Abstract
Cephamycin C production was blocked in wild-type cultures of the clavulanic acid-producing organism Streptomyces clavuligerus by targeted disruption of the gene (lat) encoding lysine ɛ-aminotransferase. Specific production of clavulanic acid increased in the lat mutants derived from the wild-type strain by 2- to 2.5-fold. Similar beneficial effects on clavulanic acid production were noted in previous studies when gene disruption was used to block the production of the non-clavulanic acid clavams produced by S. clavuligerus. Therefore, mutations in lat and in cvm1, a gene involved in clavam production, were introduced into a high-titer industrial strain of S. clavuligerus to create a double mutant with defects in production of both cephamycin C and clavams. Production of both cephamycin C and non-clavulanic acid clavams was eliminated in the double mutant, and clavulanic acid titers increased about 10% relative to those of the parental strain. This represents the first report of the successful use of genetic engineering to eliminate undesirable metabolic pathways in an industrial strain used for the production of an antibiotic important in human medicine.
Actinomycetes are rich sources of a variety of bioactive natural products, including antibacterial and antifungal antibiotics and immunosuppressive agents. To date, strain improvement of these organisms to obtain high-titer cultures more suitable for industrial fermentations has depended largely on random mutagenesis and selection techniques. However, recent advances in actinomycete molecular biology and knowledge of antibiotic biosynthetic pathways and the gene clusters that encode them offer genetic engineering as an alternative approach to the improvement of strains in a targeted manner. Engineered strains of Streptomyces spp. have been constructed in which gene dosage has been increased by the introduction of additional copies of genes encoding pathway enzymes (7), or gene expression has been increased by the introduction of positive-acting, pathway-specific regulatory genes on high-copy-number plasmids (11, 15). In each of these cases, genetic engineering has resulted in significant overproduction of the product, demonstrating the utility of this recombinant approach for strain improvement, at least when wild-type strains are used as model systems. However, applications of such methods to industrial high-titer strains to improve their production characteristics further have not yet been reported.
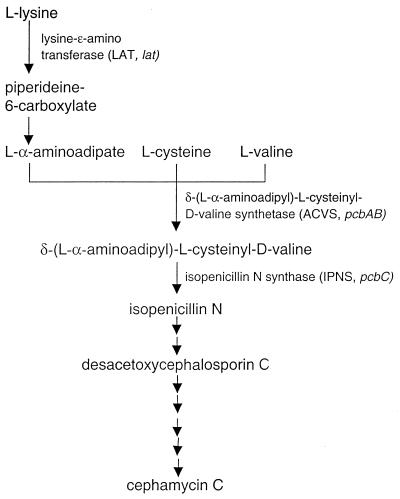
Streptomyces clavuligerus is an actinomycete well known for its ability to produce a variety of β-lactam antibiotics. Of these, isopenicillin N, desacetoxycephalosporin C, and cephamycin C are all derived from the cephamycin C pathway, which draws upon l-lysine, l-cysteine, and l-valine as precursors (Fig. 1). The gene cluster encoding the cephamycin C pathway enzymes has been well characterized (1, 5, 12). The same organism produces another group of structurally related β-lactam compounds, the clavams, in which oxygen replaces sulfur as the heteroatom in the fused bicyclic ring system. The best known of these compounds, clavulanic acid, possesses weak antibiotic activity but is a very potent β-lactamase inhibitor. Because of its β-lactamase inhibitory properties, clavulanic acid is used clinically in combination with conventional β-lactam antibiotics to treat infections caused by bacteria that would otherwise be resistant to these antibiotics. Augmentin, a mixture of amoxicillin and clavulanic acid, is the most widely prescribed of these combinations, with a market value in excess of one billion dollars yearly.
FIG. 1.
Cephamycin biosynthetic pathway. Abbreviated names of biosynthetic enzymes and their corresponding genes are given in parentheses.
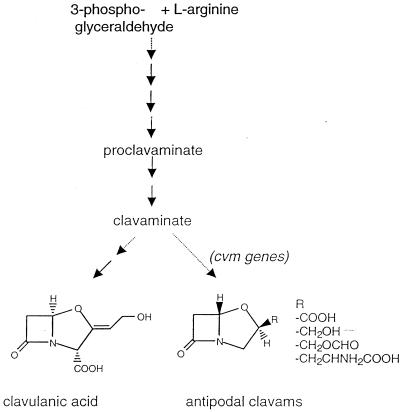
Clavulanic acid is biosynthesized by a pathway that begins by condensation of 3-phosphoglyceraldehyde with l-arginine and proceeds through a number of steps to form proclavaminic acid, then clavaminic acid, and, finally, clavulanic acid (Fig. 2). Although there are no biosynthetic enzymes shared by the cephamycin C and clavulanic acid pathways, the genes encoding clavulanic acid biosynthetic enzymes are located in a cluster adjacent to the cephamycin C gene cluster (17), and the two biosynthetic pathways are coregulated by a single transcription activation protein, CcaR (1, 11). In addition to clavulanic acid, this organism produces a number of other clavams that are structurally related to clavulanic acid, but have an inverted stereochemistry in their ring system. These antipodal clavams, some of which possess antifungal and antibacterial activities, have been proposed to share a common biosynthetic pathway with clavulanic acid up to the level of the late intermediate, clavaminic acid (3). Clavaminic acid, therefore, is a branch point and can be converted to either clavulanic acid or the antipodal clavams. We have recently reported the identification and characterization of several genes specifically involved in the production of the antipodal clavams (8).
FIG. 2.
Clavulanic acid-antipodal clavam biosynthetic pathway.
The industrial production of clavulanic acid is carried out by large-scale fermentation of S. clavuligerus. Strains capable of supporting a high level of clavulanic acid productivity have been derived from the wild-type S. clavuligerus through a conventional strain development program extending over many years. Biochemical analysis of these strains has revealed that they still have the genetic capability to produce both cephamycin C and antipodal clavams. However, these are undesirable products in an industrial clavulanic acid-producing strain. Therefore, it was of interest to determine if elimination of the pathway for the biosynthetically unrelated metabolite cephamycin C and the pathway for the biosynthetically related antipodal clavams would have beneficial effects on clavulanic acid productivity in these high-titer industrial strains. In this paper, we report the construction of mutants of wild-type S. clavuligerus blocked in the earliest step of the cephamycin C biosynthetic pathway. Furthermore, we have constructed and characterized double mutants derived from the high-titer strain that are blocked in the production of both cephamycin C and the antipodal clavams.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Streptomyces clavuligerus NRRL 3585 was obtained from the Northern Regional Research Laboratory. Streptomyces lividans TK24 was kindly provided by D. A. Hopwood (John Innes Institute, Norwich, United Kingdom). The indicator organism, Escherichia coli ESS, was a gift from A. L. Demain (Massachusetts Institute of Technology, Cambridge, Mass.). The E. coli vector pUC119 was obtained from J. Vieira (Waksman Institute of Microbiology, Rutgers University, Piscataway, N.J.), and the Streptomyces vector pIJ486 was a gift from D. A. Hopwood. E. coli strains were grown as described by Sambrook et al. (14), while Streptomyces strains were cultured as described previously (4). The S. clavuligerus wild type and related mutant strains were cultivated on Trypticase soy broth containing 1% starch (TSBS) medium for quantitation of total antibiotic production, starch asparagine (SA) medium for quantitation of clavulanic acid production, and soy medium for quantitation of both clavulanic acid production and antipodal clavam production (9).
Cultivation of high-titer strains.
The high-titer production strain S. clavuligerus SB1918 and the lat::apr-cvm::apr mutants derived from it were cultivated in 1-liter microfermentors on a proprietary soy-based fermentation medium for assessment of clavulanic acid production. S. clavuligerus SB1918 is a high-level producer of clavulanic acid that resulted from the conventional strain improvement program at Glaxo SmithKline.
Recombinant DNA techniques.
All recombinant DNA techniques involving E. coli were carried out according to the method of Sambrook et al. (14), while those involving Streptomyces spp. were carried out as described by Hopwood et al. (4).
Construction of a plasmid carrying the lat::apr-disrupted gene construct.
A 1.08-kb EcoRI-KpnI fragment carrying the 5′ end of lat and a 688-bp KpnI fragment carrying the 3′ end of lat were inserted upstream and downstream, respectively, of an apramycin resistance gene (apr) cassette that had previously been cloned into the multiple cloning site of pUC119 (9). The apr cassette was positioned in the opposite transcriptional orientation relative to lat. This generated the plasmid pCF002, which carried the entire lat gene disrupted by the presence of the apr cassette inserted at the KpnI site within the gene. The disrupted lat gene was then subcloned into pIJ486 as an EcoRI-HindIII fragment to create pLATAP.
Construction of a plasmid carrying the cvm1::apr-disrupted gene construct.
The cvm1 gene disruption plasmid was created by inserting the apr cassette into the plasmid pCEC026 at the unique BsaBI site located within cvm1 636 bp from the translational start codon (8). The disrupted cvm1 gene, carrying the apr cassette inserted in the same orientation relative to cvm1, was then subcloned into pIJ486 as a BamHI-HindIII fragment to create pCEC081.
Gene disruption technology.
Gene disruptions and replacements were carried out as described previously (9).
LAT assay.
Lysine ε-aminotransferase (LAT) activity was measured by monitoring the conversion of α-aminoadipate to 1-piperideine-6-carboxylate after derivitization with o-aminobenzaldehyde as described by Madduri et al. (6).
Quantification of production of β-lactam compounds.
Total β-lactam antibiotics were detected by a bioassay procedure with E. coli ESS as an indicator strain. Zones of inhibition were measured, and the amount of antibiotic in a sample was estimated by comparison with a standard curve constructed with cephalosporin C as a standard.
Clavulanic acid and the other antipodal clavams were quantified by high-performance liquid chromatography analysis after derivatization with imidazole as described by Paradkar et al. (10).
RESULTS
Disruption of lat in the wild-type strain.
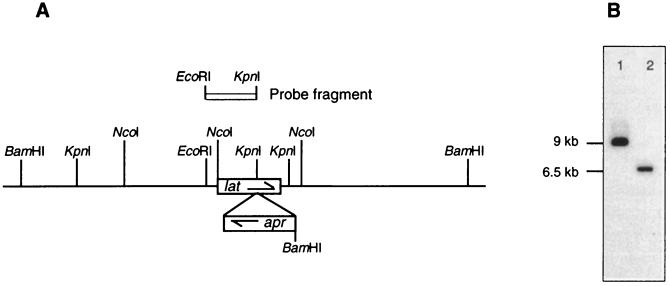
A gene replacement technique was used to introduce a mutation early in the cephamycin C pathway in order to block the formation of any products from this pathway. The gene encoding LAT, which converts l-lysine to α-aminoadipic acid, was chosen as the target (Fig. 1). The lat gene was sequenced and characterized previously (16), and a cloned copy of the gene was disrupted by insertion of an apr cassette encoding an apramycin resistance gene that is functional in both E. coli and Streptomyces spp. The DNA fragment carrying the disrupted lat gene was then transferred to the Streptomyces plasmid pIJ486 to give pLATAP. pIJ486 is segregationally unstable in S. clavuligerus in the absence of antibiotic selection and is therefore a suitable vector for the delivery of gene replacement constructs (9). The plasmid pLATAP was introduced into S. clavuligerus NRRL 3585 by protoplast transformation, and apramycin-resistant, thiostrepton-sensitive colonies were obtained from the transformants following growth and sporulation in the absence of antibiotic selection. The estimated frequency of recovery of apramycin-resistant, thiostrepton-sensitive colonies was 5 to 10%. Four such presumptive mutants were studied further, and gene replacement in these strains was confirmed by Southern analysis. The details of the gene disruption mutation are represented diagramatically in Fig. 3A, and Southern analysis of one of the mutants, lat#2, is shown in Fig. 3B. When genomic DNA isolated from both the wild type and the lat#2 mutant was digested with BamHI and separated by agarose gel electrophoresis, the lat-specific probe hybridized to a 9-kb fragment in the wild type and a 6.5-kb fragment in the lat#2 mutant. The smaller size of the hybridizing BamHI fragment in the mutant was attributable to the introduction of a BamHI site as part of the apr cassette used for the gene disruption procedure.
FIG. 3.
Disruption of the lat gene. (A) Diagram of the chromosome of a lat::apr gene disruption mutant in the region of the lat gene. Open boxes represent the lat and apr genes. Arrows indicate the direction of transcription. (B) Southern analysis of one of the lat::apr mutants (lat#2). Lanes 1 and 2, BamHI-digested genomic DNA from wild-type S. clavuligerus and the lat#2 mutant, respectively. The lat-specific probe fragment is shown in panel A. The sizes of hybridizing bands were determined by comparison with HindIII-digested lambda DNA (not shown).
Analysis of the lat::apr mutants.
As expected, all four of the lat::apr mutants failed to produce cephamycin C (Table 1). One representative lat::apr mutant, lat#2, was also examined biochemically to compare its LAT activity with that of the wild-type strain. No significant LAT activity could be detected in the mutant, whereas under the same conditions, the wild-type strain showed normal amounts of cephamycin C production and LAT activity. When the lat::apr mutants were characterized for clavulanic acid production, the specific production in the mutants was approximately 2 to 2.5 times higher than that seen in the wild-type strain. Growth of the mutants was comparable to that of the wild-type strain on both SA and TSBS media. The cephamycin C and clavulanic acid production data presented in Table 1 represent a survey of the four mutants and are taken from single cultures harvested at a single time point. However, clavulanic acid production in the lat#2 mutant was also reevaluated in comparison to that of the wild type by using triplicate cultures harvested at three time points, and the same general trend was observed. Specific clavulanic acid production was 3.8-fold higher in the mutant at 48 h, 1.7-fold higher at 72 h, and 2.0-fold higher at 99 h (data not shown).
TABLE 1.
Antibiotic, LAT, and clavulanic acid production characteristics of lat::apr mutants
| Strain | Total antibiotic concn (μg/ml/OD600)a | LAT activity (U/mg of protein)b | Clavulanic acid titer (μg/ml/OD600)c |
|---|---|---|---|
| Wild type | 1.12 | 1.9 | 4.5 |
| lat#2 | 0 | 0.1 | 11.7 |
| lat#5 | 0 | NDd | 11.5 |
| lat#7 | 0 | ND | 11.8 |
| lat#11 | 0 | ND | 11.7 |
Total antibiotic concentrations were determined with supernatants of single cultures grown in TSBS medium for 66 h at 28°C. OD600, optical density at 600 nm.
LAT activity was measured with cell extracts prepared from duplicate TSBS cultures grown for 48 h at 28°C.
Clavulanic acid titers were determined with supernatants of single cultures grown in SA medium for 66 h at 28°C.
ND, not determined.
In order to confirm that the loss of cephamycin C-producing ability was attributable to the mutation in lat, lat::apr mutants were fermented in medium supplemented with increasing amounts of α-aminoadipic acid. Cephamycin C production was restored to lat::apr mutants with the addition of α-aminoadipate. The degree of restoration of antibiotic production increased with increasing dosage of α-aminoadipate, but restoration of production never surpassed about 25 to 30% of the wild-type levels (Table 2). Subsequent studies have shown that the lat::apr mutation has a polar effect on the expression of the downstream pcbAB and pcbC genes such that α-aminoadipyl-cysteinyl-valine synthetase and isopenicillin N synthase enzyme levels are reduced to about 12 and 25% of wild-type levels, respectively, in the lat::apr mutant (data not shown). In a related study, Alexander et al. (2) recently created a lat disruption mutant designed to maintain transcription of downstream genes and showed that the mutant can be restored to wild-type levels of cephamycin C production by complementation with a lat gene under the control of the ermE* promoter.
TABLE 2.
Effect of α-aminoadipate supplementation on antibiotic production by a lat::apr mutant
| α-Aminoadipate concn (mM) | Total antibiotic concn (μg/ml/OD600)a
|
|
|---|---|---|
| Wild type | lat#2 | |
| 0 | 1.01 | 0.02 |
| 0.5 | 0.76 | 0.14 |
| 1.0 | 1.05 | 0.13 |
| 2.0 | 0.90 | 0.23 |
| 4.0 | 1.09 | 0.31 |
Total antibiotic concentrations were determined by averaging the values obtained for supernatants of duplicate cultures grown in TSBS medium supplemented with α-aminoadipate for 48 h at 28°C. OD600, optical density at 600 nm.
Disruption of cvm1 in the wild-type strain.
We recently reported that the genes cvm1, cvm4, and cvm5, located adjacent to cas1, the gene encoding the clavaminate synthase1 isoenzyme, are involved in the biosynthesis of the antipodal clavams. Mutants disrupted in cvm1 (cvm1::apr) and cvm4/5 were shown to produce elevated levels of clavulanic acid and to be specifically blocked in the production of the antipodal clavams (8).
Creation of a double mutant blocked in both lat and cvm1.
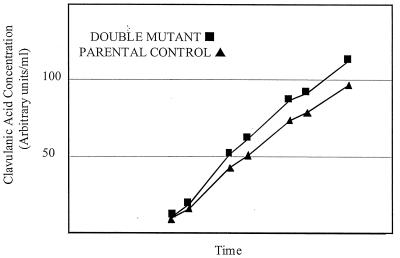
Because both the lat::apr and cvm1::apr mutants showed desirable changes in clavulanic acid production (increased), we created a double mutant in a high-titer strain by combining these two mutations. Using the same plasmid and protocols described above for the introduction of the lat::apr mutation into the wild-type strain of S. clavuligerus, the mutation was recreated in an S. clavuligerus clavulanic acid high-titer strain, SB1918. From these experiments, 10 mutant strains were isolated and characterized genetically to confirm that they contained a disrupted lat gene. One representative mutant, SB1989, was then used as the host strain for a second round of mutation to disrupt the cvm1 gene. This was accomplished by introducing a cvm1 gene disruption plasmid, pCEC081 (8), into SB1989. Transformants were selected with thiostrepton as the plasmid marker; apramycin could not be used for selection because the SB1989 strain already carried the apr gene as part of the lat disruption. After growth of the transformants in the absence of thiostrepton to promote loss of free plasmid, putative double mutants were selected based on their reduced production of clavams. Four double disruptant mutants were isolated, and their identities were confirmed by Southern analysis. The four double mutants were analyzed for clavulanic acid production, antipodal clavam production, and antibiotic production. As expected, the mutants produced no cephamycin or antipodal clavams, but in small-scale fermentor studies, clavulanic acid was produced at levels averaging about 10% higher than that of the parental SB1918 isolate (Fig. 4). The data shown here represent the average productivity of three independent fermentations for each strain, which were all carried out at the same time. Single assays were taken from each fermentor at each time point during the fermentations except for the final time point, at which we have four assay values for each fermentor, for a total of 12 assay values for each strain. The standard deviations for the final time point are 0.836% for the double mutant and 3.2% for the parental strain. The entire experiment was also repeated more than five times with similar results. To assess growth, viscosity measurements were taken throughout the fermentations, and no differences between the two strains were observed (data not shown). For operational reasons, cells within these 1-liter fermentors were grown for a set period of time and terminated when clavulanic acid production of SB 1989 was typically shown to level off in this system. However, when the double mutant strain was fermented for longer periods at pilot plant scale (1,000 liters), it still demonstrated a 10% productivity improvement over the parental strain (data not shown).
FIG. 4.
Clavulanic acid production by the SB1918 lat::apr-cvm1::apr double mutant compared to that of parental strain SB1918. Clavulanic acid accretion of both the parental strain (▴) and the double mutant strain (▪) when fermented in 1-liter fermentors is shown. Each time point represents an average clavulanic acid productivity from three parallel fermentations for each strain (single assays from each fermentor), except for the final time point, which represents the average of four determinations for each fermentor. Standard deviations for this final time point were 0.836% for the double mutant and 3.2% for the parental strain. Due to the commercially sensitive nature of the data, clavulanic acid productivity and fermentation times are represented in arbitrary units per milliliter.
A detailed fermentation analysis comparing the clavulanic acid productivity of the high-titer strain containing only the lat::apr mutation with that of the double mutant has not yet been performed. Therefore, from the results presented here, it is not possible to determine the contributions of each mutation to the overall improvement demonstrated for the high-titer strain. How these two mutations interact is clearly of interest and will be the subject of further experimentation.
DISCUSSION
Traditionally within the antibiotic industry, improvements in yield in antibiotic fermentations have been achieved by classical mutagenesis and high-throughput screening of the antibiotic-producing microorganisms. For many of the antibiotics commonly used for humans today, the genetics and biochemistry of their biosynthesis are known, so pharmaceutical companies have also been running programs to apply genetic engineering techniques to improve antibiotic yields based on this knowledge. However, reports of successful application of techniques that improve antibiotic productivity in wild-type strains to high-titer industrial strains are rare. (In fact, we cannot think of one example applied to an antibiotic in regular use in humans.) In this paper, we have demonstrated that by using gene replacement technology to eliminate competing and noncompeting biosynthetic pathways, it is possible to apply observations seen with a wild-type strain to a high-titer industrial strain.
In particular, using S. clavuligerus, we have demonstrated that elimination of cephamycin C biosynthesis by disruption of the lat gene produces a strain with increased clavulanic acid productivity. Since the biosynthetic pathways for clavulanic acid and cephamycin C are biochemically distinct, the effect must be indirect. Possibly the metabolic resources normally used to make lysine, cysteine, and valine for cephamycin C biosynthesis are spared and can now be directed towards production of arginine and 3-phosphoglyceraldehyde for clavulanic acid biosynthesis. Alternatively, it is known that both the clavulanic acid and cephamycin C biosynthetic pathways are controlled by a single regulatory gene, ccaR (1, 11). Elimination of metabolites arising from the cephamycin C pathway may have a regulatory influence upon expression of the clavulanic acid biosynthetic genes.
Studies from other groups have shown that mutations that impair cephamycin C production have variable effects on clavulanic acid production in S. clavuligerus. While some mutants not producing cephamycin C showed no change in clavulanic acid production compared to the wild type and others showed a loss of clavulanic acid production (presumed now to have mutations in the ccaR gene) (11), in at least one case, clavulanic acid production was improved (13). The basis for these changes in productivity is difficult to analyze, however, since the nature of the mutations was not determined. On the other hand, in a separate study in which cephamycin C productivity of strains was increased by addition of a second copy of lat, no information was provided as to the effect of this increased gene dosage on clavulanic acid production (1). Answers to these questions could shed light on the nature of the interactions between these two pathways.
In a previous study, we demonstrated the beneficial effects of a mutation in cvm1, a gene involved in the production of the antipodal clavams, on the production of clavulanic acid. The underlying reason for beneficial effects on clavulanic acid production resulting from blockage of clavam biosynthesis is more directly apparent than was the case for cephamycin biosynthesis. A block in the pathway between clavaminic acid and the antipodal clavams should alter the normal flux of the intermediates at this branch point, allowing more clavaminic acid to be channeled into clavulanic acid. Since secondary metabolites are typically produced as families of structurally related compounds, this approach of blocking competing pathways not only results in strains producing a less complex mixture of products, but may also improve the yield of the desired compound.
In the present study, using a well-defined system, we have demonstrated a positive effect of the lat and cvm1 mutations on clavulanic acid productivity in a wild-type strain and have shown that the mutations result in a similar phenotype in a high-titer industrial strain. Although the percentage increase in the high-titer strain is only about 10%, the level of clavulanic acid production is already orders of magnitude greater than that in the wild-type strain, so a 10% increase represents a significant gain. As far as we are aware, this is the first reported example of improvement in the productivity of a secondary metabolic pathway by the elimination of an apparently unrelated secondary metabolic pathway. Although further experimentation will be required to determine if this is a general effect that can be applied to strain improvement for other microorganisms, which make a number of structurally unrelated secondary metabolites, this approach opens up a number of new strategies that should be considered for improving the production of secondary metabolites. In particular with the advent of microbial genome sequencing, working from the sequence of an antibiotic-producing microorganism, it may be possible to tailor or “design” the strain for optimal productivity of the desired metabolite by eliminating all unrelated secondary metabolic pathways identified by sequence analysis.
In addition to the titer benefits already noted, elimination of cephamycin C and antipodal clavam production from industrial strains may also simplify the subsequent extraction and purification of clavulanic acid from the fermentation broth by eliminating contaminating metabolites with similar chromatographic properties. Furthermore, one of the antipodal clavams produced by S. clavuligerus, clavam-2-carboxylate, is known to have toxic properties. As a consequence, its levels are tightly regulated by the U.S. Pharmacopoeia guidelines for clavulanic acid, and it would be desirable to remove it as a contaminant in fermentation. From an industrial perspective, simplification of the downstream purification process can be as beneficial to the overall efficiency of the process as enhancement of titer.
ACKNOWLEDGMENTS
This work was supported by Glaxo SmithKline Pharmaceuticals and by a grant to S.E.J. from the National Sciences and Research Council of Canada.
REFERENCES
- 1.Alexander D C, Jensen S E. Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein. J Bacteriol. 1999;180:4068–4079. doi: 10.1128/jb.180.16.4068-4079.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander D C, Brumlik M J, Lee L, Jensen S E. Early cephamycin biosynthetic genes are expressed from a polycistronic transcript in Streptomyces clavuligerus. J Bacteriol. 2000;182:348–356. doi: 10.1128/jb.182.2.348-356.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egan L A, Busby R W, Iwata-Reuyl D, Townsend C A. Probable role of clavaminic acid as the terminal intermediate in the common pathway to clavulanic acid and the antipodal clavam metabolites. J Am Chem Soc. 1997;119:2348–2355. [Google Scholar]
- 4.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 5.Liras P. Biosynthesis and molecular genetics of cephamycins: cephamycins produced by actinomycetes. Antonie Leeuwenhoek. 1999;75:109–124. doi: 10.1023/a:1001804925843. [DOI] [PubMed] [Google Scholar]
- 6.Madduri K, Stuttard S, Vining L C. Lysine catabolism in Streptomyces spp. is primarily through cadaverine: β-lactam producers also make α-aminoadipate. J Bacteriol. 1989;171:299–302. doi: 10.1128/jb.171.1.299-302.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malmberg L-H, Hu W-S, Sherman D H. Precursor flux control through targeted chromosomal insertion of the lysine ε-aminotransferase (lat) gene in cephamycin C biosynthesis. J Bacteriol. 1993;175:6916–6924. doi: 10.1128/jb.175.21.6916-6924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosher R H, Paradkar A S, Anders C, Barton B, Jensen S E. Genes specific for the biosynthesis of clavam metabolites antipodal to clavulanic acid are clustered with the gene for clavaminate synthase 1 in Streptomyces clavuligerus. Antimicrob Agents Chemother. 1999;43:1215–1224. doi: 10.1128/aac.43.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paradkar A S, Jensen S E. Functional analysis of the gene encoding the clavaminate synthase 2 isoenzyme involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. J Bacteriol. 1995;177:1307–1314. doi: 10.1128/jb.177.5.1307-1314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paradkar A S, Aidoo K A, Jensen S E. A pathway-specific transcriptional activator regulates late steps of clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol Microbiol. 1998;27:831–843. doi: 10.1046/j.1365-2958.1998.00731.x. [DOI] [PubMed] [Google Scholar]
- 11.Pérez-Llarena F J, Liras P, Rodríguez-García A, Martín J F. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both β-lactam compounds. J Bacteriol. 1997;179:2053–2059. doi: 10.1128/jb.179.6.2053-2059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez-Llarena F J, Rodríguez-García A, Enguita F J, Martín J F, Liras P. The pcd gene encoding piperideine-6-carboxylate dehydrogenase involved in biosynthesis of α-aminoadipic acid is located in the cephamycin cluster of Streptomyces clavuligerus. J Bacteriol. 1998;180:4753–4756. doi: 10.1128/jb.180.17.4753-4756.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romero J, Liras P, Martin J F. Isolation and biochemical characterization of Streptomyces clavuligerus mutants in the biosynthesis of clavulanic acid and cephamycin C. Appl Microbiol Biotechnol. 1988;27:510–516. [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Stutzman-Engwall K J, Otten S L, Hutchinson C R. Regulation of secondary metabolism in Streptomyces spp. and overproduction of daunorubicin in Streptomyces peucetius. J Bacteriol. 1992;174:144–154. doi: 10.1128/jb.174.1.144-154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tobin B M, Kovacevic S, Madduri K, Hoskins J A, Skatrud P L, Vining L C, Stuttard C, Miller J R. Localization of the lysine ɛ-aminotransferase (lat) and δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase (pcbAB) genes from Streptomyces clavuligerus and production of lysine ɛ-aminotransferase activity in Escherichia coli. J Bacteriol. 1991;173:6223–6229. doi: 10.1128/jb.173.19.6223-6229.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward J M, Hodgson J E. The biosynthetic genes for clavulanic acid and cephamycin production occur as a ‘super-cluster’ in three Streptomyces. FEMS Microbiol Lett. 1993;110:239–242. doi: 10.1111/j.1574-6968.1993.tb06326.x. [DOI] [PubMed] [Google Scholar]