Abstract
Marine classification schemes based on abiotic surrogates often inform regional marine conservation planning in lieu of detailed biological data. However, these schemes may poorly represent ecologically relevant biological patterns required for effective design and management strategies. We used a community‐level modeling approach to characterize and delineate representative mesoscale (tens to thousands of kilometers) assemblages of demersal fish and benthic invertebrates in the Northwest Atlantic. Hierarchical clustering of species occurrence data from four regional annual multispecies trawl surveys revealed three to six groupings (predominant assemblage types) in each survey region, broadly associated with geomorphic and oceanographic features. Indicator analyses identified 3–34 emblematic taxa of each assemblage type. Random forest classifications accurately predicted assemblage distributions from environmental covariates (AUC > 0.95) and identified thermal limits (annual minimum and maximum bottom temperatures) as important predictors of distribution in each region. Using forecasted oceanographic conditions for the year 2075 and a regional classification model, we projected assemblage distributions in the southernmost bioregion (Scotian Shelf‐Bay of Fundy) under a high emissions climate scenario (RCP 8.5). Range expansions to the northeast are projected for assemblages associated with warmer and shallower waters of the Western Scotian Shelf over the 21st century as thermal habitat on the relatively cooler Eastern Scotian Shelf becomes more favorable. Community‐level modeling provides a biotic‐informed approach for identifying broadscale ecological structure required for the design and management of ecologically coherent, representative, well‐connected networks of Marine Protected Areas. When combined with oceanographic forecasts, this modeling approach provides a spatial tool for assessing sensitivity and resilience to climate change, which can improve conservation planning, monitoring, and adaptive management.
Keywords: climate change adaptation, community‐level modeling, conservation planning, ecological coherence, ocean warming, representativity
INTRODUCTION
Characterizing the diversity of distinct assemblages in marine ecosystems and their distribution is key to successful implementation of regional conservation planning (Foster et al., 2017; Roberts et al., 2003), ecosystem‐based management of fisheries (Koen‐Alonso et al., 2019), and marine spatial planning processes (Foley et al., 2010) that collectively aim to maintain or restore biodiversity and ecosystem functions that underpin services and benefits to society. From a conservation perspective, designing networks of marine protected areas (MPAs) that capture the full breadth of ecosystem and habitat types is a keystone principle (representativity) that maximizes their effectiveness and ecological coherence (Airamé et al., 2003; Roberts et al., 2003). Ecological coherence describes the integrity of the collective sites in a network, the processes, functions, and structures protected therein, and the network with the wider environment under changing conditions and is achieved by incorporating representativity and other ecological principles (i.e., connectivity, adequacy, replication) in their design (Ardron, 2008). MPA networks designed to be representative of taxonomic assemblages and habitat types at a regional scale are likely to conserve a greater range of biodiversity and ecosystem functions than those approaches focused on species richness alone (Roberts et al., 2003). Moreover, representative networks can serve as an insurance policy, natural reference, and seed stock to adjacent non‐protected areas (Rice & Houston, 2011), and thus confer simultaneous benefits to conservation and fisheries (Gaines et al., 2010; Roberts et al., 2003). Addressing representativity is a logical precursor to incorporating other ecological design principles into MPA networks (Roberts et al., 2003; Smith et al., 2009). However, operationalizing this criterion requires the development of regional marine classification schemes that delineate the main biogeographic subdivisions of MPA network planning areas (Rice & Houston, 2011; Roff et al., 2003). These subdivisions ideally define spatially contiguous units of relatively homogeneous community composition along important axes of environmental variation. Although larger‐scale marine ecoregions are available (e.g., Spalding et al., 2007), their spatial scale often abstracts them from the scale by which regional MPA networks are developed.
Several approaches have been adopted to develop marine classification schemes across multiple spatial scales. These approaches can be broadly characterized as classifications based on (1) abiotic habitat surrogates or (2) biologically informed predictive mapping. The first strategy leverages widely available physiographic and oceanographic data to delineate units of the classification with the assumption that distinct abiotic conditions will represent underlying community composition (e.g., Douglass et al., 2014; Roff et al., 2003). Although these abiotic surrogates can provide an adequate first‐order approximation of biological patterns (Kostylev & Hannah, 2007; Sutcliffe et al., 2015), they often rely on expert judgments of the relative biological importance of abiotic variables and assumptions on boundary locations. Furthermore, abiotic surrogates alone fail to capture the level of representativity achieved by biologically informed classifications (Ban, 2009; Rubidge et al., 2016; Sutcliffe et al., 2015), and therefore may lead to larger (inefficient) MPA network footprints to achieve adequate protection of biodiversity features (Ferrari et al., 2018). The second strategy applies one of three broad community‐level modeling approaches that identify key environmental correlates with more sparse biological survey data to extrapolate multispecies patterns (e.g., assemblage types, species groups, compositional turnover) across a planning region (Ferrier & Guisan, 2006). A “predict‐then‐assemble” approach reconstructs assemblages from independently modeled species distributions (e.g., Leathwick et al., 2006). An “assemble‐then‐predict” approach models assemblage types identified directly from the biological survey data (e.g., Moritz et al., 2013; Rubidge et al., 2016), whereas an “assemble‐and‐predict‐together” approach models multispecies responses within a single integrated process (e.g., Leathwick et al., 2012; Murillo et al., 2018; Sutcliffe et al., 2015). Each approach confers advantages and disadvantages; the choice hinges on the data at hand and intended application (Ferrier & Guisan, 2006).
Marine classifications derived using an assemble‐then‐predict approach offer multiple benefits for conservation applications. Compared to approaches that aggregate separate species distribution models, this approach offers faster processing of a large number of taxa, integrating patterns from all taxa in the data set. In this way, the modeling approach can include rare species recorded too infrequently to be reliably modeled alone (Ferrier & Guisan, 2006) and those species with weak environmental responses (Pitcher et al., 2012); improving power to detect emergent properties. The regional subdivisions identified by these classifications also correspond with more distinct assemblages compared to habitat surrogates (Cooper et al., 2019; Rubidge et al., 2016), improving the basis on which to evaluate MPA network representativity. Although it is thought addressing representativity inherently includes some degree of network connectivity and climate resilience (McLeod et al., 2009; Smith et al., 2009), it is rare in practice for demographic and genetic connectivity (Balbar & Metaxas, 2019), ecosystem wide climate change vulnerability (Wilson et al., 2020), and climate adaptation considerations (Tittensor et al., 2019) to be empirically implemented in MPA design and management (Balbar et al., 2020). Classification techniques also can allow for the identification of indicator taxa strongly associated with their respective assemblages (Murillo et al., 2018; Rubidge et al., 2016), which may facilitate selection of emblematic species for operationalizing the connectivity criterion or for selective monitoring approaches. Predictive models, including those developed using an assemble‐then‐predict approach, can be used to assess which assemblage types may persist under future climate conditions (Ferrier & Guisan, 2006). Therefore, biologically informed classification schemes could provide a pragmatic tool for implementing a recent recommendation by Tittensor et al. (2019) to incorporate climate‐smart objectives by default into MPA design and management plans (e.g., maintenance of representativity under changing climate conditions).
Researchers, MPA practitioners, and decision‐makers in Atlantic Canada are uniquely positioned to realize the extended benefits of a community‐level modeling approach to biophysical classification for marine conservation planning. MPA network planning is coordinated by the federal Department of Fisheries and Ocean (DFO) on a regional level (DFO, 2009), prompting the need for marine classification schemes suitable at that scale. However, earlier efforts to develop such classifications have been based either on mainly abiotic habitat surrogates (Kostylev & Hannah, 2007; Park & Mercier, 2014; Roff et al., 2003) or taxonomically restricted biological data sets (Chouinard & Dutil, 2011; Moritz et al., 2013). Existing classification schemes set up at global (Spalding et al., 2007), continental (Wilkinson et al., 2009), and national scales (DFO, 2009) provide useful context for setting the scale of bioregional networks, but do not resolve local ecological features identified by regional network design processes (e.g., DFO, 2018a). Regional fishery‐independent multispecies trawl surveys on the continental shelf and adjacent areas conducted annually since the 1970s in Atlantic Canada have greatly expanded in taxonomic scope in the last decade (Chadwick et al., 2007), meeting the data requirements for predictive mapping of bottom communities from a broader ecosystem perspective. With wide continental shelves covering a broad latitudinal gradient, complex bathymetry, estuarine to marine conditions, and the confluence of cold polar and warm tropical currents, the diverse oceanographic and physiographic context requires the identification of key environmental gradients driving compositional patterns. Atlantic Canada also is considered an ocean warming hotspot (Hobday & Pecl, 2014), with a projected overall loss of suitable thermal habitat for fish and benthic invertebrates predicted over the 21st century under a high carbon emissions scenario (Morley et al., 2018; Stanley et al., 2018), although, for some species, suitable habitat is expected to expand (Greenan et al., 2019). Consequently, there is increased motivation to provide scale‐appropriate ecosystem forecasts and management guidance that could bridge the gap between theory and practice (Tittensor et al., 2019) for implementing climate‐smart networks in a vulnerable area.
Here, we apply a unified community‐level (assemble‐then‐predict) modeling approach to characterize and delineate the major assemblages of demersal fish and benthic invertebrates in four regions of Atlantic Canada. First, we use clustering techniques to identify the predominant assemblage types from species occurrence data derived from ecosystem surveys conducted with bottom trawls within each of four survey regions. We perform indicator analyses to identify emblematic taxa of each assemblage type. Next we combine oceanographic and physiographic data layers with regional coverage and random forest classification to identify key environmental correlates with assemblage distributions. We use modeled assemblage–environment relationships to predictively map their distribution and produce regional, biologically informed classification schemes. Finally, we evaluate the vulnerability of assemblages in the southernmost bioregion (Scotian Shelf‐Bay of Fundy) to ocean warming using the regional distributional model to (1) hindcast spatial compositional changes associated with recent warming and (2) forecast assemblage distributions to 2075 with predictions of a general circulation model under a high‐emissions warming scenario. Our study expands on previous approaches to develop ecologically relevant regional classifications in the marine realm (Rubidge et al., 2016; Sutcliffe et al., 2015) by demonstrating the utility across multiple regions and for predicting assemblage vulnerability to climate change. Our method focusing on assemblage types also offers a complementary approach to previous work to project climate‐associated distributional changes in marine ecosystems, which are typically evaluated on a species‐by‐species basis.
METHODS
Study areas
We replicated our analyses in each of four administrative regions delineated by the Department of Fisheries and Oceans in Atlantic Canada: Quebec (NGSL); Gulf (SGSL); Newfoundland and Labrador (NL); and Scotian Shelf‐Bay of Fundy, that is, Maritimes (MAR; Figure 1) regions. These regions correspond roughly with distinct biogeographic divisions distinguished by their oceanographic and bathymetric features and are recognized as the appropriate scale for MPA network design in Canada (DFO, 2009). The NGSL region spans the lower St. Lawrence estuary and northern Gulf of St. Lawrence and the SGSL includes the southern Gulf of St. Lawrence. In the NGSL region, three deep troughs (Laurentian, Anticosti, and Esquiman Channels) dominate the bathymetry; while an expansive, shallow plateau, the Magdalen Shallows, occupies much of the SGSL region. Freshwater inputs from the St. Lawrence and Saguenay rivers and connections with the Atlantic Ocean through the Cabot Strait and Strait of Belle Isle drive an estuarine circulation pattern and seasonal sea‐ice formation occurs throughout the Gulf (DFO, 2005). The NL region is characterized by relatively broad and shallow continental shelves, especially the Grand Banks to the southeast of Newfoundland. The Newfoundland shelf is intermittently cut by deeper channels and bound to the southwest by the Laurentian Channel. The oceanographic regime of the NL region is heavily influenced by the cold, southward‐flowing Labrador Current, which splits into inshore and offshore branches that skirt the coastline and continental shelf, respectively (Templeman, 2010). The MAR region encompasses the Scotian Shelf spanning the Atlantic coast of Nova Scotia, the Bay of Fundy, a section of the Laurentian Channel on the northeastern boundary, and portions of Georges Bank and the Gulf of Maine within Canadian waters. The Scotian Shelf is characterized by complex bathymetry including valleys, ridges, shallow banks, and deep basins. Cool waters carried by the southwest flowing Nova Scotia and Labrador currents have a larger influence on the Eastern Scotian Shelf, whereas occasional incursions of warmer slope water affect the Western Scotian Shelf (Maclean et al., 2013).
FIGURE 1.
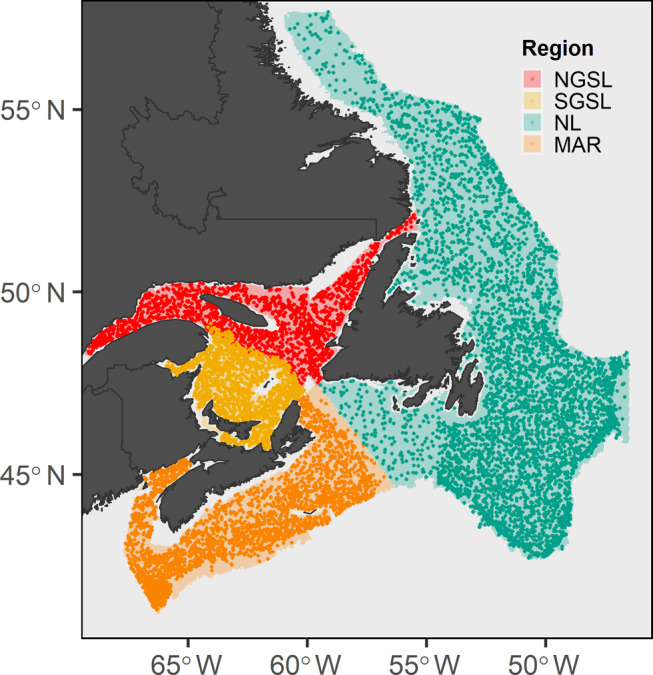
Study domain of four regions of the Northwest Atlantic: Northern Gulf of St. Lawrence (NGSL), Southern Gulf of St. Lawrence (SGSL), Newfoundland and Labrador (NL), and Maritimes (MAR). Shaded area indicates limits of regional multispecies bottom trawl surveys. Points are locations of individual trawl sets from 2007 to 2017. Surveys in the Northern and Southern Gulf of St. Lawrence partially overlap on the southern boundary of the Laurentian Trough
We delineated study area boundaries as the limits of the annual multispecies bottom trawl surveys conducted by DFO in each region. These boundaries cover depth ranges of 17–523 m (mean = 224 m, median = 224 m) for NGSL, 9–386 m (mean = 76 m, median = 59 m) for SGSL, 30–1855 m (mean = 296 m, median = 199 m) for NL, and 26–2104 m (mean = 277 m, median = 140 m) for MAR. Survey strata in the NL and MAR regions do not include the extent of Canada's Exclusive Economic Zone comprising the deepest continental slope, rise, and abyssal plain habitats. Rough bottom topography prevents sampling along part of the North Shore of the NGSL region and all surveys avoid areas with known dense aggregations of cold‐water corals (Chadwick et al., 2007). Within each study area, biological data were aggregated to cells (i.e., “sites”) in a regular grid (4‐km resolution). Therefore, observations for clustering and distribution analysis were derived as the aggregate observations of all trawls within that grid cell over the span of available data. Environmental raster layers were either aggregated or resampled (bilinear method) from their native resolutions to match this grid size. We selected the 4‐km cell size based on the average distance to nearest neighboring survey set locations within each regional data set (3–5 km) and to reflect the spatial accuracy of survey methods determined by standard tow distances (Appendix S1: Table S1). To eliminate potential bias introduced by grid cells with unequal sampling area, we also applied a 5‐km buffer around land points to exclude partial grid cells.
Biological data sources
To identify and map the distribution of the major biological assemblages in each of the four regions, we used geo‐referenced occurrence data for demersal fish and benthic invertebrate taxa collected during multispecies bottom trawl research surveys conducted annually by DFO in each region (Figure 1). We acquired positional and catch composition data from DFO regional databases and archives between the years 2007 and 2017 inclusive (only up to 2013 for NL and 2016 for MAR). Over this period, the scope of surveys expanded to improve the taxonomic resolution of non‐commercial fish species and invertebrates and survey design, gear, and protocols remained relatively consistent. Surveys in all regions follow a stratified random design with effort in each depth stratum proportional to its area (Chadwick et al., 2007). However, there are substantive regional differences with respect to survey timing, fishing vessel and gear, and fishing and sampling protocols (Appendix S1: Table S1; Chadwick et al., 2007). Therefore, we opted to treat regional data sets separately. Surveys are conducted seasonally in summer or fall in all regions, and NL and MAR regions also undertake a separate spring survey (Appendix S1: Table S1). For these regions, we elected to pool data across seasonal surveys (spring = 30%–40% of observations). In NL, the spring survey covers only the southern half of the shelf. However, ordination (nMDS) of points where the two surveys overlap indicated little separation in community structure (Appendix S1: Figure S1). From these regional data sets, we excluded invalid sets due to damaged gear, improper catch handling, or tow durations outside an acceptable range (Appendix S1: Table S1).
Surveys record the number and biomass of all fish and invertebrate taxa captured. However, data on invertebrate taxa in the NL region were available only for a handful of commercially harvested species. Taxa were identified to the highest reliable or practicable taxonomic resolution. Certain taxa, particularly invertebrates, are therefore treated in regional databases and our analyses as groups of species at the genus level or higher (e.g., Sebastes spp., Buccinidae, Pennatulacea; Appendix S1: Table S2 has complete taxa list). Many higher‐level taxa comprise a known number of species and have frequent records in the data set (Nozères et al., 2015). However, we removed any higher taxa that could result in the same species being coded to two separate taxa by different observers and therefore are too general to be of use (e.g., Annelida, Mollusca, Bivalvia). In some instances, a restricted subset of species that are easily identified are recorded separately from their parent taxon in the survey database, and represent separate taxonomic entities (e.g., Aphrodita hastata separate from all other polychaetes). Because trawl surveys are designed to specifically target benthic species, we filtered regional data sets for taxa with any of the following habitat associations listed on FishBase (Froese & Pauly, 2019), SeaLifeBase (Palomares & Pauly, 2019), or WorMS (WoRMS, 2019): benthic, benthos, sessile, reef‐associated, demersal, benthopelagic, bathydemersal, or bathypelagic.
To reduce potential biases related to differences in catchability among taxa and habitats and to increase the contribution of less common taxa, we converted abundance data to presence–absence. We counted a taxon as present if it was recorded in any trawl set located in that particular 4‐km grid cell over the period from 2007 to 2017. However, following Rubidge et al. (2016) we chose a conservative exclusion threshold and removed species that were reported in less than 1% of sampling sites in an attempt to balance the contribution of these rarer taxa with the potential distortion they may introduce to similarity‐based analyses (sensu Gauch, 1982, Legendre & Legendre, 2012). The final regional data sets consisted of 1580 sites and 162 taxa for NGSL, 1326 sites and 122 taxa for SGSL, 4908 sites and 70 taxa for NL, and 2527 sites and 113 taxa for MAR that met all inclusion criteria for catch records.
Characterizing ecological assemblages
Hierarchical clustering
To group sites in each regional data set into broad assemblage types based on their compositional similarity, we calculated pairwise distances between sites as Simpson dissimilarity (β sim) using the R package simba v. 0.3‐5 (Jurasinski & Retzer, 2012). As a measure of beta diversity, β sim captures spatial compositional turnover independent of species richness, is suitable for presence–absence data, and performs superiorly to other common metrics (Koleff et al., 2003). Dissimilarity between sites is scaled between 0 (no compositional difference) and 1 (no shared taxa) and is defined as
where a is the number of taxa shared between paired sites and b and c are the number of taxa unique to each respective site.
From the resulting dissimilarity matrices we grouped sites with similar taxonomic compositions using hierarchical agglomerative clustering with average linkage method (i.e., unweighted pair‐group method using arithmetic averages, UPGMA). Clustering was performed with the R function hclust (R version 3.6.3; R Core Team, 2020). We also evaluated our choice of dissimilarity metric and linkage method by comparing the cophenetic correlation coefficient from the resulting dendrogram to the coefficients from dendrograms produced using other dissimilarity metrics (Jaccard, Soerensen, Gower) and clustering methods (centroid, McQuitty's, median, Ward, single, complete). The cophenetic correlation coefficient indicates how well information in the original dissimilarity matrix is preserved by the dendrogram produced by clustering (Sokal & Rohlf, 1962). In all four regions, Simpson dissimilarity and UPGMA consistently outperformed other distance metrics and clustering algorithms (Appendix S1: Table S3).
To choose the similarity threshold at which to cut dendrograms, and therefore the number of clusters, in a more rigorous way, we inspected plots of several internal cluster validity indices against cluster number (k = 2–20; Appendix S1: Figure S2). These indices included the C index (Hubert & Schultz, 1976), the Calinski‐Harabasz index (Calinski & Harabasz, 1974), the Point‐Biserial index (Milligan, 1981), the Davies‐Bouldin index (Davies & Bouldin, 1979), and the Silhouette index (Roussseeuw, 1987). These indices measure the degree of within‐cluster dispersion relative to between‐cluster dispersion, and the optimal number of clusters is indicated by the maximum value of the index (minimum for Davies‐Bouldin and C indices). When multiple local optima occurred, we favored clustering solutions with more clusters, given our aim was to identify the full range of assemblage types in each region. In cases of disagreement among indices, we selected the consensus cluster number. Based on these criteria, we cut dendrograms at β sim = 0.494 (k = 7) for NGSL, β sim = 0.494 (k = 10) for SGSL, β sim = 0.512 (k = 8) for NL, and β sim = 0.587 (k = 9) for the MAR region.
The taxonomic resolution of taxa recorded at each site as well as the sampling effort (number of trawl sets) have the potential to affect the degree of similarity between sites and therefore bias clustering solutions. The majority of taxa in each regional data set were identified to at least the genus level (NGSL = 81%, SGSL = 77%, NL = 100%, MAR = 100%) and for grid cells having trawl data, few (12%–21%) contained more than one trawl set from across multiple years (mode = 1, 95th percentile = 2, max = 5–8). Furthermore, the grid cells containing multiple trawl sets were not biased toward certain clusters. Their frequency in a given cluster was proportional to cluster area (Appendix S1: Figure S10). Nonetheless, to evaluate these potential sources of bias, we re‐ran clustering analyses with a second data set in each region, filtering out taxa classified to taxonomic levels higher than genus and, for grid cells with multiple trawl sets across years, using the catch data from the most recent trawl set. In all four regions, we identified the same number of clusters (assemblage types) with the reduced data set as the original full data set and the frequency distribution of assemblage types was similar between data sets. Cramer's V indicated low (0.1 < ɸ c < 0.3) or little association (ɸ c < 0.1) between data set type (full or reduced) and cluster membership (NGSL = 0.069, SGSL = 0.020, NL = 0.088, MAR = 0.125). We present the results for the full data set to retain as much biological information as possible and because the removal of taxa higher than the genus level disproportionately removes invertebrates, which are already underrepresented.
Indicator analysis
To identify taxa emblematic of the predominant assemblage types in each region we conducted indicator analyses as developed by Dufrêne and Legendre (1997). Within each region, we calculated the indicator value of each taxon for each of the major dendrogram clusters (defined here as clusters comprising 20 or more sites) using the indval function in the labdsv R package (Roberts, 2016). The indicator value is an index scaled between 0 and 1 describing the degree to which a taxon characterizes a group of sites, the significance of which is evaluated by a randomization procedure. The value of the index increases as a taxon occurs in one particular cluster (i.e., assemblage type) with greater specificity (i.e., is rare at sites of other clusters) and fidelity (i.e., occurs frequently in sites of that cluster). We considered a taxon to be a strong indicator of an assemblage if its indicator value was maximal among all clusters, was >0.25, and significant at α = 0.05 (Dufrêne & Legendre, 1997).
Modeling distribution of assemblages based on environmental correlates
To identify environmental correlates with the distribution of the predominant assemblage types in each region, we derived raster data layers for 57 oceanographic and physiographic variables. We selected variables anticipated to form key environmental gradients driving compositional turnover on a regional scale and that were widely available in all four regions (Appendix S1: Table S4). Sources for data layers included various outputs from the BNAM ocean circulation model (Wang et al., 2018), remote‐sensing products derived from NASA satellite data and processed by the Remote Sensing Unit at the Bedford Institute of Oceanography (Fisheries and Oceans Canada, 2021a, 2021b), and mapping products obtained from spatially interpolated in situ measures including Bio‐ORACLE layers (Tyberghein et al., 2012) and GEBCO grids (GEBCO Compilation Group, 2014). Native spatial resolution varied between input sources (Appendix S1: Table S4), but we aggregated or resampled inputs to match the 4‐km resolution of the biological data. Contingent on availability, for time‐varying covariates, we constrained input data to the years 2007 to 2017 to align with biological survey data (Appendix S1: Table S4). If input sources were resolved at a monthly or more frequent time interval, we calculated multiple annual and seasonal summary variables (mean, minimum, maximum, range) that were averaged across available years (Appendix S1: Table S4). After applying a 5‐km land buffer to data layers, there were 7540 cells for NGSL, 4424 for SGSL, 39,218 for NL, and 13,326 for MAR populated with all environmental variables.
To model the relationship between the spatial distribution of assemblages and environmental predictors, we implemented random forest classifications using cluster membership as the response. We limited classifications to the major clusters in each region as minor clusters represented too few observations to be reliably modeled (generally 1 or 2 sites each; 5–10 sites in total) and were not geographically coherent enough to provide meaningful targets for conservation planners (Appendix S1: Figure S12). Major clusters were identified in dendrograms as those with ≥20 sites. Random forest is an ensemble machine learning algorithm that combines the predictions from a “forest” of decision trees to classify observations based on the majority vote among trees (Breiman, 2001). For each region, we fit a random forest model based on 10,000 trees in R using the default settings of the randomForest package (Liaw & Wiener, 2002). We evaluated model accuracy with both the out‐of‐bag (OOB) error rate (percent misclassified), and a 10‐fold cross‐validated estimate of the area under the receiver operating characteristic curve (AUC), which is an indicator of how well on average the classifier differentiates between pairwise classes (Hand & Till, 2001). We randomly partitioned data sets into 10 subsets, using each in turn to validate predictions of a model calibrated with the remaining 90% of observations by calculating the multi‐class AUC with the multiclass.roc function in R package pROC (Robin et al., 2011). We averaged AUC values across cross‐validation runs.
To improve model interpretability, we identified and removed highly correlated predictor variables prior to model fitting using an iterative variable elimination procedure similar to Knudby et al. (2013). Within each regional study area, the pair of variables with the highest absolute correlation coefficient (Pearson's r) among all cells in environmental raster layers was considered and one eliminated from the data set. We applied the following criteria when deciding which of two correlated variables should be retained. For pairs that described different summaries of the same variable: (1) annual summaries were preferred over seasonal ones with the exception of surface chlorophyll and primary production (spring summary preferred) and (2) those describing environmental extremes and, therefore, potential tolerance limits, were preferred (minimum and maximum > range > mean). For pairs describing different variables: (1) variables more closely associated with the seabed were preferred over sea surface variables (e.g., bottom temperature > sea surface temperature, depth‐integrated primary production > surface chlorophyll), (2) temporally dynamic variables were preferred over static ones to facilitate predictions under future climate scenarios (e.g., bottom salinity and temperature > depth), and (3) variables with a higher native spatial resolution and based on more contemporary data were preferred. This process continued iteratively until a subset of the original 57 variables remained for which all pairwise correlations were ≤0.7, yielding a final subset of 11–22 environmental predictors for each regional data set (Appendix S1: Table S5). Over 99% of sites with available biological data in each regional data set were assigned to a major cluster, and of these, only 23–41 (mainly nearshore) lacked associated data for the complete set of environmental predictors.
To delineate the potential distribution of each of the predominant assemblage types, we used the regional random forest classifiers to determine the predicted cluster membership for grid cells that had associated environmental data but lacked biological data from trawl surveys. We also highlighted areas of greater model uncertainty by identifying grid cells for which the probability of cluster assignment underlying the prediction was <0.70. The resulting regional biologically informed classification schemes indicated both the assemblage type and distinguishing abiotic habitat features likely to be encountered at a given location. To identify the key environmental variables associated with spatial variation in assemblage structure, we calculated the mean decrease in model accuracy resulting from randomly permuting the values of each predictor variable in the random forest classifier among observations (i.e., relative variable importance). We examined variable importance plots to assess the top predictors distinguishing individual clusters (i.e., assemblage types) and for the overall model accuracy. To further characterize the environmental conditions distinguishing assemblage types, we examined how the distribution of input grid cell values for the top predictors in each model varied among clusters.
Climate change susceptibility
We evaluated the susceptibility of modeled assemblages in the southernmost bioregion (MAR) to distributional changes associated with climatic drivers in two ways: (1) evaluating the predictive performance of the random forest classifier when hindcasting past changes, and (2) forecasting distributional changes expected under a long‐term climate projection with a regional forecasted oceanographic model developed using a high greenhouse gas emission scenario (representative concentration pathway, RCP, 8.5).
To assess the ability of our model to predict compositional changes associated with recent warming events in the MAR region, we compared hindcast predictions of the random forest classifier to observed assemblage structure for two temporal divisions of our initial biological survey data set: 2007–2011 and 2012–2016. The period 2012–2016 corresponds with an inter‐annual trend of increasing frequency and persistence of bottom water warm anomalies on the Scotian Shelf driven by incursions of warm/saline water from the continental slope (Brickman et al., 2018; Hebert et al., 2018). We limited our comparison of observed and predicted assemblage type to grid cells in the study area containing survey sets in both time periods (n = 238). We determined whether a change in the observed assemblage type occurred between time periods in cells with repeated measures by restricting the taxa presence/absence data in those cells to each period in turn prior to reclassification with hierarchical clustering as above. We then hindcast cluster membership (i.e., predicted assemblage type) from bottom temperature and salinity conditions for each time period in turn as inputs to the random forest classifier. To assess the predictive performance of model hindcasts we calculated model‐wide error rate (percent misclassified) and AUC using all hindcast predictions and sensitivity, specificity, and precision for specific classes from the confusion matrix for both time periods combined. These performance measures provide an indication of how well the model can resolve compositional changes associated with shifting climatic conditions.
To forecast how distributions of the predominant assemblage types may respond to long‐term environmental changes under a high‐emissions scenario, we compared our contemporary regional spatial classification to spatial predictions of our random forest classifier with bottom temperatures and salinities projected to 2075 under RCP scenario 8.5. We obtained projected temperature and salinity conditions for the year 2075 from the Bedford Institute of Oceanography North Atlantic Model (BNAM), a high‐resolution numerical ocean circulation model (Brickman et al., 2016). Grid cells in which the predicted assemblage type for 2075 differed from present were identified as susceptible to climate change. Finally, we compared the relative change in predicted area (from present) occupied by the major assemblage types to identify which would be most or least susceptible to change. An overall decrease (increase) in area would indicate a loss (gain) of suitable thermal habitat for a given assemblage type. This approach of modeling assemblage types based on changes to suitable thermal habitat does not account for species interactions (e.g., predation, competition, facilitation) or other factors (e.g., adaptation) that might constrain patterns of redistribution (Pinsky et al., 2020) or allow for individualistic species responses or no‐analog assemblages (Ferrier & Guisan, 2006; Nieto‐Lugilde et al., 2018). However, while the direction and rate of projected range shifts may vary regionally among taxa (Morley et al., 2018) or even between populations (Stanley et al., 2018), historical observations indicate that distributional responses to climate variability are reasonably unified among marine species within an assemblage and that, similar to single species, long‐term changes in assemblage distributions closely follow shifts in preferred thermal habitat (Kleisner et al., 2016).
RESULTS
Characterizing ecological assemblages
Hierarchical agglomerative clustering of biological survey data indicated three to six major site groupings representing the predominant assemblage types in each region (Figure 2). Of sites with available survey data, over 99% were assigned to one of these major clusters associated with discrete geomorphic and oceanographic features (Appendix S1: Figure S3). These assemblages were characterized by 3–34 indicator taxa (median = 7) that most distinguished a given assemblage from others within the same region (Appendix S1: Table S6). Of the 112 taxa we identified as strong indicators, 48 were shared across multiple regions (Appendix S1: Figure S4). The number of indicator taxa characterizing a particular assemblage was directly proportional to the taxa richness in that assemblage (Appendix S1: Figure S11).
FIGURE 2.
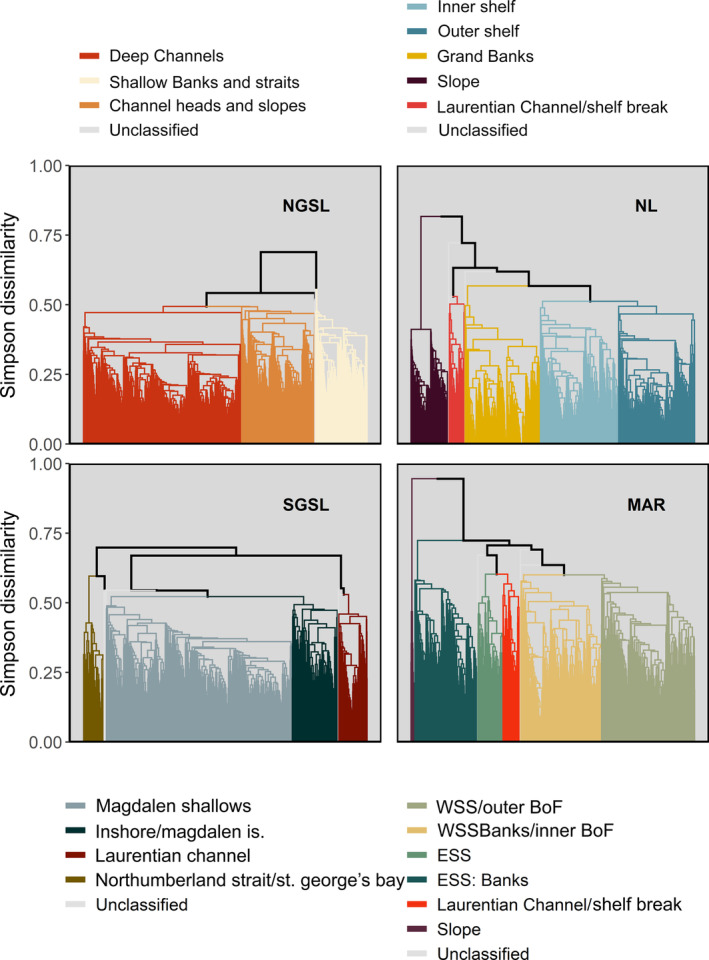
Dendrograms indicating similarity (calculated from Simpson dissimilarity) in taxonomic composition of bottom‐associated fish and benthic invertebrates between sites from annual multispecies trawl surveys in each of four regions of the Northwest Atlantic: NGSL, SGSL, NL, MAR (see Figure 1 caption for regional abbreviations). Nodes on lower branches indicate greater similarity between sites within that cluster. Colored groupings identify major clusters of sites sharing similar assemblage types (BoF, Bay of Fundy; ESS, Eastern Scotian Shelf; WSS, Western Scotian Shelf)
In the NGSL region, the majority of sites grouped in a cluster associated with deep channels (Laurentian, Anticosti, and Esquiman; Figure 2, Appendix S1: Figure S3) represented by 19 indicator taxa adapted to depth and warmer waters (Appendix S1: Table S6). This assemblage was most similar to a second cluster formed by sites concentrated at the head and along the slopes of these deeper troughs (Figure 2, Appendix S1: Figure S3). This channel‐bordering assemblage contained the fewest indicator taxa (seven) with the lowest IndVal in this region (Appendix S1: Table S6). The remainder of sites, forming the most distinct cluster, were associated with narrow straits and shallow coastal areas along the north shore of Quebec and west coast of Newfoundland, represented by a group of 34 indicator taxa largely composed of polar and subpolar invertebrate species (Figure 2, Appendix S1: Figure S3, Table S6).
Sites in the narrow and relatively shallow Northumberland Strait formed the most distinct of four clusters in the SGSL, represented by seven indicator taxa associated with shallow, coastal habitats that were often rare, but unique (high IndVal, low frequency) to this assemblage (Figure 2, Appendix S1: Figure S3, Table S6). Similar to the NGSL region, sites in the SGSL region within the Laurentian Channel also formed a unique cluster (Figure 2, Appendix S1: Figure S3) with a large overlap in the indicator taxa (14 of 19 in the SGSL) between the channel assemblages in the two regions (Appendix S1: Table S6, Figure S4). The largest cluster comprised sites on the expansive Magdalen Shallows and in Chaleur Bay (Figure 2, Appendix S1: Figure S3), with 10 of 14 indicator taxa shared with the two shallower NGSL assemblages (Appendix S1: Table S6, Figure S4). The fourth cluster of sites around the Magdalen Islands and the nearshore of SGSL shared some similarity with the Magdalen Shallows (Figure 2, Appendix S1: Figure S3) and had the fewest indicator taxa (five), which were largely soft‐bottom associated (Appendix S1: Table S6).
In NL, sites were more evenly distributed among the five major clusters (Figure 2). Sites along the continental slope formed the most distinct cluster in this region, followed by a cluster of sites occurring at the mouth of the Laurentian Channel and at the shelf break on the southwest margin of the Grand Banks (Figure 2, Appendix S1: Figure S3). The latter group had many indicator taxa in common (6 of 8) with the analogous channel assemblages in the NGSL and SGSL (Appendix S1: Table S6, Figure S4). Bathypelagic, benthopelagic, and bathydemersal fish strongly characterized the Slope assemblage, which had 16 indicator taxa (Appendix S1: Table S6). Sites occurring on the southern Grand Banks formed a unique cluster from other sites on the Newfoundland‐Labrador Shelves (Figure 2, Appendix S1: Figure S3) with only three, albeit strongly associated indicator taxa (Appendix S1: Table S6). The remaining sites were split between the Inner and Outer continental shelf corresponding roughly with the inshore and offshore branches of the Labrador Current (Figure 2, Appendix S1: Figure S3). The Inner Shelf included more polar and subpolar species (seven indicators), while temperate to boreal species (four indicators) better characterized the Outer Shelf (Appendix S1: Table S6).
The MAR region had the most clusters (n = 6), and similar to NL, the initial node of the dendrogram separated sites on the continental slope from the others (Figure 2, Appendix S1: Figure S3). A similar set of depth‐adapted indicator taxa as the Slope assemblage in NL (7 of 11 taxa in common; Appendix S1: Figure S4) characterized this assemblage (Appendix S1: Table S6). Likewise, there was also a distinct cluster formed by sites occurring in the Laurentian Channel and along the shelf break (Figure 2, Appendix S1: Figure S3) with four indicator taxa, all shared by the comparable assemblages in the other three regions (Appendix S1: Table S6, Figure S4). As with the other regions, the indicator taxa for the Slope and Laurentian Channel/Shelf Break assemblages had a strong affinity (high IndVal) to their clusters (Appendix S1: Table S6). We also observed a clear division of sites between the Eastern (ESS) and Western (WSS) Scotian Shelf (Figure 2, Appendix S1: Figure S3). Within the east–west divisions, sites associated with shallow banks and the inner Bay of Fundy (BoF) also grouped separately from deeper parts of the shelf (Figure 2, Appendix S1: Figure S3). Demersal fish largely distinguished both WSS assemblages, while the emblematic taxa for the two ESS assemblages included more invertebrates and were similar to the more northerly NGSL, SGSL, and NL regions with some subpolar species (Appendix S1: Table S6, Figure S4).
Modeling distribution of assemblages based on environmental correlates
Our regionally restricted random forest classifications produced relatively spatially contiguous predictions for the distributions of the predominant assemblage types in all four regions (Figure 3). In all regions, models performed with high accuracy as indicated by the low OOB error rates (≤20.2%) and high multiclass AUC values (>0.95) across all classes (Table 1). However, certain assemblages in each region were more frequently misclassified (Table 1), including Channel Heads and Slopes in NGSL, the Inshore/Magdalen Islands and Northumberland Strait/St. George's Bay assemblages in SGSL, Inner Shelf in NL, and ESS in the MAR region. There also was greater uncertainty (lower model assignment probabilities) in predictions for more frequently misclassified assemblages (Figure 3). We also observed a pattern of greater uncertainty in model predictions for grid cells on the boundaries between assemblages (Figure 3), likely reflecting intermediate environmental conditions. Variable importance plots indicated which specific environmental variables best distinguish between the predominant assemblages in each region (Figure 4, Appendix S1: Figure S5) and facilitate a description of the characteristic environmental habitat features occupied by those assemblages (Appendix S1: Figures S6–S9).
FIGURE 3.
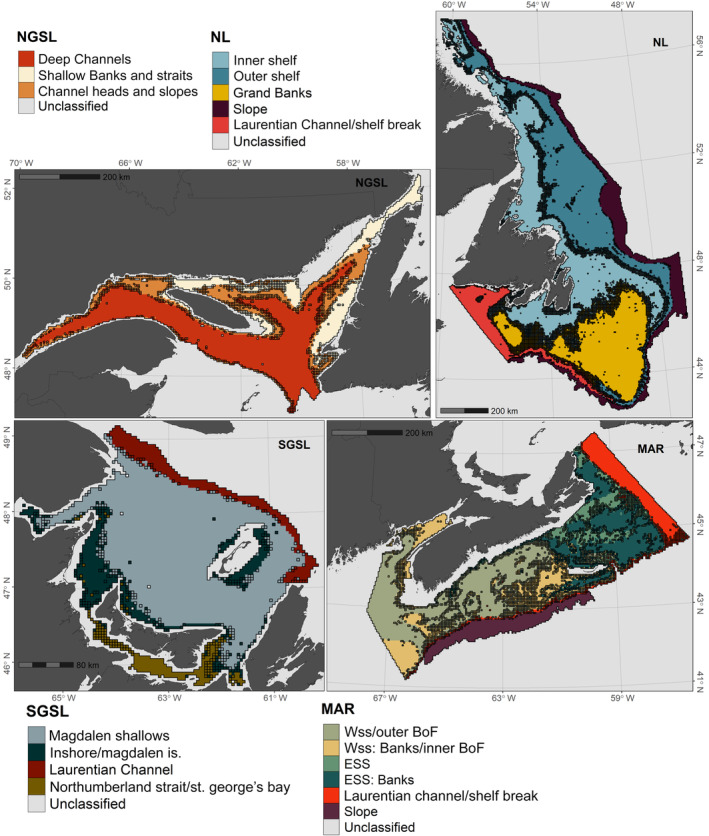
Predicted distribution of predominant assemblage types of bottom‐associated fish and benthic invertebrates in each of four regions of the Northwest Atlantic: NGSL, SGSL, NL, and MAR (see Figure 1 caption for regional abbreviations). Color denotes the expected assemblage type in a given 4‐km grid cell based on relationships with environmental correlates delineated by random forest classification (BoF, Bay of Fundy; ESS, Eastern Scotian Shelf; WSS, Western Scotian Shelf). Grid cells outlined in black indicate greater uncertainty in the model prediction (probability of assignment <0.70)
TABLE 1.
Measures of whole model and class‐specific accuracy for random forest classifications predicting cluster membership identified from biological survey data (i.e., assemblage type) based on environmental correlates
| Region and assemblage | OOB (%) | AUC |
|---|---|---|
| NGSL | ||
| All | 10.8 | 0.974 |
| Deep channels | 6.72 | |
| Shallow banks and straits | 7.43 | |
| Channel heads and slopes | 22.3 | |
| SGSL | ||
| All | 8.39 | 0.974 |
| Magdalen Shallows | 3.58 | |
| Inshore/Magdalen Is. | 23.6 | |
| Laurentian Channel | 9.09 | |
| Northumberland Strait/St. George's Bay | 19.3 | |
| NL | ||
| All | 15.2 | 0.977 |
| Inner shelf | 20.0 | |
| Outer shelf | 16.4 | |
| Grand Banks | 13.0 | |
| Slope | 7.37 | |
| Laurentian Channel/shelf break | 15.4 | |
| MAR | ||
| All | 20.2 | 0.962 |
| WSS: Outer BoF | 19.1 | |
| WSS: Banks/Inner BoF | 21.4 | |
| ESS | 30.9 | |
| ESS: Banks | 17.6 | |
| Laurentian Channel/shelf break | 22.9 | |
| Slope | 17.1 |
Note: Classifications are restricted within four regions of the Northwest Atlantic: NGSL, SGSL, NL, and MAR (see Figure 1 caption for regional abbreviations).
Abbreviations: AUC, multiclass area under the receiver operating characteristic curve (1 = perfect differentiation between classes); OOB, out‐of‐bag error rate (percent misclassified).
FIGURE 4.
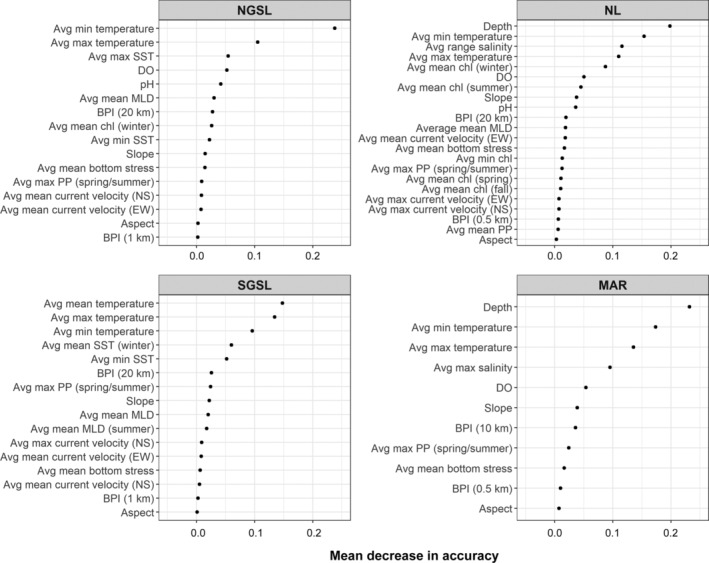
Variable importance plots indicating mean decrease in whole model accuracy resulting from randomly permuting values of environmental predictors among observations used in the four regional random forest classifiers: NGSL, SGSL, NL, and MAR (see Figure 1 caption for regional abbreviations). Variable summaries with prefix “Avg” indicate averages across years with available data. Abbreviations are SST, sea surface temperature; DO, dissolved oxygen; MLD, mixed layer depth; BPI, bathymetric position index; NS, north‐south; EW, east‐west; chl, chlorophyll; PP, primary production. Refer to Appendix S1: Table S4 for details and data sources for individual variables
In NGSL, average minimum bottom temperature was the most important environmental predictor, followed by average maximum bottom and sea surface temperature and dissolved oxygen concentration (Figure 4), but pH and a broadscale bathymetric position index (BPI, depth of a location relative to its neighbors within 20 km) also was important for distinguishing Shallow Banks and Straits (Appendix S1: Figure S5). Average minimum bottom temperature was warmer where the Deep Channel assemblage occurred relative to Shallow Banks and Straits and intermediate for Channel Heads and Slopes (Appendix S1: Figure S6). The pattern was similar, but less pronounced in average maximum bottom temperature (Appendix S1: Figure S6). Dissolved oxygen concentrations were lower but variable in Deep Channels compared to the other assemblages (Appendix S1: Figure S6). BPI values suggested, on average, the Shallow Straits and Banks assemblage occurred in elevated areas (positive BPI), the Deep Channels assemblage in depressions (negative BPI), and Channel Heads and Slopes assemblage in areas of constant slope (BPI near zero; Appendix S1: Figure S6).
As with NGSL, various metrics of bottom temperature (average mean, maximum, and minimum) were among the most important predictors in SGSL (Figure 4). Average maximum primary production in spring/summer also differentiated sites in the Northumberland Strait/St. George's Bay assemblage (Appendix S1: Figure S5). The Magdalen Shallows assemblage was characterized by low primary productivity and generally the coolest average maximum, mean, and minimum bottom temperatures, although minimum temperature was variable within this cluster (Appendix S1: Figure S7). In contrast, the Northumberland Strait/St. George's Bay assemblage was distinguished by high primary productivity, the warmest average maximum and mean bottom temperatures, and among the coolest minimum bottom temperatures (Appendix S1: Figure S7). The Inshore/Magdalen Islands assemblage occurred in areas of intermediate primary productivity, and likewise experiencing high seasonal variability in bottom temperature, though not as extreme as the Northumberland Strait (Appendix S1: Figure S7). Bottom temperatures experienced by the Laurentian Channel assemblage were less seasonally variable, and therefore, while average maximum bottom temperatures were among the lowest in this region, average minimum temperatures were warmest for this assemblage (Appendix S1: Figure S7).
Depth, average minimum and maximum bottom temperature, and average bottom salinity range were the four most important environmental predictors in the random forest classification for NL (Figure 4). Dissolved oxygen and pH also were important for distinguishing the Laurentian Channel/Shelf Break assemblage and average surface chlorophyll concentration in winter was an especially important variable for the Grand Banks assemblage (Appendix S1: Figure S5). The Grand Banks and Slope assemblages fell at opposite ends of the depth gradient, and the Inner Shelf group occurred at shallower depths on average than the Outer Shelf and Laurentian Channel/Shelf Break assemblages (Appendix S1: Figure S8). Average minimum and maximum bottom temperature was warmest for the Laurentian Channel/Shelf Break and Slope assemblages, followed by Outer Shelf, Grand Banks, and Inner Shelf (Appendix S1: Figure S8). The Slope assemblage was characterized by the smallest average range in bottom salinity; Grand Banks and Laurentian Channel/Shelf Break by the largest, albeit more variable for the latter (Appendix S1: Figure S8). This range in salinity separated conditions characterizing the Inner and Outer shelf clusters, where the former had a greater range (Appendix S1: Figure S8).
A similar suite of variables ranked as the most important environmental predictors in the MAR region (Figure 4). As expected, there was a depth gradient separating the Slope assemblage (deepest), the Laurentian Channel/Shelf Break, and assemblages occurring on the continental shelf, with a further distinction in depth for shallow banks assemblages on both the ESS and WSS (Appendix S1: Figure S9). The four assemblages on the continental shelf were further distinguished by average minimum bottom temperature (WSS > ESS; cooler for shallow banks and inner BoF), average maximum bottom temperature (WSS > ESS; warmer for shallow banks and inner BoF), and average maximum bottom salinity (WSS > ESS; less saline for shallow banks and inner BoF; Appendix S1: Figure S9). Maximum salinity was less variable and higher on average for the Slope and Laurentian Channel/Shelf Break assemblages (Appendix S1: Figure S9). Bottom temperature was seasonally consistent and less variable for the deeper assemblages, but generally warmer for the Laurentian Channel/Shelf Break compared to the Slope assemblage (Appendix S1: Figure S9). Slope and dissolved oxygen also were important variables distinguishing the two deeper assemblages (Appendix S1: Figure S5).
Climate change susceptibility
Of the 238 sites in the MAR region that were sampled both before (2007–2011) and during a recent period of warming (2012–2016) on the Scotian Shelf, 69 (29%) grouped with a different assemblage type based on taxonomic composition after the warming period began. However, changes were not proportional across the six predominant assemblage types (Figure 5). Consequently, there was a net decrease in the number of sites classified as ESS or ESS: Banks, a net increase in sites classified as Laurentian Channel/Shelf Break or WSS: Outer BoF and no net change in sites classified as Slope or WSS: Banks/Inner BoF (Figure 5). Furthermore, our regional random forest classifier was able to accurately hindcast these period‐specific cluster assignments based only on environmental conditions (error rate = 25.2%, AUC = 0.938 across both time periods). However, the classifier was better able to discern true negatives than true positives for all classes (sensitivity < specificity) and may overestimate the prevalence of the ESS, ESS: Banks, and Laurentian Channel/Banks assemblages (precision < sensitivity; Appendix S1: Table S7).
FIGURE 5.
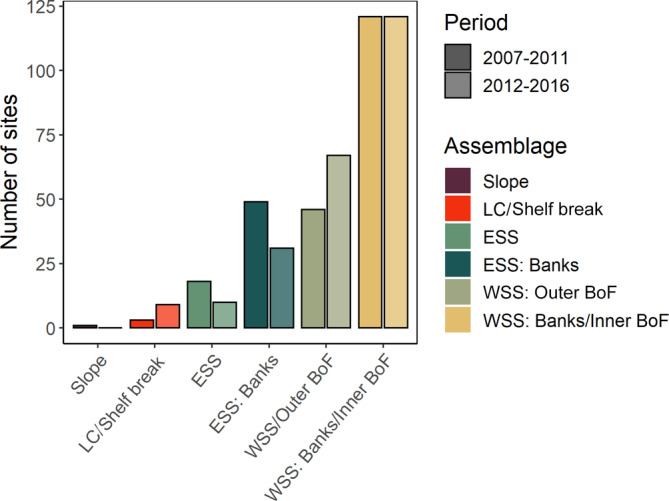
Comparison of number of sites in the Maritimes region grouping with each of the six predominant assemblage types (indicated by color hue) between two time periods (darker shading, 2007–2011; lighter shading, 2012–2016) for sites sampled for taxonomic composition in both time periods (n = 238)
Likewise, the model predicted distributional shifts of the predominant MAR assemblages with forecasted warming under RCP 8.5 in 2075 (Brickman et al., 2016). In general, the WSS/Outer BoF and WSS: Banks/Inner BoF assemblages are predicted to expand northward and east across the Scotian Shelf and to greater depths as suitable thermal habitat shifts (Figure 6b). Consequently, we projected a net increase in relative area occupied by these assemblages at the expense of decreases for the Laurentian Channel/Shelf Break, ESS, and ESS: Banks assemblages (Figure 6a). The Slope assemblage is predicted to be most resistant to change with no net change in area (Figure 6a).
FIGURE 6.
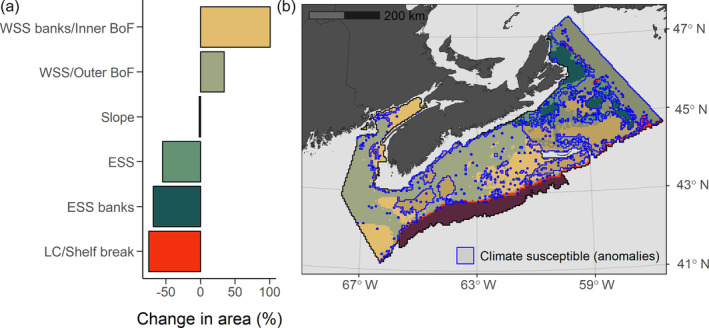
(a) Predicted relative change in area encompassed by spatial distributions of the six predominant assemblage types in Maritimes region in 2075 with bottom temperature and salinity conditions projected under RCP 8.5. Data are difference in area between 2075 and present expressed as percentage of present area. (b) Predicted 2075 assemblage distributions. Assemblage types are identified by color hue. Regions with darker shading and blue outline indicate areas susceptible to change under projected warming (i.e., predicted classification in 2075 differs from present)
DISCUSSION
Biological classifications inform ecologically coherent conservation strategies
We identified the predominant assemblages of demersal fish and benthic invertebrates in Atlantic Canada from biological survey data and developed ecologically comprehensive regional classification schemes using predictive mapping that can practically inform a number of management applications. Our classifications divide regions into mesoscale (tens to thousands of kilometers) subunits representing spatially coherent areas of similar species composition, emblematic taxa characterizing the assemblages, and the important abiotic habitat features. These subunits could be used to define management areas in an ecosystem‐based approach to fisheries management, inform the ecological objectives of marine spatial planning processes, or guide design, siting, and monitoring decisions in regional MPA network planning processes. Our results are largely consistent with previous community‐level modeling efforts in the NGSL using taxonomically restricted data sets that identified similar assemblage types associated with channels, slopes, and shallow coastal areas with indicator species and environmental correlates overlapping with our analysis (fish [Chouinard & Dutil, 2011], benthic invertebrates [Moritz et al., 2013]). However, our results differ from earlier classifications in the NL (Park & Mercier, 2014) and MAR regions that were based mainly on abiotic surrogates (DFO, 2016; Kostylev & Hannah, 2007; Roff et al., 2003). These discrepancies likely arise from differences in the identity and relative weighting of abiotic surrogates used in those classifications. Commonly available abiotic predictors do not contribute equally to explaining patterns of community composition (Pitcher et al., 2012). Our random forest classifiers indicated a restricted set of environmental axes distinguished between biotic assemblages (temperature, depth, salinity, DO), and these varied in relative importance. By integrating available biological information with abiotic predictors, our approach offers improved spatial insights for conserving the key ecological features of each region.
Our classifications provide benchmarks by which to evaluate ecological coherence in regional MPA network design and management. Studies have shown demersal fish and benthic macrofaunal assemblages identified by biologically informed classification approaches have greater homogeneity in composition, richness, and abundance within groups (Cooper et al., 2019), larger between‐group compositional differences, and indicator taxa with stronger associations (Rubidge et al., 2016) compared to abiotic habitat surrogates. Therefore, by predicting the distributions of the main assemblage types in each region at a relatively fine spatial resolution (4 km), our classifications can facilitate setting and achieving MPA network broadscale representativity targets (e.g., Government of Canada, 2011). We found multiple regions shared similar assemblages associated with the Laurentian Channel and continental slope habitats, offering additional opportunities to replicate features and implement connections among regional networks. The list of indicator taxa we identified for each assemblage type provides candidate species for evaluating regional connectivity patterns for network planning and management. Ecologically justified, quantitative conservation objectives demand a shift away from common rules‐of‐thumb approaches to connectivity (Magris et al., 2014), but demographic (e.g., individual‐based modeling, tagging studies) and genetic methods for empirically characterizing connectivity patterns require additional time, money, and resources. Indicator analyses, such as those proposed here, could help prioritize such investments. Specific indicator taxa from lengthy lists could be given priority based on conservation importance (e.g., high commercial value or cultural importance, species at risk, etc.).
Accounting for the fluid nature of ecological assemblages in space and time is required to achieve successful outcomes for conservation objectives but could be misrepresented by static maps. For example, community composition on the Scotian and Newfoundland shelves has changed over the past 40 years as a result of overexploitation and subsequent protection measures (Pedersen et al., 2017; Shackell et al., 2012). Moreover, our approach assumes a fixed relationship between species and does not account for environmental relationships in species interactions and thus non‐linear responses to climate change (e.g., thermally regulated predation rates), though our intra‐decadal comparison did show strong predictive performance. While our analysis did not account for these variables explicitly, it would be useful to repeat the same approach for earlier time periods to capture the representative range of historical and contemporary community states for conservation planning. Likewise, the boundaries between units of the classification should not be viewed as rigid features. Whereas our predictive models performed overall with high accuracy (low misclassification rate, high AUC values), the dynamic nature along boundaries was partially captured in our analysis by locations of greater model uncertainty, which were aggregated largely along margins between predicted assemblage types. These areas of higher uncertainty could represent transition zones with steep environmental gradients and rapid compositional turnover (Rubidge et al., 2016). Potentially enriched biodiversity and within‐species adaptive diversity found in transition zones increase the value of these areas as distinct conservation targets that could facilitate adaptation to changing climatic conditions (Araújo, 2002; Smith et al., 2009). The small and isolated minor clusters in our analyses were often located in these areas (Appendix S1: Figure S12), lending support to the notion of unique assemblage composition in transition zones. Further effort to characterize assemblages and environmental variability in these areas is warranted. Alternative modeling techniques that predict spatial variation in compositional dissimilarity such as Gradient Forest (Pitcher et al., 2012) and Generalized Dissimilarity Modeling (Ferrier et al., 2007) may be useful for visualizing and targeting areas of rapid compositional turnover along environmental gradients.
Temperature gradients shape ecological and evolutionary patterns relevant for climate adaptation
In our study, gradients in average minimum and maximum bottom temperature consistently emerged across regions as important variables delineating the distribution of benthic assemblages, which was reflected by the types of indicator taxa characterizing cold‐ and warm‐affinity assemblages. Metabolic constraints on oxygen supply set extremes of thermal tolerance for marine ectotherms (Pörtner & Knust, 2007), which occupy geographic ranges closer to their thermal limits than terrestrial species (Sunday et al., 2012). Temperature extremes shape the physiological, behavioral, evolutionary, and demographic responses of marine organisms and populations across wide‐ranging spatial and temporal scales (Pinsky et al., 2020). Consequently, spatial variation in biological properties across the spectrum of ecological organization are closely associated with temperature gradients in marine ecosystems including genetic structure (Stanley et al., 2018), abundance (Waldock et al., 2019), community composition (Pitcher et al., 2012, Rubidge et al., 2016, this study), diversity (Tittensor et al., 2010), and functional traits (Henriques et al., 2017). Stanley et al. (2018) documented a genetic break in population structure shared by multiple species in the Northwest Atlantic associated with a steep latitudinal gradient in seasonal temperature minima. We found a transition in community structure at a comparable location along the Atlantic coast of Nova Scotia (~44.5°–45° N) consistent with this temperature gradient, suggesting an important link between temperature stress and ecological and evolutionary processes in the region.
Our regional model predicted changes in the distribution of fish and invertebrate assemblages in the MAR region in response to recent observed and projected 21st century warming of bottom temperatures, but that changes in suitable thermal habitat varied among assemblages. Under a high‐emissions climate scenario (RCP 8.5), the two Western Scotian Shelf assemblages associated with warmer waters are projected to expand to the northeast at the expense of the deeper Laurentian Channel and cold‐affinity Eastern Scotian Shelf assemblages with little overall change for the Continental Slope assemblage. The direction and magnitude (hundreds of kilometers) of these changes are consistent with single species projections for the region (Morley et al., 2018). Similar to historical observations from bottom trawl surveys of long‐term, climate‐associated range shifts in species assemblages, our projections show a greater latitudinal response in shallower warm water assemblages compared to deeper assemblages (Kleisner et al., 2016) and an increased dominance of assemblages with affinities to warmer waters (Burrows et al., 2019). Further validation of our predictions could be achieved through comparisons to more complex community‐level models fitted to multispecies responses (reviewed by Nieto‐Lugilde et al., 2018). However, given the ability of our model to accurately hindcast compositional changes associated with warming in the last decade and that assemblage distributions are likely to track changes in extent of thermal habitats (Kleisner et al., 2016), our projections could inform climate‐smart conservation planning currently underway in Atlantic Canada. MPA networks should protect areas across a range of novel future climatic conditions (Tittensor et al., 2019). Our predictions identify potentially resilient areas that could serve as temporary thermal refugia (Keller et al., 2009; Tittensor et al., 2019) and areas more susceptible to change that may require more active monitoring and management to ameliorate the cumulative effects of other stressors (Keller et al., 2009; McLeod et al., 2009).
Community‐level modeling facilitates monitoring and adaptive management
Biologically informed classifications such as ours can promote more efficient and cost‐effective monitoring of MPA networks in other ways. Effective monitoring programs are key to adaptive management (WCPA/IUCN, 2007), but collecting additional ecological data can be prohibitively expensive. The lower within‐habitat variability associated with biologically informed classification schemes relative to abiotic surrogates translates to reduced sampling intensity required to detect changes in species richness and abundance (Cooper et al., 2019). In addition, failure to account for biogeographic variation in monitoring programs may reduce power to detect reserve effects for target and non‐target species (Hamilton et al., 2010). Evaluating network efficacy could be further simplified by targeting monitoring programs toward the indicator taxa we identified. A number of indicator taxa from our study were previously identified as more vulnerable to climate change (Stortini et al., 2015) and could therefore serve as sentinel species (e.g., mustache sculpin, snow crab). Indicators shared by multiple regions and that are already experiencing temperature‐mediated declines, like snow crab (Zisserson & Cook, 2017), should be further prioritized. We also identified the environmental surrogates most closely associated with biological patterns (e.g., temperature, depth), which can further prioritize sampling efforts to the most relevant variables especially if collection of physical data (e.g., through the Atlantic Zonal Monitoring Program; Therriault et al., 1998) is a more feasible option than a comprehensive biological survey.
Despite the extensive benefits of community‐level modeling approaches for marine conservation, practical application is limited not by an analytical framework or computational power, but by the availability of comprehensive biological data. Biological data from long‐term, spatially and taxonomically comprehensive regional surveys, such as the data sets from this study, are biased toward temperate latitudes, especially the north Atlantic Ocean (Poloczanska et al., 2016). In areas where data are sparse, ‘assemble‐then‐predict’ approaches such as ours may be more attainable compared to approaches requiring model fitting to separate species because they have greater power to detect shared patterns by pooling data across rare species (Ferrier & Guisan, 2006). Emerging techniques for quantifying marine biodiversity such as eDNA metabarcoding (Djurhuus et al., 2020) could help bridge the gap in data poor regions, provided that reference databases are sufficiently developed for confident species identifications.
Regions that have started conservation planning without or with little biological data need not necessarily revise existing networks. MPA networks designed using abiotic surrogates or incomplete biological data capture conservation features better than randomly placed reserves (Ban, 2009; Sutcliffe et al., 2015) and may provide an adequate first‐order approximation of biological patterns (Rubidge et al., 2016; Sutcliffe et al., 2015). We found that sites grouped solely based on compositional similarity were broadly associated with geomorphic and oceanographic features, suggesting that protecting such features captures representative species assemblages to some degree. However, updated classifications based on biological data can inform gap analyses to prioritize site selection for network expansion while minimizing conflicts with other stakeholders (Douglass et al., 2014; Geange et al., 2017). We suggest prioritizing assemblage types underrepresented in existing networks based on their distinctiveness from other assemblages. Distinctiveness can be inferred with the hierarchical clustering approach used here from the topological relationships between assemblage clusters in the dendrogram. Gap analyses that maximize conservation benefits while reducing user conflicts will become more critical as parties to the Convention on Biological Diversity recalibrate to more ambitious targets under a post‐2020 biodiversity agreement (Dinerstein et al., 2019; Visconti et al., 2019).
More ambitious conservation targets exacerbate the need to improve shared stakeholder buy‐in by demonstrating the efficacy and benefits of protection measures. In this study, we demonstrated how community‐level modeling can be consistently applied across regions to summarize ecologically relevant patterns of spatial variation in marine assemblages and better support the practices and principles that advance the ecological objectives of marine conservation planning (e.g., ecological coherence, climate adaptation, monitoring and adaptive management). However, a framework to align ecological objectives with positive socioeconomic outcomes that would better translate the societal benefits of ecologically coherent MPA networks has lagged behind (Rees et al., 2018). Predictive mapping that moves beyond species composition to characterize spatial variation in functional trait diversity, functional redundancy, and representative functional groups is an important extension of our work that will help bridge this connection. This can be accomplished with emerging community‐level modeling techniques that flexibly incorporate functional trait variation (Nieto‐Lugilde et al., 2018). Functional trait diversity may be tightly linked with ecosystem functions and processes that in turn support the flow of ecosystem goods and services that bolster human well‐being (Cadotte et al., 2011). Therefore, a predictive mapping application directed on functional traits would complement our results focused on taxonomic composition. Together these could support other formal processes to identify and communicate tangible societal consequences and benefits associated with levels of protection and representativity within MPA networks (Rees et al., 2018). Community‐level modeling combined with a framework that emphasizes social‐ecological coherence of MPA networks may help build the social capital needed to ensure comprehensive and lasting protection of marine ecosystems and the benefits they provide.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
We are grateful to Emily Rubidge for her invaluable input on the conceptual framework for this research and Katie Gale for software code. Lydia Stevens assisted with data collation and quality control. This work would not have been possible without data contributions from Carla Caverhill and the Bedford Institute of Oceanography Remote Sensing group, Donald Clark, and Fisheries and Oceans Canada in the Quebec, Gulf, and Newfoundland and Labrador Regions. David Brickman provided helpful comments on a previous draft. This work was supported by funding from Fisheries and Oceans Canada's Strategic Program for Ecosystem‐Based Research and Advice (SPERA).
O'Brien, John M. , Stanley Ryan R. E., Jeffery Nicholas W., Heaslip Susan G., DiBacco Claudio, and Wang Zeliang. 2022. “Modeling Demersal Fish and Benthic Invertebrate Assemblages in Support of Marine Conservation Planning.” Ecological Applications 32(3): e2546. 10.1002/eap.2546
Handling Editor: Julian D. Olden
Funding information Fisheries and Oceans Canada
DATA AVAILABILITY STATEMENT
Trawl survey data from Brodie et al. 2013; DFO 2018b, DFO 2019; DFO 2021. BIO North Atlantic Model data from Wang et al. (2018). Primary production data from Fisheries and Oceans Canada (2021a) and satellite‐derived chlorophyll data from Fisheries and Oceans Canada (2021b). Bathymetry data from GEBCO Compilation Group (2014). Bio‐ORACLE data from Tyberghein et al. (2012). R code (O'Brien & Stanley, 2021) to generate results and figures is archived on Zenodo at: https://doi.org/10.5281/zenodo.5576982.
REFERENCES
- Airamé, S. , Dugan J. E., Lafferty K. D., Leslie H., McArdle D. A., and Warner R. R.. 2003. “Applying Ecological Criteria to Marine Reserve Design: A Case Study from the California Channel Islands.” Ecological Applications 13: 170–84. [Google Scholar]
- Araújo, M. B. 2002. “Biodiversity Hotspots and Zones of Ecological Transition.” Conservation Biology 16: 1662–3. [Google Scholar]
- Ardron, J. A. 2008. “The Challenge of Assessing whether the OSPAR Network of Marine Protected Areas Is Ecologically Coherent.” Hydrobiologia 606: 45–53. [Google Scholar]
- Balbar, A. C. , Daigle R. M., Heaslip S. G., Jeffery N. W., Proudfoot B., Robb C. K., Rubidge E. & Stanley R.. 2020. “Approaches for Assessing and Monitoring Representation, Replication, and Connectivity in Marine Conservation Networks.” Canadian Science Advisory Secretariat Research Document 2020/050. vii + 57 p. Ottawa, ON: Fisheries and Oceans Canada.
- Balbar, A. C. , and Metaxas A.. 2019. “The Current Application of Ecological Connectivity in the Design of Marine Protected Areas.” Global Ecology and Conservation 17: e00569. [Google Scholar]
- Ban, N. C. 2009. “Minimum Data Requirements for Designing a Set of Marine Protected Areas, Using Commonly Available Abiotic and Biotic Datasets.” Biodiversity and Conservation 18: 1829–45. [Google Scholar]
- Breiman, L. 2001. “Random forests.” Machine Learning 45: 5–32. [Google Scholar]
- Brickman, D. , Hebert D., and Wang Z.. 2018. “Mechanism for the Recent Ocean Warming Events on the Scotian Shelf of Eastern Canada.” Continental Shelf Research 156: 11–22. [Google Scholar]
- Brickman, D. , Wang Z. & DeTracey B.. 2016. “High Resolution Future Climate Ocean Model Simulations for the Northwest Atlantic Shelf Region.” Canadian Technical Report of Hydrography and Ocean Sciences 315: xiv + 143 pp. Ottawa, ON: Fisheries and Oceans Canada (DFO).
- Brodie, B. , Mowbray F., and Power D.. 2013. DFO Newfoundland and Labrador Region Ecosystem Trawl Surveys. Version 1 in OBIS Canada Digital Collections. Dartmouth: Bedford Institute of Oceanography. Published by OBIS, Digital http://www.iobis.org/ [Google Scholar]
- Burrows, M. T. , Bates A. E., Costello M. J., Edwards M., Edgar G. J., Fox C. J., Halpern B. S., et al. 2019. “Ocean Community Warming Responses Explained by Thermal Affinities and Temperature Gradients.” Nature Climate Change 9: 1–5. [Google Scholar]
- Cadotte, M. W. , Carscadden K., and Mirotchnick N.. 2011. “Beyond Species: Functional Diversity and the Maintenance of Ecological Processes and Services.” Journal of Applied Ecology 48: 1079–87. [Google Scholar]
- Calinski, T. , and Harabasz J.. 1974. “A Dendrite Method for Cluster Analysis.” Communications in Statistics 3: 1–27. [Google Scholar]
- Chadwick, E. M. P. , Brodie W., Colbourne E., Clark D., Gascon D., and Hurlbut T.. 2007. “History of Annual Multi‐Species Trawl Surveyson the Atlantic Coast of Canada.” Atlantic Zonal Monitoring Program Bulletin 6: 25–42. [Google Scholar]
- Chouinard, P.‐M. , and Dutil J.‐D.. 2011. “The Structure of Demersal Fish Assemblages in a Cold, Highly Stratified Environment.” ICES Journal of Marine Science 68: 1896–908. [Google Scholar]
- Cooper, K. M. , Bolam S. G., Downie A. L., and Barry J.. 2019. “Biological‐Based Habitat Classification Approaches Promote Cost‐Efficient Monitoring: An Example Using Seabed Assemblages.” Journal of Applied Ecology 56: 1085–98. [Google Scholar]
- Davies, D. , and Bouldin D.. 1979. “A Cluster Separation Measure.” IEEE Transactions on Pattern Analysis and Machine Intelligence PAMI‐1: 224–7. [PubMed] [Google Scholar]
- DFO . 2005. “The Gulf of St. Lawrence: A Unique Ecosystem. The Stage for the Gulf of St. Lawrence Intergrated Management (GOSLIM).” Cat. No. F:1–30. Ottawa, ON: DFO.
- DFO . 2009. “Development of a Framework and Principles for the Biogeographic Classification of Canadian Marine Areas.” DFO Canadian Science Advisory Secretariat Advisory Report 2009/056. Ottawa, ON: DFO.
- DFO . 2016. “Evaluation of Hierarchical Marine Ecological Classification Systems for Pacific and Maritimes Regions.” DFO Canadian Science Advisory Secretariat Advisory Report 2016/003. Ottawa, ON: DFO.
- DFO . 2018a. “Design Strategies for a Network of Marine Protected Areas in the Scotian Shelf Bioregion.” DFO Canadian Science Advisory Secretariat Advisory Report 2018/006. Ottawa, ON: DFO.
- DFO . 2018b. DFO Quebec Region Multispecies bottom trawl surveys. Version 5 In OBIS Canada Digital Collections. Dartmouth: Bedford Institute of Oceanography. Published by OBIS, Digital http://www.iobis.org/ [Google Scholar]
- DFO . 2019. DFO Gulf Region Groundfish Research Vessel Surveys. Version 4 In OBIS Canada Digital Collections. Dartmouth: Bedford Institute of Oceanography. Published by OBIS, Digital http://www.iobis.org/ [Google Scholar]
- DFO . 2021. “Maritimes Research Vessel Surveys [Data sets]”. https://open.canada.ca/data/en/dataset/8ddcaeea-b806-4958-a79f-ba9ab645f53b.
- Dinerstein, E. , Vynne C., Sala E., Joshi A. R., Fernando S., Lovejoy T. E., Mayorga J., et al. 2019. “A Global Deal for Nature: Guiding Principles, Milestones, and Targets.” Science Advances 5: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djurhuus, A. , Closek C. J., Kelly R. P., Pitz K. J., Michisaki R. P., Starks H. A., Walz K. R., et al. 2020. “Environmental DNA Reveals Seasonal Shifts and Potential Interactions in a Marine Community.” Nature Communications 11: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass, L. L. , Turner J., Grantham H. S., Kaiser S., Constable A., Nicoll R., Raymond B., Post A., Brandt A., and Beaver D.. 2014. “A Hierarchical Classification of Benthic Biodiversity and Assessment of Protected Areas in the Southern Ocean.” PLoS One 9: e10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne, M. , and Legendre P.. 1997. “Species Assemblages and Indicator Species: The Need for a Flexible Asymmetrical Approach.” Ecological Monographs 67: 345–66. [Google Scholar]
- Ferrari, R. , Malcolm H., Neilson J., Lucieer V., Jordan A., Ingleton T., Figueira W., Johnstone N., and Hill N.. 2018. “Integrating Distribution Models and Habitat Classification Maps into Marine Protected Area Planning.” Estuarine, Coastal and Shelf Science 212: 40–50. [Google Scholar]
- Ferrier, S. , and Guisan A.. 2006. “Spatial Modelling of Biodiversity at the Community Level.” Journal of Applied Ecology 43: 393–404. [Google Scholar]
- Ferrier, S. , Manion G., Elith J., and Richardson K.. 2007. “Using Generalized Dissimilarity Modelling to Analyse and Predict Patterns of Beta Diversity in Regional Biodiversity Assessment.” Diversity and Distributions 13: 252–64. [Google Scholar]
- Fisheries and Oceans Canada 2021a. “Primary Production Maps of the North Atlantic [Dataset].” https://gcgeo.gc.ca/geonetwork/metadata/eng/08ea557c-5fc1-4f31-b0cd-863e614ef1a2
- Fisheries and Oceans Canada 2021b. “Satellite‐derived Chlorophyll Maps of the North Atlantic [Dataset].” https://gcgeo.gc.ca/geonetwork/metadata/eng/fadc644c-673344a3-9275-151df550c23c
- Foley, M. M. , Halpern B. S., Micheli F., Armsby M. H., Caldwell M. R., Crain C. M., Prahler E., et al. 2010. “Guiding Ecological Principles for Marine Spatial Planning.” Marine Policy 34: 955–66. [Google Scholar]
- Foster, N. L. , Rees S., Langmead O., Griffiths C., Oates J., and Attrill M. J.. 2017. “Assessing the Ecological Coherence of a Marine Protected Area Network in the Celtic Seas.” Ecosphere 8: e01688. [Google Scholar]
- Froese, R ., and Pauly D.. 2019. “FishBase. World Wide Web Electronic Publication.” www.fishbase.org
- Gaines, S. D. , White C., Carr M. H., and Palumbi S. R.. 2010. “Designing Marine Reserve Networks for both Conservation and Fisheries Management.” Proceedings of the National Academy of Sciences USA 107: 18286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauch, H. G. 1982. Multivariate Analysis in Community Ecology. Cambridge: Cambridge University Press. [Google Scholar]
- Geange, S. W. , Leathwick J., Linwood M., Curtis H., Duffy C., Funnell G., and Cooper S.. 2017. “Integrating Conservation and Economic Objectives in MPA Network Planning: A Case Study from New Zealand.” Biological Conservation 210: 136–44. [Google Scholar]
- GEBCO Compilation Group . 2014. “The GEBCO_2014 Grid, Version 201503318.” www.gebco.net
- Government of Canada . 2011. National Framework for Canada's Network of Marine Protected Areas. Ottawa, ON: Fish Ocean Canada. [Google Scholar]
- Greenan, B. J. W. , Shackell N. L., Ferguson K., Greyson P., Cogswell A., Brickman D., Wang Z., Cook A., Brennan C. E., and Saba V. S.. 2019. “Climate Change Vulnerability of American Lobster Fishing Communities in Atlantic Canada.” Frontiers in Marine Science 6: 1–18. [Google Scholar]
- Hamilton, S. L. , Caselle J. E., Malone D. P., and Carr M. H.. 2010. “Incorporating Biogeography into Evaluations of the Channel Islands Marine Reserve Network.” Proceedings of the National Academy of Sciences USA 107: 18272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand, D. J. , and Till R. J.. 2001. “A Simple Generalisation of the Area under the ROC Curve for Multiple Class Classification Problems.” Machine Learning 45: 171–86. [Google Scholar]
- Hebert, D. , Pettipas R., Brickman D., and Dever M.. 2018. “Meteorological, Sea Ice and Physical Oceanographic Conditions on the Scotian Shelf and in the Gulf of Maine during 2016.” DFO Canadian Science Advisory Secretariat Science Advisory Report 2018/016.
- Henriques, S. , Guilhaumon F., Villéger S., Amoroso S., França S., Pasquaud S., Cabral H. N., and Vasconcelos R. P.. 2017. “Biogeographical Region and Environmental Conditions Drive Functional Traits of Estuarine Fish Assemblages Worldwide.” Fish and Fisheries 18: 752–71. [Google Scholar]
- Hobday, A. J. , and Pecl G. T.. 2014. “Identification of Global Marine Hotspots: Sentinels for Change and Vanguards for Adaptation Action.” Reviews in Fish Biology and Fisheries 24: 415–25. [Google Scholar]
- Hubert, L. , and Schultz J.. 1976. “Quadratic Assignment as a General Data‐Analysis Strategy.” British Journal of Mathematical and Statistical Psychology 29: 190–241. [Google Scholar]
- Jurasinski, G. , and Retzer V.. 2012. “Simba: A Collection of Functions for Similarity Analysis of Vegetation Data.” R Package Version 0.3‐5. https://CRAN.R-project.org/package=simba.
- Keller, B. D. , Gleason D. F., McLeod E., Woodley C. M., Airamé S., Causey B. D., Friedlander A. M., et al. 2009. “Climate Change, Coral Reef Ecosystems, and Management Options for Marine Protected Areas.” Environmental Management 44: 1069–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleisner, K. M. , Fogarty M. J., McGee S., Barnett A., Fratantoni P., Greene J., Hare J. A., et al. 2016. “The Effects of Sub‐Regional Climate Velocity on the Distribution and Spatial Extent of Marine Species Assemblages.” PLoS One 11: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudby, A. , Kenchington E., and Murillo F. J.. 2013. “Modeling the Distribution of Geodia Sponges and Sponge Grounds in the Northwest Atlantic.” PLoS One 8: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koen‐Alonso, M. , Pepin P., Fogarty M. J., Kenny A., and Kenchington E.. 2019. “The Northwest Atlantic Fisheries Organization Roadmap for the Development and Implementation of an Ecosystem Approach to Fisheries: Structure, State of Development, and Challenges.” Marine Policy 100: 342–52. [Google Scholar]
- Koleff, P. , Gaston K. J., and Lennon J. J.. 2003. “Measuring Beta Diversity for Presence‐Absence Data.” Journal of Animal Ecology 72: 367–82. [Google Scholar]
- Kostylev, V. E. , and Hannah C. G.. 2007. “Process‐Driven Characterization and Mapping of Seabed Habitats.” In Mapping the Seafloor for Habitat Characterization: Geological Association of Canada, Special Paper 47, edited by Todd B. and Greene H., 171–84. St. John's, NL: Geological Association of Canada. [Google Scholar]
- Leathwick, J. , Francis M., and Kathryn J.. 2006. “Development of a Demersal Fish Community Classification for New Zealand's Exclusive Economic Zone.” In NIWA Client Report: Ham2006‐062. 21. Wellington: Department of Conservation. [Google Scholar]
- Leathwick, J. R. , Rowden A., Nodder S., Gorman R., Bardsley S., Pinkerton M., Baird S. J., Hadfield M., Currie K., and Goh A.. 2012. “A Benthic‐Optimised Marine Environment Classification (BOMEC) for New Zealand Waters.” In New Zealand Aquatic Environment and Biodiversity Report No. 88. 54. Wellington: Ministry of Fisheries. [Google Scholar]
- Legendre, P. , and Legendre L.. 2012. Numerical Ecology. Developments in Environmental Modelling, 3rd ed., Vol 24. Amsterdam: Elsevier. [Google Scholar]
- Liaw, A. , and Wiener M.. 2002. “Classification and Regression by Random Forest.” R News 2: 18–22. [Google Scholar]
- Maclean, M ., Breeze H., Walmsley J., and Corkum J.. 2013. “State of the Scotian Shelf Report.” Canadian Technical Report of Fisheries and Aquatic Sciences 3074.
- Magris, R. A. , Pressey R. L., Weeks R., and Ban N. C.. 2014. “Integrating Connectivity and Climate Change into Marine Conservation Planning.” Biological Conservation 170: 207–21. [Google Scholar]
- McLeod, E. , Salm R., Green A., and Almany J.. 2009. “Designing Marine Protected Area Networks to Address the Impacts of Climate Change.” Frontiers in Ecology and the Environment 7: 362–70. [Google Scholar]
- Milligan, G. 1981. “A Monte Carlo Study of Thirty Internal Criterion Measures for Cluster Analysis.” Psychometrika 46: 187–99. [Google Scholar]
- Moritz, C. , Lévesque M., Gravel D., Vaz S., Archambault D., and Archambault P.. 2013. “Modelling Spatial Distribution of Epibenthic Communities in the Gulf of St. Lawrence (Canada).” Journal of Sea Research 78: 75–84. [Google Scholar]
- Morley, J. W. , Selden R. L., Latour R. J., Frölicher T. L., Seagraves R. J., and Pinsky M. L.. 2018. “Projecting Shifts in Thermal Habitat for 686 Species on the North American Continental Shelf.” PLoS One 13: 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo, F. J. , Kenchington E., Tompkins G., Beazley L., Baker E., Knudby A., and Walkusz W.. 2018. “Sponge Assemblages and Predicted Archetypes in the Eastern Canadian Arctic.” Marine Ecology Progress Series 597: 115–35. [Google Scholar]
- Nieto‐Lugilde, D. , Maguire K. C., Blois J. L., Williams J. W., and Fitzpatrick M. C.. 2018. “Multiresponse Algorithms for Community‐Level Modelling: Review of Theory, Applications, and Comparison to Species Distribution Models.” Methods in Ecology and Evolution 9: 834–48. [Google Scholar]
- Nozères, C. , Bourassa M., Gendron M., Plourde S., Savenkoff C., Bourdages H., Benoît H., and Bolduc F.. 2015. “Using Annual Ecosystemic Multispecies Surveys to Assess Biodiversity in the Gulf of St. Lawrence.” Canadian Technical Report of Fisheries and Aquatic Sciences 3149.
- O'Brien, J. M. , and Stanley R. R. E.. 2021. “NW‐Atl‐Bioclassification: Identifying and Delineating Assemblages of Fish and Benthic Invertebrate in the Northwest Atlantic (v1.0.0).” Zenodo. 10.5281/zenodo.5576982. [DOI]
- Palomares, M. , and Pauly D.. 2019. SeaLifeBase. World Wide Web Electronic Publication. www.sealifebase.org
- Park, L. , and Mercier F.. 2014. “Incorporating Representativity into Marine Protected Area Network Design in the Newfoundland‐Labrador Shelves Bioregion.” Ecosystem Management Publication Series Newfoundland Labrador Reg 0010.
- Pedersen, E. J. , Link H., Thompson P. L., Taranu Z. E., Gonzalez A., Ball R. A., Moritz C., et al. 2017. “Signatures of the Collapse and Incipient Recovery of an Overexploited Marine Ecosystem.” Royal Society Open Science 4: 170215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky, M. L. , Selden R. L., and Kitchel Z. J.. 2020. “Climate‐Driven Shifts in Marine Species Ranges: Scaling from Organisms to Communities.” Annual Review of Marine Science 12: 153–79. [DOI] [PubMed] [Google Scholar]
- Pitcher, R. C. , Lawton P., Ellis N., Smith S. J., Incze L. S., Wei C. L., Greenlaw M. E., Wolff N. H., Sameoto J. A., and Snelgrove P. V. R.. 2012. “Exploring the Role of Environmental Variables in Shaping Patterns of Seabed Biodiversity Composition in Regional‐Scale Ecosystems.” Journal of Applied Ecology 49: 670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poloczanska, E. S. , Burrows M. T., Brown C. J., Molinos J. G., Halpern B. S., Hoegh‐Guldberg O., Kappel C. V., et al. 2016. “Responses of Marine Organisms to Climate Change across Oceans.” Frontiers in Marine Science 3: 1–21. [Google Scholar]
- Pörtner, H. O. , and Knust R.. 2007. “Climate Change Affects Marine Fishes through the Oxygen Limitation of Thermal Tolerance.” Science 315: 95–7. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2020. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. https://www.R-project.org/ [Google Scholar]
- Rees, S. E. , Pittman S. J., Foster N., Langmead O., Griffiths C., Fletcher S., Johnson D. E., and Attrill M.. 2018. “Bridging the Divide: Social–Ecological Coherence in Marine Protected Area Network Design.” Aquatic Conservation: Marine and Freshwater Ecosystems 28: 754–63. [Google Scholar]
- Rice, J. , and Houston K.. 2011. “Representativity and Networks of Marine Protected Areas.” Aquatic Conservation: Marine and Freshwater Ecosystems 21: 649–57. [Google Scholar]
- Roberts, C. M. , Branch G., Bustamante R. H., Castilla J. C., Dugan J., Halpern B. S., Lafferty K. D., et al. 2003. “Application of Ecological Criteria in Selecting Marine Reserves and Developing Reserve Networks.” Ecological Applications 13: 215–28. [Google Scholar]
- Roberts, D. W. 2016. “Labdsv: Ordination and Multivariate Analysis for Ecology. R Package Version 1.8‐0.” https://CRAN.R-project.org/package=labdsv
- Robin, X. , Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.‐C., and Müller M.. 2011. “PROC: An Open‐Source Package for R and S+ to Analyze and Compare ROC Curves.” BMC Bioinformatics 12: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff, J. C. , Taylor M. E., and Laughren J.. 2003. “Geophysical Approaches to the Classification, Delineation and Monitoring of Marine Habitats and Their Communities.” Aquatic Conservation: Marine and Freshwater Ecosystems 13: 77–90. [Google Scholar]
- Roussseeuw, P. 1987. “Silhouettes: A Graphical Aid to the Interpretation and Validation of Cluster Analysis.” Journal of Computational and Applied Mathematics 20: 53–65. [Google Scholar]
- Rubidge, E. M. , Gale K. S. P., and Curtis J. M. R.. 2016. “Community Ecological Modelling as an Alternative to Physiographic Classifications for Marine Conservation Planning.” Biodiversity and Conservation 25: 1899–920. [Google Scholar]
- Shackell, N. L. , Fisher J. A. D., Frank K. T., and Lawton P.. 2012. “Spatial Scale of Similarity as an Indicator of Metacommunity Stability in Exploited Marine Systems.” Ecological Applications 22: 336–48. [DOI] [PubMed] [Google Scholar]
- Smith, J , Patterson M., Alidina H. M. & Ardron J.. 2009. “Criteria and Tools for Designing Ecologically Sound Marine Protected Area Networks in Canada's Marine Regions.” Toronto, ON: WWF‐Canada.
- Sokal, R. R. , and Rohlf F. J.. 1962. “The Comparison of Dendrograms by Objective Methods.” Taxon 11: 33–40. [Google Scholar]
- Spalding, M. D. , Fox H. E., Allen G. R., Davidson N., Ferdaña Z. A., Finlayson M., Halpern B. S., et al. 2007. “Marine Ecoregions of the World: A Bioregionalization of Coastal and Shelf Areas.” Bioscience 57: 573–83. [Google Scholar]
- Stanley, R. R. , DiBacco C., Lowen B., Beiko R. G., Jeffery N. W., Van Wyngaarden M., Bentzen P., et al. 2018. “A Climate‐Associated Multispecies Cryptic Cline in the Northwest Atlantic.” Science Advances 4: eaaq0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stortini, C. H. , Shackell N. L., Tyedmers P., and Beazley K.. 2015. “Assessing Marine Species Vulnerability to Projected Warming in the Scotian Shelf, Canada.” ICES Journal of Marine Science 72: 1731–43. [Google Scholar]
- Sunday, J. M. , Bates A. E., and Dulvy N. K.. 2012. “Thermal Tolerance and the Global Redistribution of Animals.” Nature Climate Change 2: 686–90. [Google Scholar]
- Sutcliffe, P. R. , Klein C. J., Pitcher C. R., and Possingham H. P.. 2015. “The Effectiveness of Marine Reserve Systems Constructed Using Different Surrogates of Biodiversity.” Conservation Biology 29: 657–67. [DOI] [PubMed] [Google Scholar]
- Templeman, N. D. 2010. “Ecosystem Status and Trends Report for the Newfoundland and Labrador Shelf.” DFO Canadian Science Advisory Secretariat Research Document 2010/26.
- Therriault J.‐C., Petrie B. D., Pepin P., Gagnon J., Gregory D., Helbig J., Herman A., et al. 1998. “Proposal for a Northwest Atlantic Zonal Monitoring Program.” Canadian Technical Report on Hydrography and Ocean Science 194. [Google Scholar]
- Tittensor, D. P. , Beger M., Böerder K., Boyce D., Cavanagh R., Cosandey‐Godin A., Ortuño G. C., et al. 2019. “Integrating Climate Adaptation and Biodiversity Conservation in the Global Protected Ocean.” Science Advances 5: eaay9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittensor, D. P. , Mora C., Jetz W., Lotze H. K., Ricard D., vanden Berghe E., and Worm B.. 2010. “Global Patterns and Predictors of Marine Biodiversity across Taxa.” Nature 466: 1098–101. [DOI] [PubMed] [Google Scholar]
- Tyberghein, L. , Verbruggen H., Pauly K., Troupin C., Mineur F., and de Clerck O.. 2012. “Bio‐ORACLE: A global environmental dataset for marine species distribution modelling.” Global Ecology and Biogeography 21: 272–81. 10.1111/j.1466-8238.2011.00656.x [DOI] [Google Scholar]
- Visconti, B. P. , Butchart S. H. M., Brooks T. M., Langhammer P. F., Marnewick D., Vergara S., Yanosky A., and Watson J. E. M.. 2019. “Protected Area Targets Post‐2020.” Science 364: 239–41. [DOI] [PubMed] [Google Scholar]
- Waldock, C. , Stuart‐Smith R. D., Edgar G. J., Bird T. J., and Bates A. E.. 2019. “The Shape of Abundance Distributions across Temperature Gradients in Reef Fishes.” Ecology Letters 22: 685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Lu Y., Greenan B., Brickman D., and Detracey B.. 2018. “BNAM: An Eddy‐Resolving North Atlantic Ocean Model to Support Ocean Monitoring.” Canadian Technical Report of Hydrography and Ocean Sciences 327.
- WCPA/IUCN 2007. “Establishing Networks of Marine Protected Areas: A Guide for Developing National and Regional Capacity for Building MPA Networks.” WCPA IUCN Non‐Technical Summary Report .
- Wilkinson, T. , Wiken E., Creel J., Hourigan T. F., Agard T., Herrmann H., Janishevski L., Madden C., Morgan L., and Padilla M.. 2009. Marine Ecoregions of North America. Montreal: Commision for Environmental Cooperation. [Google Scholar]
- Wilson, K. L. , Tittensor D. P., Worm B., and Lotze H. K.. 2020. “Incorporating Climate Change Adaptation into Marine Protected Area Planning.” Global Change Biology 26: 3251–67. [DOI] [PubMed] [Google Scholar]
- WoRMS Editorial Board 2019. “ World Register of Marine Species .” http://www.marinespecies.org
- Zisserson, B. , and Cook A.. 2017. “Impact of Bottom Water Temperature Change on the Southernmost Snow Crab Fishery in the Atlantic Ocean.” Fisheries Research 195: 12–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Data Availability Statement
Trawl survey data from Brodie et al. 2013; DFO 2018b, DFO 2019; DFO 2021. BIO North Atlantic Model data from Wang et al. (2018). Primary production data from Fisheries and Oceans Canada (2021a) and satellite‐derived chlorophyll data from Fisheries and Oceans Canada (2021b). Bathymetry data from GEBCO Compilation Group (2014). Bio‐ORACLE data from Tyberghein et al. (2012). R code (O'Brien & Stanley, 2021) to generate results and figures is archived on Zenodo at: https://doi.org/10.5281/zenodo.5576982.


