Abstract
Mannases have industrial uses in food and pulp industries, and their regulation may influence development of the mushrooms of commercially important basidiomycetes. We expressed an Agaricus bisporus cel4 cDNA, which encodes a mannanase, in Saccharomyces cerevisiae and Pichia pastoris. CEL4 had no detectable activity on cellulose or xylan. This gene is the first isolated from this economically important fungus to encode a mannanase. P. pastoris secreted about three times more CEL4 than S. cerevisiae. The removal of the cellulose-binding domain of CEL4 lowered the secreted specific activity by P. pastoris by approximately 97%. The genomic sequence of cel4 was isolated by screening a cosmid library of A. bisporus C54-carb8. The open reading frame was interrupted by 12 introns. The level of extracellular CEL4 increases dramatically at the postharvest stage in compost extracts of A. bisporus fruiting cultures. In laboratory liquid cultures of A. bisporus, the activity of CEL4 detected in the culture filtrate reached a maximum after 21 days. The levels of CEL4 broadly mirrored the levels of enzyme activity. In the Solka floc-bound mycelium, CEL4 protein showed a maximum after 2 to 3 weeks of culture and then declined. Changes in CEL4 activity during fruiting-body development suggest that hemicellulose utilization plays an important role in sporophore formation. The availability of the cloned gene will further studies of compost decomposition and the extracellular enzymes that fungi deploy in this process.
β-Mannanase enzymes (endo-1,4-β-d-mannanase, mannan endo-1,4-β-mannosidase; EC 3.2.1.78) are endohydrolases that catalyze the random hydrolysis of β-1,4-mannosidic linkages in the main chain of galactomannan, glucomannan, galactoglucomannan, and mannan (20). These enzymes are of particular interest in ligninolytic fungi, where there is a complex interplay between the processes of degradation of lignin, cellulose, and hemicellulose (including the mannans). There are possible biotechnological applications. β-Mannanase can be used to bleach pulp, to reduce the viscosity of instant coffee, and for the clarification of fruit juices and wines (35). At the same time, the role of hemicellulases in wood decay and the breakdown of leaf litter remains poorly understood. The cultivated mushroom Agaricus bisporus, a ligninolytic leaf litter degrader (8), is the source of the mannanase described in this paper.
The deduced CEL4 amino acid sequence shows that the encoded protein has a modular structure (40). There is a signal peptide at the N terminus, typical of proteins that are secreted into the medium, a catalytic domain, and a linker rich in serine, proline, and threonine that separates a cellulose-binding domain from the catalytic domain. The catalytic domain of CEL4 had the most amino acid sequence similarity with ascomycete mannanases from Aspergillus aculeatus (5) and Trichoderma reesei (28), which belong to glycosyl hydrolase family 5 (43 and 42%, respectively). Therefore, based on amino acid similarities of the catalytic domains, CEL4 also belongs to family 5 (11). Both CEL4 and the mannanase from T. reesei contain a cellulose-binding domain but at opposite ends (N proximal and C terminal, respectively, of the protein). cel4 expression is regulated at the transcriptional level (40).
The preferred substrate for growth of A. bisporus is compost. Utilization of the compost requires the production of a number of extracellular enzymes that degrade lignocellulosic components. Studies of axenic compost cultures during growth and fruiting of A. bisporus found large changes in laccase, cellulase, and xylanase levels during fruit body development (34, 38).
Our objectives in this study were (i) to determine the genomic sequence of cel4, (ii) to express CEL4 in Saccharomyces cerevisiae and Pichia pastoris to determine its enzymatic activity, (iii) to determine the effect of the cellulose-binding domain on CEL4 activity, and (iv) to determine if CEL4 is regulated during fungal development. This work may lead to the utilization of CEL4 in the food or pulp industries and to its manipulation for crop improvement.
MATERIALS AND METHODS
Strains and culture conditions.
We isolated cel4 from a genomic library of the carboxin-resistant A. bisporus strain C54-carb8 (18, 26). cel4 cDNA from A. bisporus strain D649 (40) was used for the yeast heterologous expression experiments.
Mycelia growing on compost harvested at different stages of the fruiting cycle of A. bisporus strain U3 were obtained from Horticulture Research International (Wellsbourne, United Kingdom). The cultures were grown axenically on sterilized compost as described by Burton et al. (3). The mycelium-compost samples were stored at −70°C. Compost samples were taken from the following stages: stage 1, colonization, during which the mycelium has fully colonized the compost; stage 2, initial aggregation, the beginning of the fruiting stage with sporophores or fruit bodies being 2 to 5 mm in diameter; stage 3, the pinning stage, in which fruit bodies are about 1 cm in height; stage 4, the button stage, in which fruit bodies are about 2.5 cm in height but still closed; stage 5, the veil break, during which the veil opens and the spores are shed; stage 6, postharvest, taken after fruit bodies have been harvested from the first flush and before the emergence of the second flush; stage 7, senescence, an alternative to stage 6 during which fruit bodies are allowed to complete their development in situ and none are harvested.
Soluble protein was extracted from compost-mycelium by immersing 10 g of frozen sample in 100 ml of ice-cold sterile deionized water and stirred for 1 h at 4°C. The solution was centrifuged twice for 20 min at 2,700 × g at 4°C. The supernatant was stored in aliquots at −70°C.
For the laboratory liquid cultures, static cultures of A. bisporus D649 in 2% (wt/vol) malt extract medium (50 ml in 250-ml conical flasks) were inoculated with agar cubes (0.5 by 0.5 by 0.5 cm) cut from malt extract plate cultures to include the colony margin and incubated at 25°C in the dark. After 7 days of growth, the A. bisporus mycelium was homogenized by shaking the samples with sterile glass beads (4-mm diameter); 1-ml amounts were used to inoculate 50 ml (in 250-ml flasks) of Treschow's minimal medium (29) containing 0.5 g of d-fructose per liter (19), and the flasks were incubated at 25°C in the dark. After 14 days of growth, 1 ml of fructose-grown fragmented mycelium inoculant was transferred into 100 ml (in Roux bottles) of either fresh d-fructose medium (0.5 g/liter) or Solka floc BW40 (0.5 g/liter) (International Filler Corporation, North Tonawanda, N.Y.) as the sole carbon source. Cultures were harvested for analysis (typically two or three bottles for each time point) at weekly intervals for 10 weeks. Mycelium was harvested by filtration through nylon gauze (pore size, 2 mm), washed three times with 25 mM Tris-HCl, pH 6.5, and recovered by centrifugation at 20,000 × g for 5 min. The wet weights of the mycelial samples were determined. Both the supernatant culture filtrates and mycelium were stored at −70°C until required.
S. cerevisiae Invsc1 (MATα his-Δ1 leu2 trp1-289 URA3-520 [Invitrogen, San Diego, Calif.]) was used as the host for heterologous expression of cel4. The expression vector used was derived from pYES2 (Invitrogen), which contains the URA3 marker gene and the 2μm circle origin of replication. Growth conditions were according to the work of Chow et al. (4). The P. pastoris host strain was KM71 (Muts Arg+ His−) (Invitrogen). The expression vector was pPICZαA (supplied with the Pichia expression kit; Invitrogen), which has an α-factor secretion signal for the efficient secretion of recombinant enzymes. P. pastoris recombinants were grown on minimal medium with histidine (MMH), containing, per liter, 13.4 g of yeast nitrogen base with ammonium sulfate but without amino acids (Difco, Detroit, Mich.), 0.4 mg of biotin, 15 g of agar, 5 ml of methanol, and 4 mg of histidine; buffered glycerol complex (BMGY), containing 10 g of yeast extract, 20 g of peptone, 100 mM potassium phosphate buffer (pH 6.0), 13.4 g of yeast nitrogen base, 0.4 mg of biotin, and 10 ml of glycerol; and buffered minimal methanol medium (BMMH), containing the same ingredients as BMGY except that 5 ml of methanol per liter was used in place of glycerol. Both P. pastoris and S. cerevisiae were maintained on YPD (per liter, 10 g of yeast extract, 20 g of peptone, 20 g of dextrose). P. pastoris cultures were grown in BMGY according to the manufacturer's instructions to an optical density of 2 and then transferred into BMMY expression medium and grown for another 2 days. The culture filtrates were aliquoted and stored at −70°C.
Escherichia coli strains XL-1 Blue (Stratagene, La Jolla, Calif.), Top 10F′ (Invitrogen), and BL21(DE3) (Invitrogen) were used for recombinant-DNA manipulations.
Recombinant-DNA techniques and enzymes.
cel4 cDNA (pEYc1200) was previously isolated from a library in λZAP (40). Standard DNA manipulations were carried out essentially as described by Sambrook et al. (25). Restriction enzymes and other enzymes used for DNA manipulations were purchased from Promega (Madison, Wis.) or New England Biolabs (Beverly, Mass.).
Sequencing of cel4 genomic sequence.
Screening of cel4 from A. bisporus strain C54-carb8 (26) was according to the method of Yagüe et al. (39). A 32P-random primer-labeled HindIII-NotI 1.3-kb cel4 cDNA fragment from pEYc1200 consisting of only the catalytic domain was used as a probe. The cel4 genomic sequence was obtained by directly sequencing from the cosmid (ABI sequencer 377; University of Durham, Durham, United Kingdom). Cosmid clone 31C1 was the only clone that hybridized to the cel4 cDNA probe. The sequence from 553 bp upstream of the start codon to 308 bp downstream of the stop codon (2,853 bp) was sequenced (both strands) by using specific primers, with the following exception. The sequence from 974 to 2853 bp was subcloned into pBluescript KS(+) (Stratagene) as a 1.8-kb XbaI fragment, which enabled standard primers (T7 and T3) to be used for sequencing of the end regions of this fragment.
Prediction of O-glycosylation sites.
Putative O-glycosylation sites in the predicted CEL4 amino acid sequence were found by using the program NetOGly 2.0 (10), which was obtained from the internet site http://www.cbs.dtu.dk/databases/OGLYCBASE/.
Production of CEL4 polyclonal antibody.
We also expressed cel4 in an E. coli expression vector, pT79 (a derivative of pMW172 [33]), so that the polypeptide could be used as an immunogen. A 1.1-kb cel4 cDNA fragment specifying only the catalytic domain (252 to 1,342 bp) was synthesized by PCR. The cellulose-binding domain and linker region were not included since exclusion of this part of the protein, which is very similar to CEL1, CEL2, and CEL3, was expected to elicit an antiserum that was more specific to CEL4. The primers were the Sfi-cel4 primer, 5′-CGATCGGCCGACGTGGCCGTGTCGACCGGATTT-3′, and the Not-cel4 primer, 5′-CGTTCGCGGCCGCCGTGATCAACAATA-3′. The restriction sites are underlined, and the nucleotides (in bold) are complementary to the cel4 catalytic domain sequence. The recombinant protein was produced in E. coli BL21(DE3) as inclusion bodies that allowed a one-step purification by step gradient centrifugation (2). Antibodies against CEL4 were raised in a New Zealand White rabbit by immunization with 600 μg (500 μl) of the purified inclusion body proteins, which were emulsified with an equal volume of Speecol adjuvant (Id-dlo; Institute for Animal Science and Health, Lelystad, The Netherlands). Boosters of 150 μg of protein were given approximately every 2 weeks. The maximum titer (by enzyme-linked immunosorbent assay) was found at 13 weeks. A high dilution of the antiserum was required in the enzyme-linked immunosorbent assay, which indicates that the antiserum has a high affinity for the antigen (CEL4 secreted in P. pastoris). Serum collected prior to immunization was used as a control.
Construction of the CEL4 yeast recombinants.
Splicing by overlap extension PCR (13) was used to splice an S. cerevisiae triose phosphate isomerase promoter and SUC2 invertase secretion peptide to cel4. The yeast promoter and secretion peptide were subcloned into pBluescript (4). These clones include the full-length sequence of cel4, except that the sequence up to and including most of the secretion peptide was removed. The flanking primers used were T3 (Stratagene) and CM5 5′-CGTGATCATCTAGACTAATTCAAGCCCGG-3′, which incorporates an XbaI restriction site (underlined) and is cel4 specific (in bold). The overlap primers used were CM1, 5′-GCAGCCAAAATAGCCGATGTTCCAGTCTGG-3′, and CM2, 3′-CGTCGGTTTTATAGACGGCTACAAGGTCAGACC-5′ (the cel4 sequence is highlighted in bold). The T3 and CM2 primers were used to amplify the yeast promoter and secretion peptide fragment (1 kb), and the CM1 and CM5 primers were used to amplify the cel4 sequence (1.3 kb, nucleotides 63 to 1341). The PCR conditions used were 95°C for 20 s, 52°C for 20 s, and 74°C for 90 s for 20 cycles; 10 ng of cDNA template was used in each reaction mixture. The splicing by overlap extension PCR conditions used were 95°C for 1 min, 58°C for 30 min, and 74°C for 30 s for 20 cycles; 100 ng of each of the two PCR products was used with the flanking primers. An XhoI site upstream of the yeast promoter in the multiple cloning site of pBluescript and the XbaI restriction site (incorporated into the primer) were used to directionally clone the spliced construct into the pYES expression vector. For the cloning of cel4 into the P. pastoris shuttle vector, the cel4 cDNA (nucleotides 63 to 1320) was amplified with the following primers: XhPcel4F (5′-GGCGCGCTCGAGAAGAGAGATGTTCCAGTCTGGGG-3′) and XhPcel4R (5′-GGAGGGCGTTCTAGAGCCCGGTTTTTCATTGCAG-3′) (cel4 sequence is in bold). Primer XhPcel4F introduces an XhoI site (underlined) upstream of the cel4 sequence and primer XhPcel4R introduces an XbaI site (underlined) downstream of cel4. P. pastoris was the host used to express truncated cel4 (i.e., without the cellulose-binding domain). The 1.1-kb fragment (nucleotides 171 to 1320) encoding the linker and catalytic domain was amplified using the following primers: CEL4FCBD (5′-GGCGCGCTCGAGAAGAGACCTGGATCAACAACT-3′) (the XhoI restriction site is underlined, and the cel4 sequence is in bold) and the XhPcel4R primer.
Activity measurements and Western analysis.
Qualitative activity determinations by Congo Red staining were according to the method of Pentilä et al. (22), and the hydrolysis halos were quantified with an image analyzer (EASY Plus enhanced analysis system; Herolab, Wiesloch, Germany) running EASY Plus version 4.16 software. Culture filtrates were dialyzed against 50 mM sodium acetate, pH 5, overnight prior to enzymatic assays. The Nelson-Somogyi assay was used to make the quantitative measurements (as increases in reducing groups) essentially according to the methods of Nelson (21) and Somogyi (27). CEL4 activity was assayed with the following substrates: 1% (wt/vol) locust bean gum (LBG) (Sigma, St. Louis, Mo.), 1% (wt/vol) xylan (Sigma), 1% (wt/vol) carboxymethyl cellulose (CMC) (BDH, Toronto, Canada), and 0.1% (wt/vol) barley β-1,3-glucan (Sigma). Protein concentrations were determined using the modified Lowry assay (12) and the Bradford assay according to the instructions of the manufacturer (Bio-Rad, Hercules, Calif.). Protein samples were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis using 10% (wt/vol) polyacrylamide gels containing 0.1% (wt/vol) SDS according to the method of Laemmli (16). Either the gels were stained directly using Coomassie brilliant blue R250 or separated proteins were electroblotted onto nitrocellulose (Hybond-C; Amersham, Little Chalfont, Buckinghamshire, United Kingdom) as described by Zamanian and Mason (42). Standard protein molecular mass markers were obtained from Novex (San Diego, Calif.). Western blots were probed with anti-CEL4 antiserum and horseradish peroxidase-labeled anti-rabbit F(ab′)2 goat secondary antibody (Sigma). CEL4 was visualized with a horseradish peroxidase conjugate substrate detection kit as described by the supplier (Bio-Rad). The CEL4 antiserum was used to detect CEL4 protein (i) secreted by S. cerevisiae and P. pastoris recombinants, (ii) in the compost extracts, and (iii) in the mycelium and culture filtrates of A. bisporus grown in minimal medium containing fructose or Solka floc. The intensities of the bands from the Western blots from the compost extracts and minimal medium experiments were quantified by using an image analyzer.
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the EMBLNEW database under accession number AJ271862.
RESULTS
Isolation and sequencing of the cel4 gene.
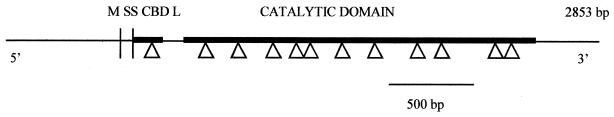
We isolated one recombinant clone from the cosmid genomic DNA library from A. bisporus after screening with a cel4 cDNA low-stringency probe. This clone, when digested with HindIII, gave a single positive band of 1.8 kb, suggesting that A. bisporus contains a single copy of cel4 and no other mannanases with similar sequences. We designed primers to sequence out from the flanking regions of the cel4 cDNA. Primers were then designed based on those sequences but reading inwards. For the sequence reported, both strands were analyzed. We found 12 short (46- to 65-bp) introns (Fig. 1). These introns had the consensus GTNN(G/T/A)-5′ and (A/C/T)AG-3′ sequences that are commonly found in fungi (32). The fifth and sixth introns delimited an exon of only 12 bp. A putative TATA box sequence, TATAAAA, was found 52 bp upstream from the start codon. The TATA box region is not GC rich. There are five putative CAAT boxes in both orientations in the 550 bp upstream of the start codon of cel4 286 and in the 382 bp (sense strand) and 206, 291, and 532 bp (antisense strand) upstream. Putative CAAT boxes are present in all A. bisporus genes sequenced to date. No sequence resembling the consensus eukaryotic polyadenylation sites was found (14), but the transcribed 3′ noncoding region may extend beyond the 308 bp sequenced from this region.
FIG. 1.
Genomic organization of cel4. The predicted signal peptide (SS) is at Met-1 to Ala-19. The putative cellulose-binding domain (CBD) is at Asp-20 to Leu-55, and the putative catalytic domain is at Gly-86 to Ala-439 (thick lines). The domains are separated by a proline/serine/threonine-rich linker (L). M represents the start codon. The presence of 12 introns (triangles) was deduced by comparison of cDNA and genomic sequences.
There were 20 predicted O-glycosylation sites in CEL4. The region from amino acid residues 60 to 80 is predicted to be very heavily O glycosylated, which is consistent with the hypothesis that this region functions as a linker. There also are predicted glycosylation sites around amino acid residues 160 and 260 in the catalytic core region. Glycosylation of cel4 has not been verified; however, Western blots detecting CEL4 in A. bisporus laboratory culture filtrates and in A. bisporus-colonized compost have shown that its molecular weight is significantly higher than the calculated value for the polypeptide alone (data not shown). O glycosylation probably accounts for this difference, as there are no N-glycosylation motifs in CEL4. The sequence of cel4 cDNA and genomic DNA differed by 16 nucleotides, which result in seven changes to the amino acid sequence of CEL4.
Heterologous expression of cel4 in S. cerevisiae and P. pastoris.
S. cerevisiae containing the cel4 gene produced clear halos on LBG plates, indicating that β-mannanase was secreted and was enzymatically active. The parent vector (control) did not produce a clear halo, and no halos were seen on plates containing 1% (wt/vol) xylan or 1% (wt/vol) CMC in place of LBG. Therefore, no xylanase or endoglucanase activities were detected. With P. pastoris, the host-vector controls secreted significant endoglucanase activity, such that any minor activity of the CEL4 protein would have been masked. The P. pastoris strains transformed with cel4 produced clear halos on the LBG plate, however, and the parent vector (control) did not. We made reducing sugar assays of the mannanase activity of CEL4 in the P. pastoris and S. cerevisiae culture filtrates and pellets (Table 1). CEL4 from which the cellulose-binding domain had been removed was expressed in P. pastoris. Its activity was less than for the unmodified protein (Table 1).
TABLE 1.
Mannanase activity of CEL4 secreted by S. cerevisiae and P. pastoris recombinants
| Mannanase source from vector host filtrate or pellet | Total vol (ml) | Total amt of protein (mg) | Mannanase activity (glucose equivalents [μmol/h/ml]) | Mannanase sp act (μmol/h/mg of protein) |
|---|---|---|---|---|
| Parent vector in S. cerevisiae culture filtratec | 100 | 20 | NDd | |
| CEL4 in S. cerevisiae culture filtratec | 100 | 30 | 0.15 ± 0.03 | 0.48 |
| CEL4 in S. cerevisiae pelleta | 0.35 ± 0.01 | |||
| Parent vector in P. pastoris culture filtratebc | 10 | 20 | ND | |
| CEL4 in P. pastoris culture filtratebc | 10 | 20 | 3.3 ± 0.44 | 1.7 |
| CEL4 in P. pastoris pelleta | 0.54 ± 0.01 | |||
| CEL4 lacking the cellulose-binding domain in P. pastoris culture filtratebc | 10 | 20 | 0.10 ± 0.014 | 0.05 |
Cell pellets were broken with a French press, and measurements were done in triplicate.
P. pastoris cultures were initially 100 ml and were resuspended in 10 ml of fresh medium prior to induction.
Culture filtrate activities were measured in duplicate from two independent experiments.
ND, not detectable because the mannose activity was less than 70 nmol/h/ml.
Regulation of CEL4 in fruiting cultures of A. bisporus compost extracts.
Western blots of the various compost extract stages were probed with CEL4 antiserum, with laccase antiserum used as a control (equal volumes of extracts containing approximately 0.3 μg of protein per μl were used). Mannanase activities from Nelson-Somogyi assays and Congo Red assays of the compost extracts from different developmental stages were also measured (Table 2). The activities from the Congo Red plate assays were determined from the areas of the hydrolysis halos using an image analyzer (Table 2).
TABLE 2.
Levels of CEL4 protein and mannanase activity detected in the developmental stages of A. bisporus compost extractsc
| Developmental stage | Mannanase activity from reducing sugar assays (μmol/h/ml of reducing sugar released) | Normalized intensity valuea for:
|
||
|---|---|---|---|---|
| Mannanase activity from plate assays | Bound CEL4 antibody | Bound laccase antibody (control) | ||
| Colonization | 0.60 ± 0.12 | 0.23 ± 0.05 | NDb | 1.0 ± 0.0 |
| Initial aggregation | 0.52 ± 0.08 | 0.23 ± 0.09 | 0.07 ± 0.02 | 1.08 ± 0.09 |
| Pinning | 0.68 ± 0.0 | 0.22 ± 0.11 | 0.08 ± 0.02 | 0.97 ± 0.06 |
| Button | 0.72 ± 0.08 | 0.23 ± 0.06 | 0.19 ± 0.12 | 1.03 ± 0.05 |
| Veil break | 1.3 ± 0.48 | 0.88 ± 0.24 | 0.41 ± 0.07 | 0.46 ± 0.13 |
| Postharvest | 2.4 ± 0.48 | 1.0 ± 0.15 | 1.0 ± 0.0 | ND |
| Senescence | 1.4 ± 0.36 | 0.57 ± 0.09 | 0.83 ± 0.11 | ND |
For comparative purposes, normalized values have been used; each data set has its largest value set to 1.0.
ND, not detectable.
Experiments were repeated on three separate occasions. Values are means ± standard errors.
The mannanase activity (Table 2) broadly correlated with the level of CEL4 protein throughout the fruiting cycle. The mycelium-bound compost used in this assay was from axenic cultures, so the activity measured was solely from A. bisporus. The level of CEL4 protein was very low until the veil-break stage, reaching a maximum at the postharvest stage. Less activity accumulated if mushrooms were not harvested (senescent sample). The presence of significant mannanase activity in the initial developmental stages prior to detection of an approximately 60-kDa predominant band on Western blots suggests that at least one other mannanase is produced by A. bisporus during growth on compost.
Regulation of CEL4 secretion in A. bisporus laboratory cultures.
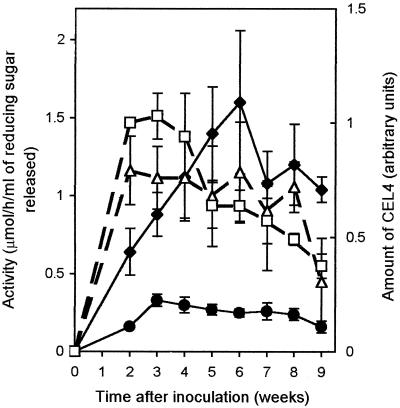
We compared cellulase and mannanase activities from the laboratory liquid culture filtrates of A. bisporus grown in the presence of Solka floc (Fig. 2). These data were compiled from the averages of four (cellulase) and five (mannanase) measurements made on separate occasions. Time zero was the point at which mycelium was transferred from fructose-containing medium. The levels of cellulase and mannanase activity rose to a peak and subsequently declined. In the case of cellulase, the activity rose steadily to a maximum at 42 days and then dropped steadily, which is consistent with previously published data (19). Mannanase activity peaked at 21 days and then dropped more gradually. The highest level of cellulase activity is about five times more than the highest level of mannanase activity.
FIG. 2.
Levels of CEL4 protein and mannanase activity in liquid cultures of A. bisporus (D649 strain) containing 0.05% (wt/vol) Solka floc grown at 25°C. Nelson-Somogyi assays were used to measure cellulase (⧫, CMC as the substrate) and mannanase (●, LBG as the substrate) activities. The levels of CEL4 in Solka floc-bound mycelium (□) and in culture filtrates (▵) were detected by CEL4 antiserum in Western blots. Error bars indicate standard errors of activities measured on five and four separate occasions for mannanase and cellulase, respectively. Western blots were repeated four and seven times for mycelium and culture filtrates, respectively. The amount of CEL4 protein from culture fluid was approximately 10-fold more than that found in the mycelium- and cellulose-bound fraction in the 3-week-old sample.
We also compared the amount of CEL4 in Solka floc-bound mycelium with that secreted into the culture filtrate (Fig. 2). The amount of CEL4 detected in the Solka floc-bound mycelium was about 20% of the total activity except in the later samples (where a large proportion of the cellulose would have been degraded).
DISCUSSION
The cel4 genomic DNA and the cel4 cDNA (accession number Z50095) differ at 16 of 1,432 nucleotides. These differences change seven amino acid residues, of which two were not changes in amino acids with functional differences. These differences might be because different alleles were cloned but may also represent differences at more than one locus. cel4 has 12 introns ranging from 46 to 65 bp. The other hemicellulase gene sequenced from A. bisporus, xylanase (accession number X78330 [7]), also has a large number of introns (11), while the cellulase genes do not (4, 24). The Congo Red plate assays and reducing sugar assays showed that CEL4 protein is a specific mannanase that, when expressed in S. cerevisiae, has no detectable activity against CMC, xylan, or barley β-glucan. The specific activity of CEL4 secreted by P. pastoris was approximately three times that secreted by S. cerevisiae. About 2.5 times more of the mannanase activity was found in the S. cerevisiae recombinant cell extracts than in the culture fluid. In P. pastoris, about six times more activity was found in the culture filtrate than in the cell extracts. Therefore, P. pastoris is much better at secreting CEL4 into the culture medium, presumably because P. pastoris has a more powerful inducible promoter and secretion peptide (α-factor pre-pro secretion signal from S. cerevisiae) than does S. cerevisiae (6).
The molecular mass of mature CEL4 deduced from the nucleotide sequence is 49 kDa; however, the apparent molecular mass of CEL4 of both the S. cerevisiae and P. pastoris cultures from the Western blot was approximately 60 kDa, probably due to glycosylation (results not shown). Hyperglycosylation has been observed with CEL3 of A. bisporus and two endoglucanases of T. reesei when they were expressed in S. cerevisiae (4, 22).
There is no evidence that CEL4 is a low-level- or mixed-specificity enzyme, and we do not know why some hemicellulases, e.g., CEL4, contain specific cellulose-binding domains and others, e.g., xylanase from A. bisporus (7), do not. Removal of the cellulose-binding domain of CEL4 reduced the specific activity significantly relative to that of full-length CEL4. Removal of the cellulose-binding domain has little influence on the activities of cellulases towards soluble substrates but decreases their activities towards insoluble cellulose (17).
A. bisporus is cultivated on composted wheat straw and can degrade lignin, cellulose, hemicellulose, protein, and microbial cell wall polymers (1, 3, 4, 7, 8, 9, 19, 23, 34, 41). Laccase accumulates in compost during its colonization by mycelium of A. bisporus, and the level of laccase rapidly decreases at the time of fruiting (30). Cellulase (31) increases during fruiting. The large increase in mannanase activity in both postharvest and senescent cultures contrasts with the changes observed in other extracellular enzymes and has no obvious explanation. Mannanase may be required to replenish stored carbohydrate levels in the mycelium. Mannanase activity is detectable at all stages, however, so mannan hydrolysis may be a source of carbon and energy even during colonization, although it is unlikely that this activity is due to CEL4 (Fig. 2). During fruit body development, CEL4 follows a profile similar to that of cellulase activity (38) and endoxylanase activity (7, 34).
In laboratory liquid cultures, the amount of CEL4 detected in the culture filtrate does not increase over time after an initial rapid increase (Fig. 2). We expected that CEL4 would remain bound to the mycelium until the cellulose was used up. The mycelium- and cellulose-bound fraction declines in later samples, but CEL4 in the medium does not increase correspondingly. Although the CEL4 activity drops after 21 days, the amount of protein remains fairly constant, consistent with CEL4 inactivation and similar to the behavior of both laccase (36) and cellulase (19). Around 80% of CEL4 remains in the culture filtrate throughout the time course.
Our results show large changes in the level of CEL4 activity during fruiting-body development, suggesting that hemicellulose degradation contributes to the large carbon and energy demand during sporophore formation and that hemicellulose degradation parallels the mobilization of cellulose. The cellulose-binding domain in CEL4 might function in some manner in the coordination of these functions. The expression of CEL4 in P. pastoris provides a baseline for future studies of compost degradation and the extracellular enzymes that fungi deploy in this process.
ACKNOWLEDGMENTS
Portions of this work were funded by a NEDO grant from MITI, Japan.
We thank Mike Challen and S. Sreenvasaprasad (Horticulture Research International, Wellsbourne, United Kingdom) for providing A. bisporus strains and compost samples, James Whiteford for providing the protein samples extracted from compost, Phil Marsh (KCL) for advice on PCR, and Phil Cunningham (KCL) for help with bioinformatics.
REFERENCES
- 1.Bonnen A M, Anton L H, Orth A B. Lignin-degrading enzymes of the commercial button mushroom, Agaricus bisporus. Appl Environ Microbiol. 1994;60:960–965. doi: 10.1128/aem.60.3.960-965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowden G A, Paredes A M, Georgiou G. Structure and morphology of protein inclusion bodies in Escherichia coli. Bio/Technology. 1991;9:725–730. doi: 10.1038/nbt0891-725. [DOI] [PubMed] [Google Scholar]
- 3.Burton K S, Partis M D, Wood D A, Thurston C F. Accumulation of serine proteinase in senescent sporophores of the cultivated mushroom, Agaricus bisporus. Mycol Res. 1997;101:146–152. [Google Scholar]
- 4.Chow C M, Yagüe E, Raguz S, Wood D A, Thurston C F. The cel3 gene of Agaricus bisporus codes for a modular cellulase and is transcriptionally regulated by the carbon source. Appl Environ Microbiol. 1994;60:2779–2785. doi: 10.1128/aem.60.8.2779-2785.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christgau S, Kauppinen S, Vind J, Kofod L V, Dalbge H. Expression cloning, purification and characterisation of a β-1,4-mannanase from Aspergillus aculeatus. Biochem Mol Biol Int. 1994;33:917–925. [PubMed] [Google Scholar]
- 6.Cregg J M, Vedvick T S, Raschke W C. Recent advances in the expression of foreign genes in Pichia pastoris. Bio/Technology. 1993;11:905–909. doi: 10.1038/nbt0893-905. [DOI] [PubMed] [Google Scholar]
- 7.DeGroot P W J, Basten D E J W, Sonnenberg A S M, Van Griensven L J L D, Visser J, Schaap P J. An endo-1,4-β-xylanase encoding gene from Agaricus bisporus is regulated by compost specific factors. J Mol Biol. 1998;277:273–284. doi: 10.1006/jmbi.1997.1605. [DOI] [PubMed] [Google Scholar]
- 8.Durrant A J, Wood D A, Cain P B. Lignocellulose degradation by Agaricus bisporus during solid substrate fermentation. J Gen Microbiol. 1991;137:751–755. [Google Scholar]
- 9.Fermor T R, Wood D A. Degradation of bacteria by Agaricus bisporus and other fungi. J Gen Microbiol. 1981;126:377–387. [Google Scholar]
- 10.Gupta R, Birch H, Rapacki K, Brunak S, Hansen J E. O-GLYCBASE version 4.0: a revised database of O-glycosylated proteins. Nucleic Acids Res. 1999;27:370–372. doi: 10.1093/nar/27.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hess H H, Lees M B, Derr J E. A linear-Folin assay for both water-soluble and sodium dodecyl sulphate-solubilised proteins. Anal Biochem. 1978;85:295–300. doi: 10.1016/0003-2697(78)90304-4. [DOI] [PubMed] [Google Scholar]
- 13.Horton R M, Cai Z, Ho S N, Pease L R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–533. [PubMed] [Google Scholar]
- 14.Humphrey T, Proudfoot N J. A beginning to the biochemistry of polyadenylation. Trends Genet. 1988;3:243–245. doi: 10.1016/0168-9525(88)90028-5. [DOI] [PubMed] [Google Scholar]
- 15.Kubicek C P. The cellulase proteins of Trichoderma reesei: structure, mutiplicity, mode of action and regulation of formation. Adv Biochem Eng Biotechnol. 1992;45:1–27. [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Linder M, Teeri T T. The roles and function of cellulose-binding domains. J Biotechnol. 1997;57:15–28. doi: 10.1016/s0168-1656(97)00088-6. [DOI] [PubMed] [Google Scholar]
- 18.Loftus M G, Moore D, Elliott T J. DNA polymorphisms in commercial and wild strains of the cultivated mushroom, Agaricus bisporus. Theor Appl Genet. 1988;76:712–718. doi: 10.1007/BF00303517. [DOI] [PubMed] [Google Scholar]
- 19.Manning K, Wood D A. Production and regulation of extracellular endocellulase by Agaricus bisporus. J Gen Microbiol. 1983;129:1839–1847. [Google Scholar]
- 20.McCleary B V. β-d-Mannanases. Methods Enzymol. 1988;160:596–610. [Google Scholar]
- 21.Nelson N. A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem. 1944;153:375–380. [Google Scholar]
- 22.Penttilä M E, Andre L, Salheimo M, Lehtovaara P, Knowles J K C. Expression of two Trichoderma reesei endoglucanases in the yeast Saccharomyces cerevisiae. Yeast. 1987;3:175–185. doi: 10.1002/yea.320030305. [DOI] [PubMed] [Google Scholar]
- 23.Perry C R, Smith M, Britnell C, Wood D A, Thurston C F. Identification of two laccase genes in the cultivated mushroom Agaricus bisporus. J Gen Microbiol. 1993;139:1209–1218. doi: 10.1099/00221287-139-6-1209. [DOI] [PubMed] [Google Scholar]
- 24.Raguz S, Yagüe E, Wood D A, Thurston C F. Isolation and characterisation of a cellulose-growth-specific gene from Agaricus bisporus. Gene. 1992;119:183–190. doi: 10.1016/0378-1119(92)90270-y. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sodhi H S. Enzyme mutants of Coprinus bilantus and recombinant DNA technology for strain improvement in Agaricus bisporus. Ph.D. thesis. London, United Kingdom: University of London; 1992. [Google Scholar]
- 27.Somogyi M. Notes on sugar determination. J Biol Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- 28.Stålbrand H, Saloheimo A, Vehmaanperä J, Henrissat B, Penttilä M. Cloning and expression in Saccharomyces cerevisiae of a Trichoderma reesei β-mannanase gene containing a cellulose binding domain. Appl Environ Microbiol. 1995;61:1090–1097. doi: 10.1128/aem.61.3.1090-1097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treschow C. Nutrition of the cultivated mushroom. Dan Bot Ark. 1944;11:1–180. [Google Scholar]
- 30.Turner E M. Phenoloxidase activity in relation to substrate and development stage in the mushroom, Agaricus bisporus. Trans Brit Mycol Soc. 1974;63:542–547. [Google Scholar]
- 31.Turner E M, Wright M, Ward T, Osbourne D J, Self R. Production of ethylene and other volatiles and changes in cellulase and laccase activities during the life cycle of the cultivated mushroom Agaricus bisporus. J Gen Microbiol. 1975;91:167–176. doi: 10.1099/00221287-91-1-167. [DOI] [PubMed] [Google Scholar]
- 32.Unkles S E. Gene organisation in industrial filamentous fungi. In: Kinghorn J R, Turner G, editors. Applied molecular genetics of filamentous fungi. London, United Kingdom: Chapman & Hall; 1992. pp. 28–53. [Google Scholar]
- 33.Way M. Identification of a region in segment-1 of gelsolin critical for actin binding. EMBO J. 1990;9:4103–4109. doi: 10.1002/j.1460-2075.1990.tb07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteford J R. Characterisation of xylanases from the cultivated mushroom Agaricus bisporus. Ph.D. thesis. London, United Kingdom: University of London; 1998. [Google Scholar]
- 35.Wong K K Y, Saddler J N. Applications of hemicellulases in the food, feed, and pulp and paper industries. In: Coughlan M P, Hazlewood G P, editors. Hemicellulose and hemicellulases. London, United Kingdom: Portland Press Ltd.; 1993. pp. 127–143. [Google Scholar]
- 36.Wood D A. Inactivation of extracellular laccase during fruiting of Agaricus bisporus. J Gen Microbiol. 1980;117:339–345. [Google Scholar]
- 37.Wood D A, Claydon N, Dudley K J, Stephens S K, Allan M. Cellulase production in the life cycle of the cultivated mushroom, Agaricus bisporus. In: Aubert P, Beguin P, Millet J, editors. Biochemistry and genetics of cellulose degradation. London, United Kingdom: Academic Press; 1988. pp. 53–70. [Google Scholar]
- 38.Wood D A, Goodenough P. Fruiting of Agaricus bisporus. Changes in extracellular enzyme activities during growth and fruiting. Arch Microbiol. 1977;114:161–165. [Google Scholar]
- 39.Yagüe E, Chow C M, Challen M P, Thurston C F. Correlation of exons with functional domains and folding regions in a cellulase from Agaricus bisporus. Curr Genet. 1996;30:56–61. doi: 10.1007/s002940050100. [DOI] [PubMed] [Google Scholar]
- 40.Yagüe E, Mehak-Zunic M, Morgan L, Wood D A, Thurston C F. Expression of CEL2 and CEL4, two proteins from Agaricus bisporus with similarity to fungal cellobiohydrolase I and β-mannanase, respectively, is regulated by the carbon source. Microbiology. 1997;143:239–244. doi: 10.1099/00221287-143-1-239. [DOI] [PubMed] [Google Scholar]
- 41.Yagüe E, Wood D A, Thurston C F. Regulation of transcription of the cell gene in Agaricus bisporus. Mol Microbiol. 1994;12:41–47. doi: 10.1111/j.1365-2958.1994.tb00993.x. [DOI] [PubMed] [Google Scholar]
- 42.Zamanian M, Mason J R. Benzene dioxygenase in Pseudomonas putida: subunit composition and immuno-cross-reactivity with aromatic dioxygenases. Biochem J. 1987;244:611–616. doi: 10.1042/bj2440611. [DOI] [PMC free article] [PubMed] [Google Scholar]