Abstract
Scope
An imbalance of the gut microbiota (“dysbiosis”) is associated with numerous chronic diseases, and its modulation is a promising novel therapeutic approach. Dietary supplementation with soluble fiber is one of several proposed modulation strategies. This study aims at confirming the impact of the resistant dextrin NUTRIOSE (RD), a soluble fiber with demonstrated beneficial health effects, on the gut microbiota of healthy individuals.
Methods and results
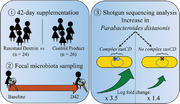
Fifty healthy women are enrolled and supplemented daily with either RD (n = 24) or a control product (n = 26) during 6 weeks. Characterization of the fecal metagenome with shotgun sequencing reveals that RD intake dramatically increases the abundance of the commensal bacterium Parabacteroides distasonis. Furthermore, presence in metagenomes of accessory genes from P. distasonis, coding for susCD (a starch‐binding membrane protein complex) is associated with a greater increase of the species. This suggests that response to RD might be strain‐dependent.
Conclusion
Supplementation with RD can be used to specifically increase P. distasonis in gut microbiota of healthy women. The magnitude of the response may be associated with fiber‐metabolizing capabilities of strains carried by subjects. Further research will seek to confirm that P. distasonis directly modulates the clinical effects observed in other studies.
Keywords: gut microbiota, resistant dextrin, Parabacteroides distasonis, shotgun metagenomics, soluble fiber
Dietary supplement with the resistant dextrin NUTRIOSE, a soluble fiber, increases the abundance of Parabacteroides distasonis in the gut microbiota of healthy women. Magnitude of P. distasonis increase is related to the presence of a pair of genes annotated as susCD, a starch‐binding membrane protein complex, in the specific strains carried by individuals.
1. Introduction
Over the past two decades, implication of the gut microbiota in human and animal health has become well recognized. Through the production of metabolites such as short chain fatty acids (SCFA), and via interactions with host cells, gut microorganisms are a key component of the host's homeostasis.[ 1 , 2 ] Imbalance in gut microbial community, also known as dysbiosis, can have adverse repercussions on the host's health. For instance, in the dysbiotic gut the concentration of lipopolysaccharide can increase and be absorbed, leading to an inflammatory response. Furthermore, specific microbial signatures (at the species or strain level) have been associated with a broad range of chronic diseases, from metabolic diseases such as obesity or diabetes[ 2 ] to psychological disorders such as anxiety and depression.[ 3 ] Gut microbiota modulation is therefore of great interest as novel therapeutic approach.
Several strategies to modulate gut microbiota currently exist and more are being developed. Fecal microbiota transplantation,[ 4 ] medication,[ 5 , 6 ] or whole‐diet modification[ 7 , 8 ] can all lead to significant changes in gut microbiota composition. However, implementing these strategies may be challenging or have adverse effects of their own.
A more convenient approach consists in supplementing the usual diet with probiotics (i.e, living microorganisms) or prebiotics. While the definition of prebiotics evolved a lot over the past decades, it is now accepted that fibers are considered to be prebiotics when they meet defined conditions: “A substrate that is selectively utilized by host microorganisms conferring a health benefit.” [ 9 ] Indeed, fibers, which are mainly carbohydrates (e.g., inulin), nonstarch polysaccharides (e.g., pectin), cellulose, resistant starch, or resistant dextrin (RD), are not (fully) digested and/or absorbed in the human small intestine. Therefore, they reach the colon where they can be hydrolyzed and fermented by bacteria.[ 10 ] The impact of various fibers on the gut microbiota and on host health continues to be described and elucidated. To mention only a few examples of apparent modulation by dietary fibers on health conditions: inulin had a protective action against cardio‐vascular disease, and restored gut homeostasis.[ 11 ] Immunomodulatory effects on allergies seen with pectin intake were related to increased concentrations of SCFA produced by the gut microbiota.[ 12 ] Supplementation with fructo‐oligosaccharides and galacto‐oligosaccharides changed the gut microbiota while also having antidepressant effects on mice.[ 13 ] According to a recent meta‐analysis, these dietary fibers could also increase Bifidobacterium and Lactobacillus spp. in the gut microbiota of healthy adults.[ 14 ] In a recent review, resistant starch and resistant dextrins (RD) were proven to have positive effects of body mass index (BMI), total body fat, and markers of metabolic disorders in healthy volunteers.[ 15 ]
However, not all individuals respond to prebiotics in the same ways. Baseline composition of the gut microbiota is often found to be one of the factors explaining response differences within study‐groups. Several studies showed that response to inulin supplementation was driven by baseline levels of various taxa, such as Coprococcus [ 16 ] or Akkermansia.[ 17 ] Supplementation with wheat bran arabinoxylan‐oligosaccharides had effects on gut microbiota modification that were directly influenced by baseline levels of Prevotella.[ 18 ] These results highlight the importance of defining and characterizing baseline microbiota profiles in responders and nonresponders when conducting prebiotic supplementation studies.
RD has already been shown to impact gut microbiota and display several health benefits through in vitro[ 19 ] and clinical studies,[ 20 ] confirming the prebiotic properties of this fiber. Modification of gut microbiota included an increase in Bacteroides spp, a decrease in Clostridium perfringens and the production of several SCFAs. In other studies performed on either animal models, healthy volunteers or specific human populations (diabetic women), RD showed different beneficial health responses, such as antiobesogenic,[ 21 , 22 , 23 , 24 , 25 , 26 ] cholesterol‐lowering,[ 27 ] anti‐inflammatory,[ 28 , 29 , 30 ] or positive influence on the immune system response.[ 29 ]
Most of the interventional (human) studies with fibers focused on specific taxa, targeting only Bifidobacterium and Lactobacillus, thus following the first prebiotic definition.[ 31 ] In the current study we expand the list of taxa being examined by performing shotgun metagenomics to precisely characterize the effects of RD consumption on the whole gut microbiota.
2. Experimental Section
2.1. Ethical Statement
This study was conducted according to Good Clinical Practice Guidelines, the Declaration of Helsinki (2000). Signed written informed consent for participation in the study was obtained from all subjects before protocol‐specific procedures were carried out. An independent ethics committee approved the protocol (IntegReview IRB, October 2015).
2.2. Study Population
For this pilot study, a total of 56 subjects in the United States (Chicago, IL) were recruited. The sample size was chosen according to sample size of other gut microbiota analyses in the context of dietary fiber intervention.[ 14 ] The inclusion criteria were as follows:
‐ female,
‐ 18–50 years of age,
‐ BMI between 18 and 29.9 kg m− 2,
‐ daily liquid intake of at least 1.5 L (self‐defined: water, tea, coffee, soft drinks, juice, etc.),
‐ non menopausal with reliable contraception, good general, and mental health.
The exclusion criteria were as follows:
‐ pregnant or lactating,
‐ high fiber consumer (self‐defined: five or more pieces of fruit or vegetables per day),
‐ smoker,
‐ suffering from ongoing or diagnosed gastrointestinal pathology, neurological pathology, metabolic/endocrine diseases, any severe chronic disease,
‐ suffering from an alternation of constipation and diarrhea,
‐ subject who had undergone gastrointestinal surgery in the last 2 years,
‐ subject taking medication or dietary supplements that could affect bowel movement / gut motility, or with a history of systemic disease that might affect gut motility according to the investigator; or which could otherwise impact the results of the study.
2.3. Dosage Information
The active product was NUTRIOSE FB06 (ROQUETTE, France) and the control product (CP) was a maltodextrin (GLUCIDEX 21, ROQUETTE, France). NUTRIOSE FB06 was a nonviscous RD obtained from wheat with a fiber content of 85% and a mono‐ and disaccharide content of ≤0.5%. The dosage of the two products was increased during the study: 5 g d−1 (week 1), 10 g d−1 (week 2), and 20 g d−1 (weeks 3–6). The products were presented in packets of 5, 10, and 20 g, labeled and packaged in one container. After a run‐in period of about 2 weeks, each randomized subject consumed one packet daily of either active product or placebo during 6 weeks. They consumed the packet in 250 mL of water, at breakfast.
2.4. Procedure
The clinical trial was a randomized, double‐blind design with a dietary supplementation using increasing doses as described in Figure 1A, conducted in 2016/2017.
Figure 1.
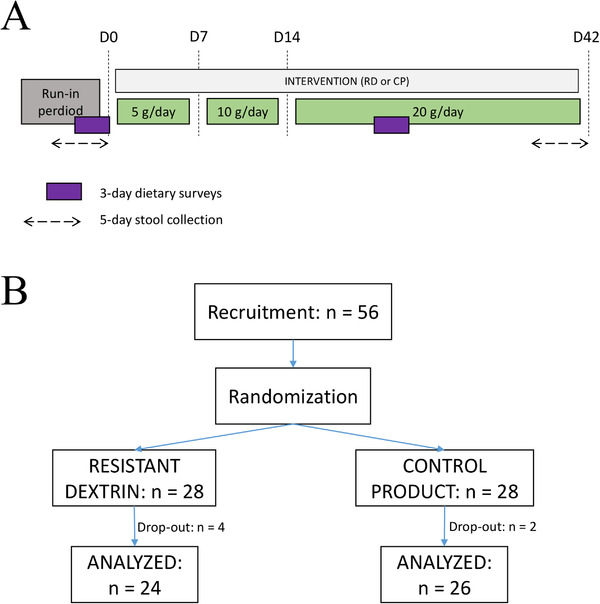
Clinical trial. A) Study design. Stool samples are collected and analyzed at baseline and D42. B) Clinical trial flowchart.
To detect effects on gut microbiota, fecal samples were collected at baseline and after 6 weeks of study product consumption (RD or CP).
Product compliance was defined by the ratio between the amount of product actually consumed and the quantities of products to be consumed per the protocol. Compliances were evaluated at each visit after baseline in order to verify that there was no difference between the two groups. To evaluate the product intake compliance, subjects were asked to bring back the nonconsumed packets at D7, D14, and D42.
2.5. Data Collection
Fecal samples were collected at the beginning of the study (baseline) and 6 weeks later (D42), and stored in a stabilizing solution (RNAlater) following the International Human Microbiome Standards (IHMS) procedure, SOP 05 (http://www.human‐microbiome.org/).[ 32 ]
Potential food intake modifications on the part of the subjects during the intervention were monitored using food diaries at two different time points (before entering the study (D‐2/D0) and on the last days of supplementation (D40/D42)). Daily energy, fibers, lipids, proteins, carbohydrates, alcohol, and water intakes were evaluated based on this data.
2.6. DNA Extraction and Shotgun Sequencing
Fecal DNA was extracted following the IHMS procedure, SOP 07 V2 (http://www.human‐microbiome.org/).[ 32 ] The DNA preparation was subjected to quality control using Qubit Fluorometric (ThermoFisher Scientific, Waltham, MA, USA) and qualified using DNA size profiling on a Fragment Analyzer (Agilent Technologies, Santa Clara, CA, USA). 3 μg of high molecular weight DNA (>10 kbp) was used to build the sequencing library. Shearing of DNA into fragments of approximately 150 bp was performed using an ultrasonicator (Covaris, Woburn, MA, USA) and DNA fragment library construction was performed using the Ion Plus Fragment Library and Ion Xpress Barcode Adapters Kits (ThermoFisher Scientific, Waltham, MA, USA). Purified and amplified DNA fragment libraries were sequenced using the Ion Proton Sequencer (ThermoFisher Scientific, Waltham, MA, USA), with a minimum of 20 million high‐quality reads of 150 bp (on average) generated per library. A mean of 22.6 million (± 1.8 million) reads per sample was generated.
2.7. Reads Mapping
To create the gene count table, the METEOR software was used (https://forgemia.inra.fr/metagenopolis/meteor)[ 33 ]: First, low‐quality reads were trimmed or filtered out with AlienTrimmer.[ 34 ] Reads that aligned to the human genome (accession number: GCF_000001405.39, nucleotide identity ≥90%) were also discarded. Then, remaining reads were trimmed to 80 bases and mapped to the Integrated Gut Catalogue 2 (IGC2),[ 35 , 36 ] comprising 10.4 million genes, using Bowtie2.[ 37 ] Uniquely mapped reads (reads mapped to a single gene in the catalogue) were attributed to their corresponding genes. Shared reads (reads that mapped with the same alignment score to multiple genes in the catalogue) were attributed according to the ratio of their unique mapping counts of the captured genes. The resulting count table was further processed using the R package MetaOMineR v1.31.[ 38 ] It was downsized to 14.5 million mapped reads to take into account differences in sequencing depth and in mapping rate across samples. Then, the downsized matrix was normalized for gene length and transformed into a frequency matrix (fragments per kilobase of exon model per million reads mapped: FPKM normalization).
2.8. Metagenomic Species (MGS) Profiles
Using MSPminer,[ 39 ] the IGC2 catalogue was previously organized into 1990 MGS,[ 36 ] which were clusters of coabundant genes corresponding to the same microbial species and organized as core genes and accessory genes. MGS taxonomy was assigned with the Genome Taxonomy Database.[ 40 ] Relative abundance of an MGS was computed as the mean abundance of 100 “marker” genes (i.e., core genes that correlated the most altogether). If less than 10% of “marker” genes were seen in a sample, the abundance of the MGS was set to 0. Relative abundances at higher taxonomical ranks were computed as the sum of the MGS that belong to a given taxon. MGS count was assessed as the number of MGS present in a sample (i.e., whose abundance was strictly positive).
2.9. Microbiome Functional Potential
Genes from the IGC2 catalogue were previously mapped with[ 41 ] onto KEGG orthologs (KO) from the KEGG database (version 8.9).[ 42 ] Each gene was assigned to the best‐ranked KO among hits with e‐value <10–5 and a bit score >60. For a given sample, the abundance of a KEGG module or a Gut‐Metabolic Modules (GMMs)[ 43 ] was assessed with the following procedure:
i) restriction of the gene content of each MGS detected in the sample to the MGS core genes and the accessory genes actually detected in the sample;
ii) calculation of the fraction of the module present in the restricted set of genes;
iii) assessment that an MGS carried a module in a given sample if the computed fraction was above 90%;
iv) sum of the abundance of all MGSs that carried the module for a given sample.
Carbohydrate‐active enzymes (CAZymes) annotation of the IGC2 catalog was performed according to the Carbohydrate‐active enzymes database CAZy (http://www.cazy.org).[ 44 ]
2.10. Statistical Analysis
Statistical analyses were performed and data were plotted using the R software v3.6.0.[ 45 ] Comparisons between two groups were performed using (paired) Wilcoxon tests. Correlations between variables were performed using Spearman's correlations. False Discovery Rate was controlled by correcting all p‐values for multiple testing with the Benjamini Hochberg Procedure. Unless stated otherwise, a corrected p‐value (q‐value) is considered significant if less than 0.1. Quantitative values were expressed as mean ± standard deviation (SD).
Bray‐Curtis dissimilarity was assessed with R package vegan v2.5.7.[ 46 ]
Impact of RD was assessed using the R package nparLD v2.1[ 47 ] for nonparametric longitudinal data (function f1.ld.f1 for experimental design with one whole‐plot factor and one subplot factor). Log fold‐change (log2FC) of clinical, dietary, and metagenomic features were computed for each individual as the log2‐transformed ratio between the value at baseline (denominator) and the value at D42 (numerator). For metagenomic features, zero abundance was replaced by a pseudoabundance equal to half the minimum abundance of detected MGS.
3. Results
3.1. Population Characteristics
Fifty six women entered the study and were randomly assigned to two groups: the RD) group (n = 28) and the CP group (n = 28). Six individuals dropped out before the end of the study because they started on medication, an excluding criteria since it could impact the results of the study. This medication was unrelated to the intervention, as RD is known to be well tolerated at the dosage used.[ 48 ] This left a total of 100 fecal samples that could be further analyzed (RD: n = 24*2; CP: n = 26*2, Figure 1B).
There was no difference at baseline between both groups regarding demographic data or dietary data (q ≥ 0.1, Table 1 ). In particular, the two groups were similar regarding age and BMI, two variables with major impact on gut microbiota.[ 38 , 49 ] Diet (assessed by food diaries) remained stable during the intervention apart from the dietary fiber intake that increased in RD group and decreased in CP group (from 15 ± 7.3 g d−1 to 18 ± 8.2 and from 19 ± 9.4 g d−1 to 16 ± 9.5, respectively, q = 0.003, Table S1, Supporting Information). However, the mean increase of dietary fiber in the RD group (3.0 ± 6.3 g) was considered to be negligible as compared to the dosage of RD during the intervention (20 g d during the last 4 weeks, with a fiber content of 85%).
Table 1.
Characteristics of the study population at baseline
| RD | CP | p a) | Q b) | |
|---|---|---|---|---|
| BMI ± SD [kg m− 2] | 25 ± 3.5 | 26 ± 3 | 0.51 | 0.64 |
| Age ± SD [years] | 38 ± 7.8 | 36 ± 8.7 | 0.64 | 0.64 |
| Energy ± SD [kcal d−1] | 1800 ± 440 | 1800 ± 440 | 0.95 | 0.95 |
| Carbohydrates ± SD [g d−1] | 200 ± 70 | 210 ± 68 | 0.68 | 0.89 |
| Proteins ± SD [g d−1] | 76 ± 18 | 79 ± 20 | 0.72 | 0.89 |
| Lipids ± SD [g d−1] | 80 ± 19 | 74 ± 19 | 0.17 | 0.41 |
| Percentage of energy from alcohol ± SD [%] | 1.3 ± 2.6 | 1.2 ± 2 | 0.76 | 0.89 |
| Water ± SD [mL d−1] | 3500 ± 670 | 4100 ± 1100 | 0.031 | 0.22 |
| Fiber ± SD [g d−1] | 15 ± 7.3 | 19 ± 9.4 | 0.086 | 0.30 |
P‐value associated with the non‐parametric longitudinal data test.
P‐value corrected for multiple testing with the Benjamini‐Hocherg procedure.
resistant dextrin (RD); control product (CP); standard deviation (SD)
Throughout the intervention the participants in the RD and CP groups observed a high compliance to protocol (99% ± 1.7 and 99% ± 2.2, respectively, see Methods).
3.2. RD Consumption does not Alter Global Structure of Gut Microbiota
MGS richness at baseline was similar in the RD and CP groups (217 ± 71 vs 198 ± 74, p = 0.29, Wilcoxon test). Its evolution between baseline and D42 also showed no difference in both groups (p = 0.32, Figure 2A).
Figure 2.
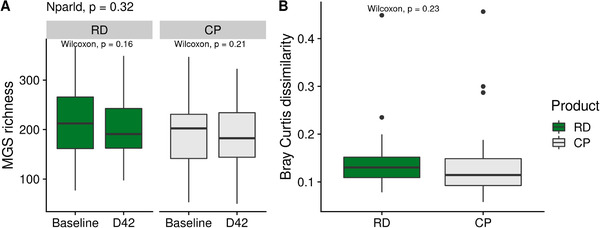
Evolution in gut microbiota global composition between RD and CP. A) Comparison of MGS richness between baseline and D42 in the RD group or in the CP group. P‐values associated with paired Wilcoxon test are displayed. P‐value associated with the test for nonparametric longitudinal data (nparld, see Methods) is displayed at the top. B) Comparison of intraindividuals Bray–Curtis dissimilarity between the RD and the CP group. P‐value associated with Wilcoxon test is displayed. CP indicates control product; MGS, MetaGenomic species; RD, resistant dextrin.
To further examine the effect of RD on microbiota composition, we computed Bray Curtis dissimilarity based on MGS abundances between samples from the same individual. There was no significant difference between the two groups (p = 0.23, Wilcoxon test, Figure 2B), confirming that RD did not alter the global composition of gut microbiota.
3.3. RD Increases the Abundance of P. distasonis
Out of 491 MGS present in at least 10% samples, P. distasonis showed a distinct evolution in the RD group as compared to the CP group (q = 0.004, Figure 3A, Table S2, Supporting Information). It increased in the RD group with a mean log2FC of 2.1 ± 1.7 between baseline and D42 (p = 1.5e–4, paired Wilcoxon test), while it remained stable in the CP group (mean log2FC: ‐0.1 ± 2.4, p = 0.53, paired Wilcoxon test). In all the subjects in this study, P. distasonis had a high prevalence (80%) and accounted for 0.92% ± 0.97 of the gut microbiota at baseline (in both study groups combined). With RD intake, its mean relative abundance raised to 7.2% ± 6.8, while it remained stable in CP group (0.68% ± 0.89). Accordingly, whereas it was only the 15th most abundant species at baseline, it became the most dominant species at D42 in gut microbiota of individuals taking RD.
Figure 3.
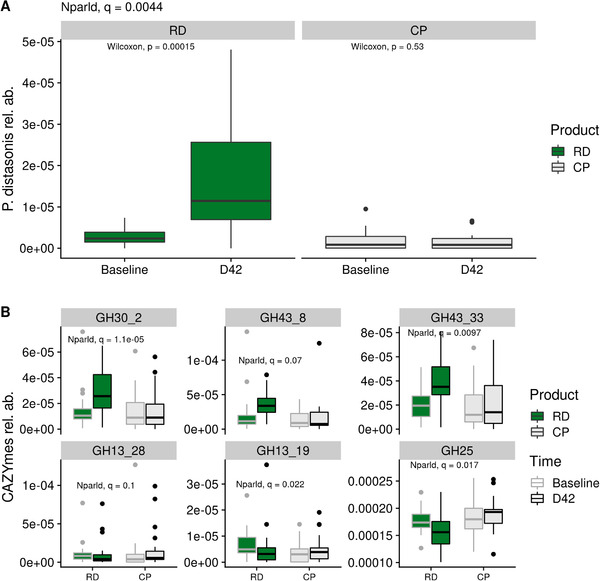
Metagenomic features altered by RD intake. A) Evolution of the relative abundance of Parabacteroides distasonis in the RD and the CP group. P‐values associated with paired Wilcoxon test are displayed. Q‐value associated with the test for nonparametric longitudinal data (nparld, see Methods) is displayed at the top. B) Evolution if the six CAZymes impacted by RD intake. Q‐value associated with the test for nonparametric longitudinal data (nparld, see Methods) is displayed for each CAZymes. CP indicates control product; RD, resistant dextrin; rel. ab., relative abundance.
We further assessed the impact of RD on gut microbiota functional potential using three databases: two functional module databases (the nonspecific database KEGG and the gut‐microbiota‐specific database GMM), and CAZy. There was no evolution in modules from KEGG or GMM, which is consistent with the low number of impacted species (Table S2, Supporting Information). In particular, abundance of modules related to SCFA did not change along the intervention. On the other hand, three CAZYmes increased with RD while three other CAZYmes decreased (q ≤ 0.1, Figure 3B). All of them belong to glycoside hydrolase families (GH). Consistently, the three increasing GH (GH30_2, GH43_8 and GH43_33) were carried by P. distasonis, whereas the three decreasing GH (GH13_28, GH13_19, and GH25) were not.
3.4. Response to RD Might Depend on Specific P. distasonis Strains
P. distasonis log2FC positively correlated with Bray–Curtis dissimilarity (rho = 0.48, p = 0.027, Figure 4A), showing that global response of gut microbiota to RD was closely related to the response of P. distasonis itself. We further defined the responder status based on P. distasonis log2FC. Out of the 24 individuals of the RD group, P. distasonis increased in 18 individuals (the responder group, log2FC > 0), slightly decreased in three individuals (the nonresponder group, log2FC < 0), and was absent from three individuals at both time points (the noncarrier group, Figure 4B).
Figure 4.
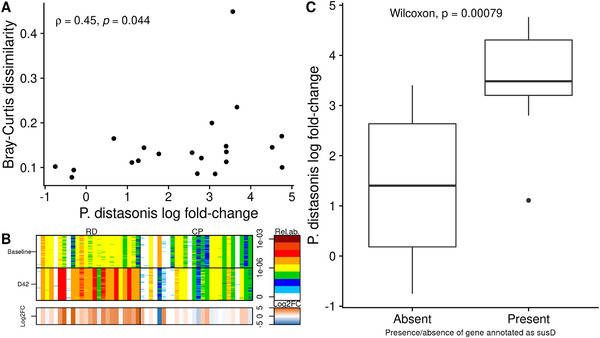
Focus on Parabacteroides distasonis. A) Relation between P. distasonis log‐fold change and intraindividuals Bray–Curtis dissimilarity, considering only individuals from the RD group carrying P. distasonis (n = 21). Spearman's coefficient along with the associated p‐value are displayed. B) Barcode of P. distasonis in the RD group (left) or in the CP group (right), at baseline (top) or at D42 (bottom). The 50 “tracer” genes are in rows, abundance is indicated by color gradient (white, not detected; red, most abundant); individuals, ordered by increasing MGS richness, are in columns. The lowest bar gives the log‐fold change between baseline and D42 associated with each individual (i.e., each column). C) Difference in P. distasonis log‐fold change when comparing individuals from the RD group whose P. distasonis strain carries a specific gene annotated as SusD (n = 10) with individuals whose strain does not have this gene (n = 11). CP indicates control product; log2FC, log‐fold change; MGS, MetaGenomic species; RD, resistant dextrin.
The three nonresponders had similar profile as the responders regarding compliance to protocol, global microbiota structure, richness, baseline P. distasonis abundance, demographic and dietary data (Figure S1, Supporting Information). Thus, these variables were not able to explain the difference in response to RD. We further hypothesized that response to RD was related to the accessory genes of P. distasonis strains carried by individuals.
Mean coverage of P. distasonis genome in the 21 RD individuals carrying the species was 6.2X ± 3.9 at baseline (min: 0.8X), and 38.0X ± 28.6 at D42 (min: 2.3X). In particular, coverage was above 3X in all individuals at one time point at least, which enables us to perform strain analysis.
Clustering individuals based on presence/absence of P. distasonis genes did not reveal any clade associated with a high or low P. distasonis log2FC (Figure S2, Supporting Information). Out of the 10 104 total genes of the P. distasonis pangenome described in our catalogue, we considered 2583 genes present in minimum five individuals and maximum 16 individuals. We then tested the association between P. distasonis log2FC and presence/absence of these genes, and found that 217 genes had a significant association (8.4%, uncorrected p ≤ 0.05, Wilcoxon test, Table S3, Supporting Information). Yet, no association remained significant after multiple test corrections (q ≤ 0.1). Only 16% and 5% of these genes had a KO or a CAZY annotation, respectively. The two most significant genes were present in the same individuals (n = 10). They were present in individuals showing a high P. distasonis log2FC (mean 3.5 ± 1.1, n = 10) and absent in individuals showing a low P. distasonis log2FC (mean = 1.4 ± 1.5, n = 11, p = 0.0008, Wilcoxon test, Figure 4C). Interestingly, one of them was annotated as susD (K21572), a starch‐binding outer membrane protein.[ 50 , 51 ] The second gene had no annotation. They both aligned on the complete genome of P. distasonis strain CBBP‐1 (accession number CP051672.1) 3250 nucleotides apart, suggesting that they might be part of the same operon. Available information on the complete genome shows that both genes are separated by a gene assigned to susC, known to associate with susD to form the complex SusCD.[ 52 ] This gene was present in IGC2 (blast, evalue = 0) but absent from automatic clustering of P. distasonis performed with MSPminer. However, susC and susD displayed highly similar abundance profiles, showing that susC was also part of P. distasonis pangenome (Figure S3, Supporting Information). SusC was present in 12 individuals, among which all but two also carry susD. Consistently, they were also present in individuals with higher P. distasonis log2FC (n = 12, p = 0.028, Figure S4, Supporting Information).
4. Discussion
In the present study, shotgun metagenomic sequencing revealed that RD specifically promoted the substantial growth of P. distasonis. This confirms and extends the results from a previous clinical trial that found (Para)bacteroides group to be increased by RD using real‐time polymerase chain reaction.[ 20 ] In line with the same study we consistently found an increase in β‐glucosidase potential activity (GH30_2, carried by P. distasonis) and no change in SCFA potential production. However, an in vitro study with RD found an increase in Roseburia as well as acetate, propionate, and butyrate.[ 19 ] These differential results may be due to difference in methods and techniques that were used (i.e., in vivo vs in vitro, shotgun vs 16S rRNA‐based fluorescence).
Interestingly, P. distasonis was also found to be increased in other studies of diverse dietary fibers,[ 53 ] such as inulin,[ 54 , 55 ] xyloglucan,[ 56 ] or galactooligosaccharide/polydextrose.[ 57 ] This suggests that P. distasonis is able to use a wide range of substrates for fermentation.
P. distasonis, highly prevalent in the gut microbiota,[ 58 ] seems to display conflicting importance regarding its potential effects on human health, as outlined in a recent review.[ 59 ] For example, it is thought to be detrimental in inflammatory bowel disease (IBD) patients, as well as in Crohn's disease. However, the species may have antitumorigenic and anti‐inflammatory potential in colorectal cancer patients. On the beneficial side, in obesity its role seems clearly protective. Notably, a study in mice found that P. distasonis and its excreted metabolites (succinate, lithocholic acid, and ursodeoxycholic acid) were associated with decreased weight gain,[ 60 ] decreased hyperglycemia, and reduced hyperlipidemia. Consistently, several RD‐related studies showed similar beneficial health effects in overweight men[ 21 ] and in hypercholesterolemic hamsters,[ 27 ] strongly suggesting that benefits could be due to gut microbiota modulation. Clearly, much more research is needed in order to better understand any cause‐and‐effect relationships between P distasonis and health outcomes. The data generated in the current study will surely help to advance such understanding.
Another significant, if not critical, factor is the basal composition of the microbiota, with regards to P. distasonis. Firstly, if P. distasonis is absent from the colon, RD might have a limited action. In our study, only three individuals did not carry P. distasonis, which effectively removed them from statistical consideration given the very small “N.” Secondly, our results suggest that response to supplementation depends on P. distasonis strains carried by individuals. Indeed, the individuals carrying a strain with specific genes coding for SusCD, a starch‐binding membrane protein complex, showed a more pronounced increase in P. distasonis, as compared to the individuals without these genes. In Bacteroides, the SusCD complex is known to enable the uptake of large nutrients through the outer membrane, and recently the mechanism of β2,6 fructo‐oligosaccharide import by the SusCD complex from Bacteroides thetaiotamicron has been characterized.[ 52 ] Besides, in the same study, authors proposed to redefine “sus” as “saccharide uptake system” rather than “starch utilization system,” as these proteins seem not to be starch‐specific. Thus, the genes annotated as susC and susD might enhance RD metabolism by P. distasonis. However, the low number of nonresponder individuals (n = 3) makes it difficult to determine true marker‐genes of RD response. Examining closely the strain carried by an individual is all the more important since several in vitro and in vivo studies showed that P. distasonis properties (such as anti‐inflammatory, promotion of GLP‐1 production or barrier protection) were highly strain‐specific.[ 61 , 62 ] The relation between P. distasonis and GLP‐1 production is consistent with three clinical trials showing that RD improves satiety in healthy adults.[ 22 , 24 , 25 ]
Given the impact of RD on specific strains of P. distasonis, we foresee that this fiber could be used to extend our understanding of its growth, function, and interaction with the host. However, this requires to apply the presented analysis on larger and more diverse datasets. Currently, the observed effects on host function may not be due exclusively to changes in P. distasonis, since the detection of taxa which might show a smaller effect size than that of P. distasonis would require a larger cohort size, for statistical reasons. Also, related to the cohort size, the low number of nonresponder individuals (n = 3) makes it difficult to determine true marker‐genes of RD response. Moreover, this pilot study focused on a specific population (healthy women from 18 to 50 years old) and further research including different populations, notably in terms of age, gender, or health states (obesity for example), might highlight signatures that could not be seen in the current study. Finally, it should be noted that physical activity was not recorded: since computed mean calorie intake per day (1800 kcal ± 440) corresponded to the estimated needs for sedentary women, the participants may have under‐reported their diet intake, which potentially biased our results. Besides those limitations, such studies could also benefit from additional datasets such as fecal and serum metabolites measurements, notably P. distasonis related metabolites (e.g., acetate, succinate, and bile acids),[ 59 ] as they could impact host metabolism. Nevertheless, this pilot study will help in designing further studies based on known P. distasonis properties.
In conclusion, RD strongly promoted the growth of P. distasonis, a highly prevalent species of the gut microbiota[ 58 ] which is of particular interest for individuals suffering from obesity. It is notable that a small proportion of subjects did not carry P. distasonis in their fecal pattern. Response to RD was a function of baseline microbiota composition, especially on the P. distasonis strain dominant in certain individuals. In vitro studies on diverse P. distasonis strains would help to further understand the differences between responders versus nonresponders to RD. Overall, our study highlights the importance of a personalized nutrition, that is, a nutrition adapted to the specificity of an individual (here, the bacterial strains carried in its gut or its health status) when trying to promote health benefits by modulation of the gut microbiota.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
F.T. and K.D.S. contributed equally to this work. C.T., L.G.D., C.V., and H.C. designed the clinical trial. C.V. and H.C. conducted the clinical trial. J.D. and S.D.E. designed the microbiota research. N.G. and F.L. generated the metagenomics sequences. K.D.S. and F.T. analyzed the sequencing data. E.L.C., M.B., N.P., F.P.O., and A.S.A. contributed to data interpretation. F.T., K.D.S., and C.T. wrote the manuscript. All authors read and approved the manuscript.
Supporting information
Supporting Information
Acknowledgements
The study was funded by Roquette, France. The authors thank Benoit Quinquis for his help in generating sequences, Bernard Henrissat for the catalogue annotation, and David Madsen for his suggestions for text improvements and English proofreading of the manuscript.
Thirion F., Da K. Silva, Plaza Oñate F., Alvarez A.‐S., Thabuis C., Pons N., Berland M., Le E. Chatelier, Galleron N., Levenez F., Vergara C., Chevallier H., Guérin‐Deremaux L., Doré J., Ehrlich S. D., Diet Supplementation with NUTRIOSE, a Resistant Dextrin, Increases the Abundance of Parabacteroides distasonis in the Human Gut. Mol. Nutr. Food Res. 2022, 66, 2101091. 10.1002/mnfr.202101091
Data Availability Statement
The data that support the findings of this study are openly available in the European Nucleotide Archive (ENA) at https://www.ebi.ac.uk reference number PRJEB42906.
References
- 1. Valdes A. M., Walter J., Segal E., Spector T. D., BMJ 2018, 361, k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fan Y., Pedersen O., Nat. Rev. Microbiol. 2021, 19, 55. [DOI] [PubMed] [Google Scholar]
- 3. Nikolova V. L., Smith M. R. B., Hall L. J., Cleare A. J., Stone J. M., Young A. H., JAMA Psychiatry 2021, 78, 1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanssen N. M. J., de Vos W. M., Nieuwdorp M., Cell Metab. 2021, 33, 1098. [DOI] [PubMed] [Google Scholar]
- 5. Maier L., Goemans C. V., Wirbel J., Kuhn M., Eberl C., Pruteanu M., Müller P., Garcia‐Santamarina S., Cacace E., Zhang B., Gekeler C., Banerjee T., Anderson E. E., Milanese A., Löber U., Forslund S. K., Patil K. R., Zimmermann M., Stecher B., Zeller G., Bork P., Typas A., Nature 2021, 599, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ke H., Li F., Deng W., Li Z., Wang S., Lv P., Chen Y., Front. Pharmacol. 2021, 12, 726707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meslier V., Laiola M., Roager H. M., De Filippis F., Roume H., Quinquis B., Giacco R., Mennella I., Ferracane R., Pons N., Pasolli E., Rivellese A., Dragsted L. O., Vitaglione P., Ehrlich S. D., Ercolini D., Gut 2020, 69, 1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cotillard A., Kennedy S. P., Kong L. C., Prifti E., Pons N., Chatelier E. Le, Almeida M., Quinquis B., Levenez F., Galleron N., Gougis S., Rizkalla S., Batto J.‐M., Renault P., Doré J., Zucker J.‐D., Clément K., Ehrlich S. D., Nature 2013, 500, 585. [DOI] [PubMed] [Google Scholar]
- 9. Gibson G. R., Hutkins R., Sanders M. E., Prescott S. L., Reimer R. A., Salminen S. J., Scott K., Stanton C., Swanson K. S., Cani P. D., Verbeke K., Reid G., Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491. [DOI] [PubMed] [Google Scholar]
- 10. Slavin J., Nutrients 2013, 5, 1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu H., Chiou J., Nutrients 2021, 13, 2878.34445037 [Google Scholar]
- 12. Blanco‐Pérez F., Steigerwald H., Schülke S., Vieths S., Toda M., Scheurer S., Curr. Allergy Asthma Rep. 2021, 21, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burokas A., Arboleya S., Moloney R. D., Peterson V. L., Murphy K., Clarke G., Stanton C., Dinan T. G., Cryan J. F., Biol. Psychiatry 2017, 82, 472. [DOI] [PubMed] [Google Scholar]
- 14. So D., Whelan K., Rossi M., Morrison M., Holtmann G., Kelly J. T., Shanahan E. R., Staudacher H. M., Campbell K. L., Am. J. Clin. Nutr. 2018, 107, 965. [DOI] [PubMed] [Google Scholar]
- 15. Włodarczyk M., Śliżewska K., Nutrients 2021, 13, 3808,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leyrolle Q., Cserjesi R., Mulders M. D. G. H., Zamariola G., Hiel S., Gianfrancesco M. A., Portheault D., Amadieu C., Bindels L. B., Leclercq S., Rodriguez J., Neyrinck A. M., Cani P. D., Lanthier N., Trefois P., Bindelle J., Paquot N., Cnop M., Thissen J.‐P., Klein O., Luminet O., Delzenne N. M., Brain Behav. Immun. 2021, 94, 289. [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez J., Hiel S., Neyrinck A. M., Roy T. Le, Pötgens S. A., Leyrolle Q., Pachikian B. D., Gianfrancesco M. A., Cani P. D., Paquot N., Cnop M., Lanthier N., Thissen J.‐P., Bindels L. B., Delzenne N. M., Gut 2020, 69, 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chung W. S. F., Walker A. W., Bosscher D., Garcia‐Campayo V., Wagner J., Parkhill J., Duncan S. H., Flint H. J., BMC Microbiol. 2020, 20, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hobden M. R., Martin‐Morales A., Guérin‐Deremaux L., Wils D., Costabile A., Walton G. E., Rowland I., Kennedy O. B., Gibson G. R., PLoS One 2013, 8, e77128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lefranc‐Millot C., Guérin‐Deremaux L., Wils D., Neut C., Miller L. E., Saniez‐Degrave M. H., J. Int. Med. Res. 2012, 40, 211. [DOI] [PubMed] [Google Scholar]
- 21. Li S., Guerin‐Deremaux L., Pochat M., Wils D., Reifer C., Miller L. E., Appl. Physiol. Nutr. Metab. = Physiol. Appl. Nutr. Metab. 2010, 35, 773. [DOI] [PubMed] [Google Scholar]
- 22. Hobden M. R., Commane D. M., Guérin‐Deremaux L., Wils D., Thabuis C., Martin‐Morales A., Wolfram S., Dìaz A., Collins S., Morais I., Rowland I. R., Gibson G. R., Kennedy O. B., Eur. J. Nutr. 2021, 60, 4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hobden M. R., Guérin‐Deremaux L., Rowland I., Gibson G. R., Kennedy O. B., Proc. Nutr. Soc. 2015, 74, 258. [DOI] [PubMed] [Google Scholar]
- 24. Guérin‐Deremaux L., Pochat M., Reifer C., Wils D., Cho S., Miller L. E., Nutr. Res. 2011, 31, 665. [DOI] [PubMed] [Google Scholar]
- 25. Guerin‐Deremaux L., Li S., Pochat M., Wils D., Mubasher M., Reifer C., Miller L. E., Int. J. Food Sci. Nutr. 2011, 62, 628. [DOI] [PubMed] [Google Scholar]
- 26. Guérin‐Deremaux L., Pochat M., Reifer C., Wils D., Cho S., Miller L. E., Glob. Epidemic Obes. 2013, 1, 2. [Google Scholar]
- 27. Juhel C., Tosini F., Steib M., Wils D., Guerin‐Deremaux L., Lairon D., Cara L., Indian J. Exp. Biol. 2011, 49, 219. [PubMed] [Google Scholar]
- 28. Breton J., Plé C., Guerin‐Deremaux L., Pot B., Lefranc‐Millot C., Wils D., Foligné B., Biomed. Res. Int. 2015, 2015, 162398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farhangi M. A., Javid A. Z., Sarmadi B., Karimi P., Dehghan P., Clin. Nutr. 2018, 37, 1216. [DOI] [PubMed] [Google Scholar]
- 30. Pouillart P. R., Dépeint F., Abdelnour A., Deremaux L., Vincent O., Mazière J.‐C., Madec J.‐Y., Chatelain D., Younes H., Wils D., Saniez M.‐H., Dupas J.‐L., Inflamm. Bowel Dis. 2010, 16, 783. [DOI] [PubMed] [Google Scholar]
- 31. Gibson G. R., Roberfroid M. B., J. Nutr. 1995, 125, 1401. [DOI] [PubMed] [Google Scholar]
- 32. Costea P. I., Zeller G., Sunagawa S., Pelletier E., Alberti A., Levenez F., Tramontano M., Driessen M., Hercog R., Jung F.‐E., Kultima J. R., Hayward M. R., Coelho L. P., Allen‐Vercoe E., Bertrand L., Blaut M., Brown J. R. M., Carton T., Cools‐Portier S., Daigneault M., Derrien M., Druesne A., de Vos W. M., Finlay B. B., Flint H. J., Guarner F., Hattori M., Heilig H., Luna R. A., van Hylckama Vlieg J., et al., Nat. Biotechnol. 2017, 35, 1069. [DOI] [PubMed] [Google Scholar]
- 33. Cotillard A., Kennedy S. P., Kong L. C., Prifti E., Pons N., Le Chatelier E., Almeida M., Quinquis B., Levenez F., Galleron N., Gougis S., Rizkalla S., Batto J.‐M., Renault P., Doré J., Zucker J.‐D., Clément K., Ehrlich S. D., Nature 2013, 500, 585. [DOI] [PubMed] [Google Scholar]
- 34. Criscuolo A., Brisse S., Genomics 2013, 102, 500. [DOI] [PubMed] [Google Scholar]
- 35. Wen C., Zheng Z., Shao T., Liu L., Xie Z., Chatelier E. L.e, He Z., Zhong W., Fan Y., Zhang L., Li H., Wu C., Hu C., Xu Q., Zhou J., Cai S., Wang D., Huang Y., Breban M., Qin N., Ehrlich S. D., Genome Biol. 2017, 18, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Plaza Onate F., Pons N., Gauthier F., Almeida M., Ehrlich S. D., Chatelier E. L.e 2021, 10.15454/FLANUP [DOI]
- 37. Langmead B., Salzberg S. L., Nat. Methods 2012, 9, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chatelier E. Le, Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., Almeida M., Arumugam M., Batto J.‐M., Kennedy S., Leonard P., Li J., Burgdorf K., Grarup N., Jørgensen T., Brandslund I., Nielsen H. B., Juncker A. S., Bertalan M., Levenez F., Pons N., Rasmussen S., Sunagawa S., Tap J., Tims S., Zoetendal E. G., Brunak S., Clément K., Doré J., Kleerebezem M., et al., Nature 2013, 500, 541. [DOI] [PubMed] [Google Scholar]
- 39. Plaza Oñate F., Chatelier E. L.e, Almeida M., Cervino A. C. L., Gauthier F., Magoulès F., Ehrlich S. D., Pichaud M., Bioinformatics 2019, 35, 1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parks D. H., Chuvochina M., Waite D. W., Rinke C., Skarshewski A., Chaumeil P.‐A., Hugenholtz P., Nat. Biotechnol. 2018, 36, 996. [DOI] [PubMed] [Google Scholar]
- 41. Buchfink B., Xie C., Huson D. H., Nat. Methods 2015, 12, 59. [DOI] [PubMed] [Google Scholar]
- 42. Kanehisa M., Goto S., Nucleic Acids Res. 2000, 28, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vieira‐Silva S., Falony G., Darzi Y., Lima‐Mendez G., Garcia Yunta R., Okuda S., Vandeputte D., Valles‐Colomer M., Hildebrand F., Chaffron S., Raes J., Nat. Microbiol. 2016, 1, 16088. [DOI] [PubMed] [Google Scholar]
- 44. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B., Nucleic Acids Res. 2014, 42, D490 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. R Core Team , R: A Language and Environment for Statistical Computing 2019, https://www.R-project.org.
- 46. Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P. R., O'Hara R. B., Simpson G. L., Solymos P., Stevens M. H. H., Szoecs E., Wagner H. 2019, https://CRAN.R-project.org/package=vegan.
- 47. Noguchi K., Gel Y. R., Brunner E., Konietschke F., J. Stat. Softw. 2012, 50, 1.25317082 [Google Scholar]
- 48. Pasman W., Wils D., Saniez M.‐H., Kardinaal A., Eur. J. Clin. Nutr. 2006, 60, 1024. [DOI] [PubMed] [Google Scholar]
- 49. de la Cuesta‐Zuluaga J., Kelley S. T., Chen Y., Escobar J. S., Mueller N. T., Ley R. E., McDonald D., Huang S., Swafford A. D., Knight R., Thackray V. G., mSystems 2019, 4, e00261–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reeves A. R., Wang G. R., Salyers A. A., J. Bacteriol. 1997, 179, 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shipman J. A., Berleman J. E., Salyers A. A., J. Bacteriol. 2000, 182, 5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gray D. A., White J. B. R., Oluwole A. O., Rath P., Glenwright A. J., Mazur A., Zahn M., Baslé A., Morland C., Evans S. L., Cartmell A., Robinson C. V., Hiller S., Ranson N. A., Bolam D. N., van den Berg B., Nat. Commun. 2021, 12, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang S., Zhang X., Li H., Ren Y., Geng Y., Lu Z., Shi J., Xu Z., Appl. Microbiol. Biotechnol. 2021, 105, 7475. [DOI] [PubMed] [Google Scholar]
- 54. Kiewiet M. B. G., Elderman M. E., El Aidy S., Burgerhof J. G. M., Visser H., Vaughan E. E., Faas M. M., de Vos P., Mol. Nutr. Food Res. 2021, 65, 2000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Benítez‐Páez A., Hess A. L., Krautbauer S., Liebisch G., Christensen L., Hjorth M. F., Larsen T. M., Sanz Y., consortium M., Mol. Nutr. Food Res. 2021, 65, 2000996. [DOI] [PubMed] [Google Scholar]
- 56. Moro Cantu‐Jungles T., do Nascimento G. E., Zhang X., Iacomini M., Cordeiro L. M. C., Hamaker B. R., Carbohydr. Polym. 2019, 206, 389. [DOI] [PubMed] [Google Scholar]
- 57. Thompson R. S., Gaffney M., Hopkins S., Kelley T., Gonzalez A., Bowers S. J., Vitaterna M. H., Turek F. W., Foxx C. L., Lowry C. A., Vargas F., Dorrestein P. C., Wright K. P. J., Knight R., Fleshner M., Brain. Behav. Immun. 2021, 97, 150. [DOI] [PubMed] [Google Scholar]
- 58. Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D. R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J.‐M., Hansen T., Paslier D. Le, Linneberg A., Nielsen H. B., Pelletier E., Renault P., et al., Nature 2010, 464, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ezeji J. C., Sarikonda D. K., Hopperton A., Erkkila H. L., Cohen D. E., Martinez S. P., Cominelli F., Kuwahara T., Dichosa A. E. K., Good C. E., Jacobs M. R., Khoretonenko M., Veloo A., Rodriguez‐Palacios A., Gut Microbes 2021, 13, 1922241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang K., Liao M., Zhou N., Bao L., Ma K., Zheng Z., Wang Y., Liu C., Wang W., Wang J., Liu S.‐J., Liu H., Cell Rep. 2019, 26, 222.e5. [DOI] [PubMed] [Google Scholar]
- 61. Cuffaro B., Assohoun A. L. W., Boutillier D., Súkeníková L., Desramaut J., Boudebbouze S., Salomé‐Desnoulez S., Hrdý J., Waligora‐Dupriet A.‐J., Maguin E., Grangette C., Cells 2020, 9, 2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cuffaro B., Assohoun A. L. W., Boutillier D., Peucelle V., Desramaut J., Boudebbouze S., Croyal M., Waligora‐Dupriet A.‐J., Rhimi M., Grangette C., Maguin E., Microorganisms 2021, 9, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are openly available in the European Nucleotide Archive (ENA) at https://www.ebi.ac.uk reference number PRJEB42906.


