae localize to cell‐cell junctions in sprouts
To characterize EHD2 localization in sprout structures, we employed a 3‐dimensional (3D) sprouting assay 25 , 37 (Figure 3A). In this assay, ECs undergo collective migration making multicellular sprouts that branch and lumenize, faithfully mimicking in vivo processes. 38 , 39 , 40 PH‐Dll4 robustly localized to adherens junctions, namely VE‐cadherin (Figure 3B,C). Likewise, we observed that both EHD2 and Dll4 localized to cell‐cell junctions in 3D sprouting structures (Figure 3D‐G). These results support the notion that EHD2 localizes at junctions, which are areas of Dll4/Notch transcytosis. 41 , 42 Staining in the mouse retina showed a similar pattern. Mouse EHD2 does not have a working antibody; however, staining for caveolin‐1, which highly colocalizes with EHD2, and Dll4 in the mouse retina showed a strong colocalization pattern consistent with our in vitro results (Figure 3H,I). This data suggests that EHD2 localizes to cell‐cell junctions in sprouts actively undergoing angiogenesis.
FIGURE 3.
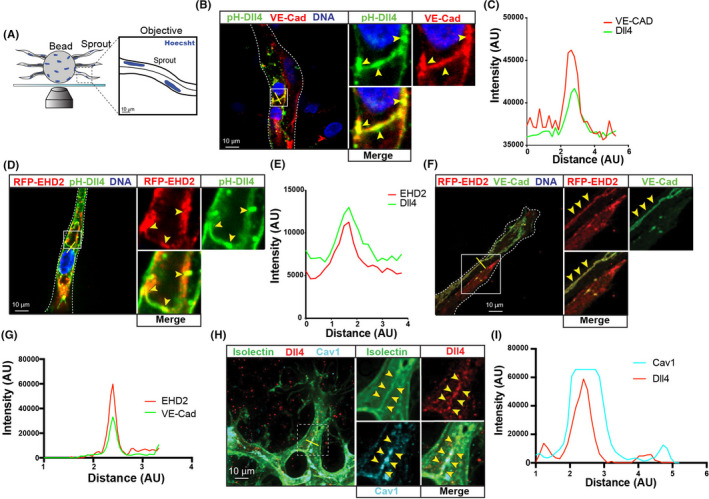
EHD2 and caveolae localize to adherens junctions in sprouts. (A) Schematic of 3‐dimensional sprout growth in fibrin‐bead assay (FBA) using Human umbilical vein endothelial cells. (B) Representative sprout stained for VE‐cadherin (VE‐Cad) expressing pHluorin‐Dll4 (pH‐Dll4). Yellow line marks line scan area. (C) Line scan of pH‐Dll4 and VE‐Cad of image in panel B. (D) Representative sprout expressing pH‐Dll4 and red fluorescence protein (RFP)‐EHD2. Yellow line marks line scan area. (E) Line scan of pH‐Dll4 and RFP‐EHD2 of image in panel D. (F) Representative sprout stained for VE‐Cad expressing RFP‐EHD2. Yellow line marks line scan area. (G) Line scan of pH‐Dll4 and VE‐Cad of image in panel F. (H) Representative image of retinal blood vessels in P6 mouse stained for isolectin, Dll4, and caveolin‐1 (Cav1). Boxes are magnified images on right. Yellow line marks line scan area. (I) Line scan of Cav1 and Dll4 in panel H. Yellow arrowheads show areas of Dll4 puncta. All experiments were done at minimum in triplicate. All images are a single confocal slice
3.4. EHD2 is required for Dll4/Notch transcytosis
With evidence of EHD2 at sites of membranous Dll4, we tested if EHD2 affected Dll4 endocytosis. To do so, we relied on a Dll4‐antibody covalently linked to pHrodo, a pH sensitive dye that fluoresces only at an acidic endosomal pH. 43 This allowed us to specifically monitor live Dll4 endocytosis (Figure 4B, Figure S2A). We first ensured the surface‐bound pHrodo label was distinguishable from the internalized pool by inhibiting endocytosis with either dynasore or cold treatment. Both treatments significantly reduced pHrode‐Dll4 signal intensity compared to control (Figure S2B). Additionally, surface‐bound pHrodo‐Dll4 signal could be rescued when the media pH was lowered to that of endosomes (~pH5; Figure S2B). Next, we pulse‐chased with the pHrodo‐Dll4 in control ECs; there was a sharp peak in fluorescent intensity at the 10‐min time point, indicating an increase in Dll4 endocytosis (Figure 4A,C). A pHrodo‐labelled IgG control was added to monitor non‐specific uptake. Dll4 internalization was significantly reduced in EHD2 siRNA‐treated groups (Figure 4C). Equally, internalization of pHrodo‐Dll4 in ECs treated with the pan‐endocytosis inhibitor Dynasore or caveolae cholesterol inhibitor MβCD also significantly reduced Dll4 internalization as compared with DMSO control (Figure 4D). These results indicate that internalization of Dll4 requires EHD2 and is dynamin and caveolin dependent.
FIGURE 4.
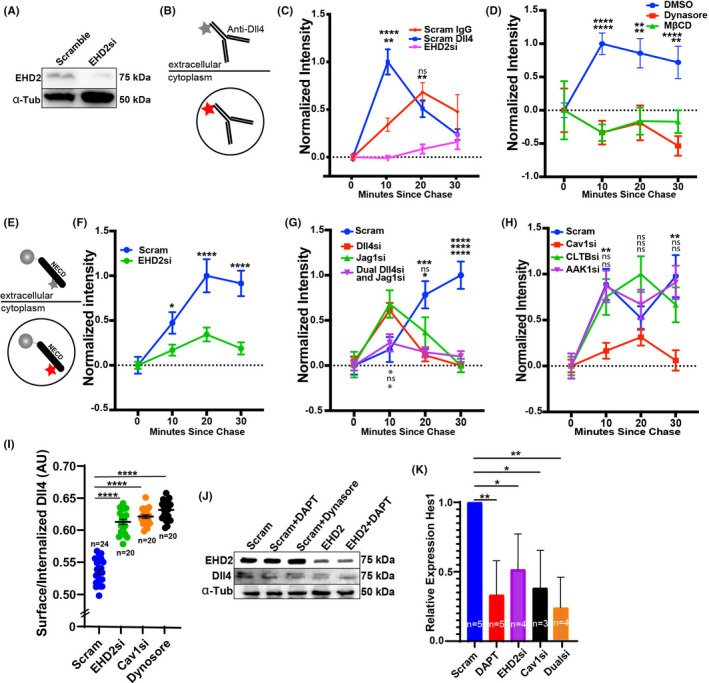
Loss of EHD2 blunts Dll4 endocytosis. (A) Western blot of EHD2 after siRNA (si) treatment in comparison to scramble (Scram) control in Human vein endothelial cells. (B) Schematic of pHrodo‐labeled delta‐like ligand 4 (Dll4) antibody. PHrodo gains fluorescent intensity with increasing endosomal pH, thus used as a metric of endocytosis. (C) Relative internalization of pHrodo‐Dll4 pulse‐chase over time between indicated siRNA groups. IgG was used as a non‐specific internalization control. A minimum of 50 cells were used per group. Order of comparisons (top to bottom): Scram si vs. EHD2 si; Scram IgG vs. pHrodo‐Dll4 with EHD2 si. (D) Relative internalization of pHrodo‐Dll4 pulse‐chase over time between indicated groups. A minimum of 50 cells were used per group. Order of comparisons (top to bottom): DMSO vs. Dynosore; DMSO vs. MβCD. (E) Schematic of recombinant Notch intracellular domain (NECD) functionalized to microbead and labeled with pHrodo. (F) Relative internalization of functionalized NECD pulse‐chase over time between indicated siRNA groups. A minimum of 50 cells were used per group. Comparisons: Scram si vs. EHD2 si treated cells. (G) Relative internalization of functionalized NECD pulse‐chase over time between indicated siRNA groups. A minimum of 50 cells were used per group. Order of comparisons (top to bottom): Scram si vs. Dll4 si; Scram si vs. Jagged1 (Jag1) si; Scram si vs. Dll4 and Jag1 si treated cells. (H) Relative internalization of functionalized NECD pulse‐chase over time between indicated siRNA groups. A minimum of 50 cells were used per group. Order of comparisons (top to bottom): Scram si vs. caveolin1 (Cav1) si; Scram si vs. clathrin light chain B (CLTB) si; Scram si vs. AP2 Associated Kinase 1 (AAK1) si treated cells. (I) Antibody feeding assay comparing internalized to surface‐bound ratio Dll4 between indicated groups. N, number of cells. (J) Western blot of Dll4 levels across indicated treatment groups. (K) Relative expression of Hes1 compared to GAPDH control in indicated si treatment groups. N, number of replicates. *p < .05, **p < .01, ***p < .001, ****p < .0001. ns, non‐significant. Error bars are SEM. All experiments were done at minimum in triplicate
To more unambiguously test Notch/Dll4 transcytosis, we next pHrodo‐labelled recombinant NECD protein. During physiological Dll4/Notch signaling, NECD binds to the adjacent Dll4‐presenting cell. Thereafter, both the bound NECD and Dll4 receptor are transcytosed by the original Dll4‐presenting cell. To best reproduce this complex, we functionalized the pHrodo‐labeled NECD to a microbead as previously reported 12 (Figure 4E). The analysis revealed a delay in the internalization kinetics in scramble siRNA‐treated ECs compared to the Dll4‐antibody internalization, likely due to the presence of a bead tether (Figure 4F). Nonetheless, siRNA knockdown against EHD2 led to a significant impairment of NECD internalization compared with control (Figure 4F). We observed the same internalization defect in Dll4, Jag1, and dual Dll4 and Jag1 siRNA‐treated ECs, suggesting NECD is specific to these Notch ligands (Figure 4G). To again confirm Dll4 uptake depends on caveolae, we knocked down both caveolin and clathrin‐related endocytic components. Knockdown of caveolin‐1 significantly reduced NECD internalization, while knockdown of clathrin‐related proteins CLTB or AP2 did not affect Dll4 internalization compared with control (Figure 4H). To further confirm our findings, we also compared Dll4 endocytosis using an antibody feeding assay. 44 Briefly, ECs were incubated with Dll4‐antibody and allowed to endocytose the ligand after a cold block was removed. Thereafter, the proportion of surface bound to internalized Dll4 was evaluated. Similar to results using pHrodo labeling, loss of EHD2, Caveolin‐1 or dynamin all produced a significant impairment in Dll4 endocytosis (Figure 4I). Overall, these data indicate that EHD2 enhances caveolin‐mediated NECD/Dll4 transcytosis.
We considered that the disruptions in Dll4 endocytosis may be due to reduced Dll4 bioavailability. However, we observed that EHD2 siRNA knockdown, Notch inhibition via treatment with DAPT or Dynasore did not affect endogenous Dll4 protein levels (Figure 4J). Therefore, the reduced internalization of Dll4 is a direct result of loss of EHD2, supporting its role as an endocytic mediator of Dll4/Notch1 transcytosis. To visualize this endocytic impairment in the absence of EHD2 we employed transmission electron microscopy imaging. EHD2 knockdown greatly increased the number of small endocytic vacuoles near the plasma membrane (Figure S2C), an observation consistent with previous reports investigating EHD2 in which caveolae are unable to be stabilized through actin anchoring and accumulate near the plasma membrane. 14 , 45 Lastly, we confirmed that knockdown of EHD2, Caveolin‐1 or both proteins resulted in a significant reduction in Hes1 expression, indicating a decrease in Notch activation (Figure 4K).
3.5. Loss of EHD2 affects development of zebrafish blood vessels
To investigate whether EHD2 played a role in angiogenesis, we first characterized how the loss of EHD2 affected intersomitic blood vessel (ISV) development in Danio rerio (zebrafish). This vessel bed requires tightly regulated tip/stalk cell specification and demonstrates stereotyped morphodynamics, making aberrations in normal blood vessel development relatively obvious. 46 Due to a gene duplication event in teleosts, EHD2 has two paralogs in zebrafish: EHD2a and EHD2b. We targeted each paralog individually, as well as in combination, using a 4‐guide CRISPR knockout (KO) approach 22 to create F0 KOs in the EHD2a/b loci (Table S2). For each KO, we evaluated one of the four CRISPR cut sites for indel formation. Sequencing revealed 100% of the putative target sites contained substantial indels in 3 random samples from each condition as well as significantly reduced mRNA expression (Figure 5A‐C). Quantification of the proportion of fish with dysmorphic ISVs using a vascular reporter line tg(kdrl:eGFP) 47 in each condition revealed a significant increase in EHD2a/b knockout fish (21.76%) compared to a scrambled single‐guide RNA control at 48 h post fertilization (hpf) (Figure 5D,E). These results suggest that EHD2a/b are necessary for normal sprouting behaviors in vivo.
FIGURE 5.
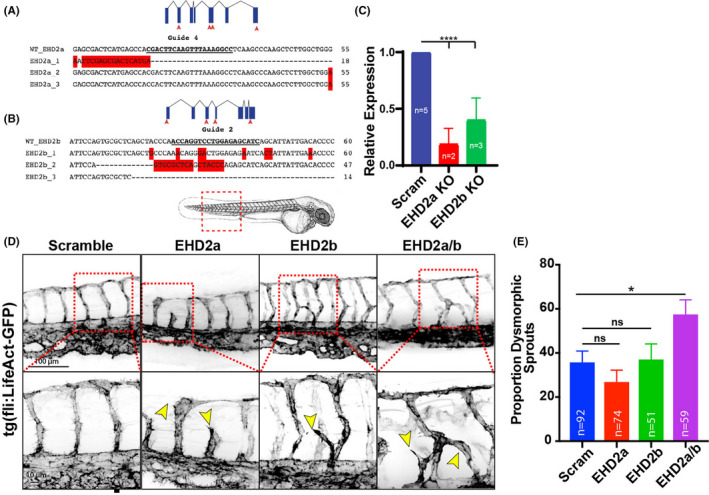
Loss of EHD2 promotes dysmorphic sprouting in zebrafish blood vessels. (A,B) DNA sequence alignments of EHD2a or EHD2b CRISPR injected embryos to a wild‐type (WT) sequence (n = 3). (C) Message RNA levels of either EHD2a or EHD2b relative to a scramble control after CRISPR injections. RNA was collected at 72 h post fertilization (hpf). 10–20 fish were pooled per repeat. N, number of repeats. (D) Representative images of intersomtic blood vessels (ISVs) on vascular reporter tg(kdrl:GFP)+/+ background at 48 hpf injected with indicated CRISPR guides. Yellow arrowheads denote abnormal vascular growth. Red dashed box denotes area of higher magnification. Larvae cartoon denotes location of imaging. (E) Quantification of the proportion of Crispant fish with dysmorphic ISVs. Dysmorphic was defined as a vessel projection emerging from the ISV distinct from the central stalk. Error bars represent SEM. N, number of fish quantified unless indicated. *p < .05, ****p < .0001. ns, non‐significant. All experiments were done at minimum in triplicate
We next tested if vascular abnormalities in the EHD2a/b KO lines were related to Notch activity, as loss of Notch signaling promotes hypersprouting both in developing zebrafish and mouse blood vessels. 5 , 48 , 49 , 50 , 51 , 52 , 53 Treatment with the small molecule Notch inhibitor LY‐411575 phenocopied the increase in dysmorphic sprouts observed in the EHD2a/b KOs (Figure 6A,B). To further explore Notch activation, we monitored expression of Hey2, a downstream Notch target, across groups in reference to a GAPDH control. We observed significantly diminished expression of Hey2 in EHD2b, and EHD2a/b KO groups in comparison with the scramble control (Figure 6C). The minimal effect of the EHD2a KO is likely due to the lack of expression (Figure S3). Overall, these results support a Notch loss‐of‐function phenotype in the absence of EHD2b in zebrafish blood vessels.
FIGURE 6.
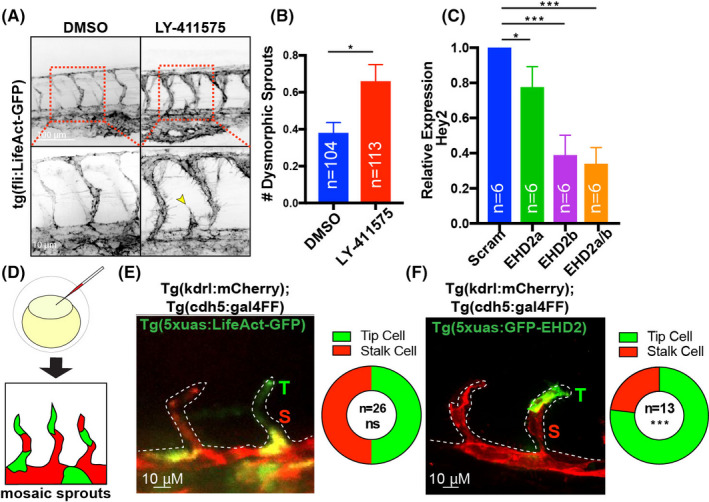
EHD2 knockout phenocopies Notch loss of function. (A) Intersomic vessels (ISVs) of fish treated with either DMSO or 2 µM LY‐411575 on tg(fli:LifeAct)+/+ background at 48 h post fertilization (hpf). Yellow arrowheads point to dysmorphic sprouts. (B) Number of dysmorphic ISVs between indicated groups. (C) Relative expression of Hey2 transcript in 48hpf Crispant groups normalized to GAPDH. 10–20 fish were pooled per repeat. N, number of repeats. (D) Cartoon of method to produce mosaic expression in zebrafish blood vessels. (E,F) Representative images of ISVs expressing mCherry (tg(kdrl:mCherry), LifeAct‐GFP or GFP‐EHD2 at 24 hpf. Quantification of the proportion of expressing endothelial cells in either the tip (T, green) or stalk (S, red) cell positions in the vascular sprouts shown to the left. N, number of fish. Significance *p < .05, ***p < .001. ns, non‐significant. Error bars are SEM. All experiments were done at minimum in triplicate
In canonical tip/stalk cell specification, tip cells exhibit elevated Dll4 levels that, in turn, elicit repressive Notch activation in the trailing stalk cells. 5 , 6 , 51 , 52 , 54 , 55 Given that KO of EHD2a/b phenocopied the Notch loss‐of‐function sprouting defects as well as its effect on Dll4 transcytosis in vitro, we hypothesized that EHD2 may influence hierarchical tip/stalk cell positioning. To determine how EHD2 functions during tip/stalk cell specification, we developed a GFP‐tagged EHD2 fusion protein that was injected into a WT (tg(kdrl:mCherry; tg(cdh5:gal4ff)) 21 vascular reporter line to produce mosaic ISVs. This approach allowed us to visualize individual ECs in the sprout collective (Figure 6D). Zebrafish EHD2a/b proteins are approximately 70% identical to the human ortholog, thus predicted to work similarly (Figure S4). We reasoned that if EHD2 did not affect Notch activation there would be an equal hierarchical EC contribution in tip or stalk cell positions. Confirming this, we injected a control tg(5xUAS:LifeAct‐GFP) construct and observed mosaic integration with an even 50/50 distribution between tip and stalk ECs in growing ISVs at 24hpf (Figure 6E). However, ECs expressing GFP‐EHD2 (tg(5xUAS:GFP‐EHD2)) demonstrated a significant bias toward the tip cell position with 76.9% of ISVs exhibiting EHD2‐overexpressing ECs in the tip cells (Figure 6F). Our interpretation of this result is that Dll4 is most highly expressed in tip cells 56 ; thus EHD2 overexpression would preferentially localize to this position given its potential role in Dll4/NECD transcytosis. Overall, these results support a role for EHD2 in Notch/Dll4 signaling during blood vessel development in vivo.
4. DISCUSSION
Although Notch signaling is critical for blood vessel development, endocytic mechanisms that regulate both Dll4 cell surface expression and Notch receptor activation remain elusive. We report that EHD2 can alter transcytosis of the Dll4/Notch1 complex. Importantly, this is the first characterization of Dll4 being influenced by caveolin‐mediated endocytosis. In a broader context, our results demonstrate a novel endocytic pathway that directly impacts Dll4/Notch signaling which is required for proper blood vessel development.
Caveolae have complex roles in regulating both endocytosis events and maintaining tissue integrity. Caveolae are 50–80 nm flask shaped invaginations of the plasma membrane fashioned through integral membrane caveolins and their associated proteins. 57 , 58 In addition to endocytosis, caveolae more recently have been shown to be a membrane reservoir that can safeguard against mechanical stress. 59 For instance, caveolin 1,3 are essential for notochord integrity in developing zebrafish. 59 Similarly, caveolae protect against membrane rupture in ECs and skeletal muscle. 60 , 61 Interestingly, global deletion of caveolin‐1 is not embryonic lethal, 62 suggesting that caveolins are not essential and/or other redundant factors are at play. In our investigation, loss of EHD2 was not lethal in zebrafish. Additionally, EHD2 knockdown did not affect caveolin formation on the plasma membrane, although, it did impact the number of caveolar pits adjacent to the plasma membrane. Others have shown that loss of EHD2 does not affect the number of caveolar pits formed in non‐endothelial tissues, but drastically increases their dynamics by not being anchored to the underlying actin network. 14 It would be prudent to predict that caveolae and by association, EHD2, likely have multiple cellular roles given the broad range of functions that have been reported for each protein.
Our work suggests that in the absence of EHD2, the caveolar pit will be destabilized and remains in the subapical space unavailable for binding. Others have shown that EHD2 can also help anchor caveolae to the actin cytoskeleton. Thus, another potential mechanism could be a lack of pit anchoring through loss of EHD2. This would preclude the force necessary to initiate Dll4/NECD pulling and subsequent S2 cleavage and transcytosis. Without NECD transcytosis, γ‐secretase is unable to cleave at the S3 intracellular site, effectively blocking Notch signaling.
In mice, global EHD2 deletion does not affect viability, in contrast with the embryonic lethality of Notch1 or Dll4 knockout models. 63 , 64 However, loss of EHD2 has been shown to increase the number of caveolae that were detached from the membrane and significantly reduced production of endothelial nitric oxide, potentially indicating a preference in endothelial function. 65 Loss of Notch function has also been associated with reduced endothelial nitric oxide synthase activity. 66 In line with these reports, we observed that EHD2 knockdown resulted in an elevated number of detached caveolae in ECs. In zebrafish, we show that EHD2 knockdown closely phenocopies loss of Notch signaling in terms of sprouting defects. Additionally, loss of EHD2 also reduces Notch activation, similar to in vivo results. However, it is important to note that these results are correlative and require additional experiments to truly demonstrate that EHD2 is directly interacting with Dll4/Notch in vivo.
Dll4 is unquestionably vital for Notch signaling and blood vessel morphogenesis. In non‐endothelial cell types, Dll1 endocytosis generates the mechanical pulling force on the NECD to expose the S2 domain for cleavage. 11 After S2 cleavage, the Dll1/NECD complex internalizes and, presumably, undergoes subsequent lysosomal degradation. In the adjacent Notch presenting cell, following S2 cleavage and NECD release, the S3 cleavage by γ‐secretase and NICD release can proceed. 7 Others have reported that the Dll1 pulling force is derived from clathrin‐dependent endocytosis 11 ; however, our results suggest a different pathway. In our investigation, we equally explored the idea that EHD2 uses clathrin‐dependent or independent programs to aid in Dll4 internalization. To our surprise, we did not observe any significant Dll4 or EHD2 colocalization with clathrin in ECs, indicating this was not the operative pathway for Dll4 endocytosis. Moreover, ablation of clathrin itself or related protein AP2 did not affect Dll4 endocytosis. With these results, we determined that Dll4 strongly relied on caveolar endocytosis for NECD transcytosis and subsequent Notch activation. To our knowledge, this is the first report indicating an association between Dll4 and caveolin‐mediated endocytosis and it contrasts reports investigating Dll1. 11 , 12 , 35 , 36 The reasons for this disparity could be due to both receptor‐type and/or tissue source differences. For instance, global Dll1 deletion in mice does not affect viability, while global loss of Dll4 is embryonic lethal. In the same vein, our results only determine that EHD2 is associated with Dll4/NECD transcytosis and does not provide a concrete mechanism of action of how EHD2 physically interacts with the Dll4 receptor.
In aggregate, our results characterize EHD2’s role in NECD/Dll4 transcytosis. Our analysis uncovers two major findings: (1) Dll4 uses a non‐clathrin‐mediated endocytic program; and (2) EHD2 is required for Dll4 internalization during Notch receptor engagement. These results raise questions pertaining to the role of other endocytic proteins in the basal‐ and/or bound‐state of Dll4. With regard to EHD2, what is the precise pulling force contribution required for Dll4 endocytosis and how much is controlled by anchoring to the cytoskeleton potentially through EHD2 is not known. It is also interesting to speculate how the caveolar machinery may interface with modifiers of Dll4, such as fringe proteins known to glycosylate Dll4's extracellular domain. Overall, we believe that in addition to transcriptional regulation of Dll4/Notch proteins, the endocytic machinery involved in their signaling may add yet another level of regulation important for blood vessel development.
5. PERSPECTIVES
EHD2 is a caveolae related protein that is enriched in blood vessels.
EHD2 associates with Dll4 and caveolin‐1 in endothelial cells.
Loss of EHD2 blunts Dll4 endocytosis and Notch extracellular domain transcytosis in vitro.
Knockout of EHD2 leads to dysmorphic blood vessel development in zebrafish.
CONFLICT OF INTERESTS
None.
AUTHOR CONTRIBUTIONS
A.M.W, H.K, K.M.L and E.J.K created zebrafish and cell line constructs. A.M.W and E.J.K conceived experiments. R.J.J. performed mouse retinal experiments. A.M.W and C.R.F performed fibrin‐bead experiments. D.E.W and S.M.M performed EHD2 TEM analysis and in situ hybridization experiments. A.M.W. and E.J.K wrote the manuscript.
Supporting information
Appendix S1
ACKNOWLEDGEMENTS
We like to thank Jennifer Bourne and the Electron Microscopy Center at the University of Colorado Anschutz Medical Campus for assistance with transmission electron micrograph collection.
Webb AM, Francis CR, Judson RJ, et al. EHD2 modulates Dll4 endocytosis during blood vessel development. Microcirculation.2022;29:e12740. doi: 10.1111/micc.12740
Funding information
Work was supported by funding from the National Heart Lung Blood Institute (Grant R15HL156106‐01A1, 1R56HL148450‐01, R00HL124311; A.M.W, C.R.F, and E.J.K)
REFERENCES
- 1. Chappell JC, Taylor SM, Ferrara N, Bautch VL. Local guidance of emerging vessel sprouts requires soluble Flt‐1. Dev Cell. 2009;17(3):377‐386. 10.1016/j.devcel.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chappell JC, Wiley DM, Bautch VL. How blood vessel networks are made and measured. Cells Tissues Organs. 2012;195(1–2):94‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gerhardt H, Golding M, Fruttiger M, et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161(6):1163‐1177. Epub 2003 Jun 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ninov N, Borius M, Stainier DY. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139(9):1557‐1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benedito R, Roca C, Sörensen I, et al. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137(6):1124‐1135. [DOI] [PubMed] [Google Scholar]
- 6. Blanco R, Gerhardt H. VEGF and Notch in tip and stalk cell selection. Cold Spring Harb Perspect Med. 2013;3(1):a006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mumm JS, Schroeter EH, Saxena MT, et al. A ligand‐induced extracellular cleavage regulates gamma‐secretase‐like proteolytic activation of Notch1. Mol Cell. 2000;5(2):197‐206. [DOI] [PubMed] [Google Scholar]
- 8. Gordon WR, Arnett KL, Blacklow SC. The molecular logic of Notch signaling–a structural and biochemical perspective. J Cell Sci. 2008;121(Pt 19):3109‐3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramasamy SK, Kusumbe AP, Wang L, Adams RH. Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature. 2014;507(7492):376‐380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pitulescu ME, Schmidt I, Giaimo BD, et al. Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat Cell Biol. 2017;19(8):915‐927. [DOI] [PubMed] [Google Scholar]
- 11. Shergill B, Meloty‐Kapella L, Musse AA, et al. Optical tweezers studies on Notch: single‐molecule interaction strength is independent of ligand endocytosis. Dev Cell. 2012;22(6):1313‐1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meloty‐Kapella L, Shergill B, Kuon J, et al. Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev Cell. 2012;22(6):1299‐1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gessler M. Dll1 and Dll4: similar, but not the same. Blood. 2009;113(22):5375‐5376. [DOI] [PubMed] [Google Scholar]
- 14. Moren B, Shah C, Howes MT, et al. EHD2 regulates caveolar dynamics via ATP‐driven targeting and oligomerization. Mol Biol Cell. 2012;23(7):1316‐1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoernke M, Mohan J, Larsson E, et al. EHD2 restrains dynamics of caveolae by an ATP‐dependent, membrane‐bound, open conformation. Proc Natl Acad Sci USA. 2017;114(22):E4360‐e4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Torrino S, Shen W‐W, Blouin CM, et al. EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription. J Cell Biol. 2018;217(12):4092‐4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kovtun O, Tillu VA, Ariotti N, Parton RG, Collins BM. Cavin family proteins and the assembly of caveolae. J Cell Sci. 2015;128(7):1269‐1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen J‐N. FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol. 2007;304(2):735‐744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hen G, Nicenboim J, Mayseless O, et al. Venous‐derived angioblasts generate organ‐specific vessels during zebrafish embryonic development. Development. 2015;142(24):4266‐4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Proulx K, Lu A, Sumanas S. Cranial vasculature in zebrafish forms by angioblast cluster‐derived angiogenesis. Dev Biol. 2010;348(1):34‐46. [DOI] [PubMed] [Google Scholar]
- 21. Bussmann J, Wolfe SA, Siekmann AF. Arterial‐venous network formation during brain vascularization involves hemodynamic regulation of chemokine signaling. Development. 2011;138(9):1717‐1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu RS, Lam II, Clay H, Duong DN, Deo RC, Coughlin SR. A rapid method for directed gene knockout for screening in G0 Zebrafish. Dev Cell. 2018;46(1):112‐125. e4. [DOI] [PubMed] [Google Scholar]
- 23. Thisse C, Thisse B. High‐resolution in situ hybridization to whole‐mount zebrafish embryos. Nat Protoc. 2008;3(1):59‐69. [DOI] [PubMed] [Google Scholar]
- 24. He TC, Zhou S, da Costa LT, Yu J, Kinzler K w, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95(5):2509‐2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakatsu MN, Davis J, Hughes CC. Optimized fibrin gel bead assay for the study of angiogenesis. J Vis Exp. 2007;3:186. 10.3791/186. Epub 2007 Apr 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fredriksson S, Gullberg M, Jarvius J, et al. Protein detection using proximity‐dependent DNA ligation assays. Nat Biotechnol. 2002;20(5):473‐477. [DOI] [PubMed] [Google Scholar]
- 27. Schindelin J, Arganda‐Carreras I, Frise E, et al. Fiji: an open‐source platform for biological‐image analysis. Nat Methods. 2012;9(7):676‐682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(Pt 3):213‐232. [DOI] [PubMed] [Google Scholar]
- 29. Stahl A, Connor KM, Sapieha P, et al. The mouse retina as an angiogenesis model. Invest Ophthalmol Vis Sci. 2010;51(6):2813‐2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suchting S, Freitas C, le Noble F, et al. The Notch ligand Delta‐like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc Natl Acad Sci USA. 2007;104(9):3225‐3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alam MS. Proximity Ligation Assay (PLA). Curr Protoc Immunol. 2018;123(1):e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guilherme A, Soriano NA, Bose S, et al. EHD2 and the novel EH domain binding protein EHBP1 couple endocytosis to the actin cytoskeleton. J Biol Chem. 2004;279(11):10593‐10605. [DOI] [PubMed] [Google Scholar]
- 33. Sankaranarayanan S, De Angelis D, Rothman JE, Ryan TA. The use of pHluorins for optical measurements of presynaptic activity. Biophys J. 2000;79(4):2199‐2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Galletta BJ, Cooper JA. Actin and endocytosis: mechanisms and phylogeny. Curr Opin Cell Biol. 2009;21(1):20‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heuss SF, Ndiaye‐Lobry D, Six EM, Israel A, Logeat F. The intracellular region of Notch ligands Dll1 and Dll3 regulates their trafficking and signaling activity. Proc Natl Acad Sci USA. 2008;105(32):11212‐11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okano M, Matsuo H, Nishimura Y, et al. Mib1 modulates dynamin 2 recruitment via Snx18 to promote Dll1 endocytosis for efficient Notch signaling. Genes Cells. 2016;21(5):425‐441. [DOI] [PubMed] [Google Scholar]
- 37. Francis CR, Kushner EJ. Capturing membrane trafficking events during 3D angiogenic development in vitro. Microcirculation. 2021;28(6):e12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kushner EJ, Ferro LS, Liu J‐Y, et al. Excess centrosomes disrupt endothelial cell migration via centrosome scattering. J Cell Biol. 2014;206(2):257‐272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kushner EJ, Ferro LS, Yu Z, Bautch VL. Excess centrosomes perturb dynamic endothelial cell repolarization during blood vessel formation. Mol Biol Cell. 2016;27(12):1911‐1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mouillesseaux KP, Wiley DS, Saunders LM, et al. Notch regulates BMP responsiveness and lateral branching in vessel networks via SMAD6. Nat Commun. 2016;7:13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Batchuluun K, Azuma M, Yashiro T, Kikuchi M. Notch signaling‐mediated cell‐to‐cell interaction is dependent on E‐cadherin adhesion in adult rat anterior pituitary. Cell Tissue Res. 2017;368(1):125‐133. [DOI] [PubMed] [Google Scholar]
- 42. Benhra N, Lallet S, Cotton M, et al. AP‐1 controls the trafficking of Notch and Sanpodo toward E‐cadherin junctions in sensory organ precursors. Curr Biol. 2011;21(1):87‐95. [DOI] [PubMed] [Google Scholar]
- 43. Kamen L, Myneni S, Langsdorf C, et al. A novel method for determining antibody‐dependent cellular phagocytosis. J Immunol Methods. 2019;468:55‐60. [DOI] [PubMed] [Google Scholar]
- 44. Boucher JM, Clark R, Chong DC, Citrin KM, Wylie LA, Bautch V. Dynamic alterations in decoy VEGF receptor‐1 stability regulate angiogenesis. Nat Commun. 2017;8:15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matthaeus C, Lahmann I, Kunz S, et al. EHD2‐mediated restriction of caveolar dynamics regulates cellular fatty acid uptake. Proc Natl Acad Sci USA. 2020;117(13):7471‐7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gore AV, Monzo K, Cha YR, Pan W, Weinstein BM. Vascular development in the zebrafish. Cold Spring Harb Perspect Med. 2012;2(5):a006684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248(2):307‐318. [DOI] [PubMed] [Google Scholar]
- 48. Therapontos C, Vargesson N. Zebrafish notch signalling pathway mutants exhibit trunk vessel patterning anomalies that are secondary to somite misregulation. Dev Dyn. 2010;239(10):2761‐2768. [DOI] [PubMed] [Google Scholar]
- 49. Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445(7129):781‐784. [DOI] [PubMed] [Google Scholar]
- 50. Arima S, Nishiyama K, Ko T, et al. Angiogenic morphogenesis driven by dynamic and heterogeneous collective endothelial cell movement. Development. 2011;138(21):4763‐4776. [DOI] [PubMed] [Google Scholar]
- 51. Ehling M, Adams S, Benedito R, Adams RH. Notch controls retinal blood vessel maturation and quiescence. Development. 2013;140(14):3051‐3061. [DOI] [PubMed] [Google Scholar]
- 52. Eilken HM, Adams RH. Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr Opin Cell Biol. 2010;22(5):617‐625. [DOI] [PubMed] [Google Scholar]
- 53. Jakobsson L, Bentley K, Gerhardt H. VEGFRs and Notch: a dynamic collaboration in vascular patterning. Biochem Soc Trans. 2009;37(Pt 6):1233‐1236. [DOI] [PubMed] [Google Scholar]
- 54. Gridley T. Notch signaling in vascular development and physiology. Development. 2007;134(15):2709‐2718. [DOI] [PubMed] [Google Scholar]
- 55. Hellstrom M, Phng L‐K, Hofmann JJ, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445(7129):776‐780. [DOI] [PubMed] [Google Scholar]
- 56. Jakobsson L, Franco CA, Bentley K, et al. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol. 2010;12(10):943‐953. [DOI] [PubMed] [Google Scholar]
- 57. Lamaze C, Tardif N, Dewulf M, Vassilopoulos S, Blouin CM. The caveolae dress code: structure and signaling. Curr Opin Cell Biol. 2017;47:117‐125. [DOI] [PubMed] [Google Scholar]
- 58. Lamaze C, Torrino S. Caveolae and cancer: a new mechanical perspective. Biomed J. 2015;38(5):367‐379. [DOI] [PubMed] [Google Scholar]
- 59. Garcia J, Bagwell J, Njaine B, et al. Sheath cell invasion and trans‐differentiation repair mechanical damage caused by loss of Caveolae in the Zebrafish notochord. Curr Biol. 2017;27(13):1982‐1989. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cheng JP, Mendoza‐Topaz C, Howard G, et al. Caveolae protect endothelial cells from membrane rupture during increased cardiac output. J Cell Biol. 2015;211(1):53‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lo HP, Nixon SJ, Hall TE, et al. The caveolin‐cavin system plays a conserved and critical role in mechanoprotection of skeletal muscle. J Cell Biol. 2015;210(5):833‐849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Razani B, Lisanti MP. Caveolin‐deficient mice: insights into caveolar function human disease. J Clin Invest. 2001;108(11):1553‐1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Krebs LT, Xue Y, Norton CR, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14(11):1343‐1352. [PMC free article] [PubMed] [Google Scholar]
- 64. Huppert SS, Le A, Schroeter EH, et al. Embryonic lethality in mice homozygous for a processing‐deficient allele of Notch1. Nature. 2000;405(6789):966‐970. [DOI] [PubMed] [Google Scholar]
- 65. Matthaeus C, Lian X, Kunz S, et al. eNOS‐NO‐induced small blood vessel relaxation requires EHD2‐dependent caveolae stabilization. PLoS One. 2019;14(10):e0223620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Patenaude A, Fuller M, Chang L, et al. Endothelial‐specific Notch blockade inhibits vascular function and tumor growth through an eNOS‐dependent mechanism. Cancer Res. 2014;74(9):2402‐2411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


