Abstract
Historical contingency has long figured prominently in the conceptual frameworks of evolutionary biology and community ecology. Evolutionary biologists typically consider the effects of chance mutation and historical contingency in driving divergence and convergence of traits in populations, whereas ecologists instead are often interested in the role of historical contingency in community assembly and succession. Although genetic differences among individuals in populations can influence community interactions, variability among populations of the same species has received relatively little attention for its potential role in community assembly and succession. We used a community‐level study of experimental evolution in two compositionally different assemblages of protists and rotifers to explore whether initial differences in species abundances among communities attributed to differences in evolutionary history, persisted as species that continued to evolve over time. In each assemblage, we observed significant convergence between two invaded treatments initially differing in evolutionary history over an observation period equal to ~40–80 generations for most species. Nonetheless, community structure failed to converge completely across all invaded treatments within an assemblage to a single structure. This suggests that whereas the species in the assemblage represent a common selective regime, differences in populations reflecting their evolutionary history can produce long‐lasting transient alternative community states. In one assemblage, we also observed increasing within‐treatment variability among replicate communities over time, suggesting that ecological drift may be another factor contributing to community change. Although subtle, these transient alternative states, in which communities differed in the abundance of interacting species, could nonetheless have important functional consequences, suggesting that the role of evolution in driving these states deserves greater attention.
Keywords: biological invasions, convergence and divergence, eco‐evolutionary dynamics, experimental evolution, historical contingency, intraspecific variability
INTRODUCTION
Historical contingency is of long‐standing interest to both evolutionary biologists and ecologists (e.g., Fukami, 2015; Gillespie, 2004; Lenski, 2017; Travisano et al., 1995), albeit from widely different perspectives. Evolutionary biologists typically consider whether historical contingency, mostly in the form of chance mutations, will influence divergence or convergence of populations of a single species, often under identical conditions imposed by selection experiments (Lenski, 2017; Simões et al., 2008). Ecologists, conversely, have more frequently focused on the role of historical contingency in community dynamics by considering how chance differences in the timing of species arrivals can have effects on community properties (e.g., Li et al., 2016). These approaches diverge in the level of biological organization on which they focus. Ecologists concentrate on the interspecific level, typically ignoring the role of historical contingency at the intraspecific level, whereas evolutionary biologists frequently begin with a single genetic clone of an asexually reproducing organism, but rarely consider the consequences of historical contingency in a larger community context (but see Meyer et al., 2012 for a two‐species community).
Interest in historical contingency among evolutionary biologists concerns whether the sequence and identity of mutations over time result in evolutionary changes that are path dependent (and consequently, divergent), or whether mutations interact with a common selective regime to produce largely repeatable (convergent) changes in phenotypes (e.g., Herron & Doebeli, 2013). Specifically, even if every sequence of evolutionary change is unique, it is still possible to reach the same peak in a fitness landscape via different underlying mutations, resulting in phenotypic convergence, despite different evolutionary pathways (Lenormand et al., 2009). Historical contingency may, however, result in evolutionary trajectories that reach different fitness peaks (Lenski & Travisano, 1994); divergence can arise from factors such as different mutations or epistatic interactions among mutations (Meyer et al., 2012). Convergence has been shown for traits directly related to fitness (i.e., adaptation) (Colosimo et al., 2005; Gilchrist et al., 2004), whereas historical contingency may be more important for traits that are less tightly tied to fitness (Travisano et al., 1995).
In community ecology, historical contingency is primarily of interest for its role in generating priority effects during community assembly (Chase, 2003; Fukami, 2015) and successional dynamics (Meiners et al., 2015), and for its role in determining species coexistence (Grainger et al., 2019; Letten et al., 2017). Priority effects do not always have long‐lasting effects on community assembly, but can persist for multiple generations in some situations (Toju et al., 2018). They can result in the divergence of community structure and function, including alternative transient or stable states, or compositional cycles (Fukami, 2015). Similarly, successional dynamics can result in community divergence (e.g., Taylor & Chen, 2011), convergence (e.g., Alday et al., 2011), or more complex patterns (e.g., del Moral & Lacher, 2005).
Historical contingency in community ecology has traditionally focused on how the different timing (order) of species arrivals affects community patterns, but not the potential consequences of genetic variation within species (Chase, 2003; Fukami, 2015; Violle et al., 2012). Only more recently have ecologists begun to investigate the importance of intraspecific diversity (e.g., Jung et al., 2010; Laughlin et al., 2012; Siefert, 2012; Zee & Fukami, 2018) or eco‐evolutionary dynamics in community assembly (e.g., Knope et al., 2012; Kremer & Klausmeier, 2017; Lee et al., 2012; Urban & De Meester, 2009), species coexistence (e.g., Klauschies et al., 2016; Kremer & Klausmeier, 2013), or community properties such as resilience (e.g., Barabás & D'Andrea, 2016). Nonetheless, work with foundation species (i.e., plant species that define habitats sensu Whitham et al., 2012) suggests that different genotypes of the same species can be an important determinant of community assembly (e.g., Keith et al., 2017; Lamit et al., 2016).
Few studies have examined the possibility that differing evolutionary histories of interacting species within a community may produce communities that differ in terms of the abundance of species (and therefore, community composition) (but see Zee & Fukami, 2018 for work with competing pairs of Pseudomonas fluorescens strains in which evolutionary history was manipulated). Even if communities containing populations of species that differ in evolutionary history eventually converge on a single community state (again, in terms of the abundance of species within the community as opposed to species richness or the identity of species present), evolutionary history may be one factor that could govern the emergence of transient alternative states. Transient alternative states can persist for many generations (Fukami & Nakajima, 2013), frequently for longer than the interval between disturbances that restart successional dynamics (Fukami & Nakajima, 2011). The emergence of transient alternative states may consequently constitute an important mechanism that maintains regional species diversity (Fukami & Nakajima, 2013).
Here we take advantage of an experiment that assessed the importance of evolutionary history in driving the outcomes of biological invasions (Faillace & Morin, 2016) to examine whether communities with differences in the recent evolutionary history of populations would exhibit persistent divergence or convergence among communities over time. We constructed communities of two assemblages of protists and rotifers (from this point forwards termed Assemblages A and B) with combinations of species that differed in their exposure to, and evolutionary history with, a designated invading species. To determine if post‐invasion evolutionary history had the ability to alter the trajectory of community development we compared species performance over time (measured as mean abundance) from treatments with naïve (i.e., with no previous history of evolutionary experience with an invader) or evolved (i.e., with a history of potential post‐invasion evolution) populations of invaders and residents. In this way we determined whether abundances of populations from initially divergent treatments became more similar over time, regardless of the initial differences in evolutionary history of invaders and residents. We reasoned that if post‐invasion interactions represent a common selective regime across all invaded treatments within each assemblage, early differences among invaded treatments might become less pronounced over time as initially naïve populations evolved following invasion during our observation period, due to a tendency to converge toward a single community end state under a shared, post‐invasion, selective regime. We predicted that if post‐invasion evolution occurred in a rapid and mostly deterministic, repeatable fashion, then naïve populations of residents and invaders would evolve to effectively become less naïve over time. In this case, we would expect to see increased similarity among invaded communities after tens of generations of interactions, compared with the initial post‐invasion dynamics assessed when species had just experienced different selective regimes (before naïve populations might be expected to undergo much evolution). Stochastic processes (i.e., drift) could also lead to increased variation among replicates within treatments depending on the degree of repeatability in evolutionary outcomes following invasion, such that drift could conceivably lead to the blurring of differences among treatments, as a result of increased within‐treatment variance relative to the among‐treatments variance. Alternatively, the evolutionary history of interacting species could instead dictate divergent trajectories of community development, leading to the formation of alternative transient or stable states differing in the abundance of species. In that case, we would expect communities that were composed of different combinations of evolved and naïve invaders and residents to remain different, or even become more divergent over time.
METHODS
Experimental design
We implemented our experimental protocol in three phases (Appendix S1: Figure S1). Phase 1 began in August 2013, during which we established five replicates of each of two compositionally different assemblages of ciliate protists and rotifers, which were feeding on the same four bacterial resource species (Bacillus cereus, Bacillus subtilis, Proteus vulgaris, and Serratia marcescens) to establish uninvaded and invaded lines of each assemblage. Assemblage A contained five ciliates, Blepharisma americanum, Euplotes patella, Paramecium bursaria, Prorodon niveus, Spirostomum teres, and one rotifer, Lecane sp. Assemblage B contained three ciliates, Euplotes daidaleos, Paramecium caudatum, Stentor coeruleus, and a second rotifer, Monostyla sp. These assemblages were both the source and the target of experimentally contrived invasions.
The protists and rotifers were originally collected from Bamboo Pond on the Rutgers University New Brunswick, NJ, campus and were maintained in the laboratory for 3 years before the start of the experiment, equal to ~550–1095 protist generations. For the years prior to the start of Phase 1 of the experiment, five species from Assemblage A, B. americanum, E. patella, Lecane sp., P. niveus, and S. teres were maintained together, whereas three species from Assemblage B were originally maintained together, Monostyla sp., P. caudatum, and S. coeruleus. Protists and rotifers were a mix of bacterivores and predators. During this time, the designated invaders, P. bursaria from Assemblage A and E. daidaleos from Assemblage B, were grown continuously as single species cultures. They were introduced to each of their source assemblages several months before the initiation of Phase 1 of the experiment to maximize their acclimation time in their source assemblage.
Assemblages grew in replicated microcosms (n = 5), that is, loosely lidded 250‐ml jars with 100 ml of protist pellet medium (1 Carolina Biological protist pellet, 2.8 L water, plus 0.14 g Herptivite) previously autoclave sterilized and then bacterized with our four bacterial resource species plus two sterile wheat seeds for additional nutrients. Identical laboratory conditions (spatial positions randomized in a Percival incubator at 22°C with a 5 h : 19 h, light : dark photoperiod) allowed resident species in each assemblage to additionally acclimate to laboratory conditions and each other during Phase 1, lasting an additional ~6 months (late August 2013 to early March 2014). Replicate microcosms in Phases 1 and 2 were intended to prevent any catastrophic loss of uninvaded and invaded lines of the two assemblages. For this reason, replicates were homogenized at each subculturing event during these phases to maintain a single uninvaded and an invaded line of each assemblage. Because our design considered a single uninvaded and a single invaded line of each assemblage, comparisons of the effects that might emerge as a result of differing mutations or other evolutionary changes were outside the scope of this experiment (i.e., had we instead maintained multiple invaded and uninvaded lines of each assemblage throughout Phases 1 and 2 to be used in Phase 3). We subcultured organisms every 3 weeks by placing ~5 ml of well mixed culture into new sterile microcosms with 100 ml of fresh medium. Although this procedure does create a population bottleneck for the protists and rotifers, it was essential to ensure that any observed evolutionary responses resulted from the interactions between the protists and rotifers, and not in response to changes within the bacterial community. Therefore, fresh sterile protist pellet medium used during subculturing was always inoculated before use with the same four bacterial taxa continuously maintained individually on agar slants, effectively resetting the bacterial composition at each subculturing event. This protocol makes it highly unlikely that the protists and rotifers had experienced consistent directional selection resulting from long‐term evolutionary changes in bacterial traits (i.e., we provided a regular replenishment of ungrazed bacteria throughout the experiment), or that any differences among treatments were driven by changes to the bacterial community itself.
At the start of Phase 2 (March 2014), we invaded the replicates of each assemblage by designating one resident species from each assemblage as the experimental invader of the other assemblage. Therefore, the invading species were naïve to species identity in newly invaded communities, but not to previous interactions with other species, a design that closely matches natural invasions. An inoculum of 15–20 individuals of Assemblage A resident, P. bursaria, invaded each of five replicates of Assemblage B. Similarly, an inoculum of 15–20 individuals of E. daidaleos, a resident from Assemblage B, invaded five replicates of Assemblage A. Invaders were functionally similar mixotrophic bacterivores and each assemblage contained a congener with which the invader might be expected to interact strongly. Replicates of invaded and uninvaded control lineages of each assemblage then were grown under identical laboratory conditions (with periodic subculturing during which we again homogenized each uninvaded and invaded line of both assemblages) for an additional 200–400 protist generations (13 months) to provide an opportunity for populations of residents and invaders to evolve following invasion. Uninvaded resident assemblages therefore experienced a total of ~300–550 protist generations during the course of the experiment (18 months), ensuring a prolonged recent evolutionary history of residents, whereas, in invaded replicates, invaders and residents experienced a recent interaction history of ~200–400 protist generations.
After 13 months of interaction, in Phase 3 we created our final treatment combinations (n = 5) of evolved (denoted with a “+”) or naïve (−) invaders (I) and residents (R) with a second round of community assembly. Following a subculturing event in which the uninvaded and invaded lines of each assemblage were again each homogenized, we established our final treatments over a 3‐week period. The treatments for these test communities included: (1) invaders and residents both evolved (coevolved treatment: +I/+R), (2) evolved invaders and naïve residents (evolved invader treatment: +I/−R), (3) naïve invaders and evolved residents (evolved residents treatment: −I/+R), (4) invaders and residents both naïve (naïve invasion treatment: −I/−R), and (5) solely naïve residents (−R) as uninvaded controls. After subculturing, replicates of the invaded line became the coevolved “end state” treatment against which other invaded treatments and the uninvaded control treatment could be compared. We then individually invaded each of five replicates of communities with 15–20 individuals of the appropriate invader (either evolved or naïve), depending on treatment. Evolved invaders (+I) originated in invaded lines of each assemblage (in which the invader had a recent evolutionary history in the novel assemblage), whereas naïve invaders (−I) originated in their source assemblage (i.e., the assemblage in which each invader was considered a resident: Assemblage A for P. bursaria and Assemblage B for E. daidaleos). Similarly, evolved residents (+R) originated in the invaded (coevolved) line of each assemblage, whereas naïve residents (−R) originated from the uninvaded (control) line of each assemblage (i.e., Assemblage A grown continuously without the invader, E. daidaleos, present, and Assemblage B grown continuously without the invader, P. bursaria). For instance, the evolved residents treatment (−I/+R) contained naïve invaders (−I) that originated from their source assemblages, invading previously reassembled communities of evolved residents from the invaded line of each assemblage (+R). For the evolved residents treatment (−I/+R), it was necessary to first reassemble resident communities without invaders by isolating evolved residents from the invaded line of each assemblage and allowing them to reach detectable equilibrium abundances before introducing naïve invaders to the community. To reassemble these resident communities, we isolated ~50 individuals of each evolved resident from the invaded line and allowed them to grow to equilibrium abundances over a period of several weeks, before introducing 15–20 naïve invaders. Treatments with naïve invaders and/or residents enabled us to make comparisons of ecological effects between ancestral (unevolved, no recent history of interaction) and evolved (recent history of interaction) states for both invaders and resident species.
After invaders exceeded a detection threshold, over a 12‐week observation period, we estimated the species densities (number per milliliter) from periodic counts (nine observations total) of the number of individuals of each species in a well‐mixed subsample of known volume (~0.3 ml, with volume measured precisely by sample mass) sampled without replacement from each replicate from all communities. All species were visually distinct and easily identified using a Nikon SMZ microscope.
Statistical analyses
Evolutionary history and community convergence
To assess whether communities tended to converge over time, we analyzed community trajectories using the framework developed by De Cáceres et al. (2019). For each experimental assemblage, we established patterns of community dissimilarity using non‐metric multidimensional scaling (NMDS) based on Bray–Curtis distances in the R package vegan (Oksanen et al., 2007). Then, we calculated community centroids for each of the invaded treatment groups at each time point throughout the experiment (n = 9) to track the central tendency of community trajectories. For all pairwise comparisons between treatments, we next calculated the Euclidean distance between their community centroids (i.e., dissimilarity between communities) at each time point and assessed whether the distance between community centroids decreased (convergence) or increased (divergence) over time. We used Mann–Kendall trend tests in the trend R package (Pohlert, 2020) to detect monotonic trends of the distance between communities over time. This test focuses on the signal of these relationships in which a tendency to observe a decrease in distance between communities over time would indicate convergence (i.e., communities are becoming more similar over time yielding negative tau [τ] values) and an increase in distance between communities indicates divergence (i.e., communities are becoming more dissimilar over time yielding positive tau values). When communities show variable periods of convergence and divergence such that no clear trend is observed, tau values are non‐significant.
For each assemblage, a first community convergence analysis tested for the effects of evolutionary history on invaded communities. These analyses excluded the uninvaded control (−R) communities, to avoid spurious significant differences in community structure due to the addition of the invader (i.e., resulting solely from the experimentally imposed treatments). Furthermore, one resident, P. niveus, in Assemblage A was excluded because it failed to establish in the evolved residents treatment (−I/+R), which was likely due to its small starting propagule size. In an additional analysis for each assemblage, we also tested whether resident community composition converged or diverged in response to invasion. To test for the effects of invasion solely on residents, we included the uninvaded control communities, but removed the invader from the analyses.
The effects of invasion and evolutionary history on population and community dynamics
To evaluate the effects of evolution over time on population and community dynamics, we modeled species abundances using multivariate generalized linear models (function “manyglm”) with a negative binomial distribution in the mvabund package (Wang et al., 2012). These models included interactions between time and (1) invader evolution, (2) resident evolution, and (3) coevolution (i.e., the interaction between resident and invader evolution). This analysis assessed both community‐level and species‐level differences among experimental microcosms. For individual species, we used univariate tests that were adjusted for multiple comparisons through resampling based on the Holm step‐down procedure (Wang et al., 2012). In our models, we accounted for repeated sampling by restricting permutations within blocks that correspond to the identity of microcosm jars using the “bootID” argument (n permutations = 999). Test statistics were based on likelihood ratio tests. Using this analysis, we conducted two separate tests of these data: (1) the effects of invasion on residents and (2) the overall effects of the evolutionary treatments in invaded communities, which excluded the uninvaded control (−R). In test 1, we removed the invader from the analysis to avoid spurious differences based on the addition of the invader in invaded communities.
Ecological drift
Stochastic processes (i.e., drift) could also lead to increased variation among replicates within treatments, depending on the degree of repeatability in evolutionary outcomes following invasion, such that drift could conceivably lead to declining differences among treatments as a result of increased within‐treatment variance relative to the among‐treatments variance. We tested for drift using linear mixed effects models that assessed whether the distance from each replicate to the treatment centroid increased over time. Our model included invader and resident evolution and their interaction as fixed effects and microcosm ID as a random effect. All analyses and graphics were produced in R v.4.0.2 (R Core Team, 2021).
RESULTS
Evolutionary history and community convergence
Despite starting at different community states, communities containing coevolved species (+I/+R) and only the evolved invader (+I/−R) converged toward a similar community composition over time in Assemblage A (tau = −0.72, p = 0.009; Figure 1; Appendix S1: Figure S2). All other community trajectory comparisons examining the effects of evolution over time in invaded communities in Assemblage A were not significant (p > 0.05; Appendix S1: Figure S2). Community trajectories showed that initially the evolved invader communities more strongly favored the resident Lecane sp. and P. bursaria than did communities from the coevolved treatment (Figure 1).
FIGURE 1.
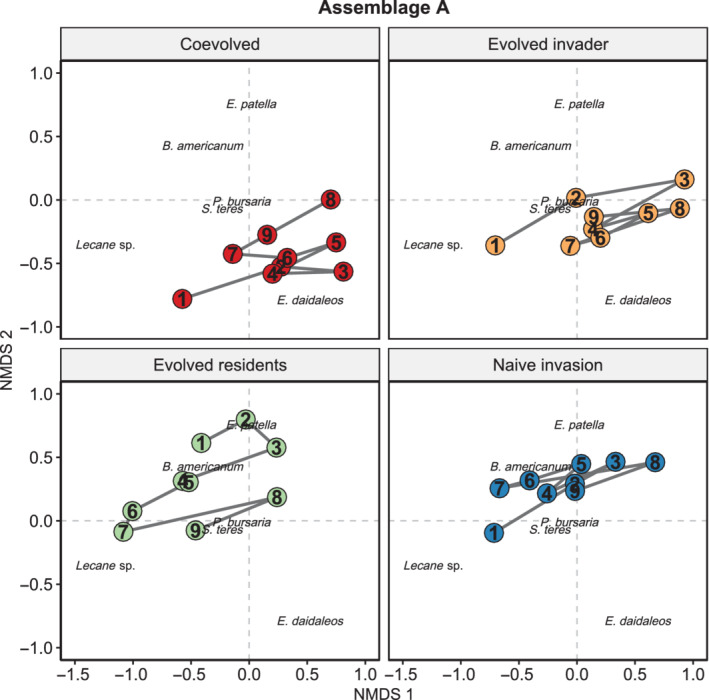
The effects of evolutionary history on invaded community trajectories in Assemblage A. Significant trends of convergence were only observed between coevolved and evolved invader communities. The position of constituent species and community centroids are plotted and, within these points, numbers indicate the sequential community survey events. Non‐metric multidimensional scaling (NMDS) stress = 0.16
Although we observed community‐level effects evolution and coevolution by invaders and residents in both assemblages (Table 1), when directly examining the effects over time of evolutionary history on species abundances in Assemblage A, we observed the effects of coevolution and invader evolution in some species (Table 2). Specifically, the abundance of Lecane sp. declined over time in communities with invader evolution (coef = −0.002, p = 0.028) and coevolution (coef = −0.054, p = 0.009; Figure 2). Abundance of P. bursaria generally declined over time (coef = −0.007, p = 0.008; Figure 2), and this decline was greater in communities with evolved invaders (coef = −0.014 p = 0.019; Figure 2). Although overall the abundance of E. daidaleos slightly increased over time (coef = 1.00, p = 0.073; Figure 2), it declined in the coevolved treatment (coef = −0.054, p = 0.003; Figure 2) and was an apparent driver of increasing similarity among communities from the coevolved and evolved invader treatments (Figure 1). B. americanum, P. bursaria, and S. teres remained more prominent within the compositions of the naïve invasion (−I/−R) and evolved residents (−I/+R) communities, relative to the coevolved and evolved invader treatments, although E. patella was relatively rare in all treatments (Figure 1). Both S. teres and E. patella declined over time (p = 0.001; Figure 2), however their population abundances were not impacted by coevolution, invader, or resident evolution (p > 0.05; Table 2). Ultimately, toward the end of the experiment, communities from both coevolved and evolved invader treatments favored the invader, E. daidaleos, and Lecane sp., relative to the naïve invasion and evolved residents communities (Figure 1). Nonetheless, the observed convergence between coevolved and evolved invader communities appeared to be driven by similar declines in the abundance of the invader E. daidaleos and the resident Lecane sp. Additionally, declines in E. patella and S. teres in all treatments contributed to the observed convergence, although this effect was unrelated to the evolutionary treatments.
TABLE 1.
Community‐level response to invader and resident evolution following invasion in Assemblages A and B
| Assemblage A | Assemblage B | |||
|---|---|---|---|---|
| Source of Variation | Deviance | p | Deviance | p |
| Time | 183.00 | 0.001 | 50.88 | 0.001 |
| Invader evolution × time | 25.87 | 0.006 | 23.94 | 0.002 |
| Resident evolution × time | 18.18 | 0.015 | 12.49 | 0.040 |
| Coevolution × time | 35.47 | 0.001 | 26.13 | 0.003 |
Note: Deviance and p‐values are reported from multivariate tests with generalized linear models that are modeled with a negative binomial distribution. Statistically significant results at α = 0.05 are denoted with p‐values in bold font.
TABLE 2.
Species‐level response to invader and resident evolution following invasion within Assemblage A
| Source of Variation | B. americanum | S. teres | E. patella | E. daidaleos | Lecane sp. | P. bursaria | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deviance | p | Deviance | p | Deviance | p | Deviance | p | Deviance | p | Deviance | p | |
| Time | 4.474 | 0.101 | 42.489 | 0.001 | 118.867 | 0.001 | 5.341 | 0.073 | 0.128 | 0.783 | 11.696 | 0.008 |
| Invader evolution × time | 0.001 | 0.406 | 0.117 | 0.899 | 2.689 | 0.222 | 4.369 | 0.094 | 8.878 | 0.028 | 9.815 | 0.019 |
| Resident evolution × time | 1.234 | 0.477 | 5.464 | 0.083 | 0.199 | 0.620 | 2.884 | 0.244 | 3.342 | 0.244 | 5.058 | 0.091 |
| Coevolution × time | 3.973 | 0.129 | 0.396 | 0.596 | 0.841 | 0.596 | 16.021 | 0.003 | 10.453 | 0.009 | 3.790 | 0.120 |
Note: Deviance and p‐values are reported from univariate tests with generalized linear models that are modeled with a negative binomial distribution. p‐values are adjusted for multiple comparisons through resampling based on the Holm step‐down procedure. Statistically significant results at α = 0.05 are denoted with p‐values in bold font.
FIGURE 2.
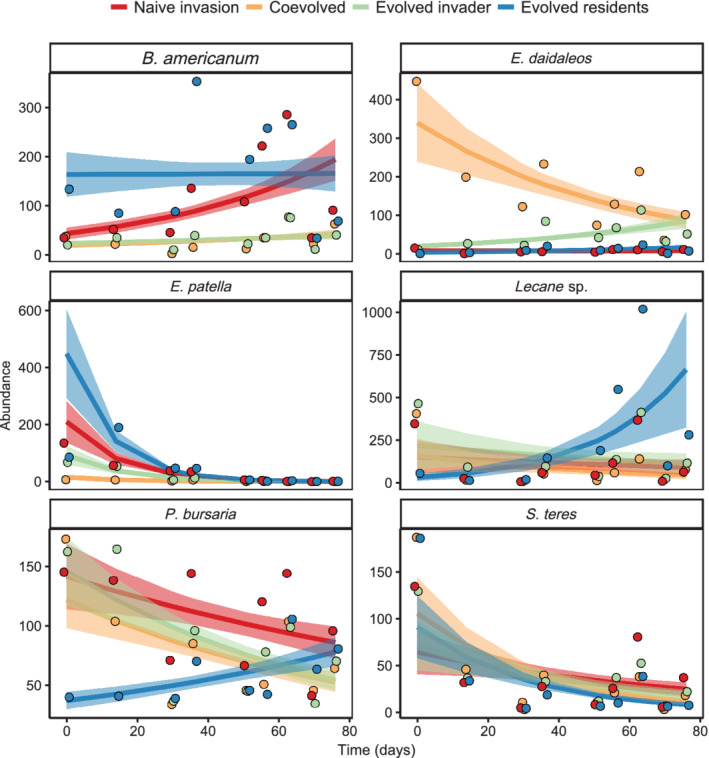
Population dynamics of species within invaded communities of Assemblage A. Means and fitted lines with standard error are plotted. Model estimates were back transformed from a negative binomial glm. Points indicate the observed raw abundances, offset to help visualize the data
When examining the changes over time due directly to invasion, we observed significant divergence between the naïve invasion treatment and the uninvaded control (−R) treatment (Appendix S1: Figures S3 and S4). Upon examining resident abundances, changes in community composition between the invaded communities and the uninvaded control were attributed to Lecane sp., which increased over time in the naïve invasion treatment (p = 0.001; Appendix S1: Table S1, Figure S5). There was a weak, marginally significant positive effect of invasion on B. americanum abundance (p = 0.095; Appendix S1: Table S1, Figure S5).
In Assemblage B, communities containing evolved residents converged with communities from the naïve invasion treatment (tau = −0.61, p = 0.03; Figure 3; Appendix S1: Figure S6). There was no notable signal of divergence among communities differing in evolutionary history within this assemblage (tau range: 0.05 to −0.61), and all other comparisons among communities were not significant (p > 0.05; Appendix S1: Figure S6). Naïve invasion communities had a community trajectory that initially favored the invader, P. bursaria, and one resident, Monostyla sp., but over time the structure became more balanced between these two species and two additional residents, E. daidaleos and P. caudatum (Figure 3). In contrast, despite converging to a similar end point, the communities with evolved residents instead initially favored three residents, E. daidaleos, P. caudatum, and S. coeruleus. Evolved residents and evolved invader communities also showed signals of convergence, but this trend was marginally significant (tau = −0.50, p = 0.07). For the evolved invader communities, the trajectory initially favored Monostyla sp. and P. bursaria (the invader) before again ending with a community balancing these two species as well as E. daidaleos and P. caudatum.
FIGURE 3.
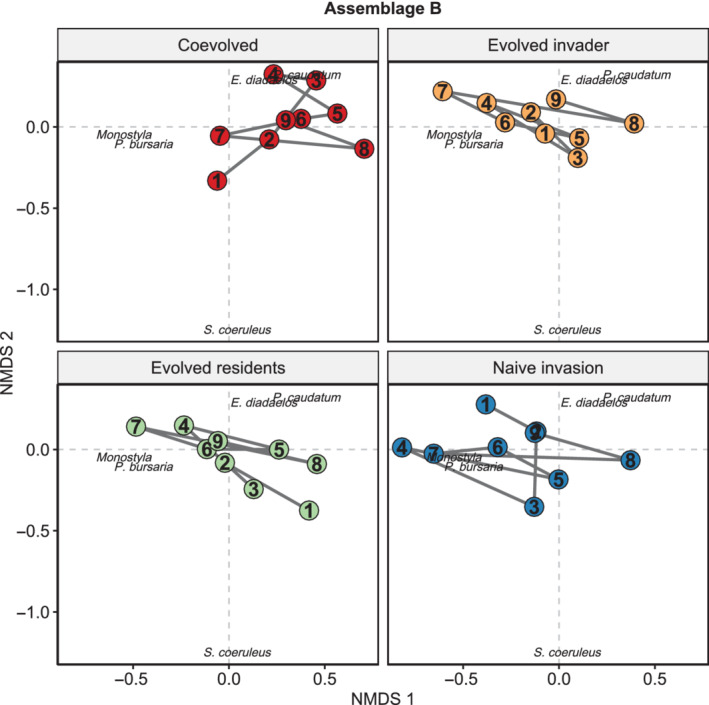
Effects of evolutionary history on invaded community trajectories in Assemblage B. Significant trends of convergence were only observed between evolved residents and naïve invasion communities. The position of constituent species and the community centroids are plotted and, within these points, numbers indicate the sequential community survey events. Non‐metric multidimensional scaling (NMDS) stress = 0.14
Examining the effects of evolutionary history on species abundances over time (Table 3), showed that the abundance of the invader, P. bursaria, decreased in the coevolved treatment (coef = −0.050, p = 0.002) and the evolved invader communities (coef = −0.009, p = 0.001; Figure 4), whereas Monostyla sp. only declined in the coevolved treatment (coef = −0.023, p = 0.051; Figure 4). The abundance of E. daidaleos increased similarly in the evolved invader (coef = 0.024, p = 0.018) and evolved residents (coef = 0.024, p = 0.014) communities (Figure 4). Finally, abundance of S. coeruleus declined similarly among all invaded treatments over time (coef = −0.041, p = 0.001), but the relationship was independent of evolutionary history (p > 0.05; Figure 4). There was no effect of time or evolution on P. caudatum population abundance (p > 0.05). Overall, convergence between the naïve invasion and evolved residents communities in Assemblage B was driven by the decline in S. coeruleus across all treatments and increase in E. daidaleos in the evolved residents treatment.
TABLE 3.
Species‐level response to invader and resident evolution following invasion within Assemblage B
| S. coeruleus | P. caudatum | Monostyla sp. | E. daidaleos | P. bursaria | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Source of Variation | Deviance | p | Deviance | p | Deviance | p | Deviance | p | Deviance | p |
| Time | 24.341 | 0.001 | 0.850 | 0.492 | 0.358 | 0.506 | 23.453 | 0.001 | 1.877 | 0.244 |
| Invader evolution × time | 0.340 | 0.810 | 0.503 | 0.810 | 0.575 | 0.810 | 8.627 | 0.018 | 13.891 | 0.001 |
| Resident evolution × time | 0.019 | 0.993 | 0.852 | 0.751 | 0.759 | 0.993 | 8.911 | 0.014 | 2.702 | 0.327 |
| Coevolution × time | 0.587 | 0.522 | 1.702 | 0.410 | 6.363 | 0.051 | 3.456 | 0.217 | 14.026 | 0.002 |
Note: Deviance and p‐values are reported from univariate tests with generalized linear models that are modeled with a negative binomial distribution. p‐values are adjusted for multiple comparisons through resampling based on the Holm step‐down procedure. Statistically significant results at α = 0.05 are denoted with p‐values in bold font.
FIGURE 4.
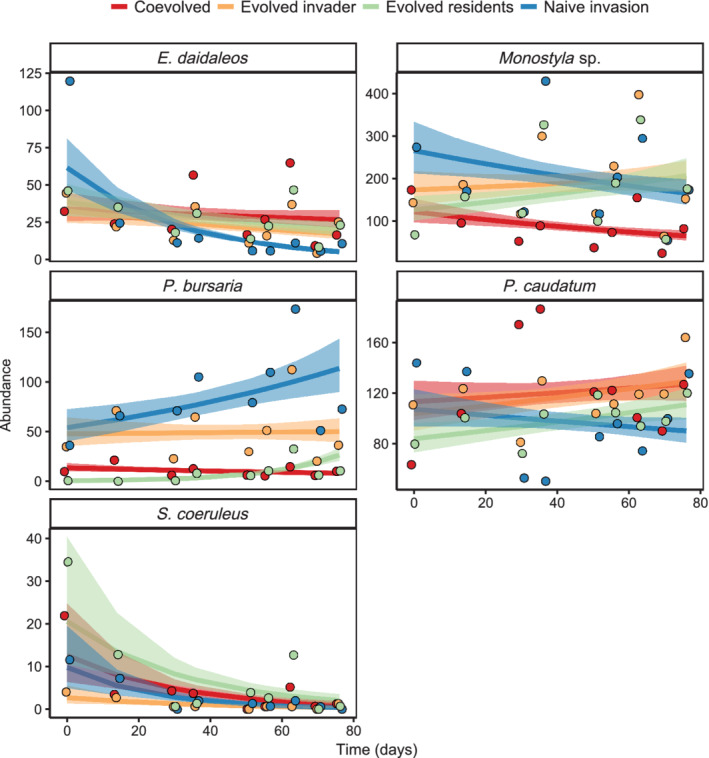
Population dynamics of species within invaded communities of Assemblage B. Means and fitted lines with standard error are plotted. Model estimates were back transformed from a negative binomial glm. Points indicate the observed raw abundances, offset to help visualize the data
When comparing the invaded resident communities to the uninvaded controls to examine the effects of invasion, we observed significant convergence between the naïve invasion treatment and the uninvaded control treatment (Appendix S1: Figures S7 and S8). E. daidaleos abundance was positively affected by invasion (p = 0.032) and the other residents did not show a main effect of invasion on their abundance (p > 0.05; Appendix S1: Table S2, Figure S9).
Drift
The contribution of drift to community patterns varied between assemblages. In Assemblage A, the distance to the within‐treatment community centroid increased over time (p < 0.0001), which is indicative of drift. However, this increase in within‐community variation over time was lower in the naïve invasion communities (Appendix S1: Figures S10 and S11). In contrast, the distance to the within‐treatment community centroid decreased over time in Assemblage B (p = 0.02), and the overall distance to the centroid was higher in the evolved residents community (resident evolution, p = 0.021), indicating that communities in this treatment had more variable compositions than those from other treatments (Appendix S1: Figures S12 and S13).
DISCUSSION
We evaluated the course of community development in two different species assemblages to understand whether populations differing in evolutionary history could produce transient alternative ecological community states. Our results demonstrated that the evolutionary history of interacting species can drive differences in community development following biological invasions. The differences in converging community structure observed in the two assemblages seemed to be related to whether the invader or resident species evolved. For Assemblage A, in which we previously demonstrated ecological effects related to evolution in the invader (Faillace & Morin, 2016, 2020), we observed increasing similarity among communities from the coevolved and evolved invader treatments. In contrast, for Assemblage B, in which we had shown that evolution of resident species drove differences in the community (Faillace & Morin, 2016, 2020), we found that communities with naïve invasions and evolved residents became more similar over time. Here, the evolved resident communities showed little directional change over time from the initial to final community state, whereas the naïve invasion communities appeared to ultimately become more similar to the communities with evolved residents.
The increase in similarity between the naïve invasion and evolved residents communities in Assemblage B was consistent with our expectation that post‐invasion evolution occurs in a rapid and mostly repeatable fashion with the species present in each assemblage representing a common selective regime, such that naïve populations would evolve to effectively become less naïve over time. Additionally, across both assemblages, communities from naïve treatments showed some trends of convergence with coevolved treatments, however the convergence was not statistically significant. It seems likely that these effects might have become stronger and statistically significant had we continued our observations beyond the tens of generations that we observed. These results agree with a recent field experiment with native and invasive grass species occurring in sympatry and allopatry, in which evolution in populations of the native species appeared to produce repeatable responses to competition with the invasive species (Germain et al., 2020). Nonetheless, in our experiment, the lack of complete convergence to a single uniform community state within each assemblage suggested that the differences in populations that we attributed to evolutionary history could produce long‐lasting transient alternative community states.
When considering the patterns of community similarity over time with the population‐level results, we observed that, for Assemblage A, signals of convergence among treatments appeared to be driven by increases in the invader, E. daidaleos, across all except the coevolved treatment, for which it declined, as well as declines in abundance in all treatments of E. patella and S. teres. Nonetheless, we also observed evidence of drift occurring among communities within treatments. Therefore, the observed pattern appears to be driven, at least in part, by increasing within‐treatment dispersion, implying an additional role for stochastic processes, specifically ecological drift, in community development for this assemblage.
For Assemblage B, the increase in among‐treatment similarity between the evolved residents and naïve invasion treatments appeared to be driven by a community structure increasingly balanced among the invading species, P. bursaria, as well as residents Monostyla sp., E. daidaleos, and P. caudatum. Additionally, the abundance of S. coeruleus declined across all invaded treatments over time. Here, although we observed changes to the degree of drift over time, the evolved resident treatment exhibited consistently greater within‐treatment dispersion compared with the remaining treatments. This elevated dispersion suggests that the abundances of interacting species in the treatment with evolved residents remained more variable over time compared with those in the other treatments. One possible explanation for this result is that the effects in this treatment are potentially driven by evolution occurring in multiple interacting species, as opposed to evolution in a single species, as seen in Assemblage A, possibly increasing variability in those interactions.
Historical contingency is known to influence the properties of both populations and communities (Fukami, 2015; Lenski, 2017). When considering interactions among species, historical contingency can determine the composition and functioning of communities through priority effects and eco‐evolutionary dynamics to influence community assembly and coexistence, as well as succession (Fukami, 2015; Grainger et al., 2019; Kremer & Klausmeier, 2017; Meiners et al., 2015). Transient alternative states can differ from alternative stable states in the conditions under which they occur and the patterns of diversity in which they result; because transient states may be common in many natural communities, they could be particularly important in maintaining biodiversity (Fukami & Nakajima, 2011, 2013). For instance, different genotypes of foundation species can promote the development of measurably different communities of associated dependent species (e.g., Keith et al., 2017; Lamit et al., 2016). Because chance mutations can cause divergence in genotypes (and therefore phenotypes) under identical selective regimes (Lenski, 2017; Meyer et al., 2012), it follows that the dominant phenotypes present among allopatrically evolving populations or in metacommunities may differ in important ways (e.g., Urban, 2010), even when the populations evolve under similar conditions. In fact, Brockhurst et al. (2006) demonstrated that independently evolved populations of wrinkly spreader (WS) phenotypes of P. fluorescens can differ in aspects of the WS phenotype with important consequences for communities, despite evolving under identical conditions. Sympatrically coevolved pairs of strains exhibited greater character displacement, yielding both greater productivity and reduced invasibility compared with randomly assembled (allopatric) pairs. Pairs of sympatrically and allopatrically evolved strains of P. fluorescens also differ in the strength of the priority effects that governed competitive outcomes (Zee & Fukami, 2018). Taken together, even in cases in which eventual convergence of phenotypes might be expected, historical contingency resulting in phenotypic variability among populations could cause important long‐term differences in community structure or function.
The importance of ecological and evolutionary processes interacting at contemporary timescales is increasingly recognized (Ellner, 2013; Koch et al., 2014; Schoener, 2011). Some studies of both laboratory and natural systems have clearly demonstrated ongoing contemporary evolution (e.g., Bassar et al., 2012; Farkas et al., 2016; Hiltunen & Becks, 2014), highlighting the need to consider ecological and evolutionary dynamics as simultaneous and interacting processes that drive community dynamics. Such eco‐evolutionary dynamics (Fussmann et al., 2007; Kinnison & Hairston, 2007) can have effects on the phenotypic traits of species (Grant & Grant, 2002; Stuart et al., 2014), population and community dynamics (Becks et al., 2010, 2012; Faillace & Morin, 2016, 2020; Yoshida et al., 2003), and even ecosystem functioning (Palkovacs et al., 2009). Eco‐evolutionary feedbacks (such as the ecology → evolution → ecology feedback observed here) may be of particular importance in governing the divergence of populations through their ability to amplify intraspecific phenotypic trait variation (Bailey et al., 2013), and may be crucial for our understanding of how species diversity arises (Post & Palkovacs, 2009) and is maintained. Despite their potential importance, few studies have attempted to identify the community‐level effects of eco‐evolutionary dynamics in complex communities. Nonetheless, we clearly need to improve our understanding of evolution in a community context (terHorst et al., 2018).
We did not identify specific molecular genetic targets of evolution, but have previously argued that our design explicitly disentangled possible induced plastic phenotypic responses from heritable changes (Faillace & Morin, 2020). Not only did the identity of species in each community remain constant with all other conditions maintained under common garden conditions, but also an additional 3 weeks passed before we began our observation period, representing the passage of ~21–42 generations for the protists. This is well after the period of maximum induction of plastic changes for protists (i.e., plasticity in protists is reasonably well characterized in the literature with phenotypically plastic responses typically fully induced in a population within the first 72 h, corresponding to about two or three generations after exposure to novel conditions) (Duquette et al., 2005; Fyda & Wiackowski, 1998; Wiackowski & Staronska, 1999). Finally, tracking abundances over multiple generations and turnovers of the individuals in populations ensured that any observed differences represent the ecological manifestations of heritable differences among lines of evolved and naïve populations of our species, rather than transient dynamics due to plasticity. The observed convergence among some treatments in each assemblage is consistent with selection acting on standing genetic variation, rather than on the appearance of new mutations (i.e., as for the populations of yeast studied by Burke et al., 2014). Nonetheless, similar to both the scale and time dependency of convergence and divergence observed in Escherichia coli (Lenski, 2017), new mutations could greatly alter performance and community composition, especially over longer time scales.
We argue that evolutionary history can contribute to the emergence of long‐lasting alternative community states in which the abundances of constituent species differ significantly, but it remains to be seen how important it is in natural communities. Common and rare species can differ dramatically in their respective roles in biological communities (Gaston & Fuller, 2008; Jain et al., 2014). This implies that when the relative contribution of rare and common species is shifted, these kinds of transient alternative states could be expected to have important functional consequences. In our experiment, demonstrably different alternative community states persisted for tens of generations in both assemblages, with incomplete convergence observed during the course of our observations, indicating that these transition states would be biologically relevant for community dynamics. The transient alternative states that we observed here, in which communities differed in the abundance but not in the identity of interacting species, are seemingly subtle, but could nonetheless have important functional consequences, suggesting that the role of evolution in driving these states deserves greater attention.
AUTHOR CONTRIBUTIONS
Cara A. Faillace and Peter J. Morin designed the study. Cara A. Faillace collected all data. Rita L. Grunberg conducted all statistical analyses. All authors jointly wrote the manuscript. The authors declare no competing financial interests.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
Rutgers University (Peter J. Morin) and a Rutgers University Graduate Program in Ecology and Evolution Ted Stiles Grant (Cara A. Faillace) supported this research. Thanks to members of the Morin laboratory and two anonymous reviewers for comments on the manuscript.
Faillace, Cara A. , Grunberg Rita L., and Morin Peter J.. 2022. “Historical Contingency and the Role of Post‐Invasion Evolution in Alternative Community States.” Ecology 103(7): e3711. 10.1002/ecy.3711
Handling Editor: Hideyuki Doi
Funding information Rutgers University
DATA AVAILABILITY STATEMENT
Data and code (Grunberg, 2022) are available in Zenodo at https://doi.org/10.5281/zenodo.5818017.
REFERENCES
- Alday, J. G. , Marrs R. H., and Martínez‐Ruiz C.. 2011. “Vegetation Convergence during Early Succession on Coal Wastes: A 6‐Year Permanent Plot Study.” Journal of Vegetation Science 22(6): 1072–83. [Google Scholar]
- Bailey, S. F. , Dettman J. R., Rainey P. B., and Kassen R.. 2013. “Competition Both Drives and Impedes Diversification in a Model Adaptive Radiation.” Proceedings of the Royal Society of London B: Biological Sciences 280(1766): 20131253. http://rspb.royalsocietypublishing.org/content/280/1766/20131253.short. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabás, G. , and D'Andrea R.. 2016. “The Effect of Intraspecific Variation and Heritability on Community Pattern and Robustness.” Ecology Letters 19(8): 977–86. [DOI] [PubMed] [Google Scholar]
- Bassar, R. D. , Ferriere R., López‐Sepulcre A., Marshall M. C., Travis J., Pringle C. M., and Reznick D. N.. 2012. “Direct and Indirect Ecosystem Effects of Evolutionary Adaptation in the Trinidadian Guppy (Poecilia reticulata).” The American Naturalist 180(2): 167–85. [DOI] [PubMed] [Google Scholar]
- Becks, L. , Ellner S. P., Jones L. E., and Hairston Jr N. G.. 2010. “Reduction of Adaptive Genetic Diversity Radically Alters Eco‐Evolutionary Community Dynamics.” Ecology Letters 13(8): 989–97. 10.1111/j.1461-0248.2010.01490.x. [DOI] [PubMed] [Google Scholar]
- Becks, L. , Ellner S. P., Jones L. E., and Hairston N. G.. 2012. “The Functional Genomics of an Eco‐Evolutionary Feedback Loop: Linking Gene Expression, Trait Evolution, and Community Dynamics.” Ecology Letters 15(5): 492–501. 10.1111/j.1461-0248.2012.01763.x. [DOI] [PubMed] [Google Scholar]
- Brockhurst, M. A. , Hochberg M. E., Bell T., and Buckling A.. 2006. “Character Displacement Promotes Cooperation in Bacterial Biofilms.” Current Biology 16(20): 2030–4. 10.1016/j.cub.2006.08.068. [DOI] [PubMed] [Google Scholar]
- Burke, M. K. , Liti G., and Long A. D.. 2014. “Standing Genetic Variation Drives Repeatable Experimental Evolution in Outcrossing Populations of Saccharomyces Cerevisiae.” Molecular Biology and Evolution 31(12): 3228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, J. M. 2003. “Community Assembly: When Should History Matter?” Oecologia 136(4): 489–98. [DOI] [PubMed] [Google Scholar]
- Colosimo, P. F. , Hosemann K. E., Balabhadra S., Villarreal G., Dickson M., Grimwood J., Schmutz J., Myers R. M., Schluter D., and Kingsley D. M.. 2005. “Widespread Parallel Evolution in Sticklebacks by Repeated Fixation of Ectodysplasin Alleles.” Science 307(5717): 1928–33. [DOI] [PubMed] [Google Scholar]
- de Cáceres, M. , Coll L., Legendre P., Allen R. B., Wiser S. K., Fortin M.‐J., Condit R., and Hubbell S.. 2019. “Trajectory Analysis in Community Ecology.” Ecological Monographs 89(2): e01350. 10.1002/ecm.1350. [DOI] [Google Scholar]
- del Moral, R. , and Lacher I. L.. 2005. “Vegetation Patterns 25 Years after the Eruption of Mount St. Helens, Washington, USA.” American Journal of Botany 92(12): 1948–56. [DOI] [PubMed] [Google Scholar]
- Duquette, S. L. , Altwegg R., and Anholt B. R.. 2005. “Factors Affecting the Expression of Inducible Defences in Euplotes: Genotype, Predator Density and Experience.” Functional Ecology 19(4): 648–55. 10.1111/j.1365-2435.2005.01013.x. [DOI] [Google Scholar]
- Ellner, S. P. 2013. “Rapid Evolution: From Genes to Communities, and Back Again?” Functional Ecology 27(5): 1087–99. [Google Scholar]
- Faillace, C. A. , and Morin P. J.. 2016. “Evolution Alters the Consequences of Invasions in Experimental Communities.” Nature Ecology & Evolution 1(1): 13. 10.1038/s41559-016-0013. [DOI] [PubMed] [Google Scholar]
- Faillace, C. A. , and Morin P. J.. 2020. “Evolution Alters Post‐Invasion Temporal Dynamics in Experimental Communities.” Journal of Animal Ecology 89: 285–98. [DOI] [PubMed] [Google Scholar]
- Farkas, T. E. , Mononen T., Comeault A. A., and Nosil P.. 2016. “Observational Evidence That Maladaptive Gene Flow Reduces Patch Occupancy in a Wild Insect Metapopulation.” Evolution 70(12): 2879–88. [DOI] [PubMed] [Google Scholar]
- Fukami, T. 2015. “Historical Contingency in Community Assembly: Integrating Niches, Species Pools, and Priority Effects.” Annual Review of Ecology, Evolution, and Systematics 46: 1–23. [Google Scholar]
- Fukami, T. , and Nakajima M.. 2011. “Community Assembly: Alternative Stable States or Alternative Transient States?” Ecology Letters 14(10): 973–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami, T. , and Nakajima M.. 2013. “Complex Plant‐Soil Interactions Enhance Plant Species Diversity by Delaying Community Convergence.” Journal of Ecology 101(2): 316–24. [Google Scholar]
- Fussmann, G. F. , Loreau M., and Abrams P. A.. 2007. “Eco‐Evolutionary Dynamics of Communities and Ecosystems.” Functional Ecology 21(3): 465–77. [Google Scholar]
- Fyda, J. , and Wiackowski K.. 1998. “Benefits and Costs of Predator‐Induced Morphological Changes in the Ciliate Colpidium kleini (Protozoa, Ciliophora).” European Journal of Protistology 34(2): 118–23. 10.1016/S0932-4739(98)80021-7. [DOI] [Google Scholar]
- Gaston, K. J. , and Fuller R. A.. 2008. “Commonness, Population Depletion and Conservation Biology.” Trends in Ecology & Evolution 23(1): 14–9. [DOI] [PubMed] [Google Scholar]
- Germain, R. M. , Srivastava D., and Angert A. L.. 2020. “Evolution of an Inferior Competitor Increases Resistance to Biological Invasion.” Nature Ecology & Evolution 4(3): 419–25. 10.1038/s41559-020-1105-x. [DOI] [PubMed] [Google Scholar]
- Gilchrist, G. W. , Huey R. B., Balanyà J., Pascual M., and Serra L.. 2004. “A Time Series of Evolution in Action: A Latitudinal Cline in Wing Size in South American Drosophila subobscura .” Evolution 58(4): 768–80. [DOI] [PubMed] [Google Scholar]
- Gillespie, R. 2004. “Community Assembly through Adaptive Radiation in Hawaiian Spiders.” Science 303(5656): 356–9. [DOI] [PubMed] [Google Scholar]
- Grainger, T. N. , Letten A. D., Gilbert B., and Fukami T.. 2019. “Applying Modern Coexistence Theory to Priority Effects.” Proceedings of the National Academy of Sciences 116(13): 6205–10. 10.1073/PNAS.1803122116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, P. R. , and Grant R. B.. 2002. “Unpredictable Evolution in a 30 Year Study of Darwin's Finches.” Science 296(April): 707–11. http://science.sciencemag.org/content/296/5568/707.short. [DOI] [PubMed] [Google Scholar]
- Grunberg, R. 2022. “ritagrunberg/Late‐dynamics: Late Dynamics code V1 (v1.0.0).” Zenodo. 10.5281/zenodo.5818017. [DOI]
- Herron, M. D. , and Doebeli M.. 2013. “Parallel Evolutionary Dynamics of Adaptive Diversification in Escherichia coli .” PLoS Biology 11(2): e1001490. 10.1371/journal.pbio.1001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen, T. , and Becks L.. 2014. “Consumer Co‐Evolution as an Important Component of the Eco‐Evolutionary Feedback.” Nature Communications 5: 5226. 10.1038/ncomms6226. [DOI] [PubMed] [Google Scholar]
- Jain, M. , Flynn D. F. B., Prager C. M., Hart G. M., DeVan C. M., Ahrestani F. S., Palmer M. I., Bunker D. E., Knops J. M. H., and Jouseau C. F.. 2014. “The Importance of Rare Species: A Trait‐Based Assessment of Rare Species Contributions to Functional Diversity and Possible Ecosystem Function in Tall‐Grass Prairies.” Ecology and Evolution 4(1): 104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, V. , Violle C., Mondy C., Hoffmann L., and Muller S.. 2010. “Intraspecific Variability and Trait‐Based Community Assembly.” Journal of Ecology 98(5): 1134–40. [Google Scholar]
- Keith, A. R. , Bailey J. K., Lau M. K., and Whitham T. G.. 2017. “Genetics‐Based Interactions of Foundation Species Affect Community Diversity, Stability and Network Structure.” Proceedings of the Royal Society B: Biological Sciences 284(1854): 20162703. http://rspb.royalsocietypublishing.org/content/284/1854/20162703.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnison, M. T. , and Hairston N. G.. 2007. “Eco‐Evolutionary Conservation Biology: Contemporary Evolution and the Dynamics of Persistence.” Functional Ecology 21(3): 444–54. [Google Scholar]
- Klauschies, T. , Vasseur D. A., and Gaedke U.. 2016. “Trait Adaptation Promotes Species Coexistence in Diverse Predator and Prey Communities.” Ecology and Evolution 6(12): 4141–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knope, M. L. , Forde S. E., and Fukami T.. 2012. “Evolutionary History, Immigration History, and the Extent of Diversification in Community Assembly.” Frontiers in Microbiology 2(JAN): 273. 10.3389/fmicb.2011.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, H. , Frickel J., Valiadi M., and Becks L.. 2014. “Why Rapid, Adaptive Evolution Matters for Community Dynamics.” Frontiers in Ecology and Evolution 2: 17. [Google Scholar]
- Kremer, C. T. , and Klausmeier C. A.. 2013. “Coexistence in a Variable Environment: Eco‐Evolutionary Perspectives.” Journal of Theoretical Biology 339: 14–25. [DOI] [PubMed] [Google Scholar]
- Kremer, C. T. , and Klausmeier C. A.. 2017. “Species Packing in Eco‐Evolutionary Models of Seasonally Fluctuating Environments.” Ecology Letters 20: 1158–68. 10.1111/ele.12813. [DOI] [PubMed] [Google Scholar]
- Lamit, L. J. , Holeski L. M., Flores‐Rentería L., Whitham T. G., and Gehring C. A.. 2016. “Tree Genotype Influences Ectomycorrhizal Fungal Community Structure: Ecological and Evolutionary Implications.” Fungal Ecology 24(December): 124–34. 10.1016/J.FUNECO.2016.05.013. [DOI] [Google Scholar]
- Laughlin, D. C. , Joshi C., Bodegom P. M., Bastow Z. A., and Fulé P. Z.. 2012. “A Predictive Model of Community Assembly That Incorporates Intraspecific Trait Variation.” Ecology Letters 15(11): 1291–9. [DOI] [PubMed] [Google Scholar]
- Lee, W. G. , Tanentzap A. J., and Heenan P. B.. 2012. “Plant Radiation History Affects Community Assembly: Evidence from the New Zealand Alpine.” Biology Letters 8(4): 558–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand, T. , Roze D., and Rousset F.. 2009. “Stochasticity in Evolution.” Trends in Ecology & Evolution 24(3): 157–65. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E. 2017. “Convergence and Divergence in a Long‐Term Experiment with Bacteria.” The American Naturalist 190(S1): S57–68. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E. , and Travisano M.. 1994. “Dynamics of Adaptation and Diversification: A 10,000‐Generation Experiment with Bacterial Populations.” Proceedings of the National Academy of Sciences 91(15): 6808–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letten, A. D. , Ke P.‐J., and Fukami T.. 2017. “Linking Modern Coexistence Theory and Contemporary Niche Theory.” Ecological Monographs 87(2): 161–77. [Google Scholar]
- Li, S.‐p. , Cadotte M. W., Meiners S. J., Zhichao P., Fukami T., and Jiang L.. 2016. “Convergence and Divergence in a Long‐Term Old‐Field Succession: The Importance of Spatial Scale and Species Abundance.” Ecology Letters 19(9): 1101–9. [DOI] [PubMed] [Google Scholar]
- Meiners, S. J. , Cadotte M. W., Fridley J. D., Pickett S. T. A., and Walker L. R.. 2015. “Is Successional Research Nearing Its Climax? New Approaches for Understanding Dynamic Communities.” Functional Ecology 29(2): 154–64. [Google Scholar]
- Meyer, J. R. , Dobias D. T., Weitz J. S., Barrick J. E., Quick R. T., and Lenski R. E.. 2012. “Repeatability and Contingency in the Evolution of a Key Innovation in Phage Lambda.” Science 335(6067): 428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J. , Kindt R., Legendre P., O'hara B., Stevens M. H. H., Oksanen M. J., and Suggests M. A. S. S.. 2007. “The Vegan Package.” Community Ecology Package 10: 631–7. [Google Scholar]
- Palkovacs, E. P. , Marshall M. C., Lamphere B. A., Lynch B. R., Weese D. J., Fraser D. F., Reznick D. N., Pringle C. M., and Kinnison M. T.. 2009. “Experimental Evaluation of Evolution and Coevolution as Agents of Ecosystem Change in Trinidadian Streams.” Philosophical Transactions of the Royal Society of London B: Biological Sciences 364(1523): 1617–28. 10.1098/rstb.2009.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlert, T. 2020. “Trend: Non‐Parametric Trend Tests and Change‐Point Detection. R Package Version 1.1.4.” https://cran.r-project.org/package=trend.
- Post, D. M. , and Palkovacs E. P.. 2009. “Eco‐Evolutionary Feedbacks in Community and Ecosystem Ecology: Interactions between the Ecological Theatre and the Evolutionary Play.” Philosophical Transactions of the Royal Society of London B: Biological Sciences 364(1523): 1629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2021. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Schoener, T. W. 2011. “The Newest Synthesis: Understanding the Interplay of Evolutionary and Ecological Dynamics.” Science 331(6016): 426–9. 10.1126/science.1193954. [DOI] [PubMed] [Google Scholar]
- Siefert, A. 2012. “Incorporating Intraspecific Variation in Tests of Trait‐Based Community Assembly.” Oecologia 170(3): 767–75. [DOI] [PubMed] [Google Scholar]
- Simões, P. , Santos J., Fragata I., Mueller L. D., Rose M. R., and Matos M.. 2008. “How Repeatable Is Adaptive Evolution? The Role of Geographical Origin and Founder Effects in Laboratory Adaptation.” Evolution 62(8): 1817–29. [DOI] [PubMed] [Google Scholar]
- Stuart, Y. E. , Campbell T. S., Hohenlohe P. A., Reynolds R. G., Revell L. J., and Losos J. B.. 2014. “Rapid Evolution of a Native Species Following Invasion by a Congener.” Science 346(6208): 463–6. 10.1126/science.1257008. [DOI] [PubMed] [Google Scholar]
- Taylor, A. R. , and Chen H. Y. H.. 2011. “Multiple Successional Pathways of Boreal Forest Stands in Central Canada.” Ecography 34(2): 208–19. [Google Scholar]
- terHorst, C. P. , Zee P. C., Heath K. D., Miller T. E., Pastore A. I., Patel S., Schreiber S. J., Wade M. J., and Walsh M. R.. 2018. “Evolution in a Community Context: Trait Responses to Multiple Species Interactions.” The American Naturalist 191(3): 368–80. 10.1086/695835 [DOI] [Google Scholar]
- Toju, H. , Vannette R. L., Gauthier M.‐P. L., Dhami M. K., and Fukami T.. 2018. “Priority Effects Can Persist across Floral Generations in Nectar Microbial Metacommunities.” Oikos 127(3): 345–52. 10.1111/oik.04243. [DOI] [Google Scholar]
- Travisano, M. , Mongold J. A., Bennett A. F., and Lenski R. E.. 1995. “Experimental Tests of the Roles of Adaptation, Chance, and History in Evolution.” Science 267(5194): 87–90. [DOI] [PubMed] [Google Scholar]
- Urban, M. C. 2010. “Microgeographic Adaptations of Spotted Salamander Morphological Defenses in Response to a Predaceous Salamander and Beetle.” Oikos 119(4): 646–58. [Google Scholar]
- Urban, M. C. , and De Meester L.. 2009. “Community Monopolization: Local Adaptation Enhances Priority Effects in an Evolving Metacommunity.” Proceedings of the Royal Society of London B: Biological Sciences 276(1676): 4129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violle, C. , Enquist B. J., McGill B. J., Jiang L., Albert C. H., Hulshof C., Jung V., and Messier J.. 2012. “The Return of the Variance: Intraspecific Variability in Community Ecology.” Trends in Ecology & Evolution 27(4): 244–52. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Naumann U., Wright S. T., and Warton D. I.. 2012. “Mvabund‐ an R Package for Model‐Based Analysis of Multivariate Abundance Data.” Methods in Ecology and Evolution 3: 471–4. [Google Scholar]
- Whitham, T. G. , Gehring C. A., Lamit L. J., Wojtowicz T., Evans L. M., Keith A. R., and Smith D. S.. 2012. “Community Specificity: Life and Afterlife Effects of Genes.” Trends in Plant Science 17(5): 271–81. [DOI] [PubMed] [Google Scholar]
- Wiackowski, K. , and Staronska A.. 1999. “The Effect of Predator and Prey Density on the Induced Defence of a Ciliate.” Functional Ecology 13(1): 59–65. 10.1046/j.1365-2435.1999.00282.x. [DOI] [Google Scholar]
- Yoshida, T. , Jones L. E., Ellner S. P., Fussmann G. F., and Hairston N. G.. 2003. “Rapid Evolution Drives Ecological Dynamics in a Predator – Prey System.” Nature 424(6946): 303–6. 10.1038/nature01767. [DOI] [PubMed] [Google Scholar]
- Zee, P. C. , and Fukami T.. 2018. “Priority Effects Are Weakened by a Short, but Not Long, History of Sympatric Evolution.” Proceedings of the Royal Society B: Biological Sciences 285(1871): 20171722. 10.1098/rspb.2017.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Data Availability Statement
Data and code (Grunberg, 2022) are available in Zenodo at https://doi.org/10.5281/zenodo.5818017.


