Abstract
Gut microbiota, a collection of microorganisms that live within gastrointestinal tract, provides crucial signaling metabolites for the physiological of hosts. In healthy state, gut microbiota metabolites are helpful for maintaining the basic functions of hosts, whereas disturbed production of these metabolites can lead to numerous diseases such as metabolic diseases, cardiovascular diseases, gastrointestinal diseases, neurodegenerative diseases, and cancer. Although there are many reviews about the specific mechanisms of gut microbiota metabolites on specific diseases, there is no comprehensive summarization of the functions of these metabolites. In this Opinion, we discuss the knowledge of gut microbiota metabolites including the types of gut microbiota metabolites and their ways acting on targets. In addition, we summarize their physiological and pathologic functions in health and diseases, such as shaping the composition of gut microbiota and acting as nutrition. This paper can be helpful for understanding the roles of gut microbiota metabolites and thus provide guidance for developing suitable therapeutic strategies to combat microbial-driven diseases and improve health.
Keywords: gut microbiota metabolites, intestinal barrier, energy metabolism, immune response, circadian rhythm, short-chain fatty acids
The gut microbiota is a community of microorganisms that dwell in a mutualistic relationship with hosts in gastrointestinal tract. Although the importance of gut microbiota has been recognized for a long time, it is only in recent decades that our understanding of gut microbiota began to surge because of the progresses in genomics, metabolomics, and other technologies such as culturomics [1]. Integrative analysis of gut microbiota of human subjects, antibiotic-treated, germ-free, or gnotobiotic animals have revealed that the gut microbiota plays a causal, or at least a correlative relationship in the development of diseases such as non-alcoholic fatty liver disease, obesity, inflammatory bowel disease, Alzheimer’s disease, Parkinson’s disease, allergy, and depression [2,3]. In addition to the direct roles in development of diseases, the gut microbiota can interact with the drugs that are taken orally and impact their efficacy and toxicity [4]. Because of the importance of gut microbiota in disease development and modification of drug efficacy, the gut microbiota has become an indispensable target that needs to be considered in medical area.
In normal condition, the gut microbiota is enclosed within the gastrointestinal lumen by intestinal barrier, a structure composed of epithelial cells, mucus, commensal bacteria, immune cells, and antibodies [5]. To circumvent this spatial limitation, the gut microbiota evolves a strategy of releasing different classes of metabolites to exert the effects on hosts and other gut bacteria. In this event, comprehensive identification of gut microbiota metabolites and investigation of their roles have naturally become important for researchers. With the help of advanced metabolomic tools such as ultra-performance liquid chromatography-tandem mass spectrometry, a large group of metabolites have been identified, such as short-chain fatty acids (SCFAs), bile acids, and choline metabolites [6,7]. These metabolites can induce a series of physiological and pathological functions on hosts and other bacteria, such as modulation of energy metabolism, nutrition absorption, and regulation of gut microbiota composition [8]. These microbiota-host and bacteria-bacteria metabolic dialogs are essential for understanding the roles of gut microbiota in maintenance of health and promotion of diseases.
Currently, there are many reviews about the specific functions and mechanisms of gut microbiota metabolites on specific diseases, such as the mechanisms of SCFAs on inflammatory bowel diseases and their functions on host metabolism [9-11]. However, there is no comprehensive summarization of the functions of gut microbiota metabolites, which has limited our understanding of the gut microbiota and our combat against microbial-associated diseases. In this Opinion, focusing on the functions of gut microbiota metabolites, we first give brief introduction on the classification and production of gut microbiota metabolites. Then, we emphasize the functions of gut microbiota-derived metabolites, especially the functions of regulating energy metabolism, local and systemic immune system, and neural activity. At last, the perspectives on future research directions are discussed.
2. Gut microbiota metabolites
2.1. Classification of gut microbiota metabolites
It is estimated that the gut microbiota contains about 1015 microbial cells and more than 22 million microbial genes, both of which exceed the cells and genes of human [12]. With these genes, the gut microbiota can synthesize a myriad of enzymes with versatile capabilities to ferment a variety of compounds that have escaped from the digestion of human enzymes or compounds that are indigestible by human enzymes such as fibers [13]. As a result, the gut microbiota can produce a battery of metabolites with wide spectrum of bioactivities. According to the origination, the gut microbiota metabolites can be broadly divided into three types: (1) metabolites that are produced by gut microbiota directly from diets, such as SCFAs and indole derivatives; (2) metabolites that are generated by the host and modified by gut microbiota, such as secondary bile acids; (3) metabolites that are produced de novo, such as polysaccharide A [14]. Many of the gut microbiota metabolites share similar chemical structure, and they showed similar functions on hosts. According to the chemical structures and their functions, we have listed the major groups of metabolites, the typical metabolites and their targets inTable 1. It should be noted that because the chemical structure of gut microbiota metabolites is diverse and thus the classification here is not very strict. In addition, a specific gut microbiota metabolite can act on multiple organs or tissues, and thus possesses multiple functions. For example, butyrate can act as energy resource and nutrition for colonocyte, modulate the intestinal barrier and systemic immune response [20]. Therefore, the functions of gut microbiota metabolites are not isolated, and should be treated systemically. Among all the gut microbiota metabolites, the most extensively studied metabolites are SCFAs, bile acids, and amino acid-derived metabolites.
Table 1.
Typical gut microbiota metabolites and their roles in health and diseases.
| Groups | Typical metabolites | Typical targets | Specific functions | Typical diseases associated | Ref. |
|---|---|---|---|---|---|
| Short-chain fatty acids | Acetate, propionate, butyrate, hexanoate, isovalerate, isobutyrate, 2-methylpropionate, valerate |
Directly act on GPR41, GPR43, GPR109A, GPR81, GPR91, HDAC1 and HDAC3 | Regulation of gut microbiota composition, gut barrier integrity, appetite, energy homeostasis, gut hormone production, circadian clocks; inhibit proinflammatory cytokines; stimulate water and sodium absorption; modulate systemic immune response | Diabetes, obesity, pancreatitis, nonalcoholic fatty liver disease, hypertension, atherosclerosis, chronic kidney disease, ulcerative colitis, radiation proctitis, Crohn’s disease, colorectal cancer, autism spectrum disorder, sclerosis, Parkinson’s disease, asthma, diarrhea | [15-21] |
| Bile acids | Cholate, hyocholate, deoxycholate, taurohyocholate, ursodeoxycholate, taurocholate, tauro- α-muricholate, glycocholate, hyodeoxycholate, tauro- β-muricholate, lithocholate, taurodeoxylcholate |
Directly act on FXR, VDR, PXR/SXR, constitutive androstane receptor (CAR), TGR5, sphingosine 1-phosphate receptor 2 (S1PR2), formyl-peptide receptor (FPR), muscarinic acetylcholine receptor (mAChR) | Facilitate lipid and vitamin absorption; regulation of gut microbiota composition, gut hormones, intestinal immunity, intestinal electrolyte and fluid balance, gut motility, lipid homeostasis, glucose homeostasis, amino acid homeostasis, circadian clocks; influence neurotransmission and physiology | Primary biliary cholangitis, primary sclerosing cholangitis, obesity, nonalcoholic fatty liver disease, non-alcoholic steatohepatitis, atherosclerosis, ulcerative colitis, cancer, hepatic encephalopathy, multiple sclerosis, Alzheimer's disease, Parkinson's disease, traumatic brain injury, stroke and amyotrophic lateral sclerosis | [8,22-25] |
| Gases | H2S, H2, CO2, CH4, NO | NO targets soluble guanylate cyclase, H2S cause conformational changes of target proteins by sulfhydration | CH4 slows gut motility; H2S regulates gut inflammation, motility, epithelial secretion and susceptibility to infections; NO mediates gastric mucosal protection and regulate mucosal blood flow | Parkinson’s disease, colitis, ulcer | [26-31] |
| Tryptophan and indole derivatives | Indole-3-lactic acid, indole acetic acid, indole-3-acetamide, indole pyruvic acid, indoxyl sulfuric acid, indole, serotonin | Directly targeting on AhR and PXR | Influence the gut microbial spore formation, drug resistance, biofilm formation, and virulence; regulate intestinal barrier functions, gut hormone secretion, gut motility, systemic immune response | Ulcerative colitis, Crohn’s disease, obesity, stroke, mucosal candidiasis, autism spectrum disorder, Alzheimer’s disease, Parkinson's disease, migraine, schizophrenia, irritable bowel syndrome | [32-36] |
| Choline metabolites | TMA, methylamine, dimethylglycine, dimethylamine, | Direct target unknown, but can activate NF-кB, protein kinase C (PKC), NLRP3 inflammasome | Inhibits bile acid synthesis; promote inflammation, thrombosis; affects myocardial hypertrophy and fibrosis; exacerbates mitochondrial dysfunction | Nonalcoholic fatty liver disease, obesity, atherosclerosis, diabetes, heart failure, hypertension | [37-39] |
| Vitamins | Vitamin B2, Vitamin B3, Vitamin B5, Vitamin B6, Vitamin B9, Vitamin B12, vitamin K | Vitamin receptors | Involved in cellular metabolism; modulate immune function and cell proliferation; supply vitamins for hosts | Vitamin associated diseases such as schizophrenia, autism, and dementia | [40,41] |
| Neurotransmitters | Dopamine, catecholamines, 5-HT, and GABA | Adrenergic receptors, 5-HT receptors, GABA receptors | Regulate gut motility, memory and stress responses, immune function of nervous system | Parkinson's disease, autism | [27,42,43] |
| Lipids | Conjugated fatty acids, cholesterol, phosphatidylcholines, triglycerides, LPS |
LPS targets directly on TLR4 | LPS triggers systemic inflammation; conjugated fatty acids regulate hyperinsulinemia, immune system, lipoprotein profiles; cholesterol acts as material bases for bile acid synthesis. | Diabetes, obesity, nonalcoholic fatty liver disease, hyperinsulinemia, hypercholesterolemia, chronic hepatitis C. | [7,44,45] |
| Others | Ethanol; triphosadenine; lantibiotic such as ruminococcin A and cytolysin; microcin such as microcin B17; organic acids such as benzoate and hippurate; polyamines such as cadaverine, and spermidine | Triphosadenine activate P2X and P2Y receptors | Enhance or damage gut barrier; regulate intestinal or systemic immune response; act as antibiotics to modulate gut microbiota composition; supply the nutrients; be toxic to host cells | Fatty liver disease, C. difficile and H. pylori infections, irritable bowel syndrome, ulcerative colitis | [7,46-48] |
2.2. Typical gut microbiota metabolites in a nutshell
During the past decade, a wealth of literatures has paid attention to SCFAs and SCFAs have become the cynosure of all the gut microbiota metabolites. SCFAs are saturated aliphatic acids with number of carbons ranges from one to six. Acetate, propionate, and butyrate are the most common SCFAs produced by gut microbiota. Most SCFA are generated by fermenting of carbohydrates that have escaped the digestion of human enzymes in stomach and small intestine, and in a much lesser extent by fermenting undigested protein-derived branched chain amino acids (BCAAs) [49]. The undigested proteins and peptides that contain BCAAs can be metabolized typically to branched-chain fatty acids such as 2-methylbutyrate and iso-valerate [49]. The SCFAs are rapidly absorbed by colonocytes in large intestine via hydrogen-dependent or sodium-dependent monocarboxylate transporters following their production [50]. In colonocytes, a large part of SCFAs function as energy source, and the remaining SCFAs can be transported into the circulation system and other tissues such as brain, heart, and lung [51]. The targets of SCFAs include G protein-coupled receptor (GPR) 41, GPR43, GPR109A, GPR81, and GPR91 which are also named as free fatty acid receptor 3 (FFAR3), FFAR2, hydroxycarboxylic acid receptor (HCA) 2, HCA1, and succinate receptor 1, correspondingly [20]. SCFAs can also target nuclear class I histone deacetylases (HDACs) including HDAC1 and HDAC3, which are mainly correlated to the anti-inflammatory phenotype [52].
Bile acids are cholesterol derived amphipathic and water-soluble metabolites of saturated hydroxylated C-24 sterols that are originally synthesized by hepatocytes and transformed by gut microbiota [53]. In the liver, primary bile acids are produced from cholesterol by the classic or alternative pathway, the former of which plays major role [54]. In the classic pathway, 7 α-hydroxylase is the rate-limiting enzyme for synthesis of primary bile acids [55]. Chenodeoxycholic acid and cholic acid are the main hepatic bile acids synthesized in human and can conjugate to taurine or glycine to form bile salts [56]. Of note is that bile acids show obvious species-specific differences. For example, the primary bile acids generated by human are chenodeoxycholic acid and cholic acid, whereas rodents mainly produce cholic acid, chenodeoxycholic acid and muricholic acids [56]. Primary bile salts are synthesized in the liver, deposited in the gallbladder and discharged into the duodenum. Primary bile acids in intestinal tract can be reabsorbed by host and delivered back to liver, a process termed enterohepatic circulation, to maintain the bile acid pool [23]. In terminal ileum and colon, the primary bile acids that escape enterohepatic circulation can be transformed by gut microbiota into secondary bile acids mainly via bile salt hydrolases and 7 α-dehydroxylases [22,23]. The bile acids nuclear receptors include farnesoid X receptor (FXR), vitamin D3 receptor (VDR), pregnane X receptor/steroid and xenobiotic-sensing receptor (PXR/SXR), constitutive androstane receptor, and the membrane receptors include Takeda G-protein receptor 5 (TGR5), sphingosine 1-phosphate receptor 2, formyl-peptide receptor, and muscarinic acetylcholine receptor [8].
The proteins and peptides that escape digestion of hosts can undergo the metabolism of gut microbiota, and produce various bioactive compounds that includes, but not limited to, ammonia, amines, sulfides, nitrogen compounds, indoles, phenols, p-cresol sulfate, and precursors to branched-chain fatty acids [57]. Among all these compounds, tryptophan and tryptophan derivatives have caught the eyes of a large group of researchers. In the gastrointestinal tract, unabsorbed tryptophan can be transformed by the gut microbiota into tryptamine, skatole, indole and indole derivatives, such as indole-3-propionic acid, 3-methyl-indole, and indoxyl sulfate [36]. Many of these metabolites can act as ligands for aryl hydrocarbon receptor (AhR), a protein that can trigger a wide range of effects on the hosts such as modulation of metabolism, immunity, and social behavior [34,35]. Some amino acids can be sequentially metabolized by gut microbiota and hosts, as epitomized by carnitine, an amino acid that is abundant in red meat. Carnitine can be metabolized to trimethylamine (TMA) in the gut lumen by gut microbiota, and then be converted into trimethylamine-N-oxide (TMAO) by flavin-containing monooxygenase 1 and 3 in the liver [37]. Elevated circulating levels of TMAO increase the risk of cardiovascular diseases via promoting atherosclerotic lesion development; however, there is still controversy over it [58]. Some amino acids metabolites can serve as neurotransmitters. The gut microbiota can synthesize phenylalanine and tyrosine derivative dopamine via decarboxylation of L-DOPA by tyrosine decarboxylase [59]. Dopamine can be converted into norepinephrine and epinephrine via hydroxylation and methylation, respectively. Other neurotransmitters produced by gut microbiota include serotonin (5-hydroxytryptamine (5-HT)), norepinephrine, and γ-aminobutyric acid (GABA) [27].
In addition to metabolites mentioned above, the gut microbiota is capable of producing a series of other metabolites that are important for physiological functions of hosts. The gut microbiota can produce gases including hydrogen (H2), methane (CH4), carbon dioxide (CO2), hydrogen sulfide (H2S), nitric oxide (NO) that can modulate the physiology of hosts [28]. Lipids play important roles in the health of hosts, and the gut microbiota can produce lipopolysaccharides (LPS), conjugated fatty acids and other lipids to modulate the functions of hosts [7]. Assessment of the genomes of human gut bacteria for biosynthesis pathways of B-vitamins showed that 40-65% of the bacteria have the capability to synthesize vitamins, and the gut microbiota is an important producer of B-vitamins [60]. It should be noted that, although the importance of gut microbiota metabolites has been well recognized, the number of them is so huge that many of them remain uncharacterized. Therefore, mining of metabolites with special functions from the gut microbiota have become an important territory [61], and many methods for discovery of these metabolites have been developed, such as functional metagenomics, sequence-based (meta)genomics, and metabolomics [6,62,63].
2.3. Ways gut microbiota metabolites act on targets
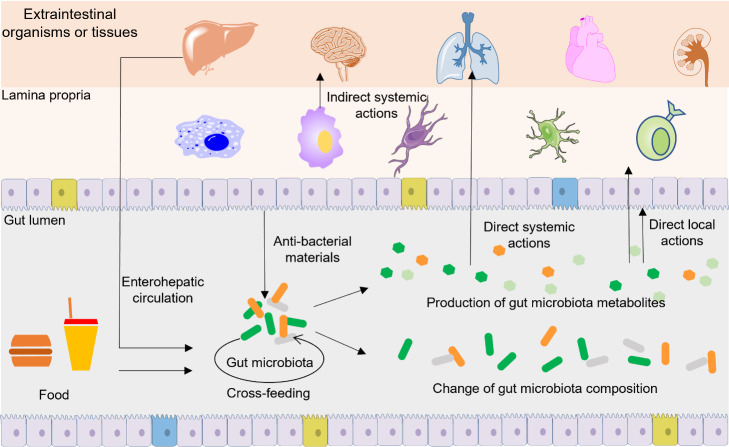
Gut microbiota metabolites act in multiple ways to directly or indirectly influence the functions of hosts (Fig. 1). The composition of gut microbiota is a key determinant in human health and disease, and the metabolism of gut microbiota is closely linked to the composition of gut microbiota. The gut microbiota metabolites can target on gut bacteria or hosts to regulate the composition and function of gut microbiota directly or indirectly. For example, gut microbiota metabolites SCFAs can serve as energy source for gut microbiota, and a high concentration of SCFAs can inhibit the growth of some gut bacteria [64]. In addition, SCFAs can modulate the production of secretory immunoglobulin A (sIgA), a non-inflammatory antibody synthesized by hosts, to prevent the invasive of pathogens [65]. Correspondingly, gut microbiota metabolites can modulate the composition of gut microbiota to indirectly influence the functions of hosts. Gut microbiota metabolites can also act on targets of host that are near or distant from gastrointestinal tract to directly modulate the functions of hosts. For example, upon their release into the gastrointestinal tract, SCFAs are directly sensed by the intestinal epithelial cells to influence the function of gut barrier [66]. SCFAs can also be delivered to tissues and organs that are remote from the gut, and then directly sensed by target tissues and organs to trigger extensive physiological changes of hosts [67]. In addition to SCFAs, other gut microbiota metabolites can also act directly or indirectly on targets that are within, near, or distant from gastrointestinal tract. We will discuss these specific mechanisms in the following section.
Figure 1.
Ways gut microbiota metabolites act on targets. Within gut lumen, gut microbiota metabolites serve as the nutrients for some bacteria and change the composition of gut microbiota. Locally, gut microbiota metabolites can act on intestinal epithelium and immune cells in the lamina propria, and the local effects can further induce downstream systemic functions. Systemically, gut microbiota metabolites can be absorbed and transported to remote organs and tissues to exert diverse functions. Some gut microbiota metabolites can indirectly regulate the composition and function of gut microbiota via inducing hosts to synthesize and release anti-bacterial materials into gut lumen. Some gut microbiota metabolites may undergo enterohepatic circulation.
Bacterial cross-feeding refers to the process that one bacteria uptake or exchange the bacterial products with another [68]. Lactate, an end product of Bifidobacterium, is the typical bacterial product of cross-feeding. Eubacterium hallii cannot grow in pure starch condition. When E. hallii and Bifidobacterium adolescentis are co-cultured, significant reduction of lactate can be observed whereas butyrate shows an increase of concentration [69]. Thus, E. hallii can synthesis butyric acid using the lactose released by B. adolescentis. Cross-feeding provides significant advantages for bacteria in poor nutrient conditions and modulates antibiotic tolerance in bacterial communities, and thus influences the composition and function of gut microbiota [68,70,71].
3. Functions of gut microbiota metabolites
3.1. Regulation of the composition and function of gut microbiota
In gastrointestinal tract, a normal gut microbiota composition act as a barrier to defend the invasion of pathogens by stimulating hosts to secrete compounds with antimicrobial effects, competitive consumption of nutrients, and occupation of attachment sites [72]. On the contrary, dysbiosis, an abnormal change of gut microbiota composition, is associated with the development of a large number of diseases [73]. In addition to aforementioned SCFAs, many gut microbiota metabolites can directly modulate the composition and function of gut microbiota. For example, indole and indole derivatives can function as interspecies signaling molecules with the capability to regulate virulence, antibiotic resistance, biofilm formation, motility, and sporulation of gut bacteria [74]. Due to the detergent properties, bile acids can reshape the composition of gut microbiota by inhibiting the growth of gut bacteria via destroying the structure of bacterial membranes [22]. Among all the gut microbiota metabolites, of particular interest are antibiotics such as ribosomally synthesized, posttranslationally modified peptides such as lantibiotics, bacteriocins, microcins [46,48]. These metabolites are often toxic for a limited spectrum of bacterial species and are therefore hoped to be developed as new generation of antibiotics in the event of global abuse of antibiotics [75]. Besides these direct roles, gut microbiota metabolites can modulate the immune systems of hosts to indirectly affect the composition and function of gut microbiota (Fig. 1). For example, sIgA is a non-inflammatory antibody specialized in mucosal protection via maintenance of non-invasive commensal bacteria and neutralization of invasive pathogens [76]. SCFAs can improve the production of cecal sIgA and thus modulate the composition of gut microbiota [77]. Similarly, bile acids can act on FXR, thereby inducing the transcription of antimicrobial agents such as inducible nitric oxide synthase and interleukin (IL)-18 to regulate the composition of gut microbiota [78].
3.2. Serving as nutrition and influencing nutrition absorption
Gut microbiota such as Bifidobacteria can de novo synthesize a series of vitamins especially vitamin K and B group vitamins [79]. These vitamins can not only maintain the basic functions of bacterial metabolism, but also act as complementary endogenous sources of vitamins to maintain the metabolic and physiological functions of mammals. SCFAs can be directly degraded to provide energy or be used as resources for gluconeogenesis and lipid biosynthesis [20]. Gut microbiota amino acid metabolites including tryptophan, leucine, valine, and isoleucine are essential amino acids required for protein synthesis and are precursor of metabolites that can significantly affect mammalian physiology [80]. Minerals have many important functions in human, such as being a part and parcel of enzymes, hemoglobin, and bones. The factors influencing the bioavailability of minerals include synergism and antagonism between different minerals, presence of complexing or chelating compounds in the food, acidity of the colon, processing methods of food, health state of the organisms, environmental pollution, and gut microbiota [81]. Gut microbiota metabolites especially SCFAs can affect the bioavailability of minerals by lowering the acidity of intestinal luminal contents and changing the intestinal tissue morphology and transport proteins [82]. As the amphipathic and water-soluble molecules, bile acids emulsify dietary lipids and fat-soluble vitamins to facilitate the absorption of these nutrients after their release into the duodenum from the gall bladder [8].
3.3. Modulation of host metabolism
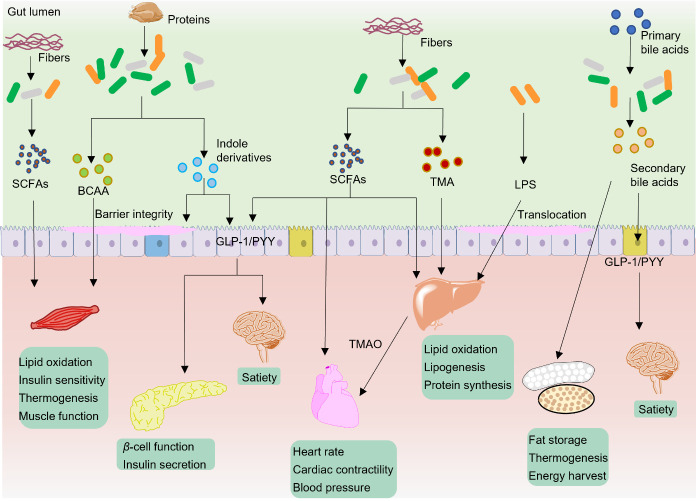
Energy homeostasis, or the balance of energy storage and release, is vital for the survival and health of organisms. In adult mammals, long-term positive energy balance can result in obesity and increase the risks for various diseases such as type 2 diabetes, hypercholesterolemia, asthma, arthritis, liver diseases, and colon cancer [83,84]. On the contrary, long-term negative energy balance can lead to a number of diseases or physical disorders, including loss of bone mass, decline of thyroid hormones, reduction of physical performance and fertility [85,86]. Nutrients such as lipids and glucose have critical roles in energy metabolism and disease development. Over the past two decades, numerous studies have demonstrated that gut microbiota and gut microbiota metabolites play crucial roles in host metabolism via regulation of non-shivering thermogenesis, nutrition metabolism, satiety, motility function of organs, insulin synthesis and secretion, and insulin sensitivity (Fig. 2).
Figure 2.
Typical gut microbiota metabolites in modulation of host metabolism. The major ways of this modulation include regulation of nutrition metabolism (lipids, proteins, glucose), non-shivering thermogenesis (browning of WAT and BAT), satiety (by secretion of hormone GLP-1 and PYY), motility function of organs (muscle and heart), insulin synthesis and secretion, and insulin sensitivity. By these ways, gut microbiota metabolites can maintain the homeostasis of energy.
Adipose tissues play important roles in energy homeostasis. In general, adipose tissues are made up of three different tissues, i.e., brown adipose tissue (BAT), white adipose tissue (WAT) and beige adipose tissue. BAT and beige adipose tissue are major contributors to non-shivering thermogenesis in mammals while WAT is the key adipose tissue compartment to energy storage [87]. SCFAs, especially acetate, can inhibit lipolysis, increase adipogenesis, and thus improve the capacity of lipid storage of adipose tissues [88]. In cold environment, butyrate can partially rescue the impaired thermogenesis induced by antibiotics via improving the thermogenesis of BAT and browning of WAT [89]. The functions of insulin and β-cell are important for maintaining the blood glucose and the development of type II diabetes mellitus (T2DM). SCFAs has beneficial effects on β-cell function and insulin secretion via GPR43 [90,91]. The skeletal muscle is the largest organ in the human body and occupies a proportion of approximate 40% body mass, 30% resting energy expenditure, and 80% insulin-related glucose uptake [92,93]. SCFAs can influence the lipid, carbohydrate, and protein metabolism of skeletal muscle via acting on GPR41, GPR43, HDACs [94]. In the heart, SCFAs can act as an energy resource and acutely reduce the heart rate, cardiac contractility, and blood pressure [95,96]. Liver is an important metabolic organ for lipid and glucose metabolism. SCFAs in the liver can directly act as sources of energy. Propionate can be used for synthesis of glucose in liver while acetate can be used as substrates to synthesize cholesterol and long-chain fatty acids [97]. SCFAs can increase energy expenditure and decrease hepatic steatosis via inducing a switch from hepatic lipogenesis to hepatic beta-oxidation thereby protect against high-fat diet-induced obesity [98].
In addition to SCFAs, other gut microbiota metabolites also play important roles in regulation of energy metabolism. Activation of bile acid targets FXR and TGR5 can increase liver glycogen synthesis and insulin sensitivity, promote pancreas insulin secretion, facilitate energy metabolism in the liver, brown adipose tissue and muscles [99-101]. Like SCFAs, bile acids can act on enteroendocrine cells and facilitate the release of gut hormones such as glucagon-like peptide-1 (GLP-1) and peptide YY (PYY), which are involved in regulation of appetite and gut motility [8,102]. In the liver, bile acids can regulate triglyceride metabolism, especially the production of very low-density lipoprotein and lipogenesis, whose dysbolism further promote obesity, T2D as well as atherosclerosis and other cardiovascular diseases [103]. The branched-chain amino acids (BCAAs) are essential amino acids synthesized by gut microbiota. In the 1960s, researchers have already found out that BCAAs have direct roles on stimulation of insulin secretion and elevated levels of BCAAs are correlated with obesity and serum insulin [104,105]. One study showed that feeding mice a diet with specifically reduced BCAAs can improve glucose tolerance and body composition [106]. The mechanism of BCAAs in increasing energy expenditure is associated with chronic phosphorylation of mammalian target of rapamycin, c-Jun NH2-terminal kinase, and insulin receptor substrate 1 at 307 residue and by accumulating multiple acylcarnitines in the muscles [107]. Many studies have demonstrated the important roles of BCAAs in regulating protein synthesis, glucose and lipid metabolism, insulin resistance, hepatocyte proliferation, and thermogenesis of BAT [108]. Other gut microbiota metabolites such as TMAO, LPS, tryptophan and indole-derivative metabolites are closely linked with energy and nutrients metabolism [99]. Disturbed production of these gut microbiota metabolites is linked to a series of diseases such as obesity, T2DM, dyslipidemia, nonalcoholic fatty liver disease, atherosclerosis, and heart failure [109-112].
3.4. Influencing the intestinal barrier and gut motility
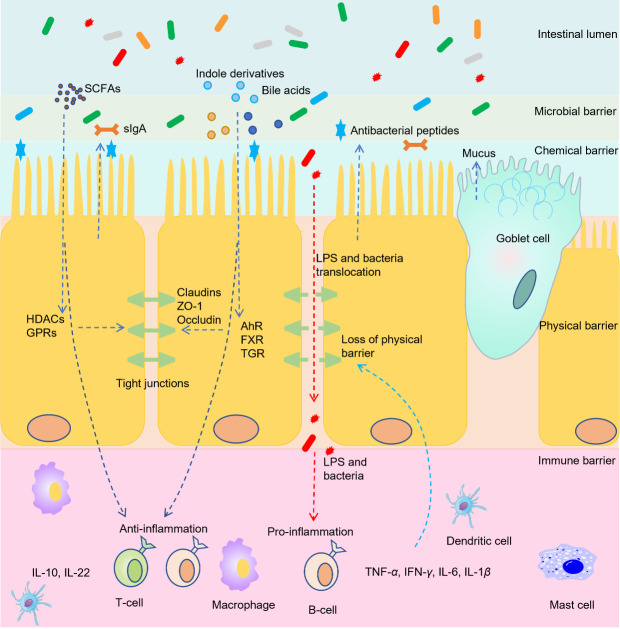
With a total surface of 200 m2, the gastrointestinal tract is the most exposed body system to the outside world [113]. Intestinal barrier is a layer of microbial, chemical, physical and immune barrier between the gut lumen and mucosal tissues with the critical functions of nutrient absorption and immune modulation. Specifically, the structure of intestinal barrier includes the outer microbial layer with gut commensals, mucus layer with the antimicrobial peptides and secretory immunoglobulin A (sIgA), the central single cell layer with epithelial cells (absorptive enterocytes, Goblet cells, enteroendocrine cells and Paneth cells), and the inner lamina propria layer where innate and adaptive immune cells dwell, such as T cells, B cells, dendritic cells, macrophages, and IgA producing plasma cells [5,114,115]. The permeability of the epithelium is determined by the tight junctions that reside near the apical surface of adjacent epithelial cells [116]. Increased intestinal epithelial permeability, also called “leaky gut”, facilitates the translocation of harmful substances such as LPS and pathogens to the inner layer of intestinal barrier and bloodstream [117]. Translocation of these materials can further affect the immune and metabolomic status of hosts and is associated with a wide range of diseases, including celiac disease, colorectal cancer, irritable bowel syndrome, inflammatory bowel disease and other extraintestinal diseases such as chronic liver diseases, T1DM, obesity and food allergy [5,118,119].
The function of intestinal barrier is largely affected by gut microbiota metabolites (Fig. 3). SCFAs especially butyrate can orchestrate the genes encoding tight-junction proteins and regulate the redistribution of occludin to prevent abnormal intestinal permeability [120]. SCFAs can also regulate intestinal barrier by stimulating the hosts to secrete antimicrobial peptides, sIgA and mucins to prevent the adherence or invasion of harmful bacteria [121,122]. The immune cells in intestinal barrier secrete inflammatory cytokines such as IL-10, IL-1 β, IL-6, IL-22, tumor necrosis factor α (TNF- α) to maintain the normal functions of immune system. On the contrary, excessive production of these cytokines can lead to systemic inflammation and diseases. SCFAs exert anti-inflammatory effects by regulating cytokine production and immune cell functions [123]. Indole and indole derivatives can act as ligands of epithelial nuclear receptors such as AhR, PXR and retinoid-related orphan receptor gamma-t to affect the functions of intestinal barrier [33]. For example, indole can enhance the trans-epithelial resistance of epithelial cells, induce the expression of intestinal tight junction proteins, decrease the inflammatory cytokines IL-1 α, IL-1 β, TNF- α and IL-6 [33,36]. Activation of bile acid target TGR5 by BAR501 shifted the macrophage phenotypes from M1 (pro-inflammatory) to M2 (tissue-protective), decreased the expression of proinflammatory cytokines including TNF- α, IFN- γ, IL-1 β, IL-6, and enhanced the expression of anti-inflammatory cytokines including transforming growth factor β and IL-10 [124].
Figure 3.
Gut microbiota metabolites modulation of intestinal barrier. Intestinal barrier consists of microbial barrier, chemical barrier, physical barrier, and immune barrier. SCFAs can enhance the chemical barrier by stimulating the secretion of antimicrobial peptides, sIgA and mucins to prevent harmful bacteria. SCFAs, bile acids, and indole derivatives can enhance physical barrier via increasing tight junction proteins such as cludins, occluden-1, and occludin. The epithelial cross of SCFAs and indole derivatives can act on immune cells and lead to release of anti-inflammatory cytokines such as IL-10 and IL-22. During chronic diseases, disturbance of tights junctions can lead to destruction of physical barrier, and further lead to translocation of LPS and bacteria. This translocation triggers the activation of immune cells and lead to production of pro-inflammatory cytokines. The release of pro-inflammatory cytokines can act on local epithelial cells to worsen physical barrier or can act on extraintestinal organs to trigger other diseases.
The proper function of intestinal motility is important for digestion, absorption, and secretion of nutrients and wastes whereas dysmotility of intestine can lead to infections, malabsorption of nutrients, diarrhea, and constipation [125,126]. Slow colonic transit is associated with increased bacterial production of methane, protein catabolism, carbohydrate deprivation, and an increase of detrimental metabolites such as ammonia and aromatic derivatives of amino acids [127]. On the contrary, fast transit is associated with increased gut health and reduced low-grade intestinal inflammation [128]. Gut microbiota metabolites such as SCFAs, LPS, secondary bile acids and methane can interact with enteric nerves and smooth muscles via neural and humoral pathways such as GLP-1, PYY, motilin, and serotonin to modulate gastric emptying and colonic motility [129,130].
3.5. Impacting the systemic immune response
In addition to modulate the local intestinal immunity, gut microbiota can also influence both adaptive and innate immune responses in multiple extraintestinal organs via acting on a plethora of immune cells such as B cells, T cells, and macrophages. In spleen B cells, SCFAs support antibody production via increasing acetyl-CoA and regulating metabolic sensors [77]. In addition, mice with low SCFA production showed defects in homeostatic and pathogen-specific antibody responses, leading to bigger susceptibility to pathogens whereas SCFA intake can restore this immune deficiency [77]. In the lung, gut microbiota metabolites predominately SCFAs are responsible for the protection against allergy and asthma. Antibiotics treatment leads to elevated allergic lung inflammation whereas administration of SCFAs reduces this inflammation by manipulation of T helper type 2 cells and decreasing the circulating immunoglobulin E [67,131]. In pregnant mice, administration of acetate in drinking water protected offspring from asthma during adulthood, and the mechanism is associated with regulating mucosal immune responses via stimulation of GPCRs such as GPR109A and GPR41 [132], and inhibition of HDACs to enhance the number and function of Treg cells [133]. Butyrate protected the lungs of influenza-infected mice via regulating the metabolism of influenza-specific CD8+ T cells and shaping Ly6c- patrolling monocyte hematopoiesis [134].
Autoimmune cholestatic liver diseases, including primary sclerosing cholangitis and primary biliary cholangitis, are a type of diseases that show impairment of bile flow and excessive accumulation of toxic bile acids [135]. In the liver, high concentrations of bile acids contribute to autoimmune liver injury and can result in activation of pro-inflammatory programs and production of NF- κB dependent mediators [136-138]. In addition, bile acids can cause direct injury on biliary epithelial cells and lead to production of IL-6 and IL-1 β, which will further result in augmentative inflammation [139]. Non-alcoholic fatty liver disease is a metabolic liver disease characterized by hepatic steatosis and can progress to non-alcoholic steatohepatitis that shows obvious hepatic inflammatory damage [140]. SCFAs such as acetate can alleviate hepatic inflammation by suppression of the macrophage proinflammatory activation [141].
Increased level of TMAO is positively correlated with atherosclerosis and cardiovascular diseases such as myocardial infarction, hypertension, and myocardial fibrosis [142]. TMAO can impair endothelial self-repair capacity and enhancing monocyte adhesion via activating NF- кB, protein kinase C and nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing 3 (NLRP3) inflammasome [143]. In mice, voluntary exercise inhibited cardiac dysfunction in western diet-induced obesity via preventing myocardial inflammation and fibrosis [144]. However, TMAO supplementation abolished this cardioprotective effects of voluntary exercise via inducing myocardial inflammation by increasing TNF- α and IL-10 [144]. In mouse models of doxorubicin-induced cardiac fibrosis, TMAO aggravated cardiac fibrosis by activating NLRP3 inflammasome. On the contrary, silencing of NLRP3 protected the mice from cardiac fibrosis as evidence by amelioration of cellular proliferation, migration and collagen deposition [145].
Osteoporosis is a skeletal disease that is characterized by low bone mineral density and microstructural destruction of bone tissues. In the bone, gut microbiota metabolites such as LPS, SCFAs, and bile acids can influence the immune function to modulate the bone metabolism [146]. For example, increased permeability of intestinal barrier causes more LPS into the circulation system and reduction of bone mineral density [147]. The mechanisms are associated with activation of TLR2 and TLR4 in mesenchymal stromal cells to inhibit osteoblastic differentiation, activation of NF- κB and MAPK signaling pathways to increase the differentiation of macrophages into osteoclasts [148]. Plasma metabolome study demonstrated the existence of gut microbiota-derived uraemic toxins, including LPS, tryptophan derivative indoxyl sulfate, tyrosine or phenylalanine derivatives p-cresol sulfate and phenylacetylglutamine [149]. In the kidney, these metabolites can accumulate and contribute to pathogenesis and progression of chronic kidney disease (CKD) via triggering renal damage, inflammation and fibrosis [150].
In addition to the effects mentioned above, gut microbiota metabolites can also affect other organs or tissues to influence the development of diseases such as systemic lupus erythematosus, rheumatoid arthritis, human immunodeficiency virus, and inflammatory skin diseases [151-153]. For example, rheumatoid arthritis is a disease caused by the malfunction of white blood cells and it affects approximately 1% of the population worldwide [154]. Butyrate supplementation ameliorates arthritis in a regulatory B cells-dependent way via elevating the content of the serotonin derivative 5-hydroxyindole-3-acetic acid, a compound that can further activate AhR, a transcriptional marker for regulatory B cell function [154]. Gut microbiota metabolites can also influence the immunity of nervous system, which we will discuss in the next section.
3.6. Influencing the nervous system
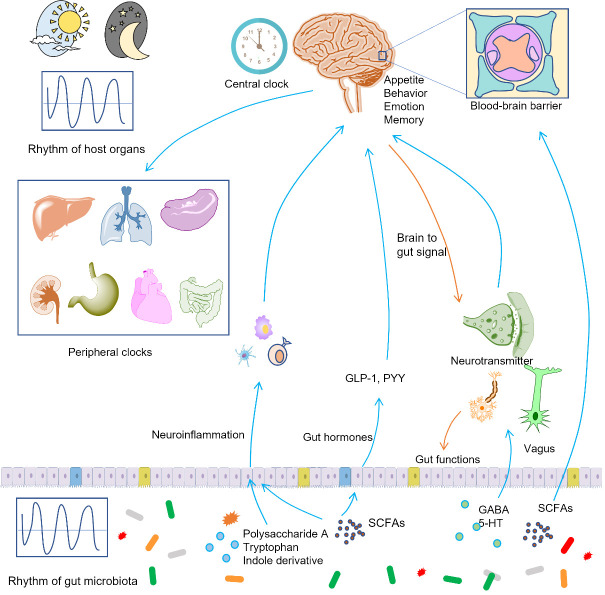
It has long been established that central nervous system (CNS) can modulate the functions of gut such as gut motility, digestive juice secretion, immune function, blood flow, and nociception [155,156]. In addition, the signaling from brain to gut can further affect the composition and function of gut microbiota via intestinal immune system [157]. On the contrary, a growing body of evidence emerged in recent years suggests that the gut microbiota metabolites exert significant influences on brain (Fig. 4), and thus affect the development of neurological and psychiatric disorders such as multiple sclerosis, major depressive disorder, anxiety disorder, Parkinson’s disease, Alzheimer’s disease, and autism [155]. For example, bacteria-derived LPS can act as the mediator to trigger inflammatory bowel disease-related psychosocial disturbances [158].
Figure 4.
Gut microbiota metabolites modulate the nervous system and circadian rhythm. Gut microbiota metabolites can modulate the cerebral inflammation, the production of gut hormones, the transmission of nervous impulse, the functions of blood-brain barrier to regulate the functions of brains such as the emotion and appetite. On the contrary, the brain can modulate the functions of gut such as the gut motility and secretion of digestive juice. The production of gut microbiota metabolites shows day-night rhythm, and this rhythm can be transported to the central clocks and peripheral clocks to modulate the systemic functions of hosts. Similarly, the central clock can transfer the light and dark cues to peripheral organs and gut microbiota, and thus synchronize the functions of gut microbiota and host clocks.
The blood-brain barrier (BBB) is part of a neurovascular unit that comprise brain microvascular endothelial cells, astrocytes, neurons, pericytes, extracellular matrix, and microglia. BBB plays essential roles in preventing the entrance of therapeutic drugs to brain and protecting the brain from injuries and diseases by tightly segregating the brain from the circulating blood. Infections, autoimmune diseases, and brain injury can change the integrity of BBB, rendering the increased accessibility of the brain to harmful products in the circulatory system [159]. SCFAs play important roles in maintaining the integrity of BBB as evidenced by the fact that intravenous or intraperitoneal administration of sodium butyrate can inhibit BBB breakdown following the traumatic brain injury [160]. SCFAs can cross BBB via transporter H+-dependent or sodium-dependent monocarboxylate transporters that are abundantly expressed in endothelial cells [50]. In the brain, SCFAs can directly regulate learning, memory, behavior, and disease progress [161]. In addition to SCFAs, many other gut microbiota metabolites such as BCAAs, GABA, and indole derivatives can cross BBB to exert the extensive effects on brain [162].
The CNS contains a variety of innate and adaptive immune cells including microglia, astrocytes, perivascular macrophages, CD4+ T and CD8+ T cells, and mast cells that can affect cerebral inflammation [155]. Usually, physiological production of cytokines by activation of CNS immune cells causes minimal impact on the CNS. On the contrary, chronic systemic inflammation can lead to significant behavioral alterations and cognitive dysfunctions [163,164]. Administration of mice with SCFAs reduced experimental autoimmune encephalomyelitis and axonal damage through increasing Treg differentiation by suppression of the c-Jun NH2-terminal kinase 1 and p38 pathway [165]. Similarly, treatment with polysaccharide A, a gut bacterial product, protected the wild-type mice against CNS demyelination and inflammation by a Toll-like receptor (TLR)-2-dependent pathway [166]. Astrocytes are one type of glial cells that present abundantly in the brain and can modulate neural inflammatory responses by cytokine production and antigen presentation [167]. The gut microbiota metabolites tryptophan and indole derivative can activate AhR to modulate astrocyte activity, and thus influence the multiple sclerosis in animals [168].
Feelings of hunger and satiety are important motivations for feeding behavior. Gut microbiota metabolites can influence the appetite, and thus maintain the metabolic health of hosts or cause a number of metabolic disorders. In the colon, gut microbiota metabolites can promote enteroendocrine cells to produce anorexigenic hormones (PYY, GLP-1, and cholecystokinin), neurotransmitter 5-HT, and peripheral hormones (leptin, ghrelin, and insulin) [169]. For example, SCFAs can binding to GRP43 and thus lead to the release of GLP-1, PYY, insulin, and leptin [170,171]. The anorexigenic hormones including GLP-1 and PYY can cross the BBB and activate proopiomelanocortin in the brain [172,173]. In addition, some gut microbiota metabolites such as GABA can act as neurotransmitter to regulate appetite. This is supported by the fact that disruption of GABA signaling pathways suppressed postweaning feeding, blunted neuropeptide Y-triggered hyperphagia, and hunger-associated appetite [174,175]. Furthermore, gut microbiota can synthesis protein sequences that are identical to peptides with appetite-regulating effects, such as caseinolytic protease, a mimic of alpha-melanocyte-stimulating hormone to induce anorexigenic effects [176].
3.7. Modulation of circadian rhythm
Human is evolved to adapt to a circadian rhythm of ~24 h that is in concert with the light/dark cycle on the earth. Circadian rhythms affect the microscopic molecular oscillations in genes, proteins, and metabolites and macroscopic aspects of biology and physiology such as behavior, sleep-wake cycles, gastrointestinal digestion, absorption, motility, and hormone secretion [177,178]. The ‘central’ circadian clock is located in the suprachiasmatic nucleus of hypothalamus. It receives environmental light and dark cues and synchronizes this information to ‘peripheral’ clocks in peripheral tissues to keep the body functioning in a same rhythm [179]. Circadian rhythms can be disrupted by lifestyle factors such as shift work, jet lag, sleep deprivation, artificial light at night, food type, and timing of food consumption [180]. Disruption of circadian rhythms is implicated in a number of diseases such as neurodegenerative diseases, cardiovascular diseases, gastrointestinal diseases, metabolic diseases, sleep and psychiatric disorders, and cancer [181-185].
Recent studies showed that gut microbiota can regulate or be regulated by the central and peripheral circadian clocks (Fig. 4). Like mammals, gut microbiota exhibits circadian rhythm and shows compositional and functional oscillation, and this oscillation further programs host transcriptome oscillations that eventually affect hepatic drug detoxification and drug hepatotoxicity [186]. The mechanisms that gut microbiota can affect host circadian rhythms include contact-dependent and contact-independent mechanisms [187]. The contact-dependent mechanisms need a direct contact between gut bacteria and gastrointestinal cells and the following activation of pattern recognition receptors such as NOD-like receptors and TLRs [187]. The contact-independent mechanisms mainly require small molecular gut microbiota metabolites such as bile acids and SCFAs to act as mediators [179]. For example, the levels of butyrate and propionate show obvious diurnal oscillation, and this oscillation is lost under high-fat feeding [188]. Treatment of murine and hepatic-derived organoids with individual SCFAs significantly changed the oscillations of expression of major hepatic circadian genes including Bmal1 and Per2[188]. On the contrary, deletion of Bmal1abolished the rhythmicity of fecal SCFA levels, supporting that circadian rhythm of hosts can regulate the rhythmicity of gut microbiota [189].
The effects of gut microbiota metabolites on circadian rhythm are vast and are necessarily interconnected with other functions of gut microbiota metabolites such as modulation of energy metabolism and immune response. These interactions between different functions via the nexus of gut microbiota metabolites are important for understanding the functions of gut microbiota metabolites holistically. For example, gut microbiota programs diurnal oscillations of a SCFAs receptor HDAC3 and thus produce synchronized diurnal oscillations in histone acetylation [190]. The oscillations of histone acetylation further regulate metabolic gene expression and nutrient uptake in a diurnally oscillatory manner. In addition, the oscillations of HDAC3 induced rhythmic transcription of the lipid transporter gene Cd36and high-fat-diet-induced obesity [190]. The study demonstrated the tight relationships of different functions of gut microbiota metabolites.
3.8. Affecting drug efficacy and toxicity
Gut microbiota plays important roles in modification of the toxicity and efficacy of drugs and herbal compounds such as metformin, berberine, aconitine [4]. Gut microbiota metabolites can compete with drug-metabolizing enzymes, affect the expression of drug transporters and hepatic drug-metabolizing enzymes to modulate the efficacy and toxicity of drugs. p-Cresol is a microbial product of tyrosine and phenylalanine by organisms that belong to Firmicutes, Bacteroidetes, Actinobacteria and Fusobacteria phyla [191,192]. Acetaminophen is a widely used non-steroidal anti-inflammatory drug to treat pain and fever. Overdose of acetaminophen can cause severe and sometimes fatal hepatotoxicity in clinic [193]. In the liver, the toxicity of acetaminophen can be reduced by transforming it into inactive acetaminophen sulfate, whereas a small portion of it can be transformed into toxic compound N-acetyl- p-benzoquinone imine by cytochrome P450 (CYP) 2E1 and CYP3A4 [194,195]. p-cresol and acetaminophen can compete for human cytosolic sulfotransferase 1A1, an enzyme responsible for transforming p-cresol to inactive compounds. This competition hampers the capability of liver to reduce the toxicity of acetaminophen [196]. Drug transporters are membrane proteins that transport a wide range of materials into and out of cells and play significant functions in the drug absorption, distribution, and excretion. They are expressed in many tissues such as the intestine, liver, kidney, and brain [197]. Butyrate can downregulate the expression and function of drug transporter P-glycoprotein via inhibiting HDAC/NF- κB pathways [198]. Hepatic metabolism of xenobiotics is mainly regulated by nuclear receptors such as constitutive androstane receptor and PXR [199]. Modulation of AhR by tryptophan and indole derivatives many further modulate the transcription of hepatic and duodenal CYP450 1a genes [200]. The modulation of metabolic enzymes and drug transporters may further impact the metabolism of drugs. However, more research is needed to support this hypothesis.
4. Considerations and perspectives
In addition to the metabolites we have mentioned, the gut microbiota has the potential to synthesize a large number of other structurally distinct metabolites. However, the structures and functions of these microbial metabolites, for the most part, remain unknown. For this reason, it is needed to mine these metabolites systemically. Currently, a variety of methods such as culture-based, genome/metagenomics-based, and metabolomics-based methods have been developed to mine the metabolites with potent bioactivities, and these methods have found plenty exciting achievements [61]. These metabolites discovered are hoped to exhibit super pharmacological effects on hosts and can be developed as novel antibiotics, immunoregulators, anti-inflammatory and anti-obesity drugs [201]. For example, traditional antibiotics can cause extensive damage to the gut microbiota composition and even lead to secondary infections [202]. On the contrary, the antimicrobial metabolites synthesized by gut microbiota such as bacteriocins can be developed as novel antibiotics because they can targeted inhibit or eliminate a certain strain [203]. In addition, the drugs mined from gut microbiota exhibit the advantage over other drugs since they can escape the chemical conversion of gut microbiota, a process that can strongly influence the drug efficacy and toxicity [204,205].
Precision medicine (personalized medicine, stratified medicine, person-centered medicine) is a field of medicine that aims to optimize the medical diagnosis and disease treatment by taking into account an individual's genes, microbiomes, environments, etc. Because of their significance in health and disease, gut microbiota has become an important consideration in the way toward precision medicine [206]. To achieve the goal of precision medicine, it is necessary to find suitable biomarkers for patient stratification and treatment decision at first. The biomarkers can be gut microbiota species, metabolites, microbial genes and enzymes that are responsible for the production of metabolites [4]. For example, SCFAs can be a potential biomarker for screening of patients with celiac disease, adenomatous polyposis, and colorectal cancer [207,208]. When the biomarkers are found, suitable methods are needed to modulate the gut microbiota. The conventional methods include fecal microbiota transplantation (FMT), diet, prebiotics, probiotics, synbiotics, and antibiotics. Recently, supplement of microbial enzyme inhibitors, gut microbiota metabolites, and probiotics have been developed as new methods for precision medicine. For example, butyrate-rich diets have recently been used for improving the redox status and fibrin lysis in Behçet’s Syndrome [209]. Targeted inhibition of microbial TMA production by 3,3-dimethyl-1-butanol non-lethally reduced the level of plasma TMAO in mice supplemented with a high-choline or L-carnitine diet [210]. More importantly, administration of 3,3-dimethyl-1-butanol can reduce the progression of atherosclerosis in Apoe-/- mice, suggesting of the use of enzyme inhibitors in precision medicine [210].
Special attention should be paid to the biomarkers of precision medicine considering that the progresses of diseases are dynamic and so does the gut microbiota. During the disease development, the levels of biomarkers are dynamic and should be monitored repeatedly. Correspondingly, the methods for manipulation of gut microbiota, the dosage of drugs and enzyme inhibitors should be adjusted according to the levels of the biomarkers. More importantly, gut microbiota and hosts are highly interactive since some gut microbiota metabolites, such as TMA, can be further metabolized by hosts. Therefore, microbial-based biomarkers may lead to the failure of precision medicine. In order to develop more precise biomarkers, integrating information about hosts, gut microbiota and other external factors as integrative biomarkers is perhaps more important. For example, vedoNet, a comprehensive neural network algorithm incorporating clinical and microbiome-related data, is useful for predict patients’ response to inflammatory bowel disease treatment [211].
5. Conclusion
The gut microbiota and the host are involved in a complex crosstalk that is influenced by environmental conditions, and they interact with each other both locally and systemically. Owing to the diversity of gut microbiota and the gut microbial genes, the gut microbiota produces many structurally distinct metabolites. These metabolites shuttle between gut microorganisms and between gut microorganisms and hosts, thereby exert a wide range of bioactivities to modulate the functions of both gut microbiota and hosts. Because of the variability of gut microbiota, targeting of gut microbiota metabolites is a necessary procedure toward precision medicine. To achieve the aim of precision medicine, diagnostic microbial biomarkers or dynamic integrative biomarkers should be developed. Of note, although a large numbers of gut microbiota metabolites have been identified, systematic discovery of more microbial metabolites is needed, and these metabolites are hoped to be developed as new drugs for disease treatment.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 82104409, 81891012, 81891010, U19A2010, China), China Postdoctoral Science Foundation (No. 2021M690490, China), Sichuan Science and Technology Program (No. 2021YJ0466, China), and the "Xinglin Scholar" Plan of Chengdu University of Traditional Chinese Medicine (No. BSH2020017, China). We thank the support from Open Research Fund of Chengdu University of Traditional Chinese Medicine Key Laboratory of Systematic Research of Distinctive Chinese Medicine Resources in Southwest China.
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- [1].Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, et al. (2018). Culturing the human microbiota and culturomics. Nat Rev Microbiol, 16:540-550. [DOI] [PubMed] [Google Scholar]
- [2].Sekirov I, Russell SL, Antunes LC, Finlay BB (2010). Gut microbiota in health and disease. Physiol Rev, 90:859-904. [DOI] [PubMed] [Google Scholar]
- [3].Morais LH, Schreiber HL, Mazmanian SK (2021). The gut microbiota-brain axis in behaviour and brain disorders. Nat Rev Microbiol, 19:241-255. [DOI] [PubMed] [Google Scholar]
- [4].Feng WW, Liu J, Ao H, Yue SJ, Peng C (2020). Targeting gut microbiota for precision medicine: Focusing on the efficacy and toxicity of drugs. Theranostics, 10:11278-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vancamelbeke M, Vermeire S (2017). The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol, 11:821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Milshteyn A, Colosimo DA, Brady SF (2018). Accessing Bioactive Natural Products from the Human Microbiome. Cell Host Microbe, 23:725-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S (2012). Host-gut microbiota metabolic interactions. Science, 336:1262-1267. [DOI] [PubMed] [Google Scholar]
- [8].Perino A, Demagny H, Velazquez-Villegas L, Schoonjans K (2021). Molecular physiology of bile acid signaling in health, disease, and aging. Physiol Rev, 101:683-731. [DOI] [PubMed] [Google Scholar]
- [9].Lavelle A, Sokol H (2020). Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol, 17:223-237. [DOI] [PubMed] [Google Scholar]
- [10].Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol, 10:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fan Y, Pedersen O (2021). Gut microbiota in human metabolic health and disease. Nat Rev Microbiol, 19:55-71. [DOI] [PubMed] [Google Scholar]
- [12].Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 464:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xu J, Chen HB, Li SL (2017). Understanding the molecular mechanisms of the interplay between herbal medicines and gut microbiota. Med Res Rev, 37:1140-1185. [DOI] [PubMed] [Google Scholar]
- [14].Postler TS, Ghosh S (2017). Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab, 26:110-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L (2014). The role of short-chain fatty acids in health and disease. Adv Immunol, 121:91-119. [DOI] [PubMed] [Google Scholar]
- [16].Felizardo RJF, Watanabe IKM, Dardi P, Rossoni LV, Câmara NOS (2019). The interplay among gut microbiota, hypertension and kidney diseases: The role of short-chain fatty acids. Pharmacol Res, 141:366-377. [DOI] [PubMed] [Google Scholar]
- [17].Canfora EE, Meex RC, Venema K, Blaak EE (2019). Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol, 15:261-273. [DOI] [PubMed] [Google Scholar]
- [18].Tahara Y, Yamazaki M, Sukigara H, Motohashi H, Sasaki H, Miyakawa H, et al. (2018). Gut microbiota-derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci Rep, 8:1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dalile B, Van Oudenhove L, Vervliet B, Verbeke K (2019). The role of short-chain fatty acids in microbiota-gut-brain communication. Rev Gastroenterol Hepatol, 16:461-478. [DOI] [PubMed] [Google Scholar]
- [20].Feng WW, Ao H, Peng C (2018). Gut Microbiota, Short-Chain Fatty Acids, and Herbal Medicines. Front Pharmacol, 9:1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pan LL, Li BB, Pan XH, Sun J (2021). Gut microbiota in pancreatic diseases: possible new therapeutic strategies. Acta Pharmacol Sin, 42:1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wahlström A, Sayin SI, Marschall HU, Bäckhed F (2016). Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab, 24:41-50. [DOI] [PubMed] [Google Scholar]
- [23].Poland JC, Flynn CR (2021). Bile acids, their receptors, and the gut microbiota. Physiology, 36:235-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Govindarajan K, MacSharry J, Casey PG, Shanahan F, Joyce SA, Gahan CG (2016). Unconjugated bile acids influence expression of circadian genes: a potential mechanism for microbe-host crosstalk. PLoS One, 11:e0167319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McMillin M, DeMorrow S (2016). Effects of bile acids on neurological function and disease. FASEB J, 30:3658-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ostojic SM (2018). Inadequate production of H2 by gut microbiota and Parkinson disease. Trends Endocrinol Metab, 29:286-288. [DOI] [PubMed] [Google Scholar]
- [27].McCarville JL, Chen GY, Cuevas VD, Troha K, Ayres JS (2020). Microbiota metabolites in health and disease. Annu Rev Immunol, 38:147-170. [DOI] [PubMed] [Google Scholar]
- [28].Kalantar-Zadeh K, Berean KJ, Burgell RE, Muir JG, Gibson PR (2019). Intestinal gases: influence on gut disorders and the role of dietary manipulations. Nat Rev Gastroenterol Hepatol, 16:733-747. [DOI] [PubMed] [Google Scholar]
- [29].Pacher P, Beckman JS, Liaudet L (2007). Nitric oxide and peroxynitrite in health and disease. Physiol Rev, 87:315-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Singh SB, Lin HC (2015). Hydrogen sulfide in physiology and diseases of the digestive tract. Microorganisms, 3:866-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sen N (2017). Functional and molecular insights of hydrogen sulfide signaling and protein sulfhydration. J Mol Biol, 429:543-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Roth W, Zadeh K, Vekariya R, Ge Y, Mohamadzadeh M (2021). Tryptophan metabolism and gut-brain homeostasis. Int J Mol Sci, 22:2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang J, Zhu SW, Ma N, Johnston LJ, Wu CD, Ma X (2021). Metabolites of microbiota response to tryptophan and intestinal mucosal immunity: A therapeutic target to control intestinal inflammation. Med Res Rev, 41:1061-1088. [DOI] [PubMed] [Google Scholar]
- [34].Modoux M, Rolhion N, Mani S, Sokol H (2021). Tryptophan metabolism as a pharmacological target. Trends Pharmacol Sci, 42:60-73. [DOI] [PubMed] [Google Scholar]
- [35].Roager HM, Licht TR (2018). Microbial tryptophan catabolites in health and disease. Nat Commun, 9:3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Agus A, Planchais J, Sokol H (2018). Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe, 23:716-724. [DOI] [PubMed] [Google Scholar]
- [37].Wang Z, Bergeron N, Levison BS, Li XS, Chiu S, Jia X, et al. (2019). Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur Heart J, 40:583-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang YX, Wang Y, Ke BB, Du J (2021). TMAO: how gut microbiota contributes to heart failure. Transl Res, 228:109-125. [DOI] [PubMed] [Google Scholar]
- [39].Yang SJ, Li XY, Yang F, Zhao R, Pan XD, Liang JQ, et al. (2019). Gut microbiota-dependent marker TMAO in promoting cardiovascular disease: inflammation mechanism, clinical prognostic, and potential as a therapeutic target. Front Pharmacol, 10:1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rudzki L, Stone T, Maes M, Misiak B, Samochowiec J, Szulc A (2021). Gut microbiota-derived vitamins-underrated powers of a multipotent ally in psychiatric health and disease. Prog Neuropsychopharmacol Biol Psychiatry, 107:110240. [DOI] [PubMed] [Google Scholar]
- [41].Stacchiotti V, Rezzi S, Eggersdorfer M, Galli F (2020). Metabolic and functional interplay between gut microbiota and fat-soluble vitamins. Crit Rev Food Sci Nutr, 1-22. [DOI] [PubMed] [Google Scholar]
- [42].Wang Y, Li N, Yang JJ, Zhao DM, Chen B, Zhang GQ, et al. (2020). Probiotics and fructo-oligosaccharide intervention modulate the microbiota-gut brain axis to improve autism spectrum reducing also the hyper-serotonergic state and the dopamine metabolism disorder. Pharmacol Res, 157:104784. [DOI] [PubMed] [Google Scholar]
- [43].Morais LH, Schreiber HL, Mazmanian SK (2021). The gut microbiota-brain axis in behaviour and brain disorders. Nat Rev Microbiol, 19:241-55. [DOI] [PubMed] [Google Scholar]
- [44].Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes, 56:1761-1772. [DOI] [PubMed] [Google Scholar]
- [45].Serino M, Luche E, Gres S, Baylac A, Bergé M, Cenac C, et al. (2012). Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut, 61:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Donia MS, Fischbach MA (2015). Small molecules from the human microbiota. Science, 349:1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Perruzza L, Gargari G, Proietti M, Fosso B, D’Erchia AM, Faliti CE, et al. (2017). T follicular helper cells promote a beneficial gut ecosystem for host metabolic homeostasis by sensing microbiota-derived extracellular ATP. Cell Rep, 18:2566-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mousa WK, Athar B, Merwin NJ, Magarvey NA (2017). Antibiotics and specialized metabolites from the human microbiota. Nat Prod Rep, 34:1302-1331. [DOI] [PubMed] [Google Scholar]
- [49].Russell WR, Gratz SW, Duncan SH, Holtrop G, Ince J, Scobbie L, et al. (2011). High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr, 93:1062-1072. [DOI] [PubMed] [Google Scholar]
- [50].Vijay N, Morris ME (2014). Role of monocarboxylate transporters in drug delivery to the brain. Curr Pharm Des, 20:1487-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT (1987). Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut, 28:1221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L (2014). The role of short-chain fatty acids in health and disease. Adv Immunol, 121:91-119. [DOI] [PubMed] [Google Scholar]
- [53].Gérard P (2014). Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens, 3:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jia W, Wei M, Rajani C, Zheng X (2021). Targeting the alternative bile acid synthetic pathway for metabolic diseases. Protein Cell, 12:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ikegami T, Honda A (2020). Reciprocal interactions between bile acids and gut microbiota in human liver diseases. J Hepatol, 72:558-577. [DOI] [PubMed] [Google Scholar]
- [56].Winston JA, Theriot CM (2020). Diversification of host bile acids by members of the gut microbiota. Gut Microbes, 11:158-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Smith EA, Macfarlane GT (1996). Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol, 81:288-302. [DOI] [PubMed] [Google Scholar]
- [58].Ussher JR, Lopaschuk GD, Arduini A (2013). Gut microbiota metabolism of L-carnitine and cardiovascular risk. Atherosclerosis, 231:456-461. [DOI] [PubMed] [Google Scholar]
- [59].van Kessel SP, Frye AK, El-Gendy AO, Castejon M, Keshavarzian A, van Dijk G, et al. (2019). Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nat Commun, 10:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Magnúsdóttir S, Ravcheev D, de Crécy-Lagard V, Thiele I (2015). Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet, 6:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wang L, Ravichandran V, Yin Y, Yin J, Zhang Y (2019). Natural products from mammalian gut microbiota. Trends Biotechnol, 37:492-504. [DOI] [PubMed] [Google Scholar]
- [62].Behsaz B, Bode E, Gurevich A, Shi YN, Grundmann F, Acharya D, et al. (2021). Integrating genomics and metabolomics for scalable non-ribosomal peptide discovery. Nat Commun, 12:3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kenshole E, Herisse M, Michael M, Pidot SJ (2021). Natural product discovery through microbial genome mining. Curr Opin Chem Biol, 60:47-54. [DOI] [PubMed] [Google Scholar]
- [64].Sun Y, O’Riordan MX (2013). Regulation of bacterial pathogenesis by intestinal short-chain fatty acids. Adv Appl Microbiol, 85:93-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Takeuchi T, Ohno H (2021). Reciprocal regulation of IgA and the gut microbiota: a key mutualism in the intestine. Int Immunol, dxab049. [DOI] [PubMed] [Google Scholar]
- [66].Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, et al. (2015). Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun, 6:6734. [DOI] [PubMed] [Google Scholar]
- [67].Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR, et al. (2018). Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol, 11:785-795. [DOI] [PubMed] [Google Scholar]
- [68].Smith NW, Shorten PR, Altermann E, Roy NC, McNabb WC (2019). The classification and evolution of bacterial cross-feeding. Front Ecol Evol, 7:153. [Google Scholar]
- [69].Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, et al. (2006). Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol, 72:3593-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Adamowicz EM, Flynn J, Hunter RC, Harcombe WR (2018). Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J, 12:2723-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR (2015). Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci U S A, 112:6449-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Maynard CL, Elson CO, Hatton RD, Weaver CT (2012). Reciprocal interactions of the intestinal microbiota and immune system. Nature, 489:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wilkins LJ, Monga M, Miller AW (2019). Defining dysbiosis for a cluster of chronic diseases. Sci Rep, 9:12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lee JH, Wood TK, Lee J (2015). Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol, 23:707-718. [DOI] [PubMed] [Google Scholar]
- [75].Garcia-Gutierrez E, Mayer MJ, Cotter PD, Narbad A (2019). Gut microbiota as a source of novel antimicrobials. Gut microbes, 10:1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Gutzeit C, Magri G, Cerutti A (2014). Intestinal IgA production and its role in host-microbe interaction. Immunol Rev, 260:76-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kim M, Qie Y, Park J, Kim CH (2016). Gut microbial metabolites fuel host antibody responses. Cell Host Microbe, 20:202-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, et al. (2006). Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A, 103:3920-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hill MJ (1997). Intestinal flora and endogenous vitamin synthesis. Eur J Cancer Prev, 6:S43-S45. [DOI] [PubMed] [Google Scholar]
- [80].Zeng SL, Li SZ, Xiao PT, Cai YY, Chu C, Chen BZ, et al. (2020). Citrus polymethoxyflavones attenuate metabolic syndrome by regulating gut microbiome and amino acid metabolism. Sci Adv, 6: eaax6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Skrypnik K, Suliburska J (2018). Association between the gut microbiota and mineral metabolism. J Sci Food Agric, 98:2449-2460. [DOI] [PubMed] [Google Scholar]
- [82].Whisner CM, Castillo LF (2018). Prebiotics, bone and mineral metabolism. Calcif Tissue Int, 102:443-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Mokdad AH, Ford ES, Bowman BA, Dietz WH, Vinicor F, Bales VS, et al. (2003). Prevalence of obesity, diabetes, and obesity-related health risk factors. JAMA, 289:76-79. [DOI] [PubMed] [Google Scholar]
- [84].Must A, Spadano J, Coakley EH, Field AE, Colditz G, Dietz WH (1999). The disease burden associated with overweight and obesity. JAMA, 282:1523-1529. [DOI] [PubMed] [Google Scholar]
- [85].Pollock N, Grogan C, Perry M, Pedlar C, Cooke K, Morrissey D, et al. (2010). Bone-mineral density and other features of the female athlete triad in elite endurance runners: a longitudinal and cross-sectional observational study. Int J Sport Nutr Exerc Metab, 20:418-426. [DOI] [PubMed] [Google Scholar]
- [86].Kurpad AV, Muthayya S, Vaz M (2005). Consequences of inadequate food energy and negative energy balance in humans. Public Health Nutr, 8:1053-1076. [DOI] [PubMed] [Google Scholar]
- [87].Moreno-Navarrete JM, Fernandez-Real JM (2019). The gut microbiota modulates both browning of white adipose tissue and the activity of brown adipose tissue. Rev Endocr Metab Disord, 20:387-397. [DOI] [PubMed] [Google Scholar]
- [88].Jocken JW, González Hernández MA, Hoebers NT, van der Beek CM, Essers YP, Blaak EE, et al. (2018). Short-chain fatty acids differentially affect intracellular lipolysis in a human white adipocyte model. Front Endocrinol, 8:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Li B, Li L, Li M, Lam SM, Wang G, Wu Y, et al. (2019). Microbiota depletion impairs thermogenesis of brown adipose tissue and browning of white adipose tissue. Cell Rep, 26:2720-2737. [DOI] [PubMed] [Google Scholar]
- [90].Priyadarshini M, Villa SR, Fuller M, Wicksteed B, Mackay CR, Alquier T, et al. (2015). An acetate-specific GPCR, FFAR2, regulates insulin secretion. Mol Endocrinol, 29:1055-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Pingitore A, Chambers ES, Hill T, Maldonado IR, Liu B, Bewick G, et al. (2017). The diet-derived short chain fatty acid propionate improves beta-cell function in humans and stimulates insulin secretion from human islets in vitro. Diabetes Obes Metab, 19:257-265. [DOI] [PubMed] [Google Scholar]
- [92].Frontera WR, Ochala J (2015). Skeletal muscle: a brief review of structure and function. Calcif Tissue Int, 96:183-195. [DOI] [PubMed] [Google Scholar]
- [93].Zurlo F, Larson K, Bogardus C, Ravussin E (1990). Skeletal muscle metabolism is a major determinant of resting energy expenditure. J Clin Invest, 86:1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Frampton J, Murphy KG, Frost G, Chambers ES (2020). Short-chain fatty acids as potential regulators of skeletal muscle metabolism and function. Nat Metab, 2:840-848. [DOI] [PubMed] [Google Scholar]
- [95].Carley AN, Maurya SK, Fasano M, Wang Y, Selzman CH, Drakos SG, et al. (2021). Short-chain fatty acids outpace ketone oxidation in the failing heart. Circulation, 143:1797-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Poll BG, Xu J, Jun S, Sanchez J, Zaidman NA, He X, et al. (2021). Acetate, a short-chain fatty acid, acutely lowers heart rate and cardiac contractility along with blood pressure. J Pharmacol Exp Ther, 377:39-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].den Besten G, Lange K, Havinga R, van Dijk TH, Gerding A, van Eunen K, et al. (2013). Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am J Physiol Gastrointest Liver Physiol, 305:G900-G910. [DOI] [PubMed] [Google Scholar]
- [98].Den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, et al. (2015). Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes, 64:2398-2408. [DOI] [PubMed] [Google Scholar]
- [99].Agus A, Clément K, Sokol H (2021). Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut, 70:1174-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Shapiro H, Kolodziejczyk AA, Halstuch D, Elinav E (2018). Bile acids in glucose metabolism in health and disease. J Exp Med, 215:383-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Molinaro A, Wahlström A, Marschall HU (2018). Role of bile acids in metabolic control. Trends Endocrinol Metab, 29:31-41. [DOI] [PubMed] [Google Scholar]
- [102].Cani PD, Van Hul M, Lefort C, Depommier C, Rastelli M, Everard A (2019). Microbial regulation of organismal energy homeostasis. Nat Metab, 1:34-46. [DOI] [PubMed] [Google Scholar]
- [103].Jonsson AL, Bäckhed F (2017). Role of gut microbiota in atherosclerosis. Nat Rev Cardiol, 14:79-87. [DOI] [PubMed] [Google Scholar]
- [104].Floyd JC, Fajans SS, Conn JW, Knopf RF, Rull J (1966). Stimulation of insulin secretion by amino acids. J Clin Invest, 45:1487-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Felig P, Marliss E, Cahill Jr GF (1969). Plasma amino acid levels and insulin secretion in obesity. N Engl J Med, 281:811-6. [DOI] [PubMed] [Google Scholar]
- [106].Fontana L, Cummings NE, Apelo SI, Neuman JC, Kasza I, Schmidt BA, et al. (2016). Decreased consumption of branched-chain amino acids improves metabolic health. Cell Rep, 16:520-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab, 9:311-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Tajiri K, Shimizu Y (2018). Branched-chain amino acids in liver diseases. Transl Gastroenterol Hepatol, 3:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chávez-Talavera O, Tailleux A, Lefebvre P, Staels B (2017). Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology, 152:1679-94. [DOI] [PubMed] [Google Scholar]
- [110].Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, et al. (2013). Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab, 17:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Zhu Y, Shui X, Liang Z, Huang Z, Qi Y, He Y, Chen C, et al. (2020). Gut microbiota metabolites as integral mediators in cardiovascular diseases. Int J Mol Med, 46:936-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Ðanić M, Stanimirov B, Pavlović N, Goločorbin-Kon S, Al-Salami H, Stankov K, et al. (2018). Pharmacological applications of bile acids and their derivatives in the treatment of metabolic syndrome. Front Pharmacol, 9:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Scaldaferri F, Pizzoferrato M, Gerardi V, Lopetuso L, Gasbarrini A (2012). The gut barrier: new acquisitions and therapeutic approaches. J Clin Gastroenterol, 46:S12-7. [DOI] [PubMed] [Google Scholar]
- [114].Liu W, Luo X, Tang J, Mo Q, Zhong H, Zhang H, et al. (2021). A bridge for short-chain fatty acids to affect inflammatory bowel disease, type 1 diabetes, and non-alcoholic fatty liver disease positively: By changing gut barrier. Eur J Nutr, 60:2317-30. [DOI] [PubMed] [Google Scholar]
- [115].Natividad JM, Verdu EF (2013). Modulation of intestinal barrier by intestinal microbiota: pathological and therapeutic implications. Pharmacol Res, 69:42-51. [DOI] [PubMed] [Google Scholar]
- [116].Wells JM, Brummer RJ, Derrien M, MacDonald TT, Troost F, Cani PD, et al. (2017). Homeostasis of the gut barrier and potential biomarkers. Am J Physiol Gastrointest Liver Physiol, 312:G171-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Camilleri M (2019). Leaky gut: mechanisms, measurement and clinical implications in humans. Gut, 68:1516-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Meddings J (2008). The significance of the gut barrier in disease. Gut, 57:438-40. [DOI] [PubMed] [Google Scholar]
- [119].König J, Wells J, Cani PD, García-Ródenas CL, MacDonald T, Mercenier A, et al. (2016). Human intestinal barrier function in health and disease. Clin Transl Gastroenterol, 7:e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Wang HB, Wang PY, Wang X, Wan YL, Liu YC (2012). Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig Dis Sci, 57:3126-35. [DOI] [PubMed] [Google Scholar]
- [121].Sunkara LT, Jiang W, Zhang G (2012). Modulation of antimicrobial host defense peptide gene expression by free fatty acids. PLoS One, 7:e49558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Jung TH, Park JH, Jeon WM, Han KS (2015). Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr Res Pract, 9:343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. (2015). Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol, 8:80-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Biagioli M, Carino A, Cipriani S, Francisci D, Marchianò S, Scarpelli P, et al. (2017). The bile acid receptor GPBAR1 regulates the M1/M2 phenotype of intestinal macrophages and activation of GPBAR1 rescues mice from murine colitis. J Immunol, 199:718-733. [DOI] [PubMed] [Google Scholar]
- [125].Di Lorenzo C, Youssef NN (2010). Diagnosis and management of intestinal motility disorders. Semin Pediatr Surg, 19:50-8. [DOI] [PubMed] [Google Scholar]
- [126].Rao M (2020). An increasingly complex view of intestinal motility. Nat Rev Gastroenterol Hepatol, 17:72-3. [DOI] [PubMed] [Google Scholar]
- [127].Verbeke KA, Boobis AR, Chiodini A, Edwards CA, Franck A, Kleerebezem M, et al. (2015). Towards microbial fermentation metabolites as markers for health benefits of prebiotics. Nutr Res Rev, 28:42-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Müller M, Canfora EE, Blaak EE (2018). Gastrointestinal transit time, glucose homeostasis and metabolic health: modulation by dietary fibers. Nutrients, 10:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Guarino MP, Cicala M, Putignani L, Severi C (2016). Gastrointestinal neuromuscular apparatus: An underestimated target of gut microbiota. World J Gastroenterol, 22:9871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Cherbut C, Aube AC, Blottiere HM, Galmiche JP (1997). Effects of short-chain fatty acids on gastrointestinal motility. Scand J Gastroenterol Suppl, 222:58-61. [DOI] [PubMed] [Google Scholar]
- [131].Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. (2012). Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep, 13:440-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med, 20:159-66. [DOI] [PubMed] [Google Scholar]
- [133].Thorburn AN, McKenzie CI, Shen SJ, Stanley D, Macia L, Mason LJ, et al. (2015). Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun, 6:7320. [DOI] [PubMed] [Google Scholar]
- [134].Trompette A, Gollwitzer ES, Pattaroni C, Lopez-Mejia IC, Riva E, Pernot J, et al. (2018). Dietary fiber confers protection against flu by shaping Ly6c- patrolling monocyte hematopoiesis and CD8+ T cell metabolism. Immunity, 48:992-1005. [DOI] [PubMed] [Google Scholar]
- [135].Dyson JK, Blain A, Shirley MD, Hudson M, Rushton S, Jones DE (2020). Geo-epidemiology and environmental co-variate mapping of primary biliary cholangitis and primary sclerosing cholangitis. JHEP Rep, 3:100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Allen K, Jaeschke H, Copple BL (2011). Bile acids induce inflammatory genes in hepatocytes: a novel mechanism of inflammation during obstructive cholestasis. Am J Pathol, 178:175-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Gujral JS, Farhood A, Bajt ML, Jaeschke H (2003). Neutrophils aggravate acute liver injury during obstructive cholestasis in bile duct-ligated mice. Hepatology, 38:355-63. [DOI] [PubMed] [Google Scholar]
- [138].Fiorucci S, Biagioli M, Zampella A, Distrutti E (2018). Bile acids activated receptors regulate innate immunity. Front Immunol, 9:1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].O'Brien KM, Allen KM, Rockwell CE, Towery K, Luyendyk JP, Copple BL (2013). IL-17A synergistically enhances bile acid-induced inflammation during obstructive cholestasis. Am J Pathol, 183:1498-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Bence KK, Birnbaum MJ (2021). Metabolic drivers of non-alcoholic fatty liver disease. Mol Metab, 50:101143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Deng MJ, Qu F, Chen L, Liu C, Zhang M, Ren FZ, et al. (2020). SCFAs alleviated steatosis and inflammation in mice with NASH induced by MCD. J Endocrinol, 245:425-37. [DOI] [PubMed] [Google Scholar]
- [142].Peng J, Xiao X, Hu M, Zhang X (2018). Interaction between gut microbiome and cardiovascular disease. Life Sci, 214:153-7. [DOI] [PubMed] [Google Scholar]
- [143].Ma G, Pan B, Chen Y, Guo C, Zhao M, Zheng L, Chen B (2017). Trimethylamine N-oxide in atherogenesis: impairing endothelial self-repair capacity and enhancing monocyte adhesion. Biosci Rep, 37:BSR20160244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Zhang H, Meng J, Yu H (2017). Trimethylamine N-oxide supplementation abolishes the cardioprotective effects of voluntary exercise in mice fed a western diet. Front Physiol, 8:944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Li X, Geng J, Zhao J, Ni Q, Zhao C, Zheng Y, et al. (2019). Trimethylamine N-oxide exacerbates cardiac fibrosis via activating the NLRP3 inflammasome. Front Physiol, 10:866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Li LS, Rao ST, Cheng YZ, Zhuo XY, Deng CH, Xu NN, et al. (2019). Microbial osteoporosis: The interplay between the gut microbiota and bones via host metabolism and immunity. Microbiologyopen, 8:e00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Behera J, Ison J, Tyagi SC, Tyagi N (2020). The role of gut microbiota in bone homeostasis. Bone, 135:115317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Lu LY, Chen XX, Liu Y, Yu XJ (2021). Gut microbiota and bone metabolism. FASEB J, 35:e21740. [DOI] [PubMed] [Google Scholar]
- [149].Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A, 106:3698-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Nallu A, Sharma S, Ramezani A, Muralidharan J, Raj D (2017). Gut microbiome in chronic kidney disease: challenges and opportunities. Transl Res, 179:24-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Yang W, Cong Y (2021). Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell Mol Immunol, 18:866-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Ferrus ML, Madrid N, et al. (2015). Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol, 8:760-72. [DOI] [PubMed] [Google Scholar]
- [153].Salem I, Ramser A, Isham N, Ghannoum MA (2018). The gut microbiome as a major regulator of the gut-skin axis. Front Microbiol, 9:1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Rosser EC, Piper CJ, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. (2020). Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells. Cell Metab, 31:837-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Fung TC, Olson CA, Hsiao EY (2017). Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci, 20:145-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Sharon G, Sampson TR, Geschwind DH, Mazmanian SK (2016). The central nervous system and the gut microbiome. Cell, 167:915-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Silva YP, Bernardi A, Frozza RL (2020). The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol, 11:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Carloni S, Bertocchi A, Mancinelli S, Bellini M, Erreni M, Borreca A, et al. (2021). Identification of a choroid plexus vascular barrier closing during intestinal inflammation. Science, 374:439-448. [DOI] [PubMed] [Google Scholar]
- [159].Grab DJ, Perides G, Dumler JS, Kim KJ, Park J, Kim YV, et al. (2005). Borrelia burgdorferi, host-derived proteases, and the blood-brain barrier. Infect Immun, 7:1014-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Li HX, Sun J, Wang FY, Ding GQ, Chen WQ, Fang RC, et al. (2016). Sodium butyrate exerts neuroprotective effects by restoring the blood-brain barrier in traumatic brain injury mice. Brain Res, 1642:70-78. [DOI] [PubMed] [Google Scholar]
- [161].Dalile B, Van Oudenhove L, Vervliet B, Verbeke K (2019). The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol, 16:461-478. [DOI] [PubMed] [Google Scholar]
- [162].Cryan JF, O'Riordan KJ, Cowan CS, Sandhu KV, Bastiaanssen TF, Boehme M, et al. (2019). The microbiota-gut-brain axis. Physiol Rev, 99:1877-2013. [DOI] [PubMed] [Google Scholar]
- [163].Perry VH, Newman TA, Cunningham C (2003). The impact of systemic infection on the progression of neurodegenerative disease. Nat Rev Neurosci, 4:103-12. [DOI] [PubMed] [Google Scholar]
- [164].Perry VH, Cunningham C, Holmes C (2007). Systemic infections and inflammation affect chronic neurodegeneration. Nat Rev Immunol, 7:161-7. [DOI] [PubMed] [Google Scholar]
- [165].Haghikia A, Jörg S, Duscha A, Berg J, Manzel A, Waschbisch A, et al. (2015). Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity, 43:817-29. [DOI] [PubMed] [Google Scholar]
- [166].Ochoa-Reparaz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, et al. (2010). A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol, 3:487-95. [DOI] [PubMed] [Google Scholar]
- [167].Dong Y, Benveniste EN (2001). Immune function of astrocytes. Glia, 36:180-90. [DOI] [PubMed] [Google Scholar]
- [168].Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, et al. (2016). Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med, 22:586-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Han H, Yi B, Zhong RQ, Wang MY, Zhang SF, Ma J, et al. (2021). From gut microbiota to host appetite: gut microbiota-derived metabolites as key regulators. Microbiome, 9:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, et al. (2013). The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun, 4:1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac-Varghese SE, et al. (2015). Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut, 64:1744-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Lerche S, Brock B, Rungby J, Bøtker HE, Møller N, Rodell A, et al. (2008). Glucagon-like peptide-1 inhibits blood-brain glucose transfer in humans. Diabetes, 57:325-31. [DOI] [PubMed] [Google Scholar]
- [173].Song Y, Koehler JA, Baggio LL, Powers AC, Sandoval DA, Drucker DJ (2019). Gut-proglucagon-derived peptides are essential for regulating glucose homeostasis in mice. Cell Metab, 30:976-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Kim ER, Wu Z, Sun H, Xu Y, Mangieri LR, Xu Y, et al. (2015). Hypothalamic non-AgRP, non-POMC GABAergic neurons are required for postweaning feeding and NPY hyperphagia. J Neurosci, 35:10440-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Xu J, Bartolome CL, Low CS, Yi X, Chien CH, Wang P, et al. (2018). Genetic identification of leptin neural circuits in energy and glucose homeostases. Nature, 556:505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Arnoriaga-Rodríguez M, Mayneris-Perxachs J, Burokas A, Pérez-Brocal V, Moya A, Portero-Otin M, et al. (2020). Gut bacterial ClpB-like gene function is associated with decreased body weight and a characteristic microbiota profile. Microbiome, 8:1-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Pearson JA, Wong FS, Wen L (2020). Crosstalk between circadian rhythms and the microbiota. Immunology, 161:278-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Segers A, Depoortere I (2021). Circadian clocks in the digestive system. Nat Rev Gastroenterol Hepatol, 18:239-51. [DOI] [PubMed] [Google Scholar]
- [179].Frazier K, Chang EB (2020). Intersection of the gut microbiome and circadian rhythms in metabolism. Trends Endocrinol Metab, 31:25-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Matenchuk BA, Mandhane PJ, Kozyrskyj AL (2020). Sleep, circadian rhythm, and gut microbiota. Sleep Med Rev, 53:101340. [DOI] [PubMed] [Google Scholar]
- [181].Lopez DE, Lashinger LM, Weinstock GM, Bray MS (2021). Circadian rhythms and the gut microbiome synchronize the host’s metabolic response to diet. Cell Metab, 33:873-87 [DOI] [PubMed] [Google Scholar]
- [182].Crnko S, Du Pré BC, Sluijter JP, Van Laake LW (2019). Circadian rhythms and the molecular clock in cardiovascular biology and disease. Nat Rev Cardiol, 16:437-47. [DOI] [PubMed] [Google Scholar]
- [183].Leng Y, Musiek ES, Hu K, Cappuccio FP, Yaffe K (2019). Association between circadian rhythms and neurodegenerative diseases. Lancet Neurol, 18:307-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Codoñer-Franch P, Gombert M (2018). Circadian rhythms in the pathogenesis of gastrointestinal diseases. World J Gastroenterol, 24:4297-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Sancar A, Van Gelder RN (2021). Clocks, cancer, and chronochemotherapy. Science, 371: eabb0738. [DOI] [PubMed] [Google Scholar]
- [186].Thaiss CA, Levy M, Korem T, Dohnalová L, Shapiro H, Jaitin DA, et al. (2016). Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell, 167:1495-510. [DOI] [PubMed] [Google Scholar]
- [187].Bishehsari F, Voigt RM, Keshavarzian A (2020). Circadian rhythms and the gut microbiota: from the metabolic syndrome to cancer. Nat Rev Endocrinol, 16:731-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, et al. (2015). Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe, 17:681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Segers A, Desmet L, Thijs T, Verbeke K, Tack J, Depoortere I (2019). The circadian clock regulates the diurnal levels of microbial short-chain fatty acids and their rhythmic effects on colon contractility in mice. Acta Physiol, 225:e13193. [DOI] [PubMed] [Google Scholar]
- [190].Kuang Z, Wang Y, Li Y, Ye C, Ruhn KA, Behrendt CL, et al. (2019). The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science, 365:1428-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Selmer T, Andrei PI (2001). P-Hydroxyphenylacetate decarboxylase from Clostridium difficile: A novel glycyl radical enzyme catalysing the formation of p-cresol. Eur J Biochem, 268:1363-72. [DOI] [PubMed] [Google Scholar]
- [192].Bone E, Tamm A, Hill M (1976). The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am J Clin Nutr, 29:1448-54. [DOI] [PubMed] [Google Scholar]
- [193].Hodgman MJ, Garrard AR (2012). A review of acetaminophen poisoning. Crit Care Clin, 28:499-516. [DOI] [PubMed] [Google Scholar]
- [194].Laine JE, Auriola S, Pasanen M, Juvonen RO (2009). Acetaminophen bioactivation by human cytochrome P450 enzymes and animal microsomes. Xenobiotica, 39:11-21. [DOI] [PubMed] [Google Scholar]
- [195].Li J, Chiew AL, Isbister GK, Duffull SB (2021). Sulfate conjugation may be the key to hepatotoxicity in paracetamol overdose. Br J Clin Pharmacol, 87:2392-2396. [DOI] [PubMed] [Google Scholar]
- [196].Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, et al. (2006). Human sulfotransferases and their role in chemical metabolism. Toxicol Sci, 90:5-22. [DOI] [PubMed] [Google Scholar]
- [197].Raj GM, Raveendran Rs (2019). Introduction to Basics of Pharmacology and Toxicology. 1st ed. Singapore, Springer Nature. [Google Scholar]
- [198].Xie QS, Zhang JX, Liu M, Liu PH, Wang ZJ, Zhu L, et al. (2021). Short-chain fatty acids exert opposite effects on the expression and function of p-glycoprotein and breast cancer resistance protein in rat intestine. Acta Pharmacol Sin, 42:470-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Tolson AH, Wang H (2010). Regulation of drug-metabolizing enzymes by xenobiotic receptors: PXR and CAR. Adv Drug Deliv Rev, 62:1238-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Dempsey JL, Cui JY (2019). Microbiome is a functional modifier of P450 drug metabolism. Curr Pharmacol Rep, 5:481-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Shanahan F, Kiely B (2007). The gut microbiota and disease-an inner repository for drug discovery. Drug Discov Today Ther Strateg, 4:195-200. [Google Scholar]
- [202].Manohar P, Loh B, Athira S, Nachimuthu R, Hua X, Welburn SC, Leptihn S (2020). Secondary bacterial infections during pulmonary viral disease: phage therapeutics as alternatives to antibiotics? Front Microbiol, 11:1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Mousa WK, Athar B, Merwin NJ, Magarvey NA (2017). Antibiotics and specialized metabolites from the human microbiota. Nat Prod Rep, 34:1302-31. [DOI] [PubMed] [Google Scholar]
- [204].Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM (2017). Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol, 14:356-65. [DOI] [PubMed] [Google Scholar]
- [205].Cheng H, Liu J, Tan Y, Feng WW, Peng C (2021). Interactions between gut microbiota and berberine, a necessary procedure to understand the mechanisms of berberine. [J] Pharm Anal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Kuntz TM, Gilbert JA (2017). Introducing the microbiome into precision medicine. Trends Pharmacol Sci, 38:81-91. [DOI] [PubMed] [Google Scholar]
- [207].Baldi S, Menicatti M, Nannini G, Niccolai E, Russo E, Ricci F, et al. (2021). Free fatty acids signature in human intestinal disorders: Significant association between butyric acid and celiac disease. Nutrients, 13:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Niccolai E, Baldi S, Ricci F, Russo E, Nannini G, Menicatti M, et al. (2019). Evaluation and comparison of short chain fatty acids composition in gut diseases. World J Gastroenterol, 25:5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Emmi G, Bettiol A, Niccolai E, Ramazzotti M, Amedei A, Pagliai G, et al. (2021). Butyrate-rich diets improve redox status and fibrin lysis in Behçet’s syndrome. Circ Res, 12:278-80. [DOI] [PubMed] [Google Scholar]
- [210].Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, et al. (2015). Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell, 163:1585-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, et al. (2017). Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe, 21:603-10. [DOI] [PMC free article] [PubMed] [Google Scholar]